- 1State Key Laboratory of Agricultural Microbiology and Key Laboratory of Plant Pathology of Hubei Province, Huazhong Agricultural University, Wuhan, China
- 2Laboratory of Biological Processing, Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences, Changsha, China
- 3United States Department of Agriculture, Agricultural Research Service, Washington State University, Pullman, WA, United States
Botrytis cinerea is an important plant pathogenic fungus with a wide range of host. It usually produces black-colored sclerotia (BS) due to deposition of 1,8-dihydroxynaphthalene melanin in sclerotial melanogenesis. Our previous study (Zhou et al., 2018) reported six B. cinerea isolates producing orange-colored sclerotia (OS) with deficiency in sclerotial melanogenesis. Comparison of ecological fitness (conidia, mycelia, sclerotia), natural distribution, and melanogenesis of selected BS and OS isolates suggests that sclerotia play an important role in the disease cycle caused by B. cinerea. However, the molecular mechanism for formation of the OS B. cinerea remains unknown. This study was done to unravel the molecular mechanism for the sclerotial melanogenesis deficiency in the OS isolates. We found that all the five sclerotial melanogenesis genes (bcpks12, bcygh1, bcbrn1/2, bcscd1) were down-regulated in OS isolates, compared to the genes in the BS isolates. However, the sclerotial melanogenesis-regulatory gene bcsmr1 had similar expression in both types of sclerotia, suggesting the sclerotial melanogenesis deficiency is due to loss-of-function of bcsmr1, rather than lack of expression of bcsmr1. Therefore, we cloned bcsmr1 from OS (bcsmr1OS) and BS (bcsmr1BS) isolates, and found a single-nucleotide deletion in bcsmr1OS. The single-nucleotide deletion caused formation of a premature stop codon in the open reading frame of bcsmr1OS, resulting in production of a 465-aa truncated protein. The transcription activity of the truncated protein was greatly reduced, compared to that of the 935-aa full-length protein encoded by bcsmr1BS in the BS isolates. The function of bcsmr1OS was partially complemented by bcsmr1BS. This study not only elucidated the molecular mechanism for formation of orange-colored sclerotia by the spontaneous mutant XN-1 of B. cinerea, but also confirmed the regulatory function of bcsmr1 in sclerotial melanogenesis of B. cinerea.
Introduction
Botrytis cinerea is an important plant pathogenic fungus in cool and temperate regions. It can infect more than 1,400 plant species, including about 500 commercially important vegetables, fruits, and ornamentals, causing gray mold disease (Elad et al., 2016). B. cinerea usually produces gray-colored conidia and hyphae, and black-colored sclerotia on the surface of the infected plant tissues, as it can synthesize melanin and deposit that dark pigment in the cell wall of hyphae, conidia, and sclerotia (Zeun and Buchenauer, 1985; Doss et al., 2003; Zhang et al., 2015; Schumacher, 2016).
Melanins are chemically diverse and polymerized phenolic or indolic compounds occurring in various organisms such as insects, mammals, plants, and fungi (Bell and Wheeler, 1986; Butler and Day, 1998; Jacobson, 2000; Hamilton and Gomez, 2002). So far, four groups of melanins, namely allomelanins, eumelanins, neuromelanins, and pheomelanins, have been identified based on their chemical structures (Ambrico, 2016). Previous studies have characterized two groups of fungal melanins, namely allomelanins such as 1,8-dihydroxynaphthalene melanin or DHN melanin, and eumelanins such as 3,4-dihydroxyphenylalanine melanin or DOPA melanin (Sapmak et al., 2015; Li et al., 2016).
Melanins are multifunctional compounds (Cordero and Casadevall, 2017). It is well-recognized that melanin deposition in the cell wall can improve fungal survival through enhancing fungal adaptation to environmental stresses, such as UV irradiation, desiccation, enzymatic lysis, toxic chemicals, antagonists, and predators (Bell and Wheeler, 1986; Butler and Day, 1998). In some plant pathogenic fungi, such as Colletotrichum legenarium and Pyricularia oryzae, melanin is a virulence factor, as it can facilitate appressorium-mediated penetration into plant tissues (Kubo et al., 1982; Howard and Valent, 1996; Chen et al., 2004). In other plant pathogenic fungi, such as Alternaria brassicicola, however, melanin biosynthesis was found to be negatively associated with virulence (Cho et al., 2012). In B. cinerea, melanin is dispensable for pathogenicity, and instead, it may affect longevity of conidia and sclerotia as proposed in previous studies (Zhang et al., 2015; Schumacher, 2016).
B. cinerea and many other ascomycetous fungi produce DHN melanins (Zeun and Buchenauer, 1985; Bell and Wheeler, 1986; Butler and Day, 1998; Doss et al., 2003; Schumacher, 2016). They derive from the polyketide pathway, which starts with the reaction for formation of 1,3,6,8-tetrahydroxynaphthalene (T4HN) from acetate units (precursor) under the catalysis of polyketide synthase or PKS (Bell and Wheeler, 1986; Butler and Day, 1998). Then, T4HN is sequentially converted to scytalone, 1,3,8-trihydoxy-naphthalene (T3HN), vermelone, and DHN monomers through reduction and dehydration reactions. Finally, the DHN monomers are polymerized through oxidative reactions to form the end product DHN melanin.
Previous studies have identified several genes related to melanogenesis in B. cinerea (Schumacher et al., 2014; Zhang et al., 2015; Cohrs et al., 2016; Schumacher, 2016). The core genes for the melanogenic enzymes include bcpks12/13 (for two PKSs), bcygh1 (for a hydrolase), bcbrn1/2 (for two reductases), and bcscd1 (for a dehydratase). Moreover, Saitoh et al. (2010) reported that bcCCC2 coding for a copper-transporting ATPase affects mycelial melanogenesis in B. cinerea. The regulatory genes involving melanogenesis of B. cinerea include those for transcription factors (TFs) such as bcsmr1, bcztf1/2, bcltf1/2, bcvelA, bcvelB, and bcvel1 (Schumacher et al., 2013, 2014; Yang et al., 2013a; Cohrs et al., 2016; Schumacher, 2016), and those for signal transduction such as bcptpA, bcptpB, bcptc1, bos1, bmp3, and sak1 (Liu et al., 2011; Yang et al., 2013b,c). Among these genes, bcsmr1, bcpks12, bcbrn1/2, and bcscd1 are required for sclerotial melanogenesis (Schumacher, 2016). Single disruption of bcsmr1, bcpks12, bcscd1, and bcbrn1/2, and double disruption of bcpks12/13 and bcbrn1/2 caused deficiency in sclerotial melanogenesis. The disruption-mutants Δbcsmr1, Δbcpks12, and ΔΔbcpks12/13 produced yellow-colored sclerotia without accumulation of DHN melanin in the epidermal cell wall (Schumacher, 2016).
Sclerotia are generally regarded as the dormancy structure for survival under adverse environmental conditions. Sclerotia produced by B. cinerea can germinate to produce conidia. However, importance of the sclerotia as the primary infection source remains controversial in the life cycle of B. cinerea, as they are not frequently observed in infected tissues of many diseased plants (Williamson et al., 2007). Zhou et al. (2018) reported six field isolates of B. cinerea producing orange-colored sclerotia (OS). The sclerotia of the OS isolate XN-1 were found to be deficient in sclerotial melanogenesis, compared to the black-colored sclerotia produced by isolate B05.10 of B. cinerea (Zhou et al., 2018). The orange-colored sclerotia were greatly reduced for survival, compared to the black-colored sclerotia. Meanwhile, Zhou et al. (2018) also found that the OS B. cinerea rarely occurred in nature. Therefore, DHN melanin deposition on the sclerotia of B. cinerea plays an important role in ecological fitness of sclerotia. However, the molecular mechanism responsible for formation of the orange-colored sclerotia by the field isolates of B. cinerea remains unknown. This study was conducted to address this question. Here, we report that a single-nucleotide deletion in bcsmr1 caused formation of a premature stop codon, resulting in formation of a truncated protein with loss-of-function in regulation of sclerotial melanogenesis.
Materials and Methods
Fungal Isolates and Culture Media
A total of eight isolates of B. cinerea were used in this study (Table 1). Among these isolates, XN-1, S417, and T417 are OS isolates, whereas B05.10, HS016, S59, WXto2-2, and XN087 are BS isolates. All these fungal isolates were maintained in 20% glycerol (v/v) at −80°C for the long-term storage. Working cultures of each isolate were prepared by transferring the mycelia of that isolate in storage on potato dextrose agar (PDA) and the resulting cultures were incubated in the dark at 20°C for 5 days. Seven cultural media (MYA, PDA, PDB, PRM, YPDA, SD/-Trp, and SD/-Trp-His) were used in this study. The composition of these media was listed in Table S1.
Gene Cloning and Sequence Analysis
Six B. cinerea isolates (XN-1, S417, T417, HS016, WXt02-2, XN087) were separately incubated on cellophane film (CF)-PDA at 20°C for 3 days (Zhou et al., 2018). The mycelial mats were collected from the cultures, and used for extraction of genomic DNA (gDNA) using the CTAB method (Zeng et al., 2014) and the total RNA using the Trizol® reagents (TaKaRa Biotechnol. Co. Ltd., Dalian, China). The gDNA was used as template to PCR-amplify the DNA sequences of bcsmr1 as well as bcpks12/13, bcbrn1/2, and bcscd1 with the specific primers and thermal programs (Tables S2, S3). The PCR product was separated by agarose gel electrophoresis. The target DNA band in the gel was purified and inserted into the pMD18-T vector (TaKaRa). The resulting re-combined plasmid was subsequently transformed into the competent cells of E. coli DH5α. The positive clones harboring the expected DNA insert were sent to AuGCT Biotechnological Company Ltd. (Beijing, China). The nucleotide (nt) sequences for bcsmr1 (including the promoter region) from different isolates and the amino acid (aa) sequences for BcSMR1 encoded by bcsmr1 were identified by BLAST searches in the public database (http://www.ncbi.nlm.nih.gov). The characteristic domains for BcSMR1 encoded by bcsmr1 were identified based on the annotation information of that protein in the reference isolate B05.10. Multiple alignments of the nt sequences of bcsmr1 or the aa sequences of BcSMR1 in the seven isolates as well as in B05.10 were done using DNAMAN version 5.2.2 (Lynnon Biosoft, Vaudreuil, Quebec, Canada).
Prokaryotic Expression of bcsmr1
The full-length cDNA of bcsmr1 was cloned by reverse transcription (RT)-PCR from the OS isolate XN-1 and the BS isolate B05.10 using the primer pair bcsmr1-etF/bcsmr1-etR (Table S2). The primers contain a Not I-restriction site. The resulting cDNA was separated by agarose gel electrophoresis and purified from the gel. Then, the cDNA was digested with Not I to release the target fragments of bcsmr1 from XN-1 (designated as bcsmr1OS) or B05.10 (designated as bcsmr1BS). The two cDNA fragments were separately inserted into the plasmid pET28a(+) (Beijing TransGen Biotechnol. Co. Ltd., Beijing China) at the Not I site and two new plasmids, namely pET28a-bcsmr1OS and pET28a-bcsmr1BS (both had a 6× His tag), were then regenerated (Figure S1). The two plasmids were separately transformed into the competent cells of E. coli BL21 (DE3) pLysS (Promega, Madison, WI, USA). Positive clones for the two plasmids were screened based on growth on LB containing kanamycin (50 μg/mL). They were shake-incubated and induced for production of the proteins BcSMR1OS and BcSMR1BS using the procedures described by Lou et al. (2015). The bacterial proteins were separated by 8% SDS-PAGE and visualized by staining with Coomassie brilliant blue G-250.
Western Blotting
The proteins in E. coli transformed with pET28a-bcsmr1OS or pET28a-bcsmr1BS were prepared using the procedures described by Lou et al. (2015) and were separated by 8% SDS-PAGE. After electrophoresis, the proteins in the gel were transferred by electrophoresis blotting (20 V, 30 min) to a piece of Bio-Rad® polyvinylidene fluoride (PVDF) membrane (6 × 4 cm, length × width) in Trans-Blot® SD Semi-Dry Electrophoretic Transfer Cell. The membrane was washed in TBST buffer (20 mmol/L Tris-base, 150 mmol/L NaCl, 0.05% Tween 20), followed by soaking for 2 h in 5% (w/v) DifcoTM skim milk solution alone and then in 5% skim milk solution amended with 0.2% (w/v) anti-His tag antibody (Sangon Biotechnol. Co. Ltd., Shanghai, China) for another 2 h. After that, the membrane was washed again in the TBST buffer for three times, 10 min each time, followed by soaking for 2 h in 5% skim milk solution amended with 0.1% (w/v) alkaline phosphatase-conjugated goat anti-mouse IgG (Sangon Biotechnol. Co. Ltd., Shanghai, China). After another washing in the TBST buffer, the membrane was soaked in 10 mL Western Blue® Stablized Substrate for Alkaline Phosphatase (Promega) to visualize the specific protein bands on the membrane. Finally, the membrane was dipped in water to stop the reaction and photographed to show the hybridzation.
Gene Transcription Activation Assay
The transcription activity of bcsmr1OS and bcsmr1BS was determined using Matchmaker™ GAL4 two-hybrid system 3 (Clontech Laboratories, 2007). The cDNA sequences for bcsmr1BS in the BS isolate B05.10 (2,080 bp long), bcsmr1OS in the OS isolate XN-1 (2,079 bp long), and bcactA (coding for actin) in B05.10 (1,128 bp long) were separately amplified by RT-PCR with the total RNA extracts from two isolates as templates using the specific RT-PCR primer sets and the specific thermal programs (Tables S2, S3). Meanwhile, the DNA sequence containing the GAL4 activation domain (ADGAL4, 408 bp long) was PCR amplified from the plasmid pGADT7 (TaKaRa) with the primer set GAL4-F/GAL4-R (Table S2). The resulting cDNA fragments for bcsmr1OS, bcsmr1BS, and bcactA, and the DNA fragment of ADGAL4 were purified from a 1% agarose gel after electrophoresis. They were separately inserted into the plasmid pGBKT7 containing the DNA binding domain of GAL4 (BDGAL4) using ClonExpress® II One Step Cloning Kit (Vazyme Biotechnol. Co. Ltd., Nanjing, China). The resulting plasmids pBDGAL4-ADGAL4 (positive control), pGBKT7 (negative control 1), pBDGAL4-BcACTA (negative control 2), pBDGAL4-BcSMR1OS, and pBDGAL4-BcSMR1BS were separately transformed into the yeast cells of strain AH109 using the LiAc/SS carrier DNA/PEG transformation method (Clontech Laboratories, 2007). Selection of the yeast mutants harboring the plasmids and detection of their transcription activity were done using the procedures described by Lou et al. (2015).
Determination of the Transcripts of the Melanogenesis-Related Genes
The transcripts of bcsmr1, bcygh1, bcpks12/13, bcbrn1/2, and bcscd1 in the sclerotia of the B. cinerea isolates were determined using qRT-PCR with the specific primers and thermal cycles (Tables S2, S3). All the investigated isolates were incubated on PDA at 20°C. The sclerotial primordia, immature sclerotia, and mature sclerotia were collected from the 6-, 8-, and 15-day-old cultures of each isolate, respectively. The total RNA in the sclerotial samples was extracted using TaKaRa RNAiso Reagent Kit (TaKaRa) and was subjected to treatment with DNase I (TaKaRa) to eliminate DNA contamination in the extracts. The treated RNA was then used as templates in qRT-PCR to determine the transcripts of bcactA (reference), bcsmr1, bcpks12/13, bcygh1, bcbrn1/2, and bcscd1. The transcript levels of each gene were calculated by the ΔΔCt method (Zeng et al., 2014). For normalization of the data, the transcript level of each gene in the sclerotial primordia of isolate B05.10 was considered as 1.0. The scale was used to calibrate the transcript level of that gene in the immature and mature sclerotia of B05.10, as well as in the sclerotial primordia, immature sclerotia, and mature sclerotia of other isolates. The qRT-PCR assay was independently repeated three times.
Complementation of the Sclerotial Melanogenesis in XN-1 with bcsmr1bs
The complementation plasmid pKS1004-BS was constructed based on the plasmid pKS1004 containing the hygromycin-resistance gene (hph) (Zeng et al., 2014). The full-length cDNA of bcsmr1BS from B05.10 was ligated at the downstream of the Aspergillus nidulans promoter Polic by fusion PCR. The resulting DNA fragment was inserted into the plasmid pKS1004 using the protocol and the reagents in ClonExpress® II One Step Cloning Kit (Vazyme Biotechnol. Co. Ltd., Nanjing, China). The plasmid was then transformed into the protoplasts of XN-1 (Zeng et al., 2014). The transformed protoplasts were regenerated at 20°C on the PRM medium (Table S1) amended with 50 μg/mL hygromycin B. The emerging colonies (isolates) were individually transferred to PDA. Meanwhile, isolates B05.10 and XN-1 were inoculated on PDA as two controls. The cultures (isolates) were incubated at 20°C for 35 days for production of sclerotia. Six complementation mutants were selected based on formation of brown- to black-colored sclerotia (indicating sclerotial melanogenesis). Integration of bcsmr1BS in the genomes of the complementation mutants was confirmed by PCR using the primer set Olic-F/bcsmr1R (Table S2). XN-1 was used as reference in the specific PCR assay. Two representative complementation mutants, namely XN-BS11 and XN-BS14, were selected for determination of the copy number of bcsmr1 by Southern blotting also with XN-1 as a reference. The bcsmr1-specific DNA probe was generated by PCR from the genomic DNA of B05.10 with the primer set bcsmr1-jcF/bcsmr1R (Table S2). In Southern blotting, the gDNA of XN-1, XN-BS11, and XN-BS14 was extracted and digested with EcoRI. The resulting DNA fragments were separated by electrophoresis and detected by the DNA probe, which was labeled with the reagents in DIG High Prime DNA Labeling and Detection Starter Kit II (Roche®, Mannheim, Germany).
Expression the six melanogenic genes (bcpks12/13, bcygh1, bcbrn1/2, bcscd1) in sclerotia produced by XN-1 (negative control), XN-BS11, XN-BS14, and B05.10 (positive control) was determined using qRT-PCR. The procedures for isolation of the total RNA from the sclerotial samples and qRT-PCR were similar to those described above.
Data Analysis
The procedure UNIVARIATE in the SAS software (SAS Institute, Cary, NC, USA, v. 8.0, 1999) was used to analyze the data on relative expression levels of each gene. Average values of each gene between the OS (3 isolates) and BS (4 isolates) groups, and between each complementation mutant and XN-1/B05.10 were compared using Student's t-test at P < 0.05 or P < 0.01.
Results
Expression of the Genes for Sclerotial Melanogenesis Is Suppressed in the Orange-Colored Sclerotia, Compared to that in the Black-Colored Sclerotia
To analyze the melanin-biosynthesis ability in orange-colored sclerotia of B. cinerea, expression of six melanogenic enzymes-coding genes, including bcpks12/13, bcygh1, bcbrn1/2, and bcscd1, in the sclerotia of seven isolates of B. cinerea (Table 1) was determined by qRT-PCR using the specific primers and thermal programs (Tables S2, S3). The results showed that all the seven investigated isolates (OS isolates: XN-1, S417, T417; BS isolates: B05.10, HS016, XN087, WXt02-2) had low expression levels of bcpks13 at all sclerotial developmental stages (Figure 1). Thus, bcpks13 did not seem to be involved in the formation of the orange-colored sclerotia.
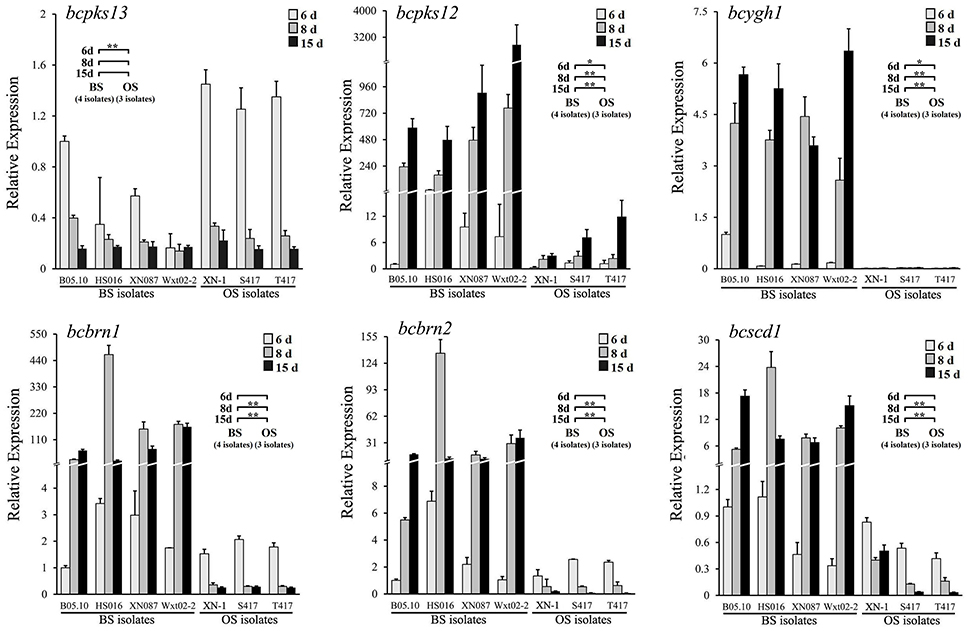
Figure 1. Relative expression values of six melanogenic enzyme-coding genes (bcpks12/13, bcygh1, bcbrn1/2, and bcscd1) in the sclerotia of B. cinerea isolates. The sclerotial primordia, immature sclerotia, and mature sclerotia of each isolate were collected from 6-, 8-, and 15-day-old PDA cultures (20°C), respectively. The results are the representative of three independent tests. Bars indicate standard deviations of the means. In each graph, “*” at P < 0.05 and “**” at P < 0.01 between the OS and BS isolates (as two groups) according to Student's t-test.
However, significant differences were found between the OS and BS isolates in expression levels of the polyketide synthase gene bcpks12 (Figure 1). Although all the seven investigated isolates showed the increased expression pattern in maturation of sclerotia, the expression levels were significantly lower in the OS isolates than in the BS isolates. In the sclerotial primordia (white-colored) from the 6-day-old PDA cultures (20°C), the expression level of bcpks12 was low with the relative expression values (REVs) ranging from 0.3 to 1.4 for the OS isolates and from 1.0 to 22.6 for the BS isolates. In the immature sclerotia (gray-colored) from the 8-day-old PDA cultures and in the mature sclerotia (black- and orange-colored in the BS and OS isolates, respectively) from 15-day-old PDA cultures, the bcpks12 expression levels slightly increased by 2.6 and 7.8-folds, respectively, in the OS isolates, whereas were largely increased by 40.4 and 121.8-folds, respectively, in the BS isolates (Figure 1).
For the other sclerotial melanogenesis genes (bcygh1, bcbrn1/2, bcscd1), the OS isolates differed from the BS isolates both in expression pattern and in expression level (Figure 1). While the BS isolates showed an increased expression pattern, the OS isolates showed a decreased expression pattern. In the sclerotial primordia of the OS isolates, the REVs ranged from 0.01 to 0.03 for bcygh1, 1.5 to 2.1 for bcbrn1, 1.3 to 2.6 for bcbrn2, and 0.4 to 0.8 for bcscd1. In the sclerotial primordia of the BS isolates, the REVs ranged from 0.1 to 1.0 for bcygh1, 1.0 to 3.4 for bcbrn1, 1.0 to 6.9 for bcbrn2, and 0.3 to 1.1 for bcscd1. In the immature sclerotia and mature sclerotia, expression of these genes was decreased by 43.2–88.5% in the OS isolates, whereas it was increased by 8 to 90-folds in the BS isolates, compared the corresponding REVs of these genes in the sclerotial primordia of B05.10.
To rule out the possibility that suppressed expression of bcpks12, bcygh1, bcbrn1/2, and bcscd1 in the sclerotia of the OS isolates is caused by mutations of these genes, the DNA sequences of these genes were cloned from the BS and OS isolates mentioned above (Table S4). Alignment analysis showed that the identity of the DNA sequences of each gene and the amino acid sequences of each protein in the seven above-mentioned isolates of B. cinerea was higher than 98% (Table S5). Single-amino acid polymorphisms were detected in the amino acid sequences of each protein (Figure S2). However, none of the polymorphic amino acids was found to be associated with formation of the orange-colored sclerotia. Therefore, suppressed expression of bcpks12, bcygh1, bcbrn1/2, and bcscd1 in the sclerotia of the OS isolates of B. cinerea is probably not caused by the point mutations in these genes. It might be caused by some regulatory genes for the sclerotial melanogenesis.
The OS Isolates Have a Single-Nucleotide Deletion in bcsmr1
To confirm whether the abnormal expression pattern of the melanin biosynthesis genes was caused by some regulatory malfunction, the transcription factor (TF) gene bcsmr1 related to the melanin biosynthesis was cloned from three OS isolates (XN-1, S417, T417) and four BS isolates (HS016, S59, WXt02-2, XN087) by PCR using the specific primers and specific thermal cycles (Tables S2, S3). The DNA sequences for bcsmr1 in these isolates were submitted to GenBank and were assigned with the accession numbers from KU743104 to KU743109, and KX098785 (Table S4). They were aligned with the DNA sequence of bcsmr1 in the reference isolate B05.10 (Accession Number: Bcin02g08760 in the website http://fungi.ensembl.org/Botrytis_cinerea). The results showed that bcsmr1 in the OS isolates (designated as bcsmr1OS) differed from bcsmr1 in other five BS isolates (designated as bcsmr1BS) in length of the open reading frame (ORF), 3,096 bp long for bcsmr1BS, whereas 3,095 bp long for bcsmr1OS (Figure 2A). The single-nucleotide deletion in bcsmr1OS is located at the position of nt 1,524, where guanine (G) is present in bcsmr1BS, but absent in bcsmr1OS. Thus, the single-nucleotide deletion was designated as G1524X. In spite of the single-nucleotide deletion, the identity of the DNA sequences in the ORF region of bcsmr1OS and bcsmr1BS in the seven isolates was higher than 98% (Table S5).
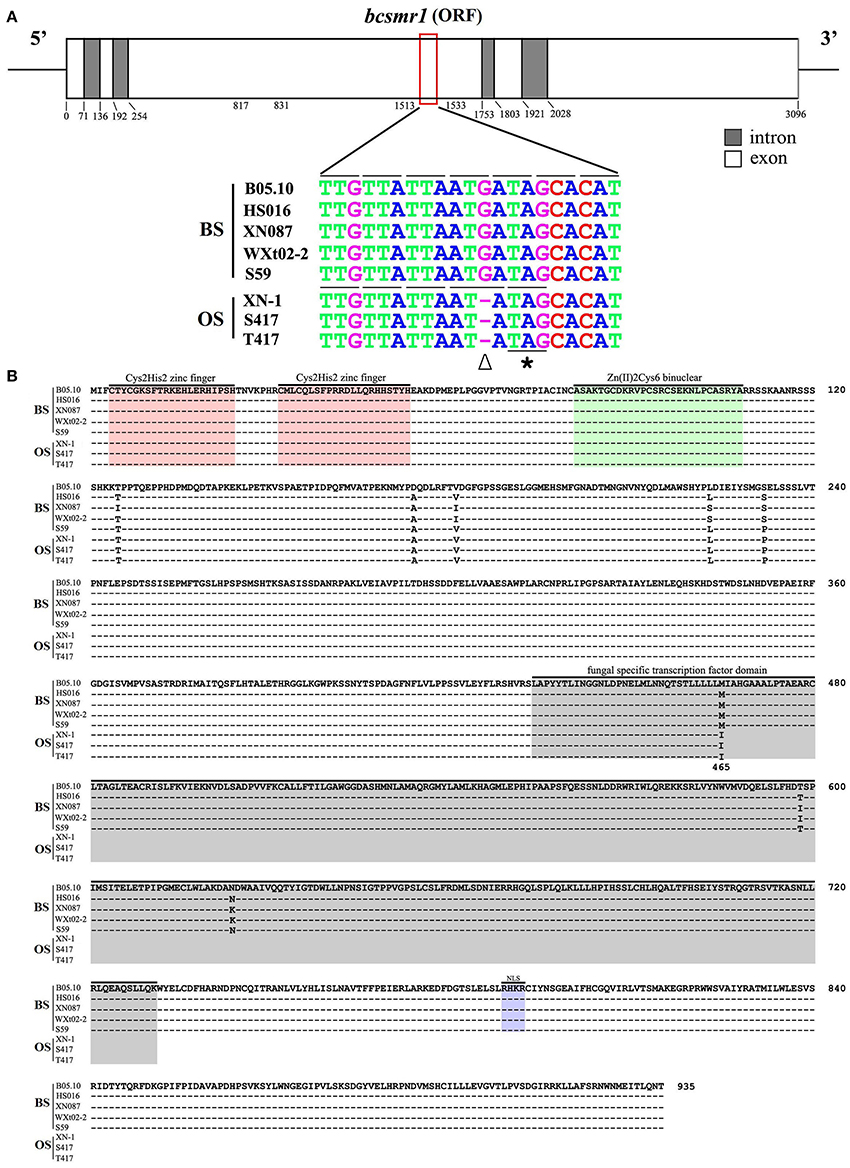
Figure 2. Comparison of the nucleotide sequences of the transcription factor gene bcsmr1 in Botrytis cinerea and the amino acids sequences encoded by bcsmr1. (A) A schematic diagram showing the structure of bcsmr1 and alignment of the partial DNA sequences of bcsmr1 in different isolates. Nucleotide positions for the exons and introns are indicated. OS, orange-colored sclerotia, BS, black-colored sclerotia. The symbols “∇” and “*” indicate the single-nucleotide deletion and the in-frame stop codon, respectively; (B) Alignment of the deduced amino acid sequences of BcSMR1 in different isolates. The symbol “–” in each column indicates the identical amino acids. Note difference in length of BcSMR1 in the OS and BS isolates, 465 aa long for BcSMR1 in the OS isolates, whereas 935 aa long for BcSMR1 in the BS isolates. NLS, Nuclear Localization Signaling region.
The single-nucleotide deletion in bcsmr1OS caused change of the triplet codon at the position nt 1,522 to 1,524 from ATG (for methionine or M) in bcsmr1BS to ATA (for isoleucine or I) in bcsmr1OS. The subsequent triplet codon at the position nt 1,525 to 1,527 was also changed from ATA (for isoleucine or I) in bcsmr1BS to TAG (stop) in bcsmr1OS (Figure 2A). As a consequence, the ORF of bcsmr1OS was deduced to encode a protein with 465 amino acid (aa) residues (designated as BcSMR1OS). It is a truncated protein of the 935-aa protein encoded by the ORF of bcsmr1BS (designated as BcSMR1BS). Alignment analysis showed that both BcSMR1465 and BcSMR1935 have the DNA binding domain at the N-terminus, consisting of two Cys2His2-zinc finger motifs and a Zn2Cys6-binuclear motif (Figure 2B). BcSMR1OS has a small portion (32 aa long) of the fungal specific transcription factor domain at the C-terminus without the nuclear localization signaling, whereas BcSMR1BS has the full fungal specific TF domain (298 aa long) with the nuclear localization signaling region. Alignment analysis also showed that there are six point mutations (T125I, A174D, V181I, L223S, S232P, M465I) in BcSMR1BS and BcSMR1OS (Figure 2B) and five of these mutations (T125I, A174D, V181I, L223S, S232P) were found in both BcSMR1BS and BcSMR1OS. Therefore, they are probably not associated with formation of the orange-colored sclerotia. The other mutation, namely M465I, was detected only in BcSMR1OS, but it was not detected in BcSMR1BS. This mutation is caused by the single-nucleotide-deletion-mediated one-nucleotide shift (Figure 2A). Therefore, the single-nucleotide deletion is probably important for formation of the orange-colored sclerotia by the OS isolates of B. cinerea. In spite of the point mutations, the identity of bcsmr1OS and bcsmr1BS is higher than 99% (Table S5).
Prokaryotic expression of bcsmr1OS and bcsmr1BS in E. coli was done to confirm formation of BcSMR1OS through the single-nucleotide deletion-mediated truncation of BcSMR1BS. The full-length cDNA of bcsmr1OS in XN-1 and bcsmr1BS in B05.10 were separately cloned by RT-PCR using the specific primers and specific thermal program (Tables S2, S3). The resulting cDNA fragments were separately introduced into the expression vector pET28a(+) (Figure S1). The resulting two plasmids, namely pET28a-bcsmr1OS and pET28a-bcsmr1BS, were then constructed and separately transformed into E. coli BL21 (DE3) pLysS. The transformed bacterial clones were selected, followed by shake-incubation in LB medium for production of BcSMR1OS and BcSMR1BS under induction by isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial proteins extracted from different E. coli cultures were detected by 8% SDS-PAGE. Results showed that two differentially-expressed proteins were consistently detected in the positive transformants of E. coli (data not show). The molecular weight was ~66 and 120 kD (both had a 6× His tag) in the transformants with pET28a-bcsmr1OS and pET28a-bcsmr1BS, respectively. The 54-kD difference in molecular weight between the two proteins is close to the 53.2-kD difference in molecular weight between the putative BcSMR1OS and BcSMR1BS, which had the predicted molecular weight of 51.3 and 104.5 kD, respectively. Western blotting analysis using the anti-His tag antibody further confirmed production of the 66- and 120-kD proteins by bcsmr1OS and bcsmr1BS, respectively, in E. coli (Figure 3).
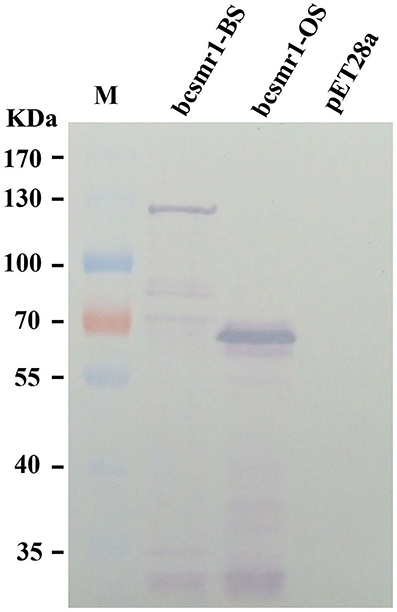
Figure 3. Western blotting detection of the E. coli proteins encoded by bcsmr1BS and bcsmr1OS using the anti-His tag antibody. The treatments “bcsmr1-BS”and “bcsmr1-OS” represent the proteins encoded by bcsmr1BS, and bcsmr1OS, respectively. The treatment “pET28a” represents the protein encoded by the blank plasmid pET28a alone without bcsmr1BS or bcsmr1OS. M = PageRulerTM prestained protein ladder.
The Transcription Activity of bcsmr1OS Is Greatly Suppressed, Compared to that of bcsmr1Bs
Both the OS isolates and the BS isolates had an increased expression pattern of bcsmr1 in sclerotia (Figure 4). We hypothesized that compared to bcsmr1BS, bcsmr1OS might be suppressed for activating transcription of the downstream genes for sclerotial melanogenesis. To test this hypothesis, the transcription activity of bcsmr1OS and bcsmr1BS was determined in Saccharomyces cerevisiae AH109 using Matchmaker™ GAL4 Two Hybrid System 3 (Clontech Laboratories, 2007). Five plasmids (pBDGAL4, pBDGAL4-BcACTA, pBDGAL4-ADGAL4, pBDGAL4-BcSMR1OS, and pBDGAL4-BcSMR1BS) were constructed. Four plasmids (pBDGAL4-BcACTA, pBDGAL4-ADGAL4, pBDGAL4-BcSMR1OS, and pBDGAL4-BcSMR1BS) contain two domains, namely the DNA-binding domain (e.g., BDGAL4) and the activation domain (e.g., ADGAL4, BcACTA, BcSMR1OS, BcSMR1BS). The remaining plasmid pBDGAL4 contains BDGAL4 alone. The plasmids were separately transformed into the yeast cells and the resulting transformants were screened on two auxotrophic media SD/-Trp and SD/Trp-His. On SD/-Trp, all the mutants showed growth. On SD/-Trp-His, however, the mutants differed in growth (Figure 5). While the transformants harboring either pBDGAL4 or pBDGAL4-BcACTA failed to grow on SD/-Trp-His, the transformants of pBDGAL4-ADGAL4, pBDGAL4-BcSMR1OS, and pBDGAL4-BcSMR1BS grew on this medium and produced blue color on the colonies when X-α-gal was added. This result indicates that ADGAL4, bcsmr1BS, and bcsmr1OS can activate transcription of the MEL1 gene coding for α-galactosidase. Relatively, the transformants of pBDGAL4-ADGAL4 and pBDGAL4-BcSMR1BS grew better than the transformant harboring pBDGAL4-BcSMR1OS. The blue color on the colonies of the transformants of pBDGAL4-ADGAL4 and pBDGAL4-BcSMR1BS was darker than that on the colonies of the transformant of pBDGAL4-BcSMR1OS. Therefore, compared to bcsmr1BS, bcsmr1OS is greatly suppressed for transcription of MEL1.
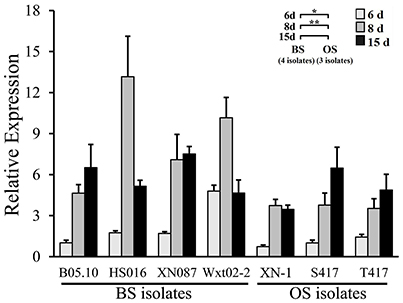
Figure 4. Relative expression values of the transcription factor gene bcsmr1 in the sclerotia of B. cinerea. The sclerotial primordia, immature sclerotia, and mature sclerotia of each isolate were collected from 6-, 8-, and 15-day-old PDA cultures (20°C), respectively. The results are the representative of the three independent tests. Bars indicate standard deviations of the means. “*” at P < 0.05 and “**” at P < 0.01 between the OS and BS isolates (as two groups) according to Student's t-test.
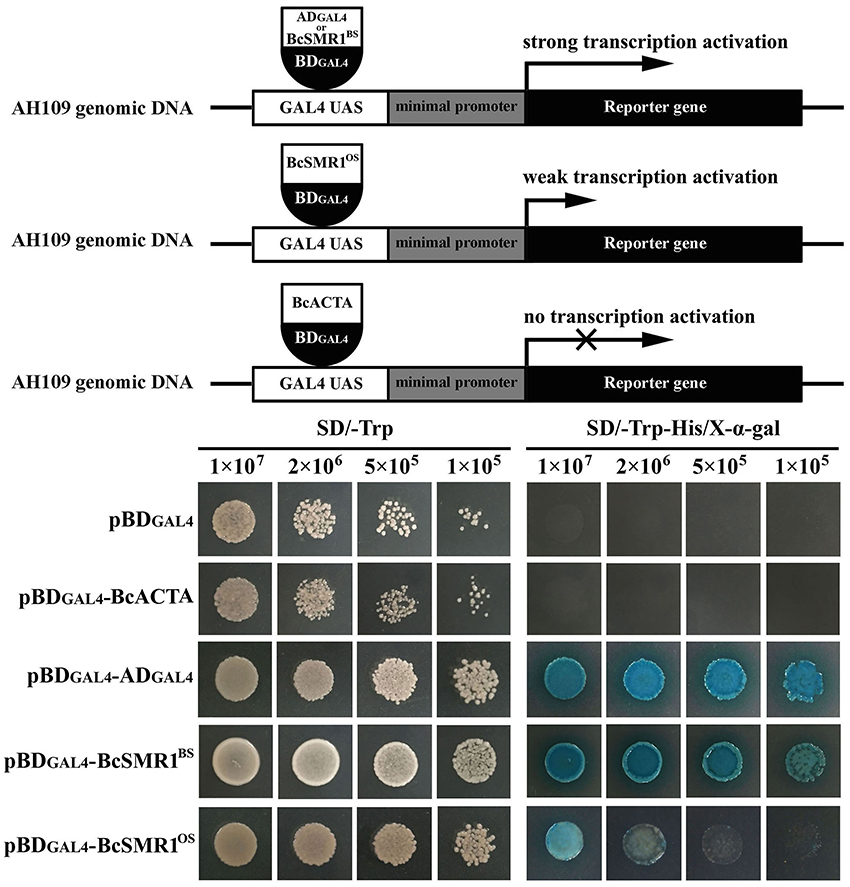
Figure 5. Transcription activation of bcsmr1 in Saccharomyces cerevisiae AH109. (Top) Three schematic diagrams depicting the experimental principle. ADGAL4 and BDGAL4 stand for the GAL4 activation domain and the GAL4 DNA-binding domain, respectively. BcSMR1BS and BcSMR1OS stand for the proteins encoded by bcsmr1BS from the BS isolate B05.10 and bcsmr1OS from the OS isolate XN-1, respectively. BcACTA stands for the protein encoded by bcactA from B05.10. BcSMR1BS, BcSMR1OS, and ADGAL4 can initiate transcription of the reporter genes HIS3, ADE2, and MEL1; (Bottom) Difference between bcsmr1BS and bcsmr1OS in transcription activity. Compared to ADGAL4, BcSMR1BS initiated strong gene transcription, whereas BcSMR1OS initiated weak gene transcription. Growth of the yeast mutants on SD/-Trp (left panel, 30°C, 1 day) indicates complementation of the auxotrophic mutant AH109. Growth of the yeast mutants on SD/-Trp-His (right panel, 30°C, 1 day) and production of blue color on the yeast colonies in the presence of X-α-Gal indicate the positive activity of gene transcription.
Complementation of bcsmr1OS with bcsmr1BS Restored Sclerotial Melanogenesis in XN-1
To confirm responsibility of the single-nucleotide deletion in bcsmr1OS for sclerotial melanogenesis deficiency, the full-length cDNA of bcsmr1BS from B05.10 was used to complement the function of bcsmr1OS in XN-1. A complementation plasmid pKS1004-BS containing the cDNA of bcsmr1BS under the control of the constitutive A. nidulans PoliC promoter was successfully constructed on the basis of the plasmid pKS1004 harboring the hygromycin-resistance gene hph (Zeng et al., 2014). The plasmid was introduced into XN-1 by protoplast-mediated transformation. Ten transformants were obtained based on hygromycin resistance. After incubation at 20°C on PDA for 35 days, XN-1 formed orange-colored sclerotia, as it has defect in sclerotial melanogenesis, whereas B05.10 formed black-colored sclerotia, as it has normal sclerotial melanogenesis (Figure 6). Four of these transformants formed orange-colored sclerotia like those formed by XN-1, indicating that the sclerotial melanogenesis in these transformants was not restored at all. In contrast, six other transformants (XN-BS1, XN-BS2, XN-BS11, XN-BS12, XN-BS14, XN-BS16) formed partially-melanized sclerotia, which were characterized by formation of color mosaic on the sclerotial surface (Figure 6). These six transformants could be classified into two groups based on the color mosaic on the sclerotia. Three transformants (XN-BS11, XN-BS12, XN-BS16) showed a brown-gray mosaic pattern, whereas the other three transformants (XN-BS1, XN-BS2, XN-BS14) showed a gray-black mosaic pattern. This result indicates that the sclerotial melanogenesis in these transformants was at least partially restored, compared to the complete sclerotial melanogenesis in B05.10 and the defective sclerotial melanogenesis in XN-1. Meanwhile, sporogenic germination (indicating lack of dormancy) was observed on the orange sclerotia produced by XN-1 and on the brown-grayish sclerotia produced by the transformants XN-BS11, XN-BS12, and XN-BS16 (Figure 6). The sporulation appeared to be vigorous on the sclerotia of XN-1, whereas was sparse on the sclerotia of the three transformants. In contrast, the black-colored sclerotia produced by B05.10 and the gray-colored sclerotia produced by the transformants XN-BS1, XN-BS2, and XN-BS14 did not germinate at all. This result indicates that the dormancy deficiency of the orange sclerotia (germination-active) of XN-1 due to lack of melanin was partially restored in the partially-melanized sclerotia produced by the complementation mutants.
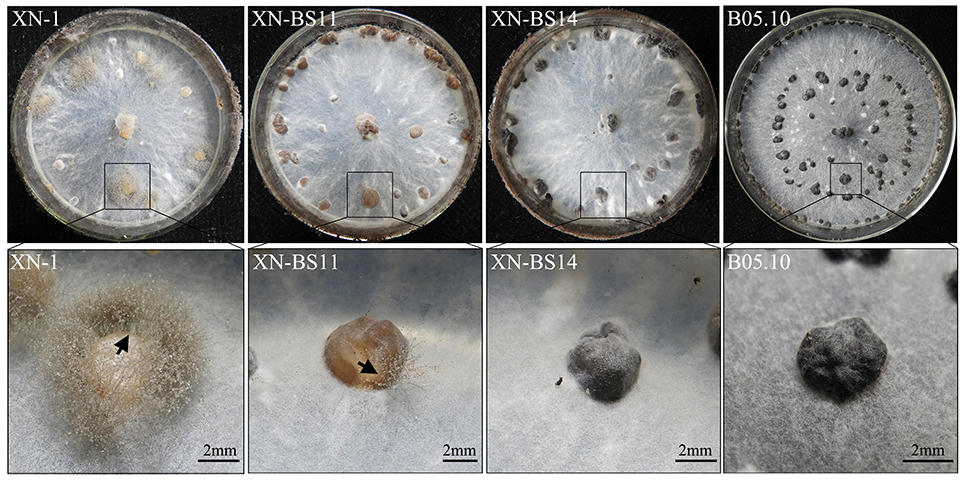
Figure 6. Colony morphology of Botrytis cinerea isolates B05.10 (BS), XN-1 (OS) and two complementation mutants of XN-1 (XN-BS11, XN-BS14) on potato dextrose agar after incubation at 20°C for 35 days. Note color of the sclerotia formed by different isolates. The arrow mark indicates sporogenic germination on the sclerotia.
The six complementation transformants were confirmed for integration of bcsmr1BS in their genomes by PCR using the primer set Olic-F/bcsmr1R (Table S2) (data not shown). Two of these transformants (XN-BS11, XN-BS14) were further determined for bcsmr1BS integration in their genomes by Southern blotting using the bcsmr1-specific DNA probe (Figure S3A). XN-1 was used as the reference isolate in Southern blotting. The result showed that the three isolates had one to three hybridization bands, one band in XN-1, two bands in XN-BS11, and three bands in XN-BS14 (Figure S3B). This result indicates that the genomes of XN-1, XN-BS11, and XN-BS14 harbor one (bcsmr1OS), two (bcsmr1OS + bcsmr1BS), and three (bcsmr1OS + bcsmr1BS + bcsmr1BS) copies of bcsmr1, respectively.
Expression of the six melanogenic genes (bcpks12/13, bcygh1, bcbrn1/2, bcscd1) in the sclerotia of isolates XN-1, XN-BS11, and XN-BS14 was determined by qRT-PCR. The two transformants were similar to XN-1 both in expression pattern and in expression level of bcpks13, which is not required for sclerotial melanogenesis according to Schumacher (2016). All the three isolates showed a decreased expression pattern and low expression levels of bcpks13 in sclerotial maturation.
In contrast, the two transformants differed greatly from XN-1 either in expression pattern or in expression level of the other five sclerotial melanogenesis genes (bcpks12, bcygh1, bcbrn1/2, bcscd1). For bcpks12 and bcygh1, the two transformants were similar to XN-1 in expression pattern, but differed from XN-1 in expression level (Figure 7). The two genes showed an increased expression pattern in maturation of the sclerotia produced by the three isolates. In the sclerotial primordia, the REVs ranged from 1.0 to 49.8 for bcpks12 and from 1.0 to 10.0 for bcygh1. In the immature sclerotia and mature sclerotia, the REVs of the two genes were largely increased by 12 to 350-folds in the two transformants, whereas were slightly increased by 3 to 12-folds in XN-1.
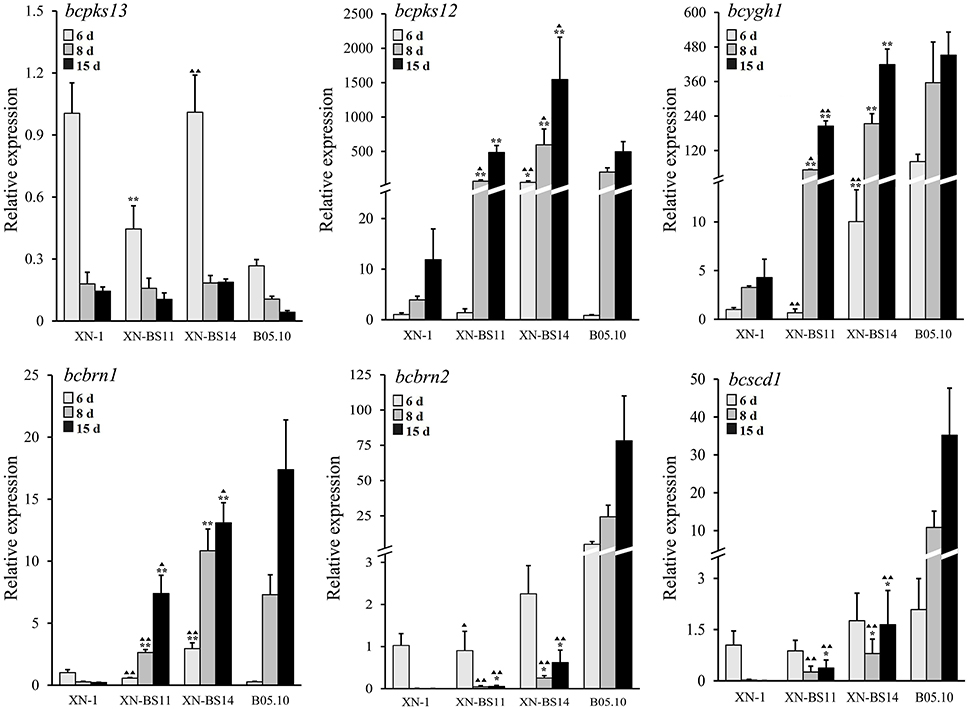
Figure 7. Relative expression values of the six melanogenic enzyme-coding genes (bcpks12/13, bcygh1, bcbrn1/2, bcscd1) in the sclerotia of isolate XN-1 of Botrytis cinerea, two complemented mutants of XN-1 (XN-BS11, XN-BS14) and B05.10. The sclerotial primordia, immature sclerotia, and mature sclerotia of each isolate were collected from 6-, 8-, and 15-day-old PDA cultures (20°C), respectively. The results are the representative of three independent tests. Bars indicate standard deviations of the means. *P < 0.05 and **P < 0.01 (Student's t-test) between each mutant and XN-1;  P < 0.05 and
P < 0.05 and  P < 0.01 (Student's t-test) between each mutant and B05.10.
P < 0.01 (Student's t-test) between each mutant and B05.10.
For bcbrn1, the two transformants differed from XN-1 both in expression pattern and in expression level (Figure 7). In XN-1, the REV was reduced from 1.0 in the sclerotial primordia to < 0.3 in the immature sclerotia/mature sclerotia. In XN-BS11 and XN-BS14, however, the REVs were increased in maturation of the sclerotia. XN-BS11 had the REV increase from 0.6 in the sclerotial primordia to 2.6 in the immature sclerotia, and 7.4 in mature sclerotia. XN-BS14 had a REV increase from 2.9 in the sclerotial primordia to 10.8 in the immature sclerotia, and 13.1 in the mature sclerotia.
For bcbrn2 and bcscd1, the two transformants were also similar to XN-1 in expression pattern, but differed from XN-1 in expression level (Figure 7). All the three isolates showed a decreased expression pattern in maturation of the sclerotia. In the sclerotial primordia, the three isolates had the REVs of 0.9–2.3 and 0.9–1.8 for bcbrn2 and bcscd1, respectively. In the immature sclerotia and mature sclerotia, while XN-1 had negligible expression of both genes, XN-BS11 had the REVs of 0.1–0.6 for bcbrn2 and 0.4–1.7 for bcscd1. In general, XN-BS14 had higher REVs of bcpks12, bcygh1, bcbrn1/2, and bcscd1 than the corresponding REVs for XN-BS11 (Figure 7). Meanwhile, the expression pattern of bcpks12, bcygh1, and bcbrn1/2 in the mutants XN-BS11 and XN-BS14 resembled that in the BS isolate B05.10, although the expression levels of these genes at each stage were significantly (P < 0.05 or P < 0.01) lower than those of B05.10 (Figure 7). This result further confirmed that the single-nucleotide deletion in bcsmr1 is the major reason for formation of the orange-colored sclerotia.
Discussion
Zhou et al. (2018) first reported natural occurrence of orange-colored sclerotial (OS) isolates of B. cinerea infecting strawberry and tomato in central China. The OS isolates are natural mutants or variants of B. cinerea, compared to the normal isolates with formation of black-colored sclerotia. Microscopic observation showed that both orange- and black-colored sclerotia of B. cinerea are quite similar in structure, consisting of the cortex layer (outside) with compact parenchymatous cells and the medulla (inside) with loose prosenchymatous cells. However, the sclerotia of the two types differ greatly in melanin deposition in the wall of the epidermal cells. While the BS isolates have normal melanin deposition in the black-colored sclerotia, the OS isolates are deficient in melanin deposition in the orange-colored sclerotia (Zhou et al., 2018). Our previous study also showed that the OS isolates occur rarely in nature, as the sclerotia produced by the OS isolates of B. cinerea survived poorly, compared to the sclerotia produced by the BS isolates (Zhou et al., 2018). Therefore, mutation of the sclerotial color from black to orange greatly changed the ecological fitness of B. cinerea.
Zeun and Buchenauer (1985) reported that B. cinerea produces DHN melanin, as tricyclazole, an inhibitor for DHN melanin biosynthesis, can delay sclerotial melanogenesis in B. cinerea. Schumacher (2016) elucidated the molecular pathway for biosynthesis of DHN melanin in B. cinerea. The result showed that bcpks12 is required for sclerotial melanogenesis, bcpks13 is required for conidial melanogenesis, and bcbrn1/2 and bcscd1 are required for both sclerotial melanogenesis and conidial melanogenesis (Schumacher, 2016). The present study revealed that the OS isolates differed greatly from the BS isolates either in expression pattern or in expression level of bcpks12, bcygh1, bcbrn1/2, and bcscd1. The expression levels of these genes in the immature and mature sclerotia of the three OS isolates were significantly lower than those of the four BS isolates (Figure 1). This molecular evidence corroborates our previous study in microscopic observation that the orange-colored sclerotia was due to lack of melanin deposition (Zhou et al., 2018).
A recent study by Schumacher (2016) showed that the transcription factor (TF) gene bcsmr1 regulates the sclerotial melanogenesis of B. cinerea. This gene has only one copy in isolate B05.10 of B. cinerea (Schumacher, 2016). The Δbcsmr1 mutant formed yellow-colored sclerotia on agar media, indicating lack of sclerotial melanogenesis (Schumacher, 2016). Bcsmr1 is an ortholog of CMR1 (Colletotrichum Melanin Regulation 1) in Collectrichum lagenarium (Tsuji et al., 2000). It is also homologous to other orthologs of CMR1, including Amr1 and CmrA in Alternaria species (Cho et al., 2012; Fetzner et al., 2014), BMR1 in Bipolaris oryzae (Kihara et al., 2008), ChCMR1 in Cochliobolus heterostrophus (Eliahu et al., 2007), and PIG1 in Magnaporthe grisea (Sweigard et al., 1998).
Bcsmr1 and other CMR1 orthologs are similar in structure. They contain a DNA-binding domain with two Cys2His2 zinc finger motifs and one Zn(II)2Cys6 binuclear cluster motif at the N-terminus, and a fungal specific transcription factor (TF) domain and a nuclear localization signaling (NSL) region at the C-terminus (Figure 2B). Tsuji et al. (2000) found that the DNA-binding domain in CMR1 is necessary for melanization in C. lagenarium. Fetzner et al. (2014) reported that CmrA encoded by CmrA can bind to the promoter region of pksA (coding for polyketide synthase), thereby initiating expression of pksA and the subsequent melanogenesis in A. alternata.
This study demonstrated that both BS and OS isolates of B. cinerea had an increased expression pattern of bcsmr1 in the sclerotia (Figure 4). The result is consistent with the finding of Schumacher (2016), who found that expression of bcsmr1 is associated with sclerotial development of B. cinerea. It implies that failure to regulate the sclerotial melanogenesis by bcsmr1OS in the OS isolates of B. cinerea is not due to expression of this gene. Instead, it may be caused by loss-of-function of bcsmr1OS.
In order to elucidate the mechanism for the loss-of-function of bcsmr1OS, we cloned bcsmr1OS and bcsmr1BS from selected isolates in this study, and compared the DNA sequences of bcsmr1OS with those of bcsmr1BS. Three OS isolates (XN-1, S417, T419) have a single-nucleotide deletion in bcsmr1OS (Figure 2B). Occurrence of single-nucleotide deletions has been frequently reported in animals (Persuy et al., 1999; Yang et al., 2003) and plants (Takano et al., 2014; Lou et al., 2015), whereas it has been rarely reported in fungi, including B. cinerea (Pihet et al., 2009).
Previous studies readily indicated that B. cinerea can mutate in response to extreme environments such as presence of UV irradiation and fungicides (Leroux et al., 2002; Kretschmer et al., 2009). These mutagens usually cause formation of single-nucleotide polymorphisms (SNPs) in populations of B. cinerea (Kretschmer et al., 2009; Schumacher et al., 2012, 2013). Most SNPs in certain genes such as mbc1 coding for β-tubulin have been found to be closely associated with sensitivity or resistance to fungicides (Leroux et al., 2002; Kretschmer et al., 2009). A few SNPs in other genes such as bcvel1 coding for the VELVET protein cause formation of a premature stop codon, thereby resulting in production of truncated proteins and change of some important phenotypes such as virulence and sclerotial formation (Schumacher et al., 2012, 2013). Whether or not these mutagens can cause formation of the single-nucleotide deletion in bcsmr1 remains unknown. Taking together all the information about the low occurrence frequency of the OS isolates of B. cinerea in nature and the reduced ecological fitness of the OS isolates, we can draw a conclusion that the single-nucleotide deletion observed in this study is important for B. cinerea. Understanding of the mutagens for formation of that single-nucleotide deletion in bcsmr1 will be helpful for designing a novel strategy for control of B. cinerea.
This study demonstrated that the single-nucleotide deletion in bcsmr1OS caused formation of a premature stop codon, which results in production of a truncated 465-aa protein BcSMR1OS (Figures 2, 3). In contrast, the full-length bcsmr1BS in the BS isolates codes for a 935-aa protein BcSMR1BS (Figures 2, 3). BcSMR1OS contains the complete DNA-binding domain, but has only the partial transcription factor domain without the nuclear signaling localization region. In S. cerevisiae AH109, bcsmr1OS had reduced activity in activating transcription of the MEL1 gene, compared to bcsmr1BS (Figure 5). This result suggests that bcsmr1OS cannot be fully functional in activating transcription of the sclerotial melanogenesis genes in B. cinerea. Like other transcription factors, bcsmr1 is transcribed in the nuclei and the resulting transcripts are translocated into the cytoplasm, where they are translated to the protein BcSMR1. Without the nuclear signaling localization domain, BcSMR1OS may not be so easy as BcSMR1BS in re-entrance into the nuclei from the cytoplasm. Without the full transcription factor domain, BcSMR1OS may not be so fully functional as BcSMR1BS in activating transcription of the downstream genes for sclerotial melanogenesis even it enters into the nuclei somehow. As a consequence, transcription of the sclerotial melanogenesis genes regulated by BcSMR1OS is reduced or blocked, compared to BcSMR1BS. Thus, the sclerotial melanin biosynthesis in the OS isolates of B. cinerea is suppressed, thereby causing formation of the orange-colored sclerotia.
Introduction of the full-length bcsmr1BS from the BS isolate B05.10 partially complemented the loss-of-function of bcsmr1OS in the OS isolate XN-1. Six complementation mutants formed partially-melanized sclerotia (Figure 6). The mutants XN-BS11 and XN-BS14 showed enhanced expression of bcpks12, bcygh1, bcbrn1/2, and bcscd1 in the immature and mature sclerotia in comparison to their progenitor XN-1(Figure 7), confirming the previous finding by Schumacher (2016) that bcsmr1 has the regulatory function for the sclerotial melanogenesis in B. cinerea. Ectopic complementation with the full-length bcsmr1BS increased expression of bcpks12, bcygh1, and bcbrn1 in sclerotia and the expression pattern was similar to that found in the BS isolate B05.10. However, this study also found that the complementation had negligible effects on expression of bcbrn1/2 and bcscd1 (Figure 7). These results suggest that bcsmr1 has different regulatory effects on expression of these genes. Coincidently, bcbrn2 and bcscd1 along with bcpks13 form one gene cluster (in chromosome No. 3) located at a different chromosomal position from the other sclerotial melanogenesis genes. Bcsmr1, bcpks12, and bcygh1 form a gene cluster located at chromosome No. 2, and bcbrn1 is located at chromosome No. 4 (Schumacher, 2016). Difference in chromosomal position for bcsmr1, bcbrn2, and bcscd1 may be responsible for the low regulatory efficiency of bcsmr1 for bcbrn2 and bcscd1. Further studies are necessary to determine the activity of bcsmr1BS in activating transcription of different sclerotial melanogenesis genes.
This study used the full-length bcsmr1 from the BS isolate B05.10, instead of the deleted nucleotide, to restore the phenotype of formation of black-colored sclerotia by the OS isolate XN-1. This complementation strategy may mask the possible effects of the point mutations of bcsmr1 on formation of orange-colored sclerotia. A few point mutations were identified in bcsmr1 among the eight isolates of B. cinerea (5 BS isolates and 3 OS isolates) (Figure 2). However, none of the point mutations was found to be associated with formation of the orange-colored sclerotia. Therefore, the result from the complementation experiment in this study at least strengthened importance of the single-nucleotide deletion in loss-of-function of bcsmr1 for sclerotial melanogenesis. Future studies are necessary to complement bcsmr1OS with the deleted nucleotide.
Schumacher (2016) reported that bcygh1 involves in conidial melanogenesis of B. cinerea, as BcYGH1 encoded by bcygh1 can convert the product catalyzed by BcPKS13 encoded by bcpks13, which is required only for conidial melanogenesis. Fan et al. (2017) reported that Vayg1, an ortholog of bcygh1, is required for microsclerotia formation and melanogenesis of microsclerotia in Verticillium dahlia, the causal agent of verticillium wilt of cotton. In this study, we found that like the essential genes for sclerotial melanogenesis (bcpks12, bcbrn1/2, bcscd1), bcygh1 was greatly suppressed for expression in the orange sclerotia of XN-1, compared to expression of bcygh1 in the black sclerotia of B05.10 (Figure 1). However, when bcsmr1BS was introduced into XN-1, bcygh1 expression was greatly increased (Figure 7). This result implies that besides Bcltf1/2 (two transcription factors), bcsmr1 may regulate expression of bcygh1. It also suggests that the expression of bcygh1 is closely associated with sclerotial melanogenesis in B. cinerea. Further studies are necessary to clarify the interaction between bcsmr1 and bcygh1 and to elucidate how the interaction affects the sclerotial melanogenesis in B. cinerea.
Author Contributions
YZ and JZ: designed research; LY and MW: provided new agents and analyzed the data; YZ, WC, and GL: wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (Grant No. 31571955) and China's Special Fund for Agro-Scientific Research in the Public Interest (Grant No. 201303025). We appreciate the kind help from Prof. Zhonghua Ma (Zhejiang University, Hangzhou, China) for providing isolate B05.10 of B. cinerea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02492/full#supplementary-material
Figure S1. A schematic diagram showing the procedure for construction of the plasmids used for prokaryotic expression of bcsmr1OS and bcsmr1BS from Botrytis cinerea isolates XN-1 and B05.10, respectively.
Figure S2. A schematic diagram showing the structure of the five melanogenic enzymes. (A) The proposed pathway for biosynthesis of DHN-melanin in B. cinerea. It was modified on the basis of Figure S8 prepared by Schumacher et al. (2014); (B) Five schematic diagrams showing differential amino acids in the five melanogenic enzymes among the investigated isolates of B. cinerea. Two polyketide synthases: BcPKS12 and BcPKS13 encoded by bcpks12 and bcpks13, respectively; two reductases: BcBRN1 and BcBRN2 encoded by bcbrn1 and bcbrn2, respectively; and one dehydratase: BcSCD1 encoded by bcscd1. Abbreviations for the conserved domains: KS, β-ketoacyl synthase domain; AT, acyl transferase domain; ACP, acyl carrier protein domain; TE, thioesterase domain; ENR, enoyl-ACP reductase doamin; SD, scytalone dehydratase domain. The GenBank accession numbers for the polypeptides-coding genes were listed in Table S4.
Figure S3. Southern blotting detection of bcsmr1 in isolate XN-1 (OS) of Botrytis cinerea and two complementation mutants of XN-1 (XN-BS11, XN-BS14). (A) Two schematic diagrams showing the position of the bcsmr1-specific DNA probe on the DNA sequence of bcsmr1OS and on the complemented cDNA sequence of bcsmr1BS; (B) Southern blotting detection of bcsmr1 in different isolates.
Table S1. Cultural media used in this study and their compositions.
Table S2. Oligonucleotides PCR primers used in this study.
Table S3. Thermal programs for PCR and qRT-PCR in this study.
Table S4. The GenBank accession numbers for the melanin biosynthesis-related genes and their promoter regions in different isolates of B. cinerea.
Table S5. Identity of bcsmr1 (open reading frame) and the polypeptide BcSMR1 encoded by bcsmr1 in black-colored sclerotial (or BS) or orange-colored sclerotial (or OS) isolates of Botrytis cinerea.
References
Ambrico, M. (2016). Special issue: melanin, along lasting history bridging natural pigments and organic bioelectronics. Polym. Int. 65, 1249–1250. doi: 10.1002/pi.5239
Bell, A. A., and Wheeler, M. H. (1986). Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451. doi: 10.1146/annurev.py.24.090186.002211
Butler, M. J., and Day, A. W. (1998). Fungal melanins: a review. Can. J. Microbiol. 44, 1115–1136. doi: 10.1139/w98-119
Chen, Z. J., Nunes, M. A., Silva, M. C., and Rodrigues, C. J. Jr. (2004). Appressorium turgor pressure of Colletotrichum kahawae might have a role in coffee cuticle penetration. Mycologia 96, 1199–1208. doi: 10.1080/15572536.2005.11832868
Cho, Y., Srivastava, A., Ohm, R. A., Lawrence, C. B., Wang, K. H., Grigoriev, I. V., et al. (2012). Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola. PLoS Pathog. 8:e1002974. doi: 10.1371/journal.ppat.1002974
Clontech Laboratories (2007). Matchmaker™ GAL4 Two Hybrid System 3 and Libraries User Manual. Protocol No. PT3247-1, Version No. PR742219, Mountain View, CA.
Cohrs, K. C., Simon, A., Viaud, M., and Schumacher, J. (2016). Light governs asexual differentiation in the grey mould fungus Botrytis cinerea via the putative transcription factor BcLTF2. Environ. Microbiol. 18, 4068–4086. doi: 10.1111/1462-2920.13431
Cordero, R. J. B., and Casadevall, A. (2017). Functions of fungal melanin beyond virulence. Fung. Biol. Rev. 31, 99–112. doi: 10.1016/j.fbr.2016.12.003
Doss, R. P., Deisenhofer, J., von Nidda, H. A. K., Soeldner, A. H., and McGuire, R. P. (2003). Melanin in the extracellular matrix of germlings of Botrytis cinerea. Phytochemistry 63, 687–691. doi: 10.1016/S0031-9422(03)00323-6
Elad, Y., Pertot, I., Cotes Prado, A. M., and Stewart, A. (2016). “Plant hosts of Botrytis spp.” in Botrytis-The Fungus, the Pathogen and Its Management Agricultural Systems, eds S. Fillinger and Y. Elad (Cham: Springer International Publishing), 413–486.
Eliahu, N., Igbaria, A., Rose, M. S., Horwitz, B. A., and Lev, S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 6, 421–429. doi: 10.1128/EC.00264-06
Fan, R., Klosterman, S. J., Wang, C., Subbarao, K. V., Xu, X., Shang, W., et al. (2017). Vayg1 is required for microsclerotium formation and melanin production in Verticillium dahliae. Fung. Genet. Biol. 98, 1–11. doi: 10.1016/j.fgb.2016.11.003
Fetzner, R., Seither, K., Wenderoth, M., Herr, A., and Fischer, R. (2014). Alternaria alternata transcription factor CmrA controls melanization and spore development. Microbiology 160, 1845–1854. doi: 10.1099/mic.0.079046-0
Hamilton, A. J., and Gomez, B. L. (2002). Melanins in fungal pathogens. J. Med. Microbiol. 51, 189–191. doi: 10.1099/0022-1317-51-3-189
Howard, R. J., and Valent, B. (1996). Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. doi: 10.1146/annurev.micro.50.1.491
Jacobson, S. (2000). Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13, 708–717. doi: 10.1128/CMR.13.4.708-717.2000
Kihara, J., Moriwaki, A., Tanaka, N., Tanaka, C., Ueno, M., and Arase, S. (2008). Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 281, 221–227. doi: 10.1111/j.1574-6968.2008.01101.x
Kretschmer, M., Leroch, M., Mosbach, A., Walker, A. S., Fillinger, S., Mernke, D., et al. (2009). Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 5:e1000696. doi: 10.1371/journal.ppat.1000696
Kubo, Y., Suzuki, K., Furusawa, I., and Yamamoto, M. (1982). Effect of tricyclazole on appressorial pigmentation and penetration from appressoria of Colletotrichum lagenarium. Phytopathology 72, 1198–1200. doi: 10.1094/Phyto-72-1198
Leroux, P., Fritz, R., Debieu, D., Alberini, C., Lanen, C., Bach, J., et al. (2002). Mechanisms of resistance to fungicides in field strains of Botrytis cinerea. Pest. Manag. Sci. 58, 876–888. doi: 10.1002/ps.566
Li, X. Q., Guo, B. L., Cai, W. Y., Zhang, J. M., Huang, H. Q., Zhan, P., et al. (2016). The role of melanin pathways in extreme tolerance and virulence of Fonsecaea revealed by de novo assembly transcriptomics using illumina paired-end sequencing. Stud. Mycol. 83, 1–18. doi: 10.1016/j.simyco.2016.02.001
Liu, W., Soulié, M. C., Perrino, C., and Fillinger, S. (2011). The osmosensing signal transduction pathway from Botrytis cinerea regulates cell wall integrity and MAP kinase pathways control melanin biosynthesis with influence of light. Fung. Genet. Biol. 48, 377–387. doi: 10.1016/j.fgb.2010.12.004
Lou, Y., Han, Y. C., Yang, L., Wu, M. D., Zhang, J., Cheng, J. S., et al. (2015). CmpacC regulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungus Coniothyrium minitans. Environ. Microbiol. 17, 4711–4729. doi: 10.1111/1462-2920.13018
Persuy, M. A., Printz, C., Medrano, J. F., and Mercier, J. C. (1999). A single nucleotide deletion resulting in a premature stop codon is associated with marked reduction of transcripts from a goat β-casein null allele. Anim. Genet. 30, 444–451. doi: 10.1046/j.1365-2052.1999.00547.x
Pihet, M., Vandeputte, P., Tronchin, G., Renier, G., Saulnier, P., Georgeault, S., et al. (2009). Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 9:177. doi: 10.1186/1471-2180-9-177
Saitoh, Y., Izumitsu, K., Morita, A., and Tanaka, C. (2010). A copper-transporting ATPase BcCCC2 is necessary for pathogenicity of Botrytis cinerea. Mol. Genet. Genom. 284, 33–43. doi: 10.1007/s00438-010-0545-4
Sapmak, A., Boyce, K. J., Andrianopoulos, A., and Vanittanakom, N. (2015). The pbrB gene encodes a laccase required for DHN-melanin synthesis in conidia of Talaromyces (Penicillium) marneffei. PLoS ONE 10:e0122728. doi: 10.1371/journal.pone.0122728
Schumacher, J. (2016). DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 99, 729–748. doi: 10.1111/mmi.13262
Schumacher, J., Gautier, A., Morgant, G., Studt, L., Ducrot, P. H., Le Pêcheur, P., et al. (2013). A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLoS ONE 8:e53729. doi: 10.1371/journal.pone.0053729
Schumacher, J., Pradier, J. M., Simon, A., Traeger, S., Moraga, J., Collado, I. G., et al. (2012). Natural variation in the VELVET Gene bcvel1affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS ONE 7:e47840. doi: 10.1371/journal.pone.0047840
Schumacher, J., Simon, A., Cohrs, K. C., Viaud, M., and Tudzynski, P. (2014). The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 10:e1004040. doi: 10.1371/journal.pgen.1004040
Sweigard, J. A., Carroll, A. M., Farrall, L., Chumley, F. G., and Valent, B. (1998). Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant Microbe Interact. 11, 404–412. doi: 10.1094/MPMI.1998.11.5.404
Takano, S., Matsuda, S., Kinoshita, N., Shimoda, N., Sato, T., and Kato, K. (2014). Genome-wide single nucleotide polymorphisms and insertion-deletions of Oryza sativa L. subsp. japonica cultivars grown near the northern limit of rice cultivation. Mol. Breed. 34, 1007–1021. doi: 10.1007/s11032-014-0093-4
Tsuji, G., Kenmochi, Y., Takano, Y., Sweigard, J., Farrall, L., Furusawa, I., et al. (2000). Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38, 940–954. doi: 10.1046/j.1365-2958.2000.02181.x
Williamson, B., Tudzynsk, B., Tudzynski, P., and van Kan, J. A. L. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Yang, Q. Q., Chen, Y. F., and Ma, Z. H. (2013a). Involvement of BcVeA and BcVelB in regulating conidiation, pigmentation and virulence in Botrytis cinerea. Fung. Genet. Biol. 50, 63–71. doi: 10.1016/j.fgb.2012.10.003
Yang, Q. Q., Jiang, J. H., Mayr, C., Hahn, M., and Ma, Z. H. (2013b). Involvement of two type 2C protein phosphatases BcPtc1 and BcPtc3 in the regulation of multiple stress tolerance and virulence of Botrytis cinerea. Environ. Microbiol. 15, 2696–2711. doi: 10.1111/1462-2920.12126
Yang, Q. Q., Yu, F. W., Yin, Y. N., and Ma, Z. H. (2013c). Involvement of protein tyrosine phosphatases BcPtpA and BcPtpB in regulation of vegetative development, virulence and multi-stress tolerance in Botrytis cinerea. PLoS ONE 8:e61307. doi: 10.1371/journal.pone.0061307
Yang, T. Y., McNally, B. A., Ferrone, S., Liu, Y., and Zheng, P. (2003). A single-nucleotide deletion leads to rapid degradation of TAP-1 mRNA in a melanoma cell line. J. Biol. Chem. 278, 15291–15296. doi: 10.1074/jbc.M300954200
Zeng, L. M., Zhang, J., Han, Y. C., Yang, L., Wu, M. D., Jiang, D. H., et al. (2014). Degradation of oxalic acid by the mycoparasite Coniothyrium minitans plays an important role in interacting with Sclerotinia sclerotiorum. Environ. Microbiol. 16, 2591–2610. doi: 10.1111/1462-2920.12409
Zeun, R., and Buchenauer, H. (1985). Effect of tricyclazole on production and melanin contents of sclerotia of Botrytis cinerea. J. Phytopathol. 112, 259–267. doi: 10.1111/j.1439-0434.1985.tb00802.x
Zhang, C. H., He, Y. F., Zhu, P. K., Chen, L., Wang, Y. W., Ni, B., et al. (2015). Loss of bcygh1 and bcpks13 in Botrytis cinerea not only blocks melanization but also increases vegetative growth and virulence. Mol. Plant Microbe Interact. 28, 1091–1101. doi: 10.1094/MPMI-04-15-0085-R
Keywords: Botrytis cinerea, orange-colored sclerotia, sclerotial melanogenesis, bcsmr1, single-nucleotide deletion
Citation: Zhou Y, Yang L, Wu M, Chen W, Li G and Zhang J (2017) A Single-Nucleotide Deletion in the Transcription Factor Gene bcsmr1 Causes Sclerotial-Melanogenesis Deficiency in Botrytis cinerea. Front. Microbiol. 8:2492. doi: 10.3389/fmicb.2017.02492
Received: 29 September 2017; Accepted: 30 November 2017;
Published: 12 December 2017.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Eugenio Mancera, Centro de Investigación y de Estudios Avanzados Irapuato, MexicoLuis Antonio Pérez-García, Universidad Autónoma de San Luis Potosí, Mexico
Copyright © 2017 Zhou, Yang, Wu, Chen, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhang, zhangjing1007@mail.hzau.edu.cn
 Yingjun Zhou1,2
Yingjun Zhou1,2 Weidong Chen
Weidong Chen Guoqing Li
Guoqing Li Jing Zhang
Jing Zhang