- 1Institute of Applied Synthetic Chemistry, TU Wien, Vienna, Austria
- 2Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, United States
This paper describes the measurement and analysis of in vivo activity and stability of cyclohexanone monooxygenase from Acinetobacter sp. NCIMB 9871 (CHMO), a model Baeyer–Villiger monooxygenase, in the recombinant host Escherichia coli. This enzyme was often described as poorly stable in vitro, and has recently been found to deactivate rapidly in the absence of its essential cofactors and antioxidants. Its stability in vivo was scarcely studied, so far. Under conditions common for the overexpression of CHMO we investigated the ability of the host to support these properties using metabolomics. Our results showed that E. coli failed to provide the intracellular levels of cofactors required to functionally stabilize the enzyme, although the biocatalyst was produced in high concentration, and was invariably detected after protein synthesis had stopped. We thus infer that biotechnological applications of CHMO with this host relied on a residual activity of approximately 5-10%. Other microorganisms might offer a more efficient solution for recombinant production of CHMO and related enzymes.
Introduction
Flavin-dependent monooxygenases (FMOs), and especially Baeyer–Villiger monooxygenases (BVMOs), are useful biocatalysts for synthetic purposes because of their promiscuous acceptance of substrates (Torres Pazmiño et al., 2010; Bornscheuer et al., 2012; Fink et al., 2012; Bučko et al., 2016), and their mostly excellent stereoselectivity. Their dependence on oxygen as a terminal oxidant, and high activity at ambient temperature, can also offer technical advantages over (in)organic-synthetic methods (Bučko et al., 2016). A prominent member of this class of enzymes with a reportedly broad scope of substrates is cyclohexanone monooxygenase from Acinetobacter sp. NCIMB 9871 (CHMO) (Donoghue et al., 1976). Recently, it was found to be highly unstable in a cell-free setting (Van Beek et al., 2014). Such instability is likely common to many BVMOs, with few exceptions originating from thermophile organisms [PAMO (Fraaije et al., 2005) and TmCHMO (Romero et al., 2016)]. Genetic engineering has been used to enhance the stability and activity of CHMO in vitro: the introduction of stabilizing disulfide bonds led to an augmented thermodynamic stability by increasing the melting temperature of CHMO by 6°C (Van Beek et al., 2014), or 5°C (Schmidt et al., 2015). Still, even the improved variants showed poor kinetic stability (e.g., 33% residual activity after 24 h); the wild-type enzyme had a half-life of less than 2 min (Goncalves et al., 2017). As a consequence, even though CHMO and other BVMOs are chemically superior to other methods for the Baeyer–Villiger oxidation, their poor operational stability hinders applications on a scale relevant for the manufacture of fine or bulk chemicals (Balke et al., 2012). Only one industrial process (Bong et al., 2013) using a BVMO has been described, catalyzing the final step in the synthesis of esomeprazole (the API in the proton-pump inhibitor Nexium) on a multi-10 g scale by applying a heavily mutated CHMO variant (41 mutations).
A supposedly general solution for the lack of stability, or mitigation of its effects, was long seen in the use of recombinant whole-cell biocatalysts, e.g., Escherichia coli. Hypothetically, the maintenance of physiologically compatible conditions by the homeostatic cell would provide a stabilizing environment for fragile enzymes, such as CHMO. Failing that, the continuous re-synthesis of the enzyme, along with essential, spent cofactors, would still enable an efficient and conveniently usable system. For BVMOs, and specifically for CHMO, these assumptions are largely unchallenged, and were never quantitatively proven. Previous reports analyzed the effects of substrate and product inhibition, oxygen limitation, temperature, and pH on the operational stability of BVMOs as a whole-cell system (Doig et al., 2002, 2003; Baldwin and Woodley, 2006; Baldwin et al., 2008). It had been observed that a high intracellular concentration of NADPH was beneficial for the activity of CHMO (Walton and Stewart, 2004), but the mechanistic link between activity and operational stability of the BVMOs was hypothesized clearly, nor established experimentally.
In an in vitro study, we recently identified the following factors as influential in the deactivation of BVMOs in vitro: concentrations of (i) the BVMO itself, (ii) of the reduced cofactor NADPH, and (iii) of the coenzyme FAD (Goncalves et al., 2017). Supporting these results, we recently found by kinetic modeling that CHMO was the rate-limiting enzyme in a reaction cascade in vivo (Milker et al., 2017), even though it had the highest specific activity of the three sequential catalysts. We concluded that the low concentration of active CHMO was likely caused by an insufficient supply of NADPH and FAD in the cell. This study challenges the assumption that E. coli provides a beneficial environment for CHMO, or FMOs in general.
Materials and Methods
E. coli CHMO Batch Cultivation
Escherichia coli was cultivated in modified M9∗ minimal medium supplemented with 100 μg mL-1 ampicillin (For medium composition see Supplementary Information). Precultures were inoculated from late exponential phase-harvested, permanent cultures (-80°C, 1:100), and were grown for 16 h in a volume of 30 mL in non-baffled 500 mL Erlenmeyer flasks at 37°C and 350 rpm in an orbital shaker (InforsHT Multitron 2 Standard). The fermentation was performed at 37°C in a 2 L RALF fermenter (Bioengineering, Inc.) with a filling volume of 1 L. The stirring speed of the impeller turbines was set to 1000 rpm and the aeration rate kept constant at 1.5 vvm with air control by MX4/4 (DASGIP). The pH value was adjusted to 6.5 with 3 M NaOH and 3 M H3PO4, and inoculation was done at a ratio of 1:50 with the preculture. The off-gas analysis was performed with a GA4 off-gas analyzer (DASGIP). When foaming occurred antifoaming agent Antifoam O-30 (100 μL, Sigma–Aldrich) was added. At OD = 1, the bioreactor was cooled down to 20°C and the CHMO expression was induced with IPTG (100 μM, final concentration).
Cell Density
The cell density of cultures was measured via absorbance at 590 nm in a spectrophotometric cell before the culture liquid was used for subsequent assays. The first sample was taken directly after inoculation. Therefore, 1 mL of the reaction culture was sampled every hour, and if necessary, diluted in a ratio of 1:10 with the medium to stay within the linear range (up to 0.7) of the apparatus (WPA colorwave CO7500 Colorimeter). We used the following relation: 1 OD = 0.43 g DCW-1 to determine the dry cell weight (Walton and Stewart, 2004).
Cell Physiology
Samples for the analysis of the cell physiology were taken every hour. One mL of culture was transferred to a 1.5 mL Eppendorf reaction tube on ice and centrifuged (5000 × g, 10 min). The supernatant was filtered with a 0.2 μm syringe filter and stored at -20°C until HPLC analysis (Nexera Shimadzu). The supernatant was analyzed with a refractive index (RI) detector and photodiode array (PDA) detector for quantification of the analytes and an electrospray ionization (ESI) ion source with a quadrupole mass analyzer for additional confirmation of the substances (LC–MS 2020 Shimadzu). Separation was performed with an ROA-Organic Acid H+ (8%) column (300 mm × 7.8 mm, Phenomenex) with an isocratic flow of 0.4 mL min-1 5 mM formic acid in water.
Quantification of Soluble CHMO
The quantification of soluble CHMO was performed as single measurements per time point. The volume of the sample was adjusted, so that after centrifugation and the washing steps, the re-suspended cells (0.5 mL) would have an OD at 590 nm of 7.0 to later on have a quantifiable amount of CHMO. The sample was at first centrifuged (5000 × g, 10 min) and the resulting pellet was washed with 1 mL 50 mM Tris-HCl buffer, pH 7.5, then centrifuged again (5000 × g, 10 min), and stored at -20°C until further workup.
For cell lysis, the pellets were re-suspended in 0.5 mL 50 mM Tris-HCl, pH 7.5, containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were placed on ice and sonicated in nine cycles (5 s pulse, 55 s break, amplitude 50%, Bandelin KE76 sonotrode connected to a Bandelin Sonopuls HD 3200 wave generator). Precipitates were removed by centrifugation (15000 × g, 15 min) and the clear supernatants containing the soluble proteome, including CHMO, were analyzed by SDS-PAGE (Laemmli, 1970). Every SDS-PAGE gel was additionally loaded with at least three, differently concentrated samples of pure CHMO (range: 0.4–2.5 μg) in order to quantify the soluble CHMO concentration in every sample (for preparation of purified enzyme: see Supplementary Information). The gels were stained with SimplyBlue SafeStain (Thermo Fisher) using the microwave method according to the manufacturer’s protocol, scanned with the Molecular Imager Gel Doc XR System (BioRad), and quantified with the Image Lab Software (BioRad).
E. coli Metabolomics
The sampling procedure was repeated every hour as a single point, and every 2 h in triplicates unless stated otherwise. Metabolomics samples were taken using a fast filtration sampling procedure, as described elsewhere (Link et al., 2012) (for details see Supplementary Information).
LC–MS/MS Measurement
Dry extracts were re-suspended in water (100 μL) and centrifuged (12000 × g, 3 min). The supernatants were used for subsequent analyses. Separation was achieved with a Luna-NH2 HPLC column (150 mm × 2 mm, 3 μm particle size, 100 Å pore size, Phenomenex) using a binary gradient method (Solvent A: acetonitrile/Solvent B: 10 mM ammonium acetate, pH 9.9). Gradient parameters were as follows: 0–24 min: 20–100% B; 25–34 min: 100% B; 35 min: 20% B. Detection was performed with a tandem mass spectrometry detector with an ESI ion source (Shimadzu LCMS-8040) in multiple reaction monitoring (MRM) mode (Lorenz et al., 2011). Peak areas were normalized to fully 13C-labeled internal standards and absolute quantification of metabolites was achieved with linear calibration curves of the standards. Finally, concentrations were normalized to the amount of biomass. For MRM fragments see Supplementary Table S2.
Activity Measurements in Vivo
Bicyclo[3.2.0]hept-2-en-6-one was purchased from Sigma–Aldrich and used as obtained. The activity measurements of CHMO in the batch culture were performed every 3 h. The first measurement was done 3 h post-induction with IPTG. For the activity measurement 5 mL of fermentation broth were transferred to a 250 mL non-baffled Erlenmeyer flask, and bicyclo[3.2.0]hept-2-en-6-one (8 mM, 1 M solution in dioxane) was added as substrate. The culture was placed on an orbital shaker at 20°C and 350 rpm to mimic the fermentation conditions in terms of aeration, mixing, and temperature. The first sample was taken immediately after substrate addition. Therefore, 100 μL of the culture broth were extracted with 500 μL ethyl acetate containing methyl benzoate (1 mM) as internal standard. The activity test was performed for 1 h; samples were taken every 15 min. The consumption of substrate as well as formation of the corresponding lactone was monitored by calibrated GC. The activity was normalized to the concentration of biomass, and to the soluble CHMO content to obtain the specific activity.
Results
Hypotheses and Experimental Design
We investigated the levels of and changes in concentrations of the following metabolites in a whole-cell overexpression system, which had been identified as crucial for CHMO’s activity and stability in earlier studies in vitro and in silico: CHMO itself, NADPH, FAD (Goncalves et al., 2017; Milker et al., 2017). We based our study on the following hypotheses:
(A) CHMO, produced by over-expression, is stable in vivo over the duration of a preparative experiment.
(B) CHMO is produced by E. coli to a sufficient concentration to achieve high specific activity of the whole-cell catalyst.
(C) The cofactors FAD and NADP(H) are produced by E. coli to sufficiently high concentrations to efficiently maintain a high specific activity of CHMO.
(D) E. coli is a suitable organism to act as an efficient whole-cell biocatalyst with FAD- and NADP(H)-dependent enzymes.
To test these hypotheses, we designed and conducted an experiment for the production of CHMO under controlled conditions in a bioreactor (pH, temperature, oxygen saturation) and performed the following analyses: we measured the intracellular concentration of CHMO (via SDS-PAGE and densitometry) and determined the activity of CHMO in a Baeyer–Villiger oxidation in a satellite culture to characterize the performance of the whole-cell biocatalyst. We determined the concentrations of the cofactors FAD and NADPH and their respective precursors (GXP, ATP), plus several indicative metabolites: amino acids (building blocks for the synthesis of CHMO) and compounds of the central carbon metabolism and tricarboxylic acid cycle (TCA) to monitor the physiology of E. coli. The sampling schedule is described in Supplementary Table S1.
Activity, Stability, and Concentration of CHMO
We induced the production of CHMO with IPTG in the exponential phase, and held the culture stationary with >60 mM glucose for 15 h after reaching a limitation by the nitrogen (Supplementary Figure S1). The maximum concentration of soluble CHMO was measured after 19 h (20.9 μg mg DCW-1, 11 h after induction). This value decreased toward the end of the experiment to 15.2 μg mg DCW-1 (Figure 1) with a small exponential decay constant of approximately 0.04 (assuming first-order deactivation of the enzyme in the stationary phase). We thus observed a loss of 27% of the soluble CHMO. This observation led us to reject hypothesis A: CHMO was not stable over a time span common for a preparative experiment.
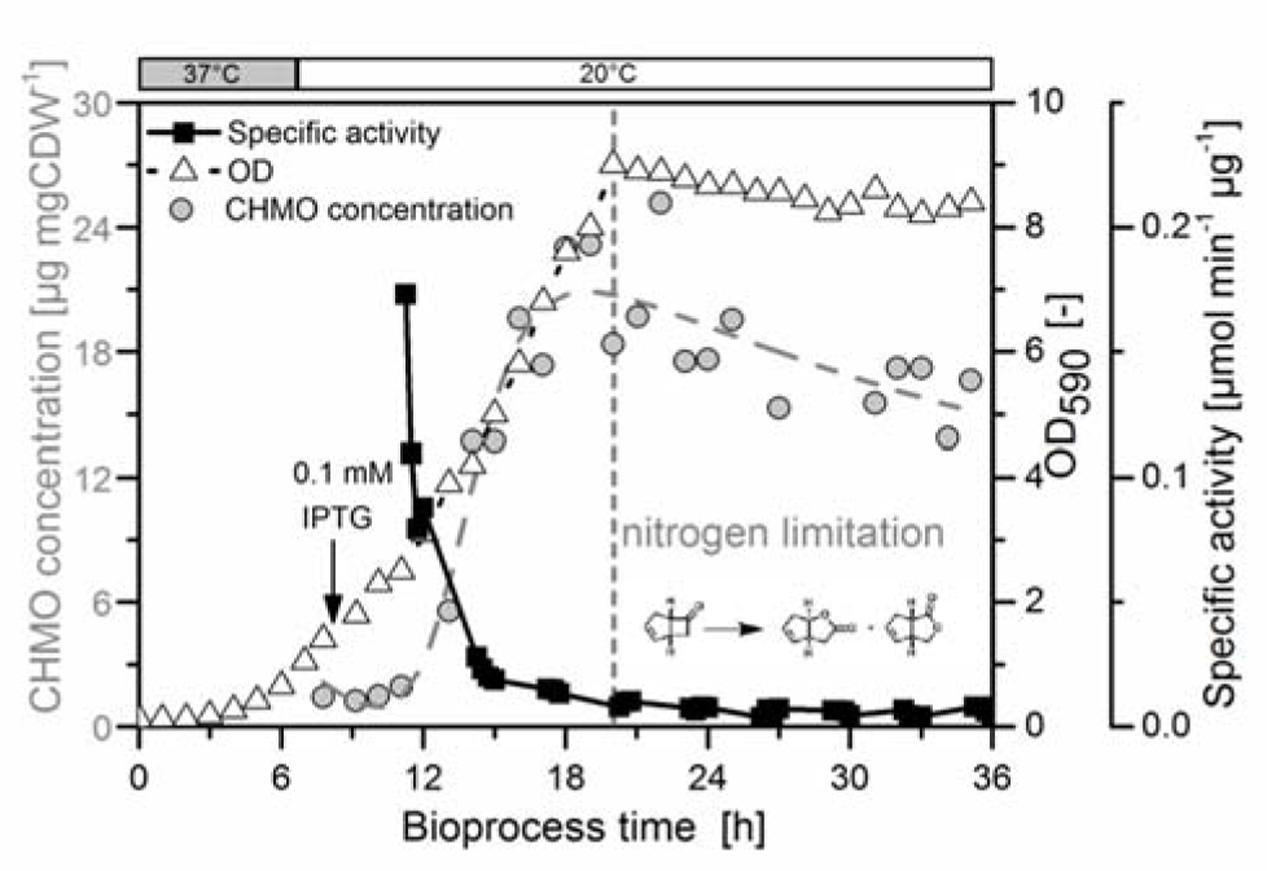
FIGURE 1. The specific activity of CHMO decayed rapidly to a basal level. E. coli BL21(DE3) pET22b::chmo was grown in minimal medium with sufficient aeration (see Supplementary Information, Supplementary Figure S2). The initial temperature of 37°C (exponential growth, growth rate at 37°C: 0.44 h-1) was lowered to 20°C after 8 h, shortly prior to induction with IPTG at a biomass concentration of 0.6 g dry cell weight per liter (g DCW L-1; OD590= 1.40). The culture reached nitrogen limitation after 20 h (maximum biomass concentration: 3.87 g DCW L-1). In the stationary phase the biomass decreased slightly to the final value of 3.61 g DCW L-1. The concentration of CHMO was determined by SDS-PAGE (Supplementary Figure S3) with a maximum value of 20.9 μg mg DCW-1 after 19 h, and was empirically interpolated using a composite function (exponential growth–exponential decay, see Supplementary Information). The activity of the cells in a Baeyer–Villiger oxidation of bicyclo[3.2.0]hept-2-en-6-one was measured in a satellite culture, which was incubated at the same temperature and comparable aeration/mixing conditions as in the bioreactor (Klöckner and Büchs, 2012). The specific activity was obtained by GC analysis, and normalized to the dry cell weight and to the protein concentration.
We also observed a fast decrease in the specific activity with rising concentration of soluble CHMO: the highest activity was observed at the first measurement (3 h after induction). The specific activity dropped by approximately 90% within the first 5 h, and then remained constant for the next 21 h (Figure 1). Despite the inverse relationship of the concentration of CHMO and its specific activity, E. coli produced enough CHMO to theoretically reach high specific activity, thus supporting hypothesis B.
Concentration of Cofactors NADPH and FAD
Based on our analytical results from metabolomics (quantification by 13C-standards in MS analysis), we estimated the intracellular concentrations of NADPH and FAD, using a published value for the volume of 3.2 ± 1.2 × 10-15 L per cell (Volkmer and Heinemann, 2011).
We estimated the cytosolic concentration of CHMO, but the propagated error from the estimated volume did not allow the identification of a significant trend (see Figure 2C); the mean value was still used for calculations. We observed stable intracellular concentrations of FAD (40 ± 16 μM, Figure 2A) and NADPH (63 ± 29 μM, Figure 2B) after adaption to 20°C (increase in the uptake of oxygen, Supplementary Figure S2). Since the cells were extracted vigorously in sampling procedure, the measurement could not differentiate between the free and bound form of FAD (or of any other metabolite that would form non-covalent complexes), preventing the exact calculation of equilibrium concentrations. That notwithstanding, and given the uncertainty in the estimated values, we made the following, drastically simplifying assumptions to obtain an extreme boundary condition: (i) the total concentration of FAD in the cell is equal to the concentration of unbound FAD, and (ii) CHMO is the only FAD-dependent enzyme in the cell. This way, a hypothetical, but optimum scenario was created for the population of the CHMO⋅FAD complex (i.e., loading 𝜃), calculated using a plain logistic function:
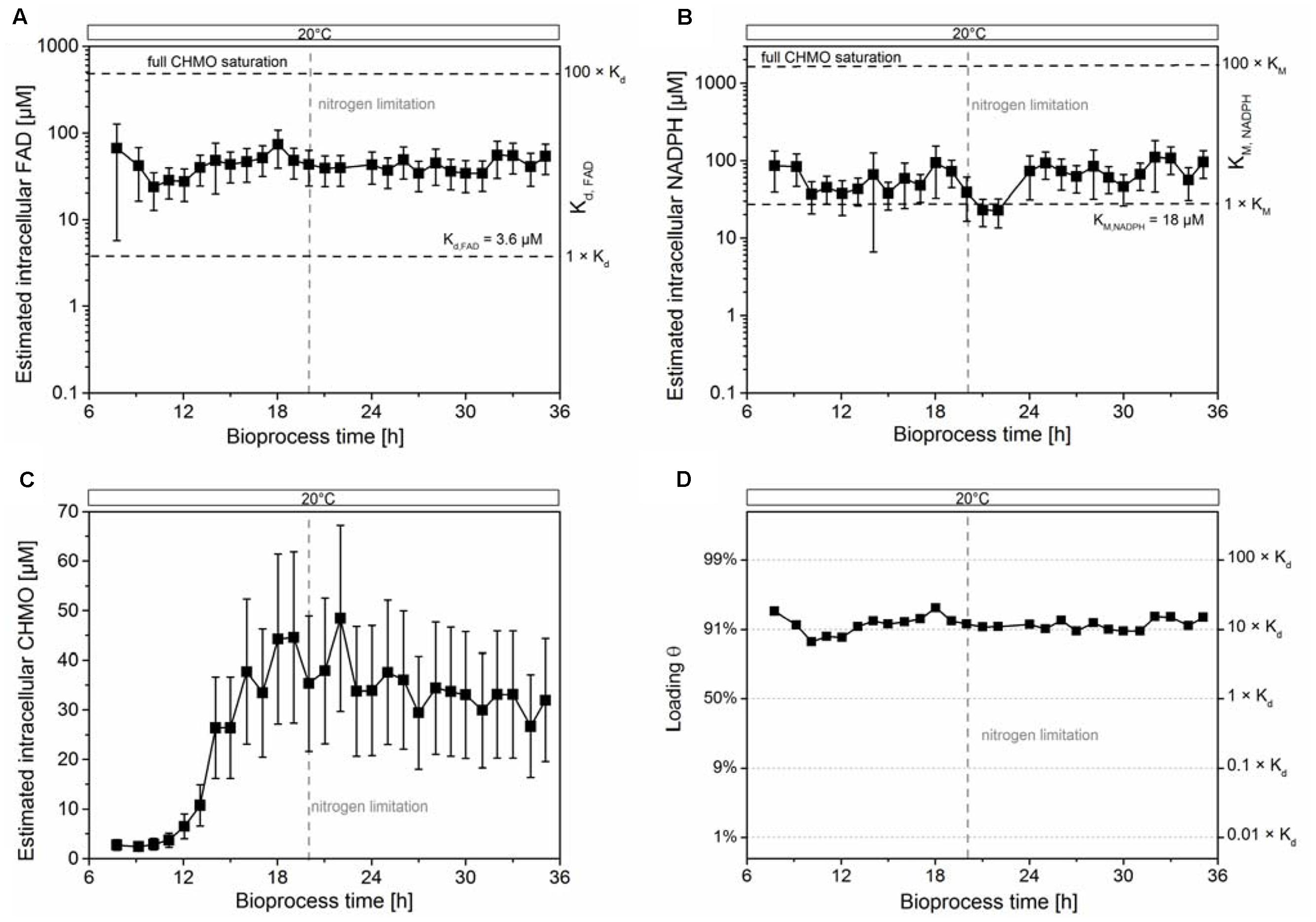
FIGURE 2. The intracellular concentrations of FAD and NADPH were insufficient to stabilize CHMO. The estimated intracellular concentrations of FAD and NADPH were stable (A,B). The estimated intracellular concentration of CHMO (C) and the estimated saturation of CHMO with FAD (D) was stable under nitrogen limiting conditions within experimental error. The compounded errors were propagated from the uncertainty of the single components (see Supplementary Methods). Data are reported as mean ± propagated error (1 SD; see Supplementary Information).
with [CHMO⋅FAD] as the concentration of the CHMO⋅FAD complex, [CHMOapo] as the concentration of the CHMO apoenzyme, Kd as the dissociation constant of FAD (3.5 ± 0.6 μM; Goncalves et al., 2017), and [FAD] as the total intracellular concentration of FAD. Even under these theoretical conditions, the intracellular concentration of FAD was only sufficient to saturate on average 90–95% of CHMO with the coenzyme, far from thermodynamic saturation (>>99% at [FAD] > 100 × Kd; Figure 2D). Given the large compounded uncertainty, and our extreme assumptions, we speculated that most of CHMO was actually present in its apo form. The decay of specific activity to a residual non-zero value corroborated the insufficient loading, indicating a rising lack of FAD with rising CHMO concentrations in the cell.
The estimated intracellular concentration of NADPH was sufficiently high to largely saturate the enzyme (0.73 × vmax, KM,NADPH = 18 μM; Figure 2B). Nevertheless, the concentration of NADPH was much lower than the level required for a strong stabilization, as earlier determined in vitro. There, a titer of >1 mM was found to have a large, beneficial effect on the thermodynamic and kinetic stability of CHMO (Goncalves et al., 2017). Together, these results could not clearly disprove hypothesis C on the basis of FAD; still, our data clearly showed that the cells could not provide enough of the coenzyme to saturate most CHMO molecules.
Biosynthesis of the Cofactors
We then analyzed the metabolic network (Figure 3) to identify potential bottlenecks in the metabolic upkeep of active CHMO [specifically FAD, NADPH, and key metabolites that are required for their synthesis: guanosine and adenosine mono-, di-, and triphosphates (GMP, GDP, GTP, AMP, ADP, and ATP)]. We measured the concentrations of these biosynthetic precursors (Figure 4) to determine if a shortage thereof was limiting the availability of the cofactor to CHMO. GTP decreased already during the exponential growth phase of the cells (Figure 4A). After the culture had reached limitation by nitrogen, the concentration of the GXP pool was stable at approximately 0.300 ± 0.01 μmol gCDW-1; the three compounds of this pool can be interconverted by the host metabolism. The concentration of ATP was stable over the entire observed period (approximately 2.0 ± 0.2 μmol gCDW-1; Figure 4B); the concentrations of AMP and ADP decreased until the onset of the nitrogen limitation, and were then stable. We could not discern any apparent, meaningful correlation between the concentrations of ATP or GTP and FAD, or a bottleneck in their supply. The lack in FAD was thus not directly, or not obviously, caused by a poor supply of its precursors.
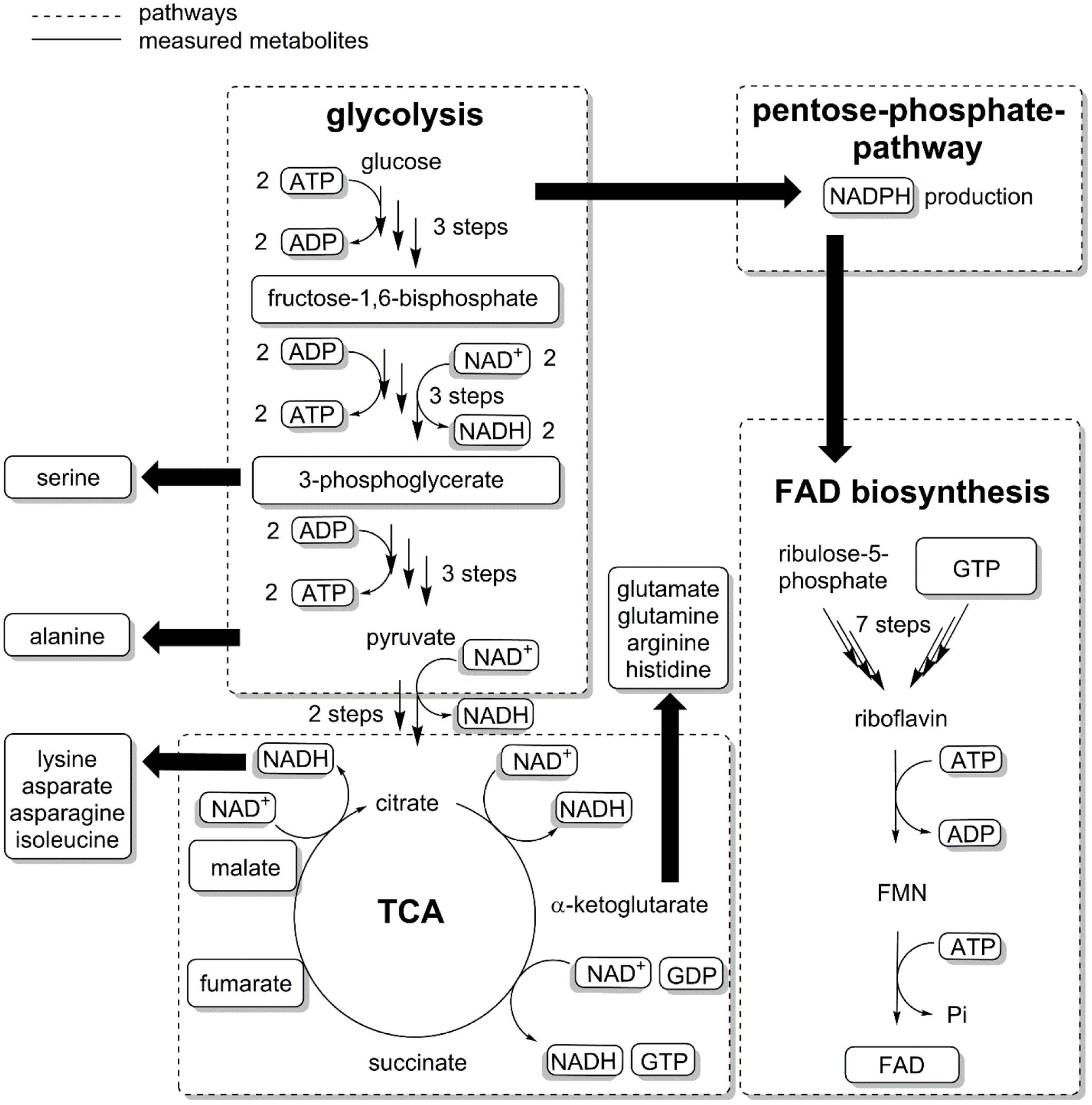
FIGURE 3. The biosynthesis of FAD requires GTP, ATP and ribulose-5-phosphate. GTP and Ribulose-5-phosphate are precursors for riboflavin, which after phosphorylation and reaction with ATP give FMN (flavin mononucleotide) and subsequently FAD. Metabolites were extracted from E. coli samples and measured with LC–MS/MS (see Supplementary Methods). Some metabolites were not quantified either because of low solubility (riboflavin) or insufficient resolution of the chromatographic method (ribulose-5-phosphate).
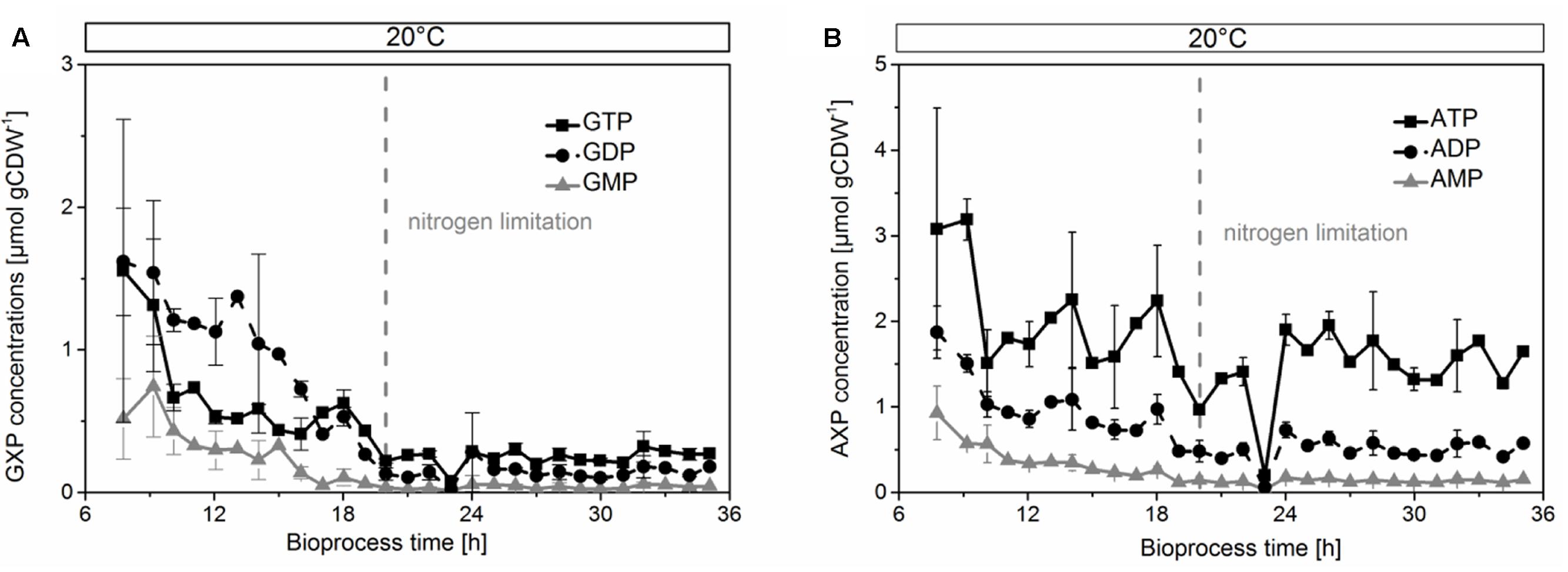
FIGURE 4. Concentrations of GXP (A) and AXP (B) decreased in the growing phase, but were stable during limitation by nitrogen. The concentrations were normalized by cell dry weight.
To assess the proper function of the host organism, we investigated indicative parameters for growth, physiology, and biocatalytic performance of E. coli. We found an overall decrease of the glycolytic intermediates fructose-1,6-bisphosphate and 3-phosphoglycerate (Figure 5A), and the TCA components fumarate and malate (Figure 5B) after the onset of the nitrogen limitation. These metabolites remained constant within experimental error after physiological adaption of the culture to 20°C. The heavy metabolic burden of heterologous overexpression became apparent in an unfavorable value for the adenylate energy charge (Figure 5C) of 0.65 at the beginning of protein overproduction. It later stabilized to 0.83 under nitrogen limitation. This value reflected a balanced pool of adenosine phosphates (Chapman et al., 1971) and cell viability until the end of the cultivation. The protein production stopped when the culture reached the nitrogen limitation, which was reflected in the concentration curves of most amino acids. Specifically, the low concentrations of glutamine, glutamate, and aspartate after the nitrogen limitation prevented the organism from further synthesizing CHMO (or other proteins, Supplementary Figures S4D–F).
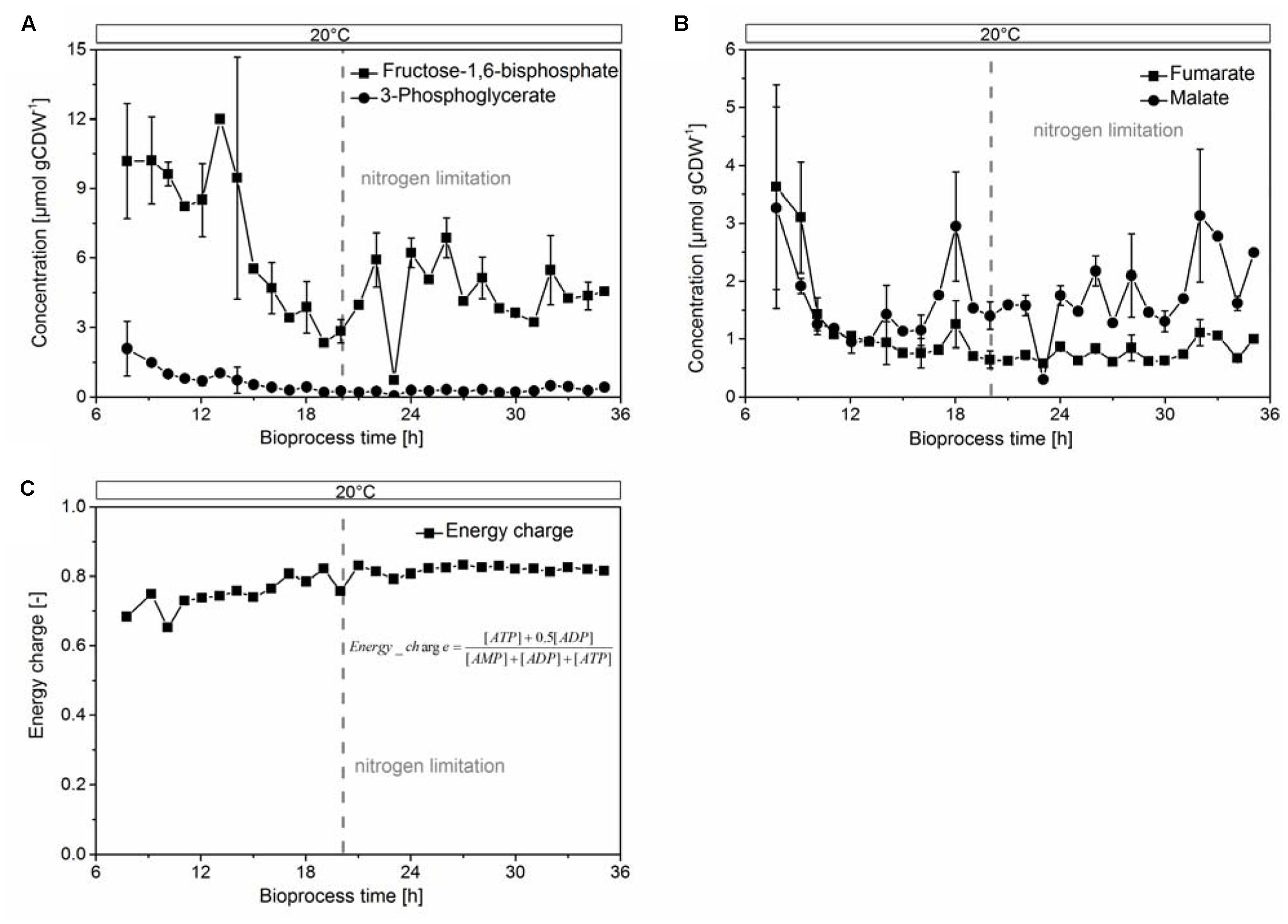
FIGURE 5. Glycolytic intermediates (A) fructose-1,6-bisphosphate, 3-phosphoglycerate; TCA intermediates (B) fumarate, malate, and the energy charge, calculated from the concentrations of ATP, ADP and AMP (C), were stable under nitrogen-limited conditions.
In summary, the host organism E. coli was found capable of maintaining the NADPH requirements of CHMO, providing a stable environment with sufficiently high levels of precursors for the protein itself, as well as for essential cofactors. However, due to the low concentration of FAD, it failed to fully saturate CHMO in overexpression. Although the experimental evidence cannot lead us to directly reject hypothesis D — all necessary compounds were invariably present to some extent—, it does not support the assumption that E. coli is a suitable host for the efficient maintenance of CHMO.
Discussion
Our findings suggest that the paradigm of placing a poorly stable biocatalyst, such as CHMO, into a living cell to provide for a stabilizing environment lacks specific experimental evidence. In regard of demanding requirements to stabilize CHMO (high concentrations of the cofactors NADPH and FAD), we compared in vitro (Goncalves et al., 2017) and in vivo reaction modes, and provided substantial evidence that generic in vivo stabilization utterly failed with CHMO; we additionally infer that this host organism would also fail with similar enzymes. Under our controlled reaction conditions, the peak concentration of soluble CHMO decreased slightly, but without any meaningful correlation to the significant loss in activity (Figure 1). This finding is in contrast to a previous study, which identified the (unspecified) decay of CHMO itself as main reason for the loss of activity (Walton and Stewart, 2004). We found that the supply of NADPH in this standard E. coli strain was sufficiently high to allow the enzyme to operate efficiently (approximately 3 × KM), but it was still orders of magnitude below a stabilizing concentration (approximately 1 mM). Similarly, the intracellular concentration of FAD was insufficient to completely saturate the enzyme. We speculate that the processes of deactivation and synthesis of the target enzyme were competing, as long as the culture was growing, as a plausible reason for the drop in specific activity during that period. When synthesis ceased, in the final phase of the experiment, a constant residual activity of CHMO in the range of 5–10% was observed, likely resulting from a small fraction of holo-CHMO.
We followed the biosynthetic pathway for FAD, starting from GTP and ribulose-5-phosphate, an intermediate of the pentose phosphate pathway (PPP) (Abbas and Sibirny, 2011) (Figure 3). Nitrogen limitation caused a decrease in PPP metabolites (Brauer et al., 2006), and thus interfered with the supply of building blocks for FAD. These results were corroborated by our previous study on an enzymatic cascade, where the presence of a second, overexpressed flavin-dependent enzyme in E. coli was found likely to decrease the availability of FAD (Milker et al., 2017). Even under the extreme assumption that CHMO was the only FAD-dependent enzyme in E. coli, the host failed to provide enough of the cofactor to reach maximum efficiency. Moreover, the genome of E. coli encodes for approximately 80 flavin-dependent enzymes (Macheroux et al., 2011). They presumably all are essential for growth, or homeostasis, to unknown relative proportions, and are thus constantly competing for the pool of available, free FAD. Given the low stability of the CHMO⋅FAD complex, even an FAD-overproducing organism might not provide sufficient amounts for complete functional stabilization.
These results strongly suggest that the investigated organism E. coli BL21 (DE3) is incapable of efficiently maintaining the activity of CHMO, an important FMO, under commonly used conditions for overexpression. Overall, its performance as a stabilizing host would only be sufficient for NADPH-dependent enzymes, but not if they also rely on FAD, with an affinity to that of CHMO. Analysis of the central carbon metabolism and the biosynthetic pathway for FAD indicated that the scarcity was a deep-rooted and widely distributed problem created by the metabolic network. Other well-explored species for heterologous expression, but with reportedly better flavin production (e.g., Pichia pastoris, Bacillus subtilis, Candida famata) (Abbas and Sibirny, 2011) might thus offer a more suitable artificial habitat for CHMO.
Author Contributions
MF and FR conceived the research and planned the experiments. All authors performed the culturing experiment and analyzed the data. SM performed the instrumental analyses. LG performed the protein purification. All authors co-wrote the manuscript and approved of the final version.
Funding
This work was supported by the Austrian Science Fund FWF (SM and FR; grant nos. I723-N17 and P24483-B20), by the COST Action Systems Biocatalysis CM1303 WG2 (SM and FR), by TU Wien (ABC-Top Anschubfinanzierung), and by the Coordination for the Improvement of Higher Education Personnel CAPES (LG; grant no. 2505-13-4).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Dr. Oliver Spadiut (TU Wien) provided access to and assistance with densitometry for the quantification of CHMO. We thank Dr. Roland Ludwig (BOKU Wien) and Prof. Marko D. Mihovilovic (TU Wien) for valuable discussions of the data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02201/full#supplementary-material
References
Abbas, C. A., and Sibirny, A. A. (2011). Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 75, 321–360. doi: 10.1128/MMBR.00030-10
Baldwin, C. V. F., Wohlgemuth, R., and Woodley, J. M. (2008). The first 200-L scale asymmetric Baeyer-Villiger oxidation using a whole-cell biocatalyst. Org. Process Res. Dev. 12, 660–665. doi: 10.1021/op800046t
Baldwin, C. V. F., and Woodley, J. M. (2006). On oxygen limitation in a whole cell biocatalytic Baeyer–Villiger oxidation process. Biotechnol. Bioeng. 95, 362–369. doi: 10.1002/bit.20869
Balke, K., Kadow, M., Mallin, H., Sass, S., and Bornscheuer, U. T. (2012). Discovery, application and protein engineering of Baeyer-Villiger monooxygenases for organic synthesis. Org. Biomol. Chem. 10, 6249–6265. doi: 10.1039/c2ob25704a
Bong, Y. K., Clay, M. D., Collier, S. J., Mijts, B., Vogel, M., Zhang, X., et al. (2013). Synthesis of prazole compounds. European Patent No. EP2010836590.
Bornscheuer, U. T., Huisman, G. W., Kazlauskas, R. J., Lutz, S., Moore, J. C., and Robins, K. (2012). Engineering the third wave of biocatalysis. Nature 485, 185–194. doi: 10.1038/nature11117
Brauer, M. J., Yuan, J., Bennett, B. D., Lu, W., Kimball, E., Botstein, D., et al. (2006). Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. U.S.A. 103, 19302–19307. doi: 10.1073/pnas.0609508103
Bučko, M., Gemeiner, P., Schenkmayerová, A., Krajčovič, T., Rudroff, F., and Mihovilovič, M. D. (2016). Baeyer-Villiger oxidations: biotechnological approach. Appl. Microbiol. Biotechnol. 100, 6585–6599. doi: 10.1007/s00253-016-7670-x
Chapman, A. G., Fall, L., and Atkinson, D. E. (1971). Adenylate energy charge in Escherichia coli during growth and starvation. J. Bacteriol. 108, 1072–1086.
Doig, S. D., Avenell, P. J., Bird, P. A., Gallati, P., Lander, K. S., Lye, G. J., et al. (2002). Reactor operation and scale-up of whole cell Baeyer-Villiger Catalyzed lactone synthesis. Biotechnol. Prog. 18, 1039–1046. doi: 10.1021/bp0200954
Doig, S. D., Simpson, H., Alphand, V., Furstoss, R., and Woodley, J. M. (2003). Characterization of a recombinant Escherichia coli TOP10 [pQR239] whole-cell biocatalyst for stereoselective Baeyer–Villiger oxidations. Enzyme Microb. Technol. 32, 347–355. doi: 10.1016/S0141-0229(02)00317-4
Donoghue, N. A., Norris, D. B., and Trudgill, P. W. (1976). The purification and properties of cyclohexanone oxygenase from Nocardia globerula CL1 and Acinetobacter NCIB 9871. Eur. J. Biochem. 63, 175–192. doi: 10.1111/j.1432-1033.1976.tb10220.x
Fink, M. J., Rial, D. V., Kapitanova, P., Lengar, A., Rehdorf, J., Cheng, Q., et al. (2012). Quantitative comparison of chiral catalysts selectivity and performance: a generic concept illustrated with cyclododecanone monooxygenase as Baeyer–Villiger biocatalyst. Adv. Synth. Catal. 354, 3491–3500. doi: 10.1002/adsc.201200453
Fraaije, M. W., Wu, J., Heuts, D. P. H. M., Van Hellemond, E. W., Spelberg, J. H. L., and Janssen, D. B. (2005). Discovery of a thermostable Baeyer–Villiger monooxygenase by genome mining. Appl. Microbiol. Biotechnol. 66, 393–400. doi: 10.1007/s00253-004-1749-5
Goncalves, L., Kracher, D., Milker, S., Rudroff, F., Fink, M. J., Ludwig, R., et al. (2017). Mutagenesis-independent, stabilization of class B flavin monooxygenases in operation. Adv. Synth. Catal. 359, 2121–2131. doi: 10.1002/adsc.201700585
Klöckner, W., and Büchs, J. (2012). Advances in shaking technologies. Trends Biotechnol. 30, 307–314. doi: 10.1016/j.tibtech.2012.03.001
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Link, H., Buescher, J. M., and Sauer, U. (2012). “Chapter 5 - Targeted and quantitative metabolomics in bacteria,” in Methods in Microbiology, eds H. Colin and W. Anil (Cambridge, MA: Academic Press), 127–150.
Lorenz, M. A., Burant, C. F., and Kennedy, R. T. (2011). Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 83, 3406–3414. doi: 10.1021/ac103313x
Macheroux, P., Kappes, B., and Ealick, S. E. (2011). Flavogenomics - a genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x
Milker, S., Fink, M. J., Oberleitner, N., Ressmann, A. K., Mihovilovic, M. D., Bornscheuer, U. T., et al. (2017). Kinetic modeling of an enzymatic redox cascade in vivo reveals cofactor-caused bottlenecks. ChemCatChem 9, 3420–3427. doi: 10.1002/cctc.201700573
Romero, E., Castellanos, J. R. G., Mattevi, A., and Fraaije, M. W. (2016). Characterization and crystal structure of a robust cyclohexanone monooxygenase. Angew. Chem. Int. Ed. Engl. 55, 15852–15855. doi: 10.1002/anie.201608951
Schmidt, S., Genz, M., Balke, K., and Bornscheuer, U. T. (2015). The effect of disulfide bond introduction and related Cys/Ser mutations on the stability of a cyclohexanone monooxygenase. J. Biotechnol. 214, 199–211. doi: 10.1016/j.jbiotec.2015.09.026
Torres Pazmiño, D. E., Dudek, H. M., and Fraaije, M. W. (2010). Baeyer–Villiger monooxygenases: recent advances and future challenges. Curr. Opin. Chem. Biol. 14, 138–144. doi: 10.1016/j.cbpa.2009.11.017
Van Beek, H. L., Wijma, H. J., Fromont, L., Janssen, D. B., and Fraaije, M. W. (2014). Stabilization of cyclohexanone monooxygenase by a computationally designed disulfide bond spanning only one residue. FEBS Open Bio 4, 168–174. doi: 10.1016/j.fob.2014.01.009
Volkmer, B., and Heinemann, M. (2011). Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLOS ONE 6:e23126. doi: 10.1371/journal.pone.0023126
Keywords: biotechnology, Escherichia coli, redox biocatalysis, Baeyer–Villiger monooxygenase, flavin, enzyme stability
Citation: Milker S, Goncalves LCP, Fink MJ and Rudroff F (2017) Escherichia coli Fails to Efficiently Maintain the Activity of an Important Flavin Monooxygenase in Recombinant Overexpression. Front. Microbiol. 8:2201. doi: 10.3389/fmicb.2017.02201
Received: 19 September 2017; Accepted: 26 October 2017;
Published: 13 November 2017.
Edited by:
Selin Kara, Technical University of Hamburg, GermanyReviewed by:
Dirk Tischler, Freiberg University of Mining and Technology, GermanyDominique Böttcher, University of Greifswald, Germany
Copyright © 2017 Milker, Goncalves, Fink and Rudroff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Fink, mfink@gmwgroup.harvard.edu Florian Rudroff, florian.rudroff@tuwien.ac.at
 Sofia Milker
Sofia Milker Leticia C. P. Goncalves
Leticia C. P. Goncalves Michael J. Fink
Michael J. Fink Florian Rudroff
Florian Rudroff