- 1Dental School, Oral Health Centre of Western Australia, The University of Western Australia, Perth, WA, Australia
- 2Laboratory of Oral Microbiology, Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
- 3Department of Prosthodontics, School of Dentistry, University of São Paulo, São Paulo, Brazil
- 4Dental Division Research, Guarulhos University, Guarulhos, Brazil
Probiotics are live microorganisms that confer benefits to the host health. The infection rate of potentially pathogenic organisms such as Candida albicans, the most common agent associated with mucosal candidiasis, can be reduced by probiotics. However, the mechanisms by which the probiotics interfere with the immune system are largely unknown. We evaluated the effect of probiotic bacteria on C. albicans challenged human macrophages. Macrophages were pretreated with lactobacilli alone (Lactobacillus rhamnosus LR32, Lactobacillus casei L324m, or Lactobacillus acidophilus NCFM) or associated with Escherichia coli lipopolysaccharide (LPS), followed by the challenge with C. albicans or LPS in a co-culture assay. The expression of pattern-recognition receptors genes (CLE7A, TLR2, and TLR4) was determined by RT-qPCR, and dectin-1 reduced levels were confirmed by flow cytometry. The cytokine profile was determined by ELISA using the macrophage cell supernatant. Overall probiotic lactobacilli down-regulated the transcription of CLEC7A (p < 0.05), resulting in the decreased expression of dectin-1 on probiotic pretreated macrophages. The tested Lactobacillus species down-regulated TLR4, and increased TLR2 mRNA levels in macrophages challenged with C. albicans. The cytokines profile of macrophages challenged with C. albicans or LPS were altered by the probiotics, which generally led to increased levels of IL-10 and IL-1β, and reduction of IL-12 production by macrophages (p < 0.05). Our data suggest that probiotic lactobacilli impair the recognition of PAMPs by macrophages, and alter the production of pro/anti-inflammatory cytokines, thus modulating inflammation.
Introduction
Candida albicans is the most common yeast isolated from the human mucosal surfaces (Scully et al., 1994). Despite its commensal nature, Candida causes oral and genital candidiasis that may evolve toward a systemic dissemination. For some decades, antifungals such as nystatin, amphotericin B, and fluconazole have been the drugs of choice for a successful treatment of oral candidiasis (McCullough and Savage, 2005). However, the administration of these agents may result in a temporary gastrointestinal disorder such as nausea, vomiting, and diarrhea (Lewis et al., 1991). The limited spectrum and toxicity of available antifungals, in addition to the gradual emergence of drug-resistant Candida strains (Niimi et al., 2010), bring urgency to alternative therapies against fungal infections, such as probiotic bacteria.
Probiotics are defined as live microorganisms that when administered in adequate amounts confer benefits to the host (Sanders and Klaenhammer, 2001). Bacteria belonging to the genus Lactobacillus have been used as probiotics due to their association with the healthy gastrointestinal tract in humans (Dunne et al., 1999). Animal (Elahi et al., 2005; Matsubara et al., 2012) and human studies (Hatakka et al., 2007; Ishikawa et al., 2015) have shown that the daily consumption of probiotic bacteria may reduce the oral colonization of C. albicans. The available literature supports the use of probiotics in the management of mucosal candidiasis not only in the oral cavity but also in the urogenital system and gastrointestinal tract, both as adjuvant therapy and as a prophylactic agent (Matsubara et al., 2016a). In vivo and in vitro studies suggest the potential use of probiotics for preventing recurrent vulvovaginal candidiasis (Falagas et al., 2006a), and recurrent urinary tract infections in women (Falagas et al., 2006b). Despite these promising findings, further investigation based on clinical trials is needed to confirm these beneficial effects of probiotics for the human being before its widespread use.
The restoration of a natural healthy microbiome on mucosal surfaces seems to be the major effect of probiotics (Matsubara et al., 2016a). These beneficial microorganisms may control the establishment of opportunistic pathogens, such as C. albicans, on mucosal surfaces. This control might be possible by directly interfering with pathogen survival and virulence expression (Ribeiro et al., 2016; Wang et al., 2017), competing for nutrients and adhesion sites or modulating the host immune response (Saarela et al., 2000; Reid et al., 2011; Matsubara et al., 2016b). Probiotic lactobacilli were also found to affect C. albicans biofilm formation and filamentation (Matsubara et al., 2016b). The effect of probiotic bacteria can occur by direct interactions between cell surfaces or indirectly through their products (Rizzo et al., 2015), such as lactate, a major fermentation product of lactic acid bacteria (LAB), that regulates critical functions of macrophages and dendritic cells, and also modulates the inflammatory response of epithelial cells (Garrote et al., 2015).
The innate immune system is the first line of defense against pathogens in the mucosa beyond the epithelial barrier (Romani, 2004), and the most important mechanism of protection against disseminated candidiasis (Soloviev et al., 2011). In this scenario, macrophages link the innate and the adaptive immune responses, due to their multifunctional characteristics that include phagocytic and microbicidal activities, a capacity of cytokines production, and antigen presentation (Auwerx, 1991; Romani, 2004).
The capacity of lactobacilli to induce the release of proinflammatory cytokines by macrophages was found to be strain-specific, with lactobacilli cells promoting different responses compared to probiotic cell-free supernatants (Quinteiro-Filho et al., 2015). On the other hand, another study showed that both Lactobacillus crispatus cells and its supernatant enhanced the anti-inflammatory response of macrophages and epithelial cell by increasing the production of the regulatory cytokine IL-10 and reducing the proinflammatory cytokines TNF-α and IL-6 (Rizzo et al., 2015).
The recognition of C. albicans pathogen-associated molecular patterns (PAMPs) by macrophages is mediated by an array of membrane-bound and soluble pattern-recognition receptors (PRRs) (Brown, 2011), such as Toll-like receptors (TLRs), C-type lectin receptors, including dectin-1, and the galectin family proteins (Taylor et al., 2002). Dectin-1 is the primary receptor for C. albicans in phagocytic cells, which recognizes β-glucans of Candida cell wall. This recognition leads to an orchestrated signaling that culminates in phagocytosis, antimicrobial effector functions, and production of defensins, chemokines, cytokines and reactive oxygen species (ROS) (Brown, 2011; Romani, 2011). TLR4 in turn not only recognizes mannan of fungi cell wall but also LPS from Gram-negative bacteria (Brown, 2011).
Lactobacilli were found to attenuate the adaptive and innate immune responses in macrophages challenged by LPS or lipoteichoic acid of other bacteria (Pradhan et al., 2016). In turn, specific Lactobacillus rhamnosus and Lactobacillus casei strains enhanced the phagocytic and microbicidal activities of peritoneal macrophages in mice challenged with C. albicans (Marranzino et al., 2012). Despite these evidences, the effects of probiotics on human immune systems affected by Candida are is still unclear.
Hence, the present study sought to evaluate the influence of probiotics on the human response to C. albicans by determining the effect of three Lactobacillus strains on PRR (dectin-1, TLR4, and TLR2) expression, and measuring the impact of these probiotics on inflammatory cytokines production by human macrophages challenged with C. albicans and LPS.
Materials and Methods
Microorganisms and Culture Conditions
Candida albicans ATCC SC5314, isolated from human clinical infection (Jones et al., 2004), and the probiotic strains L. rhamnosus LR32, Lactobacillus acidophilus NCFM (Danisco, Madison, WI, United States) and L. casei L324m (clinical isolate, Institute of Biomedical Sciences, University of São Paulo, Brazil) were used (Matsubara et al., 2016b). All strains were stored in 20% glycerol at -80°C prior to the experiments.
Candida albicans was cultivated in Sabouraud dextrose broth (SDB; Difco Laboratories, Detroit, MI, United States) at 37°C, for 18 h in an orbital shaker. Probiotics were cultivated in De Man, Rogosa and Shape broth (MRS; Difco Laboratories) at 37°C, in an anaerobic incubator (85% N2, 10% CO2, and 5% H2) for 18 h.
Probiotic bacteria and yeasts were harvested by centrifugation (2,000 × g/5 min), washed twice with phosphate-buffered saline (PBS, pH 7.2), and resuspended at ∼2 × 108 and 6 × 106 CFU/ml, respectively, in RPMI-1640 medium (Sigma–Aldrich, St. Louis, MO, United States) without fetal bovine serum (FBS) and antibiotics.
Cell Culture
The human macrophage-like cell line THP-1 was grown in RPMI-1640 medium (Sigma–Aldrich, St. Louis, MO, United States) enriched with 10% heat-inactivated FBS (Sigma–Aldrich), 0.05 mM b-mercaptoethanol (Sigma–Aldrich), 11 mM sodium bicarbonate (NaHCO3), 1% sodium-pyruvate (Sigma–Aldrich), 10 mM Hepes (Sigma–Aldrich), 1% glutamine (Sigma–Aldrich), and 1% penicillin/streptomycin (Sigma–Aldrich). Cells were incubated at 37°C in a humidified atmosphere containing 10% CO2.
Co-culture Assay
THP-1 cells were plated at 1 × 106 cells/well, in 6-well plates (Corning-Costar, Lowell, MA, United States). The differentiation into macrophages was achieved by treating THP-1 cells with 100 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma–Aldrich) for 72 h (Figure 1). After incubation, plastic-adherent cells were washed twice and culture medium was replaced by RPMI 1640 medium with no FBS and antibiotics, and incubated for 24 h (37°C, 10% CO2) (Chanput et al., 2012).
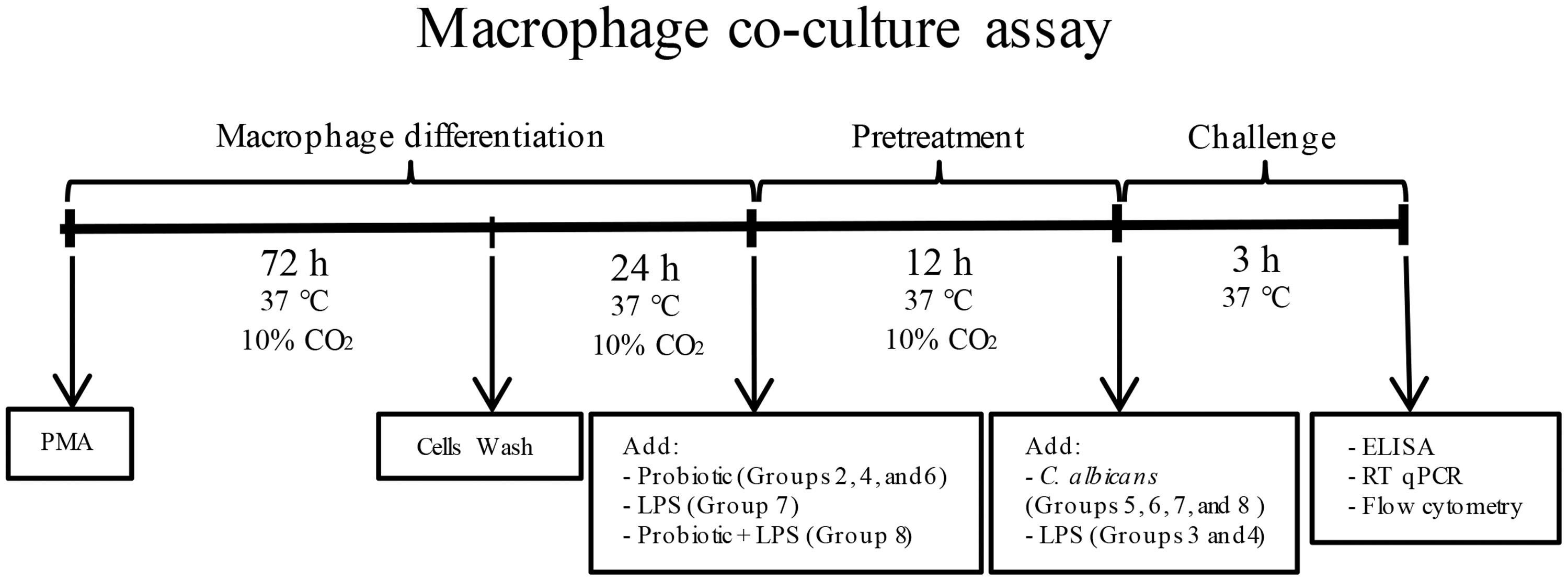
FIGURE 1. Flowchart of macrophage co-culture assay. Macrophages were pretreated with probiotic lactobacilli alone (Groups 2, 4, and 6), LPS alone (Group 7) or both probiotic and LPS (Group 8). After 12 h of incubation (37°C, 10% CO2), macrophages were challenged with Candida albicans (Groups 5, 6, 7, and 8) or LPS (Groups 3 and 4). Groups 3 and 5 had no pretreatment, Group 2 received no challenge, and Group 1 was the negative control.
Macrophages were pretreated with probiotic bacteria and/or Escherichia coli lipopolysaccharide (LPS) before C. albicans or LPS challenge, comprising eight groups of study for each probiotic bacteria as follows: Group 1 – THP-1 (negative control); Group 2 – THP-1 + Probiotic pretreatment; Group 3 – THP-1 + LPS challenge; Group 4 – THP-1 + Probiotic pretreatment + LPS challenge; Group 5 – THP-1 + Candida challenge; Group 6 – THP-1 + Probiotic pretreatment + Candida challenge; Group 7 – THP-1 + LPS pretreatment + Candida challenge; and Group 8 – THP-1 + LPS and Probiotic pretreatments + Candida challenge. Macrophages were pretreated with probiotic lactobacilli alone (1 × 107 CFU/well) (Groups 2, 4, and 6), LPS alone (Sigma–Aldrich, 100 ng/ml) (Group 7) or both LPS and probiotic bacteria (Group 8) (Yoon et al., 2013), for 12 h, at 37°C. The cell supernatant of all groups was replaced by fresh medium after pretreatments. Afterward, macrophages were challenged with 50 μl of C. albicans suspension (6 × 106 yeast/ml) (Groups 5, 6, 7, and 8) or LPS (100 ng/ml) (Groups 3 and 4) for 3 h, at 37°C. Negative control macrophages received neither pretreatment nor challenge (Group 1). Equivalent total volume was obtained in all groups by the addition of fresh culture medium (RPMI 1640 with no FBS and antibiotics).
Viability of THP-1 cells was maintained after co-culture of macrophages in any of the studied conditions and it was assessed by trypan blue exclusion assay (Barker et al., 2005) (Supplementary Figure S1). Relative transcription of CLEC7A, TLR4 and TLR2, expression of dectin-1 on macrophage surface, and cytokines production were evaluated after the final incubation. All assays were carried out in triplicate in three independent assays.
Relative Transcription of CLEC7A, TLR4, and TLR2
Macrophages were lysed and total RNA was extracted using RNeasy KIT (QIAGEN, Valencia, CA, United States). The concentration, purity, and quality of the isolated RNA samples were determined using a Nano DropOne/One Spectrophotometer (Thermo Scientific, Waltham, MA, United States). The RNA from each sample was immediately reverse transcribed into cDNA using SuperScript VILO MasterMix (Invitrogen, Waltham, MA, United States). The conditions for reverse transcription were 15 min at 37°C, 5 s at 85°C. Relative expression levels of CLEC7A, TLR4, and TLR2 were evaluated by quantitative real-time reverse transcription PCR (RT-qPCR), using TaqMan Gene Expression Master Mix (Applied Biosciences, Foster City, CA, United States), TaqMan primers and probes for CLEC7A (Hs01902549_sl), TLR4 (Hs01060206_m1), TLR2 (Hs00152932_m1), and GAPDH (Hs02758991_gl) and 100 ng of cDNA in each reaction. The RT-qPCR comprised an initial step of 50°C for 2 min, 95°C for 10 min followed by 40 cycles at 95°C for 15 s, and 50°C for 1 min, using Step One Plus System (Applied Biosciences). All data were normalized to GAPDH transcripts levels in the same cDNA set (Pfaffl, 2001; Gantner et al., 2005).
Expression of Dectin-1 on Macrophages Surface
After the final incubation, macrophages were washed with PBS and treated with AccutaseTM cell detachment solution (BD Biosciences, San Jose, CA, United States). Cells were incubated with Fc BlockTM (Human BD Fc Block, BD Biosciences) and stained with either an anti-human Clec7a – Dectin-1 antibody or an isotype control (5 μl/million cells, both labeled with PE from BD Biosciences) (30 min, 4°C in the dark) (Supplementary Figure S2). After washing with PBS, stained cells were analyzed by fluorescence-activated cell sorting (FACS) for at least 10,000 events (Guava easyCyteTM Flow Cytometers – Merck Millipore, Merck KgaA, Darmstadt, Germany).
Cytokine Profile
The concentrations of TNF-α, IL-10, IL-12 (p70), and IL-1β secreted by macrophages after 3 h of co-culture were determined in macrophage cells supernatants by ELISA (BD Biosciences, San Jose, CA, United States) following the manufacturer’s recommendation. The cytokines levels were determined by comparison with a standard calibration curve.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD) from three independent experiments. One-way ANOVA test followed by Tukey’s Multiple Comparison test was used for statistical analyses. Statistical significance was set at p < 0.05 (GraphPad Prism® Version 6.0c – GraphPad Software, La Jolla, CA, United States).
Results
Probiotics Alter the Cytokine Profile of Macrophages Challenged with LPS and/or Candida albicans
In order to evaluate the effect of probiotic bacteria on macrophages challenged with C. albicans and/or LPS, we first investigated the impact of C. albicans and LPS challenges on cytokine production by macrophages (Figure 2). The challenge with C. albicans alone was not able to induce a significant production of TNF-α, IL-10, and IL-12 (p > 0.01) as compared to control non-challenged macrophages, although the stimulus with C. albicans increased the level of IL-1β significantly (p < 0.01). In contrast, TNF-α and IL-10 levels were dramatically increased (p > 0.01) in the supernatants of macrophages challenged with LPS, but the same effect was not observed for IL-12 and IL-1β levels. The pretreatment with LPS before the challenge with C. albicans promoted a synergistic effect on macrophages when compared to macrophages challenged with C. albicans or LPS alone (except for TNF-α), inducing the highest levels of proinflammatory cytokines among the given groups. This was associated with an increase in the expression of clec7a (Figure 2E) in macrophages pretreated with LPS before C. albicans challenge.
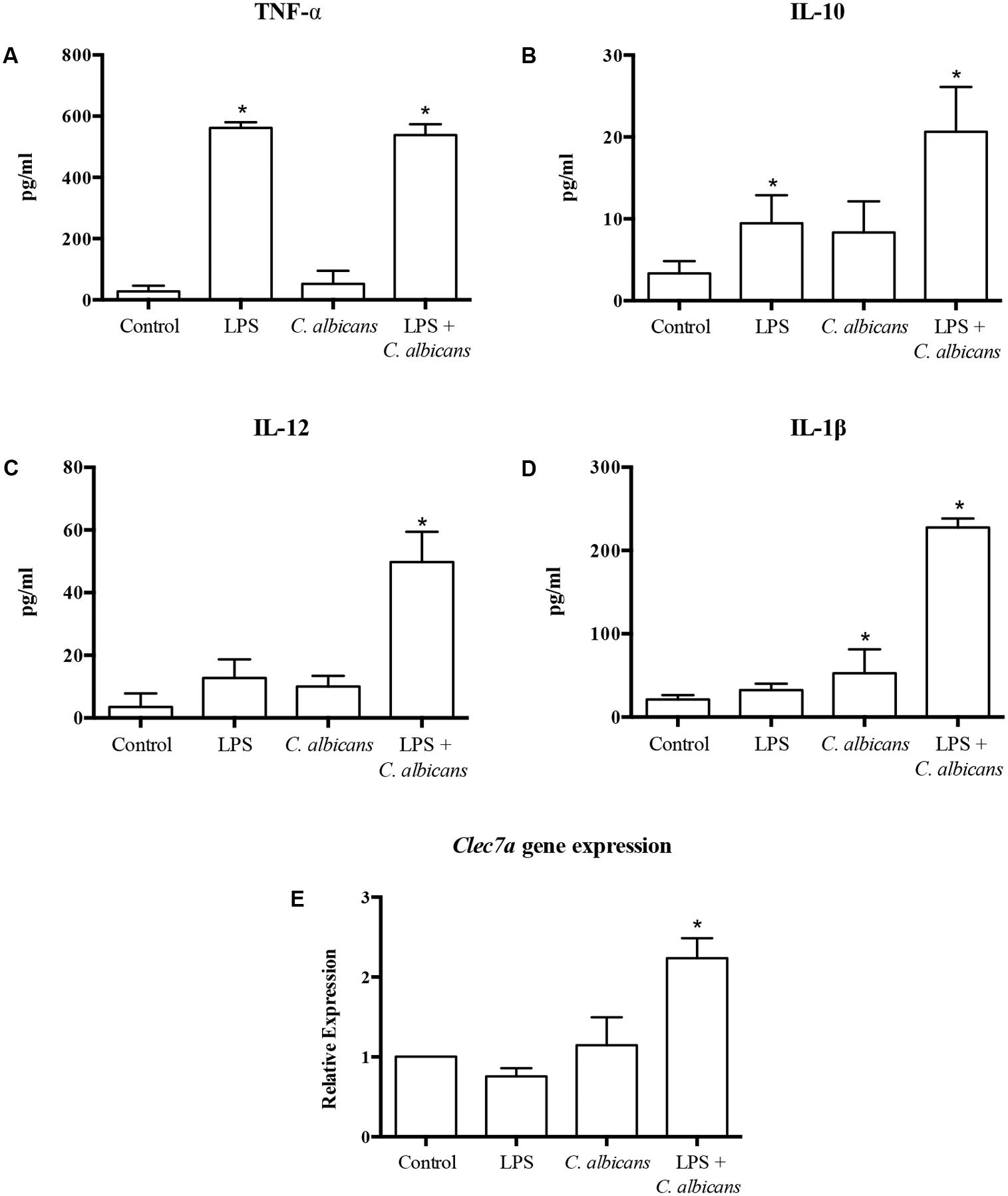
FIGURE 2. Cytokine production [TNF-α (A), IL-10 (B), IL-12 (C), and IL-1β (D)], and transcription of CLEC7A (E) by macrophages challenged with LPS, C. albicans, or pretreated with LPS and challenged by C. albicans (LPS + C. albicans). Control group had neither challenge nor pretreatment. Data are presented as mean ± SD of three independent experiments. ∗p < 0.05.
The treatment of macrophages with probiotics affected the production of cytokines significantly, with similar performances for all three tested probiotic strains (Figure 3). The pretreatment with probiotic lactobacilli (Group 2) induced a significant production of TNF-α and IL1-β as compared to non-treated macrophages (Group 1). The pretreatment with probiotics on LPS challenged macrophages caused a substantial increase in the levels of IL1-β and IL-10, except for L. rhamnosus pretreated macrophages. On the contrary, the production of TNF-α and IL-12 by LPS stimulated macrophages was not affected by probiotic bacteria.
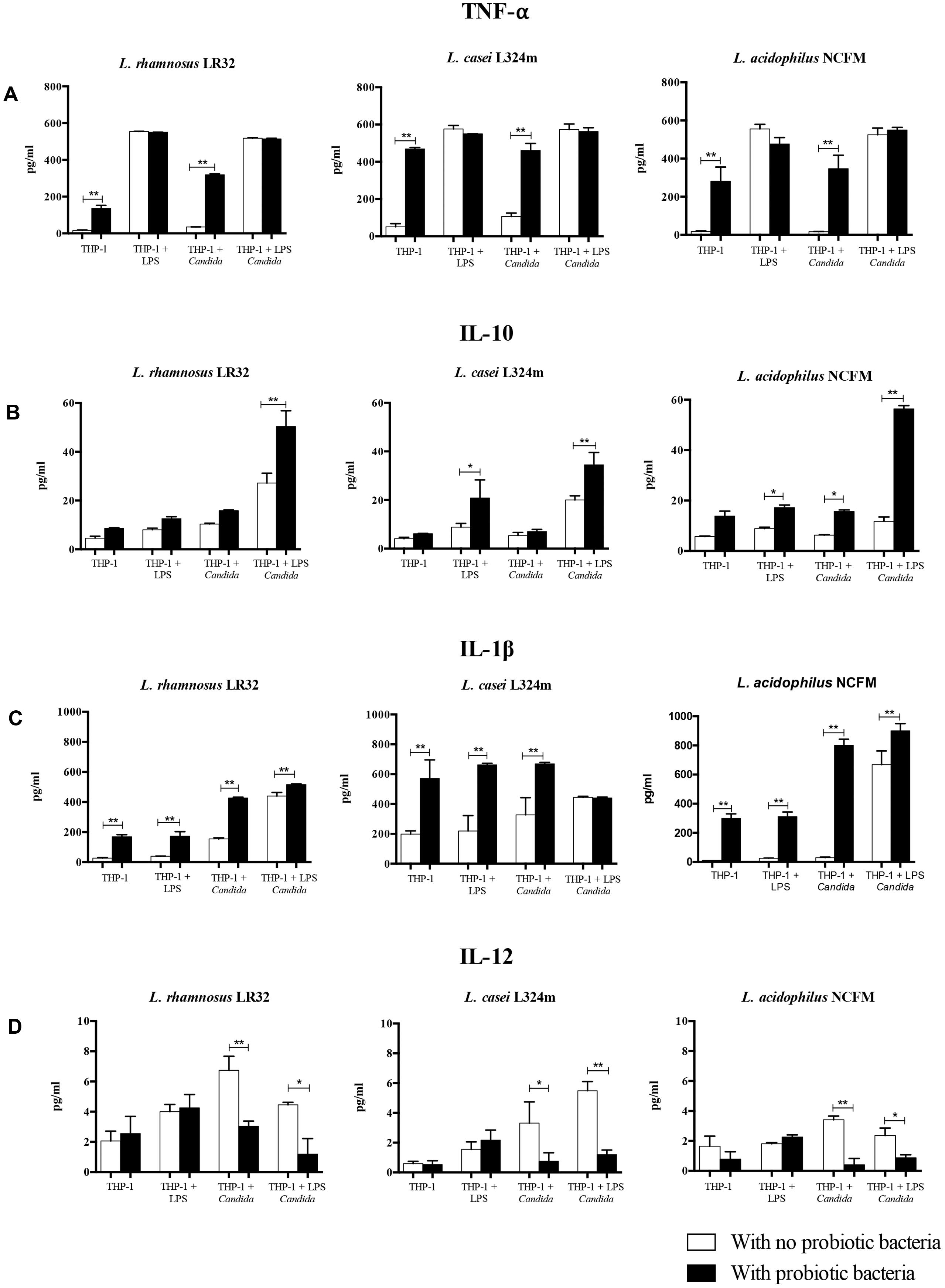
FIGURE 3. Effect of probiotic bacteria pretreatment on the production of TNF-α (A), IL-10 (B), IL-1β (C), and IL-12 (D) by macrophages. The quantification of cytokines in the macrophage supernatants was determined by ELISA. Eight groups of study were performed for each Lactobacillus strain tested: Group 1 – THP-1 (negative control); Group 2 – THP-1 + Probiotic pretreatment; Group 3 – THP-1 + LPS challenge; Group 4 – THP-1 + Probiotic pretreatment + LPS challenge; Group 5 – THP-1 + Candida challenge; Group 6 – THP-1 + Probiotic pretreatment + Candida challenge; Group 7 – THP-1 + LPS pretreatment + Candida challenge; and Group 8 – THP-1 + LPS and Probiotic pretreatments + Candida challenge. Three different strains of probiotic bacteria were tested: Lactobacillus rhamnosus LR32, Lactobacillus casei L324m, and Lactobacillus acidophilus NCFM. Data are presented as mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01.
In turn, the probiotic pretreatment of macrophages challenged with C. albicans (Group 6) led to increased levels of TNF-α and IL-1β (p < 0.01), while the secretion of IL-10 was increased significantly only by L. acidophilus pretreated macrophages. In contrast, L. rhamnosus, L. acidophilus, and L. casei pretreatments reduced significantly (p < 0.05) the levels of IL-12. In the most complex situation simulated in our study, the effects of probiotics on increasing the production of IL-10 and IL-1β (except for L. casei), and decreasing IL-12 were observed in cells pretreated with LPS and challenged by C. albicans, in different levels according to the probiotic strain.
Probiotics Reduce Transcription of CLEC7A and Dectin-1 Expression in Macrophages
The pretreatment of macrophages with all three tested probiotics down-regulated the transcription of CLEC7A (p < 0.01) (Figure 4), which codifies dectin-1 receptors. L. rhamnosus was the most effective in down-regulating CLEC7A (p < 0.01) in macrophages pretreated with LPS and challenged with C. albicans compared to the remaining Lactobacillus strains. However, both L. acidophilus and L. casei induced higher down-regulation of CLEC7A than L. rhamnosus in macrophages with no additional challenge (Figure 4).
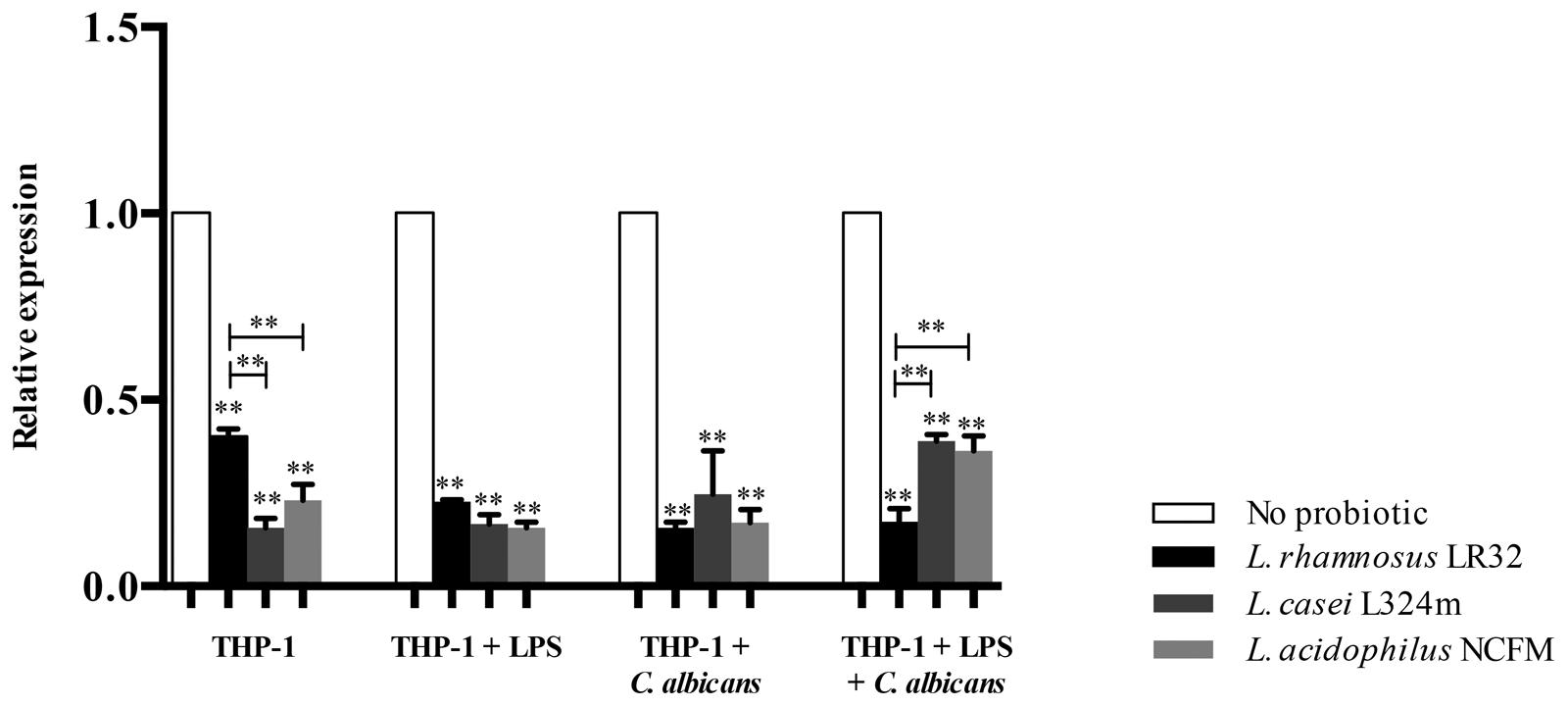
FIGURE 4. Relative transcription of CLEC7A in macrophages pretreated with probiotics alone or with both LPS and probiotics for 12 h before being challenged with C. albicans or LPS for 3 h. L. rhamnosus LR32, L. casei L324m, and L. acidophilus NCFM were tested in these different situations. The transcription of CLEC7A was normalized to GAPDH reference gene (internal control). Groups with no probiotic bacteria pretreatment were set at 1. Data are presented as mean ± SD. ∗∗p < 0.01.
The reduced expression of dectin-1 receptor on macrophage surfaces promoted by probiotics was confirmed by flow cytometry (Figure 5). L. rhamnosus and L. acidophilus presented similar percentage of cells expressing dectin-1, whereas L. casei was found to interfere to a lesser extent under all simulated conditions. The used antibody had no non-specific binding to either C. albicans or Lactobacillus cells (Supplementary Figure S3).
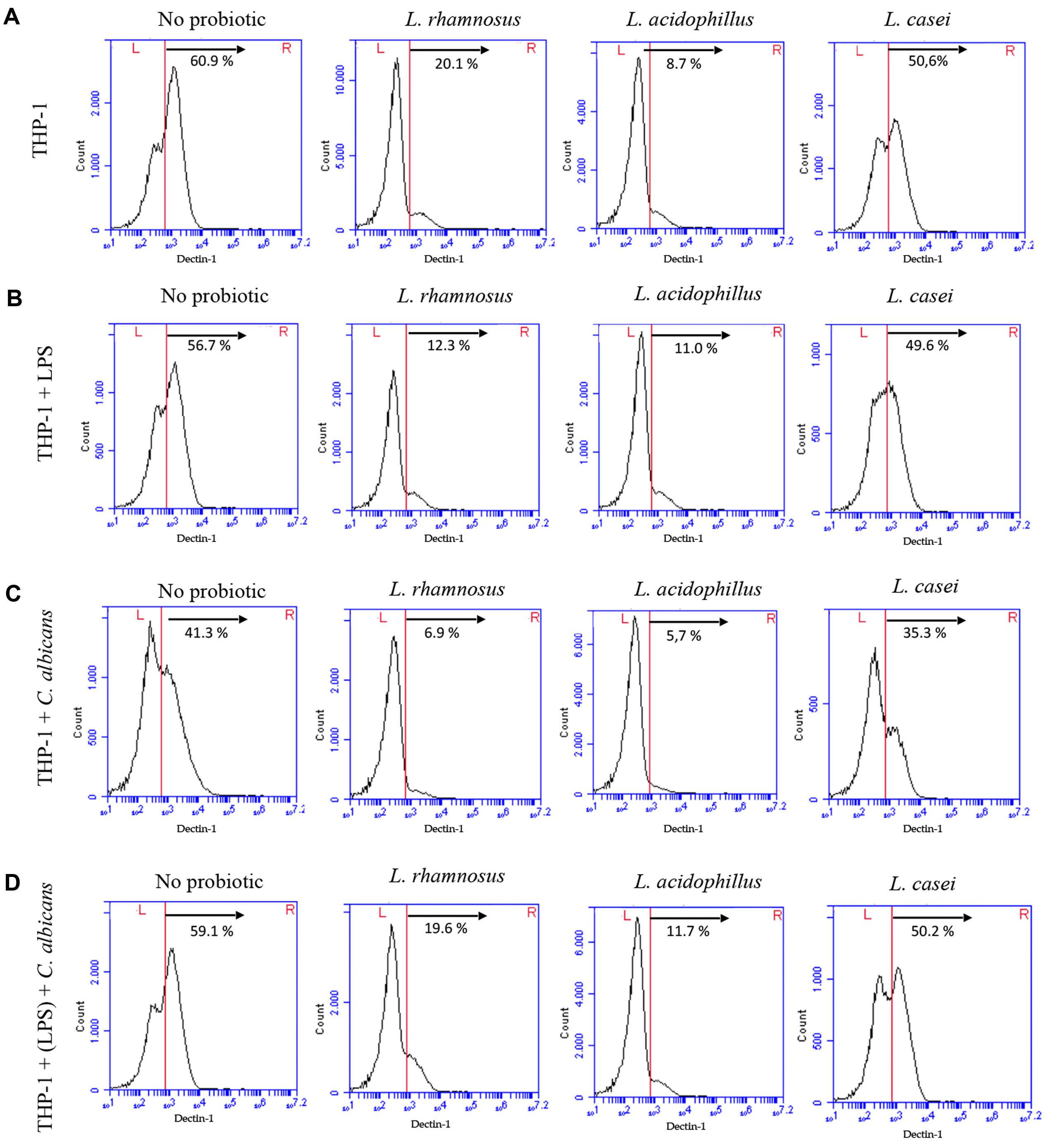
FIGURE 5. Expression of dectin-1 receptors by THP-1 macrophages in control groups (no probiotic pretreatment) and test groups (pretreated with L. rhamnosus LR32, L. acidophilus NCFM, or L. casei L324m) determined by flow cytometry. (A) Macrophages pretreated with probiotic bacteria; (B) macrophages pretreated with probiotic bacteria and challenged with LPS; (C) macrophages pretreated with probiotic and challenged with C. albicans; (D) macrophages pretreated with both LPS and probiotic before C. albicans challenge. Percentage of cells expressing high levels of dectin-1 is shown for each situation.
Probiotics Down-Regulate TLR4 and Up-Regulate TLR2 in Macrophages Challenged with C. albicans
The challenge of macrophages with C. albicans was found to down-regulate TLR4 (Figure 6A). The tested Lactobacillus strains further reduced the transcription of this gene in macrophages stimulated by C. albicans, except for L. casei that increased TLR4 mRNA levels when the macrophages were pretreated with LPS and challenged by C. albicans (Figure 6C). In turn, the LPS challenge alone was not able to alter the transcription of TLR4 after 3 h of incubation (Figure 6A), but a significant decrease in the transcription of TLR4 was observed when these macrophages were pretreated with probiotics (Figure 6C).
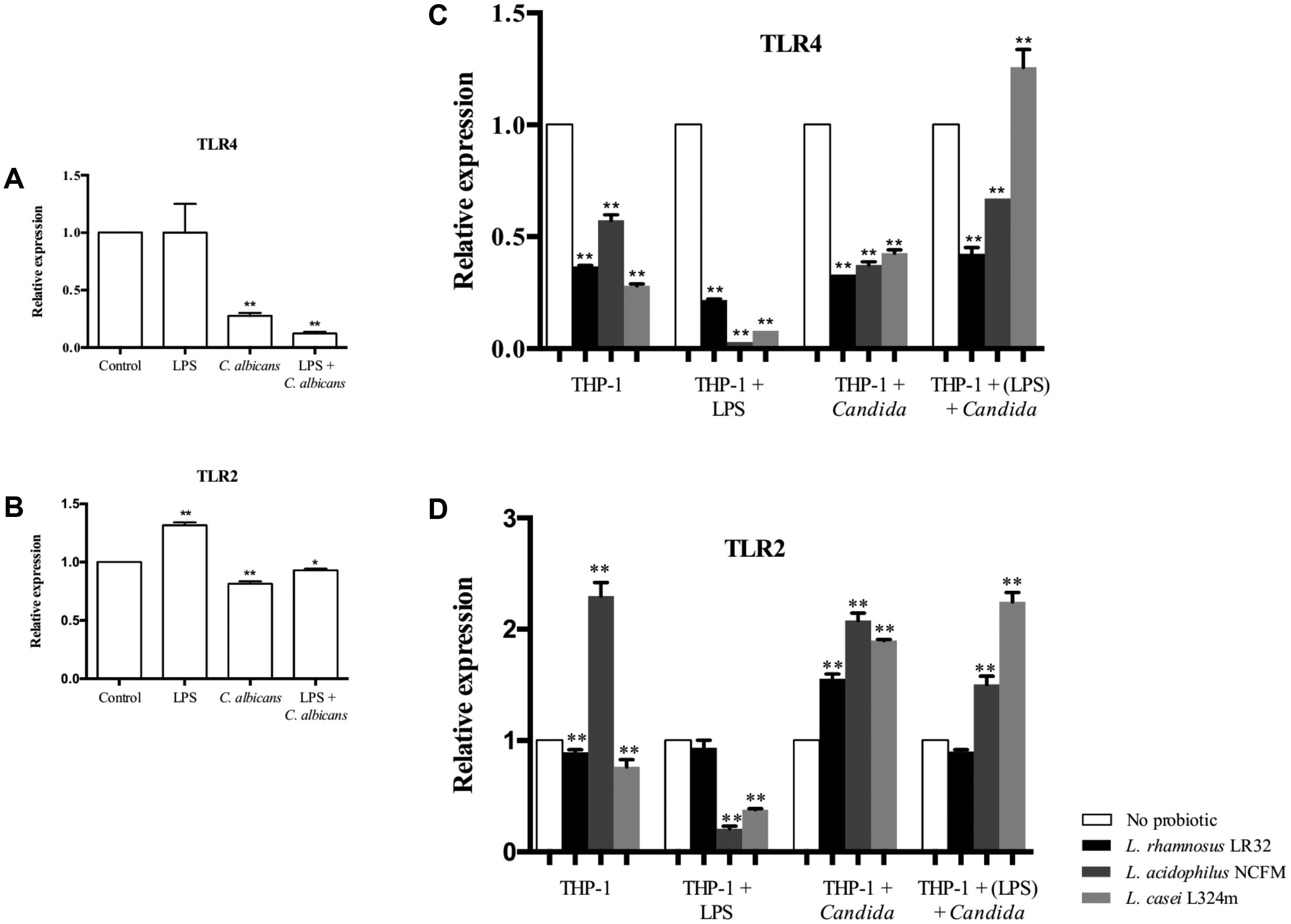
FIGURE 6. Relative transcription of TLR4 (A) and TLR2 (B) in macrophages challenged with LPS, C. albicans, or pretreated with LPS before C. albicans challenge. Transcription of TLR4 (C) and TLR2 (D) in macrophages pretreated with probiotics alone or both LPS and probiotics before C. albicans or LPS challenge. Gene expression was normalized to GAPDH transcripts levels in the same cDNA set. Groups with no probiotic bacteria pretreatment were set at 1. Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
Macrophages challenged with LPS alone presented up-regulation of TLR2, while a reduced expression of this gene was observed when macrophages were challenged with C. albicans (Figure 6B). However, the expression of TLR2 was entirely changed by probiotic pretreatments as the transcription of TLR2 was up-regulated in C. albicans challenged macrophages for all three Lactobacillus strains tested (Figure 6B). Probiotics also promoted a significant (p < 0.01) decrease in TLR2 mRNA levels when the macrophages were challenged with LPS, except for macrophages pretreated with L. rhamnosus. It is noteworthy that the pretreatment of macrophages with L. acidophilus alone showed a unique effect increasing TLR2, while the other probiotic bacteria (L. rhamnosus and L. casei) reduced the transcription of this gene.
Discussion
The reduction of C. albicans oral colonization promoted by LAB, such as Lactobacillus species, has already been demonstrated in animal models (Matsubara et al., 2012) and clinical trials (Ishikawa et al., 2015). However, the mechanisms underlying this beneficial effect are still poorly understood, especially from the human immune system perspective. In the present study, we investigated how Lactobacillus species influence the immune response of human macrophages challenged by C. albicans, analyzing two important factors for the macrophages activity: production of pro- and anti-inflammatory cytokines and expression of receptors for Candida recognition.
Our study was designed to simulate different conditions encountered in the human mucosa. The LPS, during both pretreatment or challenge stimulus, represented Gram-negative bacteria colonizing the mucosal surfaces. The pretreatment of macrophages with LPS aimed to enhance the response of macrophages to C. albicans as demonstrated previously (Rogers et al., 2013). The LPS challenge, in turn, acted as a positive control to the C. albicans challenge groups. Groups 7 and 8 simulated the clinical situation where macrophages are continuously stimulated by endogenous bacteria before the overgrowth of C. albicans.
Macrophages with no probiotic pretreatment (Groups 1, 3, 5, and 7), challenged with C. albicans alone, produced only significant levels of IL-1β (Figure 2D). However, the pretreatment of macrophages with LPS enhanced the response to C. albicans, which not only expanded the production of TNF-α, IL-1β and IL-12, but also increased the secretion of the regulatory cytokine IL-10 (Figures 2A–D). It has already been demonstrated that the presence of bacterial LPS induces the expression of dectin-1 on macrophages, which in turn leads to an enhanced immune response against C. albicans (Rogers et al., 2013). This evidence corroborates with our results of up-regulation of transcription of CLEC7A in LPS challenged macrophages (Figure 2E).
In lactobacilli pretreated macrophages, the probiotics supplied an additional ‘reprogramming’ stimulus suggesting their immunomodulatory effect, due to decreased production of IL-12, and increased levels of IL-10. The interference with the release of pro- and anti-inflammatory cytokines by human macrophages is probiotic-strain dependent, since strains of the same species may show opposite effects (Drago et al., 2010). Other lactobacilli species, such as L. crispatus, an important urogenital species routinely found in the vagina of healthy women, also enhanced the production of the IL-10, and reduced the proinflammatory cytokine production in macrophages and epithelial cells challenged with Chlamydia trachomatis (Rizzo et al., 2015). This immunomodulatory effect induced by probiotic bacteria may lead to an enhanced capacity of macrophages to combat C. albicans infections. A previous in vivo study using a C. albicans infection murine model showed significant increase in the phagocytic and microbicidal activities of peritoneal macrophages of mice treated with L. rhamnosus and L. casei, which resulted in decreased levels of the pathogen in infected organs (Marranzino et al., 2012).
IL-10 is one of the most important immunosuppressive cytokines, aiming the maintenance of homeostasis (Ma et al., 2015), and its production can be enhanced by probiotics (Lucas et al., 2005). In turn, during pathogen infections, IL-12 produced by macrophages/monocytes and dendritic cells (Trinchieri, 1995) influences the production of interferon-gamma (IFN-γ) by natural killer (NK) and T cells, which favor phagocytic cells activation and inflammation. IL-12 also plays an important role by favoring a T helper type 1 (Th1) response (Trinchieri, 1993). In our study, the reduction of IL-12 promoted by probiotics may be also associated with the increase in IL-10 levels, since IL-10 is a potent inhibitor of IL-12 production by human peripheral blood mononuclear cells (D’Andrea et al., 1993), by suppressing IL-12 production at the transcriptional level (Aste-Amezaga et al., 1998).
The significant increase of IL1-β production by lactobacilli-treated macrophages after C. albicans challenge (Figure 3C) may suggest that probiotic bacteria are activating inflammasome, as shown for L. rhamnosus (Miettinen et al., 2012). Since inflammasomes contribute to an anti-Candida response (Martinon et al., 2002), probiotic treatment may increase Candida elimination.
In our co-culture assay, L. rhamnosus LR32, L. casei L324m, and L. acidophilus NCFM induced similar cytokine profile in human macrophages, although the level of inhibition (e.g., IL-12) or stimulation (e.g., IL-10) in cytokines production varied according to the tested bacterial strain. These differences may be associated with their mechanisms to modulate TLR expression in phagocytic cells (Kotzamanidis et al., 2010). In fact, the tested L. rhamnosus strain down-regulated the expression of TLR4, L. casei up-regulated TLR4 and TLR2, whereas L. acidophilus decreased TLR4 and increased TLR2 transcription levels in C. albicans challenged macrophages.
The decreased expression of TLR4 in LAB pretreated macrophages observed in our study is in accordance with previous in vivo data reporting that probiotic lactobacilli reduce TLR4 mRNA levels and decrease TLR2 responses in blood mononuclear cells (Forsberg et al., 2014). In another study, the pretreatment with L. crispatus also downregulated the expression of TLR4 and TLR2 by epithelial cells challenged with C. albicans, suggesting that L. crispatus may act as an anti-inflammatory agent through TLR2/4 (Rizzo et al., 2013). Since TLR4 signaling culminates in the expression of genes encoding inflammatory molecules (Aksoy et al., 2012), the down-regulation of TLR4 transcription by L. rhamnosus and L. acidophilus favors an anti-inflammatory effect, reducing the production of proinflammatory cytokines such IL-12 (Re and Strominger, 2001).
Our data also indicated that probiotics increased TLR2 mRNA levels in C. albicans challenged macrophages (Figure 6D). C. albicans cell wall mannan and phospholipomannan are able to trigger TLR2-signaling (Brown, 2011), but the role of TLR2 signaling in controlling C. albicans infections is still contradictory. Increased susceptibility or resistance to infection with C. albicans were both showed in TLR2-deficient mice, due to reduced proinflammatory cytokines production or decreased IL-10/increased IL-12 and IFN-γ production, respectively (Brown, 2011). TLR2 and TLR4 signaling are not equivalent as TLR2 stimulus elicits less IL-12 (p70) than TLR4 (Re and Strominger, 2001), but induces abundant IL-10, favoring Th2 and T cytotoxic responses (Dillon et al., 2004). Thus, the up-regulation of TLR2 by Lactobacillus spp. in the presence of C. albicans (Figure 6D) may partially explain the cytokine profile observed in our study (Figures 3B,D), with high IL-10 and low IL-12 levels after probiotics treatment.
All three tested probiotics down-regulated dectin-1 expression in Candida challenged macrophages. Even though bacterial LPS induces dectin-1 expression in macrophages (Rogers et al., 2013), the probiotic pretreatments were able to reduce dectin-1 expression even in LPS challenged macrophages (Figure 4), by down-regulating the transcription of CLEC7A. The importance of dectin-1 signaling to control commensal C. albicans levels in the gastrointestinal tract is still controversial. Individuals with genetic deficiencies associated with decreased expression of dectin-1 are more susceptible to fungal diseases (Romani, 2011). On the other hand, in an animal model, dectin-1 was found to be essential for controlling infection of the gastrointestinal tissues during systemic candidiasis, but it had no influence on gastrointestinal colonization of C. albicans in mice (Vautier et al., 2012).
The decrease of dectin-1 expression in macrophages promoted by L. rhamnosus and L. acidophilus, and to a lesser extent by L. casei, showed in our study, may indicate that these beneficial bacteria may limit an exacerbated inflammatory response originated from the recognition of C. albicans yeasts by macrophages. This inflammatory control may avoid tissue destruction that facilitates the establishment of C. albicans infections, but still allowing host defense mechanisms.
This hypothesis is reinforced by a previous study showing that the dectin-1 level reduction is associated with an increased regulatory T cells differentiation and a local homeostasis, thus leading to ameliorate colitis (Tang et al., 2015). The down-regulation of pro-inflammatory genes was also observed in murine macrophages treated with L. acidophilus (Pradhan et al., 2016). Furthermore, the capacity of probiotics to reduce yeast-to-hyphae transition of C. albicans, as demonstrated previously by our group (Matsubara et al., 2016b), may also contribute to the reduction of local inflammation, as Candida remains in the commensal yeast form.
The alteration of C. albicans recognition by immune cells in the presence of lactobacilli may involve other mechanisms. For instance, the exposure of C. albicans to lactate, produced by LAB, induces β-glucan masking in C. albicans by changing the expression of cell-wall-related genes (Ballou et al., 2016). As the fungal cell wall architecture play an essential role in the competitive colonization of Candida species in mucosal surfaces (Sem et al., 2016), the effect of probiotic bacteria on fungal cell wall needs to be further investigated.
Conclusion
Lactobacilli may contribute in the response against C. albicans colonization and infection by affecting the expression of dectin-1, TLR2, and TLR4 at the transcription level, thus altering the recognition of C. albicans and the cytokine profile of macrophages. This immunomodulation favors an anti-inflammatory response to C. albicans colonization (Figure 7). Our novel findings open new possibilities for the study of probiotics against C. albicans infections. Further investigations are necessary to determine whether the immune modulation is caused by a direct or an indirect effect of probiotic bacteria on macrophages. More studies testing other immune cells and analyzing different inflammatory signaling pathways will help to clarify the role of probiotic bacteria on Candida-host interactions.
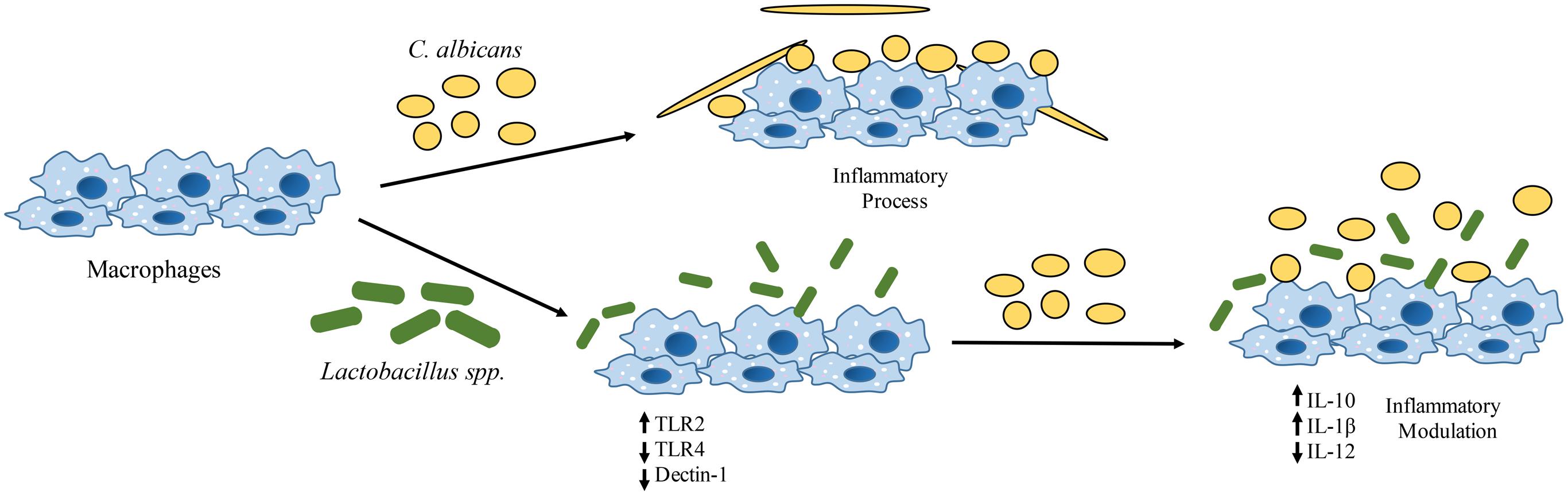
FIGURE 7. Effects of Lactobacillus spp. on human macrophages challenged with C. albicans. There is an alteration in the transcription of pattern-recognition receptors (PRRs) genes (CLEC7A, TLR2, and TLR4) with a significant reduction of C. albicans receptors (dectin-1) on macrophages. These changes have an impact on the cytokine profile that leads to an anti-inflammatory effect.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author Contributions
VM designed and conducted experiments, analyzed data and wrote the article; KI helped with flow cytometer assay; EA-S helped with RT-qPCR assay; BB-S helped with cell culture experiments; AN gave a critical review of the manuscript; MM supervised the project and the article writing.
Funding
Part of this study was supported by FAPESP grant 2015/18273-9 and 2006/1315-7.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank the Coordination for the Improvement of Higher Education Personnel (CAPES) Foundation for supporting VM.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02280/full#supplementary-material
FIGURE S1 | Number of viable macrophage cells after the final incubation in the different groups. Data are presented as cells/well.
FIGURE S2 | (A) Macrophages under different stimulation (cell with no challenge or LPS pretreated cells challenged with C. albicans) incubated with isotype control antibody. (B) Flow cytometry gating on macrophages.
FIGURE S3 | Flow cytometry data from control wells containing C. albicans or Lactobacillus strains submitted to anti-Human CLEC7A – Dectin-1 antibody. (A) macrophages alone with no antibody; (B) C. albicans + antibody; (C) L. rhamnosus + antibody; (D) L. acidophilus + antibody; (E) L. casei + antibody. Percentage of cells expressing the antibody, shown in each situation, reveals no non-specific binding in any control group.
References
Aksoy, E., Taboubi, S., Torres, D., Delbauve, S., Hachani, A., Whitehead, M. A., et al. (2012). The p110delta isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat. Immunol. 13, 1045–1054. doi: 10.1038/ni.2426
Aste-Amezaga, M., Ma, X., Sartori, A., and Trinchieri, G. (1998). Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J. Immunol. 160, 5936–5944.
Auwerx, J. (1991). The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia 47, 22–31. doi: 10.1007/BF02041244
Ballou, E. R., Avelar, G. M., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal beta-glucan masking and immune evasion. Nat. Microbiol. 2:16238. doi: 10.1038/nmicrobiol.2016.238
Barker, K. S., Liu, T., and Rogers, P. D. (2005). Coculture of THP-1 human mononuclear cells with Candida albicans results in pronounced changes in host gene expression. J. Infect. Dis. 192, 901–912. doi: 10.1086/432487
Brown, G. D. (2011). Innate antifungal immunity: the key role of phagocytes. Annu. Rev. Immunol. 29, 1–21. doi: 10.1146/annurev-immunol-030409-101229
Chanput, W., Reitsma, M., Kleinjans, L., Mes, J. J., Savelkoul, H. F., and Wichers, H. J. (2012). β-Glucans are involved in immune-modulation of THP-1 macrophages. Mol. Nutr. Food Res. 56, 822–833. doi: 10.1002/mnfr.201100715
D’Andrea, A., Aste-Amezaga, M., Valiante, N. M., Ma, X., Kubin, M., and Trinchieri, G. (1993). Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 178, 1041–1048. doi: 10.1084/jem.178.3.1041
Dillon, S., Agrawal, A., Van Dyke, T., Landreth, G., McCauley, L., Koh, A., et al. (2004). A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J. Immunol. 172, 4733–4743. doi: 10.4049/jimmunol.172.8.4733
Drago, L., Nicola, L., Iemoli, E., Banfi, G., and De Vecchi, E. (2010). Strain-dependent release of cytokines modulated by Lactobacillus salivarius human isolates in an in vitro model. BMC Res. Notes 3:44. doi: 10.1186/1756-0500-3-44
Dunne, C., Murphy, L., Flynn, S., O’Mahony, L., O’Halloran, S., Feeney, M., et al. (1999). Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Van Leeuwenhoek 76, 279–292. doi: 10.1023/A:1002065931997
Elahi, S., Pang, G., Ashman, R., and Clancy, R. (2005). Enhanced clearance of Candida albicans from the oral cavities of mice following oral administration of Lactobacillus acidophilus. Clin. Exp. Immunol. 141, 29–36. doi: 10.1111/j.1365-2249.2005.02811.x
Falagas, M. E., Betsi, G. I., and Athanasiou, S. (2006a). Probiotics for prevention of recurrent vulvovaginal candidiasis: a review. J. Antimicrob. Chemother. 58, 266–272. doi: 10.1093/jac/dkl246
Falagas, M. E., Betsi, G. I., Tokas, T., and Athanasiou, S. (2006b). Probiotics for prevention of recurrent urinary tract infections in women: a review of the evidence from microbiological and clinical studies. Drugs 66, 1253–1261.
Forsberg, A., Abrahamsson, T. R., Bjorksten, B., and Jenmalm, M. C. (2014). Pre- and postnatal administration of Lactobacillus reuteri decreases TLR2 responses in infants. Clin. Transl. Allergy 4:21. doi: 10.1186/2045-7022-4-21
Gantner, B. N., Simmons, R. M., and Underhill, D. M. (2005). Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24, 1277–1286. doi: 10.1038/sj.emboj.7600594
Garrote, G. L., Abraham, A. G., and Rumbo, M. (2015). Is lactate an undervalued functional component of fermented food products? Front. Microbiol. 6:629. doi: 10.3389/fmicb.2015.00629
Hatakka, K., Ahola, A. J., Yli-Knuuttila, H., Richardson, M., Poussa, T., Meurman, J. H., et al. (2007). Probiotics reduce the prevalence of oral candida in the elderly–a randomized controlled trial. J. Dent. Res. 86, 125–130. doi: 10.1177/154405910708600204
Ishikawa, K. H., Mayer, M. P., Miyazima, T. Y., Matsubara, V. H., Silva, E. G., Paula, C. R., et al. (2015). A multispecies probiotic reduces oral Candida colonization in denture wearers. J. Prosthodont. 24, 194–199. doi: 10.1111/jopr.12198
Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., et al. (2004). The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101, 7329–7334. doi: 10.1073/pnas.0401648101
Kotzamanidis, C., Kourelis, A., Litopoulou-Tzanetaki, E., Tzanetakis, N., and Yiangou, M. (2010). Evaluation of adhesion capacity, cell surface traits and immunomodulatory activity of presumptive probiotic Lactobacillus strains. Int. J. Food Microbiol. 140, 154–163. doi: 10.1016/j.ijfoodmicro.2010.04.004
Lewis, M. A., Samaranyake, L. P., and Lamey, P. J. (1991). Diagnosis and treatment of oral candidosis. J. Oral Maxillofac. Surg. 49, 996–1002. doi: 10.1016/0278-2391(91)90066-U
Lucas, M., Zhang, X., Prasanna, V., and Mosser, D. M. (2005). ERK activation following macrophage FcgammaR ligation leads to chromatin modifications at the IL-10 locus. J. Immunol. 175, 469–477. doi: 10.4049/jimmunol.175.1.469
Ma, X., Yan, W., Zheng, H., Du, Q., Zhang, L., Ban, Y., et al. (2015). Regulation of IL-10 and IL-12 production and function in macrophages and dendritic cells. F1000Res 4:1465. doi: 10.12688/f1000research.7010.1
Marranzino, G., Villena, J., Salva, S., and Alvarez, S. (2012). Stimulation of macrophages by immunobiotic Lactobacillus strains: influence beyond the intestinal tract. Microbiol. Immunol. 56, 771–781. doi: 10.1111/j.1348-0421.2012.00495.x
Martinon, F., Burns, K., and Tschopp, J. (2002). The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426. doi: 10.1016/S1097-2765(02)00599-3
Matsubara, V. H., Bandara, H. M., Mayer, M. P., and Samaranayake, L. P. (2016a). Probiotics as antifungals in mucosal candidiasis. Clin. Infect. Dis. 62, 1143–1153. doi: 10.1093/cid/ciw038
Matsubara, V. H., Silva, E. G., Paula, C. R., Ishikawa, K. H., and Nakamae, A. E. (2012). Treatment with probiotics in experimental oral colonization by Candida albicans in murine model (DBA/2). Oral Dis. 18, 260–264. doi: 10.1111/j.1601-0825.2011.01868.x
Matsubara, V. H., Wang, Y., Bandara, H. M., Mayer, M. P., and Samaranayake, L. P. (2016b). Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 100, 6415–6426. doi: 10.1007/s00253-016-7527-3
McCullough, M. J., and Savage, N. W. (2005). Oral candidosis and the therapeutic use of antifungal agents in dentistry. Aust. Dent. J. 50(4 Suppl. 2), S36–S39. doi: 10.1111/j.1834-7819.2005.tb00383.x
Miettinen, M., Pietila, T. E., Kekkonen, R. A., Kankainen, M., Latvala, S., Pirhonen, J., et al. (2012). Nonpathogenic Lactobacillus rhamnosus activates the inflammasome and antiviral responses in human macrophages. Gut Microbes 3, 510–522. doi: 10.4161/gmic.21736
Niimi, M., Firth, N. A., and Cannon, R. D. (2010). Antifungal drug resistance of oral fungi. Odontology 98, 15–25. doi: 10.1007/s10266-009-0118-3
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pradhan, B., Guha, D., Ray, P., Das, D., and Aich, P. (2016). Comparative analysis of the effects of two probiotic bacterial strains on metabolism and innate immunity in the RAW 264.7 murine macrophage cell line. Probiotics Antimicrob. Proteins 8, 73–84. doi: 10.1007/s12602-016-9211-4
Quinteiro-Filho, W. M., Brisbin, J. T., Hodgins, D. C., and Sharif, S. (2015). Lactobacillus and Lactobacillus cell-free culture supernatants modulate chicken macrophage activities. Res. Vet. Sci. 103, 170–175. doi: 10.1016/j.rvsc.2015.10.005
Re, F., and Strominger, J. L. (2001). Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J. Biol. Chem. 276, 37692–37699. doi: 10.1074/jbc.M105927200
Reid, G., Younes, J. A., Van der Mei, H. C., Gloor, G. B., Knight, R., and Busscher, H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. doi: 10.1038/nrmicro2473
Ribeiro, F. C., de Barros, P. P., Rossoni, R. D., Junqueira, J. C., and Jorge, A. O. (2016). Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 122, 201–211. doi: 10.1111/jam.13324
Rizzo, A., Fiorentino, M., Buommino, E., Donnarumma, G., Losacco, A., and Bevilacqua, N. (2015). Lactobacillus crispatus mediates anti-inflammatory cytokine interleukin-10 induction in response to Chlamydia trachomatis infection in vitro. Int. J. Med. Microbiol. 305, 815–827. doi: 10.1016/j.ijmm.2015.07.005
Rizzo, A., Losacco, A., and Carratelli, C. R. (2013). Lactobacillus crispatus modulates epithelial cell defense against Candida albicans through Toll-like receptors 2 and 4, interleukin 8 and human beta-defensins 2 and 3. Immunol. Lett. 156, 102–109. doi: 10.1016/j.imlet.2013.08.013
Rogers, H., Williams, D. W., Feng, G. J., Lewis, M. A., and Wei, X. Q. (2013). Role of bacterial lipopolysaccharide in enhancing host immune response to Candida albicans. Clin. Dev. Immunol. 2013:320168. doi: 10.1155/2013/320168
Romani, L. (2011). Immunity to fungal infections. Nat. Rev. Immunol. 11, 275–288. doi: 10.1038/nri2939
Saarela, M., Mogensen, G., Fonden, R., Matto, J., and Mattila-Sandholm, T. (2000). Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84, 197–215. doi: 10.1016/S0168-1656(00)00375-8
Sanders, M. E., and Klaenhammer, T. R. (2001). Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84, 319–331. doi: 10.3168/jds.S0022-0302(01)74481-5
Scully, C., el-Kabir, M., and Samaranayake, L. P. (1994). Candida and oral candidosis: a review. Crit. Rev. Oral Biol. Med. 5, 125–157. doi: 10.1177/10454411940050020101
Sem, X., Le, G. T., Tan, A. S., Tso, G., Yurieva, M., Liao, W. W., et al. (2016). β-Glucan exposure on the fungal cell wall tightly correlates with competitive fitness of Candida species in the mouse gastrointestinal tract. Front. Cell. Infect. Microbiol. 6:186. doi: 10.3389/fcimb.2016.00186
Soloviev, D. A., Jawhara, S., and Fonzi, W. A. (2011). Regulation of innate immune response to Candida albicans infections by alphaMbeta2-Pra1p interaction. Infect. Immun. 79, 1546–1558. doi: 10.1128/IAI.00650-10
Tang, C., Kamiya, T., Liu, Y., Kadoki, M., Kakuta, S., Oshima, K., et al. (2015). Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe 18, 183–197. doi: 10.1016/j.chom.2015.07.003
Taylor, P. R., Brown, G. D., Reid, D. M., Willment, J. A., Martinez-Pomares, L., Gordon, S., et al. (2002). The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J. Immunol. 169, 3876–3882. doi: 10.4049/jimmunol.169.7.3876
Trinchieri, G. (1993). Interleukin-12 and its role in the generation of TH1 cells. Immunol. Today 14, 335–338. doi: 10.1016/0167-5699(93)90230-I
Trinchieri, G. (1995). Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 13, 251–276. doi: 10.1146/annurev.iy.13.040195.001343
Vautier, S., Drummond, R. A., Redelinghuys, P., Murray, G. I., MacCallum, D. M., and Brown, G. D. (2012). Dectin-1 is not required for controlling Candida albicans colonization of the gastrointestinal tract. Infect. Immun. 80, 4216–4222. doi: 10.1128/IAI.00559-12
Wang, S., Wang, Q., Yang, E., Yan, L., Li, T., and Zhuang, H. (2017). Antimicrobial compounds produced by vaginal Lactobacillus crispatus are able to strongly inhibit Candida albicans growth, hyphal formation and regulate virulence-related gene expressions. Front. Microbiol. 8:564. doi: 10.3389/fmicb.2017.00564
Keywords: candidiasis, Candida albicans, Lactobacillus, immune system, macrophage, pattern recognition receptor
Citation: Matsubara VH, Ishikawa KH, Ando-Suguimoto ES, Bueno-Silva B, Nakamae AEM and Mayer MPA (2017) Probiotic Bacteria Alter Pattern-Recognition Receptor Expression and Cytokine Profile in a Human Macrophage Model Challenged with Candida albicans and Lipopolysaccharide. Front. Microbiol. 8:2280. doi: 10.3389/fmicb.2017.02280
Received: 30 May 2017; Accepted: 06 November 2017;
Published: 29 November 2017.
Edited by:
Celio Geraldo Freire De Lima, Universidade Federal do Rio de Janeiro, BrazilReviewed by:
Jeanette Wagener, University of Aberdeen, United KingdomDaniel Ferreira Feijó, Gonçalo Moniz Institute (IGM), Brazil
Laura Noelia Cariddi, National University of Río Cuarto, Argentina
Lucia Helena Pinto da Silva, Universidade Federal Rural do Rio de Janeiro, Brazil
Copyright © 2017 Matsubara, Ishikawa, Ando-Suguimoto, Bueno-Silva, Nakamae and Mayer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcia P. A. Mayer, mpamayer@icb.usp.br
†Present address: Victor H. Matsubara, Oral Health Centre of Western Australia, The University of Western Australia, Perth, WA, Australia
Bruno Bueno-Silva, Praça Teresa Cristina, Guarulhos University, Guarulhos, Brazil
 Victor H. Matsubara
Victor H. Matsubara Karin H. Ishikawa
Karin H. Ishikawa Ellen S. Ando-Suguimoto
Ellen S. Ando-Suguimoto Bruno Bueno-Silva
Bruno Bueno-Silva Atlas E. M. Nakamae
Atlas E. M. Nakamae Marcia P. A. Mayer
Marcia P. A. Mayer