- 1Department of Veterinary and Animal Science, Faculty of Health and Medical Sciences, University of Copenhagen, Frederiksberg, Denmark
- 2National Food Institute, Technical University of Denmark, Søborg, Denmark
- 3DTU Biosustain, Technical University of Denmark, Kongens Lyngby, Denmark
- 4Institut für Medizinische Mikrobiologie und Hygiene, Universitätsklinikum Tübingen, Tübingen, Germany
- 5Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark
Human strains of Staphylococcus aureus commonly carry the bacteriophage ΦSa3 that encodes immune evasion factors. Recently, this prophage has been found in livestock-associated, methicillin resistant S. aureus (MRSA) CC398 strains where it may promote human colonization. Here, we have addressed if exposure to biocidal products induces phage transfer, and find that during co-culture, Φ13 from strain 8325, belonging to ΦSa3 group, is induced and transferred from a human strain to LA-MRSA CC398 when exposed to sub-lethal concentrations of commercial biocides containing hydrogen peroxide. Integration of ΦSa3 in LA-MRSA CC398 occurs at multiple positions and the integration site influences the stability of the prophage. We did not observe integration in hlb encoding β-hemolysin that contains the preferred ΦSa3 attachment site in human strains, and we demonstrate that this is due to allelic variation in CC398 strains that disrupts the phage attachment site, but not the expression of β-hemolysin. Our results show that hydrogen peroxide present in biocidal products stimulate transfer of ΦSa3 from human to LA-MRSA CC398 strains and that in these strains prophage stability depends on the integration site. Knowledge of ΦSa3 transfer and stability between human and livestock strains may lead to new intervention measures directed at reducing human infection by LA-MRSA strains.
Introduction
Staphylococcus aureus is an opportunistic pathogen in human and animals, and is one of the leading causes of acute and chronic infections. When treating staphylococcal infections, resistance to β-lactam antibiotics is an increasing problem with methicillin resistant S. aureus (MRSA) clones epidemically spreading in the hospitals as well as in the community (DeLeo et al., 2010; Dulon et al., 2011). In recent years, strains belonging to clonal complex 398 (CC398) have been widely detected in pigs and have also been found in other livestock animals such as turkey, chicken and cattle (Smith and Pearson, 2011). In general, livestock-associated MRSA (LA-MRSA) CC398 is regarded less virulent than human-associated MRSA clones due to the absence of several common virulence factors, such as enterotoxins and phage encoded Panton-Valentine leukocidin (PVL) (Schijffelen et al., 2010; Price et al., 2012; Becker et al., 2015). However, LA-MRSA CC398 causes infections in humans with livestock contact (Köck et al., 2011; Pérez-Moreno et al., 2016) and since first detected in the early 2000s (van Loo et al., 2007) the number of clinical cases in humans has increased steadily (van Cleef et al., 2011; Cuny et al., 2015). The infection of humans by LA-MRSA CC398 is primarily regarded as an occupational risk for farmers, veterinarians and meat handlers (Smith and Pearson, 2011) but a secondary risk is the spread of CC398 to the community (Smith and Pearson, 2011; Smith, 2015). In fact, infections of LA-MRSA CC398 in patients without animal contact have been reported both in Europe and the United States (Welinder-Olsson et al., 2008; Wulf et al., 2008; Bhat et al., 2009; Larsen et al., 2015, 2016; Diene et al., 2017).
The ability of S. aureus to colonize host organisms is in part determined by prophages. The majority of human-associated S. aureus isolates contain β-hemolysin negative-converting bacteriophages, which are classified as ΦSa3 (Goerke et al. 2006b). They share an integrase (Goerke et al., 2009) and an attachment core sequence (5′-TGTATCCAAACTGG-3′) in the hlb gene (Coleman et al., 1991). ΦSa3 phages contain the immune evasion cluster (IEC) encoding the chemotaxin inhibitory protein (CHIPS), staphylococcal complement inhibitor (SCIN), staphylokinase (SAK), as well as some enterotoxins (Coleman et al., 1989; van Wamel et al., 2006; Goerke et al., 2009; McCarthy and Lindsay, 2010), which contribute to adaptation to the human host (de Haas et al., 2004; Rooijakkers et al., 2005; Jung et al., 2016). Recent data suggest that ΦSa3 phages are occasionally found in livestock-associated S. aureus strains, and the acquisition of ΦSa3 is important for the host-jump of S. aureus from animals to humans (Price et al., 2012; van der Mee-Marquet et al., 2014; Larsen et al., 2015, 2016).
Induction of prophages generally occurs in response to DNA damage elicited by for example reactive oxygen species and some antibiotics, and is mediated by RecA activation and cleavage of the phage repressor (Frye et al., 2005; Goerke et al., 2006a; Nanda et al., 2015). Thus, extrinsic factors contribute to the mobility of ΦSa3 phages among S. aureus strains. Disinfection and cleaning are critical steps when maintaining a desired hygiene status in household products, in hospitals and in food animal production. In livestock production, biocides are widely applied for cleaning and disinfection of animal-associated production areas, equipment, during transport and even directly on animals to prevent skin diseases (Quinn and Markey, 2001). Major biocides applied in livestock production include hydrogen peroxide, peracetic acid, glutaric aldehyde, quaternary ammonium compounds, and isopropanol (Kjølholt et al., 2001; SCHENIHR (Scientific Committee on Emerging and Newly Identified Health Risks), 2009). Hydrogen peroxide is a common disinfectant applied in animal production. It is a powerful oxidizing agent that oxidizes thiol groups in enzymes and proteins and leads to free radical production (Kjølholt et al., 2001; Russell, 2003). Here we have applied a prophage Φ13 from strain 8325 belonging to ΦSa3 group to examine if sub-lethal concentrations of commercial biocides and biocidal compounds induce phage ΦSa3 and stimulate its transmission to LA-MRSA CC398 strains.
Materials and Methods
Strain Collection and Chemical Reagents
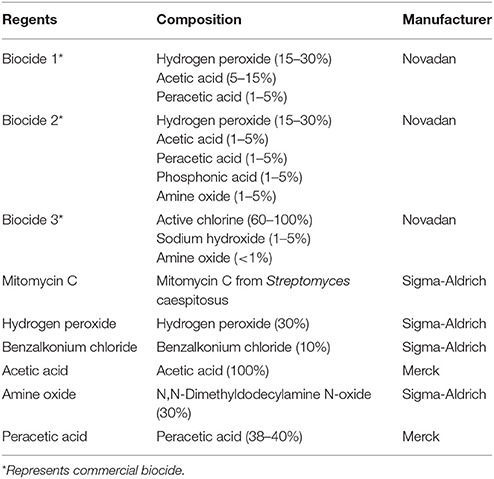
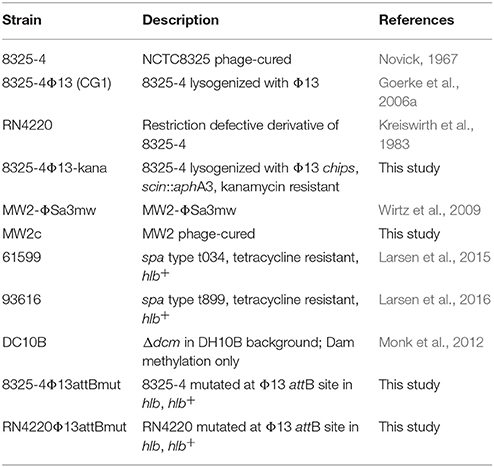
Chemicals and biocides used in this study are listed in Table 1 and strains in Table 2. Media used in this study included Mueller-Hinton broth (MHB), tryptic soy broth (TSB) and agar (TSA) from Oxoid. Sheep blood is from Department of Veterinary Disease Biology, University of Copenhagen.
Strain Construction
Phage-cured MW2c was obtained by treating MW2-ΦSa3mw (Wirtz et al., 2009) with 1 μg/ml mitomycin C for 2 h at 37°C, 200 rpm in TSB and subsequently with 0.5 mM hydrogen peroxide for 3 h at 37°C, 200 rpm. Serial dilutions were plated on TSA with 0.5 mM hydrogen peroxide and incubated at 37°C for overnight. Colonies grew on the plates were selected as phage cured bacteria (MW2c) and checked by PFGE (Goerke et al., 2004) for lacking of the phage.
Strain 8325-4Φ13-kana was obtained by replacing part of the 3′-end of Φ13 (chps and scin) in strain Φ8325-4Φ13 (Goerke et al., 2006a) with kanamycin resistance cassette aphA3. In brief, two fragments flanking the 3′-region of Φ13 and the kanamycin-resistant cassette from pDG782 (Guérout-Fleury et al., 1995) were amplified and annealed by overlapping PCR. The amplicon was cloned into KpnI restriction site into pBT2 (Brückner, 2006) to gain pCG6, which was electroporated into strain RN4220 (Kreiswirth et al., 1983) and further transduced into 8325-4Φ13. pCG6 was then used to mutagenize strain 8325-4Φ13 as described by Brückner (2006) to obtain strain 8325-4Φ13kana, and confirmed by sequencing.
To identify the Φ13 attB site in strain 8325-4 (Novick, 1967) and RN4220 (Peng et al., 1988), the original Φ13 attB site in 8325-4 was predicted by subtracting the Φ13 sequence (accession No.: NC_004617) from the NCTC 8325 genome harboring Φ13 (accession No.: NC_007795), and aligning the resulting sequence to the known hlb sequence containing the Φ13 attB site (Coleman et al., 1991) (accession No.: X61716) for confirmation. To construct a plasmid containing a mutated Φ13 attB (attBmut), 800 bp upstream sequence of Φ13 attB site in hlb gene of 8325-4 (Novick, 1967) was amplified by overlapping PCR primer pairs pIMAYhlbattfor and hlbattmutrev (Supplementary Table 1), and 800 bp sequence downstream of Φ13 attB site was amplified by overlapping PCR primer pairs hlbattmutfor and pIMAYhlbattrev, where the Φ13 attB mutation from strain 61599 was included in primer hlbattmutfor (Supplementary Table 1). The PCR fragments and pIMAY vector were digested by EcoRI, and ligated together by Gibson Assembly® Master Mixt (BioLabs®) to form the plasmid named as pIMAY_Φ13attmut. This plasmid was electroporated into RN4220 (Kreiswirth et al., 1983) resulting in the strain RN4220pIMAY_Φ13attmut, and the Φ13 attB mutation was introduced into the RN4220 chromosome by homologous recombination (Monk et al., 2012). The mutation at attB in RN4220 was confirmed by sequencing and the strain was named RN4220Φ13attBmut.
For construction of Φ13 attB mutation in strain 8325-4, plasmid pIMAY_Φ13attmut was electroporated from RN4220pIMAY_Φ13attmut into 8325-4 (Novick, 1967), and the Φ13 attB mutation from strain 61599 was introduced to 8325-4 chromosome by homologous recombination with pIMAY allelic replacement system (Monk et al., 2012). The mutation at attB in 8325-4 was confirmed by sequencing and the strain was named as 8325-4Φ13attBmut.
Determination of Minimal Inhibitory Concentrations (MICs)
MICs of biocides and chemical compounds in strain 8325-4Φ13 were determined according to the guideline of Clinical and laboratory standards institute (Clinical and Laboratory Standards Institute (CLSI), 2008). Strains from overnight TSA plate was resuspended in 0.9% NaCl to achieve the turbidity of 0.5 McFarland standard and further diluted 100-fold in MHB. The working solution of biocides and chemical compound were benzalkonium chloride (2.67 μg/ml), hydrogen peroxide (3% w/w), Biocide 1 (5% v/v), Biocide 2 (5% v/v), Biocide 3 (5% v/v), and mitomycin C (1 μg/ml). In brief, the working solution of biocides and mitomycin C were prepared with two-fold dilution series in MHB in 96-well microtiter plates with 100 μl volume. Further, 100 μl of cell suspension was added to each well. Positive growth control of wells without biocide, and negative controls of wells with only MHB were included. The microtiter plates were incubated at 37°C for 24 h. MIC values were determined as the lowest concentration of the compounds that eliminated the visible growth of bacteria.
Φ13 Induction Assay by Different Biocides and Mitomycin C
To perform the phage induction assay, strain 8325-4Φ13 was grown to the exponential phase (OD600 = 0.8) (Wirtz et al., 2009) by shaking at 37°C, 200 rpm in TSB. Then, different concentration of Biocide 1, Biocide 2, Biocide 3, benzalkonium chloride, hydrogen peroxide and mitomycin C were added into the broth culture respectively, and further incubated for 2 h at 37°C, 200 rpm. The concentration series of each biocide was determined according to MIC values of strain 8325-4 Φ13 (Table 3), which included MIC, 5X MIC, 10X MIC, 20X MIC, and 30X MIC. Supernatants were sterilized by 0.45 μm pore diameter membrane filter. The phage induction levels were evaluated by PFU determination as previously described (Goerke et al., 2006b). Briefly, 100 μl of each phage supernatant dilution was mixed 100 μl indicator strain MW2c (OD600 = 0.1) and incubated for 10 min at room temperature before mixing with top agar and pouring onto a TSA plated with 10 μM CaCl2, and further incubated overnight at 37°C.
Lysogenization Assay of Φ13 to LA-MRSA
Strain 8325-4 Φ13-kana was used as the phage donor strain. Phage lysate were obtained by growing the strain to exponential phase (OD600 = 0.8) at 37°C, 200 rpm, mixing with 1 μg/ml mitomycin C and further incubating for 4 h under the same condition. The culture was centrifuged at 4°C, 8,500 rpm for 6 min, and sterile filtered by 0.45 μm pore diameter membrane filter before determining the titer. Φ13 lysogens were obtained by mixing phage Φ13-kana and LA-MRSA CC398 strains (61599 and 93616) at MOIs of 0.001, 0.01, 0.1, 1, 10, 100, and 1,000 and incubating the mixture at 30°C for 30 min. Hundred microliter of the mixture was then spread on TSA plate containing 100 μl/ml kanamycin, 10 μg/ml tetracycline and 5% of sheep blood (Kan-Tet plate), and incubated at 37°C overnight. Lysogens were selected as colonies able to grow on Kan-Tet plate. The plates were stored at 4°C overnight to detect β-hemolysin activity. Lysogens were verified by colony morphology and hemolysin activity (Supplementary Figure 6) as well as by PCR to detect ahpA3 and sak genes. The lysogenization frequency was evaluated as the ratio of the Φ13 CC398 lysogen colony count (CFU/ml) on Kan-Tet plate (Φ13 CC398 lysogen) to the total recipient colony count (CFU/ml) on TSA plate with 10 μg/ml tetracycline (Tet plate).
PCR Analysis
Bacterial DNA was released by suspending 2-3 colonies in 100 μl Milli Q water and incubated at 95°C for 10 min. For confirming the transfer of Φ13 to LA-MRSA CC398 strain, primer pair sak-for/sak-rev for the sak gene and primer pair kanR-for/kanR-rev for the aph gene were used. For checking if the hlb gene was intact, primer pair hlbPhi13attB-for/hlbPhi13attB-rev of hlb gene was used. For primer sequences see Supplementary Table 1. PCR amplification was performed by mixing 12.5 μl DreamTaq Green PCR Master Mix (2X) (Thermo Fisher Scientific), 10.5 μl Milli Q water, 1 μl DNA template and 1 μl (0.1 μM) each primer. For determining the attB core sequence mutation in strain 8325-4 and RN4220, the PCR products of hlb gene were purified by GeneJet PCR purfication kit (Thermo Fisher Scientific) and sequenced by Macrogen Inc.
In Vitro Liquid Co-Cultivation Assay
The liquid co-cultivation assay was carried out in TSB with donor strain 8325-4Φ13-kana resistant to kanamycin and recipient strain LA-MRSA 61599 resistant to tetracycline. In brief, both donor and recipient strains were grown to exponential phase (approximately 108 cfu/ml), and mixed with a ratio of 1:1. Different concentrations of hydrogen peroxide, Biocide 1 and mitomycin C were added to the culture, and then incubated at 37°C, 200 rpm for 4 h. Phage transfer was detected by plating serial dilutions on Kan-Tet plate and Tet plate followed by incubating at 37°C for 18 h. The Φ13 CC398 lysogens were counted based on the colony morphology of Φ13 lysogens (Supplementary Figure 6) and also further confirmed the PCR program of sak and kanR genes. To confirm the lysogens were belonging to CC398 strain and avoid miscounting of donor strain 8325-4Φ13-kana, a spa-mecA multiplex PCR (Tang et al., 2017) was conducted to CC398 lysogens, which could differentiate CC398 lysogens to donor strain 8325-4Φ13-kana (Supplementary Figure 5). The transfer ratio of Φ13-kana was considered as the ratio of the Φ13 CC398 lysogen colony count on Kan- Tet plates (CFU/ml) to the recipient colony count on Tet plates (CFU/ml).
Influence of Chemical Agents to Strains and Phage
The influence of applied chemical regents to donor and recipient strains was separately evaluated by growth curve, which culture of both strains at exponential phase (approximately 108 cfu/ml) was treated with a series of concentrations of mitomycin C, hydrogen peroxide and Biocide1, and measured OD600 value every hour till 4 h at 37°C, 200 rpm. The influence of applied chemical to Φ13-kana was evaluated by treating the phage stock with a series of concentration of chemical agents and the plaque assay was conducted after 4 h at 37°C, 200 rpm, with the plaque assay mentioned above. The influence of applied chemical regents to the induction of Φ13 with the kanamycin resistant cassette was evaluated after 4 h of treatment by Φ13 induction assay mentioned above. In addition, the influence of chemical compounds present in Biocide 1 to the donor and recipient strains was evaluated individually by growth curve and Φ13 induction assays as described above in the presence of a series concentrations of peracetic acid, amine oxide and acetic acid.
Characterization of LA-MRSA CC398 Φ13 Lysogens
Stability of the prophages in CC398 Φ13 lysogens was examined in 10 Φ13 lysogens of strain 61599 and passaging them for 20 days. In brief, 5 μl overnight culture of each lysogen was diluted 1000-fold in 5 ml TSB and incubated for 24 h at 37°C, 200 rpm. After each passage, 100 μl culture was spread on TSA plate containing 10 μg/ml tetracycline and incubated overnight at 37°C. From this plate, 50 colonies were picked up and streaked on TSA plate containing both 100 μg/ml kanamycin and 10 μg/ml tetracycline and TSA plate containing only 10 μg/ml tetracycline, and then incubated overnight at 37°C. The stability of each lysogen was determined by the ratio of colonies survived on TSA plate contain both kanamycin and tetracycline to the colonies survived on TSA plate containing only tetracycline. Further, lysogens with different Φ13 stability characteristics were sequenced by paired-end sequencing (2 × 251 bp) using Nextera XT DNA Library Preparation Kit (Illumina Inc.) on a MiSeq sequencer (Illumina Inc.). Contigs were de novo assembled using CLC-bio assembler (Qiagen).
Lysogenization Assay of Φ13 attB Mutant
Stains 8325-4Φ13attBmut and RN4220Φ13attBmut, and their wild type strains were individually mixed with the Φ13-kana phage stock at a MOI of 0.1 and were incubated at 37°C for 4 hrs. After making serial dilutions, 100 μl of each dilution series was spread on TSA plates with 5% sheep blood containing 100 μl/ml kanamycin (Kan-plate) and TSA plate with 5% sheep blood (TSA plate), which were incubated at 37°C overnight. Lysogens were selected as colonies able to grow on Kan plate. The plates were stored at 4°C overnight to detect β-hemolysin activity. The lysogen frequency was determined as the ratio of colony count (CFU/ml) on Kan-plate to the total colony count (CFU/ml) on TSA plate. Further, PCR amplification of hlb gene to check if hlb gene was interrupted, PCR amplification of hlb gene including the Φ13 attB site was conducted for 16 colonies of lysogenized mutants.
Results
Induction of ΦSa3 by Biocides
Staphylococcus aureus strain 8325 harbors a ΦSa3 class of phage termed Φ13 (Iandolo et al., 2002). In this study, we applied strain 8325-4Φ13 (Goerke et al., 2006a), which is a derivative of the phage cured strain 8325-4 (Novick, 1967) lysogenized with Φ13 to evaluate if commercially available biocides can induce ΦSa3. We evaluated the susceptibility of strain 8325-4Φ13 to three commercially available biocides in addition to hydrogen peroxide, benzalkonium chloride as well as mitomycin C (Table 1) by MIC (Table 3). Further, we exposed the strain 8325-4Φ13 to these biocides and chemical reagents (Table 1) and monitored plaque formation on indicator strain MW2c. Strain MW2c is a derivative of strain MW2-ΦSa3mw in which all phages have been cured (Wirtz et al., 2009). We found that excision of Φ13 was induced by mitomycin C (≤3.75 mg/ml), by biocides containing hydrogen peroxide [Biocide 1 (≤0.2% v/v) and 2 (≤ 0.2% v/v)] as well as by hydrogen peroxide (≤0.9% w/w) in a dose-dependent manner (Figures 1A–E). No induction of Φ13 was observed by benzalkonium chloride or Biocide 3 containing sodium hypochlorite as the active ingredient (Figures 1C,F) or in the absence of stimuli indicating that less than 10 pfu/ml is released.
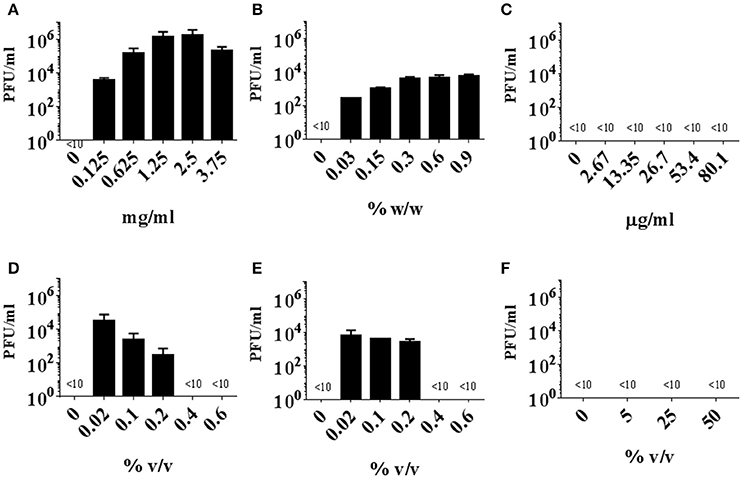
Figure 1. Induction of Φ13 from S. aureus strain 8325-4Φ13 by biocides. Strain 8325-4Φ13 was exposed to mitomycin (A), hydrogen peroxide (B), benzalkonium chloride (C), Biocide 1 (D), Biocide 2 (E), Biocide 3 (F) at the concentrations of MIC, 5X MIC, 10X MIC, 20X MIC, and 30X MIC. The induction of Φ13 was measured by PFU counts on indicator strain MW2c, and the detection limit was more than ten PFU counts. The detection limit is < 10, the error bar represents SD, n = 3.
Transfer of Φ13 to LA-MRSA CC398
To monitor integration of Φ13 into LA-MRSA CC398, a Φ13 phage stock was obtained by mitomycin C induction from strain 8325-4Φ13-kana, a derivative of 8325-4Φ13 where the 3'-end of chps and scn in Φ13 has been replaced by the kanamycin resistance gene, aphA3. This phage stock was used to infect two tetracycline-resistant LA-MRSA CC398 strains 61599 and 93616 isolated from humans in Denmark (Table 2). Strain 61599 with spa type t034 (Larsen et al., 2015) was obtained from a pig farm worker and strain 93616 with spa type t899 (Larsen et al., 2016) was recovered from a mink farmer. Neither of the strains contained ΦSa3 phage by sequence analysis (Larsen et al., 2015, 2016). After repeated attempts, only strain 61599 could be lysogenised with Φ13. At multiplicity of infection (MOI) of less than one, we observed colonies resistant to both kanamycin and tetracycline, indicating lysogenization of LA-MRSA CC398 by Φ13-kana (Supplementary Figure 1). To confirm that the resulting colonies were lysogens and not spontaneous antibiotic resistant mutants, we used PCR amplification of the aphA3 and sak genes of Φ13-kana (data not shown).
To examine if Φ13-kana could transfer directly from S. aureus 8325-4Φ13-kana (donor strain) to recipient LA-MRSA CC398 strain 61599 in the presence of biocides or mitomycin, we first assessed a series of concentrations of mitomycin C, hydrogen peroxide and Biocide 1 ranging from sub-lethal to lethal with respect to their influence on growth (Supplementary Figure 2) and the induction levels of Φ13-kana (Supplementary Figure 3). We also confirmed that these concentrations have minimum effect on Φ13-kana in plaque assays (Supplementary Figure 4). Further, we evaluated MIC of strain 8325-4Φ13 to acetic acid, amine oxide and peracetic acid (Table 1), which are present in Biocide 1 (Table 3), and exposed the strain to these chemicals to evaluate their effect on growth and Φ13 induction (Supplementary Figure 5). We found that, in addition to hydrogen peroxide (Supplementary Figure 3B), Φ13-kana can also be induced by peracetic acid, which is able to spontaneously decompose to hydrogen peroxide and acetic acid (Yuan et al., 1997; Supplementary Figure 6).
We co-cultured the donor strain 8325-4Φ13-kana with the recipient strain 61599, in the presence of sub-lethal to lethal concentrations of mitomycin C, hydrogen peroxide, and Biocide 1. Colonies of strain 61599 lysogenized with Φ13-kana were detected by plating on blood plates containing kanamycin and tetracycline. The transfer of Φ13-kana was confirmed by PCR for sak and aphA3 and the identity of the resulting colonies was confirmed by PCR amplification of spa and mecA (Supplementary Figure 7). We observed that Φ13-kana transferred from strain 8325-4Φ13-kana to 61599 with a transfer frequency between 10−5 and 10−6 in the presence of 0.002–0.1%(v/v) Biocide 1, which contains hydrogen peroxide as the active compound (Figure 2C). Exposure to mitomycin C and hydrogen peroxide also induced the transfer of Φ13-kana (Figures 2A,B). Surprisingly, in the absence of DNA damaging agents, we also observed a transfer frequency of Φ13-kana was around 2.3 × 10−7 (Figure 2). These results show that sublethal to lethal concentrations of biocides containing hydrogen peroxide and mitomycin C promote transfer of Φ13 between human and livestock-adapted strains in vitro.
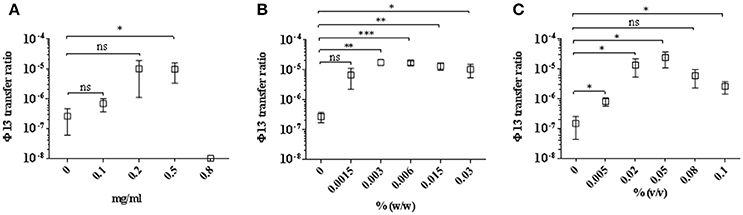
Figure 2. Transfer of Φ13 during co-cultivation of LA-MRSA CC398 with 8325-4Φ13-kana. Strains LA-MRSA CC398 and 8325-4Φ13-kana were co-cultivated and exposed for 4 h to different concentrations of (A) mitomycin C, (B) hydrogen peroxide and (C) Biocide 1. Open squares represent the Φ13 transfer ratio calculated as the number of colonies resistant to tetracycline and kanamycin relative to the number resistant to tetracycline. Error bars represent ±SD, n = 3. ***P < 0.001, **P < 0.01 and *P < 0.05, NS, none significant by t-test analysis.
Characterization of Φ13 in CC398 Lysogens
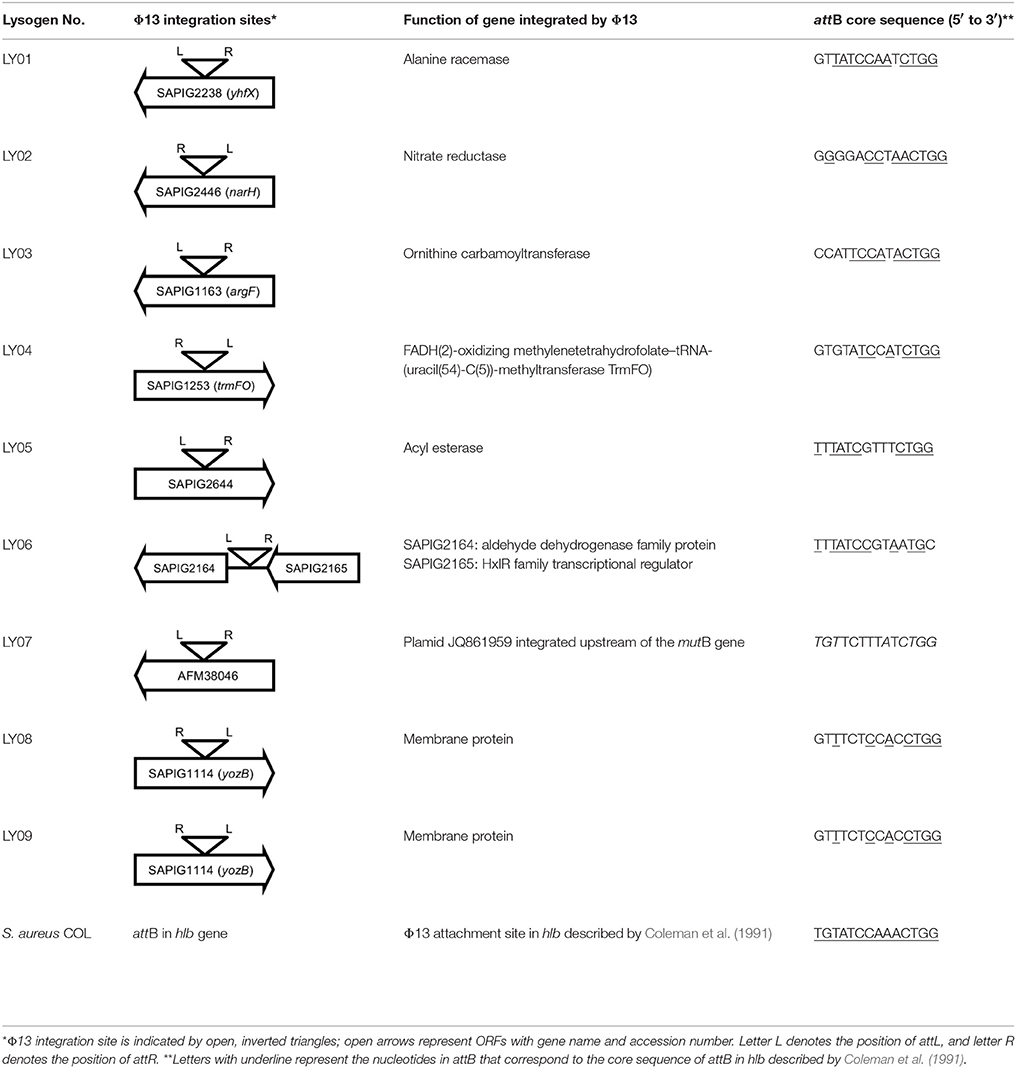
Initially, we assessed hemolysin production of nine Φ13 lysogens (LY01-09) in LA-MRSA CC398 strain 61599 on blood agar plates and observed that all were β-hemolysin positive (Supplementary Figure 8) suggesting that the phage had not integrated in the hlb attachment site commonly preferred in human strains (Goerke et al., 2006b). Subsequently, we evaluated stability of the lysogens over a period of 20 days (corresponding to approximately 960 generations) by plating on tetracycline agar plates and recording the fraction of kanamycin-resistant colonies (Figure 3). Here we observed different stability patterns with Φ13-kana being completely stabile in six lysogens (LY01-06), and being partially stable in the rest of the lysogens with 44% (LY08) and 29% (LY09) of cells retaining the phage, respectively. In LY07, the percentage of cells carrying the phage varied between 96 and 100% during the experiment indicating sporadic loss. To examine the genetic basis for the different stability patterns we sequenced the genomes of lysogens and observed eight different Φ13 integration sites in strain 61599 (Table 4). In the six lysogens (LY01-LY06) where Φ13-kana was completely stable, the phage was integrated at six different locations in the chromosome of 61599. In five of the six lysogens (LY01-05), Φ13-kana was inserted in annotated genes, whereas in LY06 the phage was integrated in an unrelated intergenic region (Table 4). In LY07, where Φ13-kana stability varied the phage was integrated in a gene encoding a hypothetical protein located on plasmid JQ861959 that had integrated in the chromosome, which may explain why recovery of the phage varied from plating to plating. In both LY08 and LY09 from which the phage was lost in the majority of cells after the 20 day period, Φ13-kana was integrated in yozB encoding a putative membrane protein, the function of which is unclear. These results strongly indicate that Φ13 integrates at alternative sites in LA-MRSA CC398 and that the overall stability of the integrated phage is influenced by its integration site in the chromosome.
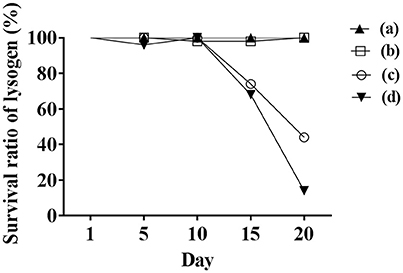
Figure 3. Φ13 stability in LA-MRSA CC398 strain 61599. Nine lysogens were grown over a 20 period with 1000 fold dilution performed daily from the previous day overnight culture. The stability of the phage was monitored by plating on agar plates with tetracycline, and scoring the number that were also kanamycin resistant. (a) filled triangle represents LY01-LY06 of lysogens, which were fully stable with Φ13 prophages during the 20-day test; (b) opened square represents LY07, which had sporadically lost Φ13 during 20 days; (c) open circle represent LY08 and (d) filled inverted triangle represents LY09, both lost Φ13 in the majority of the analyzed colonies after 20 days, respectively.
By sequence analysis, we found that all integration sites in the nine lysogens were partially similar to the attB sequence for Φ13 in hlb gene originally described by Coleman et al. (1991) (Table 4). However, we were not able to observe other similarities between the integration sites or regions. By further comparing these integration sites in lysogens to the corresponding sequences in strain 61599 and strain 8325-4, we found that only the integration site in LY03 contained one nucleotide variation between the strain 61599 and strain 8325-4, but this variation did not enhance resemblance to the attB in the hlb gene (Supplementary Table 2). In addition, sequence analysis of the hlb gene of strain 61599 revealed variation at two residues in the 14 nucleotide attB sequence when compared to attB in hlb gene of the human derived strain S. aureus COL (Projan et al., 1989). Both nucleotide substitutions are silent leaving the β-hemolysin expression intact (Figure 4). This finding suggests that point mutations in hlb of LA-MRSA may drive Φ13 to integrate elsewhere in the chromosome. To test this hypothesis, we compared the attB core sequence of ΦSa3 from strain 61599 to the 69 of genome sequenced CC398 isolates from a previous study by Price et al. (2012). We observed that 65 out of 69 CC398 isolates from Price et al.'s study showed the same substitutions of the attB core sequence for Φ13 in the hlb gene as we found in strain 61599, while four of CC398 isolates had three substitutions compared to the attB core sequence in hlb described by Coleman et al. (1991; Figure 5).
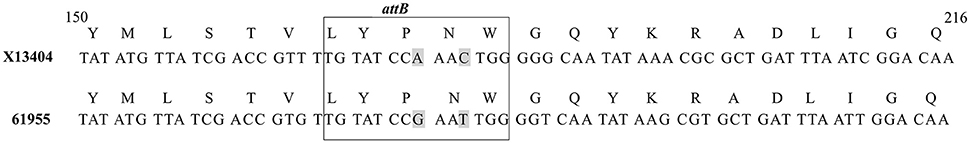
Figure 4. Core sequence of attachment site (attB) [boxed] in hlb gene in S. aureus COL (Accession No. X13404) compared to attB in the hlb gene of strain 61599. Two point mutations occurred in attB of hlb gene in strain 61599. The selected nucleotide sequence was part of the X13404 CDS region (CAA31769). The gray shaded letters represent the variations in the attB of strain 61599.

Figure 5. Variation of attB core sequence for Φ13 in hlb gene in ΦSa3-negative CC398 isolates compared to the attB in hlb gene of S. aureus COL (accession no. X13404) described by Coleman et al. (1991). CC398 isolate information was obtained from the study of Price et al. (2012). The gray shaded letters represent the variations in the attB of CC398 isolates.
To support our hypothesis that the mutation at attB in hlb of CC398 isolates influences integration of ΦSa3, we mutated the Φ13 attB site of hlb both in strain 8325-4 deviated from NCTC 8325 cured of Φ11, Φ12, and Φ13 (Novick, 1967), and in strain RN4220 derived from 8325-4 (Peng et al., 1988). After infecting both attB mutated strains with Φ13-kana at MOI of 0.1, we observed a significant decrease in lysogenization of Φ13-kana in both attB mutant strains, when compared to the respective wild type control strains (Figure 6) and confirmed by PCR amplification of hlb (Supplementary Figure 9) and aph3 (Supplementary Figure 10). These results demonstrate that point mutations of attB core sequence in hlb of CC398 isolates strongly influence the integration of ΦSa3 into the hlb gene, and the mutation favors ΦSa3 to integrate elsewhere in the bacterial genome and keeps the hlb intact.
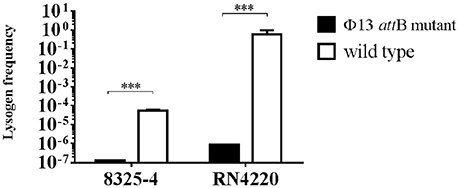
Figure 6. Lysogenization of 8325-4Φ13attBmut and RN4220Φ13attBmut by Φ13. Φ13-kana were mixed with 8325-4Φ13attBmut, RN4220Φ13attBmut, and the corresponding wild type strains with MOI of 0.1 and lysogen frequency was monitored as ratio of CFU on TSA plates with 100 μg/ml kanamycin and 5% of sheep blood (Φ13-kana LA-MRSA CC398 lysogen) relative to the total CFU count on TSA plates with 5% of sheep blood. Horizontal lines represent mean value, error bar represents ±SD, n = 3. ***P < 0.001 by t-test analysis.
Discussion
Mitomycin C and hydrogen peroxide are known to induce prophages in bacteria by causing DNA damage, which activates the SOS response and in turn cleaves the phage repressor (Frye et al., 2005; Goerke et al., 2006a; Loś et al., 2010; Nanda et al., 2015). Here, we show that commercial biocides containing hydrogen peroxide can induce phage Φ13, a phage belonging to Sa3 phage group that is known to encode human immune evasion genes and likely to be important for human colonization (Iandolo et al., 2002). Additionally, when co-culturing the human originating strain, 8325-4Φ13-kana with the livestock MRSA CC398 strain 61599 (spa type t034), the transfer frequency of Φ13 to strain 61599 was significantly increased in the presence of sublethal concentrations of hydrogen peroxide or commercial biocides containing hydrogen peroxide when compared to the transfer frequency without any treatment. Integration of Φ13 was also attempted in another LA-MRSA CC398 strain namely 93616 (spa type t899), but in this strain, we did not obtain lysogens. The inability of Φ13 to lysogenize strain 93616 could be caused by strain variations such as the restriction modification systems that restricts DNA transfer between staphylococcal lineages (Sadykov, 2016), or differences in wall teichoic acid glycopolymers that are the receptors of Φ13 and are highly strain-specific (Xia et al., 2010).
Hydrogen peroxide is a commonly applied bactericidal compound for disinfection in livestock production (Kjølholt et al., 2001). Our study indicates that this compound can act as an extrinsic factor contributing to the induction of ΦSa3 by triggering excision and propagation of the prophage, and subsequent transfer from human to livestock-associated S. aureus strains. In contrast, neither sodium hypochlorite that is known to affect the bacterial cell wall (Maillard, 2002), nor benzalkonium chloride that is a membrane-active agent (SCHENIHR (Scientific Committee on Emerging and Newly Identified Health Risks), 2009), were able to induce Φ13. In the absence of external stimuli, no spontaneous induction of Φ13 was observed in concordance with a previous study (Goerke et al., 2006a). Interestingly, we did observe transfer of Φ13 from strain 8325-4Φ13-kana to strain 61599 when the two strains were co-cultured. The most likely reason for this is that Φ13 propagates on strain 61599 resulting in more phages that in turn increase the chance of phage integration in strain 61599.
ΦSa3 group is known as hlb-converting phages that integrate in the hlb-gene at the attB attachment site (5′-TGTATCCAAACTGG-3′) recognized by the phage integrase (Coleman et al., 1991). A likely explanation for the atypical integration of ΦSa3 in strain 61599 is the two silent point mutations in the attB core sequence located within hlb. We speculate that these mutations in attB may be the reason that we did not observe phage integration in hlb but rather at numerous other locations in the LA-MRSA CC398 strain 61599. These integration sites share homology to the attB core sequence for Φ13 (Coleman et al., 1991). In particularly seven of the eight integration site sequences contain a four nucleotides sequence (5′-CTGG-3′) at the 3′-end, which is present in the Φ13 attB core sequence in hlb gene (5′-TGTATCCAAACTGG-3′) (Coleman et al., 1991), but is not in the attB core sequence in hlb from CC398 strains (5′-TGTATCCGAATTGG-3′ and 5′-TATATCCGAATTGG-3′). We speculate that this sequence similarity is important for the integration of ΦSa3 into CC398. It was previously reported that Φ13 can be integrated at different locations in S. aureus (Goerke et al., 2006b; Kraushaar et al., 2017), but the reason for the atypical integration of ΦSa3 had not been studied before. Here, we exchanged the core sequence of Φ13 attB in hlb from both strain 8325-4 and strain RN4220 to the attB core sequence from CC398 strain 61599. After infecting with Φ13, we observed a significant decrease in lysogenization frequency in the Φ13 attB mutants compared to cells carrying the intact hlb gene. This result demonstrates that the mutations at the attB core sequence in hlb gene of CC398 strain cause the low transfer frequency of Φ13 to CC398 strains and also drives the atypical integration of Φ13 in CC398 strains.
In conclusion, we show that in the presence of commercially available biocides containing hydrogen peroxide, ΦSa3 is transferred from a human-associated S. aureus donor strain to a strain, belonging to the CC398 complex. Previous studies have shown that the CC398 ancestor was adapted to humans but jumped to animals by loss of prophage Sa3 during the close human-livestock activities (Price et al., 2012). Recently, it was shown that ΦSa3 may be re-introduced into livestock-associated CC398 strains in a single horizontal gene transfer event from human-associated S. aureus and that it can be maintained stably as a prophage in the CC398 lysogens (Larsen et al., 2016). Our study highlights the importance of environmental factors in transfer of ΦSa3 between S. aureus strains and that just a four nucleotides sequence may be enough to guide integration of the phage LA-MRSA strains. Future studies will be needed to determine if and how integration frequencies vary from strain to strain and to identify factors and conditions that may prevent ΦSa3 transmission.
Author Contributions
YT, JH, LN, CWo, and HI designed the study, YT and AH conducted the microbiological analysis, CWi and YT constructed strains, YT, JL, JH and HI analyzed the results. YT wrote the manuscript and YT, LN, AH, JH, CWi, PA, JL, CWo, and HI reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by a Danish Research Council Sapere Aude grant to JH, the Deutsche Forschungsgemeinschaft (GRK1708) and the PhD Scholarship from the Faculty of Health and Medical Sciences, University of Copenhagen. We would like to thank Elvira Chapka from Department of Microbiology and Infection Control, Statens Serum Institut for their technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02418/full#supplementary-material
References
Becker, K., Ballhausen, B., Kahl, B. C., and Köck, R. (2015). The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet. Microbiol. 200, 33–38. doi: 10.1016/j.vetmic.2015.11.013
Bhat, M., Dumortier, C., Taylor, B. S., Miller, M., Vasquez, G., Yunen, J., et al. (2009). Staphylococcus aureus ST398, New York City and Dominican Republic. Emerging Infect. Dis. 15, 285–287. doi: 10.3201/eid1502.080609
Brückner, R. (2006). Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151, 1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x
Clinical and Laboratory Standards Institute (CLSI). (2008). Preformance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals; Approve Standard, 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Coleman, D., Knights, J., Russell, R., Shanley, D., Birkbeck, T. H., Dougan, G., et al. (1991). Insertional inactivation of the Staphylococcus aureus β-toxin by bacteriophage ϕ13 occurs by site-and orientation-specific integration of the ϕ13 genome. Mol. Microbiol. 5, 933–939.
Coleman, D. C., Sullivan, D. J., Russell, R. J., Arbuthnott, J. P., Carey, B. F., and Pomeroy, H. M. (1989). Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J. Gen. Microbiol. 135, 1679–1697.
Cuny, C., Wieler, L. H., and Witte, W. (2015). Livestock-associated MRSA: the impact on humans. Antibiotics 4, 521–543. doi: 10.3390/antibiotics4040521
de Haas, C. J., Veldkamp, K. E., Peschel, A., Weerkamp, F., Van Wamel, W. J., Heezius, E. C., et al. (2004). Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199, 687–695. doi: 10.1084/jem.20031636
DeLeo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. doi: 10.1128/CMR.00081-09
Diene, S. M., Corvaglia, A. R., Francois, P., van der Mee-Marquet, N., and Regional Infection Control Group of the Centre, R. (2017). Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics 18:133. doi: 10.1186/s12864-017-3516-x
Dulon, M., Haamann, F., Peters, C., Schablon, A., and Nienhaus, A. (2011). MRSA prevalence in European healthcare settings: a review. BMC Infect. Dis. 11:138. doi: 10.1186/1471-2334-11-138
Frye, J. G., Porwollik, S., Blackmer, F., Cheng, P., and McClelland, M. (2005). Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 187, 1485–1492. doi: 10.1128/JB.187.4.1485-1492.2005
Goerke, C., Matias y Papenberg, S., Dasbach, S., Dietz, K., Ziebach, R., Kahl, B. C., et al. (2004). Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J. Infect. Dis. 189, 724–734. doi: 10.1086/381502
Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191, 3462–3468. doi: 10.1128/JB.01804-08
Goerke, C., Koller, J., and Wolz, C. (2006a). Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 171–177. doi: 10.1128/AAC.50.1.171-177.2006
Goerke, C., Wirtz, C., Fluckiger, U., and Wolz, C. (2006b). Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 61, 1673–1685. doi: 10.1111/j.1365-2958.2006.05354.x
Guérout-Fleury, A. M., Shazand, K., Frandsen, N., and Stragier, P. (1995). Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167, 335–336. doi: 10.1016/0378-1119(95)00652-4
Iandolo, J. J., Worrell, V., Groicher, K. H., Qian, Y., Tian, R., Kenton, S., et al. (2002). Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289, 109–118. doi: 10.1016/S0378-1119(02)00481-X
Jung, P., Abdelbary, M. M. H., Kraushaar, B., Fetsch, A., Geisel, J., Herrmann, M., et al. (2016). Impact of bacteriophage Saint3 carriage on the immune evasion capacity and hemolytic potential of Staphylococcus aureus CC398. Vet. Microbiol. 200, 46–51. doi: 10.1016/j.vetmic.2016.02.015
Kjølholt, J., Nielsen, P. J., and Samsøe-Petersen, L. (2001). Inventory of Biocides Used in Denmark. Environmental Project No. 585. Copenhagen: Miljøstyrelsen.
Köck, R., Siam, K., Al-Malat, S., Christmann, J., Schaumburg, F., Becker, K., et al. (2011). Characteristics of hospital patients colonized with livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) CC398 versus other MRSA clones. J. Hosp. Infect. 79, 292–296. doi: 10.1016/j.jhin.2011.08.011
Kraushaar, B., Hammerl, J. A., Kienöl, M., Heinig, M. L., Sperling, N., Dinh Thanh, M., et al. (2017). Acquisition of virulence factors in livestock-associated MRSA: lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci. Rep. 7:2004. doi: 10.1038/s41598-017-02175-4
Kreiswirth, B. N., Löfdahl, S., Betley, M. J., O'Reilly, M., Schlievert, P. M., Bergdoll, M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712.
Larsen, J., Petersen, A., Sørum, M., Stegger, M., van Alphen, L., Valentiner-Branth, P., et al. (2015). Meticillin-resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro. Surveill. 20, 5–13. doi: 10.2807/1560-7917.ES.2015.20.37.30021
Larsen, J., Stegger, M., Andersen, P. S., Petersen, A., Larsen, A. R., Westh, H., et al. (2016). Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 63, 1349–1352. doi: 10.1093/cid/ciw532
Loś, J. M., Loś, M., Wegrzyn, A., and Wegrzyn, G. (2010). Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 58, 322–329. doi: 10.1111/j.1574-695X.2009.00644.x
Maillard, J. Y. (2002). Bacterial target sites for biocide action. J. Appl. Microbiol. 16S−27S. doi: 10.1046/j.1365-2672.92.5s1.3.x
McCarthy, A. J., and Lindsay, J. A. (2010). Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 10:173. doi: 10.1186/1471-2180-10-173
Monk, I. R., Shah, I. M., Xu, M., Tan, M. W., and Foster, T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3:e00277-11. doi: 10.1128/mBio.00277-11
Nanda, A. M., Thormann, K., and Frunzke, J. (2015). Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 197, 410–419. doi: 10.1128/JB.02230-14
Novick, R. (1967). Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33, 155–166. doi: 10.1016/0042-6822(67)90105-5
Peng, H. L., Novick, R. P., Kreiswirth, B., Kornblum, J., and Schlievert, P. (1988). Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988
Pérez-Moreno, M. O., Centelles-Serrano, M. J., Nogales-López, J., Domenech-Spanedda, M. F., Lozano, C., and Torres, C. (2016). Unusual presence of the immune evasion gene cluster in livestock-associated MRSA of lineage CC398 causing peridural and psoas abscesses in a poultry farmer. Enferm. Infecc. Microbiol. Clin. doi: 10.1016/j.eimc.2016.07.008. [Epub ahead of print].
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3:e00305-11. doi: 10.1128/mBio.00305-11
Projan, S. J., Kornblum, J., Kreiswirth, B., Moghazeh, S. L., Eisner, W., and Novick, R. P. (1989). Nucleotide sequence: the beta-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 17:3305. doi: 10.1093/nar/17.8.3305
Quinn, P. J., and Markey, B. K. (2001). “Disinfection and disease prevention in veterinary medicine,” in Disinfection, Sterilization and Preservation, 5th Edn. ed S. Block (Philadelphia, PA: Lea & Febiger), 1069–1103.
Rooijakkers, S. H., Ruyken, M., Roos, A., Daha, M. R., Presanis, J. S., Sim, R. B., et al. (2005). Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927. doi: 10.1038/ni1235
Russell, A. D. (2003). Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 52, 750–763. doi: 10.1093/jac/dkg422
Sadykov, M. R. (2016). “Restriction–modification systems as a barrier for genetic manipulation of Staphylococcus aureus,” in The Genetic Manipulation of Staphylococci: Methods and Protocols, ed J. L. Bose (New York, NY: Springer New York), 9–23.
SCHENIHR (Scientific Committee on Emerging and Newly Identified Health Risks) (2009). Assesssment of the Antibiotic Resistance Effects of Biocides. Brussels: SCHENIHR.
Schijffelen, M. J., Boel, C. H, van Strijp, J. A., and Fluit, A. C. (2010). Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376
Smith, T. C. (2015). Livestock-associated Staphylococcus aureus: the United States experience. PLoS Pathog. 11:e1004564. doi: 10.1371/journal.ppat.1004564
Smith, T. C., and Pearson, N. (2011). The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11, 327–339. doi: 10.1089/vbz.2010.0072
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
van Cleef, B. A., Monnet, D. L., Voss, A., Krziwanek, K., Allerberger, F., Struelens, M., et al. (2011). Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerging Infect. Dis. 17, 502–505. doi: 10.3201/eid1703.101036
van der Mee-Marquet, N. L., Corvaglia, A., Haenni, M., Bertrand, X., Franck, J. B., Kluytmans, J., et al. (2014). Emergence of a novel subpopulation of CC398 Staphylococcus aureus infecting animals is a serious hazard for humans. Front. Microbiol. 5:652. doi: 10.3389/fmicb.2014.00652
van Loo, I., Huijsdens, X., Tiemersma, E., de Neeling, A., van de Sande-Bruinsma, N., Beaujean, D., et al. (2007). Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerging Infect. Dis. 13, 1834–1839. doi: 10.3201/eid1312.070384
van Wamel, W. J., Rooijakkers, S. H., Ruyken, M., van Kessel, K. P., and van Strijp, J. A. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188, 1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006
Welinder-Olsson, C., Floren-Johansson, K., Larsson, L., Oberg, S., Karlsson, L., and Ahren, C. (2008). Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerging Infect. Dis. 14, 1271–1272. doi: 10.3201/eid1408.071427
Wirtz, C., Witte, W., Wolz, C., and Goerke, C. (2009). Transcription of the phage-encoded Panton-Valentine leukocidin of Staphylococcus aureus is dependent on the phage life-cycle and on the host background. Microbiology 155(Pt 11), 3491–3499. doi: 10.1099/mic.0.032466-0
Wulf, M. W., Markestein, A., van der Linden, F. T., Voss, A., Klaassen, C., and Verduin, C. M. (2008). First outbreak of methicillin-resistant Staphylococcus aureus ST398 in a Dutch hospital, June 2007. Euro Surveill. 13:8051. doi: 10.2807/ese.13.09.08051
Xia, G., Maier, L., Sanchez-Carballo, P., Li, M., Otto, M., Holst, O., et al. (2010). Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415. doi: 10.1074/jbc.M109.096172
Keywords: LA-MRSA CC398, biocide, prophage, ΦSa3, phage transfer
Citation: Tang Y, Nielsen LN, Hvitved A, Haaber JK, Wirtz C, Andersen PS, Larsen J, Wolz C and Ingmer H (2017) Commercial Biocides Induce Transfer of Prophage Φ13 from Human Strains of Staphylococcus aureus to Livestock CC398. Front. Microbiol. 8:2418. doi: 10.3389/fmicb.2017.02418
Received: 28 September 2017; Accepted: 22 November 2017;
Published: 07 December 2017.
Edited by:
Peter Mullany, University College London, United KingdomReviewed by:
Chad W. Euler, Hunter College (CUNY), United StatesGrzegorz Wegrzyn, University of Gdańsk, Poland
Copyright © 2017 Tang, Nielsen, Hvitved, Haaber, Wirtz, Andersen, Larsen, Wolz and Ingmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanne Ingmer, hi@sund.ku.dk
 Yuanyue Tang
Yuanyue Tang Lene N. Nielsen2
Lene N. Nielsen2 Paal S. Andersen
Paal S. Andersen Jesper Larsen
Jesper Larsen Christiane Wolz
Christiane Wolz Hanne Ingmer
Hanne Ingmer