- Department of Biochemistry, University of Otago, Dunedin, New Zealand
Extracytoplasmic function (ECF) sigma factors control expression of large numbers of genes in bacteria. Most ECF sigma factors are inhibited by antisigma proteins, with inhibition being relieved by environmental signals that lead to inactivation of the antisigma protein and consequent sigma factor activity. In cell surface signaling (CSS) systems in Gram negative bacteria antisigma activity is controlled by an outer membrane protein receptor and its ligand. In Pseudomonas aeruginosa one such system controls expression of genes for secretion and uptake of a siderophore, pyoverdine. In this system the activities of two sigma factors σFpvI and σPvdS are inhibited by antisigma protein FpvR20 that binds to the sigma factors, preventing their interaction with core RNA polymerase. Transport of ferripyoverdine by its outer membrane receptor FpvA causes proteolytic degradation of FpvR20, inducing expression of σFpvI- and σPvdS-dependent target genes. Here we show that degradation of FpvR20 and induction of target gene expression was initiated within 1 min of addition of pyoverdine. FpvR20 was only partially degraded in a mutant lacking the intracellular ClpP protease, resulting in an FpvR20 subfragment (FpvR12) that inhibited σFpvI and σPvdS. The translation inhibitor chloramphenicol did not prevent induction of an σFpvI-dependent gene, showing that degradation of FpvR20 released pre-existing σFpvI in an active form. However, chloramphenicol inhibited induction of σPvdS-dependent genes showing that active σPvdS is not released when FpvR20 is degraded and instead, σPvdS must be synthesized in the absence of FpvR20 to be active. These findings show that sigma factor activation occurs rapidly following addition of the inducing signal in a CSS pathway and requires ClpP protease. Induction of gene expression that can arise from release of active sigma from an antisigma protein but can also require new sigma factor synthesis.
Introduction
Extracytoplasmic function sigma factors are the largest and most diverse family of sigma factors in bacteria, directing expression of genes in response to a wide range of environmental stimuli (Staron et al., 2009; Ho and Ellermeier, 2012; Mascher, 2013). The activities of most ECF sigma factors are controlled by antisigma proteins that bind to and inhibit their cognate sigma factors (Campbell et al., 2007; Osterberg et al., 2011). CSS systems control the activities of a large proportion of the ECF sigma factors in Gram negative bacteria. In CSS systems antisigma protein activity (and hence that of the cognate ECF sigma factor) is controlled by an outer membrane protein receptor in response to an extracellular chemical signal, commonly a ferrisiderophore (Visca et al., 2002; Braun et al., 2006; Llamas et al., 2014). One of the best-characterized CSS systems controls expression of genes for synthesis of a siderophore pyoverdine and subsequent uptake of ferripyoverdine in the opportunistic pathogen Pseudomonas aeruginosa (Figure 1). In this system sigma factors σFpvI and σPvdS are inhibited by antisigma protein FpvR20 that is formed by cleavage of a 37 kDa precursor protein (Draper et al., 2011). FpvR20 extends from the periplasm through the cytoplasmic membrane into the cytoplasm and inhibition involves binding of the sigma factors by FpvR20, which also causes degradation of σPvdS although not σFpvI (Spencer et al., 2008; Edgar et al., 2014, 2017). Importation of ferripyoverdine results in molecular rearrangement of its receptor, FpvA (Schalk et al., 2009), initiating a proteolytic cascade that results in complete degradation of FpvR20. σFpvI and σPvdS then direct expression of genes for synthesis of FpvA and pyoverdine, respectively. σPvdS also directs expression of genes encoding a secreted exotoxin and a protease (Lamont et al., 2002). The rate of induction of target gene expression in response to the appropriate environmental signal has not been determined for this or any other CSS pathway.
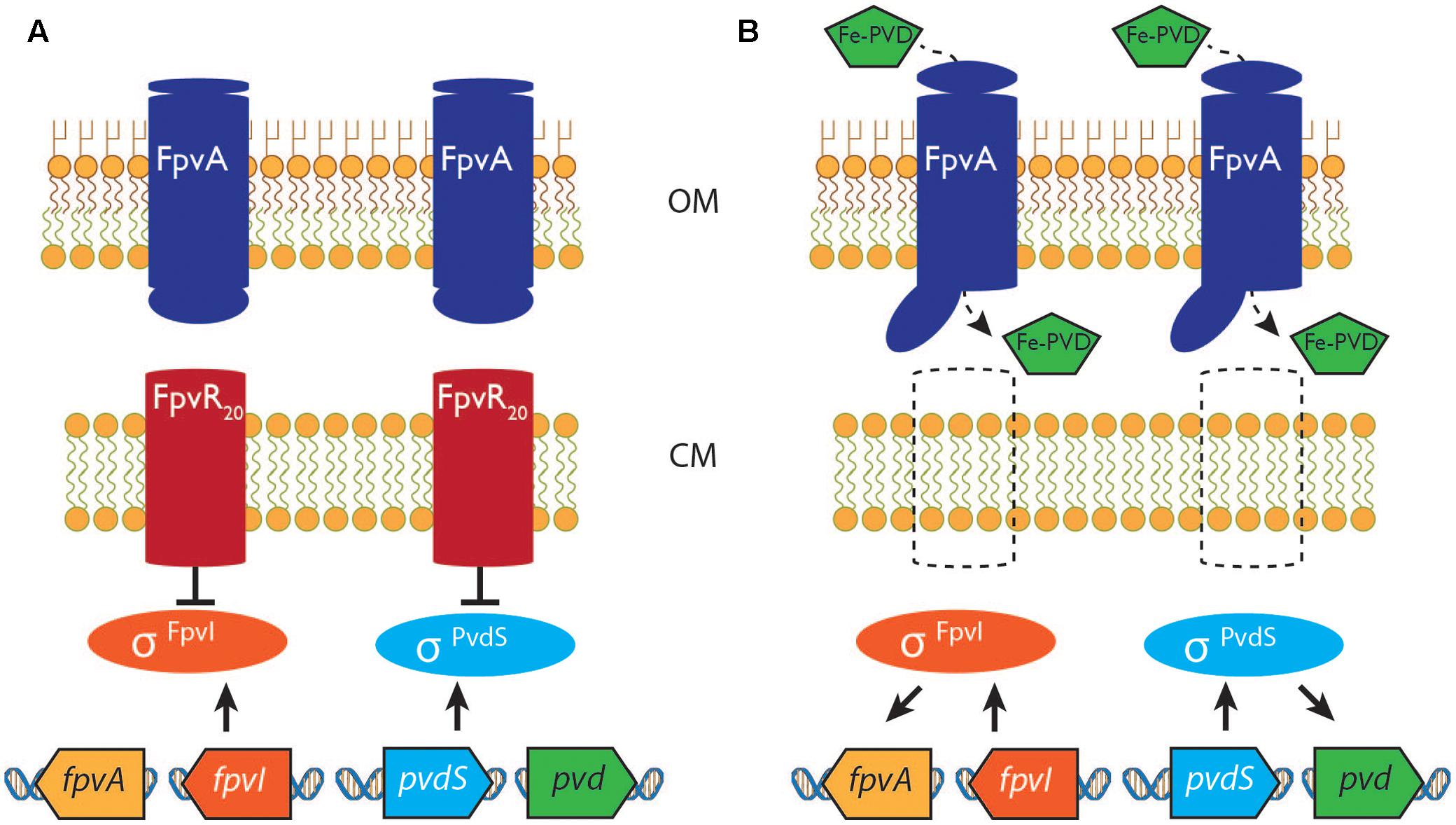
FIGURE 1. The pyoverdine signaling pathway. (A) In the absence of ferripyoverdine the activities of sigma factors σFpvI and σPvdS are inhibited by the antisigma protein FpvR20. (B) Import of ferripyoverdine (Fe-PVD) causes a molecular rearrangement of the FpvA receptor protein, triggering a proteolytic cascade that degrades FpvR20. σFpvI and σPvdS then become active, stimulating expression of the fpvA gene and of pyoverdine (pvd) synthesis genes, respectively. See text and (Llamas et al., 2014) for more detailed information. OM, outer membrane; CM, cytoplasmic membrane.
The molecular mechanisms underlying signal transduction in CSS pathways are only partially understood. The proteolytic cascade that leads to degradation of FpvR20 includes the cytoplasmic membrane protease RseP but the other proteases involved have not yet been identified (Draper et al., 2011). RseP and its homologs are also required for cleavage of other antisigma proteins that inhibit ECF sigma factors (King-Lyons et al., 2007; Draper et al., 2011; Damron and Goldberg, 2012; Barchinger and Ades, 2013). The periplasmic protease Prc is part of the proteolytic cascade in other CSS systems (Bastiaansen et al., 2014) but is not required for signal transduction in the pyoverdine system (Draper et al., 2011). The protease(s) required for degradation of the cytoplasmic antisigma component and consequent sigma factor activity are not yet known in this or any other CSS pathway.
The simplest model for induction of gene expression in sigma-antisigma systems is that degradation of antisigma protein releases active sigma factor that can then interact with core RNA polymerase to initiate transcription from target promoters. To the best of our knowledge this model has not been tested experimentally. A possible alternative mechanism, suggested by proteolysis of σPvdS in the presence of FpvR20, is that sigma factors are inactivated following binding by antisigma proteins and are only active when synthesized in the absence of the cognate antisigma.
The aims of the work described here were to investigate the time-course of degradation of FpvR20 and consequent induction of target gene expression in response to the ferripyoverdine inducing signal; to identify the protease responsible for degrading the cytoplasmic portion of FpvR20; and to investigate whether active σPvdS and σFpvI sigma factors are released following proteolysis of FpvR20, or whether sigma factors must be synthesized in the absence of FpvR20 to be active.
Materials and Methods
Growth of Bacteria
Strains of P. aeruginosa used in this study are listed in Table 1. Bacteria were routinely grown in LB medium or on LB agar at 37°C. For Western blotting and reverse transcription quantitative PCR (RT-qPCR) P. aeruginosa was grown in King’s B medium (King et al., 1954). Antibiotics were added as required at the same concentrations as described previously (Mettrick and Lamont, 2009) with chloramphenicol being added to a final concentration of 150 μg/mL that completely prevents protein synthesis (Kay and Gronlund, 1969).
Genetic Manipulations
Plasmids used in this study are listed in Table 1. Restriction of DNA molecules and DNA cloning were carried out using standard methods (Sambrook et al., 2000) with enzymes purchased from Roche Molecular Biologicals. DNA fragments required for strain construction were amplified from genomic DNA of P. aeruginosa PAO1 by PCR with FirePol DNA Polymerase (Solis Biodyne) or Taq DNA Polymerase Reddymix (ThermoPrime) using appropriate primers (Supplementary Table S1) that were designed on the basis of the P. aeruginosa PAO1 genome sequence1 (Winsor et al., 2011). DNA fragments were cloned into the required vectors and all plasmid constructs were verified by DNA sequencing.
Construction of an unmarked deletion in the P. aeruginosa clpP gene was carried out as described previously (Hoang et al., 1998; Mettrick and Lamont, 2009; Draper et al., 2011). Briefly, fragments of DNA flanking the deletion site in clpP were amplified by PCR using primer pairs listed in Supplementary Table S1, ligated together and cloned into the allele replacement vector pEX18Gm to give plasmid pEX18Gm::ΔclpP. Chromosomal allele replacement was then carried out (Hoang et al., 1998). Over 90% of clpP was deleted and the deletion was in-frame with downstream genes. Deletions and allele replacements were confirmed by PCR. An analogous method was used to create a deletion in the lon gene. For complementation of the clpP mutation, a 2.3 kb PCR product spanning the clpP gene, the upstream tig gene and the predicted promoter was cloned into the integrating plasmid miniCTX2 (Hoang et al., 2000) that was then transferred into P. aeruginosa pvdF clpP by conjugation from Escherichia coli S17-1 as described previously (Shirley and Lamont, 2009).
Western Blotting
Bacteria were grown in King’s B medium (King et al., 1954) (20 mL) to late exponential phase (OD600 between 1.8 and 2.2 [0.6 and 0.8 for PAO1 pvdF clpP]). A sample (400 μL [1000 μL for PAO1 pvdF clpP]) was centrifuged in a bench-top microcentrifuge (13,000 rpm, 20 s) and the pellet resuspended in SDS–PAGE loading buffer [2% (w/v) SDS, 5% (w/v) β-mercaptoethanol, 10% (v/v) glycerol, 0.002% bromophenol blue, 62.5 mM Tris–HCl; pH 6.8] (15 μL [8 μL for PAO1 pvdF clpP]) and PBS (85 μL [42 μL for PAO1 pvdF clpP]) at 99°C. Pyoverdine (150 μM), purified from P. aeruginosa PAO1 as described previously (Mettrick and Lamont, 2009), was added to each culture and samples were taken 1, 3, 5, 10, 20, 30, 60, 90, and 120 min after the addition of pyoverdine. The samples were centrifuged and the pellets resuspended as described above. The tubes were heated at 99°C for 20 min, centrifuged briefly and vortexed. Proteins were separated by electrophoresis on 12.5% SDS–PAGE gels and transferred to nitrocellulose membranes using standard methods (Harlow and Lane, 1999). Membranes were blocked in 15% (v/v) SeaBlock (Pierce) in TBS buffer (0.9% NaCl, 100 mM Tris–HCl; pH 7.5) containing 0.1% (v/v) Tween. Blots were probed with monoclonal antibody anti-σPvdS (Xiong et al., 2000) or anti-FpvRN (Draper et al., 2011) that was raised against a peptide corresponding to residues 62–75 within the N-terminal (cytoplasmic) region of FpvR20. Equal protein loadings were confirmed by probing the membranes with monoclonal antibody anti-RpoD (Santa Cruz Biotechnology). Detection was carried out using anti-Mouse HRP conjugates (Sigma), Super Signal ECL (Pierce), and a Fuji LAS-1000 Imager. Western blotting was carried out with bacteria from at least two independent cultures for each strain and growth condition and representative data are shown.
RT-qPCR
Bacteria were grown at 37°C in King’s B medium (King et al., 1954) (20 mL) to late exponential phase. A sample (500 μL) was taken from each culture (0 min) and transferred to a tube containing 1 mL of RNAprotect Bacteria Reagent (Qiagen). Pyoverdine (150 μM) was added to each culture. Samples (500 μL) were taken 5, 10, 30, and 60 min after the addition of pyoverdine. The samples were transferred to tubes containing 1 mL of RNAprotect Bacteria Reagent (Qiagen) and vortexed vigorously. RNA was extracted and RT-qPCR, including control reactions without template or without reverse transcriptase, was carried out as described previously (Draper et al., 2011; Konings et al., 2013). Relative quantification was performed using the second derivative maximum method corrected for primer efficiencies, with clpX and oprL as the combined reference genes, as described previously (Draper et al., 2011; Konings et al., 2013); the reference genes gave similar outcomes when used individually. qPCR was carried out twice for each cDNA sample, with three replicates each time. cDNA was prepared from at least two independent cultures for each strain and growth condition and representative data are shown.
Results
Time Course for Induction of Gene Expression
In the absence of ferripyoverdine, FpvR20 inhibits σPvdS and σFpvI. Addition of pyoverdine, which chelates iron to become ferripyoverdine, induces σPvdS-dependent expression of pyoverdine synthesis genes and the σFpvI-dependent gene fpvA (Lamont et al., 2002; Beare et al., 2003; Tiburzi et al., 2008; Draper et al., 2011). However, the rate of increase in gene expression following addition of the inducing signal has not been determined for this or any other CSS system. We therefore determined the time-course of activation of transcription for two σPvdS-dependent genes pvdH and pvdL, and the sole known σFpvI-dependent gene fpvA (Figure 2 and Supplementary Figure S1). pvdH and pvdL had very similar rates of induction, with maximal transcription 30 min after addition of pyoverdine. Expression of the fpvA gene was also maximal after 30 min although transcription of this gene was less strongly induced than that of pvdH and pvdL.
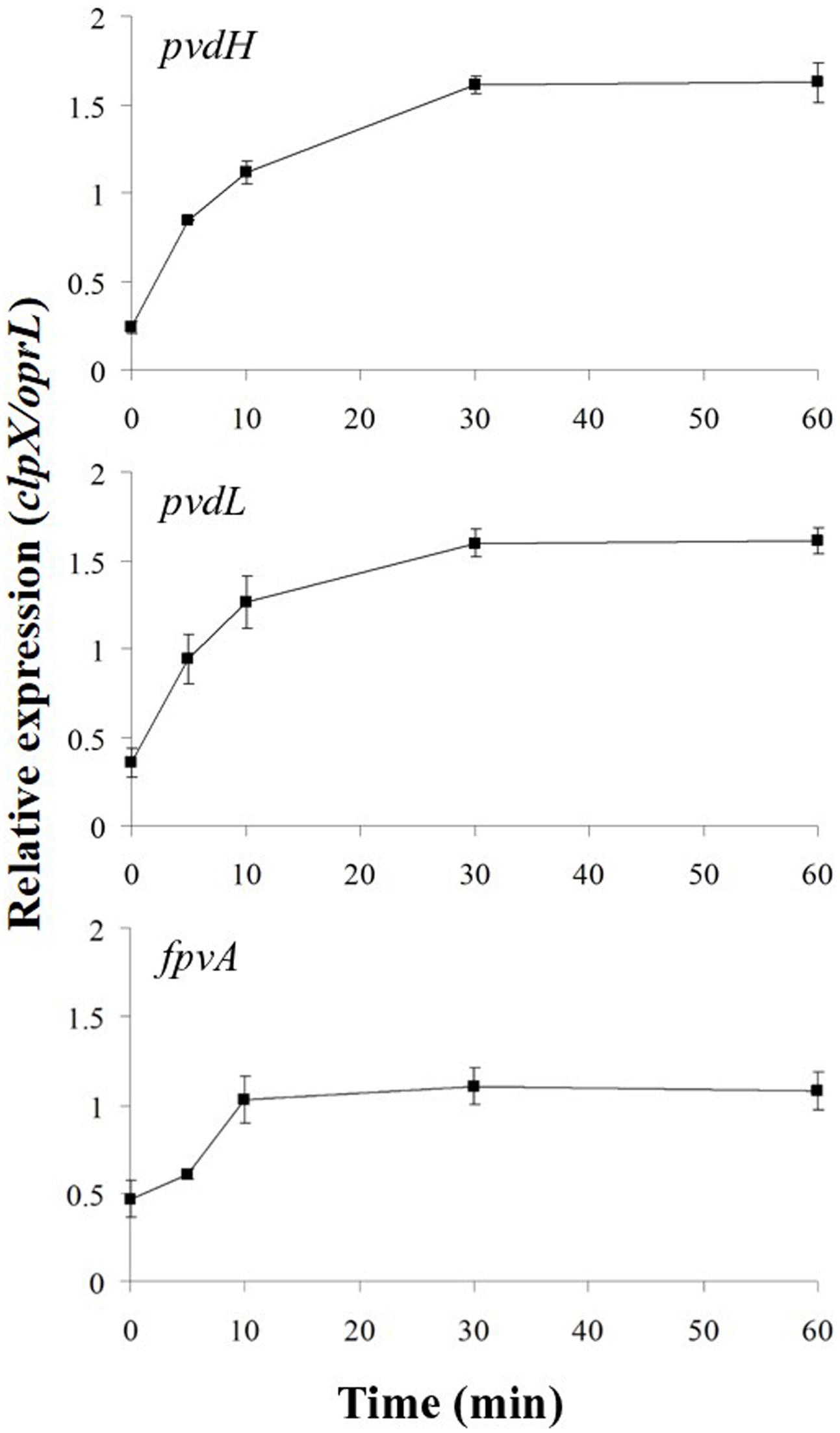
FIGURE 2. Activation of gene expression following addition of pyoverdine. Pyoverdine was added to Pseudomonas aeruginosa PAO1 pvdF bacteria (0 min) and samples were collected at intervals and analyzed by reverse transcription quantitative PCR (RT-qPCR). The amounts of pvdH, pvdL, and fpvA transcripts are shown relative to the reference genes clpX and oprL. Data are means of six technical replicates with standard deviation shown. Similar results were obtained when the experiment was repeated (Supplementary Figure S1).
Degradation of FpvR20 and σPvdS
σPvdS and σFpvI are inhibited by the FpvR20 protein and induction of gene expression following addition of pyoverdine requires degradation of FpvR20 (Draper et al., 2011). To determine the time-course of degradation of FpvR20, Western blotting was carried out using an antibody specific to the cytoplasmic (sigma factor binding) domain of the protein (Figure 3A and Supplementary Figure S2). There was significantly less FpvR20 per cell within 1 min of addition of pyoverdine and FpvR20 was almost undetectable by 30 min. This timeframe correlates well with the induction of gene expression (Figure 2). Pyoverdine-mediated induction of gene expression is dependent on the FpvA ferripyoverdine receptor protein (Shen et al., 2002; Beare et al., 2003; James et al., 2005; Draper et al., 2011). Addition of pyoverdine to an fpvA mutant did not result in degradation of FpvR20 (Figure 3B and Supplementary Figure S2) confirming the requirement for FpvA as well as pyoverdine for degradation of FpvR20.
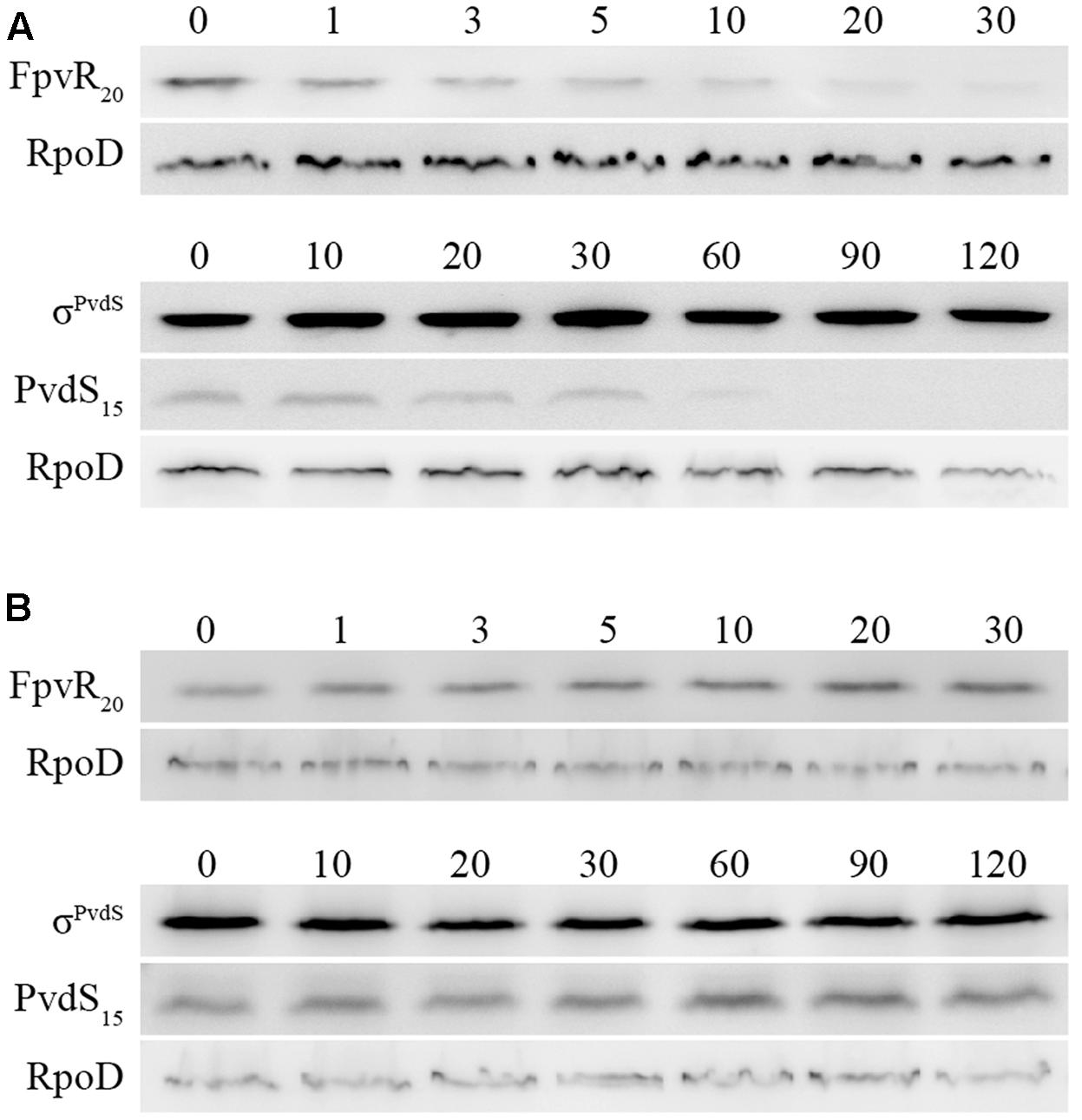
FIGURE 3. Time-course of degradation of FpvR20 and σPvdS following addition of pyoverdine. Pyoverdine was added to P. aeruginosa bacteria (0 min). Samples were collected at intervals and analyzed by Western blotting with antibodies against FpvR20, σPvdS or RpoD (loading control). FpvR20, σPvdS (full-size), PvdS15 and RpoD are indicated. Times are shown in minutes. (A) PAO1 pvdF (B) PAO1 pvdF fpvA. Similar results were obtained when the experiment was repeated (Supplementary Figure S2).
The presence of FpvR20 results in proteolysis of σPvdS to generate a subfragment (PvdS15) that is likely to be an intermediate in the proteolytic degradation of σPvdS, lowering the amount of σPvdS per cell (Spencer et al., 2008). The amount of PvdS15 decreased following the addition of pyoverdine (Figure 3A). In the absence of FpvA (and consequent presence of FpvR20) there was no change in the amount of PvdS15 following addition of pyoverdine (Figure 3B). These findings are consistent with the requirement of FpvR20 for degradation of σPvdS (Spencer et al., 2008). The amount of σFpvI per cell is not altered by the presence of FpvR20 (Edgar et al., 2017).
ClpP Protease Is Part of the Degradation Pathway
Degradation of FpvR20 requires one or more cytoplasmic proteases (Draper et al., 2011). The cytoplasmic protease ClpP contributes to the degradation of the antisigma factors RseA in E. coli (Flynn et al., 2004) and RsiW in Bacillus subtilis (Zellmeier et al., 2006). We therefore tested the hypothesis that ClpP contributes to degradation of FpvR20 and consequent activity of σPvdS and σFpvI. An in-frame deletion of the clpP (PA1801) gene was engineered in P. aeruginosa PAO1 pvdF. The effect of the clpP mutation on gene expression was then determined (Figure 4 and Supplementary Figure S3). The clpP mutation completely prevented induction of expression of pvdH, pvdL, and fpvA. Indeed, gene expression in the clpP mutant was even lower than in Clp+ bacteria in the absence of pyoverdine. Complementation of the mutant with wild-type clpP restored gene expression. These data show that ClpP protease is essential for induction of gene expression in the pyoverdine signaling pathway.
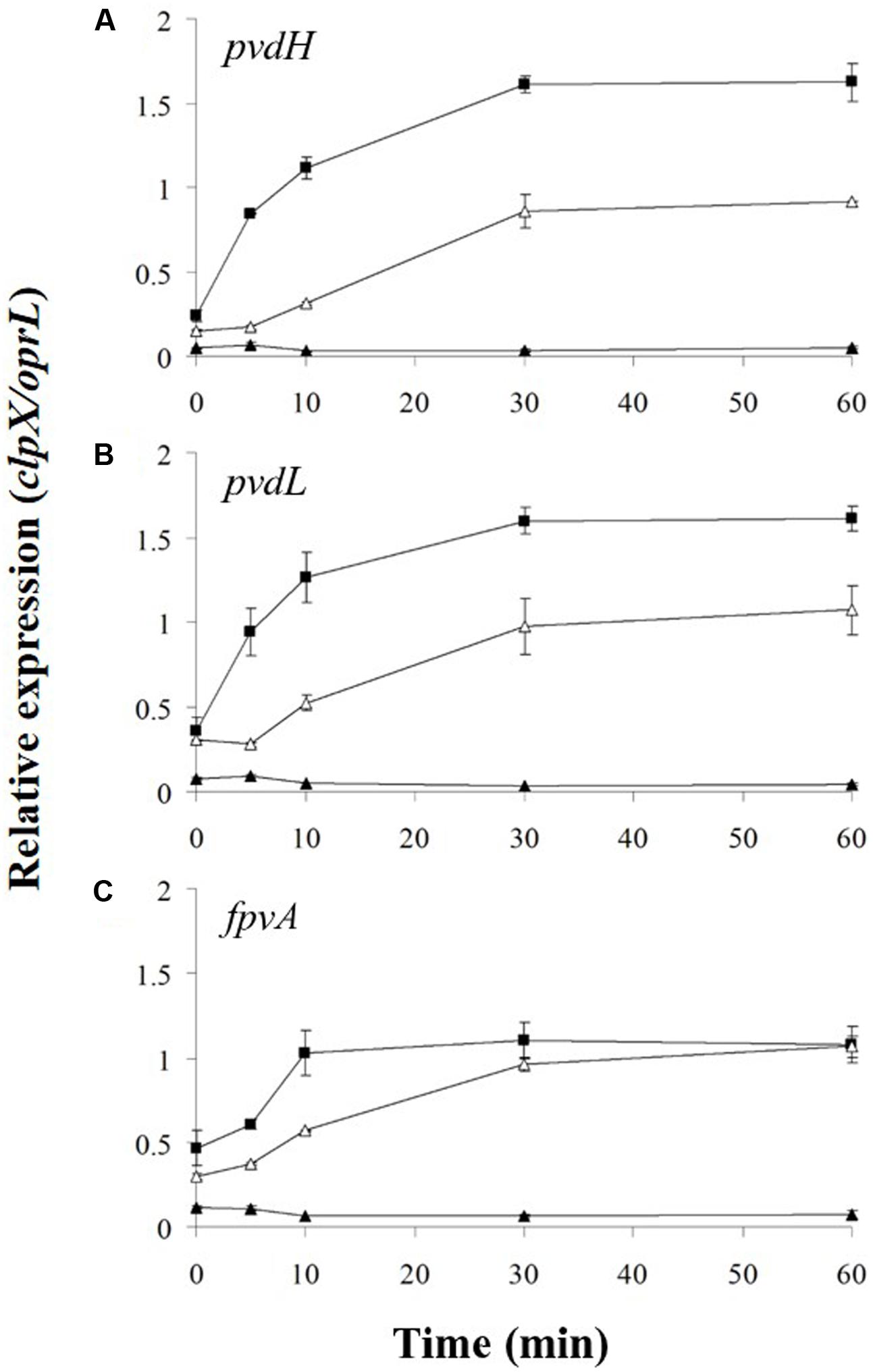
FIGURE 4. Effect of clpP mutation on induction of gene expression. Pyoverdine was added (0 min) to P. aeruginosa PAO1 pvdF clpP (black triangles) and P. aeruginosa PAO1 pvdF clpP (minictx::tig-clpP) (open triangles). Samples were collected at intervals and analyzed by RT-qPCR. Data are means of six technical replicates with standard deviation shown. Equivalent data from strain PAO1 pvdF bacteria (Figure 2) (black squares) are included for comparison. (A) pvdH. (B) pvdL. (C) fpvA.
We therefore tested the hypothesis that ClpP is required for degradation of FpvR20. Following addition of pyoverdine to clpP mutant bacteria, FpvR20 was degraded and a sub-fragment of approx. 12 kDa (FpvR12) was present that was not detected in Clp+ strains (Figure 5A and Supplementary Figure S4). The increasing amount of this fragment following addition of pyoverdine was inversely proportional to the decreasing amount of FpvR20 indicating that FpvR12 is formed as a result of proteolysis of FpvR20. The FpvR12 fragment retains the cytoplasmic sigma binding domain of FpvR20 (Edgar et al., 2014) explaining the inhibition of sigma factor activity that occurs in the clpP mutant even when pyoverdine is present (Figure 4). Complementation with wild-type clpP restored wild-type phenotype of an absence of FpvR12 (Figure 5B and Supplementary Figure S4) and largely restored expression of the pvdH, pvdL, and fpvA genes (Figure 4); incomplete restoration of gene expression was most likely a consequence of the different chromosomal context of the introduced clpP gene. In contrast to ClpP+ bacteria, reduction in the amount of PvdS15 following addition of pyoverdine did not occur in the clpP mutant (Figure 5A and Supplementary Figure S4). Collectively these data show that ClpP protease is required for, and most likely catalyzes, degradation of the cytoplasmic portion of FpvR20.
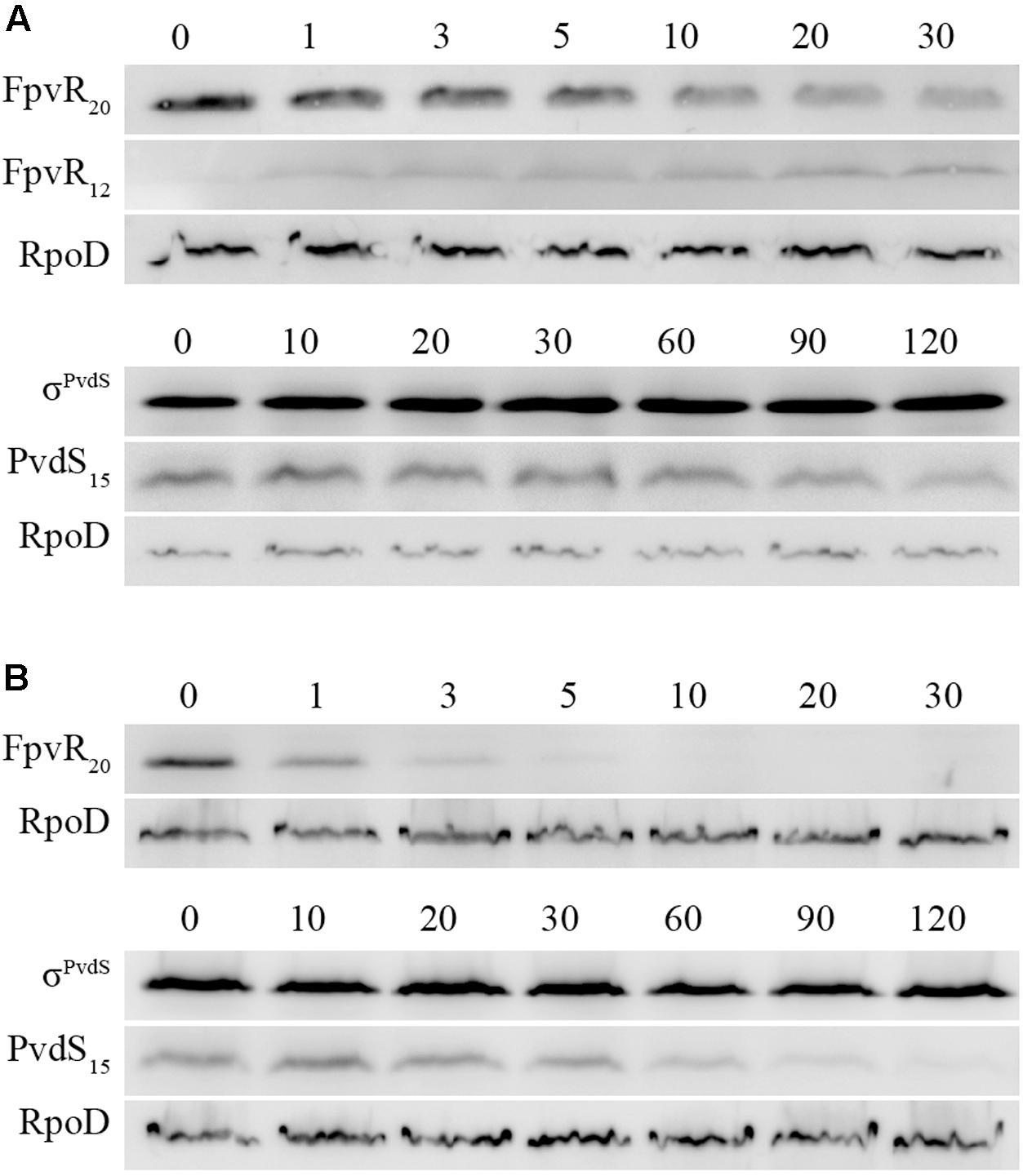
FIGURE 5. Effects of clpP mutation on degradation of FpvR20 and σPvdS. Pyoverdine was added (0 min) to P. aeruginosa PAO1 pvdF bacteria containing a mutation in the clpP gene. Samples were collected at intervals and analyzed by Western blotting. Times are shown in minutes. (A) P. aeruginosa PAO1 pvdF clpP. (B) P. aeruginosa PAO1 pvdF clpP (minictx::tig-clpP). FpvR20, FpvR12, σPvdS (full-size), PvdS15 and RpoD are indicated. FpvR12 was not detected in P. aeruginosa PAO1 pvdF clpP (minictx::tig-clpP). Similar results were obtained when the experiment was repeated (Supplementary Figure S4).
Gene Expression Requires Newly Synthesized σPvdS But Not σFpvI
The proteolytic degradation of σPvdS that occurs in the presence of FpvR20 raised the question, is active sigma factor released when FpvR20 is degraded or must the sigma factors be synthesized in the absence of FpvR20 to be active? To address this question, the effects of the translation inhibitor chloramphenicol on transcription of target genes were measured. If newly synthesized sigma factor is required for transcription of target genes, prevention of sigma factor synthesis would prevent increased target gene expression; however, if degradation of FpvR20 releases active sigma factor, chloramphenicol would not prevent induction of target gene expression.
The results are shown in Figure 6A and Supplementary Figure S5A. Addition of chloramphenicol prevented pyoverdine-mediated induction of expression of pvdH and pvdL, indicating that σPvdS must be synthesized in the absence of FpvR20 for pvd gene expression to occur. Induction of fpvA gene expression was not prevented by the presence of chloramphenicol, indicating that degradation of FpvR20 released active σFpvI that could direct transcription of fpvA. The slight delay in induction of expression of fpvA in the presence of chloramphenicol may indicate that σFpvI released from degraded FpvR20 would normally be supplemented by newly synthesized σFpvI during pyoverdine-mediated induction of gene expression.
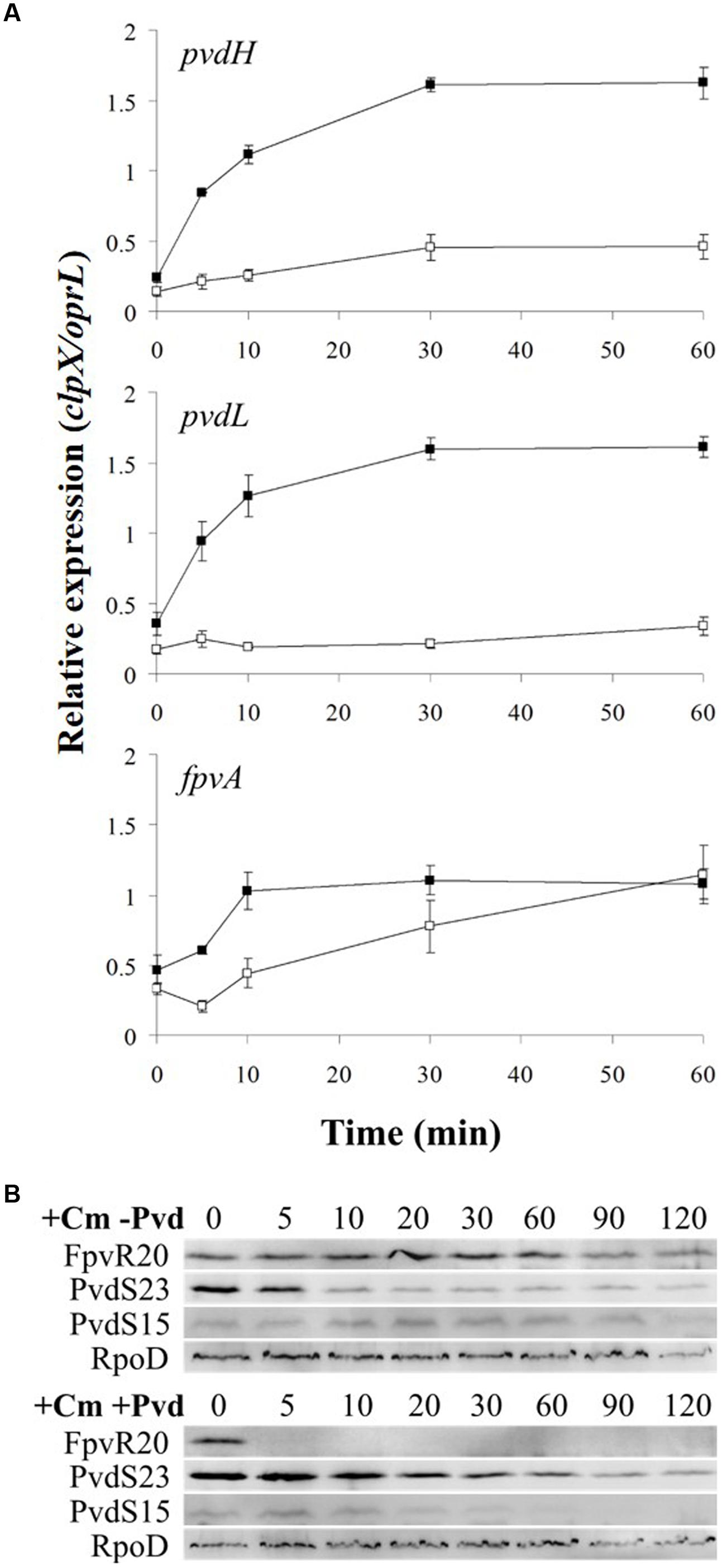
FIGURE 6. Effect of chloramphenicol on induction of gene expression. Chloramphenicol (Cm) and then pyoverdine were added to P. aeruginosa PAO1 pvdF bacteria (0 min) and samples were collected at intervals. (A) Samples were analyzed by RT-qPCR for pvdH, pvdL, and fpvA. Open squares, Cm present; Black squares, Cm absent. Data are means of six technical replicates with standard deviation shown. (B) Samples were analyzed by Western blotting for σPvdS or FpvR20 in the presence or absence of pyoverdine, as shown. FpvR20, σPvdS (full-size), PvdS15 and RpoD are indicated. Times are shown in minutes. Similar results were obtained when the experiment was repeated (Supplementary Figure S5).
The effect of chloramphenicol on protein amounts was also investigated (Figure 6B). In the presence of pyoverdine FpvR20 was rapidly degraded, as expected and as also occurred in the absence of chloramphenicol (Figure 2). In the absence of pyoverdine the amount of FpvR20 per cell did not greatly decrease for at least 60 min following addition of chloramphenicol. The continuing presence of FpvR20 in the absence of protein synthesis indicates that this protein is relatively stable under these conditions.
In the absence of pyoverdine and consequent presence of FpvR20, the amount of σPvdS decreased after the addition of chloramphenicol (Figure 6B and Supplementary Figure S5B). The amount of PvdS15 formed from σPvdS did not vary during the course of the experiment suggesting that the rates of formation and degradation of PvdS15 are similar. The decrease in the amount of σPvdS was slower following the addition of pyoverdine and consequent absence of FpvR20. These results indicate that FpvR20 accelerates, but is not essential for, degradation of σPvdS.
Discussion
In this research we show that degradation of the FpvR20 antisigma protein in the pyoverdine CSS pathway occurs within 1 min of addition of pyoverdine with expression of the pvd and fpvA target genes being maximal by 30 min. Target gene expression is dependent on ClpP protease for removal of FpvR20 antisigma activity and requires de novo synthesis of σPvdS, but not σFpvI.
The time-course of antisigma degradation has previously been examined for the E. coli stress response ECF sigma-antisigma system, in which antisigma RseA inhibits the activity of sigma factor σE. RseA has a half-life of about 8 min in uninduced bacteria and 1–2 min following heat-induced envelope stress (Ades et al., 2003; Chaba et al., 2007). In the absence of the pyoverdine inducing signal there was no detectable reduction in the amount of FpvR20 over at least 60 min when chloramphenicol was present to prevent new protein synthesis (Figure 6B), indicating that FpvR20 has a long half-life under non-inducing conditions. In contrast, FpvR20 was almost undetectable 5 min after addition of pyoverdine in the presence of chloramphenicol (Figure 6B) indicating a much shorter half-life. The rates of degradation of the FpvR20 and RseA antisigma proteins in induced and uninduced cells are therefore comparable, in each case allowing a rapid response to the relevant environmental signal. So far as we are aware the time-course of induction of target gene expression has not previously been determined for any CSS system or indeed, any ECF sigma-antisigma system.
The proteolytic cascade that leads to degradation of FpvR20 and induction of gene expression requires ClpP protease (Figures 4, 5). A CSS system is therefore part of the expanding repertoire of regulatory pathways in which Clp proteases are required for degradation of an antisigma protein and consequent sigma factor activity (Flynn et al., 2004; Zellmeier et al., 2006). Unfoldase chaperones – either ClpA or ClpX – are required to render substrate proteins susceptible to proteolysis by ClpP (Liu et al., 2014; Olivares et al., 2016) and it will be of interest to determine which unfoldase is required for complete degradation of FpvR20. In the clpP mutant a subfragment of FpvR20, FpvR12, is present that is presumably formed by proteolysis of FpvR20 and is further degraded by a ClpP-containing protease in ClpP+ bacteria. The size of FpvR12 indicates that it contains the complete cytoplasmic antisigma domain of FpvR20 and so inhibits σPvdS and σFpvI (Edgar et al., 2014, 2017). This is consistent with the finding that expression of pvdH, pvdL, and fpvA was significantly lower in the clpP mutant than in Clp+ bacteria in the absence of pyoverdine (Figure 6). Although FpvR20 has a long half-life in the absence of pyoverdine there was a gradual reduction in the amount of FpvR20 in the absence of new protein synthesis (Figure 4). These findings are consistent with proteolytic degradation of FpvR20 in wild-type bacteria even in the absence of pyoverdine. Auto-inducing systems such as the pyoverdine CSS system require a basal level of gene expression in order to produce and detect the inducing signal. In the pyoverdine system, some degradation of FpvR20 in the absence of pyoverdine is evidently necessary for low-level sigma factor activity to provide basal expression of fpvA and pvd genes that is needed for up-regulation of gene expression when pyoverdine is present.
The simplest model for sigma factor activation in sigma-antisigma systems is that degradation of the antisigma protein results in release of active sigma factor and consequent gene expression (Brooks and Buchanan, 2008; Ho and Ellermeier, 2012). So far as we are aware, this model has not been directly tested for any sigma/antisigma pair. Our data indicate that σPvdS must be synthesized in the absence of the FpvR20 antisigma in order for expression of pvd target genes to occur following addition of pyoverdine (Figure 6). This demonstrates an alternative model, in which a sigma factor must be synthesized in the absence of its cognate antisigma in order to be active. The presence of FpvR20 is associated with proteolysis of σPvdS that generates a subfragment (PvdS15), a likely intermediate in the proteolytic degradation of σPvdS, resulting in a lower amount of σPvdS per cell when FpvR20 is present (Figure 3) (Spencer et al., 2008). Proteolysis of σPvdS in the presence of FpvR20 may partially explain the requirement for sigma factor synthesis in the absence of FpvR20 in order for the σPvdS to be active and for target gene expression to occur following addition of pyoverdine. Conversely induction of the σFpvI-dependent fpvA gene did not require new protein synthesis (Figure 4A) showing that in this case, degradation of FpvR20 did result in release of active sigma factor. This is consistent with the cellular level of σFpvI being unaffected by the presence of FpvR20 (Edgar et al., 2017) indicating that FpvR20 does not trigger degradation of σFpvI. It will be of interest to determine whether other ECF sigma factors must, like σPvdS, be synthesized in the absence of the cognate anti-sigma in order to be active. A mutation in clpP or in the gene encoding Lon protease that degrades a number of regulatory proteins (Van Melderen and Aertsen, 2009) did not prevent formation of PvdS15 (Figure 5 and Supplementary Figure S6). The protease that catalyzes the formation of PvdS15 from σPvdS therefore remains to be identified.
Conclusion
Our results demonstrate the speed with which a CSS system can respond to the presence of the inducing signal and the importance of ClpP protease in this process. They also show that induction of gene expression in an ECF sigma-antisigma system can require synthesis of sigma factor in the absence of antisigma, rather than release of active sigma factor following degradation of the antisigma. The molecular mechanisms that initiate degradation of FpvR20 and presumably involve its interaction with FpvA remain to be elucidated.
Author Contributions
IL conceived the study and the project outline. TB, LM, and IL designed methods and experiments, analyzed the data and interpreted results. TB and LM carried out experiments and collected data. IL, TB, and LM wrote the manuscript. IL, TB, and LM contributed to revision of, and approved, the final manuscript.
Funding
This work was supported in part by research grant UOO0502 from the Marsden Fund administered by the Royal Society of New Zealand.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors are grateful to Dr. Rebecca Edgar for her comments on an earlier version of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02442/full#supplementary-material
Abbreviations
CSS, cell surface signaling; ECF, extracytoplasmic function.
Footnotes
References
Ades, S. E., Grigorova, I. L., and Gross, C. A. (2003). Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185, 2512–2519. doi: 10.1128/JB.185.8.2512-2519.2003
Barchinger, S. E., and Ades, S. E. (2013). Regulated proteolysis: control of the Escherichia coli σE-dependent cell envelope stress response. Subcell. Biochem. 66, 129–160. doi: 10.1007/978-94-007-5940-4_6
Bastiaansen, K. C., Ibanez, A., Ramos, J. L., Bitter, W., and Llamas, M. A. (2014). The prc and rsep proteases control bacterial cell-surface signaling activity. Environ. Microbiol. 16, 2433–2443. doi: 10.1111/1462-2920.12371
Beare, P. A., For, R. J., Martin, L. W., and Lamont, I. L. (2003). Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47, 195–207. doi: 10.1046/j.1365-2958.2003.03288.x
Braun, V., Mahren, S., and Sauter, A. (2006). Gene regulation by transmembrane signaling. Biometals 19, 103–113. doi: 10.1007/s10534-005-8253-y
Brooks, B. E., and Buchanan, S. K. (2008). Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim. Biophys. Acta 1778, 1930–1945. doi: 10.1016/j.bbamem.2007.06.005
Campbell, E. A., Greenwell, R., Anthony, J. R., Wang, S., Lim, L., Das, K., et al. (2007). A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 27, 793–805. doi: 10.1016/j.molcel.2007.07.009
Chaba, R., Grigorova, I. L., Flynn, J. M., Baker, T. A., and Gross, C. A. (2007). Design principles of the proteolytic cascade governing the σE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 21, 124–136. doi: 10.1101/gad.1496707
Damron, F. H., and Goldberg, J. B. (2012). Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 84, 595–607. doi: 10.1111/j.1365-2958.2012.08049.x
Draper, R. C., Martin, L. W., Beare, P. A., and Lamont, I. L. (2011). Differential proteolysis of sigma regulators controls cell-surface signalling in Pseudomonas aeruginosa. Mol. Microbiol. 82, 1444–1453. doi: 10.1111/j.1365-2958.2011.07901.x
Edgar, R. J., Hampton, G. E., Garcia, G. P. C., Maher, M. J., Perugini, M. A., Ackerley, D. F., et al. (2017). Integrated activities of two alternative sigma factors coordinate iron acquisition and uptake by Pseudomonas aeruginosa. Mol. Microbiol. 106, 891–904. doi: 10.1111/mmi.13855
Edgar, R. J., Xu, X., Shirley, M., Konings, A. F., Martin, L. W., Ackerley, D. F., et al. (2014). Interactions between an anti-sigma protein and two sigma factors that regulate the pyoverdine signaling pathway in Pseudomonas aeruginosa. BMC Microbiol. 14:287. doi: 10.1186/s12866-014-0287-2
Flynn, J. M., Levchenko, I., Sauer, R. T., and Baker, T. A. (2004). Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 18, 2292–2301. doi: 10.1101/gad.1240104
Harlow, E., and Lane, D. (1999). Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Ho, T. D., and Ellermeier, C. D. (2012). Extra cytoplasmic function sigma factor activation. Curr. Opin. Microbiol. 15, 182–188. doi: 10.1016/j.mib.2012.01.001
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hoang, T. T., Kutchma, A. J., Becher, A., and Schweizer, H. P. (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72. doi: 10.1006/plas.1999.1441
James, H. E., Beare, P. A., Martin, L. W., and Lamont, I. L. (2005). Mutational analysis of a bifunctional ferrisiderophore receptor and signal-transducing protein from Pseudomonas aeruginosa. J. Bacteriol. 187, 4514–4520. doi: 10.1128/JB.187.13.4514-4520.2005
Kay, W. W., and Gronlund, A. F. (1969). Amino acid pool formation in Pseudomonas aeruginosa. J. Bacteriol. 97, 282–291.
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
King-Lyons, N. D., Smith, K. F., and Connell, T. D. (2007). Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J. Bacteriol. 189, 6266–6275. doi: 10.1128/JB.00629-07
Konings, A. F., Martin, L. W., Sharples, K. J., Roddam, L. F., Latham, R., Reid, D. W., et al. (2013). Pseudomonas aeruginosa utilises multiple pathways for iron acquisition in cystic fibrosis. Infect. Immun. 81, 2697–2704. doi: 10.1128/IAI.00418-13
Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I., and Vasil, M. L. (2002). Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99, 7072–7077. doi: 10.1073/pnas.092016999
Liu, K., Ologbenla, A., and Houry, W. A. (2014). Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases. Crit. Rev. Biochem. Mol. Biol. 49, 400–412. doi: 10.3109/10409238.2014.925421
Llamas, M. A., Imperi, F., Visca, P., and Lamont, I. L. (2014). Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597. doi: 10.1111/1574-6976.12078
Mascher, T. (2013). Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 16, 148–155. doi: 10.1016/j.mib.2013.02.001
McMorran, B. J., Kumara, H. M. C. S., Sullivan, K., and Lamont, I. L. (2001). Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology 147, 1517–1524. doi: 10.1099/00221287-147-6-1517
Mettrick, K. A., and Lamont, I. L. (2009). Different roles for anti-sigma factors in siderophore signalling pathways of Pseudomonas aeruginosa. Mol. Microbiol. 74, 1257–1271. doi: 10.1111/j.1365-2958.2009.06932.x
Olivares, A. O., Baker, T. A., and Sauer, R. T. (2016). Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44. doi: 10.1038/nrmicro.2015.4
Osterberg, S., Del Peso-Santos, T., and Shingler, V. (2011). Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65, 37–55. doi: 10.1146/annurev.micro.112408.134219
Sambrook, J., Russell, D. W., and Irwin, N. (2000). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Schalk, I. J., Lamont, I. L., and Cobessi, D. (2009). Structure-function relationships in the bifunctional ferrisiderophore FpvA receptor from Pseudomonas aeruginosa. Biometals 22, 671–678. doi: 10.1007/s10534-008-9203-2
Shen, J., Meldrum, A., and Poole, K. (2002). FpvA receptor involvement in pyoverdine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 184, 3268–3275. doi: 10.1128/JB.184.12.3268-3275.2002
Shirley, M., and Lamont, I. L. (2009). Role of TonB1 in pyoverdine-mediated signaling in Pseudomonas aeruginosa. J. Bacteriol. 191, 5634–5640. doi: 10.1128/JB.00742-09
Spencer, M. R., Beare, P. A., and Lamont, I. L. (2008). Role of cell surface signaling in proteolysis of an alternative sigma factor in Pseudomonas aeruginosa. J. Bacteriol. 190, 4865–4869. doi: 10.1128/JB.01998-07
Staron, A., Sofia, H. J., Dietrich, S., Ulrich, L. E., Liesegang, H., and Mascher, T. (2009). The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74, 557–581. doi: 10.1111/j.1365-2958.2009.06870.x
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Tiburzi, F., Imperi, F., and Visca, P. (2008). Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol. Microbiol. 67, 213–227. doi: 10.1111/j.1365-2958.2007.06051.x
Van Melderen, L., and Aertsen, A. (2009). Regulation and quality control by Lon-dependent proteolysis. Res. Microbiol. 160, 645–651. doi: 10.1016/j.resmic.2009.08.021
Visca, P., Leoni, L., Wilson, M. J., and Lamont, I. L. (2002). Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45, 1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x
Winsor, G. L., Lam, D. K., Fleming, L., Lo, R., Whiteside, M. D., Yu, N. Y., et al. (2011). Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600. doi: 10.1093/nar/gkq869
Xiong, Y. -Q., Vasil, M. L., Johnson, Z., Ochsner, U. A., and Bayer, A. S. (2000). The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J. Infect. Dis. 181, 1020–1026. doi: 10.1086/315338
Keywords: ECF sigma factor, antisigma, ClpP protease, pyoverdine, siderophore, regulated proteolysis, bacterial signal transduction, cell surface signaling
Citation: Bishop TF, Martin LW and Lamont IL (2017) Activation of a Cell Surface Signaling Pathway in Pseudomonas aeruginosa Requires ClpP Protease and New Sigma Factor Synthesis. Front. Microbiol. 8:2442. doi: 10.3389/fmicb.2017.02442
Received: 18 October 2017; Accepted: 24 November 2017;
Published: 12 December 2017.
Edited by:
Daniela De Biase, Sapienza Università di Roma, ItalyReviewed by:
Kürşad Turgay, Leibniz University of Hanover, GermanyMichael L. Vasil, University of Colorado Denver School of Medicine, United States
Copyright © 2017 Bishop, Martin and Lamont. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iain L. Lamont, iain.lamont@otago.ac.nz
 Thomas F. Bishop
Thomas F. Bishop Lois W. Martin
Lois W. Martin Iain L. Lamont
Iain L. Lamont