- 1Laboratory of Clinical Microbiology, Department of Biomedical Sciences for Health, University of Milan, Milan, Italy
- 2Villa Santa Maria Institute, Como, Italy
- 3Clinical-Chemistry and Microbiology Lab, IRCCS Galeazzi Orthopedic Institute, University of Milan, Milan, Italy
The human milk is fundamental for a correct development of newborns, as it is a source not only of vitamins and nutrients, but also of commensal bacteria. The microbiota associated to the human breast milk contributes to create the “initial” intestinal microbiota of infants, having also a pivotal role in modulating and influencing the newborns’ immune system. Indeed, the transient gut microbiota is responsible for the initial change from an intrauterine Th2 prevailing response to a Th1/Th2 balanced one. Bacteria located in both colostrum and mature milk can stimulate the anti-inflammatory response, by stimulating the production of specific cytokines, reducing the risk of developing a broad range of inflammatory diseases and preventing the expression of immune-mediated pathologies, such as asthma and atopic dermatitis. The aim of the present Mini Review is to elucidate the specific immunologic role of the human milk-associated microbiota and its impact on the newborn’s health and life, highlighting the importance to properly study the biological interactions in a bacterial population and between the microbiota and the host. The Auto Contractive Map, for instance, is a promising analytical methodology based on artificial neural network that can elucidate the specific role of bacteria contained in the breast milk in modulating the infants’ immunological response.
Introduction
The human milk is a rich and complete nourishment that is essential for the correct development of the infant’s organism (Ballard and Morrow, 2013).
The first milk produced by mothers after the delivery is called colostrum and it is biochemically and functionally different from the mature milk (Castellote et al., 2011). Colostrum, indeed, contains high concentration of lactoferrin, Immunoglobulin A (IgA), leukocytes and specific developmental factors, and a low amount of lactose, potassium and calcium, underlying its immunological functions rather than nutritional (Kulski and Hartmann, 1981; Pang and Hartmann, 2007).
From 5 days to 2 weeks postpartum, there is the production of transitional milk which shares some characteristics of colostrum, although its main function is to support newborns at nutritional level (Henderson et al., 2008; Nommsen-Rivers et al., 2012).
Finally, 2 weeks after the delivery the milk can be considered as mature and its composition tends to be stable over the time, even if slight variations can occur during lactation (Ballard and Morrow, 2013). The main components of human milk are: (i) macronutrients, such as protein, fat and lactose, which concentration depends on the stage of lactation and maternal characteristics; (ii) micronutrients, including vitamins A, B1, B2, B12 and D that vary in human milk in relation to maternal diet and body stores; (iii) growth factor, which are strongly active on the endocrine system, nervous system, vasculature and intestinal tract; (iv) immunological factors, which are essential to defend the newborn from inflammation and infection, and for this reason, the early milk is rich in immune components that can support infants in the first delicate stages of their life; (v) the microbiota, which comprises more than 200 different bacterial species with a pivotal role in the formation of the newborn’s first gut microbiota (Drago et al., 2017).
The aim of the present Mini Review is to highlight the specific and fundamental role of human milk-associated bacteria in modulating and influencing the newborns’ immune system during their life.
The Milk Microbiota and the Newborn’S Immune System
The specific mechanisms that lead to the formation of the human milk microbiota are still unknown; however, there are different hypothesis that can explain the origin of milk-associated bacteria. Indeed, some microorganisms belonging to the maternal skin or infant’s oral cavity may become an integral component of the milk microbiota by means of a milk flow back into mammary ducts during lactation (Rodríguez, 2014). This mechanism may justify the presence of cutaneous and oral bacteria that are recovered in the milk microbiota, such as Streptococcus spp. and Staphylococcus spp. (Gao et al., 2007; Grice et al., 2009). Interestingly, human milk contained also a great number of intestinal bacteria, which may spread from the maternal intestinal environment by a mechanism involving dendritic cells (DCs) and CD18+ cells (Rodríguez, 2014); these cellular types would be able to capture intestinal microorganisms from the gut lumen and transfer them to lactating mammary glands by means of translocation, which results to be increased during late pregnancy and lactation (Rodríguez, 2014). Consequently, the milk microbiota can shape the initial intestinal microbiome of newborns, together with the maternal intestinal and vaginal microorganisms that are ingested by the neonate during the passage through the birth canal (Houghteling and Walker, 2015). Human milk can stimulate the proliferation of numerous Bifidobacterium and Lactobacillus strains, the main probiotic microorganisms present in the gut, creating an acidic environmental rich in short chain fatty acids (SCFAs) with a protective and nutritive role at intestinal level (Bode, 2012; Walker and Iyengar, 2015). The constant intake, during lactation, of bacteria contained in the human milk leads to the formation of a transient intestinal microbiota that deeply impacts on the newborn’s development, acting mainly on the maturation of his immune system (Houghteling and Walker, 2015). Indeed, several studies underscored the strict link between the gut microbiota signals, the mucosal host defense and the maturation of immune system, both at intestinal and systemic level (Smith et al., 2007; Sekirov et al., 2008; Walker and Iyengar, 2015). It has been showed that an altered colonization of newborns’ gut may lead to a persistent intestinal dysbiosis and, consequently, to immune-mediated and metabolic diseases during infancy and childhood (Gareau et al., 2010; Johnson and Versalovic, 2012). Moreover, breast-fed newborns have shown to possess a more stable intestinal bacterial population and a well-balanced mucosal immune response if compared to the formula-fed ones (Gronlund et al., 2000; Bezirtzoglou et al., 2011); indeed, a healthy intestinal microbiota can induce specific T cell responses and modulate substrates oxidation, decreasing the impact of autoimmune and allergic diseases not only during childhood but also in adulthood (Guaraldi and Salvatori, 2012; Palma et al., 2012). Finally, breastfeeding has been observed to have a protective role against respiratory and gastrointestinal infections between the ages of 7 and 12 months, leading to a general improvement of symptoms associated to gastrointestinal infections (Duijts et al., 2010).
Intestinal bacteria can also stimulate lymphoid elements and positively influence the maturation of both innate and adaptive immune system, as clearly demonstrated by studying germ-free animals (Cash and Hooper, 2005). It has been shown that in germ-free mice the villus capillaries develop poorly during weaning and remained in this condition also during adulthood, suggesting that the intestinal microbiota is fundamental for intestinal blood vessel to be completely developed (Martin et al., 2010). More interestingly, intestinal bacteria can promote B cell development in Peyer’s Patches and increase the production of mucosal IgA, the main antibody class in secretions that acts as first line of defense (Martin et al., 2010).
Moreover, bacterial surface-expressed or secreted ligands can interact with specific receptors on mucosal immune system and enterocytes leading to a self-limited inflammatory response for preventing pathogen mucosal penetration (Round and Mazmanian, 2009; Walker and Iyengar, 2015).
As underlined by Latuga et al. (2014), all newborns have an immature immune system and the cord blood rich in anti-inflammatory T regulatory cells; furthermore, infants have a high T helper 2 (Th2) that promotes humoral immunity with the production of IL4, IL6 and IL21, thus promoting an increased B cell response and, potentially, a higher allergic sensitization (Latuga et al., 2014). The pivotal role of milk-associated microbiota in influencing the neonates’ immune system is over-emphasized by the cytotoxic function promoted by microbial ligands in breast milk (Donnet-Hughes, 2008). Indeed, in vitro stimulation of DCs with lipopolysaccharide can lead to T-cells differentiation, supporting the hypothesis that mature milk may implement the maturation of cytotoxic Th1 cells and improve their activity against infections (M’Rabet et al., 2008). Probably, commensal colonic bacteria may stimulate the release of specific cytokines that create a balanced microenvironment suitable for naive Th0 cells to ripen toward Th1 cell type (Walker and Iyengar, 2015).
Bacteroides is a bacterial genus that is very abundant in human colostrum and it may have a main role in the early stages of newborns’ gut colonization, as reported by Mazmanian and Kasper (2006). In particular, the polysaccharide A located on the surface of Bacteroides fragilis can interact with Toll receptor 2 on intestinal DCs to stimulate cytokine production which, in turn, favor the proliferation of FOXP3 T cells in the lamina propria. FOXP3 belongs to the forhead transcription factor family bindweed in the expansion of regulatory T cells, thus having a suppressive role in the host’s immune system (Kim, 2009). Therefore, it is clear that a correct stimulation of neonates’ intestinal environment is fundamental for the physiological development of mucosal immune system and tolerance; the latter, in particular, is extremely important to avoid developing allergy or autoimmune diseases (Walker and Iyengar, 2015). Indeed, germ-free animals cannot develop tolerance due to the lack of intestinal bacteria and only the adequate colonization of newborns’ gut can lead to a complete tolerance generation (Karlsson et al., 1999; Olszak et al., 2012).
Consequently, breastfeeding is essential for oral tolerance in newborns, as it is extremely important for the establishment of local and systemic immune tolerance to antigens ingested during lactation (Verhasselt, 2010). Infants are daily exposed to specific antigens, part of which belong directly to the human milk microbiota, and that can translocate across the intestinal barrier, being involved in the presentation by antigen-presenting cells to T lymphocytes. Furthermore, bacteria located in the human milk are fundamental to correctly stimulate the Peyer’s patches, increasing the number of IgA-producing plasma cells in the intestinal environment of newborns (Gross, 2007). Consequently, IgA can trap food antigens favoring their elimination by specific enzymes, avoid the adherence of viruses and microorganisms to intestinal mucosa also counteracting the proliferation of pathogens and exert a direct immunomodulatory activity (Verhasselt, 2010).
Finally, the oral tolerance seems to be actively involved in the prevention of allergic diseases onset in babies, avoiding also the impact of respiratory and gastrointestinal infections during the early stages of their life (Lack, 2008).
Interestingly, some strains contained in the breast milk can stimulate immune responses in both animal and human models and their activity seems to have a moderate suppleness based on the gut environment (Fernández et al., 2013). Some Lactobacillus strains can enhance the production of Th1 cytokines as well as TNF-alpha even without inflammatory stimuli and activate NK cells, CD4+ and CD8+ T cells and regulatory T cells.
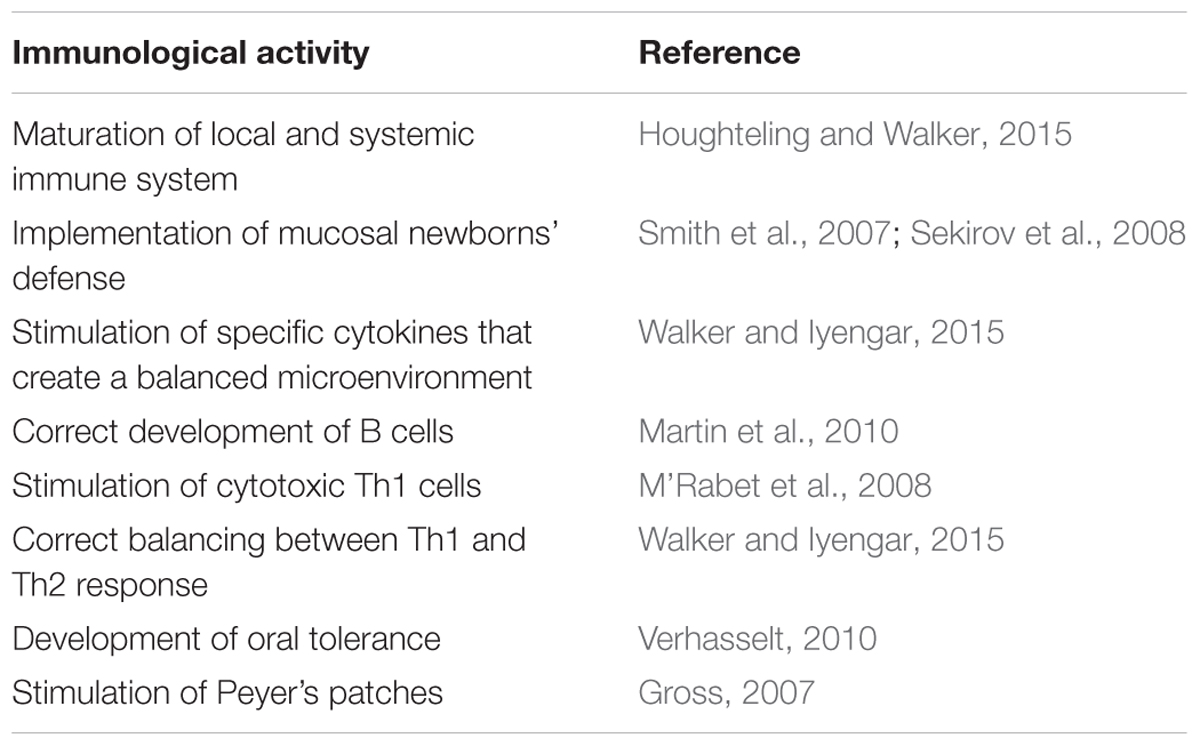
Table 1 summarizes the main immunomodulatory activities of human milk microbiota.
Microbial Network: New Insights Into Bacterial Ecology
The study of microbial interactions within a bacterial population is of extreme importance to clearly understand the specific role of microbiome. Indeed, microorganisms compete for nutrients, exchange genetic material and metabolites, being responsible of influencing the microbiota composition and the host’s health (Layeghifard et al., 2017). Due to its dynamic nature and high heterogeneity, the microbiota can be considered a complex and variable ecosystem not often well understandable. For this reason, in the last years a novel approach has been developed to study the microbiota, by using graph-theoretical, systems-oriented method able to facilitate the understanding of evolutionary and complex ecological processes (Layeghifard et al., 2017). Bacterial network is becoming essential to study microbial relationships and clarify the impact of various interactions on the host by identifying the main “hubs” that may represent the most influential member in a bacterial community (Layeghifard et al., 2017). Moreover, a central node is thought to have more links with other hubs, having a pivotal role in the stability of the whole microbial network.
Numerous methods to identify network hubs exist, but in the last years there was a great interest in using and apply the Auto-Contractive map (AutoCM). AutoCM system is a fourth-generation unsupervised artificial neural network (ANN) that can outperform numerous unsupervised algorithms (Buscema and Sacco, 2016). The system uses the minimum spanning tree (MST) theory to underline the natural connections between variables (Buscema and Grossi, 2008; Buscema et al., 2008). A MST is a spanning tree of a connected, undirected graph that links all vertices together with the minimal total pondering for its edges. The concept is that as all biological and natural systems are inclined to a minimal energetic condition, the graph delineates the essential biological information of this process. The final goal of this approach is finding out all specific correlations between variables, creating a semantic map of connections in which non-linear associations are maintained. This approach allows to both highlight the relevant nodes of the system and show the network of the main relations among variables. Nodes, also defined as “hubs,” are defined as variables with the highest number of relations in the map (Buscema and Grossi, 2008; Buscema et al., 2008; Buscema and Sacco, 2016).
AutoCM has already been used in the microbiological field to study the bacterial network of human colostrum and mature milk in Italian and Burundian populations, highlighting the main microbial hubs that represent the biological leading structure of the whole network (Drago et al., 2017). This mathematical approach could partially clarify the complexity of human-bacteria mutualism, and above all the role that some specific microorganisms have in the human milk.
Figure 1 shows a graphical representation of the human mature milk microbiota network obtained using AutoCM. The main bacterial hubs (blue lines) represent those microorganisms that may have a pivotal and leader role in the whole microbiota (Drago et al., 2017).
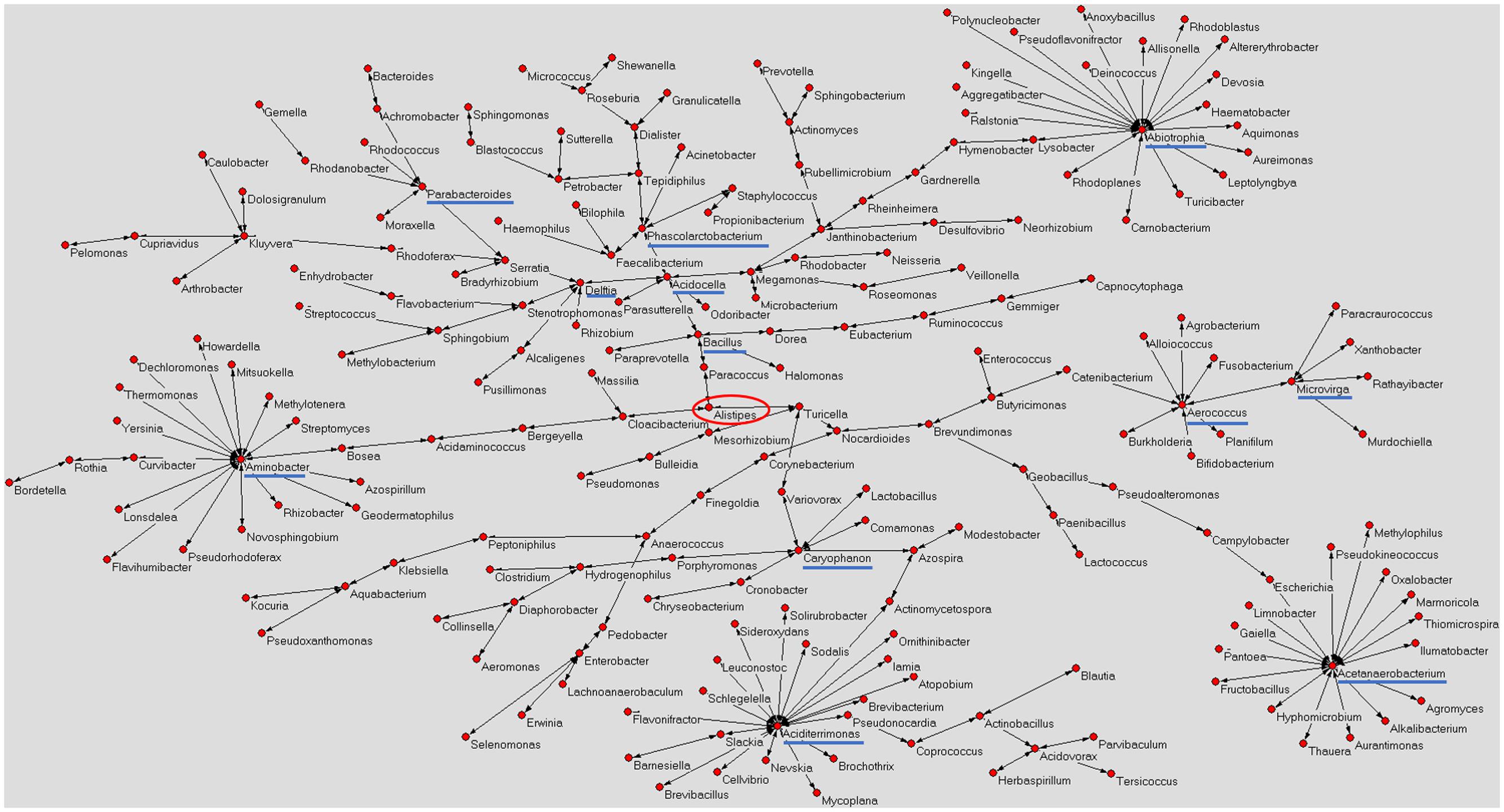
FIGURE 1. Microbiota network of Italian mature milk. The figure shows the distribution of bacterial population contained in the Italian human mature milk. The network was obtained applying the AutoCM. The main hubs of the bacterial network are underlined with a blue line; red circle shows the central node of the network (Drago et al., 2017). As authors of the previously published manuscript, we maintain the ownership of copyright as stated in the license-to-publish we signed for the Nature Publishing Group.
In the future, the AutoCM may be useful in understanding which microbe is directly involved in the stimulation and manipulation of newborns’ immune system, maybe defining a “immunomodulatory bacterial pattern” with a pivotal role in the development of infant’s immunity.
Conclusion
The human milk microbiota seems to have a central role in stimulate newborns’ immune system, contributing to create the first transient intestinal microbiota with strong immunomodulatory activities. However, further studies are needed to highlight the direct and strong connection between the human milk microbiota and the stimulation of newborns’ immune system, as to date there are no clear and specific evidence about this association.
Application of network biology will significantly improve our knowledge on bacterial interactions among the milk microbiota, with important applications for eventual targeted modification of bacterial composition, aimed to enhance the abundance of those microorganisms that may be essential not only for the modulation of the infants’ immune system, but also for improving the whole host’s health.
Author Contributions
LD provided the general concept. LD and EG drafted part of the manuscript. MT and RDG wrote the manuscript. All authors revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ballard, O., and Morrow, A. L. (2013). Human milk composition: nutrients and bioactive factors. Pediatr. Clin. North Am. 60, 49–74. doi: 10.1016/j.pcl.2012.10.002
Bezirtzoglou, E., Tsiotsias, A., and Welling, G. W. (2011). Microbiota profile in feces of breast- and formula-fed newborns by using florescence in situ hybridization (FISH). Anaerobe 17, 478–482. doi: 10.1016/j.anaerobe.2011.03.009
Bode, L. (2012). Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22, 1147–1162. doi: 10.1093/glycob/cws074
Buscema, M., and Grossi, E. (2008). The semantic connectivity map: an adapting self-organising knowledge discovery method in data bases. Experience in gastro-oesophageal reflux disease. Int. J. Data Min. Bioinform. 2, 362–404.
Buscema, M., Grossi, E., Snowdon, D., and Antuono, P. (2008). Auto-contractive maps: an artificial adaptive system for data mining. An application to Alzheimer disease. Curr. Alzheimer Res. 5, 481–498.
Buscema, M. B. M., and Sacco, P. L. (2016). MST Fitness Index and implicit data narratives: a comparative test on alternative unsupervised algorithms. Phys. A 461, 726–746.
Cash, H. L., and Hooper, L. V. (2005). Commensal bacteria shape intestinal immune system development. ASM News 71, 77–83.
Castellote, C., Casillas, R., Ramírez-Santana, C., Pérez-Cano, F. J., Castell, M., Moretones, M. G., et al. (2011). Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J. Nutr. 141, 1181–1187. doi: 10.3945/jn.110.133652
Donnet-Hughes, A. (2008). “Protective properties of human milk,” in Nutrition in Pediatrics, 4th Edn, ed. D. Duggan (Toronto, ON: Decker Publishing), 355–362.
Drago, L., Toscano, M., De Grandi, R., Grossi, E., Padovani, E. M., and Peroni, D. G. (2017). Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 11, 875–884. doi: 10.1038/ismej.2016.183
Duijts, L., Jaddoe, V. W., Hofman, A., and Moll, H. A. (2010). Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics 126, e18–e25. doi: 10.1542/peds.2008-3256
Fernández, L., Langa, S., Martín, V., Maldonado, A., Jiménez, E., Martín, R., et al. (2013). The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 69, 1–10. doi: 10.1016/j.phrs.2012.09.001
Gao, Z., Tseng, C., Pei, Z., and Blaser, M. J. (2007). Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U.S.A. 104, 2927–2932.
Gareau, M. G., Sherman, P. M., and Walker, W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514. doi: 10.1038/nrgastro.2010.117
Grice, E. A., Kong, H. H., Conlan, S., Deming, C. B., Davis, J., Young, A. C., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. doi: 10.1126/science.1171700
Gronlund, M. M., Arvilommi, H., Kero, P., Lehtonen, O. P., and Isolauri, E. (2000). Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch. Dis. Child. Fetal Neonatal Ed. 83, F186–F192.
Gross, L. (2007). Microbes colonize a baby’s gut with distinction. PLOS Biol. 5:e191. doi: 10.1371/journal.pbio.0050191
Guaraldi, F., and Salvatori, G. (2012). Effect of breast and formula feeding on gut microbiota shaping in newborns. Front. Cell. Infect. Microbiol. 2:94. doi: 10.3389/fcimb.2012.00094
Henderson, J. J., Hartmann, P. E., Newnham, J. P., and Simmer, K. (2008). Effect of preterm birth and antenatal corticosteroid treatment on lactogenesis II in women. Pediatrics 121, e92–e100. doi: 10.1542/peds.2007-1107
Houghteling, P. D., and Walker, W. A. (2015). Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 60, 294–307. doi: 10.1097/MPG.0000000000000597
Johnson, C. L., and Versalovic, J. (2012). The human microbiome and its potential importance to pediatrics. Pediatrics 129, 950–960. doi: 10.1542/peds.2011-2736
Karlsson, M. R., Kahu, H., Hanson, L. A., Telemo, E., and Dahlgren, U. I. (1999). Neonatal colonization of rats induces immunological tolerance to bacterial antigens. Eur. J. Immunol. 29, 109–118.
Kulski, J. K., and Hartmann, P. E. (1981). Changes in human milk composition during the initiation of lactation. Aust. J. Exp. Biol. Med. Sci. 59, 101–114.
Lack, G. (2008). Epidemiologic risks for food allergy. J. Allergy Clin. Immunol. 121, 1331–1336. doi: 10.1016/j.jaci.2008.04.032
Latuga, M. S., Stuebe, A., and Seed, P. C. (2014). A review of the source and function of microbiota in breast milk. Semin. Reprod. Med. 32, 68–73. doi: 10.1055/s-0033-1361824
Layeghifard, M., Hwang, D. M., and Guttman, D. S. (2017). Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 25, 217–228. doi: 10.1016/j.tim.2016.11.008
Martin, R., Nauta, A. J., Ben Amor, K., Knippels, L. M., Knol, J., and Garssen, J. (2010). Early life: gut microbiota and immune development in infancy. Benef. Microbes 1, 367–382. doi: 10.3920/BM2010.0027
Mazmanian, S. K., and Kasper, D. L. (2006). The love–hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 6, 849–858. doi: 10.1038/nri1956
M’Rabet, L., Vos, A. P., Boehm, G., and Garssen, J. (2008). Breast-feeding and its role in early development of the immune system in infants: consequences for health later in life. J. Nutr. 138, 1782S–1790S.
Nommsen-Rivers, L. A., Dolan, L. M., and Huang, B. (2012). Timing of stage II lactogenesis is predicted by antenatal metabolic health in a cohort of primiparas. Breastfeed. Med. 7, 43–49. doi: 10.1089/bfm.2011.0007
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
Palma, G. D., Capilla, A., Nova, E., Castillejo, G., Varea, V., Pozo, T., et al. (2012). Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLOS ONE 7:e30791. doi: 10.1371/journal.pone.0030791
Pang, W. W., and Hartmann, P. E. (2007). Initiation of human lactation: secretory differentiation and secretory activation. J. Mammary Gland Biol. Neoplasia 12, 211–221. doi: 10.1007/s10911-007-9054-4
Rodríguez, J. M. (2014). The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 5, 779–784. doi: 10.3945/an.114.007229
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi: 10.1038/nri2515
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736. doi: 10.1128/IAI.00319-08
Smith, K., McCoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi: 10.1016/j.smim.2006.10.002
Verhasselt, V. (2010). Oral tolerance in neonates: from basics to potential prevention of allergic disease. Mucosal Immunol. 3, 326–333. doi: 10.1038/mi.2010.25
Keywords: human milk, milk microbiota, colostrum, immunomodulation, newborn’s immune system, AutoCM
Citation: Toscano M, De Grandi R, Grossi E and Drago L (2017) Role of the Human Breast Milk-Associated Microbiota on the Newborns’ Immune System: A Mini Review. Front. Microbiol. 8:2100. doi: 10.3389/fmicb.2017.02100
Received: 21 July 2017; Accepted: 13 October 2017;
Published: 25 October 2017.
Edited by:
Gayane Manukyan, Institute of Molecular Biology (NAS RA), ArmeniaReviewed by:
Eugene Michael Dempsey, University College Cork, IrelandRyo Inoue, Kyoto Prefectural University, Japan
Copyright © 2017 Toscano, De Grandi, Grossi and Drago. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Drago, lorenzo.drago@unimi.it
 Marco Toscano
Marco Toscano Roberta De Grandi
Roberta De Grandi Enzo Grossi
Enzo Grossi Lorenzo Drago
Lorenzo Drago