- 1Department of Biochemistry and Molecular Biology, Colorado State University, Fort Collins, CO, United States
- 2Department of Biochemistry and Molecular Genetics, University of Colorado School of Medicine, Aurora, CO, United States
- 3Department of Microbiology, Ohio State University, Columbus, OH, United States
- 4Biomolecular Labeling Laboratory, Institute for Bioscience and Biotechnology Research, National Institute of Standards and Technology and the University of Maryland, Rockville, MD, United States
The initiation of DNA replication is typically tightly regulated by proteins that form initiation complexes at specific sequences known as replication origins. In Archaea and Eukaryotes, Cdc6, a near-universally conserved protein binds and facilitates the origin-dependent assembly of the replicative apparatus. TK1901 encodes Cdc6 in Thermococcus kodakarensis but, as we report here, TK1901 and the presumed origin of replication can be deleted from the genome of this hyperthermophilic Archaeon without any detectable effects on growth, genetic competence or the ability to support autonomous plasmid replication. All regions of the genome were equally represented in the sequences generated by whole genome sequencing of DNA isolated from T. kodakarensis strains with or without TK1901, inconsistent with DNA initiation occurring at one or few origins, and instead suggestive of replication initiating at many sites distributed throughout the genome. We were unable to generate strains lacking the recombination factors, RadA or RadB, consistent with T. kodakarensis cells, that are oligoploid (7–19 genomes per cell), employing a recombination-based mechanism of DNA replication. Deletion of the previously presumed origin region reduced the long-term viability of cultures supporting the possibility that retaining an origin-based mechanism of DNA initiation provides a survival mechanism for stationary phase cells with only one genome.
Introduction
DNA replication is fundamental for cellular life and although there are differences in the details, the initiation of genome replication has common features in Bacteria, Archaea, and Eurkaryotes. An initiator protein or protein complex recognizes and assembles at one (all Bacteria and some Archaea) or multiple sites (some Archaea and all Eukaryotes) that function as origins of replication (Jacob et al., 1963). Under exceptional circumstances, initiator protein-independent genome replication, termed recombination-driven DNA replication initiation (RDR; also termed inducible and constitutive stable DNA replication) has been documented in Bacteria, but such mechanisms support—at best—only minimal cell growth (Ogawa et al., 1984; Masai et al., 1994; Kogoma, 1997; Maduike et al., 2014). It was surprising then when Hawkins et al. (2013) proposed that RDR not only facilitates genome replication but supports faster-than-wild type growth of a strain of the halophilic archaeon, Haloferax volcanii from which they had genetically deleted all four of the recognized origins of genome replication. Consistent with RDR initiation, the recombination factor RadA was essential for viability of the origin-less strain but could be deleted from the genome of the parental, origins-containing H. volcanii.
Given the established and convincingly large body of evidence that archaeal genomes have defined origins recognized and bound by initiator proteins (Matsunaga et al., 2001, 2007, 2010; Norais et al., 2007; Wigley, 2009; Kawakami and Katayama, 2010; Beattie and Bell, 2011; Scholefield et al., 2011; Pelve et al., 2013), the proposal that RDR supports rapid growth in an archaeon (Hawkins et al., 2013) is unique and challenging. Most archaea encode replication initiator proteins that are homologous to eukaryotic initiation factors Orc1 and Cdc6, and one or more Cdc6-encoding genes are present in almost all sequenced archaeal genomes, usually located adjacent to a known or predicted origin(s) of replication (Robinson and Bell, 2005; Barry and Bell, 2006; Dueber et al., 2011; Bell, 2012; Makarova and Koonin, 2013; Samson et al., 2013; Arora et al., 2014; Wu et al., 2014; Cossu et al., 2015). An increase in the number of Cdc6 proteins is often positively correlated with the number of replication origins (Samson et al., 2013); H. volcanii encodes fourteen Cdc6 proteins that function at three chromosomal origins and an integrated viral origin (Norais et al., 2007). Some species are reliant on a single encoded, or only a single-functional Cdc6 protein to initiate replication, and the remaining Cdc6 isoforms are predicted to play roles in transcription regulation, recombination, replication restart, or negative regulation of replication initiation (Ausiannikava and Allers, 2017).
To address the roles of Cdc6, presumptive origin sequences, and the potential of RDR to support rapid growth of archaeal strains, we took advantage of a procedure that permits the precise deletion of non-essential genome sequences and provides strong statistical evidence for essential genes in the hyperthermophilic archaeon Thermococcus kodakarensis (Hileman and Santangelo, 2012). Employing similar techniques, several essential and some surprisingly non-essential genes have already been identified revealing unanticipated features in archaeal DNA replication (Li et al., 2010, 2014; Pan et al., 2011, 2013; Cubonova et al., 2013). Bioinformatic analysis including GC-skew and Z-curve analysis predict only one origin of replication located directly adjacent to the gene encoding Cdc6 in T. kodakarensis (Fukui et al., 2005; Ojha and Swati, 2010; Cossu et al., 2015). Most members of the Thermococcales, including T. kodakarensis, encode only one identifiable Cdc6 protein (Makarova and Koonin, 2013). We now report that Cdc6 and the adjacent previously-presumed origin of replication can be deleted from T. kodakarensis with no detectable consequences for viability, growth, genetic competence, or plasmid maintenance. Data obtained by whole genome sequence and marker frequency analyses (Xu et al., 2012), coupled with the apparent essentiality of RadA and RadB, provide strong evidence that T. kodakarensis normally employs a RDR mechanism for initiation that occurs at many sites around the genome.
Results
Construction of T. kodakarensis Δcdc6
The procedure employed to delete genes from the T. kodakarensis genome permits a statistical definition of essentiality (Hileman and Santangelo, 2012). Plasmids are constructed and used to transform a parental strain (here T. kodakarensis TS559) so that the target locus is flanked by two sets of direct repeats. Spontaneous recombination in this intermediate strain between one set of the repeats results in the markerless deletion of the target locus, whereas an equally-probable recombination between the second set of repeats regenerates the parental strain. When only the parental strain is recovered, after screening >30 isolates generated from at least two independently-constructed intermediate strains, the target locus is defined operationally as essential for T. kodakarensis viability under our laboratory conditions.
TK1901-TK1902-TK1903 (encoding Cdc6, DNA polymerase D small and large subunits, respectively) form an operon (Jager et al., 2014) and essentiality has been previously established for TK1902 and TK1903 (Figure 1; Cubonova et al., 2013). Surprisingly, this was not true for TK1901, the only gene in T. kodakarensis that encodes a recognizable Cdc6 homolog. The design of the plasmid constructed to delete TK1901 ensured retention of the upstream promotor and so continued expression of TK1902-TK1903, and avoided deletion of any sequences in the adjacent ~900 bp region predicted to contain the origin of replication based on homology with the origin region in Pyrococcus furiosus, a related member of the Thermococcales (Figure 1; Farkas et al., 2011; Cossu et al., 2015).
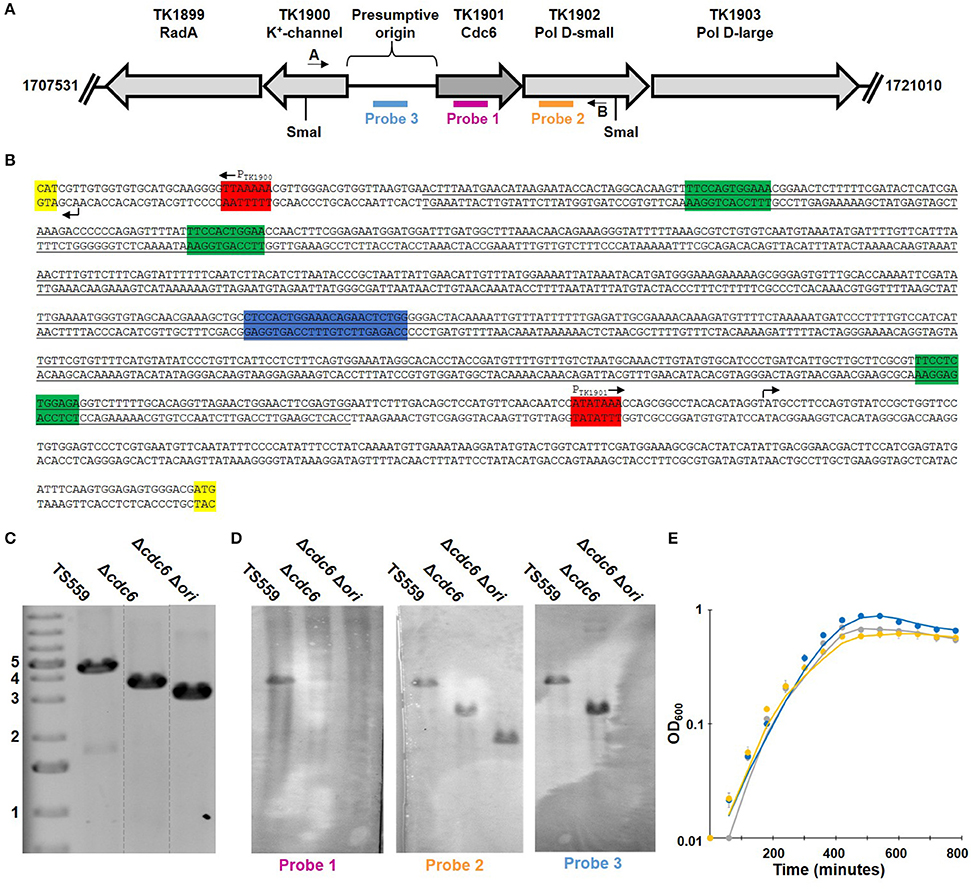
Figure 1. Deletion of TK1901 and the presumptive origin of replication from T. kodakarensis is non-phenotypic. (A) Organization of the T. kodakarensis genome surrounding TK1901. The locations of sequences used as primers in PCRs, probes in Southern blotting and SmaI recognition sites are shown. (B) Sequence of the presumptive origin region of T. kodakarensis with the ORB (blue), mini-ORBs (green), promoters (red), and the first codon of the gene (yellow) marked. The full sequence of cdc6 plus the underlined nucleotides were deleted from the genome in T. kodakarensis Δcdc6 Δori. (C) PCR generate amplicons confirm deletion of TK1901, as well as TK1901 and the presumptive origin from T. kodakarensis Δcdc6 and Δcdc6 Δori, respectively. (D) Southern blots of SmaI-digested genomic DNA from T. kodakarensis TS559, Δcdc6, and Δcdc6 Δori confirm deletion of cdc6, as well as cdc6 and the presumptive origin, respectively. (E) Deletion of cdc6 or cdc6/ori does not affect laboratory growth of T. kodakarensis TS559 (gray), Δcdc6 (blue), and Δcdc6 Δori (yellow). Error bars report standard error of the mean of three biological replicates grown in triplicate.
The presence of the TK1901 deletion was confirmed by diagnostic PCR and Southern blotting in two independent isolates (Figure 1). Amplicon sequencing confirmed that the 1,248 bp deletion extended precisely from the ATG-start codon to TGA-stop codon of TK1901, and this was subsequently re-confirmed by deep-sequencing (see below) of the entire genome of one isolate, designated T. kodakarensis Δcdc6. This isolate was phenotypically similar to the parental T. kodakarensis TS559 strain: cultures grew at similar rates and reached the same final cell densities (Figure 1) and T. kodakarensis Δcdc6 was genetically competent and supported the autonomous replication of pTN1-based plasmids (Santangelo et al., 2008).
Marker Frequency Analysis of Genomic DNA
Regions adjacent to an origin(s) are over-represented in growing cells, and marker frequency analyses comparing the number of sequencing reads across the genome has been used to identify replication origin(s) in many archaeal genomes (Andersson et al., 2010; Hawkins et al., 2013; Pelve et al., 2013). Genomic DNA was therefore isolated, fragmented, and deep-sequenced from growing and stationary phase cultures of T. kodakarensis TS559 and Δcdc6. The sequences obtained confirmed the deletion of TK1901 but, in repeated experiments, all regions of the genome were equally represented in the DNA reads from both growing and stationary phase cells of both T. kodakarensis TS559 and Δcdc6 (Figure 2 and Supplementary Figure 1). Given this unanticipated result, to provide confidence in the laboratory and computational procedures, the experiments were repeated with genomic DNAs from Escherichia coli MG1655 and P. furiosus strain JFW02(Farkas et al., 2012), species with established origins of replication. Quantification of the whole genome sequencing (WGS) reads clearly and correctly identified the origin loci established in the genomes of E. coli and P. furiosus (Supplementary Figure 2; Burland et al., 1993; Farkas et al., 2012).
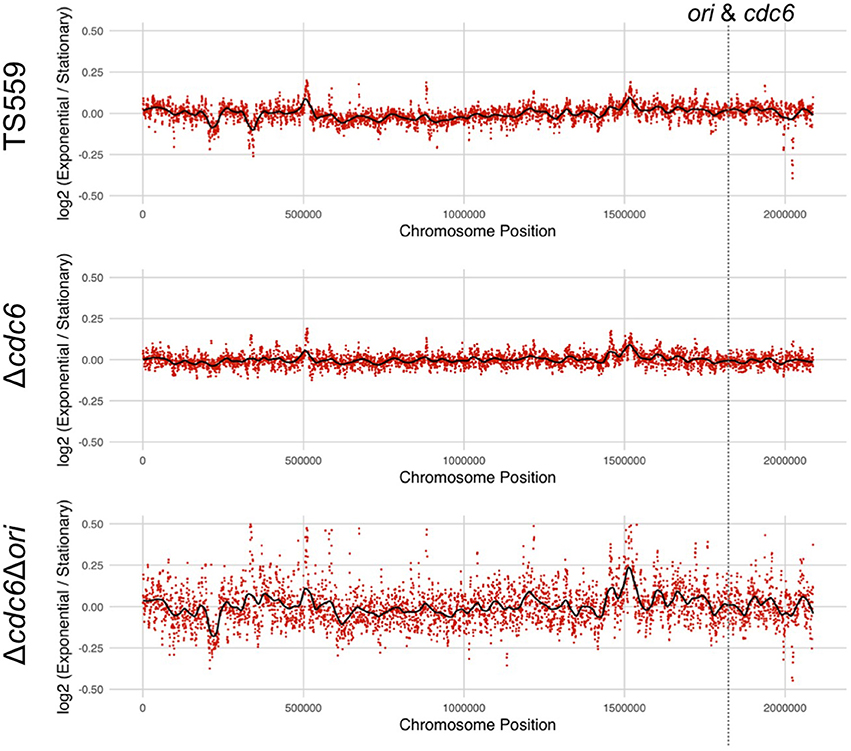
Figure 2. Marker frequency analysis of DNA sequence reads from T. kodakarensis strains fails to identify a defined origin(s) of replication. The log2 ratio of each nucleotide in sequences from exponentially growing cells divided by sequence from stationary phase cells is shown in each panel. Frequencies were calculated using 1 Kbp intervals (red dots) with a sliding window of 500 bp generating the average frequency shown in black. The location of the ori-cdc6 region is indicated.
Construction of T. kodakarensis Δcdc6 Δori
Using the same markerless-genome modification techniques, the presumed origin sequences (Ojha and Swati, 2010) were easily deleted in T. kodakarensis Δcdc6 (Figure 1). All the bioinformatically identified mini-origin recognition boxes (mini-ORBs; green) and one full ORB (blue) were deleted while retaining the promoters (red), transcription start sites (arrows), and translation start sites (yellow) for TK1902-1903, and for TK1900 (Figure 1). Both amplicon- and whole genome deep-sequencing confirmed the precision of the deletion. A representative isolate, designated T. kodakarensis Δcdc6 Δori, was phenotypically indistinguishable from T. kodakarensis TS559 and Δcdc6. All three strains grew at the same rate, achieved the same final cell densities, were genetically competent and supported plasmid replication. Quantification of WGS reads also failed to identify any preferred origin(s) sequences and indicated that replication was initiated at many sites around the genome of T. kodakarensis Δcdc6 Δori (Figure 2).
Spontaneous Genome Deletion and Inversion
Although, the WGS reads did not identify origins of replication, with >2,000x genome coverage, they did identify spontaneous recombination events at two locations in subpopulations (<10%) of T. kodakarensis TS559 cells (Figure 3). The recombinations inverted an ~150 kbp region or excised ~100 kbp, and these events resulted in small spikes and dips in the marker frequency analyses at sites previously established to contain vestigial prophage TKV2 and TKV3 genomic sequences (Figure 3; Fukui et al., 2005; Tagashira et al., 2013). Only a small number of sequences were obtained that extended across the sites of recombination, (Figure 3) but these were more prevalent in DNAs isolated from growing than from stationary phase cells. Based on PCR amplicons, these recombination events also occurred in T. kodakarensis Δcdc6 and Δcdc6 Δori. As deletion of TKV3 severely hinders growth (Tagashira et al., 2013) and several presumably essential genes are within the deleted sequences, these recombination events are likely lethal and prevent the effected cells from contributing to continued culture growth.
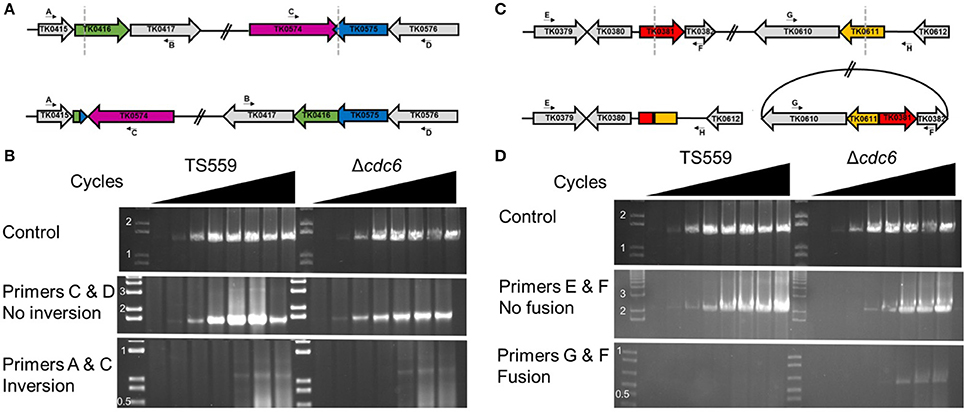
Figure 3. Semi-quantitative PCR supports large-scale genomic rearrangements. (A) Representations of TS559 (top) and inverted (bottom) genome structures. The end points of the inversion events are marked with dotted gray lines in the TS559 genome representation. (B) Semi-quantitative PCRs demonstrate the inversion genome rearrangements identified from the WGS data. The presence of the inversion was identified in <10% of the samples. (C) Representations of TS559 (top) and fusion-event (bottom) genome structures. The end points of the fusion events are marked with dotted gray lines in the TS559 genome representation. (D) Semi-quantitative PCRs demonstrate the fusion event identified from the WGS data in <10% of the samples.
Mutations Acquired by T. kodakarensis Since Isolation
T. kodakarensis KOD1 has been the focus of continuous research since its isolation in 1994 (Morikawa et al., 1994). It seemed possible therefore that the cdc6- and origin-independent replication of T. kodakarensis TS559 might result from mutations acquired and selected during laboratory culture. The genome sequence determined here for T. kodakarensis TS559 was therefore compared with that published for T. kodakarensis KOD1 (Fukui et al., 2005). All the changes known to have been intentionally introduced to generate T. kodakarensis TS559 from T. kodakarensis KOD1 were present, and although there were no large genome rearrangements, an additional 35 single nucleotide differences were identified. Some of these changes are within open reading frames, but none would be predicted to radically change DNA replication or recombination (Table 1).
Why Are Cdc6 and the Origin-Sequences Retained?
T. kodakarensis is oligoploid (7–19 genomes/cell; Spaans et al., 2015) and this is consistent with the use of RDR. Nutrient-stress, the absence of defined DNA segregation strategies, and the potential for continued cell division without DNA replication may occasionally result in T. kodakarensis cells with only one genome. Such cells would be unable to restart growth by RDR initiation but could do so if a cdc6-oriC system of replication initiation was also available. To evaluate whether retention of cdc6 or presumptive origin sequences promotes long-term viability, aliquots were taken from stationary phase cultures maintained at 85°C for extended periods without nutrient addition, and assayed for viability. Extended (several months) incubation at elevated temperatures in nutrient poor conditions was predicted to deplete energy reserves, introduce stress into the genome, and potentially reduce ploidy as genomes were consumed to provide nutrients. Cells in cultures of T. kodakarensis TS559 and Δcdc6, strains with the presumed origin (oriC) region, were viable for ~40 days longer than cells in cultures of T. kodakarensis Δori (Figure 4). Retention of Cdc6, however, did not influence long-term viability.
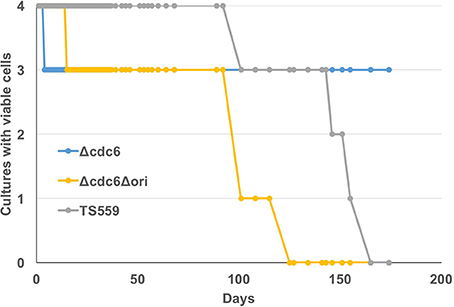
Figure 4. Presence of the origin region increases long-term viability. Four biological replicates of T. kodakarensis TS559 (gray), Δcdc6 (blue), and Δcdc6Δori (yellow) were grown to stationary phase and incubation was continued without additions to the medium for >170 days. Aliquots were removed at intervals and used to inoculate fresh growth media. The number of cultures with viable cells that generated progeny cultures is plotted against days of incubation at 85°C.
TK1899 (RadA) and TK2231 (RadB) Are Essential Genes
RadA was required for growth—presumably by RDR initiation—of the origin-deleted halophilic H. volcanii strains but non-essential in the parental, origins-containing strain (Hawkins et al., 2013). Despite repeated attempts with different transforming DNA strategies, we were unable to generate T. kodakarensis strains with RadA (TK1899) or RadB (TK2231) deleted, regardless of the retention or absence of cdc6 and/or origin sequences.
Discussion
TK1901 encodes what appears to be a fully functional Cdc6 protein, with intact Walker A and Walker B motifs, DNA, and ATP-binding domains. TK1901 is co-transcribed with TK1902 and TK1903, essential genes that encode the subunits of DNA polymerase D (Jager et al., 2014) and is located immediately adjacent to a region with sequences very similar to those of the origin of replication in P. furiosus. Nevertheless, the results reported establish conclusively that TK1901 and so Cdc6 are not required for T. kodakarensis viability and the absence of Cdc6 has no detectable effect on laboratory growth, genetic competence, or the ability to support autonomous plasmid replication. Equally surprising, the previously presumed origin of replication can also be deleted without any detectable phenotypic consequence. Consistent with Cdc6 and oriC having no essential roles, marker frequency analyses of WGS data obtained from cultures of T. kodakarensis TS559, Δcdc6, and Δcdc6 Δori provided no evidence for origin-dependent replication initiation, even when the origin and the recognition protein were both present. Given the depth of the WGS, any regional over-representation of reads, consistent with initiation at as many as 5 separate locations would have been detected, but this was not the case. In contrast, the WGS results argue for genome replication in T. kodakarensis TS559 being initiated at many sites distributed around the genome, consistent with the proposal for RDR-dependent genome replication in the H. volcanii strain with all origins of replication deleted (Hawkins et al., 2013). We extend this proposal to suggest that RDR supports growth of T. kodakarensis despite the presence of a predicted origin. In support of this assertion, despite a considerable effort, we were unable to generate T. kodakarensis strains with TK1899 (RadA) or TK2231 (RadB) deleted. In E. coli, long-lived R-loops accumulate in strains lacking RNase H, and these R-loops can facilitate initiator protein (DnaA) independent constitutive stable DNA replication, but their growth is very slow (Masai et al., 1994; Masai and Arai, 1996; Maduike et al., 2014). The T. kodakarensis strains investigated here all express TK0805, the gene that encodes RNase HII (Heider et al., 2017) and thus R-loop accumulation is unlikely to be responsible for origin-independent genome replication in T. kodakarensis.
Employing RDR for genome replication could also explain why T. kodakarensis is atypically naturally competent and so amenable to genetic manipulation. Additional features of T. kodakarensis are consistent with RDR. In genomes replicated from distinct origins, highly expressed genes are transcribed predominantly in the same direction as replication fork movement (Smith et al., 2007; Andersson et al., 2010; Paul et al., 2013; Cossu et al., 2015). But, if bidirectional replication was initiated from the previously presumed origin in T. kodakarensis, 628 transcripts would be transcribed with, and 626 would be transcribed against the direction of the replisome movement (Fukui et al., 2005; Jager et al., 2014; Cossu et al., 2015). A computational search also failed to identify any location, and so a putative origin, anywhere on the genome that would result in transcription and DNA replication occurring predominantly in the same direction (Cossu et al., 2015). Often, when cloned, an origin of genome replication will still function and can be used to construct self-replicating plasmids. This is the case for the origin of replication cloned from P. furiosus (Farkas et al., 2011) and the generated plasmids replicate not only in the cytoplasm of P. furiosus but also in T. kodakarensis. In contrast, cloning many variants of the very similar presumed origin region from the T. kodakarensis genome did not result in a replicating plasmid.
RDR initiation requires the retention of more than one genome, and it is now well-established that members of the Euryarchaeota including T. kodakarensis, are naturally oligoploid (Breuert et al., 2006; Hildenbrand et al., 2011; Spaans et al., 2015). To date, however, there is no evidence for precise genome segregation strategies suggesting that growing cultures will naturally produce cells with varying ploidy—including monoploid cells. If cells occur with only one genome, then the presence of an origin-dependent initiation module would provide a survival mechanism. With this in mind, we demonstrated that cultures of T. kodakarensis TS559 and Δcdc6 did retain viability longer than T. kodakarensis Δcdc6 Δori. The signature of an origin suggests that T. kodakarensis has relied on an origin-dependent replication strategy during its evolutionary history (Ojha and Swati, 2010). Retention of Cdc6 did not influence survival under identical conditions, suggesting that Cdc6 may not be necessary for use of the presumptive origin sequences.
The presence of ≥~20 genomes per T. kodakarensis cell (Spaans et al., 2015) raises challenging questions as to how they are all accommodated and replicated within a generation time of ~40 min. As established for the DNA clamp (PCNA1; >1,000 molecules/cell; Kuba et al., 2012; Pan et al., 2013), at minimum, the replisome components must be present at very high levels and maybe this also facilitates simultaneous replication from many sites around the T. kodakarensis genome. How the replicative apparatus is assembled and how simultaneous rounds of replication are prohibited or accommodated during rapid growth remain outstanding questions.
Materials and Methods
Growth of Microorganisms
T. kodakarensis strains were grown in artificial seawater (ASW) supplemented with 5 g/L of both yeast extract and tryptone (YT) and 2 g/L of sulfur (S°) or 5 g/L sodium pyruvate (Pyr) at 85°C. P. furiosus strain JFW02 (Farkas et al., 2012) was grown at 95°C as described using maltose as a carbon source (Adams et al., 2001). E. coli strain MG1655 was grown in Luria-Bertani (LB) broth at 37°C. The growth of cultures was measured by increases in optical density at 600 nm (OD600). T. kodakarensis cultures were harvested at an OD600 of 0.2 (early exponential) and 0.6 (late-exponential) while the stationary phase cells were harvested at an OD600 of ~1.0. In Figure 4, four independent cultures of each T. kodakarensis strain were maintained at 85°C in sealed vessels with no additions to the cultures over ~180 days. Loss of culture viability was defined as the inability of aliquots, sampled multiple times over 3 days, to initiate culture growth when inoculated into fresh medium. The inability of 9 or more individual aliquots, removed from cultures over 3 consecutive days, to support outgrowth confirmed that these cultures had lost all viable CFUs.
Strain Construction of T. kodakarensis
Standard procedures (Hileman and Santangelo, 2012) were used to construct plasmids, pOSUTK1901B and pJG4 respectively, that were used to delete TK1901 or TK1901 plus the origin sequences from T. kodakarensis TS559. In the resulting strains, T. kodakarensis Δcdc6 and Δcdc6 Δori, the Cdc6 encoding sequence [TK1901; 1,248 bp] was deleted but the promoter for the TK1901-TK1903 operon was retained to sure expression of TK1902-1903. Similarly, in T. kodakarensis Δcdc6 Δori, the putative origin (640 bp) with one origin recognition box (ORB) and three mini-ORBs were deleted, but the promoters for TK1901-1903 and TK1900 were retained (Figure 1). Use of the same procedures, and plasmids designed to precisely delete the TK1899 (RadA) and TK2231 (RadB) sequences, did not generate viable strains with the desired deletions.
Isolation of Genomic DNA
Genomic DNA was isolated from all strains as described (Santangelo et al., 2007). Cells pelleted from cultures at the designated OD600 were resuspended in 10% (w/v) sucrose, 20 mM Tris-HCl pH 8.0, and 5 mM EDTA. SDS (2% final concentration) and proteinase K (0.25 mg/ml) were added to the resulting lysate and the mixture was incubated for 1 h at 55°C. NaCl (1 M final concentration) was then added, the mixture was chilled, centrifuged, and an equal volume of isopropanol was added to the clarified supernatant. The precipitated nucleic acids were pelleted, resuspended with 10 mM Tris-HCl pH 8.0, 50 μg of RNase A added and incubation continued at 37°C for 30 min. The DNA remaining was further purified by repeated phenol/chloroform/isoamyl alcohol (25:24:1) extraction and an alcohol precipitation.
Southern Blotting
The procedure used has been previously described (Cubonova et al., 2013). In Figure 1, TK1901 (Probe 1, pink) is only detectable in strain TS559 whereas origin sequences (Probe 3, blue) are detectable in strains TS559 and Δcdc6, but not strain Δcdc6 Δori; probe 3 highlights a smaller product in Δcdc6 that reflects deletion of TK1901. Probe 2 (orange) detected the sequences encoding TK1902 in all samples and the fragment lengths identified are appropriate for the corresponding strains.
Whole Genome Sequencing (WGS)
Sequencing libraries were prepared using TruSeq DNA library preparation kits (Illumina, San Diego, CA) and were subjected to WGS (pair-ended, 2 × 150 bp per read; 1 × 125 bp reads for E. coli) on an Illumina Hi-Seq 2000 platform (University of Colorado Denver Genomics and Microarray Core Facility). Individual genome coverages ranged from 3,300× to 7,800×.
Comparison of Genome Sequences
The reference genome for Thermococcus kodakarensis KOD1 (https://www.ncbi.nlm.nih.gov/nuccore/NC_006624.1) was downloaded from Genbank and manually edited to account for the laboratory manipulations made in the lineage leading to T. kodakarensis TS559. The reference genomes for P. furiosus COM1 and Escherichia coli MG1655 were downloaded from Genbank here (https://www.ncbi.nlm.nih.gov/nuccore/CP003685 and https://www.ncbi.nlm.nih.gov/nuccore/NC_000913.3, respectively).
The T. kodakarensis TS559 and KOD1 genomes were aligned and differences identified by using Universal Genotyper (GATK version v2.1-8; Van der Auwera et al., 2013) and MUMmer (version 3.1; Kurtz et al., 2004). Low quality sequences, regions with <5x coverage and a small number of variants identified in DNA from only growing or stationary phase cells of the same isolate were not included. The coordinates of the RefSeq GFF file were updated to account for identified insertions and deletions, and the resulting variants annotated using snpEff (version 4.0e; Cingolani et al., 2012).
Alignment and Marker Frequency Analysis of Whole Genome Sequences
An index was built for each reference genome with bowtie2-build (version 2.2.9; Langmead and Salzberg, 2012) using default settings. Illumina adaptor sequences and low quality bases (quality score <10) were trimmed from the 3′-end of each read using cutadapt (version 1.11; Martin, 2011) with reads discarded if more than half the bases were trimmed. The filtered reads were aligned to the reference genome using bowtie2 (version 2.2.9; Langmead and Salzberg, 2012) selecting the best alignment for each read. The alignment statistics are reported in Supplementary Table 1. Reads that did not align as proper pairs were treated as single end reads. For copy number estimation, each reference genome was binned into 1 kb windows with a 500 bp sliding overlap between windows using bedtools (version 2.17.0; Quinlan and Hall, 2010). For each sample, the coverage for each 1 kb was calculated as the number of sequenced bases that overlap with that window. The %G+C content of each window was calculated, and corrections made for potential bias in library preparation and sequencing due to GC content. The average coverage for each GC bin was plotted against the GC content and smoothed by a Lowess regression model. A correction factor, calculated by dividing the global mean coverage by the fitted model, was applied to each sample. Source code available at http://github.com/dpastling/plethora.
Author Contributions
JR, ZK, and TS conceived and directed the project. AG, DA, JR, and TS wrote the manuscript and prepared figures. AG, RM, BB constructed and phenotypically characterized strains. AG, DA, and KJ analyzed the WGS data.
Funding
This work was supported by grants from the National Institutes of Health (R01-GM100329 to TS; R24-GM098176 to JR and TS) and from the Department of Energy (004010-00002 to TS).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Certain commercial equipment, instruments, or materials are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified is necessarily the best available for the purpose.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02084/full#supplementary-material
References
Adams, M. W., Holden, J. F., Menon, A. L., Schut, G. J., Grunden, A. M., Hou, C., et al. (2001). Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183, 716–724. doi: 10.1128/JB.183.2.716-724.2001
Andersson, A. F., Pelve, E. A., Lindeberg, S., Lundgren, M., Nilsson, P., and Bernander, R. (2010). Replication-biased genome organisation in the crenarchaeon Sulfolobus. BMC Genomics 11:454. doi: 10.1186/1471-2164-11-454
Arora, J., Goswami, K., and Saha, S. (2014). Characterization of the replication initiator Orc1/Cdc6 from the Archaeon Picrophilus torridus. J. Bacteriol. 196, 276–286. doi: 10.1128/jb.01020-13
Ausiannikava, D., and Allers, T. (2017). Diversity of DNA replication in the Archaea. Genes 8:56. doi: 10.3390/genes8020056
Barry, E. R., and Bell, S. D. (2006). DNA Replication in the Archaea. Microbiol. Mol. Biol. Rev. 70, 876–887. doi: 10.1128/MMBR.00029-06
Beattie, T. R., and Bell, S. D. (2011). Molecular machines in archaeal DNA replication. Curr. Opin. Chem. Biol. 15, 614–619. doi: 10.1016/j.cbpa.2011.07.017
Bell, S. D. (2012). Archaeal orc1/cdc6 proteins. Subcell. Biochem. 62, 59–69. doi: 10.1007/978-94-007-4572-8_4
Breuert, S., Allers, T., Spohn, G., and Soppa, J. (2006). Regulated polyploidy in halophilic archaea. PLoS ONE 1:e92. doi: 10.1371/journal.pone.0000092
Burland, V., Plunkett, G., Daniels, D. L., and Blattner, F. R. (1993). DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics 16, 551–561. doi: 10.1006/geno.1993.1230
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang le L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 80–92. doi: 10.4161/fly.19695
Cossu, M., Da Cunha, V., Toffano-Nioche, C., Forterre, P., and Oberto, J. (2015). Comparative genomics reveals conserved positioning of essential genomic clusters in highly rearranged Thermococcales chromosomes. Biochimie 118, 313–321. doi: 10.1016/j.biochi.2015.07.008
Cubonova, L., Richardson, T., Burkhart, B. W., Kelman, Z., Connolly, B. A., Reeve, J. N., et al. (2013). Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J. Bacteriol. 195, 2322–2328. doi: 10.1128/JB.02037-12
Dueber, E. C., Costa, A., Corn, J. E., Bell, S. D., and Berger, J. M. (2011). Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 39, 3621–3631. doi: 10.1093/nar/gkq1308
Farkas, J., Chung, D., DeBarry, M., Adams, M. W., and Westpheling, J. (2011). Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 77, 6343–6349. doi: 10.1128/AEM.05057-11
Farkas, J., Stirrett, K., Lipscomb, G. L., Nixon, W., Scott, R. A., Adams, M. W., et al. (2012). Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 78, 4669–4676. doi: 10.1128/AEM.00936-12
Fukui, T., Atomi, H., Kanai, T., Matsumi, R., Fujiwara, S., and Imanaka, T. (2005). Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363. doi: 10.1101/gr.3003105
Hawkins, M., Malla, S., Blythe, M. J., Nieduszynski, C. A., and Allers, T. (2013). Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547. doi: 10.1038/nature12650
Heider, M. R., Burkhart, B. W., Santangelo, T. J., and Gardner, A. F. (2017). Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J. Biol. Chem. 292, 8835–8845. doi: 10.1074/jbc.M117.783472
Hildenbrand, C., Stock, T., Lange, C., Rother, M., and Soppa, J. (2011). Genome copy numbers and gene conversion in methanogenic archaea. J. Bacteriol. 193, 734–743. doi: 10.1128/JB.01016-10
Hileman, T. H., and Santangelo, T. J. (2012). Genetics techniques for Thermococcus kodakarensis. Front. Microbiol. 3:195. doi: 10.3389/fmicb.2012.00195
Jacob, F., Brenner, S., and Cuzin, F. (1963). On the Regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 28, 329–348. doi: 10.1101/SQB.1963.028.01.048
Jager, D., Forstner, K. U., Sharma, C. M., Santangelo, T. J., and Reeve, J. N. (2014). Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genomics 15:684. doi: 10.1186/1471-2164-15-684
Kawakami, H., and Katayama, T. (2010). DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem. Cell Biol. 88, 49–62. doi: 10.1139/o09-154
Kogoma, T. (1997). Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238.
Kuba, Y., Ishino, S., Yamagami, T., Tokuhara, M., Kanai, T., Fujikane, R., et al. (2012). Comparative analyses of the two proliferating cell nuclear antigens from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells 17, 923–937. doi: 10.1111/gtc.12007
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359.
Li, Z., Huang, R. Y., Yopp, D. C., Hileman, T. H., Santangelo, T. J., Hurwitz, J., et al. (2014). A novel mechanism for regulating the activity of proliferating cell nuclear antigen by a small protein. Nucleic Acids Res. 42, 5776–5789. doi: 10.1093/nar/gku239
Li, Z., Santangelo, T. J., Cubonova, L., Reeve, J. N., and Kelman, Z. (2010). Affinity purification of an archaeal DNA replication protein network. MBio 1:e00221-10 doi: 10.1128/mBio.00221-10
Maduike, N. Z., Tehranchi, A. K., Wang, J. D., and Kreuzer, K. N. (2014). Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol. Microbiol. 91, 39–56. doi: 10.1111/mmi.12440
Makarova, K. S., and Koonin, E. V. (2013). Archaeology of Eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol. 5:a012963. doi: 10.1101/cshperspect.a012963
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Masai, H., and Arai, K. (1996). Mechanisms of primer RNA synthesis and D-loop/R-loop-dependent DNA replication in Escherichia coli. Biochimie 78, 1109–1117. doi: 10.1016/S0300-9084(97)86737-5
Masai, H., Asai, T., Kubota, Y., Arai, K., and Kogoma, T. (1994). Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 13, 5338–5345.
Matsunaga, F., Forterre, P., Ishino, Y., and Myllykallio, H. (2001). In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. U.S.A. 98, 11152–11157. doi: 10.1073/pnas.191387498
Matsunaga, F., Glatigny, A., Mucchielli-Giorgi, M.-H., Agier, N., Delacroix, H., Marisa, L., et al. (2007). Genomewide and biochemical analyses of DNA-binding activity of Cdc6/Orc1 and Mcm proteins in Pyrococcus sp. Nucleic Acids Res. 35, 3214–3222. doi: 10.1093/nar/gkm212
Matsunaga, F., Takemura, K., Akita, M., Adachi, A., Yamagami, T., and Ishino, Y. (2010). Localized melting of duplex DNA by Cdc6/Orc1 at the DNA replication origin in the hyperthermophilic archaeon Pyrococcus furiosus. Extremophiles 14, 21–31. doi: 10.1007/s00792-009-0284-9
Morikawa, M., Izawa, Y., Rashid, N., Hoaki, T., and Imanaka, T. (1994). Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566.
Norais, C., Hawkins, M., Hartman, A. L., Eisen, J. A., Myllykallio, H., and Allers, T. (2007). Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3:e77. doi: 10.1371/journal.pgen.0030077
Ogawa, T., Pickett, G. G., Kogoma, T., and Kornberg, A. (1984). RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 81, 1040–1044
Ojha, K. K., and Swati, D. (2010). Mapping of origin of replication in Themococcales. Bioinformation 5, 213–218
Pan, M., Santangelo, T. J., Li, Z., Reeve, J. N., and Kelman, Z. (2011). Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 39, 9671–9680. doi: 10.1093/nar/gkr624
Pan, M., Santangelo, T. J., Cubonová, L., Li, Z., Metangmo, H., Ladner, J., et al. (2013). Thermococcus kodakarensis has two functional PCNA homologs but only one is required for viability. Extremophiles 17, 453–461. doi: 10.1007/s00792-013-0526-8
Paul, S., Million-Weaver, S., Chattopadhyay, S., Sokurenko, E., and Merrikh, H. (2013). Accelerated gene evolution through replication–transcription conflicts. Nature 495, 512–515. doi: 10.1038/nature11989
Pelve, E. A., Martens-Habbena, W., Stahl, D. A., and Bernander, R. (2013). Mapping of active replication origins in vivo in thaum- and euryarchaeal replicons. Mol. Microbiol. 90, 538–550. doi: 10.1111/mmi.12382
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Robinson, N. P., and Bell, S. D. (2005). Origins of DNA replication in the three domains of life. FEBS J. 272, 3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x
Samson, R. Y., Xu, Y., Gadelha, C., Stone, T. A., Faqiri, J. N., Li, D., et al. (2013). Specificity and function of archaeal DNA replication initiator proteins. Cell. Rep. 3, 485–496. doi: 10.1016/j.celrep.2013.01.002
Santangelo, T. J., Cubonova, L., James, C. L., Reeve, J. N., Cubonová, L., James, C. L. et al. (2007). TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367, 344–357. doi: 10.1016/j.jmb.2006.12.069
Santangelo, T. J., Cubonova, L., and Reeve, J. N. (2008). Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74, 3099–3104. doi: 10.1128/AEM.00305-08
Scholefield, G., Veening, J. W., and Murray, H. (2011). DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 21, 188–194. doi: 10.1016/j.tcb.2010.10.006
Smith, C. E., Llorente, B., and Symington, L. S. (2007). Template switching during break-induced replication. Nature 447, 102–105. doi: 10.1038/nature05723
Spaans, S. K., van der Oost, J., and Kengen, S. W. (2015). The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles 19, 741–750. doi: 10.1007/s00792-015-0750-5
Tagashira, K., Fukuda, W., Matsubara, M., Kanai, T., Atomi, H., and Imanaka, T. (2013). Genetic studies on the virus-like regions in the genome of hyperthermophilic archaeon, Thermococcus kodakarensis. Extremophiles 17, 153–160. doi: 10.1007/s00792-012-0504-6
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11.10.1–33 doi: 10.1002/0471250953.bi1110s43
Wigley, D. B. (2009). ORC proteins: marking the start. Curr. Opin. Struct. Biol. 19, 72–78. doi: 10.1016/j.sbi.2008.12.010
Wu, Z., Liu, J., Yang, H., Liu, H., and Xiang, H. (2014). Multiple replication origins with diverse control mechanisms in Haloarcula hispanica. Nucleic Acids Res. 42, 2282–2294. doi: 10.1093/nar/gkt1214
Keywords: archaea, DNA replication, Thermococcus kodakarensis, recombination, DNA origins
Citation: Gehring AM, Astling DP, Matsumi R, Burkhart BW, Kelman Z, Reeve JN, Jones KL and Santangelo TJ (2017) Genome Replication in Thermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Front. Microbiol. 8:2084. doi: 10.3389/fmicb.2017.02084
Received: 31 July 2017; Accepted: 11 October 2017;
Published: 27 October 2017.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Thorsten Allers, University of Nottingham, United KingdomYoshizumi Ishino, Kyushu University, Japan
Copyright © 2017 Gehring, Astling, Matsumi, Burkhart, Kelman, Reeve, Jones and Santangelo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas J. Santangelo, thomas.santangelo@colostate.edu
 Alexandra M. Gehring
Alexandra M. Gehring David P. Astling
David P. Astling Rie Matsumi
Rie Matsumi Brett W. Burkhart
Brett W. Burkhart Zvi Kelman
Zvi Kelman John N. Reeve
John N. Reeve Kenneth L. Jones2
Kenneth L. Jones2 Thomas J. Santangelo
Thomas J. Santangelo