- Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
The purpose of this study was to investigate the prevalence and genetic elements of oqxAB among Escherichia coli isolates from animals, retail meat, and humans (patients with infection or colonization) in Guangzhou, China. A total of 1,354 E. coli isolates were screened for oqxAB by PCR. Fifty oqxAB-positive isolates were further characterized by pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), S1-PFGE, genetic environment analysis, plasmid replicon typing, and plasmid sequencing. oqxAB was detected in 172 (33.79%), 60 (17.34%), and 90 (18.07%) E. coli isolates from animal, food, and human, respectively. High clonal diversity was observed among oqxAB-positive isolates. In 21 oqxAB-containing transformants, oqxAB was flanked by two IS26 elements in the same orientation, formed a composite transposon Tn6010 in 19 transformants, and was located on plasmids (33.3~500 kb) belonging to IncN1-F33:A-:B- (n = 3), IncHI2/ST3 (n = 3), F-:A18:B- (n = 2), F-:A-:B54 (n = 2), or others. Additionally, oqxAB was co-located with multiple resistance genes on the same plasmid, such as aac(6′)-Ib-cr and/or qnrS, which were identified in two F-:A18:B- plasmids from pigs, and blaCTX−M−55, rmtB, fosA3, and floR, which were detected in two N1-F33:A-:B- plasmids from patients. The two IncHI2/ST3 oqxAB-bearing plasmids, pHNLDF400 and pHNYJC8, which were isolated from human patient and chicken meat, respectively, contained a typical IncHI2-type backbone, and were similar to each other with 2-bp difference, and also showed 99% identity to the Salmonella Typhimurium oqxAB-carrying plasmids pHXY0908 (chicken) and pHK0653 (human patient). Horizontal transfer mediated by mobile elements may be the primary mechanism underlying oqxAB spread in E. coli isolates obtained from various sources in Guangzhou, China. The transmission of identical oqxAB-carrying IncHI2 plasmids between food products and humans might pose a serious threat to public health.
Introduction
The efflux pump OqxAB, originally identified in the conjugative IncX1 plasmid pOLA52 from a porcine Escherichia coli isolate in 2003, belongs to the resistance-nodulation-division family, and is encoded by the oqxA and oqxB genes, which are located in the same operon (Sørensen et al., 2003; Hansen et al., 2004). OqxAB mediates resistance, or reduces susceptibility to multiple antimicrobials, including quinoxalines, chloramphenicol, trimethoprim, and quinolones, and is recognized as a plasmid-mediated quinolone resistance (PMQR) determinant (Hansen et al., 2007; Ruiz et al., 2012).
Recently, the oqxAB genes have been identified as the most prevalent PMQR genes in E. coli isolates from food-producing animals in China (Liu et al., 2012, 2013; Xu et al., 2015), as well as from animal-derived food products (Xu et al., 2014). This could be due to the widespread use of olaquindox as a growth promoter for pigs weighing below 35 kg and mequindox against enteropathogenic E. coli infections in swine and poultry (He T. et al., 2015). Our previous study demonstrated high oqxAB prevalence in E. coli isolates from animals, farm environment, and farm workers, and clonal transmission of oqxAB-carrying isolates between swine and farm workers was observed (Zhao et al., 2010). Additionally, oqxAB was observed to be relatively prevalent in E. coli isolates from pigs, ducks, chickens, meat (pork and chicken meat), and healthy humans (55.7, 40.6, 25.8, 16.2, and 7.2%, respectively; Yang T. et al., 2014). However, oqxAB has been rarely reported in human clinics and animals in Europe, where olaquindox has been banned as an animal feed additive since 1999 (Hansen et al., 2005), with detection rates of 0.46% in E. coli isolates from patients in the UK and Ireland (Ciesielczuk et al., 2013) and 1.62% from porcine E. coli strains from Denmark and Sweden (Hansen et al., 2005). Altogether, oqxAB might possibly be transmitted from animals to humans via the food chain or close contact, in China. Furthermore, IS26 plays an important role in oqxAB dissemination (Hansen et al., 2004; He T. et al., 2015). IncHI2 plasmids have been recently demonstrated to mediate oqxAB spread in S. Typhimurium and S. Indiana among animals in China, as well as in clinical S. Typhimurium isolates in Hong Kong (Li et al., 2013; Wong et al., 2016). Thus, this study was aimed at determining the distribution and genetic elements (IS26 and plasmids) of oqxAB among E. coli isolates from different sources (animals, animal-derived food products, and human clinics) in Guangzhou, China, determining the complete nucleotide sequence of two IncHI2 plasmids carrying oqxAB and comparing them with those previously reported from different sources in China, and outlining the possible routes of oqxAB transmission via the food chain.
Materials and Methods
Bacterial Strains
A total of 1,354 individual E. coli isolates were collected from Guangzhou, Guangdong province, China between July 2011 and May 2013, including 509 animal strains (372 pig and 137 chicken strains) isolated from fecal samples of healthy animals from one hog market and one live poultry market, 346 strains from retail meat (247 pork and 99 chicken meat samples) recovered from fresh or chilled pork and chicken samples purchased from supermarkets and farmers' markets, and 498 strains from patients (71 E. coli strains from urine samples from inpatients with urinary tract infection, 427 E. coli strains from fecal samples of inpatients and outpatients) from four hospitals. The samples were inoculated onto MacConkey agar, and suspected E. coli colonies (one isolate per sample) were selected and identified by standard biochemical testing. All the isolates were tested for the presence of oqxAB by PCR, and 100 amplified products were randomly selected for sequencing (Table S1).
Antimicrobial Susceptibility Testing
All oqxAB-positive E. coli isolates were tested for their MICs of ampicillin, cefotaxime, amikacin, gentamycin, neomycin, apramycin, tetracycline, florfenicol, ciprofloxacin, olaquindox, colistin, and sulfamethoxazole/trimethoprim using the agar dilution or broth microdilution method (limited to colistin). Antimicrobial susceptibility tests were performed and evaluated according to the protocols recommended by M100-S25 of the Clinical and Laboratory Standards Institute (Wayne, PA, USA) (Clinical and Laboratory Standards Institute, 2015). Colistin (>2 mg/L), neomycin (>8 mg/L), and florfenicol (>16 mg/L) were interpreted according to the clinical breakpoints or epidemiological cutoff values of the European Committee on Antimicrobial Susceptibility Testing (EUCAST; https://mic.eucast.org/Eucast2/); apramycin (>16 mg/L) and olaquindox (>64 mg/L) were analyzed according to DANMAP 2012 and DANMAP98, respectively (http://www.danmap.org/Downloads/Reports.aspx). The E. coli reference strain ATCC 25922 was used as the quality control. Epi Info version 7.2 (CDC) was used to perform statistical analysis. Comparison of prevalence of oqxAB and antimicrobials resistance rates was conducted by the χ2-test. P < 0.05 were considered statistically significant.
Pulsed-Field Gel Electrophoresis and Multilocus Sequence Typing (MLST)
The genetic diversity of 50 randomly selected oqxAB-positive E. coli isolates from animals (n = 21), food products (n = 9), and patients (n = 20) was characterized by multilocus sequence typing (MLST; http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and PFGE (Gautom, 1997). MLST data were aligned using ClustalW, and a phylogenetic tree was constructed by the neighbor joining algorithm using MEGA 7.0 (Kumar et al., 2008). PFGE patterns were compared and analyzed using BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) using a cut-off value of 90% to define PFGE clusters.
Conjugation/Transformation Experiments and Plasmid Characterization
The 50 oqxAB-positive E. coli strains subjected to PFGE were further analyzed by performing conjugation experiments with streptomycin-resistant E. coli C600 as the recipient strain following a previously described protocol (Chen et al., 2007). Transconjugants were selected using 32 μg/mL olaquindox and 3,000 μg/mL streptomycin.
Transformation was conducted via heat-shock or electroporation using E. coli strain DH5α as the recipient (Hanahan, 1983; Dower et al., 1988). Plasmid DNA was extracted using the E.Z.N.A. Plasmid DNA Midi Kit (Omega, Norcross, GA, USA). Transformants were selected using MacConkey agar plates containing 16 μg/mL olaquindox.
The presence of oqxAB in the transconjugants/transformants was confirmed via PCR and sequencing. Other resistance genes, including floR, blaCTX−M, rmtB, and fosA3 were also screened using the primers listed in Table S1. Antimicrobial susceptibility of all the transformants and E. coli DH5α recipient strain were determined using the agar dilution or broth microdilution method (limited to colistin). All the transformants were characterized by PCR-based replicon typing and were screened IncX plasmids as described previously (Carattoli et al., 2005; Johnson et al., 2012). Replicon sequence typing and plasmid double locus sequence typing were performed to further characterize IncFII and IncHI2 plasmids according to previously described protocols (García-Fernández and Carattoli, 2010; Villa et al., 2010). S1-PFGE (Barton et al., 1995), and Southern blot hybridization were performed to determine the number of plasmids and the sizes of oqxAB-carrying plasmids in all the transformants, using a non-radioactively labeled oqxAB-specific probe. The genetic context of oqxAB was determined by PCR mapping (Table S2). Ten transformants with a single oqxAB-carrying plasmid were further analyzed by restriction fragment length polymorphism (RFLP) using the endonuclease ApaLI, TZC215-1, and AHH13-1 were excluded since their sizes were significantly different from other oqxAB-carrying plasmids.
Plasmid Sequencing
Two IncHI2 plasmids, namely, pHNLDH400 from a patient with E. coli colonization and pHNYJC8 from chicken meat, were selected for sequencing. Plasmid DNA was purified from the transformants using QIAGEN® Plasmid Midi Kit according to the manufacturer's instructions (Qiagen, Hilden, Germany), and was completely sequenced by PacBio single-molecule real-time sequencing (RSII platform) (Pacific Biosciences, Menlo Park, CA, USA). Raw reads were introduced into the non-hybrid Hierarchical Genome Assembly Process. Analysis and annotation of the resulting sequences were performed using RAC (http://rac.aihi.mq.edu.au/rac/), ISfinder (https://www-is.biotoul.fr//), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), RAST server (Aziz et al., 2008), and the Gene Construction Kit 4.0 (Textco BioSoftware, Inc., Raleigh, NC, USA). Plasmids pHNLDF400 and pHNYJC8 were compared with similar oqxAB-bearing IncHI2 plasmids pHXY0908 and pHK0653 obtained from S. Typhimurium, as well as our previously reported plasmids pHNSHP45-2 (Zhi et al., 2016), and pHNAH67 from E. coli isolate AHC67 (Yang X. et al., 2014) using BLASTn and BRIG (Alikhan et al., 2011). The IncHI2 plasmid R478 (GenBank accession number BX664015) served as the reference plasmid for annotation.
Nucleotide Sequences Accession Numbers
The nucleotide sequences of plasmids pHNLDF400 and pHNYJC8 have been deposited in the GenBank database under the accession numbers KY019258 and KY019259, respectively.
Results and Discussion
Prevalence of oqxAB and Antimicrobial Susceptibility
Of 1,354 E. coli isolates examined in this study, oqxAB was detected in 322 (23.78%) E. coli isolates. Consistent with the results of our previous study (Yang T. et al., 2014), E. coli isolates from animal sources showed the highest oqxAB prevalence rate (172/509, 33.79%). However, oqxAB prevalence among the isolates of human origins (90/498, 18.07%) was slightly higher than those of food origins (60/346, 17.34%), contradictory to previous results, which was probably because we detected clinical E. coli isolates instead of strains from healthy volunteers. The oqxAB-positive isolates from patients were possibly selected under pressure exerted by antimicrobials, such as fluoroquinolones. However, oqxAB was previously reported to have low prevalence among clinical E. coli isolates in Europe (0.46%; Ciesielczuk et al., 2013), Korea (0.4%; Kim et al., 2009), Taiwan (6.05%; Kao et al., 2016), and mainland China (6.6%; Yuan et al., 2012; Zhao et al., 2015). Interestingly, the chicken isolates (51/137, 37.23%) showed a slightly higher oqxAB prevalence rate than the porcine isolates (121/372, 32.53%); similarly, the chicken meat isolates (22/99, 22.22%) showed higher oqxAB prevalence than the pork isolates (38/247, 15.38%).
As shown in Figure 1, the isolates from food-producing animals showed the highest resistance rates against multiple antimicrobials, including neomycin (57.6%), apramycin (60.5%), florfenicol (74.4%), olaquindox (98.3%), and colistin (18.0%), which are used only in veterinary medicine in China (colistin was banned from animal feed in April 2017 in China), as well as tetracycline (97.1%) and sulfamethoxazole/trimethoprim (97.7%). It was followed by the isolates from food (56.7, 51.7, 61.7, 55.0, 16.7, 81.7, and 93.3%, respectively), and humans (26.7, 17.8, 41.1, 44.4, 1.1, 58.9, and 83.3%, respectively). However, the food isolates showed the highest resistance rate of 51.7% against gentamycin, which was considerably higher than the resistance rates of animal and human isolates (37.2 and 30%, respectively). In addition, the resistance rates against cefotaxime were significantly higher in the oqxAB-carrying isolates from humans (50%) than from animals (19.8%; P < 0.01) and food products (30%; P < 0.05), which could be explained by the relatively more frequent use of cephalosporins in clinical medicine.
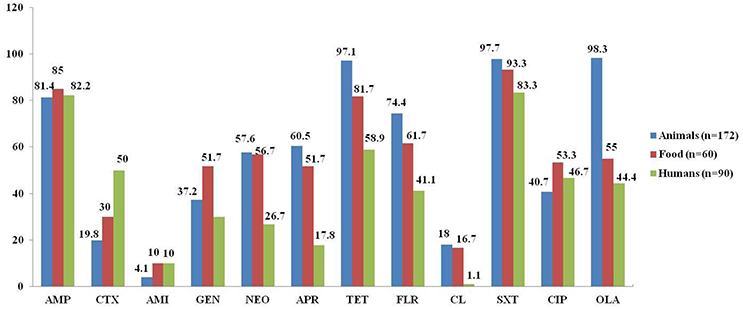
Figure 1. Comparison of antimicrobial resistance among oqxAB-positive E. coli isolates from food-producing animals, food products, and human patients. AMP, ampicillin; CTX, cefotaxime; AMI, amikacin; GEN, gentamycin; NEO, neomycin; APR, apramycin; TET, tetracycline; FLR, florfenicol; CL, colistin; SXT, sulfamethoxazole/trimethoprim; CIP, ciprofloxacin; OLA, olaquindox.
Molecular Typing of oqxAB-Positive E. coli Isolates
The MLST analysis identified 21 different sequence types (STs) among 39 oqxAB-positive E. coli isolates (Table S3). The most commonly identified genotypes were ST10 (n = 10), followed by ST410 (n = 4) and ST744 (n = 3), whereas the isolates belonging to ST93 (n = 2), ST165 (n = 2), ST542 (n = 2), ST602 (n = 2), ST48, ST57, ST58, ST178, ST206, ST224, ST301, ST359, ST453, ST746, ST847, ST1421, ST3339, and ST6697 were also identified (Table S3). The remaining 11 oqxAB-positive E. coli were assigned to 11 novel STs (Table S3). In our study, the dominant ST10 clone, which has been frequently detected in animals, humans, food products, and the environment, and is responsible for the spread of multiple antibiotic resistance genes (Belmar Campos et al., 2014; Müller et al., 2016; Röderova et al., 2017), was detected in the isolates from animals, chicken meat, and patients. Additionally, other STs described in this study, such as ST410, ST744, ST93, ST48, and ST58, have been detected in various sources (Loncaric et al., 2013; Belmar Campos et al., 2014; Abraham et al., 2015; Falgenhauer et al., 2016; Röderova et al., 2017). The results of the phylogenetic analysis revealed the genetic relatedness of oqxAB-positive isolates (Figure S1).
Among the 50 oqxAB-positive E. coli isolates from different sources analyzed using XbaI-PFGE, 44 oqxAB-carrying E. coli isolates from animals (n = 19), animal-derived food products (n = 6), and patients (n = 19) exhibited 43 distinct PFGE patterns (Figure 2). Notably, the isolates belonging to the same STs identified in our study showed different PFGE patterns; however, the ST410 E. coli isolates BYMP20 and YZHF29, which were obtained from pork and human sources from different districts in Guangzhou within 12 km, showed similar PFGE patterns (E1 and E2; Figure 2).
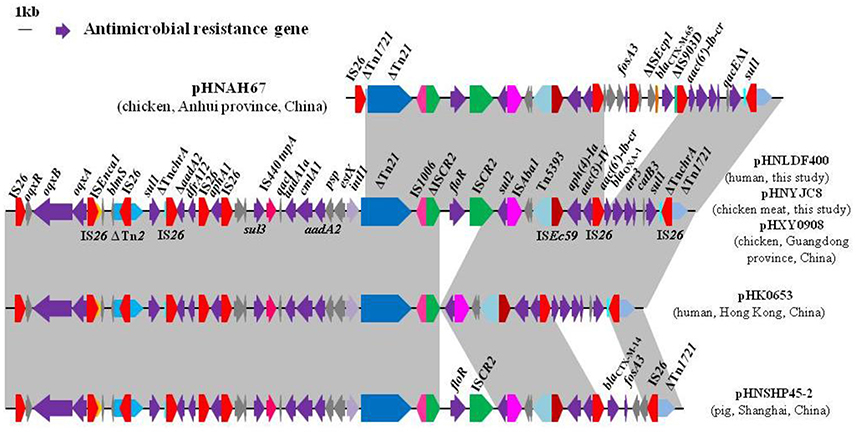
Figure 2. Comparison of the multidrug resistance regions of plasmids pHNLDF400 and pHNYJC8 with other similar IncHI2 plasmids. Arrows indicate the positions and directions of gene transcription. Regions with >99% homology are shaded in gray. ΔIndicates a truncated gene. The 1-kb distance scale is displayed in the upper left corner. Sequences were obtained from GenBank accession numbers pHNAH67, KX246266; pHXY0908, KM877269; pHK0653, KT335433; and pHNSHP45-2, KU341381.
MLST and PFGE demonstrated the molecular diversity of the oqxAB-positive E. coli isolates, and suggested that clonal transfer might not be the main mechanism underlying oqxAB dissemination among E. coli isolates in Guangzhou, China.
Analysis of oqxAB-Carrying Plasmids
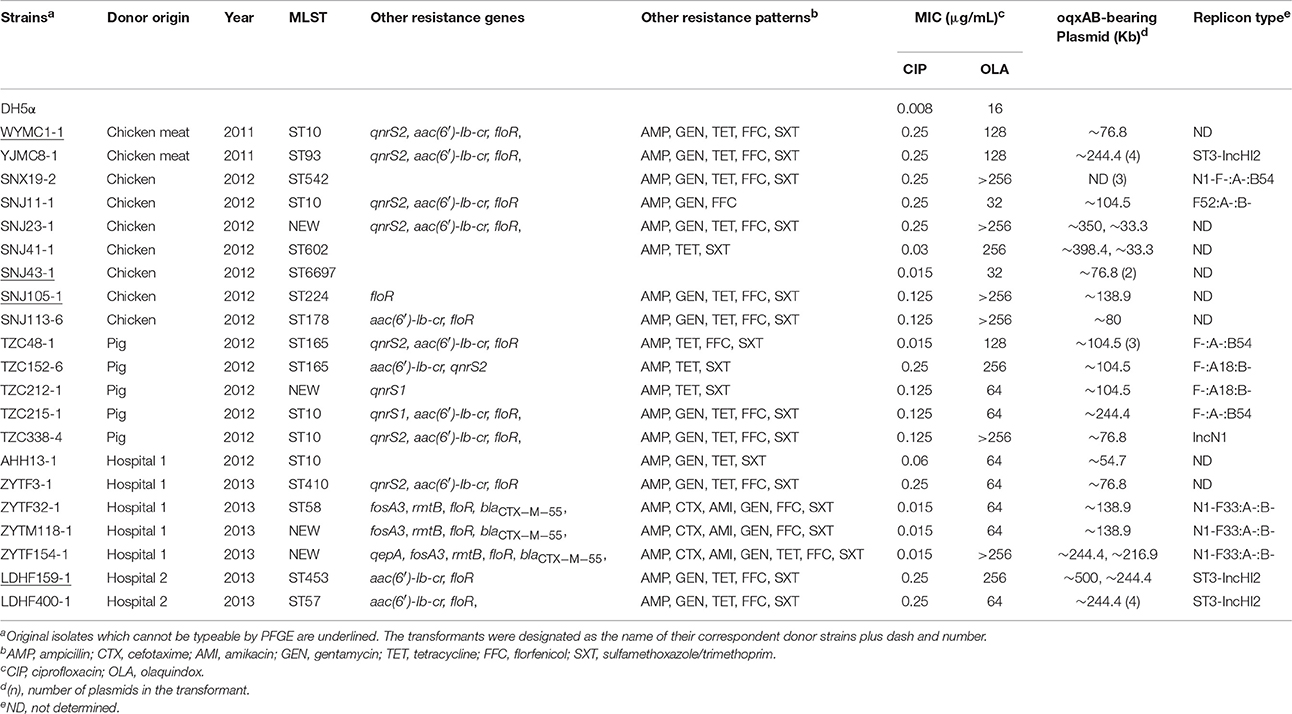
Surprisingly, none of the 50 randomly selected oqxAB-positive E. coli isolates transferred oqxAB to E. coli C600 via conjugation, which is consistent with the results reported for S. Typhimurium (Wong et al., 2014). Therefore, transformation assays were performed, and 21 transformants were obtained successfully. All the transformants were subjected to S1-PFGE and Southern blotting. The results demonstrated that the transformants carried one to four plasmids of various sizes. oqxAB was detected in plasmids with sizes ranging from ~33.3 to ~500 kb. Four transformants (SNJ23-1, SNJ41-1, ZYTM154-1, and LDHF159-1) showed two hybridization signals, whereas no hybridization signal was observed in SNX19-2 (Table 1). Thirteen transformants contained IncFII (n = 4), IncFIA (n = 2), IncFIB (n = 3), IncHI2 (n = 3), and IncN (n = 5) replicons. Notably, three transformants carried both IncN and IncFII replicons, which were classified as N1-F33: A-: B- (Table 1). Interestingly, the three transformants were obtained from three original E. coli isolates exhibiting different PFGE patterns from the same hospital, of which two carried only one oqxAB-bearing plasmid with similar sizes (Table 1 and Figure S2). Three IncHI2 plasmids from food product and patients were assigned to ST3 via plasmid double locus sequence typing (Table 1). PCR-based replicon typing could not determine the replicon types of the remaining eight transformants. Although rare, an untypable oqxAB-carrying plasmid was described previously (Liu et al., 2013). Ten transformants with single plasmid were selected for RFLP with ApaLI. Interestingly, two N1-F33:A-:B- plasmids from the patients of the same hospital (ZYTM118-1 and ZYTF32-1) showed identical patterns, as well as two F-:A18:B- plasmids from pigs (TZC152-6 and TZC212-1). Additionally, three ~76.8 oqxAB-bearing plasmids obtained from pig, chicken meat, and human, exhibited identical patterns (Figure S3).
Sequences of the regions surrounding oqxAB were determined via PCR mapping and sequencing. oqxAB was flanked by two IS26 elements in the same orientation, and formed a composite transposon Tn6010 (IS26-oqxA-oqxB-oqxR-IS26) in 19 transformants. Additionally, one IS26 element was present in upstream or downstream of oqxAB in the remaining transformants TZC338-4 and SNX19-2. Since its first identification in the plasmid pOLA52 from porcine E. coli in 2003 (Sørensen et al., 2003), Tn6010 is believed to play a vital role in oqxAB transmission among Enterobacteriaceae isolates (Liu et al., 2013; He T. et al., 2015). Our results further support that horizontal transfer mediated by mobile elements seems to be the main mechanism for oqxAB transmission in E. coli from different sources.
The MICs of ciprofloxacin and olaquindox against the transformants were 2- to 32-fold (0.015–0.25 μg/mL) and 2- to > 32-fold (32 to >256 μg/mL) higher than the recipient E. coli DH5α strain (0.008 and 16 μg/mL), respectively. Furthermore, the co-transfer of resistance to ampicillin (n = 12), gentamycin (n = 10), tetracycline (n = 9), florfenicol (n = 9), sulfamethoxazole/trimethoprim (n = 11), cefotaxime (n = 3), and amikacin (n = 3) was observed in 12 transformants containing one plasmid. Multiple resistance genes were observed to coexist with oqxAB within the same plasmid accounting for the antimicrobial resistance; such as blaCTX−M−55, rmtB, fosA3, and floR were detected in two N1-F33:A-:B- plasmids from patients; aac(6′)-Ib-cr and/or qnrS were identified in both F-:A18:B- plasmids from pigs (Table 1). This allows for the co-selection of other genes conferring resistance to antibiotics that are routinely used in clinics, such as cephalosporins, aminoglycosides, and fosfomycin, or subject to selective pressure by quinolones, as well as quinoxalines used in food-producing animals. In turn, the co-selection of multiple drug resistance genes might facilitate further oqxAB transfer among Enterobacteriaceae isolates.
Plasmid Sequencing and Comparative Analysis
Two oqxAB-bearing IncHI2 plasmids in this study, namely pHNYJC8 from YJMC8 of food origin and pHNLDF400 from LDHF400 of human origin, were selected for sequencing. Plasmids pHNLDF400 and pHNYJC8 have sizes of 249,152 and 249,153 bp, respectively, and both have a GC content of 46.51%. They contain a typical IncHI2-type backbone, which encodes genes for plasmid replication, conjugative transfer, maintenance, and stability. Similar to other IncHI2 plasmids, such as pEC5207 (KT347600, porcine E. coli, China), pHNLDF400 and pHNYJC8 also carry a set of tellurite resistance determinants (terZABCDEF).
Sequence comparisons demonstrated that pHNYJC8 and pHNLDF400, obtained from the E. coli clones ST93 and ST57, respectively, were identical with only 2-bp nucleotide difference, thereby confirming that the same oqxAB-carrying IncHI2 plasmid was transferred horizontally among E. coli isolates in human and food samples. Furthermore, both plasmids showed 99% identity to the previously sequenced oqxAB-bearing IncHI2 plasmids pHXY0908 from S. Typhimurium in chicken from China and pHK0653 from clinical S. Typhimurium in Hong Kong (Wong et al., 2016), except that a 4,284-bp fragment (floR-ISCR2) was absent in pHK0653 (Figures 2, 3), further suggesting that the same or similar oqxAB-carrying IncHI2 plasmids are transmitted among Enterobacteriaceae isolates from various sources in different geographic regions of China. Additionally, the backbones of pHNLDF400 and pHNYJC8 showed 99% identity to those of our previously sequenced colistin-resistant mcr-1-bearing plasmids pHNSHP45-2 (KU341381) obtained from porcine E. coli isolate SHP45 in Shanghai, China (Zhi et al., 2016) and pHNAH67 (KX246266) obtained from an E. coli isolate from chicken in Anhui province, China. However, the abovementioned plasmids contain distinct variable regions. pHNSHP45-2 contains a 3,376-bp fragment harboring fosA3 and blaCTX−M−14. Contrastingly, in our study, both pHNLDF400 and pHNYJC8 contain a 4,713-bp segment harboring the |aac(6′)-Ib-cr|blaOXA−1|catB3 |arr3|qacEΔ1|sul| cassette array, which is flanked by the 3′-conserved segment (3′-CS) and interrupted by a Tn21-like transposon TnchrA, itself truncated by IS26. Most importantly, pHNSHP45-2 also harbors the colistin resistance gene, mcr-1, which is associated with ISApl1, and is inserted in the plasmid backbone (Figure 3). Similarly, a 6,760-bp fragment containing fosA3 and blaCTX−M−65 is identified in pHNAH67, whereas pHNLDF400 and pHNYJC8 contain a ~28.3-kb fragment with multiple resistance genes oqxAB, sul1, ΔaadA2, dfrA12, aphA1, sul3, aadA1a, cmlA1, and aadA2, which is absent in pHNAH67 (Figure 2). The absence/presence of these regions might be explained by IS26-mediated homologous recombination or replicative transposition (He S. et al., 2015). Furthermore, the oqxAB-associated composite transposon Tn6010 and the upstream fragment (3,678-bp) containing ΔTn2, IS26 (which interrupts the Tn2 segment), and the bleomycin resistance genes blms, orf63, and ΔISEnca1 (which is truncated by IS26 of Tn6010), were identical to the corresponding regions in the originally identified IncX1 oqxAB-carrying plasmid pOLA52, as well as pHXY0908, pHK0653, and pHNSHP45-2. The above mentioned results suggest that these plasmids may have originated from the same IncHI2-type plasmid, acquiring or losing various regions containing resistance genes, and spread among Enterobacteriaceae species in livestock, food products, and humans in different regions in China.
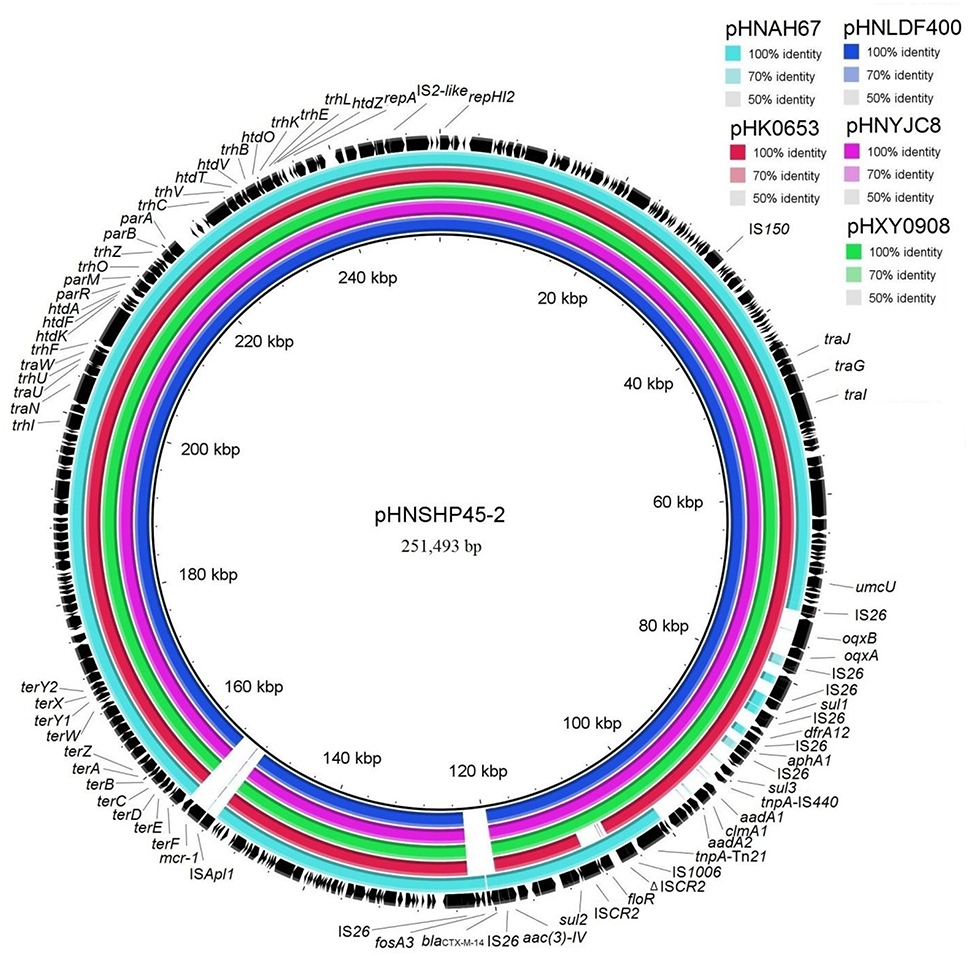
Figure 3. Sequence comparison of plasmids pHNLDF400 and pHNYJC8 with pHNSHP45-2 (GenBank accession number KU341381), pHXY0908 (KM877269), pHK0653 (KT335433), and pHNAH67 (KX246266) using BRIG. The reference sequence pHNSHP45-2 is shown in black.
Conclusion
In conclusion, our findings suggested that oqxAB dissemination among E. coli isolates from various sources could be due to horizontal transfer mediated by mobile elements, such as Tn6010, N1-F33:A-:B-, and IncHI2 plasmids. Identical oqxAB-carrying IncHI2 (ST3) plasmids were detected in the retail meat samples and human patients, similar to the previously described oqxAB-bearing IncHI2 plasmids pHXY0908 and pHK0653 from S. Typhimurium from chicken and patient. Thus, the transmission of similar oqxAB plasmids between animals, animal-derived food, and humans, and further human-to-human contact in communities and hospitals requires continued monitoring.
Ethics Statement
This study was carried out in accordance with the recommendation of ethical guidelines of South China Agricultural University. Individual written informed consent for the use of fecal or urine samples was obtained from all the patients and animal owners.
Author Contributions
JHL, ZZ, and JW conceived the study. CZ, XC, ZG, JL, JW, WL, XH, LZ, JH, YX, MY, and TH carried out the experiments. JW, CZ, and XC analyzed the data. JW wrote the manuscript. JHL and ZZ revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ni Song and Yan-Yan Liu for assistance with sample collection from patients. This work was supported in part by the grants from National Natural Science Foundation of China (No. 31272610), National Key Basic Research Program of China (No. 2013CB127200), and the National Science Fund for Distinguished Young Scholars (No. 31625026).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01982/full#supplementary-material
References
Abraham, S., Jordan, D., Wong, H. S., Johnson, J. R., Toleman, M. A., Wakeham, D. L., et al. (2015). First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J. Glob. Antimicrob. Resist. 3, 273–277. doi: 10.1016/j.jgar.2015.08.002
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 12:402. doi: 10.1186/1471-2164-12-402
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics. 9:75. doi: 10.1186/1471-2164-9-75
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Belmar Campos, C., Fenner, I., Wiese, N., Lensing, C., Christner, M., Rohde, H., et al. (2014). Prevalence and genotypes of extended spectrum beta-lactamases in Enterobacteriaceae isolated from human stool and chicken meat in Hamburg, Germany. Int. J. Med. Microbiol. 304, 678–684. doi: 10.1016/j.ijmm.2014.04.012
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chen, L., Chen, Z. L., Liu, J. H., Zeng, Z. L., Ma, J. Y., and Jiang, H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. doi: 10.1093/jac/dkm065
Ciesielczuk, H., Hornsey, M., Choi, V., Woodford, N., and Wareham, D. W. (2013). Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 62, 1823–1827. doi: 10.1099/jmm.0.064428-0
Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement. CLSI document M100-S25. Wayne, PA: CLSI.
Dower, W. J., Miller, J. F., and Ragsdale, C. W. (1988). High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids. Res. 16, 6127–6145.
Falgenhauer, L., Imirzalioglu, C., Ghosh, H., Gwozdzinski, K., Schmiedel, J., Gentil, K., et al. (2016). Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 47, 457–465. doi: 10.1016/j.ijantimicag.2016.03.019
García-Fernández, A., and Carattoli, A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. doi: 10.1093/jac/dkq101
Gautom, R. K. (1997). Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35, 2977–2980.
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hansen, L. H., Jensen, L. B., Sørensen, H. I., and Sørensen, S. J. (2007). Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60, 145–147. doi: 10.1093/jac/dkm167
Hansen, L. H., Johannesen, E., Burmølle, M., Sørensen, A. H., and Sørensen, S. J. (2004). Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 48, 3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004
Hansen, L. H., Sørensen, S. J., Jørgensen, H. S., and Jensen, L. B. (2005). The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb. Drug Resist. 11, 378–382. doi: 10.1089/mdr.2005.11.378
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio 6:e00762. doi: 10.1128/mBio.00762-15
He, T., Wang, Y., Qian, M., and Wu, C. (2015). Mequindox resistance and in vitro efficacy in animal-derived Escherichia coli strains. Vet. Microbiol. 177, 341–346. doi: 10.1016/j.vetmic.2015.04.007
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kao, C. Y., Wu, H. M., Lin, W. H., Tseng, C. C., Yan, J. J., Wang, M. C., et al. (2016). Plasmid-mediated quinolone resistance determinants in quinolone-resistant Escherichia coli isolated from patients with bacteremia in a university hosptial in Taiwan, 2001-2015. Sci. Rep. 6:32281. doi: 10.1038/srep32281
Kim, H. B., Wang, M., Park, C. H., Kim, E. C., Jacoby, G. A., and Hooper, D. C. (2009). oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. doi: 10.1128/AAC.01574-08
Kumar, S., Nei, M., Dudley, J., and Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics. 9, 299–306. doi: 10.1093/bib/bbn017
Li, L., Liao, X., Yang, Y., Sun, J., Li, L., Liu, B., et al. (2013). Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 68, 2263–2268. doi: 10.1093/jac/dkt209
Liu, B. T., Liao, X. P., Yang, S. S., Wang, X. M., Li, L. L., Sun, J., et al. (2012). Detection of mutations in the gyrA and parC genes in Escherichia coli isolates carrying plasmid-mediated quinolone resistance genes from diseased food-producing animals. J. Med. Microbiol. 61, 1591–1599. doi: 10.1099/jmm.0.043307-0
Liu, B. T., Yang, Q. E., Li, L., Sun, J., Liao, X. P., Fang, L. X., et al. (2013). Dissemination and characterization of plasmids carrying oqxAB-blaCTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE 8:e73947. doi: 10.1371/journal.pone.0073947
Loncaric, I., Stalder, G. L., Mehinagic, K., Rosengarten, R., Hoelzl, F., Knauer, F., et al. (2013). Comparison of ESBL- and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS ONE 8:e84048. doi: 10.1371/journal.pone.0084048
Müller, A., Stephan, R., and Nüesch-Inderbinen, M. (2016). Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Tot. Environ. 541, 667–672. doi: 10.1016/j.scitotenv.2015.09.135
Röderova, M., Halova, D., Papousek, I., Dolejska, M., Masarikova, M., Hanulik, V., et al. (2017). Chracteristics of quinolone resistance in Escherichia coli isolates from humans, animals, and the environment in the Czech Republic. Front. Microbiol. 7:2147. doi: 10.3389/fmicb.2016.02147
Ruiz, J., Pons, M. J., and Gomes, C. (2012). Transferable mechanisms of quinolone resistance. Int. J. Antimicrob. Agents 40, 196–203. doi: 10.1016/j.ijantimicag.2012.02.011
Sørensen, A. H., Hansen, L. H., Johannesen, E., and Sørensen, S. J. (2003). Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Chemother. 47, 798–799. doi: 10.1128/AAC.47.2.798-799.2003
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wong, M. H., Chan, E. W., Liu, L. Z., and Chen, S. (2014). PMQR genes oqxAB and aac(6')Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 5:521. doi: 10.3389/fmicb.2014.00521
Wong, M. H., Chan, E. W., Xie, L., Li, R., and Chen, S. (2016). IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella spp. Antimicrob. Agents Chemother. 60, 6911–6915. doi: 10.1128/AAC.01555-16
Xu, G., An, W., Wang, H., and Zhang, X. (2015). Prevalence and characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front. Microbiol. 6:1103. doi: 10.3389/fmicb.2015.01103
Xu, X., Cui, S., Zhang, F., Luo, Y., Gu, Y., Yang, B., et al. (2014). Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses and Ground Pork, China. Microb. Drug Resist. 20, 73–81. doi: 10.1089/mdr.2012.0224
Yang, T., Zeng, Z., Rao, L., Chen, X., He, D., Lv, L., et al. (2014). The association between occurrence of plasmid-mediated quinolone resistance and ciprofloxacin resistance in Escherichia coli isolates of different origins. Vet. Microbiol. 170, 89–96. doi: 10.1016/j.vetmic.2014.01.019
Yang, X., Liu, W., Liu, Y., Wang, J., Lv, L., Chen, X., et al. (2014). F33:A-:B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX–M–55/–14/–65 Escherichia coli from chickens in China. Front. Microbiol. 5:688. doi: 10.3389/fmicb.2014.00688
Yuan, J., Xu, X., Guo, Q., Zhao, X., Ye, X., Guo, Y., et al. (2012). Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 67, 1655–1659. doi: 10.1093/jac/dks086
Zhao, J., Chen, Z., Chen, S., Deng, Y., Liu, Y., Tian, W., et al. (2010). Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 54, 4219–4224. doi: 10.1128/AAC.00139-10
Zhao, L., Zhang, J., Zheng, B., Wei, Z., Shen, P., Li, S., et al. (2015). Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J. Clin. Microbiol. 53, 766–770. doi: 10.1128/JCM.02594-14
Keywords: antimicrobial resistance, Escherichia coli, food safety, plasmids, PMQR
Citation: Wang J, Zhi C-P, Chen X-J, Guo Z-W, Liu W-L, Luo J, Huang X-Y, Zeng L, Huang J-W, Xia Y-B, Yi M-Y, Huang T, Zeng Z-L and Liu J-H (2017) Characterization of oqxAB in Escherichia coli Isolates from Animals, Retail Meat, and Human Patients in Guangzhou, China. Front. Microbiol. 8:1982. doi: 10.3389/fmicb.2017.01982
Received: 04 June 2017; Accepted: 26 September 2017;
Published: 13 October 2017.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Liang Li, Los Angeles Biomedical Research Institute, United StatesAngela Novais, UCIBIO. Faculdade de Farmácia da Universidade do Porto, Portugal
Copyright © 2017 Wang, Zhi, Chen, Guo, Liu, Luo, Huang, Zeng, Huang, Xia, Yi, Huang, Zeng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Hua Liu, jhliu@scau.edu.cn
†Present Address: Xiao-Jie Chen, National Feed Drug Reference Laboratories, Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
Wu-Ling Liu, State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University, Guizhou, China
Li Zeng, Foshan Standard BioTech Co., Ltd., Foshan, Guangdong, China
‡These authors have contributed equally to this work.
 Jing Wang‡
Jing Wang‡ Wu-Ling Liu
Wu-Ling Liu Zhen-Ling Zeng
Zhen-Ling Zeng Jian-Hua Liu
Jian-Hua Liu