- 1School of Medical Informatics, Xuzhou Medical University, Xuzhou, China
- 2Department of Clinical Pharmacology, Xuzhou Infectious Diseases Hospital, Xuzhou, China
- 3School of Anaesthesia, Xuzhou Medical University, Xuzhou, China
- 4Clinical Laboratory of Tuberculosis, Xuzhou Infectious Diseases Hospital, Xuzhou, China
- 5School of Computer Science and Software Engineering, University of Western Australia, Perth, WA, Australia
- 6The Marshall Centre for Infectious Diseases Research and Training, University of Western Australia, Perth, WA, Australia
The intriguing sit-and-wait hypothesis predicts that bacterial durability in the external environment is positively correlated with their virulence. Since its first proposal in 1987, the hypothesis has been spurring debates in terms of its validity in the field of bacterial virulence. As a special case of the vector-borne transmission versus virulence tradeoff, where vector is now replaced by environmental longevity, there are only sporadic studies over the last three decades showing that environmental durability is possibly linked with virulence. However, no systematic study of these works is currently available and epidemiological analysis has not been updated for the sit-and-wait hypothesis since the publication of Walther and Ewald’s (2004) review. In this article, we put experimental evidence, epidemiological data and theoretical analysis together to support the sit-and-wait hypothesis. According to the epidemiological data in terms of gain and loss of virulence (+/-) and durability (+/-) phenotypes, we classify bacteria into four groups, which are: sit-and-wait pathogens (++), vector-borne pathogens (+-), obligate-intracellular bacteria (--), and free-living bacteria (-+). After that, we dive into the abundant bacterial proteomic data with the assistance of bioinformatics techniques in order to investigate the two factors at molecular level thanks to the fast development of high-throughput sequencing technology. Sequences of durability-related genes sourced from Gene Ontology and UniProt databases and virulence factors collected from Virulence Factor Database are used to search 20 corresponding bacterial proteomes in batch mode for homologous sequences via the HMMER software package. Statistical analysis only identified a modest, and not statistically significant correlation between mortality and survival time for eight non-vector-borne bacteria with sit-and-wait potentials. Meanwhile, through between-group comparisons, bacteria with higher host-mortality are significantly more durable in the external environment. The results of bioinformatics analysis correspond well with epidemiological data, that is, non-vector-borne pathogens with sit-and-wait potentials have higher number of virulence and durability genes compared with other bacterial groups. However, the conclusions are constrained by the relatively small bacterial sample size and non-standardized experimental data.
Introduction
According to the conventional theories of evolution of virulence, all pathogens will end up in a relationship of commensalism or mutualism with their hosts, that is, being avirulent to their hosts (Ewald, 1983, 1987b). However, with more experimental data collected and theoretical studies conducted, the conventional wisdom underwent critical challenges and was overturned. Anderson and May are among the pioneers to use mathematics to model population dynamics of infectious diseases and proposed basic reproductive ratio Ro to measure pathogen fitness (Anderson and May, 1979, 1980, 1982; May and Anderson, 1979; Bull and Lauring, 2014), which is written as:
#Mortality caused by the disease is defined as bacterial virulence for its simplicity (Thomas and Elkinton, 2004).
Only when Ro > 1, that is, change rate in a population Y is larger than 0 where Y is the number of the infected, can virulence (disease) be maintained in the population (Anderson and May, 1979). Fitness, defined by Ro, proposes that virulence can be achieved at a certain level with the influences of β, α, and γ individually, or interactively, and an optimal level of virulence can be reached when all parameters are appropriately balanced (Alizon et al., 2009). However, most models assume that parameters are coupled and a tradeoff exists, such as between recovery and virulence (Anderson and May, 1982), transmission and recovery (Alizon, 2008), and transmission and virulence (Ewald, 1983), etc.
Evolutionary biologist Paul W. Ewald was among the firsts to argue that bacterial transmission rate β and virulence α are linked (Ewald, 1983). In his work, costs and benefits tradeoff between transmission and virulence were systematically studied through epidemiological analysis, which validated the idea that non-vector-borne pathogens relying on direct host transmission are generally benign while those with vector-borne transmission mode are much more virulent (Ewald, 1983). From evolutionary perspective, a pathogen with direct transmission mode prefers maintaining a relatively low virulence phenotype because high virulence will lead to immobilization or death of the infected hosts, which incurs high costs by reducing contact opportunities with susceptible hosts. Without a vector, the pathogens would be trapped in dead or immobilized hosts and die out. Thus, even if a recent association between a pathogen and a host would lead to high virulence, the severity will over time reduce if the pathogen relies on host’s mobility for transmission (Ewald, 1983). On the other hand, if a pathogen were vector-borne, it would be risk-free to exploit hosts extensively without the impediment of host immobilization. Vectors such as mosquitoes, ticks, or flies can transfer pathogens from infected or immobilized hosts to a group of new susceptible population. In addition, since vector-borne pathogens rely on vectors for transmission, they tend to have lower virulence to their vectors than to their main hosts, which reinforces the virulence-transmission tradeoff theory (Elliot et al., 2003). A variety of pathogens that cause severe epidemics to human society fall into this category, such as Yersinia pestis (vector: fleas; disease: Bubonic Plague), Borrelia burgdorferi (vector: ticks; disease: Lyme disease), and Francisella tularensis (vector: ticks and flies; disease: Tularemia), etc. (Perry and Fetherston, 1997; Gubler, 2009; Conlan, 2011; Cook, 2015).
During a study of transmission modes of pathogens and their virulence, a set of non-vector-borne pathogens was identified as highly or extremely highly virulent bacteria, such as Mycobacterium tuberculosis and Corynebacterium diphtheriae, etc. (Ewald, 1983; Walther and Ewald, 2004). Careful study of these outliers showed that they are actually a special instance of vector-borne pathogens that have long-lived free-living stages during their infection cycles. The reasoning is as simple as that long-lived pathogens in the external environment can live long enough to wait for potential hosts to reach them. Thus, they are able to be highly virulent in their hosts without incurring high costs, just like the vector-borne pathogens (Ewald, 1987a). In fact, high virulence and long environmental longevity were initially studied in a Susceptible-Infected-Recovery (SIR) model by Anderson and May because they cause cyclic changes of host abundance (Anderson and May, 1980). However, interactions between the two parameters were not further explored. Ewald studied those models (Ewald, 1987a) and originally proposed that pathogenic bacteria with high durability in the external environment tend to evolve toward higher virulence phenotypes because they do not rely on the mobility of their hosts for transmission and will have lower costs and higher gains by incurring the immobility of their hosts (Ewald, 1983, 1987a, 2004; Walther and Ewald, 2004). Those bacteria are therefore termed sit-and-wait pathogens (Ewald, 1987a).
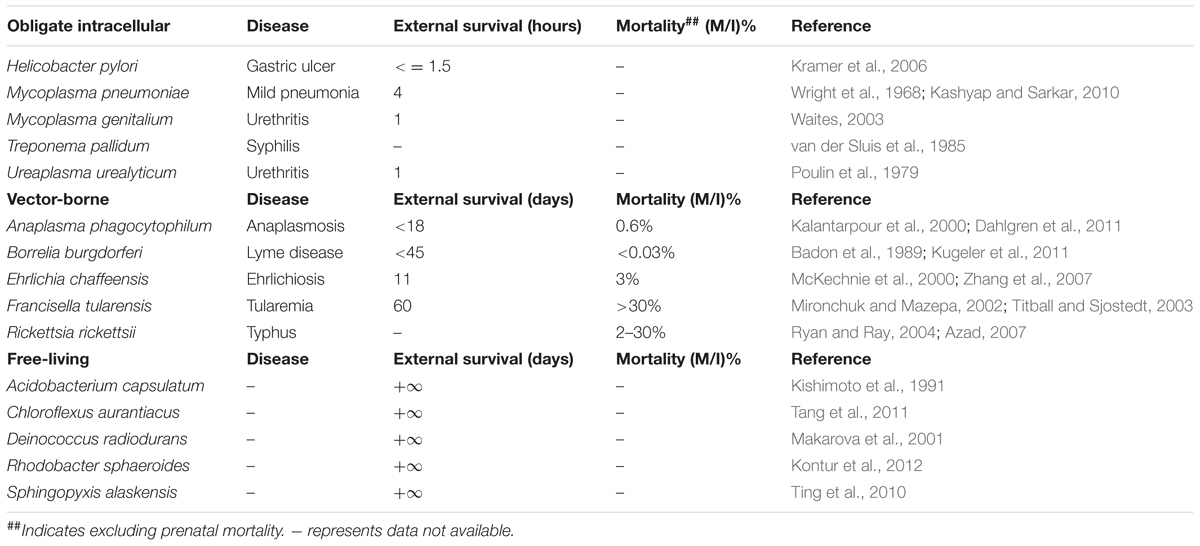
Walther and Ewald (2004) collected data about the survival time and mortality-rate of a set of human respiratory pathogens to support the sit-and-wait hypothesis. The reason to limit the study to human respiratory pathogens was to eliminate variation due to host species or site of infection (Walther and Ewald, 2004). However, considering that sit-and-wait hypothesis should be a general mechanism for bacterial virulence evolution, there is no need to restrict the investigation to respiratory pathogens only. In this study, based on the gain and loss of virulence (+/-) and durability (+/-) phenotypes, we divided bacteria into four categories (Figure 1): exclusively host-associated bacteria (-/-), sit-and-wait pathogens (+/+), vector-borne pathogens (+/-), and free-living bacteria (-/+). For each category, epidemiological data are collected for bacterial mortality and survival time based on literature mining in order to investigate the relationship between durability and virulence, that is, whether higher durability leads to higher virulence.
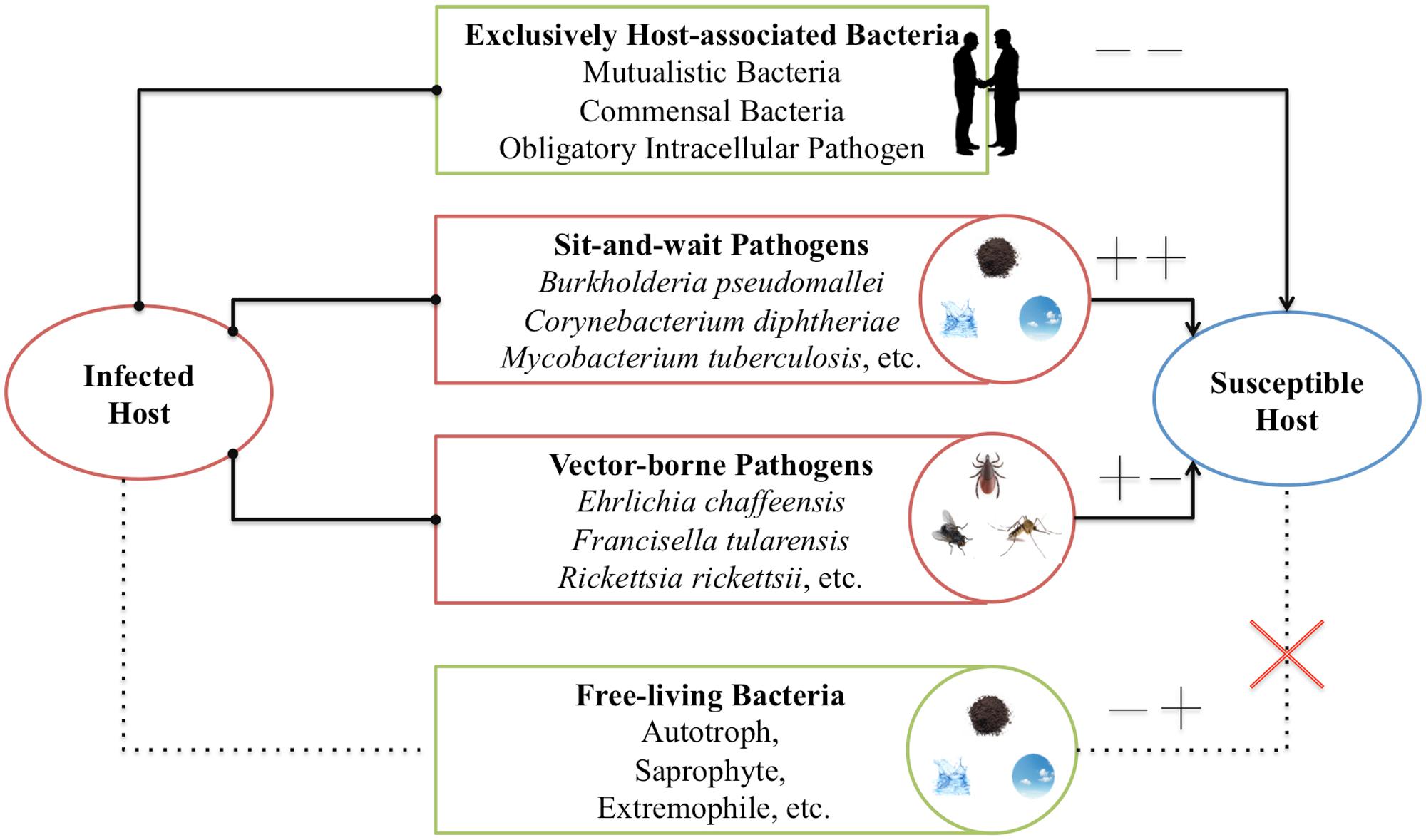
FIGURE 1. Classification of bacteria based on gain (+) or loss (+) of durability and virulence phenotypes. In this classification, four groups are included: exclusively host-associated bacteria (-), sit-and-wait pathogens (++), vector-borne pathogens (+-), and free-living bacteria (-+).
A recent experimental study found that environmental durability can diversify the population structure and virulence strategies of a fish pathogen, that is, longer survival time corresponds to higher virulence (Sundberg et al., 2014). For sit-and-wait pathogens, in order to survive hash external environment and maintain viability, they have to deal with a vast range of abiotic stresses, such as temperature fluctuation, hyperosmolarity, pH change, and hazardous radiation, etc. Thus, their survival depends on their ability to sense and respond to the ever changing environment (Boor, 2006). In addition, maintenance of viability requires careful consumption of large amount of energy storage compounds, otherwise known as ‘energy of maintenance’ (Wilkinson, 1963; Roszak and Colwell, 1987). Thus, energy reserve is another important factor for bacterial survival, especially when considering that bacteria need to adapt to extensive changes after shedding from hosts to the external environment where they are deprived of nutrients. In brief, bacterial energy storage mechanisms (ESM) and abiotic stress resistance (ASR) are two major factors for bacterial survival in the external environment. Why this is the case is discussed below.
Bacteria consume energy all the time for their regular activities, ranging from mobility to reproduction. Thus, energy storage compounds are an indispensable element for bacterial durability due to the fluctuation of nutrients in the external environment. Some bacteria have one or more types of energy reserves and can therefore be very durable in the external environment, e.g., Corynebacterium diphtheriae, that accumulates glycogen, and Mycobacterium tuberculosis (Mtb), that accumulates both wax ester and glycogen, while other obligate intracellular bacteria such as Ricketssia and Borrelia do not accumulate any energy reserves and can only survive inside hosts (Wang and Wise, 2011). Coincidentally, the former two bacteria are more highly virulent pathogens than the latter two obligate intracellular pathogens. In fact, several studies have already linked one of the major energy reserves, glycogen, with bacterial persistence, colonization, and virulence (Wilson et al., 2010). Henrissat et al. (2002) screened 55 completely sequenced bacterial genomes and found that most of the free-living bacteria accumulate glycogen as an energy reserve while those with parasitic lifestyles do not, and concluded that glycogen accumulation can be a marker for bacterial parasitic behavior. Thus, bacterial energy reserves are very much likely to be an indicator for bacterial durability, lifestyle, and probably virulence. Currently, there are five major storage compounds identified in bacteria: triacylglycerol (TAG), wax esters (WE), polyhydroxybutyrate (PHB), polyphosphate (PolyP), and glycogen (Wang and Wise, 2011), all of which will be investigated in this study.
Bacteria exist almost everywhere on the planet from deep sea (Cowellia hadaliensis) to nuclear reactors (Deinococcus radiodurans) to inside the human body (Escherichia coli) (Kato and Qureshi, 1999; Marles-Wright and Lewis, 2007). Accordingly, bacteria are experts in evolving sufficient mechanisms to deal with and/or adapt to harsh and fluctuating environments. Although many proteins have been reported to protect bacteria from harsh conditions, they are normally synthesized only when bacteria sense the corresponding stresses, which can save bacterial energy and improve working efficiency. Stress sensing is generally the first step for bacteria to react to abiotic stresses. Specifically, when sensing a stress, transcription factors, such as sigma factors, etc., will be activated to interact with RNA polymerase and expression of stress responsive genes will be unregulated. These genes encode stress proteins that are involved in dealing with stresses directly (Marles-Wright and Lewis, 2007). Thus, the former are termed general stress genes while the later are termed as specific stress genes (Schumann, 2007). Currently, there are three types of sensors: DNA, mRNA, and proteins, among which, DNA and mRNA may sense stresses by changing their conformations, which change gene expression level by influencing transcription-related protein binding (Schumann, 2007). However, these sensors cannot be detected by screening proteomes. In addition, only protein sensors are counted as major stress sensors, which include molecular chaperones, proteases, kinases, and sigma factors, etc. (Schumann, 2007). Since these protein sensors are rather diverse, forming sophisticated networks, and remote from the actual stresses, we will not review them specifically. However, a set of proteins that are more proximal to the actual stresses have been collected and used for studying the sit-and-wait pathogens, that is, proteins protecting bacteria from reactive oxygen species (ROS), UV radiation, and desiccation, etc. In addition to the three typical abiotic stresses introduced above, there are still many other abiotic stresses in the external environment, such as high salinity and extreme temperature, etc. Obviously, resistance to abiotic stress can increase the opportunities for pathogens to survive longer in unfriendly environments. For a complete list of ASR types, please refer to Supplementary Table S1.
In this study, we constructed two sets of hidden Markov models (HMMs) in terms of ESM and ASR. For each bacterial category, five representative proteomes were screened using the HMM models for distribution of ESM- and ASR-related genes. In addition, virulence factors collected from VFDB were also searched in the proteomes in order to identify homologous sequences (Chen et al., 2016). It was hypothesized that the distribution patterns of durability and virulence genes should be consistent with epidemiological data and also with the four-way classification of bacteria described above. The proposal of sit-and-wait hypothesis had spurred a debate about its validity both theoretically (Bonhoeffer et al., 1996; Walther and Ewald, 2004; Alizon et al., 2009; Roche et al., 2011) and experimentally (Tolba et al., 2007; Ayyadurai et al., 2008; Sundberg et al., 2014). While Walther and Ewald (2004) collected abundant epidemiological data on respiratory pathogens supporting the existence of a positively correlation between durability and virulence in 2004, no systematic study of this hypothesis has been conducted since then. Only sporadic scientific articles are available so far to support the hypothesis (Tolba et al., 2007; Jani and Cotter, 2010; Vezzulli et al., 2010; Sundberg et al., 2014). In this study, we searched the literature linked with sit-and-wait hypothesis tracing back through several decades. We also put experimental evidence and epidemiological data together in order to examine the validity of this intriguing hypothesis. In addition, we endeavored to correlate bacterial durability with virulence using bioinformatics methods so as to facilitate the investigation and provide a preliminary guide to test the hypothesis.
Materials and Methods
Collection of Bacterial Proteomes and Epidemiological Data
For the sit-and-wait pathogens, proteomes of eight representative bacteria were selected. For the other three categories, only proteomes of five representative bacteria were sourced. All proteomes are downloaded from UniProt (The UniProt Consortium, 2017). In order to make the investigation more persuasive, we included quantitative data, such as external survival time and host mortality, for comparison. For details, please refer to Table 1. All eight sit-and-wait bacteria were selected based on a literature search using key words such as “sit-and-wait,” “pathogenic,” and “bacteria,” in PubMed and Google Scholar databases. Representative bacteria from other categories were also selected using the same method. All absent data were further sourced from government databases, medical and biological textbooks, and also referenced papers. The collected epidemiological data are only representative and should not be considered to be comprehensive due to the large volume of literature available. It only serves to provide preliminary support for the linkage between durability and virulence.
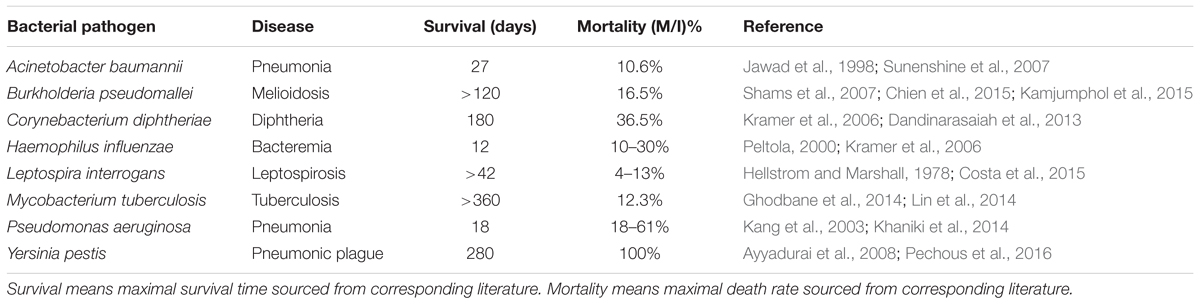
TABLE 1. Epidemiological analysis of 8 non-vector-borne environmental human pathogens with sit-and-wait potentials.
Construction of HMMs Related to Energy Storage Mechanisms
Enzymes directly linked with energy storage metabolisms in bacteria were first collected through a comprehensive up-to-date review of literature (Lopez et al., 1998; Henrissat et al., 2002; Zhang et al., 2002; Ishige et al., 2003; Kalscheuer and Steinbuchel, 2003; Brown and Kornberg, 2004, 2008; Kulaev et al., 2004; Uchino et al., 2007; Rao et al., 2009; Finkelstein et al., 2010; Kalscheuer, 2010; Achbergerova and Nahalka, 2011; Wang and Wise, 2011; Whitehead et al., 2014; Albi and Serrano, 2016). For a complete list of all the enzymes and their distributions in bacterial proteomes, please refer to Supplementary Table S1 of the review by Wang and Wise (2011). All sequences of ESM-related enzymes were downloaded from The UniProt Consortium (2017). HMMs of all enzymes were constructed by using HMMER based on multiple sequence alignments downloaded directly from Pfam database (Eddy, 2003; Finn et al., 2010). The command, hmmsearch, was used to search all proteomes for homologous sequences of ESM-related enzymes with E-value set to 1e-50. Complete gain or loss of all enzymes in the metabolism of an energy storage compound was marked as 1 and 0, respectively. Short Python scripts (Cock et al., 2009) were written and executed during the processes of file handling and text mining in order to integrate data from hmmsearch results into Supplementary Table S2.
Construction of HMMs Related to Abiotic Stress Resistance
Seed sequences of ASR related proteins were selected by using a web-based browser for Gene Ontology (GO) terms and annotations, QuickGo (provided by the UniProt-GOA group at EBI) (Binns et al., 2009). Database was filtered by designating the taxonomic group as bacteria (Taxonomy Identifier = 2). Meanwhile, 35 GO identifiers under the category of response to abiotic stimulus (GO:0009628) were selected for filtering meaningful and important proteins associated with ASR. A complete list of the 35 GO identifiers is presented below in Supplementary Table S1. In total, 1406 bacterial proteins were collected as seed proteins and used for constructing HMMs corresponding to ASR after using Perl script nrdb90.pl to remove the sequences with more than 90% similarity from the selected proteins (Holm and Sander, 1998). After obtaining sequences for all seed proteins, remote BLAST was performed to collect homologous sequences for each seed protein from the NCBI non-redundant database of protein sequences. In addition, since the sequences returned from remote BLAST were in extensible markup language (XML) file format, a Python script for parsing XML file was also written to extract the homologous sequences. The standalone command-line version of MUSCLE was used so the MSAs were done automatically (Edgar, 2004). Heads or tails of multiple sequence alignments tend to be more inconsistent (Wise, 2010). Thus all MSAs were manually edited to remove heads and tails by using JalView, a free program for MSA editing, visualization, and analysis (Waterhouse et al., 2009). HMMER was selected for the construction of HMMs through hmmbuild command by using multiple sequence alignments. Since HMMER only recognizes STOCKHOLM format, all MSAs results were converted from FASTA to STOCKHOLM format. For detailed steps and python scripts of HMM construction from scratch, please refer to Wang (2013). Python scripts and HMM model for bacterial ASR and ESM are both available under request. For searching homologs, routine procedures are performed by following HMMER User’s Guide (Eddy, 2003). In order to avoid the influence of bacterial proteome size, the number of ASR genes was converted into the percentage of ASR genes in bacterial proteome.
Homologs of Bacterial Virulence Factors
VF sequence package includes 2595 non-redundant core bacterial virulence factors sourced from VFDB (downloadable from VFDB website http://www.mgc.ac.cn/VFs/) (Chen et al., 2005). BLAST-equivalent phmmer command from HMMER package was recruited to automatically search VF homologs in all selected proteomes by using E-value of 1e-50. For usage of HMMER package please refer to the protocol document (Eddy, 2003). Results were all under further processed through a set of short python scripts (Cock et al., 2009). In order to avoid the influence of bacterial proteome size, number of VFs was converted into percentages of VFs in the respective bacterial proteomes.
Statistical Analysis
Unless otherwise indicated, Student’s t-test was used for all statistical analyses via a Python script, specifically the scipy statistical computation package. Data used in the comparisons can be found in the Tables. Where a range is given, the midpoint in the range has been used in the statistical analyses.
Results and Discussion
Through literature review and data mining, we collected bacterial durability and mortality data for representative bacteria in the previously defined four groups, which are: exclusively host-associated bacteria, sit-and-wait pathogens, vector-borne pathogens, and free-living bacteria. The results are summarized in Table 1 for sit-and-wait pathogens and in Table 2 for other categories, respectively. Correlation between durability and virulence phenotypes were calculated within and between groups. After that, HMMs were used to search for distribution patterns of enzymes related with energy storage metabolism and ASR. Homologous sequences of core virulence factors in each bacterium were also searched. The combined result is presented in Figure 2 with details summarized in Supplementary Table S2. Relationships between durability and virulence based on bioinformatics analysis are presented exclusively for each bacterial group. In addition, bacterial proteome size was also identified as an important factor for bacterial grouping and lifestyle based on our analysis.
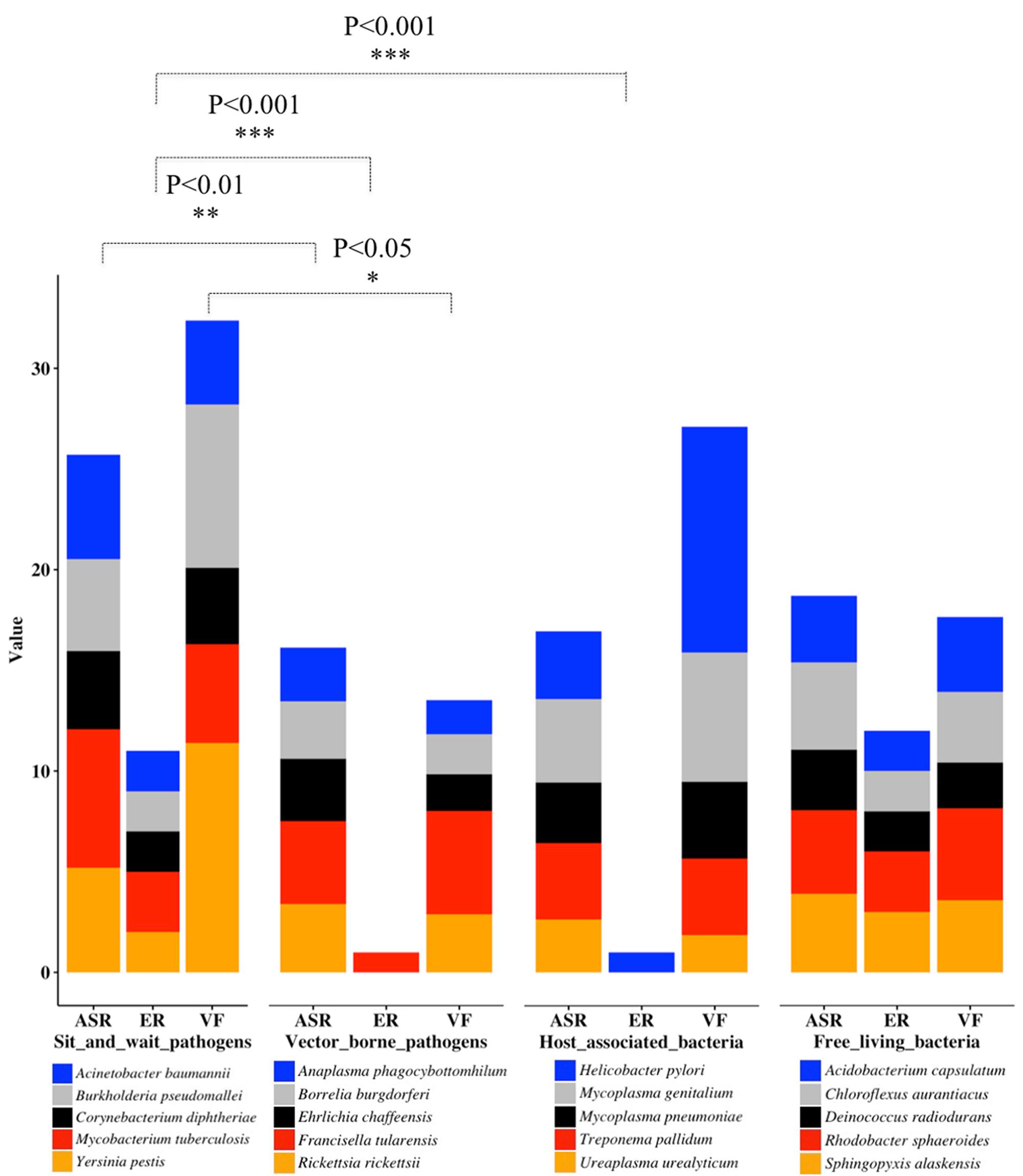
FIGURE 2. Stacked column chart for the illustration of bacterial classification based on distribution patterns of durability and virulence genes as previously defined. For each category, only five representative bacterial species are displayed. Energy Reserve (ER) includes glucose, polyphosphate, triacylglycerol, wax ester, and polyhydroxybutyrates. Value of ER represents total number of energy storage compound types. Value of ASR represents added percentages of genes related to starvation, temperature, osmolarity, pH, and desiccation, etc. Value of VF means added percentages of bacterial virulence factors. For statistical analysis, we classify the results into three levels, ∗∗∗P < 0.001, ∗∗P < 0.01, and ∗P < 0.05.
Statistical Analysis of Host Mortality versus Species Durability
Eight representative non-vector-borne human pathogens were collected based on literature review and first studied in terms of external survival time and host mortality. All these pathogens have common features, that is, long environmental survival time and high mortality (Table 1) when compared with bacteria in the other three categories (Table 2). According to the sit-and-wait hypothesis, higher durability should facilitate higher virulence. However, linear regression of host mortality (percentage) against persistence in the environment (in days) across the data in Table 1 only yielded a modest, and not statistically significant, correlation (r-value 0.387, P-value 0.344). While this may be due to the small sample size and the use of statistics from non-standardized experiments for the survival and mortality measurements, it is also understandable given the argument that durability is an enabler of virulence, but is not causally linked.
In order to better understand the selected bacteria and their sit-and-wait potentials, we dive into each species for a detailed investigation. Acinetobacter baumannii is a ubiquitous Gram-negative bacterial pathogen in the environment, which is capable of causing a wide range of infections and ranked among the top 10 pathogens causing septicemia (Jawad et al., 1998). Studies have shown that A. baumannii can persist in hospital environments for up to 3 years (Sherertz and Sullivan, 1985). However, a systematic study found out that multi-drug resistant (MDR) and susceptible strains of A. baumannii have similar survival times in distilled water, for a mean time of 27 days (Jawad et al., 1998). As for mortality, MDR A. baumannii shows a higher rate (26%) than the susceptible strain (17.6%) (Sunenshine et al., 2007), which may be caused by more virulence factors in the MDR strains rather than survival time in the environment. Since the mortality rate of uninfected control is 11.2% (Sunenshine et al., 2007), the average mortality rate for A. baumannii is re-adjusted to10.6%. Burkholderia pseudomallei is a Gram-negative bacillus and the causative agent of Melioidosis with varied severity and difficulty of treatment (Shams et al., 2007). It is normally dwelling on inmate surfaces and can survive in soil microcosms for at least 120 days (Shams et al., 2007; Kamjumphol et al., 2015). A recent study of a B. pseudomallei outbreak in Singapore between 2001 and 2010 hospital showed that the bacteria has a high mortality of 16.5% among 170 patients (Chien et al., 2015). Survival of Corynebacterium diphtheriae on dry surface is reported to be up to 6 months (Kramer et al., 2006). As a leading cause of childhood mortality in developing countries, a study of diphtheria spanning 12 years in a hospital found that the mortality rate is as high as 36.5% (Dandinarasaiah et al., 2013). Haemophilus influenzae is capable of surviving on a dry surface for 12 days and of causing a variety of clinical manifestations with a mortality rate worldwide ranging from 10 to 30% without vaccination (Peltola, 2000; Kramer et al., 2006).
Leptospira interrogans is a worldwide zoonotic pathogen and causes the emerging infectious disease Leptospirosis (Levett, 2001). Its survival in a typical New Zealand soil is at least 42 days (Hellstrom and Marshall, 1978). A systematic study showed that the worldwide mortality rate of Leptospirosis ranges from 4 to 13% between 1970 and 1999 (Costa et al., 2015). Mycobacterium tuberculosis is a typical sit-and-wait pathogen. It causes serious public health issues and its estimated overall mortality rate is 12.3%, based on case studies between 2003 and 2007 (Lin et al., 2014). Its viability and virulence in soil as Mycobacterium tuberculosis complex (MTC) lasts more than 12 months (Ghodbane et al., 2014). Pseudomonas aeruginosa is a major nosocomial pathogen with high morbidity and mortality that ranges from 18 to 61% (Kang et al., 2003) and can survive in bottled water for more than 50 days (Khaniki et al., 2014). However, the survival time heavily relies on temperature, with high temperature leading to better viability. Although Yersinia pestis has a vector-borne transmission route and causes endemic bubonic plague, it can also be transmitted from human-to-human via air droplets and causes pneumonic plague (Galimand et al., 2006). It has the ability to survive and remain virulent in the external environment for more than 280 days, which makes it one of most dangerous sit-and-wait bacterial pathogens (Ayyadurai et al., 2008). Without antibiotic treatment, the death rate would be 100% within 24-h infection, while mortality of treated patients can approach 50% (Pechous et al., 2016).
Vector-borne pathogens tend to have a comparatively shorter external survival time or no lifecycle out of hosts/vectors at all, due to their highly developed adaptation to their host environment. For example, Rickettsia rickettsii cannot survive outside hosts (Azad, 2007). However, our review of the literature did not found survival times of Anaplasma phagocytophilum, Borrelia burgdorferi, and Ehrlichia chaffeensis in the environment. The reported external survival time of the three pathogens in Table 2 is actually their viability in in vitro cells or blood products under refrigeration conditions (Badon et al., 1989; Kalantarpour et al., 2000; McKechnie et al., 2000). Thus, these pathogenic bacteria could be very sensitive to environmental stresses and can only survive within host or vector cells. F. tularensis is an exception and is able to survive in the water system for up to 2 months (Mironchuk and Mazepa, 2002), which suggests that F. tularensis may have multiple transmission routes, hence, higher virulence. The vector-borne pathogens normally have high mortality rate (Titball and Sjostedt, 2003; Ryan and Ray, 2004). Although Lyme disease is reported to be rarely fatal (Kugeler et al., 2011), if left untreated, the mortality rate of the disease can be high. A similar situation also applies to A. phagocytophilum and E. chaffeensis infections. Obligate intracellular bacteria (commensal, symbiotic, and pathogenic) are all exclusively associated with hosts, with extremely weak environmental survival ability and do not contribute to host illness and mortality or only cause mild syndromes to the host. The maximal survival time falls into the scale of several hours. Finally, free-living bacteria generally have long survival times but do not have the ability to attack humans, so there is no reported mortality. For representative examples, please refer to Table 2.
Comparative analysis of the four bacterial groups in terms of external survival and mortality shows obvious differences among each other. Sit-and-wait pathogens generally have higher mortality rate and strong durability in the environment. Although vector-borne pathogens seem to be long-lived and highly pathogenic (Table 2), the survival data come from persisting in in vitro cells and blood samples. There are insufficient reports relating to their environmental survival. As for obligate intracellular bacteria, due to their high dependence on hosts, both the durability and mortality are inconsequential. Finally, free-living bacteria display effectively infinite survival times in the environment but no host mortality. The epidemiological data clearly separate the bacteria into four groups according to the durability and mortality factors. In addition, high durability and virulence are apparently associated with sit-and-wait pathogens.
Bioinformatic and Statistical Analysis of Energy Reserves
When t-tests were used to compare the counts of durable-energy-reserve pathways in the sit-and-wait species versus the other groups of species, we noted that sit-and-wait species have significantly more than both host-associated (P-value 0.000105) and vector-born (P-value 0.000105) species, but the counts of energy reserve pathways in the sit-and-wait species are similar to those found in free-living species (P-value 0.545) (Figure 2). That is, sit-and-wait pathogens and free-living bacteria normally use more than one storage compound for energy metabolism, while vector-borne and intracellular bacteria seem to almost completely lose the ability to accumulate and utilize energy compounds. This expands the conclusion of Henrissat et al. (2002) that glycogen metabolism loss could be a marker for bacterial parasitic lifestyle.
Among the five energy storage reserves studies, wax ester (WE) is a neutral lipid that is accumulated as intracellular storage compounds by a few bacteria, such as Acinetobacter spp. and Rhodococcus opacus (Ishige et al., 2003). In addition, WE is also reported to be detected in the following bacteria: Moraxella, Micrococcus, Fundibacter, Neisseria, Marinobacter, Pseudomonas, Corynebacterium, Mycobacterium, and Nocardia (Ishige et al., 2003; Waltermann and Steinbuchel, 2005). A recent study shows that the final enzyme responsible for WE synthesis in bacteria is the bifunctional wax ester synthase/acyl-coenzyme A (acyl-CoA): diacylglycerol acyltransferase (WS/DGAT), which exhibits both acyl-CoA: fatty alcohol acyltransferase (WE synthesis) and acyl-CoA: diacylglycerol acyltransferase (TAG synthesis) activity (Kalscheuer and Steinbuchel, 2003). Thus, TAG and WE share the same final synthesis step. Both TAG and WE possess higher calorific value than carbohydrates and proteins, which means that when oxidized, more energy is released. That is why these reserves are able to support the environmental persistence of sit-and-wait bacteria. A recent study by Sirakova et al. (2012) shows that the dormancy of TAG and WE deficient Mtb strains via inactivating fatty acyl-CoA reductase genes is disrupted with increased cell wall permeability, high metabolic activity, and also impaired antibiotic tolerance (Sirakova et al., 2012), which indicates that the two energy storage compounds play essential roles in the environmental persistence of Mtb. Another bacterial energy reserve PHB acts as an osmotically neutral compound for carbon and energy storage (Uchino et al., 2007). Many bacteria accumulate it during stationery phase, especially when nitrogen is limited and a carbon source is abundant, which makes it a more widespread energy reserve than TAG and WE in bacteria. Sporadic studies have already proposed that accumulation of PHB can increase bacterial survival ability (Lopez et al., 1998). For example, B. pseudomallei, which does not store glycogen at all but is able to accumulate and utilize PHB, is rather durable in the external environment (Inglis and Sagripanti, 2006).
PolyP is a linear polymer of 100s of inorganic phosphate residues linked together by higher-energy phosphoanhydride bonds. It is ubiquitous in Nature and has been claimed to be present in all bacteria (Rashid et al., 2000a). In addition, it is labile and rapidly used up. Thus, there is no much literature studying its distribution in bacteria although it has been linked with bacterial mobility, biofilm development, long-term survival, and virulence (Rashid et al., 2000a,b; Kim et al., 2002). Last, but not the least, among the five energy reserves is glycogen, a homopolysaccharide consisting of glucosyl residues only. Currently, glycogen has been reported to exist in more than 50 bacteria across all bacterial species (Preiss, 2009). Theoretical study suggests that more than 300 bacterial proteomes harbor a complete glycogen metabolism pathway (Wang and Wise, 2011). According to numerous experimental studies, glycogen is strongly linked with bacterial lifestyle, persistence, and virulence, etc. (Strange, 1968; Bonafonte et al., 2000; Henrissat et al., 2002; Wilson et al., 2010).
Individually, each of the five energy storage compounds contributes to bacterial durability. With more than one ER in a bacterium, it would be strong advantage for the species to survive harsh conditions. This also explains why sit-and-wait pathogens normally harbor more than one type of energy reserves. However, only glycogen and PolyP metabolism have been shown experimentally to impact bacterial virulence at currently stage, which links durability and virulence together. As for WE, TAG, and PHB, more studies are required and similar tests should be performed in order to check their contributions to the virulence of pathogenic bacteria at molecular level.
Bioinformatic and Statistical Analysis of Abiotic Stress Resistance
Analysis of the second factor, ASR, provided similar groupings. The group of sit-and-wait species has a significantly larger percentage of ASR proteins compared with the host-associated species (P-value 0.0143, versus threshold of 0.05) and compared with the vector-born species (P-value 0.00871). This is understandable, as host-associated species and vector-born species are largely shielded from the external environment. Interestingly, the sit-and-wait species also have a significantly higher percentage of ASR protein when compared with the free-living species (P-value 0.0357). We speculate that this may be because sit-and-wait pathogens have not only to deal with a hostile external environment, but also host immune system attack.
Bioinformatic and Statistical Analysis of Virulence Factors
The situation for the third factor, presence of virulence factors, is a little more complicated. It can be argued that the number of virulence factors do not represent the degree of bacterial virulence due to the influences of gene transcription and post-translational modification. This is to some level true. However, the number of VFs is a sufficient indicator to reflect the diverse mechanisms that bacteria are armed with to cause harm in hosts, which is an important aspect of bacterial virulence. Analyzing the percentages of virulence factors in Supplementary Table S2 as a proxy for virulence, we found that sit-and-wait pathogens have a significantly higher percentage than vector-born species (P-value 0.0442), a higher percentage (but not significantly so) than free-living species and a similar percentage to host-associated species (P-value 0.639). The first of these results is likely due to vector-born pathogens needing to evade the vector immune systems of both the vector species and the eventual host, which may constrain the range of virulence factors. The vector-borne pathogen F. tularensis appears to be an outlier in this group with its large percentage of virulence factors, which matches its mortality very well. ESM analysis also shows that F. tularensis has the complete glycogen metabolism pathway. Although it is accepted as a vector-borne pathogen, it is also reported to have long-term survival ability, which makes it a potential pathogen similar to Yersinia pestis for multi-modal transmission (Mironchuk and Mazepa, 2002). Further investigation into the composition of the virulence factors should be performed for a better understanding of this phenomenon.
Comparison of sit-and-wait pathogens and host-associated bacteria in terms of the number of virulence factors did not find significant difference. Such a result is reasonable since host-associated bacteria also need to invade and colonize hosts, which should necessitate them being equipped with a set of VFs. As for non-pathogenic free-living bacteria, it is a little bit confusing to see that they have considerable number of virulence factors in the proteomes. The reason for the paradox is that virulence factors have also been identified in non-pathogenic bacteria (Niu et al., 2013). Keen reported that non-pathogenic bacteria are able to encode VFs like types III and IV secretion systems previously considered unique to pathogens (Keen, 2012). These findings make the classical definition of virulence factors controversial. A recent study classified VFs into two categories: pathogen-specific VFs and general VFs and general VFs are common in non-pathogenic bacteria (Niu et al., 2013). In our preliminary study, we used all VFs as a single group in order to search proteomes thoroughly for all VFs. For future work, we will make our screening subtler by using different categories of VFs, which might differentiate distribution patterns of VFs between bacterial groups.
Proteome Size and Bacterial Lifestyles
Genome reduction is the dominant theory for bacteria transiting from free-living to facultative and finally to intracellular bacteria with parasitic or symbiotic lifestyles, and has been widely observed in Nature. Examples include Mycoplasma and Rickettsia (Moran, 2002; Wolf and Koonin, 2013). For the 20 bacteria listed in Supplementary Table S2 separated by transmission modes, the analysis of bacterial proteome sizes shows obvious differences between the four groups. The average proteome sizes for sit-and-wait, vector-borne, host-associated, and free-living bacteria are 3989, 1295, 855, and 3547, respectively. Statistical analysis shows that proteome sizes of sit-and-wait pathogens are significantly different from vector-borne (P-value 0.00351) and host-associated bacteria (P-value 0.00177), but are not significantly different from those of free-living bacteria (P-value 0.543). Considering that both sit-and-wait pathogens and free-living bacteria have a long-term external survival period, they need to adapt to different niches and cope with a variety of stresses. These results are consistent with epidemiological data in terms of bacterial lifestyle. Secondly, distribution patterns of ESMs (Wilkinson, 1963) shown in Figure 2 match with transmission modes and also correlate with bacterial proteome sizes.
Potential Targets for Testing the Linkages between Durability and Virulence
Theoretical analysis in this study shed light on the relationship between durability and virulence, although the small sample volume compromises its strength. By incorporating previously experimental studies, we collected several candidates that may support this linkage, which may worth further experimental exploration. For example, urease, an enzyme that catalyzes the hydrolysis of urea into carbon dioxide and ammonia, has been reported to enhance survival of pathogens that experience acidic environments, such as Yersinia pseudotuberculosis (Hu et al., 2009), Helicobacter pylori (Stingl et al., 2002; Schoep et al., 2010), Haemophilus influenzae (Murphy and Brauer, 2011), and Edwardsiella ictaluri (Booth et al., 2009), by neutralization of the acidic environments. Interestingly, urease has also been suggested to be a virulence factor that contributes to the persistence and replication in low pH intracellular compartments of macrophages (Booth et al., 2009; Murphy and Brauer, 2011). Such a linkage shows that bacterial resistance to abiotic stresses may be a contributor to virulence.
Another abiotic stress that bacteria frequently encounter is ROS, such as superoxide anion radical, hydrogen peroxide, and hydroxyl radicals. ROS causes damages to DNA, RNA, protein, and lipids (Cabiscol et al., 2000). For example, intestinal mucosa uses epithelial NADPH oxidases to generate ROS for defending against Campylobacter jejuni infection, impairing bacterial capsule formation and decreasing C. jejuni virulence (Corcionivoschi et al., 2012). Oxidative stress has also been reported to affect the pathogenicity of E. coli by decreasing the expression of all virulence factors (Hegde et al., 2008). In addition, when invading hosts, bacterial pathogens risk being phagocytosed by phagocytes that generate superoxide and other ROS in order to kill these intruders (Hassett and Cohen, 1989). Environmental factors, such as desiccation and near UV radiation, can also generate intracellular oxygen ions (Cabiscol et al., 2000). In order to survive these harsh conditions, bacteria develop a set of mechanisms for sensing ROS acutely and maintaining ROS at a certain steady level. Once bacteria sense high-level ROS, defense mechanisms are initiated for getting rid of ROS by expressing antioxidant enzymes, such as superoxide dismutase and catalase, and repairing oxidative damages, such as initiating DNA-repair systems.
Take B. pseudomallei and Salmonella typhimurium for example. Both of the bacteria are human pathogens that are capable of intracellular survival in human macrophages and can synthesize superoxide dismutase (SOD, EC = 1.15.1.1), an enzyme that changes superoxide, a compound with superoxide anion (O2-) generated by passing a single electron to molecular oxygen (Shvinka et al., 1979), into oxygen and hydrogen peroxide (H2O2). Comparisons of wild type and SOD-deficient strains of both pathogens revealed that viabilities of SOD-deficient strains decrease and presence of SOD is required for pathogen virulence (De Groote et al., 1997; Vanaporn et al., 2011). However, hydrogen peroxide is still a comparatively strong oxidizer for bacteria. Thus, in order to avoid its damage, many bacteria possess an enzyme known as catalase, which can decompose H2O2 into H2O and O2. Mandell (1975) studied catalase functions in Staphylococcus aureus and concluded that catalase has a role in protecting intraphagocytic microbes and may be an important virulence factor. Recent investigation confirmed the potential role that catalase plays for the virulence of pathogens by studying Leptospira interrogans (Eshghi et al., 2012). However, this is not a solid relationship. A study of Salmonella typhimurium showed that catalase mutant was resistant to macrophages and retained full virulence (Buchmeier et al., 1995). Another study focusing on Staphylococcus aureus showed that catalase-negative strain retained virulence in a mouse model of chronic granulomatous disease, which contradicted with the result reported previously (Messina et al., 2002). Thus, although antioxidant enzymes are widely accepted as bacterial protectors against oxidative stresses, their role as a virulence factors should be investigated more deeply.
Desiccation is also a common abiotic stress for free-living stage bacteria in drought-like conditions. For example, many nosocomial pathogens have the ability to persist on dry inanimate surfaces, such as scissors, metal plates, and fabrics etc., in order to increase their opportunities to transfer from one patient to another, which is why inanimate surfaces are often considered as the source for outbreaks of nosocomial infections (Kramer et al., 2006). However, bacterial desiccation resistance varies. Helicobacter pylori, a Gram-negative bacterium causing non-lethal gastritis, can only survive for less than 90 min on dry surfaces while Corynebacterium diphtheriae, a pathogenic bacterium that causes diphtheria with fatality rates of 36.5%, is able to persist on dry surfaces for up to 6 months (Kramer et al., 2006). This empirical evidence also indicates a possible linkage between ASR and virulence. Currently, several desiccation resistance mechanisms have been reported for bacteria. The most well-known mechanism is to use non-reducing disaccharides, trehalose and sucrose, as desiccation stress protectants. Trehalose is a disaccharide with a α-1,1-glucosidic bond linking two glucosyl residues. Sucrose is composed of glucose and fructose. Non-reducing disaccharides increase bacterial desiccation tolerance mainly by lowering the temperature of dry membrane phase transition and maintaining general protein structure (Leslie et al., 1995). In addition, these disaccharides are responsible for the formation of vitreous cytoplasmic matrix, a virulence factor for bacterial pathogenicity (Garcia, 2011). Besides, trehalose metabolism is also interlinked with glycogen and maltose pathway, the later contributing to E. coli O157:H7 colonization in the intestine of mice hosts (Jones et al., 2008). The relationship between trehalose, maltose, and glycogen could serve a good example for tackling the sit-and-wait hypothesis experimentally by linking ER, ASR, and virulence all together.
Conclusion
In this study, we systematically explored the recent progress of the sit-and-wait hypothesis. Epidemiological data were collected to investigate the relationship between durability and virulence through representative bacterial pathogens. Our analysis provides preliminary evidence that non-vector-borne pathogens with sit-and-wait potentials investigated in this study have higher durability and virulence compared with other groups of bacteria, although the conclusion is subjected to limited bacterial sample volume. In addition, two sets of HMMs were used to scan proteomes from a sample set of bacterial species for proteins related ASR and the presence of durable energy storage pathways, while the presence of virulence factors was assessed via sequence searches. Differential patterns were identified in terms of bacterial proteome size, ESMs, ASR, and also virulence factors. All of these results support that sit-and-wait pathogens have multiple ESMs and higher number of durability and virulence genes. It is noteworthy that our results are based on a small sample volume, that is, only five species for each category. However, our study is the first theoretical support for the hypothesis at the genetic level through bioinformatics methodologies. We urge that further efforts should consider exploring the topic with an increased sample size through similar methods. In addition, this study also provides protein targets for experimentally investigating the hypothesis, such as proteins related with ESMs and also ASR. In conclusion, molecular experimental studies should be performed in order to provide evidence at the cellular and population levels for this hypothesis.
Author Contributions
LW and MW conceived the core idea and wrote the manuscript. MW also contributed to the statistical analysis. LW and ZL did all data collection and statistical analysis. SD and JY contributed to data analysis and manuscript writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Startup Fund for Excellent Researchers from Xuzhou Medical University (Project No. D2016007), Nature and Science Fund for Colleges and Universities of Jiangsu Province (Project No. 16KJB180028), Science and Technology Bureau of Xuzhou Municipal Funding (Project No. XZZDY1612), and also Jiangsu Students’ Platform for Innovation and Entrepreneurship Training Program provide financial support for the project. Part of the project was conducted at The University of Western Australia and supported by a UWA China Scholarship. A part of this paper has been publicly available in LW Ph.D. dissertation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02167/full#supplementary-material
References
Achbergerova, L., and Nahalka, J. (2011). Polyphosphate-an ancient energy source and active metabolic regulator. Microb. Cell Fact. 10:63. doi: 10.1186/1475-2859-10-63
Albi, T., and Serrano, A. (2016). Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J. Microbiol. Biotechnol. 32, 27. doi: 10.1007/s11274-015-1983-2
Alizon, S. (2008). Transmission-recovery trade-offs to study parasite evolution. Am. Nat. 172, E113–E121. doi: 10.1086/589892
Alizon, S., Hurford, A., Mideo, N., and Van Baalen, M. (2009). Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259. doi: 10.1111/j.1420-9101.2008.01658.x
Anderson, R. M., and May, R. M. (1979). Population biology of infectious diseases: Part I. Nature 280, 361–367. doi: 10.1038/280361a0
Anderson, R. M., and May, R. M. (1980). Infectious diseases and population cycles of forest insects. Science 210, 658–661. doi: 10.1126/science.210.4470.658
Anderson, R. M., and May, R. M. (1982). Coevolution of hosts and parasites. Parasitology 85(Pt 2), 411–426. doi: 10.1017/S0031182000055360
Ayyadurai, S., Houhamdi, L., Lepidi, H., Nappez, C., Raoult, D., and Drancourt, M. (2008). Long-term persistence of virulent Yersinia pestis in soil. Microbiology 154, 2865–2871. doi: 10.1099/mic.0.2007/016154-0
Azad, A. F. (2007). Pathogenic rickettsiae as bioterrorism agents. Clin. Infect. Dis. 45, S52–S55. doi: 10.1086/518147
Badon, S. J., Fister, R. D., and Cable, R. G. (1989). Survival of Borrelia burgdorferi in blood products. Transfusion 29, 581–583. doi: 10.1046/j.1537-2995.1989.29789369673.x
Binns, D., Dimmer, E., Huntley, R., Barrell, D., O’Donovan, C., and Apweiler, R. (2009). QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 25, 3045–3046. doi: 10.1093/bioinformatics/btp536
Bonafonte, M. A., Solano, C., Sesma, B., Alvarez, M., Montuenga, L., Garcia-Ros, D., et al. (2000). The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191, 31–36. doi: 10.1111/j.1574-6968.2000.tb09315.x
Bonhoeffer, S., Lenski, R. E., and Ebert, D. (1996). The curse of the pharaoh: the evolution of virulence in pathogens with long living propagules. Proc. Biol. Sci. 263, 715–721. doi: 10.1098/rspb.1996.0107
Boor, K. J. (2006). Bacterial stress responses: what doesn’t kill them can make them stronger. PLOS Biol. 4:e23. doi: 10.1371/journal.pbio.0040023
Booth, N. J., Beekman, J. B., and Thune, R. L. (2009). Edwardsiella ictaluri encodes an acid-activated urease that is required for intracellular replication in channel catfish (Ictalurus punctatus) Macrophages. Appl. Environ. Microbiol. 75, 6712–6720. doi: 10.1128/AEM.01670-09
Brown, M. R., and Kornberg, A. (2004). Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. U.S.A. 101, 16085–16087. doi: 10.1073/pnas.0406909101
Brown, M. R., and Kornberg, A. (2008). The long and short of it - polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33, 284–290. doi: 10.1016/j.tibs.2008.04.005
Buchmeier, N. A., Libby, S. J., Xu, Y. S., Loewen, P. C., Switala, J., Guiney, D. G., et al. (1995). DNA-repair is more important than catalase for Salmonella virulence in mice. J. Clin. Invest. 95, 1047–1053. doi: 10.1172/JCI117750
Bull, J. J., and Lauring, A. S. (2014). Theory and empiricism in virulence evolution. PLOS Pathog. 10:e1004387. doi: 10.1371/journal.ppat.1004387
Cabiscol, E., Tamarit, J., and Ros, J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8.
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chen, L. H., Yang, J., Yu, J., Ya, Z. J., Sun, L. L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328.
Chien, M. F. J., Wijaya, L., and tan, T. T. (2015). High mortality and relapse rates of melioidosis (Burkholderia pseudomallei infections) in Singapore. Open Forum Infect. Dis. 2, 1628. doi: 10.1093/ofid/ofv133.1181
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Conlan, J. W. (2011). Francisella tularensis: a red-blooded pathogen. J. Infect. Dis. 204, 6–8. doi: 10.1093/infdis/jir224
Cook, M. J. (2015). Lyme borreliosis: a review of data on transmission time after tick attachment. Int. J. Gen. Med. 8, 1–8. doi: 10.2147/IJGM.S73791
Corcionivoschi, N., Alvarez, L. A., Sharp, T. H., Strengert, M., Alemka, A., Mantell, J., et al. (2012). Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59. doi: 10.1016/j.chom.2012.05.018
Costa, F., Hagan, J. E., Calcagno, J., Kane, M., Torgerson, P., Martinez-Silveira, M. S., et al. (2015). Global morbidity and mortality of leptospirosis: a systematic review. PLOS Negl. Trop. Dis. 9:e0003898. doi: 10.1371/journal.pntd.0003898
Dahlgren, F. S., Mandel, E. J., Krebs, J. W., Massung, R. F., and McQuiston, J. H. (2011). Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000-2007. Am. J. Trop. Med. Hyg. 85, 124–131. doi: 10.4269/ajtmh.2011.10-0613
Dandinarasaiah, M., Vikram, B. K., Krishnamurthy, N., Chetan, A. C., and Jain, A. (2013). Diphtheria Re-emergence: problems faced by developing countries. Indian J. Otolaryngol. Head Neck Surg. 65, 314–318. doi: 10.1007/s12070-012-0518-5
De Groote, M. A., Ochsner, U. A., Shiloh, M. U., Nathan, C., McCord, J. M., Dinauer, M. C., et al. (1997). Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 94, 13997–14001. doi: 10.1073/pnas.94.25.13997
Eddy, S. R. (2003). HMMER User’s Guide. Biological Sequence Analysis Using Profile Hidden Markov Models. Available at: http://hmmer.org/
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Elliot, S., Adler, F., and Sabelis, M. (2003). How virulent should a parasite be to its vector? Ecology 84, 2568–2574. doi: 10.1890/02-8013
Eshghi, A., Lourdault, K., Murray, G. L., Bartpho, T., Sermswan, R. W., Picardeau, M., et al. (2012). Leptospira interrogans catalase is required for resistance to H2O2 and for Virulence. Infect. Immun. 80, 3892–3899. doi: 10.1128/IAI.00466-12
Ewald, P. W. (1983). Host-parasite relations, vectors, and the evolution of disease severity. Annu. Rev. Ecol. Syst. 14, 465–485. doi: 10.1146/annurev.es.14.110183.002341
Ewald, P. W. (1987a). “Pathogen-induced cycling of outbreak and insect populations,” in Insect Outbreaks, ed. P. Barbosa (London: Academic Press), 269–286. doi: 10.1016/B978-0-12-078148-5.50015-4
Ewald, P. W. (1987b). Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 503, 295–306.
Ewald, P. W. (2004). Evolution of virulence. Infect. Dis. Clin. North Am. 18, 1–15. doi: 10.1016/S0891-5520(03)00099-0
Finkelstein, D. B., Brassell, S. C., and Pratt, L. M. (2010). Microbial biosynthesis of wax esters during desiccation: adaptation for colonization of the earliest terrestrial environments? Geology 38, 247–250. doi: 10.1130/G30398.1
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Galimand, M., Carniel, E., and Courvalin, P. (2006). Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 50, 3233–3236. doi: 10.1128/AAC.00306-06
Garcia, A. H. (2011). Anhydrobiosis in bacteria: from physiology to applications. J. Biosci. 36, 939–950. doi: 10.1007/s12038-011-9107-0
Ghodbane, R., Mba Medie, F., Lepidi, H., Nappez, C., and Drancourt, M. (2014). Long-term survival of tuberculosis complex mycobacteria in soil. Microbiology 160, 496–501. doi: 10.1099/mic.0.073379-0
Gubler, D. J. (2009). Vector-borne diseases. Rev. Sci. Tech. 28, 583–588. doi: 10.20506/rst.28.2.1904
Hassett, D. J., and Cohen, M. S. (1989). Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 3, 2574–2582.
Hegde, A., Bhat, G. K., and Mallya, S. (2008). Effect of exposure to hydrogen peroxide on the virulence of Escherichia coli. Indian J. Med. Microbiol. 26, 25–28. doi: 10.4103/0255-0857.38853
Hellstrom, J. S., and Marshall, R. B. (1978). Survival of Leptospira interrogans serovar pomona in an acidic soil under simulated New Zealand field conditions. Res. Vet. Sci. 25, 29–33.
Henrissat, B., Deleury, E., and Coutinho, P. M. (2002). Glycogen metabolism loss: a common marker of parasitic behaviour in bacteria? Trends Genet. 18, 437–440.
Holm, L., and Sander, C. (1998). Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics 14, 423–429. doi: 10.1093/bioinformatics/14.5.423
Hu, Y. B., Lu, P., Wang, Y., Ding, L. S., Atkinson, S., and Chen, S. Y. (2009). OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155, 2522–2531. doi: 10.1099/mic.0.028381-0
Inglis, T. J. J., and Sagripanti, J. L. (2006). Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 72, 6865–6875. doi: 10.1128/AEM.01036-06
Ishige, T., Tani, A., Sakai, Y. R., and Kato, N. (2003). Wax ester production by bacteria. Curr. Opin. Microbiol. 6, 244–250. doi: 10.1016/S1369-5274(03)00053-5
Jani, A. J., and Cotter, P. A. (2010). Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8, 2–6. doi: 10.1016/j.chom.2010.06.012
Jawad, A., Seifert, H., Snelling, A. M., Heritage, J., and Hawkey, P. M. (1998). Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36, 1938–1941.
Jones, S. A., Jorgensen, M., Chowdhury, F. Z., Rodgers, R., Hartline, J., Leatham, M. P., et al. (2008). Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76, 2531–2540. doi: 10.1128/IAI.00096-08
Kalantarpour, F., Chowdhury, I., Wormser, G. P., and Aguero-Rosenfeld, M. E. (2000). Survival of the human granulocytic ehrlichiosis agent under refrigeration conditions. J. Clin. Microbiol. 38, 2398–2399.
Kalscheuer, R. (2010). “Genetics of wax ester and Triacylglycerol biosynthesis in bacteria,” in Handbook of Hydrocarbon and Lipid Microbiology, 1st Edn, ed. K. Timmis (Berlin: Springer), 527–535.
Kalscheuer, R., and Steinbuchel, A. (2003). A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278, 8075–8082. doi: 10.1074/jbc.M210533200
Kamjumphol, W., Chareonsudjai, P., Taweechaisupapong, S., and Chareonsudjai, S. (2015). Morphological alteration and survival of Burkholderia pseudomallei in soil microcosms. Am. J. Trop. Med. Hyg. 93, 1058–1065. doi: 10.4269/ajtmh.15-0177
Kang, C. I., Kim, S. H., Kim, H. B., Park, S. W., Choe, Y. J., Oh, M. D., et al. (2003). Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 37, 745–751. doi: 10.1086/377200
Kashyap, S., and Sarkar, M. (2010). Mycoplasma pneumoniae: clinical features and management. Lung India 27, 75–85. doi: 10.4103/0970-2113.63611
Kato, C., and Qureshi, M. H. (1999). Pressure response in deep-sea piezophilic bacteria. J. Mol. Microbiol. Biotechnol. 1, 87–92.
Keen, E. C. (2012). Paradigms of pathogenesis: targeting the mobile genetic elements of disease. Front. Cell Infect. Microbiol. 2:161. doi: 10.3389/fcimb.2012.00161
Khaniki, G. R. J., Aghaee, E. M., Alimohammadi, M., and Dehghani, M. H. (2014). Effects of environmental conditions on growth and permanence of Pseudomonas aeruginosa in bottled water. J. Appl. Biol. Sci. 8, 91–95.
Kim, K. S., Rao, N. N., Fraley, C. D., and Kornberg, A. (2002). Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U.S.A. 99, 7675–7680. doi: 10.1073/pnas.112210499
Kishimoto, N., Kosako, Y., and Tano, T. (1991). Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22, 1–7. doi: 10.1007/BF02106205
Kontur, W. S., Schackwitz, W. S., Ivanova, N., Martin, J., Labutti, K., Deshpande, S., et al. (2012). Revised sequence and annotation of the Rhodobacter sphaeroides 2.4.1 genome. J. Bacteriol. 194, 7016–7017. doi: 10.1128/JB.01214-12
Kramer, A., Schwebke, I., and Kampf, G. (2006). How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6:130. doi: 10.1186/1471-2334-6-130
Kugeler, K. J., Griffith, K. S., Gould, L. H., Kochanek, K., Delorey, M. J., Biggerstaff, B. J., et al. (2011). A review of death certificates listing lyme disease as a cause of death in the United States. Clin. Infect. Dis. 52, 364–367. doi: 10.1093/cid/ciq157
Kulaev, I. S., Vagabov, V. M., and Kulakovskaya, T. V. (2004). The Biochemistry of Inorganic Polyphosphates. Hoboken, NJ: Wiley.
Leslie, S. B., Israeli, E., Lighthart, B., Crowe, J. H., and Crowe, L. M. (1995). Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl. Environ. Microbiol. 61, 3592–3597.
Levett, P. N. (2001). Leptospirosis. Clin. Microbiol. Rev. 14, 296–326. doi: 10.1128/CMR.14.2.296-326.2001
Lin, C. H., Lin, C. J., Kuo, Y. W., Wang, J. Y., Hsu, C. L., Chen, J. M., et al. (2014). Tuberculosis mortality: patient characteristics and causes. BMC Infect. Dis. 14:5. doi: 10.1186/1471-2334-14-5
Lopez, N. I., Ruiz, J. A., and Mendez, B. S. (1998). Survival of poly-3-hydroxybutyrate-producing bacteria in soil microcosms. World J. Microbiol. Biotechnol. 14, 681–684. doi: 10.1023/A:1008857420434
Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., et al. (2001). Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79. doi: 10.1128/MMBR.65.1.44-79.2001
Mandell, G. L. (1975). Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal–leukocyte interaction. J. Clin. Invest. 55, 561–566. doi: 10.1172/JCI107963
Marles-Wright, J., and Lewis, R. J. (2007). Stress responses of bacteria. Curr. Opin. Struct. Biol. 17, 755–760. doi: 10.1016/j.sbi.2007.08.004
May, R. M., and Anderson, R. M. (1979). Population biology of infectious diseases: Part II. Nature 280, 455–461. doi: 10.1038/280455a0
McKechnie, D. B., Slater, K. S., Childs, J. E., Massung, R. F., and Paddock, C. D. (2000). Survival of Ehrlichia chaffeensis in refrigerated, ADSOL-treated RBCs. Transfusion 40, 1041–1047. doi: 10.1046/j.1537-2995.2000.40091041.x
Messina, C. G. M., Reeves, E. P., Roes, E., and Segal, A. W. (2002). Catalase negative Staphylococcus aureus retain virulence in mouse model of chronic granulomatous disease. FEBS Lett. 518, 107–110. doi: 10.1016/S0014-5793(02)02658-3
Mironchuk, I., and Mazepa, A. V. (2002). Viability and virulence of Francisella tularensis subsp. Holarctica in water ecosystems (experimental study). Zh. Mikrobiol. Epidemiol. Immunobiol. 2, 9–13.
Moran, N. A. (2002). Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586. doi: 10.1016/S0092-8674(02)00665-7
Murphy, T. F., and Brauer, A. L. (2011). Expression of urease by Haemophilus influenzae during human respiratory tract infection and role in survival in an acid environment. BMC Microbiol 11:183. doi: 10.1186/1471-2180-11-183
Niu, C., Yu, D., Wang, Y., Ren, H., Jin, Y., Zhou, W., et al. (2013). Common and pathogen-specific virulence factors are different in function and structure. Virulence 4, 473–482. doi: 10.4161/viru.25730
Pechous, R. D., Sivaraman, V., Stasulli, N. M., and Goldman, W. E. (2016). Pneumonic plague: the darker side of Yersinia pestis. Trends Microbiol. 24, 190–197. doi: 10.1016/j.tim.2015.11.008
Peltola, H. (2000). Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13, 302–317. doi: 10.1128/CMR.13.2.302-317.2000
Perry, R. D., and Fetherston, J. D. (1997). Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66.
Poulin, S. A., Kundsin, R. B., and Horne, H. W. Jr. (1979). Survival of Ureaplasma urealyticum on different kinds of swabs. J. Clin. Microbiol. 10, 601–603.
Preiss, J. (2009). “Glycogen Biosynthesis,” in Encyclopedia of Microbiology, 3rd Edn, ed. M. Schaechter (Oxford: Elsevier), 145–158. doi: 10.1016/B978-012373944-5.00085-7
Rao, N. N., Gomez-Garcia, M. R., and Kornberg, A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647. doi: 10.1146/annurev.biochem.77.083007.093039
Rashid, M. H., Rao, N. N., and Kornberg, A. (2000a). Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182, 225–227.
Rashid, M. H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H., et al. (2000b). Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 9636–9641.
Roche, B., Drake, J. M., and Rohani, P. (2011). The curse of the Pharaoh revisited: evolutionary bi-stability in environmentally transmitted pathogens. Ecol. Lett. 14, 569–575. doi: 10.1111/j.1461-0248.2011.01619.x
Roszak, D. B., and Colwell, R. R. (1987). Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51, 365–379.
Ryan, K. J., and Ray, C. G. (2004). Sherris Medical Microbiology: An Introduction to Infectious Diseases. New York, NY: McGraw-Hill.
Schoep, T. D., Fulurija, A., Good, F., Lu, W., Himbeck, R. P., Schwan, C., et al. (2010). Surface properties of Helicobacter pylori urease complex are essential for persistence. PLOS ONE 5:e15042. doi: 10.1371/journal.pone.0015042
Schumann, W. (2007). “Bacterial stress sensors,” in Protein Reviews, Vol. 7, ed. S. K. Calderwood (New York, NY: Springer), 36–56.
Shams, A. M., Rose, L. J., Hodges, L., and Arduino, M. J. (2007). Survival of Burkholderia pseudomallei on environmental surfaces. Appl. Environ. Microbiol. 73, 8001–8004. doi: 10.1128/AEM.00936-07
Sherertz, R. J., and Sullivan, M. L. (1985). An outbreak of infections with Acinetobacter calcoaceticus in burn patients: contamination of patients’ mattresses. J. Infect. Dis. 151, 252–258. doi: 10.1093/infdis/151.2.252
Shvinka, J. E., Toma, M. K., Galinina, N. I., Skards, I. V., and Viesturs, U. E. (1979). Production of superoxide radicals during bacterial respiration. J. Gen. Microbiol. 113, 377. doi: 10.1099/00221287-113-2-377
Sirakova, T. D., Deb, C., Daniel, J., Singh, H. D., Maamar, H., Dubey, V. S., et al. (2012). Wax ester synthesis is required for Mycobacterium tuberculosis to enter in vitro dormancy. PLOS ONE 7:e51641. doi: 10.1371/journal.pone.0051641
Stingl, K., Altendorf, K., and Bakker, E. P. (2002). Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10, 70–74. doi: 10.1016/S0966-842X(01)02287-9
Sundberg, L. R., Kunttu, H. M., and Valtonen, E. T. (2014). Starvation can diversify the population structure and virulence strategies of an environmentally transmitting fish pathogen. BMC Microbiol. 14:67. doi: 10.1186/1471-2180-14-67
Sunenshine, R. H., Wright, M. O., Maragakis, L. L., Harris, A. D., Song, X., Hebden, J., et al. (2007). Multidrug-resistant acinetobacter infection mortality rate and length of hospitalization. Emerg. Infect. Dis. 13, 97–103. doi: 10.3201/eid1301.060716
Tang, K. H., Barry, K., Chertkov, O., Dalin, E., Han, C. S., Hauser, L. J., et al. (2011). Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 12:334. doi: 10.1186/1471-2164-12-334
The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Thomas, S. R., and Elkinton, J. S. (2004). Pathogenicity and virulence. J. Invertebr. Pathol. 85, 146–151. doi: 10.1016/j.jip.2004.01.006
Ting, L., Williams, T. J., Cowley, M. J., Lauro, F. M., Guilhaus, M., Raftery, M. J., et al. (2010). Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 12, 2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x
Tolba, O., Loughrey, A., Goldsmith, C. E., Millar, B. C., Rooney, P. J., and Moore, J. E. (2007). Survival of epidemic strains of nosocomial- and community-acquired methicillin-resistant Staphylococcus aureus on coins. Am. J. Infect. Control. 35, 342–346. doi: 10.1016/j.ajic.2006.10.015
Uchino, K., Saito, T., Gebauer, B., and Jendrossek, D. (2007). Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 189, 8250–8256. doi: 10.1128/JB.00752-07
van der Sluis, J. J., ten Kate, F. J., Vuzevski, V. D., Kothe, F. C., Aelbers, G. M., and van Eijk, R. V. (1985). Transfusion syphilis, survival of Treponema pallidum in stored donor blood. II. Dose dependence of experimentally determined survival times. Vox Sang. 49, 390–399. doi: 10.1111/j.1423-0410.1985.tb01131.x
Vanaporn, M., Wand, M., Michell, S. L., Sarkar-Tyson, M., Ireland, P., Goldman, S., et al. (2011). Superoxide dismutase C is required for intracellular survival and virulence of Burkholderia pseudomallei. Microbiology 157, 2392–2400. doi: 10.1099/mic.0.050823-0
Vezzulli, L., Pruzzo, C., Huq, A., and Colwell, R. R. (2010). Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ. Microbiol. Rep. 2, 27–33. doi: 10.1111/j.1758-2229.2009.00128.x
Waites, K. B. (2003). “Mycoplasma and Ureaplasma,” in Manual of Clinical Microbiology, 8th Edn, eds P. R. Murray, E. J. Baron, M. A. Pfaller, J. H. Jorgensen, and R. H. Yolken (Washington, DC: ASM Press), 19.
Waltermann, M., and Steinbuchel, A. (2005). Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 187, 3607–3619. doi: 10.1128/JB.187.11.3607-3619.2005
Walther, B. A., and Ewald, P. W. (2004). Pathogen survival in the external environment and the evolution of virulence. Biol. Rev. Camb. Philos. Soc. 79, 849–869. doi: 10.1017/S1464793104006475
Wang, L. (2013). Exploring the Genetic Basis for the Sit-and-Wait Hypothesis: Abiotic Stress Resistance and Energy Storage Mechanisms. Ph.D. thesis, University of Western Australia, Crawley WA.
Wang, L., and Wise, M. J. (2011). Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften 98, 719–729. doi: 10.1007/s00114-011-0832-x
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Whitehead, M. P., Eagles, L., Hooley, O., and Brown, M. R. (2014). Most bacteria synthesize polyphosphate by unknown mechanisms. Microbiology 160, 829–831. doi: 10.1099/mic.0.075366-0
Wilkinson, J. F. (1963). Carbon and energy storage in bacteria. J. Gen. Microbiol. 32, 171–176. doi: 10.1099/00221287-32-2-171
Wilson, W. A., Roach, P. J., Montero, M., Baroja-Fernández, E., Muñoz, F. J., Eydallin, G., et al. (2010). Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 34, 952–958. doi: 10.1111/j.1574-6976.2010.00220.x
Wise, M. J. (2010). No so HoT – heads or tails is not able to reliably compare multiple sequence alignments. Cladistics 26, 438–443. doi: 10.1111/j.1096-0031.2009.00292.x
Wolf, Y. I., and Koonin, E. V. (2013). Genome reduction as the dominant mode of evolution. Bioessays 35, 829–837. doi: 10.1002/bies.201300037
Wright, D. N., Bailey, G. D., and Hatch, M. T. (1968). Role of relative humidity in the survival of airborne Mycoplasma pneumoniae. J. Bacteriol. 96, 970–974.
Zhang, H., Ishige, K., and Kornberg, A. (2002). A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16678–16683. doi: 10.1073/pnas.262655199
Keywords: sit-and-wait hypothesis, pathogen, durability, virulence, energy storage mechanism, abiotic stress resistance
Citation: Wang L, Liu Z, Dai S, Yan J and Wise MJ (2017) The Sit-and-Wait Hypothesis in Bacterial Pathogens: A Theoretical Study of Durability and Virulence. Front. Microbiol. 8:2167. doi: 10.3389/fmicb.2017.02167
Received: 08 July 2017; Accepted: 23 October 2017;
Published: 03 November 2017.
Edited by:
John R. Battista, Louisiana State University, United StatesReviewed by:
Ilya R. Akberdin, San Diego State University, United StatesMichael Thomas Guarnieri, National Renewable Energy Laboratory (DOE), United States
Copyright © 2017 Wang, Liu, Dai, Yan and Wise. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Wise, michael.wise@uwa.edu.au Liang Wang, leonwang@xzhmu.edu.cn
†These authors have contributed equally to this work.
 Liang Wang1*†
Liang Wang1*† Zhanzhong Liu
Zhanzhong Liu Michael J. Wise
Michael J. Wise