- Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, Fukuoka, Japan
This review summarizes the mechanisms of the initiator protein DnaA in replication initiation and its regulation in Escherichia coli. The chromosomal origin (oriC) DNA is unwound by the replication initiation complex to allow loading of DnaB helicases and replisome formation. The initiation complex consists of the DnaA protein, DnaA-initiator-associating protein DiaA, integration host factor (IHF), and oriC, which contains a duplex-unwinding element (DUE) and a DnaA-oligomerization region (DOR) containing DnaA-binding sites (DnaA boxes) and a single IHF-binding site that induces sharp DNA bending. DiaA binds to DnaA and stimulates DnaA assembly at the DOR. DnaA binds tightly to ATP and ADP. ATP-DnaA constructs functionally different sub-complexes at DOR, and the DUE-proximal DnaA sub-complex contains IHF and promotes DUE unwinding. The first part of this review presents the structures and mechanisms of oriC-DnaA complexes involved in the regulation of replication initiation. During the cell cycle, the level of ATP-DnaA level, the active form for initiation, is strictly regulated by multiple systems, resulting in timely replication initiation. After initiation, regulatory inactivation of DnaA (RIDA) intervenes to reduce ATP-DnaA level by hydrolyzing the DnaA-bound ATP to ADP to yield ADP-DnaA, the inactive form. RIDA involves the binding of the DNA polymerase clamp on newly synthesized DNA to the DnaA-inactivator Hda protein. In datA-dependent DnaA-ATP hydrolysis (DDAH), binding of IHF at the chromosomal locus datA, which contains a cluster of DnaA boxes, results in further hydrolysis of DnaA-bound ATP. SeqA protein inhibits untimely initiation at oriC by binding to newly synthesized oriC DNA and represses dnaA transcription in a cell cycle dependent manner. To reinitiate DNA replication, ADP-DnaA forms oligomers at DnaA-reactivating sequences (DARS1 and DARS2), resulting in the dissociation of ADP and the release of nucleotide-free apo-DnaA, which then binds ATP to regenerate ATP-DnaA. In vivo, DARS2 plays an important role in this process and its activation is regulated by timely binding of IHF to DARS2 in the cell cycle. Chromosomal locations of DARS sites are optimized for the strict regulation for timely replication initiation. The last part of this review describes how DDAH and DARS regulate DnaA activity.
Introduction
The genome of Escherichia coli consists of a single circular 4.6 Mb chromosome, with a unique replication origin called oriC. Replication initiation at oriC results in construction of a pair of replisomes, which migrate bi-directionally to replicate the entire chromosome. Replication initiation at oriC is regulated to occur only once during each cell cycle, and the timing of initiation is coordinated with the cellular growth rate. Even when cells grow rapidly and the copy number of oriC increases to more than two per cell, initiation occurs at sister oriC regions simultaneously only once at a specific time during the cell cycle. As such, the time of initiation at oriC is regulated and re-initiation during the same cell cycle is strictly repressed (Skarstad and Katayama, 2013; Wolański et al., 2015; Riber et al., 2016).
The 245 bp minimal oriC region has multiple binding sites for the chromosomal replication initiator protein DnaA (DnaA boxes), and a single binding site for the integration host factor (IHF), in addition to an AT-rich duplex-unwinding element (DUE) (Figures 1A, 2A; Kaguni, 2011; Leonard and Grimwade, 2015; Wolański et al., 2015; Shimizu et al., 2016). The 9-mer DnaA box consensus sequence is 5′-TTATnCACA-3′. DnaA-initiator-associating protein DiaA is a DnaA-binding protein that stimulates ATP-bound DnaA (ATP-DnaA) assembly on oriC, and that is required for timely replication initiation (Ishida et al., 2004; Keyamura et al., 2007). Binding of IHF to DNA causes a sharp (120–180°) bend in the double helix (Swinger and Rice, 2004). A complex consisting of oriC, IHF, DiaA, and oligomeric ATP-DnaA is considered to make up the initiation complex in E. coli (Keyamura et al., 2007, 2009). This complex unwinds the oriC DUE, enabling loading of DnaB helicases onto single-stranded DNA (ssDNA) by specific protein–protein interactions and dissociations, which in turn leads to formation of replisomes (for a review, see Bell and Kaguni, 2013).
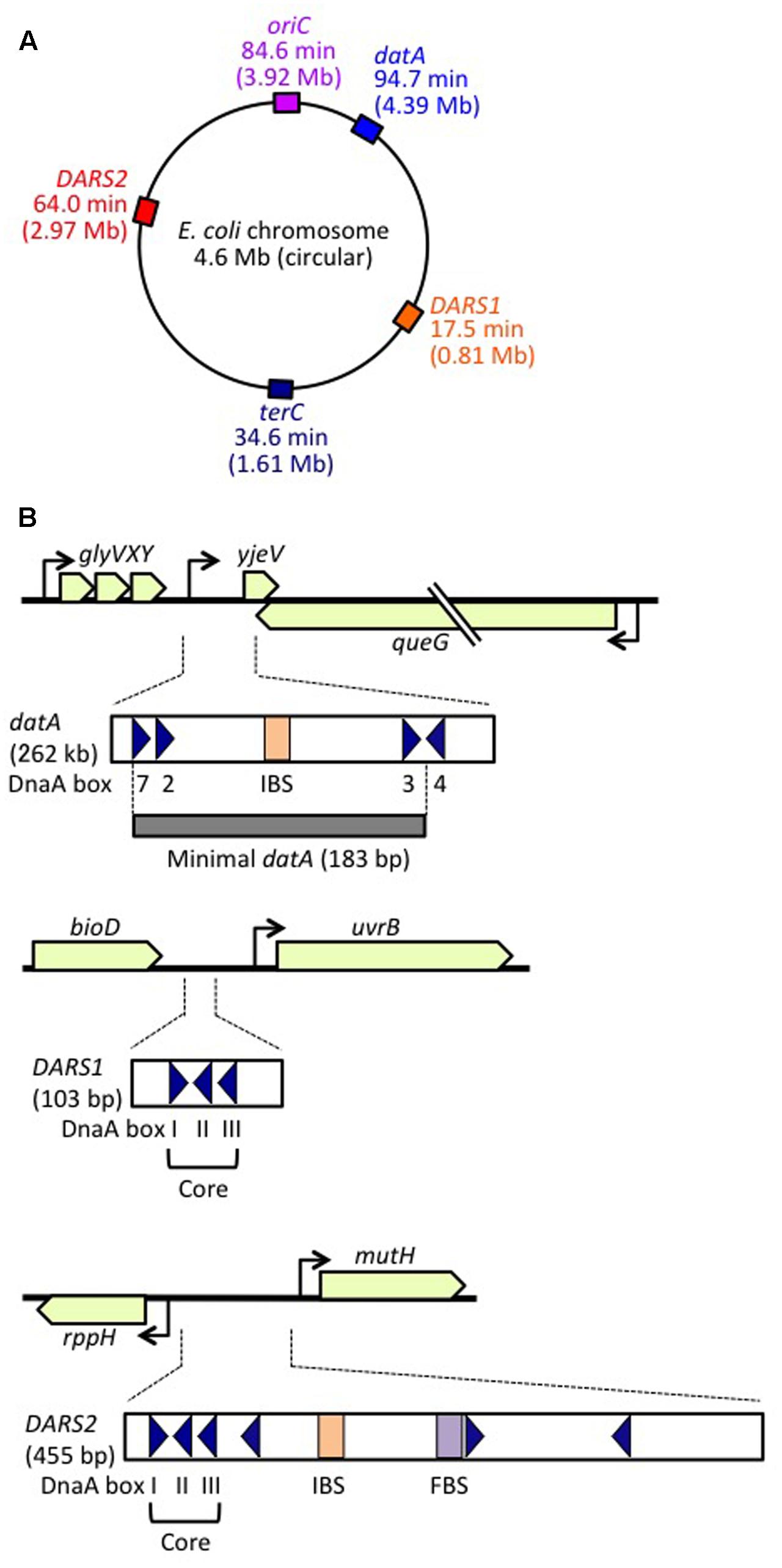
FIGURE 1. Regulatory DNA elements involved in replication initiation on the E. coli genome. (A) Schematic representation of the genomic loci of oriC, datA, DARS1, and DARS2 (and terC) in the 4.6 Mb circular E. coli genome, with positions also indicated on the scale of 0–100 min. (B) Structures of datA, DARS1, and DARS2. Open or gray bars indicate minimal regions. Triangles represent 9 bp DnaA-binding sites (DnaA boxes). Filled squares represent IHF-binding sites (IBS; shown in orange) and a Fis-binding site (FBS; shown in purple). Minimal datA consists of DnaA boxes 2, 3, and 7 and a single IBS. DnaA box 4 stimulates DDAH in vitro. DARS1 and DARS2 both have core regions containing DnaA boxes I–III. DARS2 also contains additional DnaA boxes and regulatory IBS and FBS.
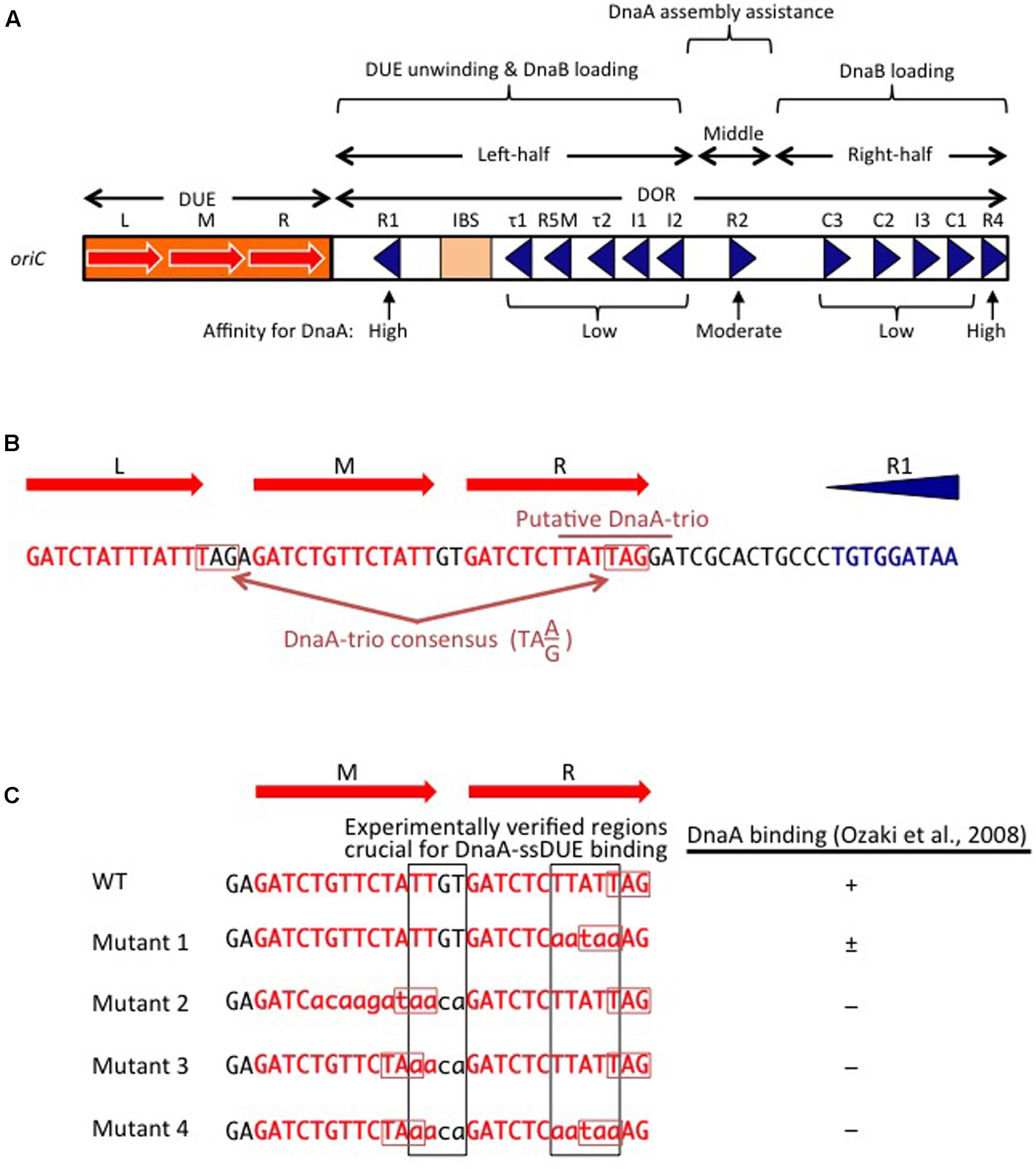
FIGURE 2. Basic features of oriC. (A) Structure of oriC. Features of the minimal 245 bp oriC sequence are shown, including DnaA boxes (triangles), IHF-binding site (IBS; rectangle), and duplex-unwinding element (DUE) AT-rich 13 bp elements (red arrows). Boxes with high, moderate, and low affinity for DnaA are indicated. (B) A portion of the oriC sequence including the DUE and R1 is shown in detail, with the putative DnaA-trio indicated (Richardson et al., 2016). Perfect DnaA-trio consensus sequences are boxed. (C) Effect of mutations on single-stranded DUE (ssDUE) DnaA binding. A portion of the DUE including the M and R 13-mers is indicated in red. Wild-type sequences and mutations are in uppercase and lowercase, respectively. Perfect DnaA-trio consensus sequences are boxed in red. DnaA binding with the indicated ssDUE sequences is summarized (Ozaki et al., 2008). Experimentally verified regions essential for DnaA-ssDUE binding are boxed in black (i.e., TTGT and TTATTT). Note that even if the DnaA-trio consensus is preserved, mutations in the boxed sequences abolish DnaA binding.
Multiple negative and positive regulatory systems target the oriC, the dnaA gene, and DnaA and work harmoniously to ensure that initiation occurs in a timely manner, in some cases via negative feedback from DNA replication. As for oriC, the minimal region contains 11 sites with 5′-GATC-3′ sequences that are specific targets of DNA adenine methylase (Dam) (Waldminghaus and Skarstad, 2009). As GATC is palindromic in duplex DNA, A residues in both strands can be methylated. In newly replicated DNA, only the sites on the parental strand are methylated, whereas those on the daughter strand are unmethylated. The hemimethylated state of oriC persists for ∼10 min in cells with a doubling time of 30 min depending on SeqA protein (Lu et al., 1994). SeqA protein has an N-terminal self-oligomerization domain and a C-terminal DNA-binding domain, and binds to the hemimethylated sites, forming self-oligomers. oriC–SeqA complexes inhibit binding of DnaA to oriC, blocking re-initiation from newly replicated DNA. SeqA sequestration is, therefore, a negative-feedback system coupled to DNA replication (Waldminghaus and Skarstad, 2009; Skarstad and Katayama, 2013). In another negative-feedback system, transcription of dnaA is autoregulated by DnaA, and is also repressed by SeqA–Dam-dependent post-replicative regulation (Campbell and Kleckner, 1990; Bogan and Helmstetter, 1997; Speck et al., 1999; Waldminghaus and Skarstad, 2009). These SeqA mechanisms have been well documented elsewhere (Waldminghaus and Skarstad, 2009), and this review will focus instead on regulation of DnaA protein.
Escherichia coli cells have at least three regulatory systems for DnaA activity (Skarstad and Katayama, 2013; Riber et al., 2016). In regulatory inactivation of DnaA (RIDA), ATP-DnaA is inactivated in a negative-feedback manner coupled to DNA replication (Katayama et al., 2010). In this system, the clamp subunit of DNA polymerase III holoenzyme has a key role; it remains on the nascent DNA strand after Okazaki-fragment completion, and the clamp–DNA complex binds to the ADP form of DNA regulatory inactivator Hda protein, an ‘ATPases associated with various cellular activities’ (AAA+) protein with an N-terminal clamp-binding site (Katayama et al., 1998; Kato and Katayama, 2001; Su’etsugu et al., 2008; Baxter and Sutton, 2012; Kim et al., 2017). The resultant ADP-Hda–clamp–DNA complex interacts with ATP-DnaA molecules catalytically, simulating ATP hydrolysis to yield ADP-DnaA. This system is predominant in the inactivation of DnaA after replication initiation, and strongly represses over-initiation of replication. RIDA has been well documented elsewhere (Katayama et al., 2010; Skarstad and Katayama, 2013; Riber et al., 2016), and this review will focus on the two other DnaA regulatory systems.
DDAH (datA-dependent DnaA-ATP hydrolysis) regulates the inactivation of DnaA (Kasho and Katayama, 2013; Riber et al., 2016) and requires the non-coding, chromosomal DNA element datA (Kitagawa et al., 1996, 1998; Figure 1). The 262 bp minimal datA region has a specific DnaA-box cluster and a single IHF-binding site (IBS) (Kasho and Katayama, 2013; Kasho et al., 2017; Figure 1B). A datA–IHF complex forms after replication initiation, and stimulates formation of specific ATP-DnaA oligomers, and hydrolysis of DnaA-bound ATP, independently of RIDA.
DnaA-reactivating sequence (DARS) sites promote reactivation of DnaA (Fujimitsu et al., 2009). The E. coli genome has at least two DARS sites (DARS1 and DARS2) (Figure 1), which contain specific DnaA-box clusters (Figure 1B), and promote oligomerization of ADP-DnaA, leading to the release of ADP (Fujimitsu et al., 2009; Kasho et al., 2014). The resultant nucleotide-free apo-DnaA preferentially dissociates from the complex, and binds ATP. Both DARS1 and DARS2 are required to sustain timely initiation in vivo. In particular, DARS2 is activated by binding of IHF and Fis (a nucleoid-associated factor); IHF binding occurs temporarily before replication initiation (Kasho et al., 2014). The RIDA, DDAH, and DARS DnaA-regulating systems are all required for timely initiation of chromosomal replication.
This review provides an up-to-date detailed synopsis of the various factors and mechanisms involved, particularly from the perspective of the mechanisms involved in the regulation of the timely activation and inactivation of the initiator protein DnaA. Sections “Basic Features of oriC and DnaA and DnaA Complex on oriC” describe mainly the principal features of oriC, DnaA and oriC-DnaA complexes, which are based on progress gained over the last 30 years. Sections “DDAH System and DARS System” mainly describe recent developments regarding DDAH and DARS systems and the salient features of DnaA complexes at datA and DARS sites. These latter sections provide information on the mechanisms of functionally different DnaA-DNA complexes. Readers who are interested only in DDAH and DARS systems might skip the Section “DnaA Complex on oriC,” while readers who are interested only in oriC-DnaA complexes might skip the Sections “DDAH System and DARS System.”
Basic Features of oriC and DnaA
oriC
The E. coli oriC is a 245 bp DNA element located at 84.6 min of the circular chromosome, and composed of two functionally distinct regions: the DUE and the DnaA-oligomerization region (DOR) (Figures 1A, 2A). The DUE has three AT-rich 13-mer sequences (L, M, and R) with the consensus 5′-GATCTnTTnTTTT-3′, where the Watson and the Crick strands are T-rich and A-rich, respectively (Bramhill and Kornberg, 1988; Messer, 2002). An additional AT-rich region flanking the L sequence assists in DUE unwinding (Messer, 2002). When the DUE is unwound, the single-stranded T-rich DUE strand binds to DnaA oligomers that are bound to the DOR (Ozaki et al., 2008) (also see below). Stable binding of the single-stranded DUE (ssDUE) requires the presence of a region spanning at least the M and R sequences; 5′-TTGT-3′ and 5′-TTATT-3′ sequences are specifically required for the binding (Ozaki et al., 2008; Figure 2B). In Bacillus subtilis oriC, a repeating trinucleotide motif (DnaA-trio) with the consensus 5′-TA(A/G)-3′ is present in the AT-rich DUE and has been proposed to sustain ssDNA binding by direct interaction with DnaA (Figure 2; Richardson et al., 2016). As requirements of specific sequences in DUE are different, E. coli and B. subtilis may differ in their mechanisms for recognition of ssDUE by DnaA (Figure 2B).
The DOR is directly connected to the right edge of the DUE, and contains 12 DnaA boxes, which are present in both orientations (Figure 2A). The DOR can be subdivided into three structurally distinct sub-regions: left-half, middle, and right-half (Leonard and Grimwade, 2015; Shimizu et al., 2016). Of these, the left-half sub-region contains six DnaA boxes (R1, τ1, R5M, τ2, I1, I2) in one orientation, and the right-half sub-region contains five boxes (C3, C2, I3, C1, R4) in the opposite orientation, whereas the middle region contains one box (R2) in the right-half orientation (McGarry et al., 2004; Rozgaja et al., 2011; Ozaki and Katayama, 2012; Ozaki et al., 2012a; Shimizu et al., 2016; Figure 2A). DnaA complexes assembled on these different sub-regions have specific roles in DUE unwinding and DnaB-helicase loading.
DnaA boxes R1 and R4 (at the left and right ends of the DOR, respectively) match the consensus 5′-TTATnCACA-3′ and have the highest affinity among DOR boxes for DnaA (in both its ATP and ADP bound forms) (Messer, 2002; McGarry et al., 2004; Kawakami et al., 2005; Figure 2A). DnaA box R2 has basically moderate affinity, and ATP-DnaA more efficiently binds to this box present in DOR than ADP-DnaA (Messer, 2002; McGarry et al., 2004; Kawakami et al., 2005; Keyamura et al., 2009). All the other DnaA boxes (τ1–I2 and R4–C1 in the left and right halves, respectively) are clustered, and each individual DnaA box only has a low affinity (McGarry et al., 2004; Kawakami et al., 2005; Rozgaja et al., 2011). Although these low-affinity boxes have only moderate similarities to the DnaA-box consensus, ATP-DnaA-specific cooperative binding does occur. This binding depends on DnaA Arg285 for recognition of DnaA-bound ATP (Kawakami et al., 2005; Keyamura et al., 2007, 2009; Kaur et al., 2014; Sakiyama et al., 2017) (for details, see below). ATP-DnaA binding to box τ1 occurs in the absence of IHF binding, but not in the presence of IHF binding, probably because of steric interference (Kawakami et al., 2005; Sakiyama et al., 2017) (see below).
The left-half DOR has a specific binding motif for the nucleoid-associated, DNA-bending protein IHF (Figure 2A). The consensus of the IBS is 5′-(A/T)ATCAAnnnnTT(A/G)-3′ (Swinger and Rice, 2004). In vitro, IHF binding stimulates DUE unwinding and replication from oriC; another nucleoid-associated protein (HU) can substitute for IHF in these reactions (Hwang and Kornberg, 1992). IHF stimulates DnaA binding to moderate-affinity (R2) and low-affinity (R5M and I1-3) sites (Grimwade et al., 2000; McGarry et al., 2004), and promotes binding to ssDUE by DnaA that is complexed with the DOR (Ozaki and Katayama, 2012). Double mutations resulting in deficiency of both IHF and HU severely inhibit cell growth, and cause synthetic lethality at high temperatures (Kano and Imamoto, 1990). Although a Fis binding consensus sequence is present between the R2 and C3 boxes, mutant cells bearing a sequence defective in Fis binding and the results of in vitro replication systems suggest that binding of Fis to oriC is not an important element in the regulation of initiation (Margulies and Kaguni, 1998; Weigel et al., 2001).
DnaA
DnaA is a 52.5 kDa basic protein composed of 473 amino acids in four domains (Ozaki and Katayama, 2009; Kaguni, 2011; Figure 3A). The largest (domain III) has an AAA+ ATPase fold that contains Walker A and B ATP/ADP-binding/hydrolysis motifs, motifs for domain III–domain III head-to-tail interaction, and motifs for ssDUE binding. Domain III AAA+ sensor 1 motifs and N-linker motif contribute to an extraordinarily high affinity for ATP/ADP (Kd = 10–100 nM) (Kawakami et al., 2006; Ozaki et al., 2012b). ATP binding causes conformational changes in DnaA, and structural differences between ATP-DnaA and ADP-DnaA are thought to occur in domains II and III (Saxena et al., 2015). Lys178, an essential residue within the Walker A motif of DnaA, is acetylated specifically in stationary-phase cells (Zhang et al., 2016).
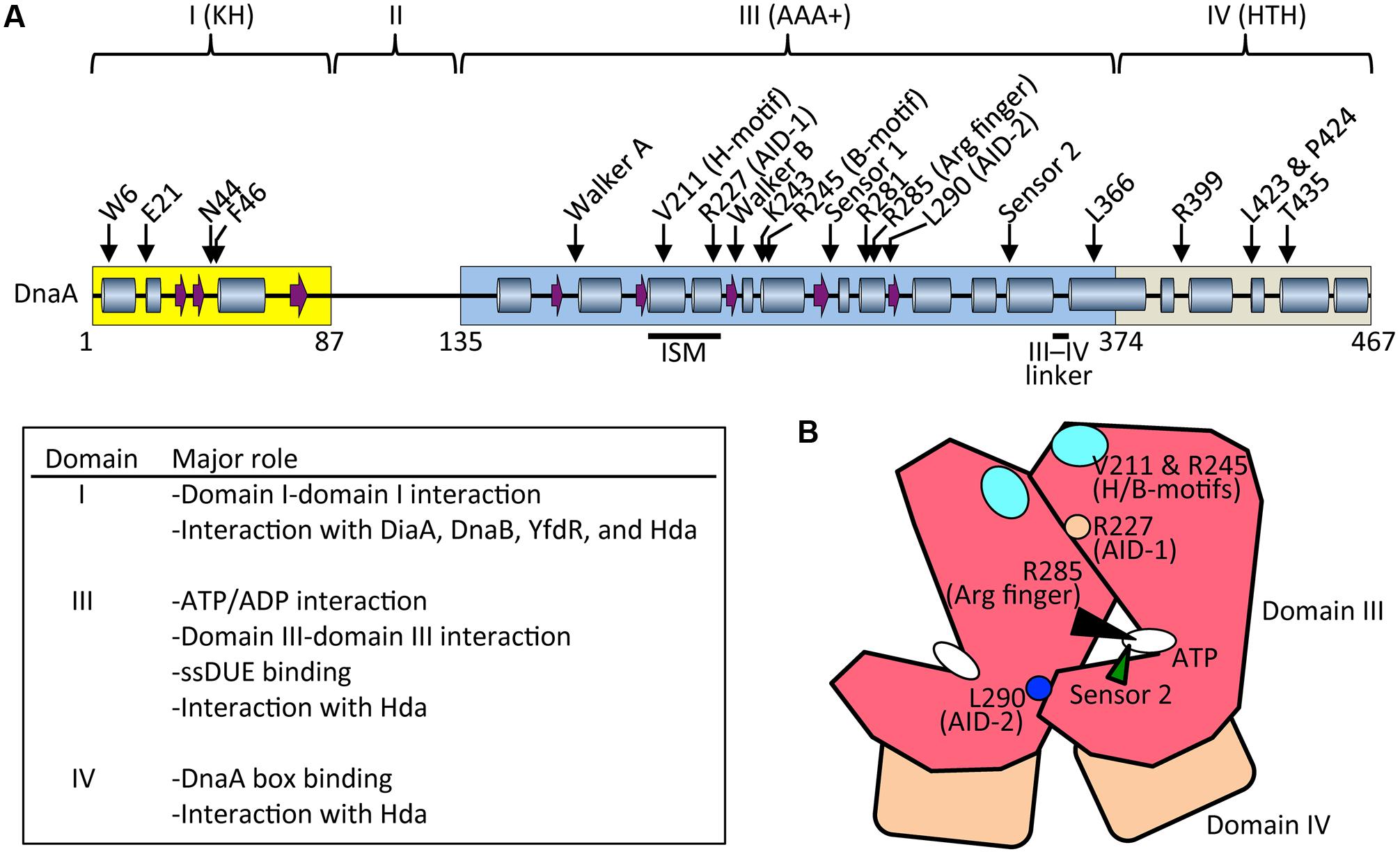
FIGURE 3. Basic features of DnaA. (A) Possible secondary structure of full length DnaA. Domains I, III, and IV are shown in yellow, blue, and gray boxes, respectively. α helices (cylinders) and β strands (arrows) were determined from the NMR structure of DnaA domain I–II (PDB: 2E0G; Abe et al., 2007) and the crystal structure of DnaA domain IV (PDB: 1J1V; Fujikawa et al., 2003), combined with a homology model of DnaA domain III generated using the crystal structure of Thermotoga maritima DnaA (PDB: 2Z4S; Ozaki et al., 2008) (SWISS-MODEL Repository: P03004). For clarity, extremely short helices in the homology model are omitted according to the crystal structure of A. aeolicus DnaA domain III (Erzberger et al., 2002). Representative motifs in the AAA+ fold and residues crucial for DnaA functions are indicated. Major roles of domains I, II, and IV are summarized. Arg finger, arginine finger (Arg285). (B) Schematic presentation of domain III (red) and IV (brown). Important motifs and residues for DnaA–DnaA interaction and ATP recognition/hydrolysis in DnaA complexes are indicated. ATP, white oval; arginine finger Arg285, black arrowhead; AID-1 Arg227, orange circle; AID-2 Leu270, blue circle; sensor 2, green arrowhead; and H/B-motifs Val211 and Arg245, cyan oval.
ATP-DnaA forms oligomers at oriC more efficiently than ADP-DnaA. The arginine finger motif (Arg285), ATP-DnaA-specific interactive locus for DUE unwinding (AID) motifs (Arg227, Leu290), box VII motif (Arg281), and Lys243 are required for functional domain III–domain III interactions (Felczak and Kaguni, 2004; Kawakami et al., 2005; Ozaki et al., 2008, 2012a; Figure 3) (see section Structure of the DnaA–IHF–oriC Complex for DUE Unwinding and DnaB Loading). In particular, Arg285 Arg finger is important for the ATP-dependent activation of DnaA complexes on oriC (Kawakami et al., 2005) and is exposed on the side opposite to that of the ATP/ADP-binding site of domain III (Figure 3B). Whereas DNA-free DnaA is monomeric in solution, domain III enables DnaA to self-oligomerize in an ATP-dependent, head-to-tail manner via interactions between bound ATP and Arg285 of adjacent protomers, resulting in the binding of multiple DnaA protomers to oriC (Kawakami et al., 2005; Ozaki et al., 2008).
During unwinding of DUE by DnaA, hydrophobic (H) and basic (B) motifs (Val211 and Arg245, respectively), which are highly conserved in DnaA orthologs, have a crucial role in binding to the T-rich ssDUE strand (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017; Figure 3) (see Section Structure of the DnaA–IHF–oriC Complex for DUE Unwinding and DnaB Loading). The H and B motifs are located in helices between the Walker A and B motifs and the Walker B and sensor 1 motifs, respectively (Ozaki and Katayama, 2009; Figure 3A). Whereas typical AAA+ proteins have only a single α helix between Walker A and B motifs, DnaA-related proteins (collectively termed the replication initiator clade) have two α helices in tandem, and the H motif is located on one of the two, giving a structure called an initiator/loader-specific motif (Iyer et al., 2004; Dueber et al., 2007; Mott et al., 2008; Ozaki et al., 2008; Ozaki and Katayama, 2009).
Certain residues in DnaA have distinct roles in its ATPase activity. Sensor 2 Arg334 is important for the intrinsic ATPase activity of DnaA, RIDA, and DDAH, probably via an interaction between the γ phosphate of ATP and Arg334. However, this residue is dispensable for the formation of functional initiation complexes on oriC (Sekimizu et al., 1987; Nishida et al., 2002; Kasho and Katayama, 2013; Figure 3). Arginine finger Arg285 is essential for intrinsic ATPase activity and for regulation by DDAH, but not by RIDA (Kawakami et al., 2005; Kasho and Katayama, 2013). Box VII Arg281 represses intrinsic ATPase activity and is dispensable for RIDA, but is required for DDAH (Kawakami et al., 2005; Kasho and Katayama, 2013) (see below).
The DnaA C-terminal domain IV has a typical helix-turn-helix (HTH) motif and binds with sequence specificity to DnaA boxes (Erzberger et al., 2002; Obita et al., 2002; Fujikawa et al., 2003; Yoshida et al., 2003; Figure 3A). This binding causes DNA bending by about 20°. An α helix consisting of amino-acid residues 434–451 is part of the HTH motif that is inserted in the major groove of the 9 bp DnaA box, recognizing the 3′ portion (5′-TnCACA-3′) of the consensus sequence (Fujikawa et al., 2003). Arg399, which is located in the turn (or loop) region of the HTH motif, is inserted in the minor groove of the DnaA box, recognizing the 5′ portion (5′-TTA-3′) of the consensus sequence (Fujikawa et al., 2003). In consistent, DnaA T435M and DnaA R399A are defective in DnaA-box binding (Sutton and Kaguni, 1997; Blaesing et al., 2000). The short linker connecting domains III and IV is a structurally flexible loop that enables swiveling of domain IV (Erzberger et al., 2002), and the results of molecular dynamics simulation indicate that it is important in the assembly of DnaA oligomers on oriC (Shimizu et al., 2016; Figure 3A). Leu366 located in an α helix downstream of the short linker is required for a conformational change in DnaA complexes constructed on oriC (Garner and Crooke, 1996; Saxena et al., 2011; Figure 3A). In addition, Leu423 and Pro424 contribute to the Hda binding that is crucial for RIDA (Keyamura and Katayama, 2011).
The N-terminal domain I of DnaA has important roles in protein–protein interaction. Weak domain I–domain I binding depends on Trp6, which contributes to DnaA self-oligomerization (Sutton et al., 1998; Simmons et al., 2003; Felczak et al., 2005; Figure 3A). Glu21 and Phe46 form a binding site for both DnaB helicase and DiaA protein (Abe et al., 2007; Keyamura et al., 2009). Phe46 contributes more to DiaA binding than Glu21, and is also required for binding to YfdR protein, a potential inhibitor of DnaA–DiaA or DnaA–DnaB interactions that is encoded by a cryptic prophage (Noguchi and Katayama, 2016). Asn44 is thought to interact with Hda to facilitate functional interaction between DnaA domain III and Hda during RIDA (Su’etsugu et al., 2013). Domain I also binds to HU, Dps (DNA protection during starvation protein), and ribosomal L2 protein, but the residues involved in these interactions have not yet been identified (Chodavarapu et al., 2008a,b, 2011). In addition, DnaA domain I contains a K-homology domain, which enables binding to ssDNA and RNA (Abe et al., 2007). However, domain I has only slight activity in ssDUE binding in vitro (Abe et al., 2007), and notably, DnaA with deletion of domains I and II retains full DUE-unwinding activity in vitro (Sutton et al., 1998), which indicates that domain III plays a predominant role in ssDUE binding sustaining DUE unwinding (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017). Domain II is an unstructured linker and the least-conserved domain among DnaA orthologs in eubacterial species (Abe et al., 2007; Nozaki and Ogawa, 2008; Figure 3A).
DnaA Complex on oriC
The Role of DiaA
The 196 amino acid DiaA protein forms a homotetramer in which each protomer has a DnaA-binding site (Ishida et al., 2004; Keyamura et al., 2007, 2009). DiaA stimulates ATP-DnaA assembly on oriC and the unwinding of DUE. A single DiaA tetramer can bind multiple DnaA molecules (theoretically up to four; experimentally at least three), potentially leading to the linkage effect for stimulating cooperative binding. With the linkage effect, it has been demonstrated that, if two proteins each with a weak affinity for DNA are stably connected by a linker molecule, the affinity of the linked molecule for DNA is increases drastically (theoretically by up to 103- to 105-fold) because it permits binding between the protein and multiple points on the DNA (Zhou, 2001; Stauffer and Chazin, 2004). A mutant that encodes a variant of DiaA with constitutive binding of just two DnaA protomers is defective in stimulation of DnaA assembly on oriC and DUE unwinding (Keyamura et al., 2007). The sequence of the diaA gene is widely conserved in the genomes of eubacterial species (Keyamura et al., 2007).
The DiaA-binding site is located in domain I of DnaA (and includes residues Glu21 and Phe46), and this site also binds DnaB helicase, as described in Section DnaA (Abe et al., 2007; Keyamura et al., 2009; Figure 3A). Both overproduction of DiaA and deletion of the diaA gene moderately inhibit initiation in vivo (Ishida et al., 2004; Flåtten et al., 2015). DiaA therefore has a positive effect on initiation through stimulation of DnaA assembly, and a negative effect probably via its ability to inhibit DnaA–DnaB binding. DiaA that is bound to the DnaA assembly on oriC is thought to dissociate (enabling DnaB binding) at (or after) the time of DUE unwinding, by an unknown mechanism (Keyamura et al., 2009). Regulation of the inhibitory activity of DiaA has not yet been determined.
Structure of the DnaA–IHF–oriC Complex for DUE Unwinding and DnaB Loading
The question of the regularity and directionality of DnaA boxes in oriC has been resolved by the identification of low-affinity DnaA boxes, and by the correction of previously obtained information. The left-half DOR is now known to contain only DnaA boxes with a leftward directionality (R1, τ1, R5M, τ2, I1, I2), whereas the middle region and right-half DOR contain only DnaA boxes with a rightward directionality (R2, C3, C2, I3, C1, R4) (Figure 2A). As explained above, head-to-tail binding of domain III leads to ATP-DnaA-dependent assembly of sub-complexes with opposite orientations in the left-half and right-half DORs (Kawakami et al., 2005; Noguchi et al., 2015; Shimizu et al., 2016; Sakiyama et al., 2017; Figures 3, 4).
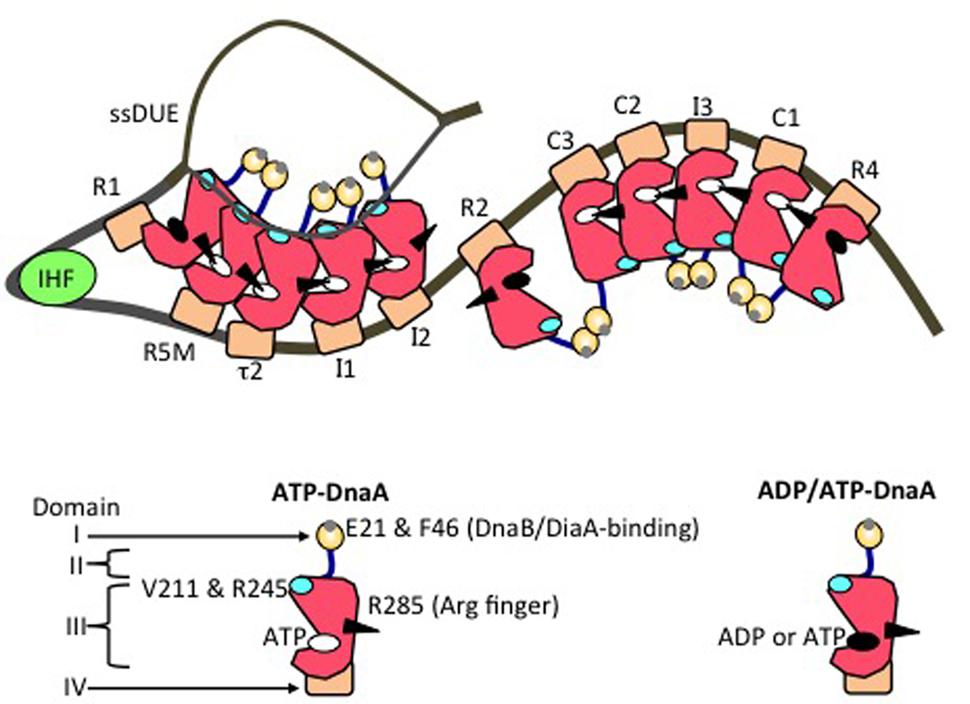
FIGURE 4. Model of an unwound state of the initiation complex. DnaA domains I (orange), II (blue), III (red), and IV (brown) are schematically presented. A small gray circle on domain I shows a patch including Glu21 and Phe46 (for binding of DiaA and DnaB helicase). A black arrowhead, cyan oval, white oval, and black oval on domain III represent the arginine finger Arg285, H/B motifs, ATP, and either ADP or ATP, respectively. IHF is shown as a light green circle. DnaA-box positions are labeled. On the left-half and right-half DnaA-oligomerization regions, head-to-tail DnaA-oligomer sub-complexes assemble, with interactions between ATP and Domain III Arg285. The T-rich ssDUE strand interacts with the DnaA H/B motifs in the left-half sub-complex.
In DnaA sub-complexes, nucleotides that are associated with R1 box- and R4 box-bound DnaA protomers are located at the outer edges of the DOR, so that those cannot interact with DnaA protomers bound to other DOR sites (Figures 2A, 4). Thus, even when ADP-DnaA is bound to R1, DUE-unwinding activity is fully sustained (Noguchi et al., 2015). ATP-DnaA assembly at the left-half DOR fundamentally depends on ATP–DnaA binding to the low-affinity R5M box (but not to the R1 box) and is stimulated by R1-bound DnaA (Sakiyama et al., 2017). Cooperative binding of ATP-DnaA molecules would operate effectively in the region from the R5M box to the I2 box (Sakiyama et al., 2017). Also, when ADP-DnaA is bound to the R4 box, assembly of the DnaA sub-complex on the right-half DOR is fully sustained with ATP-DnaA (Noguchi et al., 2015). ATP-DnaA assembly to the low-affinity sites in the right-half DOR depends on DnaA binding to the R4 box (Noguchi et al., 2015).
Evidence suggests that domain III of R2-bound DnaA does not interact with DnaA protomers bound to I2 or C3 (Rozgaja et al., 2011; Shimizu et al., 2016), as expected by the long spacing between R2 and I2 (9 bp) and between R2 and C3 (20 bp) (Figure 2A). Each box in the τ1-R5M-τ2-I1-I2 region and the R4-C1-I3-C2-C3 region is separated by 2–4 bp. However, domain I of R2-bound DnaA might interact with that of I2-bound or C3-bound DnaA, simulating cooperative DnaA binding (Miller et al., 2009; Ozaki and Katayama, 2012; Shimizu et al., 2016; Figure 4).
The existence of nucleotide-independent activities of DnaA bound to R1, R4, and R2 is consistent with data from in vivo dimethyl-sulfate footprint analysis, which suggest that DnaA binding to these boxes is stable throughout the cell cycle (Samitt et al., 1989; McGarry et al., 2004; Kawakami et al., 2005). By contrast, in in vivo experiments with DnaA fused to enhanced yellow fluorescent protein, results from photobleaching and high-resolution microscopy suggest that, overall, DnaA complexes bound to oriC are rapidly turned over with a time scale of a few seconds (Schenk et al., 2017). These results are consistent with the unstable binding of DnaA to low-affinity sites. Such dynamic features might be important for timely assembly of ATP-DnaA on oriC after the DARS system has increased in total level of ATP-DnaA.
The left-half ATP-DnaA sub-complex possesses full activity for DUE unwinding and ssDUE binding (Ozaki and Katayama, 2012; Figure 4). In addition to the Arg finger Arg285, AID1 (Arg227) and AID2 (Leu290) motifs, which are highly conserved in DnaA orthologs, are thought to be located at the domain III–domain III interface and to contribute to ATP–DnaA-specific interactions and the establishment of DnaA oligomers within the left-half DOR that are competent for DUE unwinding (Ozaki et al., 2012a; Figure 3B). Lys243 also contributes to oligomerization of ATP-DnaA on oriC and DUE unwinding (Ozaki et al., 2008). Also, Lys243 can be acetylated in vivo. Although the significance of this is unknown, it moderately affects DnaA binding to oriC in vitro (Li et al., 2017). Molecular dynamics simulation shows that sharp DNA bending (>120°) caused by IHF binding enables domain III–domain III interaction between DnaA protomers bound to R1 and R5M in the absence of DnaA–τ1 binding (Shimizu et al., 2016; Figure 4). Consistently, in vitro experiments indicate that DnaA–τ1 interaction is inhibited by IHF binding (Sakiyama et al., 2017).
DUE unwinding depends on temperature and superhelicity of oriC DNA, which contribute to destabilization of the duplex state of the DUE (Baker and Kornberg, 1988; Sekimizu et al., 1988). In addition, the unstable ssDUE binds to, and is stabilized by, DnaA domain III, enabling loading of DnaB helicase to the unwound region (Ozaki et al., 2008). In E. coli and B. subtilis, only a specific strand (the T-rich strand in E. coli) binds to DnaA (Ozaki et al., 2008; Richardson et al., 2016). Also, in E. coli, the H/B-motif residues of domain III are required for ssDUE binding (Ozaki et al., 2008; Sakiyama et al., 2017). This binding mode is consistent with that seen in the co-crystal structure of the poly-A ssDNA-bound DnaA domain III ortholog from the hyperthermophilic bacterium Aquifex aeolicus (Duderstadt et al., 2011).
Notably, when multiple DnaA molecules bind to the DOR, ssDUE binding by DnaA increases dramatically and is much higher than that of DOR-unbound DnaA molecules (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017). ssDUE directly binds to domain III of DnaA protomers that have associations through domain IV with the left-half DOR (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017; Figure 4). In particular, domain III of the R1- and R5M-bound DnaA protomers play predominant roles in ssDUE binding for replication initiation (Sakiyama et al., 2017). These and the other observations mentioned above support the idea that ssDUE directly binds to R1- and R5M-bound DnaA protomers via sharp DNA bending by IHF, which stimulates ssDUE recruitment to those DnaA molecules. This is referred to as the ssDUE-recruitment mechanism (Figure 4; Ozaki et al., 2008; Ozaki and Katayama, 2012; Noguchi et al., 2015; Shimizu et al., 2016; Sakiyama et al., 2017). This reasonably explains the role of IHF and dispensability of ATP of R1-bound DnaA in DUE unwinding, and the striking stimulation of ssDUE binding by DnaA-oligomer assembly on the DOR, as well as the strict requirements in DUE unwinding for the spacing between the DUE and the DnaA R1 box and between the DnaA R1 box and the IBS, and the requirement for H/B motifs of the DOR-bound DnaA for ssDUE binding (Figure 4) (for details, see Sakiyama et al., 2017).
Multiple DnaA molecules bound to the DOR are required for binding of DnaB helicase which is a homohexamer (Abe et al., 2007; Keyamura et al., 2009; Ozaki and Katayama, 2009). Multiple DnaA domain I molecules of the DOR-bound DnaA complexes would be allied so as to stably bind DnaB helicases by using multiple binding points (Figure 4). In consistent, Box VII Arg281 stabilizes DnaA oligomers assembled on the DOR, and is required for DnaB-helicase loading, but not for DUE unwinding (Felczak and Kaguni, 2004). This suggests that stability of DnaA complexes is more important for DnaB-helicase loading than for DUE unwinding.
In addition to the basic DnaB-binding/loading activity of the left-half-DOR DnaA sub-complex, the right-half-DOR DnaA sub-complex stimulates DnaB binding/loading (Ozaki and Katayama, 2012; Figures 2A, 4). This activity might result in binding of two DnaB helicases, one to each of the DnaA sub-complexes, enabling bi-directional replication (Ozaki and Katayama, 2012; Noguchi et al., 2015; Figure 4). The spacing between the R2 and C3 boxes is important for efficient DnaB loading, suggesting coordination of the left and right DnaA subcomplexes in a DnaB loading process (Shimizu et al., 2016; Figure 4).
DDAH System
Function
datA-dependent DnaA inactivation has been termed DDAH. Three hundred 9-mer DnaA box consensus sequences are widely distributed throughout the E. coli chromosome (Tesfa-Selase and Drabble, 1992; Roth and Messer, 1998; Hansen et al., 2007). Some of these sites regulate the activity of DnaA for replication initiation, whereas some of others function for transcriptional regulation by DnaA (Kitagawa et al., 1996; Messer and Weigel, 1997; Speck et al., 1999; Olliver et al., 2010). The datA locus includes a DnaA-box cluster and was originally identified as a potent DnaA-binding locus that can repress untimely replication initiation (Kitagawa et al., 1996; Ogawa et al., 2002; Nozaki et al., 2009; Figure 1A). DnaA binding at datA is stimulated by IHF (Nozaki et al., 2009); the datA–IHF complex stimulates hydrolysis of DnaA-bound ATP independently of RIDA to produce ADP-DnaA (Kasho and Katayama, 2013; Kasho et al., 2017; Figure 5A).
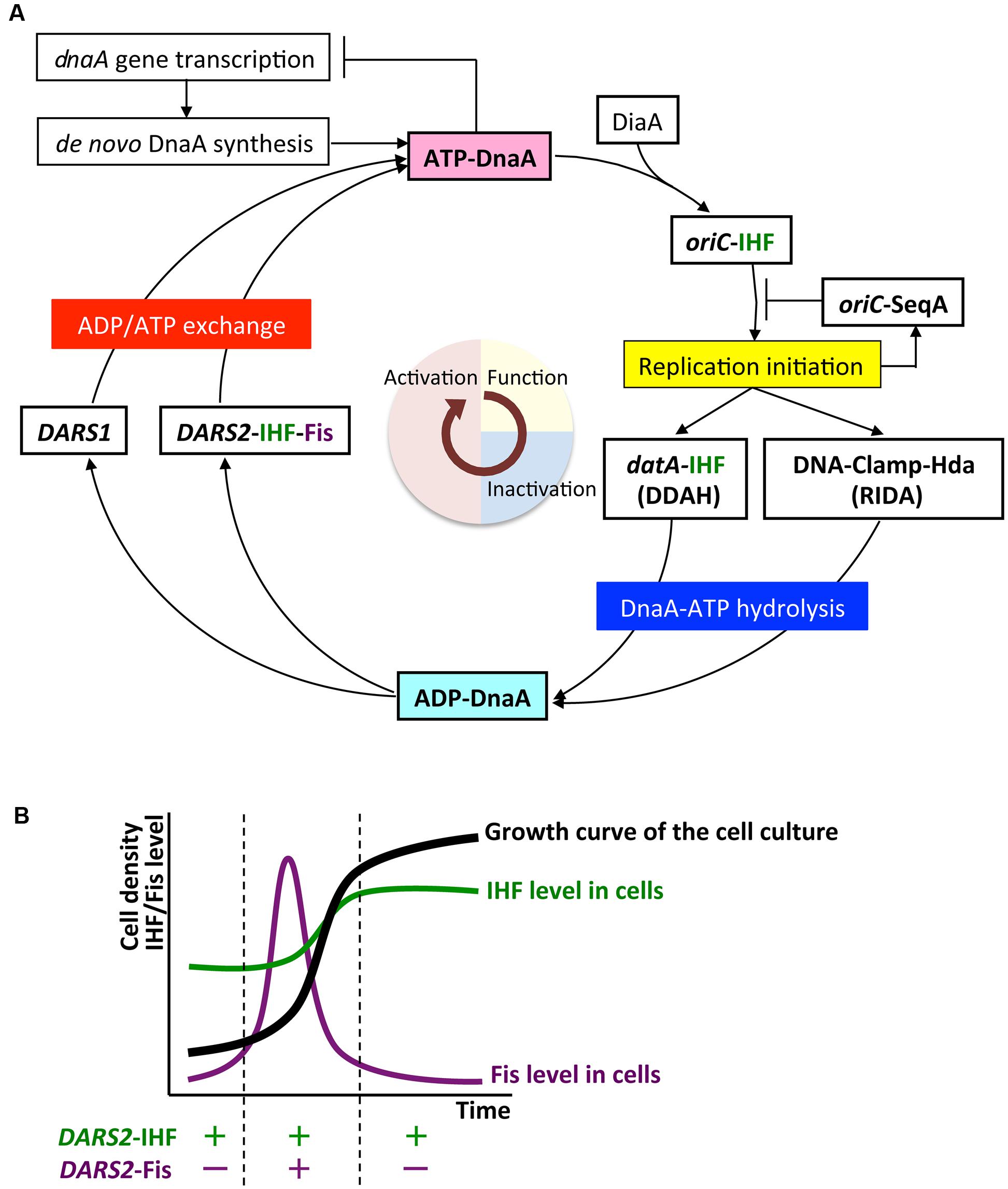
FIGURE 5. The regulatory cycle of DnaA. (A) Schematic presentation of the regulatory cycle of DnaA. ATP-DnaA forms oligomers on oriC with the aid of integration host factor (IHF) and DnaA interacting protein DiaA, and initiates replication (period in light yellow). After initiation, ATP-DnaA is converted to replication-inactive ADP-DnaA (RIDA) and datA-dependent DnaA-ATP hydrolysis (DDAH) systems (period in light blue). RIDA involves Hda protein and the clamp subunit of DNA polymerase III holoenzyme, and DDAH involves the datA locus and IHF. DnaA-reactivating sequence (DARS1 and DARS2) loci stimulate nucleotide exchange of ADP-DnaA and regenerate ATP-DnaA (period in light red). IHF binds to specific sites in oriC, datA, and DARS2 in a cell-cycle-coordinated manner. Fis binds to DARS2 in log-phase cells. (B) Growth-phase coordination of regulation of Fis expression. The cellular level of Fis varies through the growth phases, and is much higher in exponential-phase cells (enabling Fis binding to DARS2) than in stationary-phase cells. By contrast, the cellular level of IHF is highest in stationary-phase cells.
Several lines of in vivo evidence point to the important role of the datA–IHF complex in the repression of untimely replication initiation. Deletion of datA or IHF genes (ihfA or ihfB) increases cellular levels of ATP-DnaA, resulting in higher levels of ATP-DnaA than in wild-type cells and untimely replication initiation without cell growth inhibition (Kitagawa et al., 1996; Nozaki et al., 2009; Kasho and Katayama, 2013). As inhibition of RIDA causes severe over-initiation of replication and arrest of cell growth (Kato and Katayama, 2001; Nishida et al., 2002; Fujimitsu et al., 2008), DDAH is thought to play a supportive role in RIDA. Also, in DDAH-deficient mutants, replication initiation is not completely repressed in the presence of rifampicin, an inhibitor of transcription and replication initiation (von Freiesleben et al., 2000; Morigen, Skarstad and Molina, 2005), probably due to elevated levels of ATP-DnaA.
Structure of datA
The minimal datA for DDAH function is 183 bp containing high-affinity DnaA boxes 2 and 3, low-affinity DnaA box 7 and a single IBS, which are all essential for efficient ATP–DnaA binding and DDAH activity (Kitagawa et al., 1998; Ogawa et al., 2002; Nozaki et al., 2009; Kasho and Katayama, 2013; Kasho et al., 2017; Figure 1B). All the essential DnaA boxes have the same directionality, suggesting the importance of head-to-tail ATP–DnaA interactions. The results of mutation analyses indicate that, as in the oriC DORs, the orientations and interval lengths of the datA DnaA boxes 2 and 3 and the IBS are optimized for repression of untimely initiations through DDAH (Nozaki et al., 2009; Kasho and Katayama, 2013). The datA DnaA box 4 stimulates DnaA assembly and DnaA-ATP hydrolysis in vitro, but is not essential for initiation regulation in vivo (Ogawa et al., 2002; Kasho and Katayama, 2013; Kasho et al., 2017; Figure 1B). Unidentified factor(s) might stimulate DnaA assembly in vivo even in the absence of DnaA box 4 (see below).
Mechanism
In the datA region, oligomers containing three ATP-DnaA molecules form on and around the core DnaA boxes and higher oligomers form depending on IHF (Kasho and Katayama, 2013; Kasho et al., 2017; Figure 6A). IHF binding causes DNA looping, which would promote interaction between the box 2-bound DnaA and the box 3-bound DnaA, thereby stabilizing DnaA complexes in a cooperative manner for the activation of DnaA-ATP hydrolysis. The results of biochemical analyses using mutant DnaA proteins provide evidence that, in datA–DnaA complexes, DnaA–DnaA interactions depend on AAA+ domain III arginine finger Arg285, box VII Arg281, and AID2 Leu290, and stimulation of ATP hydrolysis depends on AAA+ sensor 2 motif Arg334 (Kasho and Katayama, 2013; Kasho et al., 2017). DnaA AID-2 Leu290 might facilitate dissociation of the resultant ADP-DnaA, which is unstable in datA, thereby enabling catalytic reaction of DDAH (Kasho et al., 2017). Overall, these requirements for specific residues are similar to those of the initiation complex, except for the crucial role of sensor 2 Arg334 in ATP hydrolysis. This seems reasonable because both DDAH complexes and the initiation complex are constructed mainly from head-to-tail DnaA–DnaA interactions (Figures 3B, 6A).
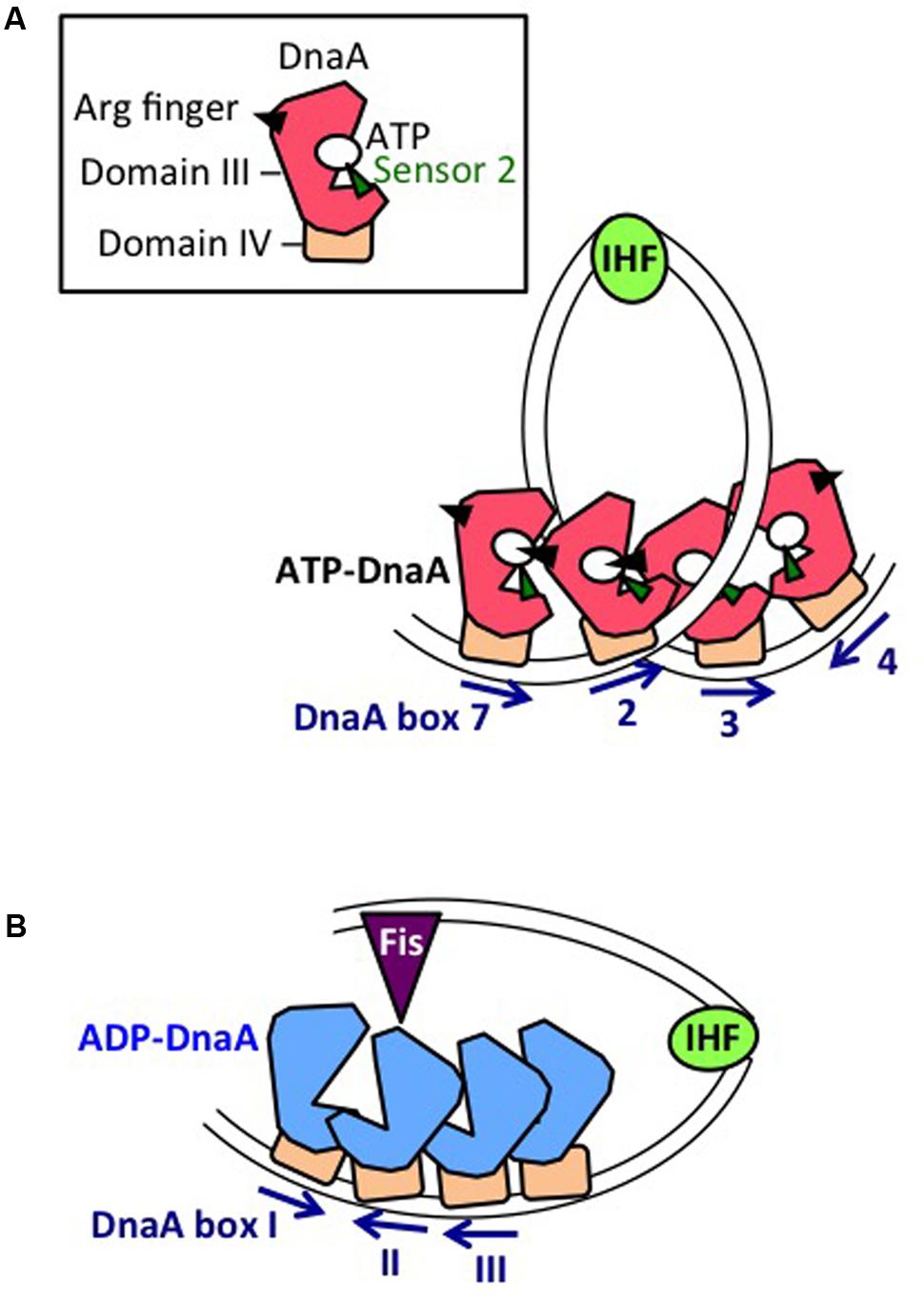
FIGURE 6. Mechanistic view of DnaA regulation by datA and DARS2. (A) Mechanistic model of datA-dependent DnaA-ATP hydrolysis (DDAH). Oligomers of ATP-DnaA (with domain III shown as a red polygon, and domain IV as an brown square) assemble on the datA minimal region containing DnaA boxes 7, 2, and 3. Sharp DNA bending by IHF could stimulate contact between DnaA bound at boxes 2 and 3. Cooperative ATP-DnaA binding on datA DnaA box 7 might induce conformational change of the DnaA to activate DDAH. (B) Mechanistic model of DARS2-mediated DnaA activation. Oligomers of ADP-DnaA (with domain III shown as a blue polygon, and domain IV as an brown square) assemble on the DARS2 core region. IHF and Fis bind to specific sites in the DARS2 regulatory region, and the resultant complex stimulates conformational changes in the core-bound DnaA oligomer, to induce structural changes in DnaA and ADP dissociation. Nucleotide-free apo-DnaA dissociates from the core region and binds ATP (which is abundant in the cytosol), yielding ATP-DnaA.
The stimulatory role of IHF in DDAH cannot be replaced by HU, which is a structural homolog of IHF and a widely conserved nucleoid-associated protein in most bacteria, although IHF can be functionally replaced by HU in an in vitro reconstituted system of replication initiation from oriC (Hwang and Kornberg, 1992; Dillon and Dorman, 2010; Kasho and Katayama, 2013). Like IHF, HU induces sharp DNA bending, but unlike IHF, HU binds to DNA without sequence specificity. At datA and oriC, the IBS is located between DnaA boxes, but the inter-DnaA box region in datA (i.e., between box 2 and box 3) is longer than that (i.e., between R1 box and R5M box) at oriC (Figures 1B, 2A), suggesting that a functional conformational change in datA for activating DDAH is strongly dependent on specific DNA bending at the datA IBS induced by IHF binding.
Regulation
Coupling of datA Dosage (Copy Number) with Replication
datA is located at 94.7 min (4.39 Mb position) in the E. coli genome, near to oriC (84.6 min, 3.92 Mb position), which means that datA is duplicated soon after replication initiation, resulting in a temporary higher dosage (copy number) of datA per cell. This is thought to be important for the repression of untimely initiations under certain growth conditions (Kitagawa et al., 1996; Figure 1A). In agreement with these observations, increasing the datA copy number via a multi-copy plasmid negatively impacts replication initiation and cell growth (Morigen et al., 2003; Kasho et al., 2017). Conversely, translocation of datA to the replication-termination locus terC at 34.6 min, which is >2 Mb apart from oriC (Figure 1A), does not result in a temporary increase in its copy number until replication is nearly completed, allowing untimely initiations in nutrition-rich medium, similar to those observed in datA-deleted cells (Kitagawa et al., 1998; Frimodt-Møller et al., 2015). The datA copy number may also be important for the regulation of cell division (Morigen et al., 2014). Notably, in B. subtilis and Streptomyces coelicolor, chromosomal DnaA-box clusters analogous to datA repress untimely replication initiation (Smulczyk-Krawczyszyn et al., 2006; Okumura et al., 2012), suggesting that the DnaA regulation by datA is a conserved feature of most eubacterial species.
Cell-Cycle-Dependent Binding of IHF
The activation of DDAH and RIDA reduces the ATP-DnaA level and represses untimely initiations (Kasho and Katayama, 2013). Regulation of IHF binding to datA is essential for the timely activation of DDAH. Cell-cycle analysis of datA–IHF binding indicates that IHF dissociates from datA before replication initiation and temporarily binds to datA after initiation (Kasho and Katayama, 2013). As both IHF binding to and dissociation from datA occur during on-going chromosome replication, DDAH is suggested to be regulated by a mechanism that couples certain cell cycle events to datA–IHF binding. Conversely, RIDA is activated by DNA replication-coupled feedback (Katayama et al., 2010; Kasho and Katayama, 2013)
The transcription inhibitor rifampicin might inhibit timely dissociation of datA–IHF complexes (Kasho and Katayama, 2013). As IHF is abundant in exponentially growing cells (Ali Azam et al., 1999), a specific factor inhibiting datA–IHF binding might be required to be transcribed at a particular moment in the cell cycle. DDAH activity is inhibited by translocation of the datA sequence to a highly transcribed region (Frimodt-Møller et al., 2016), suggesting that transcription through datA might inhibit datA–IHF binding. Consistently, the essential core region of datA (i.e., box 7-box 2-IBS-box 3) is located at an intergenic region downstream of the glyVXY operon and queG gene (Kitagawa et al., 1998; Nozaki et al., 2009; Kasho et al., 2017; Figure 1B). It remains to be determined whether transcription of these genes oscillates in a coordinated manner during the cell cycle.
Effects of DNA Superhelicity and Nutrition
DDAH is suggested to regulated also by DNA supercoiling. In vitro experiments indicate that negative DNA supercoiling, which is modulated by DNA topoisomerases such as DNA gyrase and by nucleoid protein binding (Travers and Muskhelishvili, 2005), stabilizes ATP-DnaA oligomerization and IHF binding on datA and increases DDAH activity twofold compared with datA activity on relaxed DNA (Kasho et al., 2017). Modulation of negative supercoiling by novobiocin, a DNA-gyrase inhibitor, decreases the function of datA on plasmid DNA for repressing replication initiation (Kasho et al., 2017), suggesting supercoiling-dependent regulation of DDAH in vivo. Negative supercoiling is predicted to increase under osmotic-stress conditions (Higgins et al., 1988), which might affect datA activity for repressing initiation.
Nutrition-dependent regulation of DDAH has been suggested. Untimely initiations in datA-null cells are more severely exhibited in nutrient-poor medium containing acetate or glucose than in rich medium containing glucose-casamino acid or LB (Kitagawa et al., 1998; Flåtten et al., 2015). Chromosome-conformation-capture analysis has revealed that oriC can physically interact with datA under conditions of replication stress induced by serine hydroxamate, which causes amino-acid starvation and inhibits replication initiation (Cagliero et al., 2013). Free DnaA molecules present around oriC could efficiently interact with datA under poor nutrition conditions. Conversely, in rich medium, the datA dosage could be important for the regulation of DDAH. As described above, datA translocation to terC causes excess initiations in rich medium, but not in poor medium (Kitagawa et al., 1998).
DARS System
Function
In E. coli, ATP-DnaA is produced by binding of apo-DnaA to ATP, which is abundant in the cytosol (Bochner and Ames, 1982). Replication initiation occurs when the cellular level of ATP-DnaA reaches a peak that is high enough to enable assembly on oriC (Kurokawa et al., 1999). At least three mechanisms for production of apo-DnaA have been characterized: de novo DnaA synthesis; nucleotide dissociation from ADP-DnaA by acidic phospholipids in the cell membrane; and a mechanism involving specific chromosomal DNA elements termed DARS sites (Figure 1).
In de novo synthesis, newly translated apo-DnaA binds to ATP in the cytosol (Figure 5A). In addition to autoregulation by DnaA (Speck et al., 1999), transcription of dnaA is down-regulated by the SeqA-Dam system immediately after initiation (Campbell and Kleckner, 1990; Bogan and Helmstetter, 1997). This would contribute to the reduction in de novo DnaA synthesis (i.e., production of ATP-DnaA) immediately after initiation and maintain coordination between replication initiation and the cell cycle (Riber and Løbner-Olesen, 2005).
The E. coli cell membrane contains acidic phospholipids such as cardiolipin and phosphatidylglycerol that can promote dissociation of ADP or ATP from DnaA by interaction with DnaA domain III. These phospholipids can convert ADP-DnaA to ATP-DnaA in vitro (Saxena et al., 2013). In mutant cells with reduction of levels of acidic phospholipids, initiation from oriC is repressed (Fingland et al., 2012). The possibility that acidic phospholipids regulate DnaA activity during the cell cycle deserves to be further investigated.
The DARS1 and DARS2 chromosomal DNA sequences promote dissociation of ADP from ADP-DnaA, stimulating nucleotide exchange and increasing levels of ATP-DnaA, thereby stimulating replication initiation (Fujimitsu et al., 2009; Kasho et al., 2014; Figures 1, 5A). In vitro, DARS1 can promote ADP dissociation of ADP-DnaA without additional factors, whereas DARS2 has little activity by itself, but is substantially activated by IHF and Fis, which binds site-specifically and bends DNA by <60° (Fujimitsu et al., 2009; Kasho et al., 2014; Figure 1B). In cells, DARS1 and DARS2 both have stimulatory roles in ATP-DnaA production and replication initiation, and the deletion of either or both delays initiation (Fujimitsu et al., 2009; Kasho et al., 2014). Deletion of DARS2 inhibits timely initiation more severely than deletion of DARS1, causing asynchronous initiations, and consistently, oversupply of DARS2 has a greater effect than oversupply of DARS1 on stimulation of extra initiations, which can cause inhibition of cell growth (Fujimitsu et al., 2009; Kasho et al., 2014). Deletion of DARS2, substitution of the IBS or Fis-binding site (FBS), or deletion of Fis gene decreases the cellular ATP-DnaA level more than deletion of DARS1 under RIDA-deficient conditions (Fujimitsu et al., 2009; Kasho et al., 2014; Figure 1B). These observations suggest that the complex formed by DARS2, IHF, and Fis is important for increasing the level of ATP-DnaA and determining initiation timing, and that DARS1 might maintain basal levels of ATP-DnaA.
Structure of DARS1 and DARS2
A common structural feature of DARS1 (101 bp) and DARS2 (455 bp) is the presence of a core region containing three DnaA boxes (DnaA boxes I, II, and III): the spacing between these boxes is the same at both DARS1 and DARS2, and box I has the opposite orientation to boxes II and III (Figure 1B). In addition to the core regions, DARS1 and DARS2 have regulatory regions of different lengths. The ∼50 bp regulatory region of DARS1 enhances DnaA-ADP-dissociation activity in vitro, but its role in vivo has not been determined (Fujimitsu et al., 2009). The ∼400 bp DARS2 regulatory region contains the IBS and FBS, which stimulate ADP-DnaA assembly on DARS2 and are essential for ADP dissociation from DnaA (Kasho et al., 2014).
Mechanism
Unlike oriC or datA, DARS sites preferentially bind ADP-DnaA rather than ATP-DnaA. Notably, head-to-head (but not head-to-tail) interactions would occur between the box I-bound DnaA and the box II-bound DnaA, which is a prominent feature at DARS sites (Figures 1B, 6B). Formation of oligomers of four or more ADP-DnaA molecules on the DARS2 core region depends on DnaA AID-2 Leu290, and results in dissociation of ADP (Figure 6B). Formation of similar oligomers on the DARS1 core region promotes dissociation of ADP and is dependent on DnaA AAA+ sensor 1 Asp269, which would participate in probable heat-dependent conformational change of the DnaA nucleotide-binding pocket (Fujimitsu et al., 2009). DnaA sensor 1 Asp269 is dispensable for ADP dissociation at DARS2 (Kasho et al., 2014), possibly because of still unknown functions of the activator proteins IHF and Fis (Figure 6B). Dissociation of ADP produces apo-DnaA molecules that are unstable in binding to the DARS core region, enabling ATP binding and the production of ATP-DnaA (Fujimitsu et al., 2009; Kasho et al., 2014).
Binding of IHF is essential for DARS2 activity, and, as with DDAH, IHF cannot be replaced by HU. The complex of DARS2–IHF–Fis promotes ∼10 times as much ADP dissociation as DARS1 in vitro (Kasho et al., 2014). The results of mutational analyses indicate that the spacing between the core-region DnaA boxes, IBS, and FBS is optimized for efficient activation of DARS2 (Kasho et al., 2014). The sequences and lengths of the spaces between the DARS2 core region, IBS, and FBS are highly conserved in E. coli-proximal β- or γ-proteobacterial species in which IHF and Fis homologs are conserved, suggesting that the regulatory mechanism for DARS2 is shared among proteobacterial species, including pathogenic species (Kasho et al., 2014).
Regulation
Cell-Cycle-Dependent IHF Binding
IHF binding to DARS2 is dynamically regulated in a cell-cycle-coordinated manner (Kasho et al., 2014). In contrast to datA–IHF binding, DARS2–IHF binding occurs specifically in the pre-initiation period, consistent with its role in timely production of ATP-DnaA for replication initiation (Kasho et al., 2014). Regulation of DARS2–IHF binding and dissociation is resistant to rifampicin (Kasho et al., 2014), suggesting that it is independent of transcription. The timing of DARS2–IHF binding is also independent of replication initiation at oriC or replication fork progression (Kasho et al., 2014), supporting the suggestion that functional activation of DARS2 might be controlled by specific cell cycle (but not DNA replication)-coupled events.
Growth-Phase-Dependent Fis Binding
In addition to cell-cycle regulation, growth-phase-dependent regulation contributes to timely activation of DARS2. The cellular level of Fis increases specifically in the exponential-growth phase (Ali Azam et al., 1999; Dillon and Dorman, 2010; Figure 5B). Fis binding to DARS2 occurs in the exponential phase, but not in the stationary phase (Kasho et al., 2014). The activation of DARS2 only in exponential phase is consistent with the requirement for ATP-DnaA for replication initiation.
Chromosomal-Position Effect
Escherichia coli DARS1 (101 bp) and DARS2 (455 bp) are both located in intergenic regions, at 17.5 min (0.81 Mb position) and 64.0 min (2.97 Mb position), respectively, between oriC and terC (Figure 1A), and these positions are well conserved among related bacteria (Fujimitsu et al., 2009; Kasho et al., 2014; Frimodt-Møller et al., 2015). Translocation of DARS2 to a terC-proximal locus moderately inhibits the timing of initiation, as does its translocation to an oriC-proximal locus (Frimodt-Møller et al., 2016; Inoue et al., 2016). Inhibition at the terC-proximal locus is sustained in cells lacking MatP, a DNA-binding protein involved in formation of terC-specific subchromosomal structure (Mercier et al., 2008; Inoue et al., 2016). The results of 3C (chromosome-conformation-capture) analysis suggest that oriC can physically interact with the wild-type DARS2 locus (Cagliero et al., 2013). These results suggest that the chromosomal location of DARS2 might be important for efficient interaction with oriC. In addition, a specific local chromosomal structure might be important for DARS function.
DnaA activation by DARS1 and DARS2 stimulates efficient cell growth under nutrient-poor conditions, and colonization of the large intestine of streptomycin-treated mice (Frimodt-Møller et al., 2015), suggesting a requirement for DARS1 and DARS2 for environmental adaptation of E. coli. Stimulation of replication initiation by oversupply of DARS2 has more severe effects on viability under aerobic conditions than under anaerobic conditions (Charbon et al., 2014), suggesting that over-initiation in aerobic conditions leads to lethal levels of encounters between replication forks and sites of oxidative damage (Charbon et al., 2017).
Perspectives for Coupling Between DnaA Regulation and Cell Growth in E. coli
During the E. coli cell cycle, levels of ATP-DnaA rise to a peak that induces replication initiation, after which they fall, predominantly as a result of RIDA, a replication-coupled negative-feedback mechanism (Katayama et al., 2010). In the pre-initiation period, DARS2 is predominant in the conversion of ADP-DnaA to ATP-DnaA. IHF binding to DARS2 promotes this activity in a timely manner, but IHF is abundant throughout the cell cycle, and the mechanism responsible for its timely binding and dissociation is not yet known. Similarly, the mechanism responsible for the regulation of datA–IHF binding, which occurs at a specific time after replication initiation, has not yet been determined. Further studies are required to determine which mechanisms contribute to coupling between these processes and cell-cycle progression. In addition, further studies are required to understand DnaA complex formation/dissociation, ADP dissociation, and ATP hydrolysis during DDAH and in the formation of protein complexes at DARS sites.
Chromosomal positioning of DARS1, DARS2, and datA is important for regulation of the timing of initiation under specific growth conditions. Chromosomal structural determinants such as superhelicity, co-localization of specific loci in the 3D chromosomal structure, or the timing of changes in copy number during replication that is related to the distance from oriC might all contribute to the importance of chromosomal location. Further studies can help to determine which of these mechanisms is relevant for the correct timing of replication initiation.
How DnaB helicase binding to oriC-DnaA complexes in vivo is regulated is an open question. Even in the presence of a high ATP-DnaA level and oriC DUE unwinding, DnaB helicase might still not be able to be bind to oriC-DnaA complexes because of the stable interaction between DiaA and DnaA. Further studies of the regulatory mechanisms governing the DiaA binding/dissociation to/from DnaA and DnaB helicase loading in vivo are required to understand fully how initiation is timed correctly during the cell cycle.
Intensive research over the last 40 years has increased knowledge of the factors and regulatory mechanisms involved in chromosome replication initiation in E. coli. Our knowledge has exploded, and it is possible today to provide a considerably detailed outline about the factors involved and how replication initiation is controlled. At the same time, such progress creates further important mysteries, too. Further work in this area will undoubtedly continue to surprise and inform us about how replication is controlled in this erstwhile considered simple organism and advance knowledge about how this process is controlled in other organisms as well.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by MEXT/JSPS KAKENHI Grant numbers 17H03656, 16H00775, 15K18479 and 17K07338.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, Y., Jo, T., Matsuda, Y., Matsunaga, C., Katayama, T., and Ueda, T. (2007). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. doi: 10.1074/jbc.M701841200
Ali Azam, T., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370.
Baker, T. A., and Kornberg, A. (1988). Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell 55, 113–123. doi: 10.1016/0092-8674(88)90014-1
Baxter, J. C., and Sutton, M. D. (2012). Evidence for roles of the Escherichia coli Hda protein beyond regulatory inactivation of DnaA. Mol. Microbiol. 85, 648–668. doi: 10.1111/j.1365-2958.2012.08129.x
Bell, S. P., and Kaguni, J. M. (2013). Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5:a010124. doi: 10.1101/cshperspect.a010124
Blaesing, F., Weigel, C., Welzeck, M., and Messer, W. (2000). Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 36, 557–569. doi: 10.1046/j.1365-2958.2000.01881.x
Bochner, B. R., and Ames, B. N. (1982). Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257, 9759–9769.
Bogan, J. A., and Helmstetter, C. E. (1997). DNA sequestration and transcription in the oriC region of Escherichia coli. Mol. Microbiol. 26, 889–896. doi: 10.1046/j.1365-2958.1997.6221989.x
Bramhill, D., and Kornberg, A. (1988). A model for initiation at origins of DNA replication. Cell 54, 915–918. doi: 10.1016/0092-8674(88)90102-X
Cagliero, C., Grand, R. S., Jones, M. B., Jin, D. J., and O’Sullivan, J. M. (2013). Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 41, 6058–6071. doi: 10.1093/nar/gkt325
Campbell, J. L., and Kleckner, N. (1990). E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979. doi: 10.1016/0092-8674(90)90271-F
Charbon, G., Bjørn, L., Mendoza-Chamizo, B., Frimodt-Møller, J., and Løbner-Olesen, A. (2014). Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res. 42, 13228–13241. doi: 10.1093/nar/gku1149
Charbon, G., Campion, C., Chan, S. H. J., Bjørn, L., Weimann, A., da Silva, L. C. N., et al. (2017). Rewiring of energy metabolism promotes viability during hyperreplication stress in E. coli. PLOS Genet. 13:e1006590. doi: 10.1371/journal.pgen.1006590
Chodavarapu, S., Felczak, M. M., and Kaguni, J. M. (2011). Two forms of ribosomal protein L2 of Escherichia coli that inhibit DnaA in DNA replication. Nucleic Acids Res. 39, 4180–4191. doi: 10.1093/nar/gkq120
Chodavarapu, S., Felczak, M. M., Rouvière-Yaniv, J., and Kaguni, J. M. (2008a). Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 67, 781–792. doi: 10.1111/j.1365-2958.2007.06094.x
Chodavarapu, S., Gomez, R., Vicente, M., and Kaguni, J. M. (2008b). Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67, 1331–1346. doi: 10.1111/j.1365-2958.2008.06127.x
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Dueber, E. L. C., Corn, J. E., Bell, S. D., and Berger, J. M. (2007). Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317, 1210–1213. doi: 10.1126/science.1143690
Erzberger, J. P., Pirruccello, M. M., and Berger, J. M. (2002). The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773. doi: 10.1093/emboj/cdf496
Felczak, M. M., and Kaguni, J. M. (2004). The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279, 51156–51162. doi: 10.1074/jbc.M409695200
Felczak, M. M., Simmons, L. A., and Kaguni, J. M. (2005). An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 280, 24627–24633. doi: 10.1074/jbc.M503684200
Fingland, N., Flåtten, I., Downey, C. D., Fossum-Raunehaug, S., Skarstad, K., and Crooke, E. (2012). Depletion of acidic phospholipids influences chromosomal replication in Escherichia coli. Microbiology 1, 450–466. doi: 10.1002/mbo3.46
Flåtten, I., Fossum-Raunehaug, S., Taipale, R., Martinsen, S., and Skarstad, K. (2015). The DnaA protein is not the limiting factor for initiation of replication in Escherichia coli. PLOS Genet. 11:e1005276. doi: 10.1371/journal.pgen.1005276
Frimodt-Møller, J., Charbon, G., Krogfelt, K. A., and Løbner-Olesen, A. (2015). Control regions for chromosome replication are conserved with respect to sequence and location among Escherichia coli strains. Front. Microbiol. 6:1011. doi: 10.3389/fmicb.2015.01011
Frimodt-Møller, J., Charbon, G., Krogfelt, K. A., and Løbner-Olesen, A. (2016). DNA replication control is linked to genomic positioning of control regions in Escherichia coli. PLOS Genet. 12:e1006286. doi: 10.1371/journal.pgen.1006286
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Fujimitsu, K., Senriuchi, T., and Katayama, T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. doi: 10.1101/gad.1775809
Fujimitsu, K., Su’etsugu, M., Yamaguchi, Y., Mazda, K., Fu, N., Kawakami, H., et al. (2008). Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 190, 5368–5381. doi: 10.1128/JB.00044-08
Garner, J., and Crooke, E. (1996). Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 15, 2313–2321.
Grimwade, J. E., Ryan, V. T., and Leonard, A. C. (2000). IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35, 835–844. doi: 10.1046/j.1365-2958.2000.01755.x
Hansen, F. G., Christensen, B. B., and Atlung, T. (2007). Sequence characteristics required for cooperative binding and efficient in vivo titration of the replication initiator protein DnaA in E. coli. J. Mol. Biol. 367, 942–952. doi: 10.1016/j.jmb.2007.01.056
Higgins, C. F., Dorman, C. J., Stirling, D. A., Waddell, L., Booth, I. R., May, G., et al. (1988). A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52, 569–584.
Hwang, D. S., and Kornberg, A. (1992). Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267, 23083–23086.
Inoue, Y., Tanaka, H., Kasho, K., Fujimitsu, K., Oshima, T., and Katayama, T. (2016). Chromosomal location of the DnaA-reactivating sequence DARS2 is important to regulate timely initiation of DNA replication in Escherichia coli. Genes Cells 21, 1015–1023. doi: 10.1111/gtc.12395
Ishida, T., Akimitsu, N., Kashioka, T., Hatano, M., Kubota, T., Ogata, Y., et al. (2004). DiaA, a novel DnaA-binding protein, ensures the initiation timing of E. coli chromosome replication. J. Biol. Chem. 279, 45546–45555. doi: 10.1074/jbc.M402762200
Iyer, L. M., Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31. doi: 10.1016/j.jsb.2003.10.010
Kaguni, J. M. (2011). Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613. doi: 10.1016/j.cbpa.2011.07.016
Kano, Y., and Imamoto, F. (1990). Requirement of integration host factor (IHF) for growth of Escherichia coli deficient in HU protein. Gene 89, 133–137. doi: 10.1016/0378-1119(90)90216-E
Kasho, K., Fujimitsu, K., Matoba, T., Oshima, T., and Katayama, T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. doi: 10.1093/nar/gku1051
Kasho, K., and Katayama, T. (2013). DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941. doi: 10.1073/pnas.1212070110
Kasho, K., Tanaka, H., Sakai, R., and Katayama, T. (2017). Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA-ATP hydrolysis. J. Biol. Chem. 292, 1251–1266. doi: 10.1074/jbc.M116.762815
Katayama, T., Kubota, T., Kurokawa, K., Crooke, E., and Sekimizu, K. (1998). The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94, 61–71.
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Kato, J., and Katayama, T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. doi: 10.1093/emboj/20.15.4253
Kaur, G., Vora, M. P., Czerwonka, C. A., Rozgaja, T. A., Grimwade, J. E., and Leonard, A. C. (2014). Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 91, 1148–1163. doi: 10.1111/mmi.12525
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Kawakami, H., Su’etsugu, M., and Katayama, T. (2006). An isolated Hda-clamp complex is functional in the regulatory inactivation of DnaA and DNA replication. J. Struct. Biol. 156, 220–229. doi: 10.1016/j.jsb.2006.02.007
Keyamura, K., Abe, Y., Higashi, M., Ueda, T., and Katayama, T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. doi: 10.1074/jbc.M109.002717
Keyamura, K., Fujikawa, N., Ishida, T., Ozaki, S., Su’etsugu, M., Fujimitsu, K., et al. (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP-DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099. doi: 10.1101/gad.1561207
Keyamura, K., and Katayama, T. (2011). DnaA protein DNA-binding domain binds to Hda protein to promote inter-AAA+ domain interaction involved in regulatory inactivation of DnaA. J. Biol. Chem. 286, 29336–29346. doi: 10.1074/jbc.M111.233403
Kim, J. S., Nanfara, M. T., Chodavarapu, S., Jin, K. S., Babu, V. M. P., Ghazy, M. A., et al. (2017). Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res. 45, 3888–3905. doi: 10.1093/nar/gkx081
Kitagawa, R., Mitsuki, H., Okazaki, T., and Ogawa, T. (1996). A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19, 1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x
Kitagawa, R., Ozaki, T., Moriya, S., and Ogawa, T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043. doi: 10.1101/gad.12.19.3032
Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K., and Katayama, T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18, 6642–6652. doi: 10.1093/emboj/18.23.6642
Leonard, A. C., and Grimwade, J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. doi: 10.3389/fmicb.2015.00545
Li, S., Zhang, Q., Xu, Z., and Yao, Y. (2017). Acetylation of lysine 243 inhibits the oriC binding ability of DnaA in Escherichia coli. Front. Microbiol. 8:699. doi: 10.3389/fmicb.2017.00699
Lu, M., Campbell, J. L., Boye, E., and Kleckner, N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. doi: 10.1016/0092-8674(94)90156-2
Margulies, C., and Kaguni, J. M. (1998). The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 26, 5170–5175. doi: 10.1093/nar/26.22.5170
McGarry, K. C., Ryan, V. T., Grimwade, J. E., and Leonard, A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. doi: 10.1073/pnas.0400340101
Mercier, R., Petit, M. A., Schbath, S., Robin, S., El Karoui, M., Boccard, F., et al. (2008). The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135, 475–485. doi: 10.1016/j.cell.2008.08.031
Messer, W. (2002). The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374.
Messer, W., and Weigel, C. (1997). DnaA initiator–also a transcription factor. Mol. Microbiol. 24, 1–6. doi: 10.1046/j.1365-2958.1997.3171678.x
Miller, D. T., Grimwade, J. E., Betteridge, T., Rozgaja, T., Torgue, J. J., and Leonard, A. C. (2009). Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U.S.A. 106, 18479–18484. doi: 10.1073/pnas.0909472106
Morigen, Løbner-Olesen, A., and Skarstad, K. (2003). Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50, 349–362. doi: 10.1046/j.1365-2958.2003.03695.x
Morigen, Skarstad, K., and Molina, F. (2005). Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187, 3913–3920. doi: 10.1128/JB.187.12.3913-3920.2005
Morigen, M., Flåtten, I., and Skarstad, K. (2014). The Escherichia coli datA site promotes proper regulation of cell division. Microbiology 160, 703–710. doi: 10.1099/mic.0.074898-0
Mott, M. L., Erzberger, J. P., Coons, M. M., and Berger, J. M. (2008). Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135, 623–634. doi: 10.1016/j.cell.2008.09.058
Nishida, S., Fujimitsu, K., Sekimizu, K., Ohmura, T., Ueda, T., and Katayama, T. (2002). A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277, 14986–14995. doi: 10.1074/jbc.M108303200
Noguchi, Y., and Katayama, T. (2016). The Escherichia coli cryptic prophage protein YfdR binds to DnaA and initiation of chromosomal replication is inhibited by overexpression of the gene cluster yfdQ-yfdR-yfdS-yfdT. Front. Microbiol. 7:239. doi: 10.3389/fmicb.2016.00239
Noguchi, Y., Sakiyama, Y., Kawakami, H., and Katayama, T. (2015). The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312. doi: 10.1074/jbc.M115.662601
Nozaki, S., and Ogawa, T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384. doi: 10.1099/mic.0.2008/019745-0
Nozaki, S., Yamada, Y., and Ogawa, T. (2009). Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells 14, 329–341. doi: 10.1111/j.1365-2443.2008.01269.x
Obita, T., Iwura, T., Su’etsugu, M., Yoshida, Y., Tanaka, Y., Katayama, T., et al. (2002). Determination of the secondary structure in solution of the Escherichia coli DnaA DNA-binding domain. Biochem. Biophys. Res. Commun. 299, 42–48. doi: 10.1016/S0006-291X(02)02590-1
Ogawa, T., Yamada, Y., Kuroda, T., Kishi, T., and Moriya, S. (2002). The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44, 1367–1375. doi: 10.1046/j.1365-2958.2002.02969.x
Okumura, H., Yoshimura, M., Ueki, M., Oshima, T., Ogasawara, N., and Ishikawa, S. (2012). Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 40, 220–234. doi: 10.1093/nar/gkr716
Olliver, A., Saggioro, C., Herrick, J., and Sclavi, B. (2010). DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 76, 1555–1571. doi: 10.1111/j.1365-2958.2010.07185.x
Ozaki, S., and Katayama, T. (2009). DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62, 71–82. doi: 10.1016/j.plasmid.2009.06.003
Ozaki, S., and Katayama, T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. doi: 10.1093/nar/gkr832
Ozaki, S., Kawakami, H., Nakamura, K., Fujikawa, N., Kagawa, W., Park, S.-Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. doi: 10.1074/jbc.M708684200
Ozaki, S., Noguchi, Y., Hayashi, Y., Miyazaki, E., and Katayama, T. (2012a). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. doi: 10.1074/jbc.M112.372052
Ozaki, S., Noguchi, Y., Nishimura, M., and Katayama, T. (2012b). Stable nucleotide binding to DnaA requires a specific glutamic acid residue within the AAA+ box II motif. J. Struct. Biol. 179, 242–250. doi: 10.1016/j.jsb.2012.05.001
Riber, L., Frimodt-Møller, J., Charbon, G., and Løbner-Olesen, A. (2016). Multiple DNA binding proteins contribute to timing of chromosome replication in E. coli. Front. Mol. Biosci. 3:29. doi: 10.3389/fmolb.2016.00029
Riber, L., and Løbner-Olesen, A. (2005). Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. J. Bacteriol. 187, 5605–5613. doi: 10.1128/JB.187.16.5605-5613.2005
Richardson, T. T., Harran, O., and Murray, H. (2016). The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534, 412–416. doi: 10.1038/nature17962
Roth, A., and Messer, W. (1998). High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol. 28, 395–401. doi: 10.1046/j.1365-2958.1998.00813.x
Rozgaja, T. A., Grimwade, J. E., Iqbal, M., Czerwonka, C., Vora, M., and Leonard, A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. doi: 10.1111/j.1365-2958.2011.07827.x
Sakiyama, Y., Kasho, K., Noguchi, Y., Kawakami, H., and Katayama, T. (2017). Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 45, 12354–12373. doi: 10.1093/nar/gkx914
Samitt, C. E., Hansen, F. G., Miller, J. F., and Schaechter, M. (1989). In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 8, 989–993.
Saxena, R., Fingland, N., Patil, D., Sharma, A. K., and Crooke, E. (2013). Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 14, 8517–8537. doi: 10.3390/ijms14048517
Saxena, R., Rozgaja, T., Grimwade, J., and Crooke, E. (2011). Remodeling of nucleoprotein complexes is independent of the nucleotide state of a mutant AAA+ protein. J. Biol. Chem. 286, 33770–33777. doi: 10.1074/jbc.M111.223495
Saxena, R., Vasudevan, S., Patil, D., Ashoura, N., Grimwade, J. E., and Crooke, E. (2015). Nucleotide-induced conformational changes in Escherichia coli DnaA protein are required for bacterial ORC to pre-RC conversion at the chromosomal origin. Int. J. Mol. Sci. 16, 27897–27911. doi: 10.3390/ijms161126064
Schenk, K., Hervás, A. B., Rösch, T. C., Eisemann, M., Schmitt, B. A., Dahlke, S., et al. (2017). Rapid turnover of DnaA at replication origin regions contributes to initiation control of DNA replication. PLOS Genet. 13:e1006561. doi: 10.1371/journal.pgen.1006561
Sekimizu, K., Bramhill, D., and Kornberg, A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. doi: 10.1016/0092-8674(87)90221-2
Sekimizu, K., Bramhill, D., and Kornberg, A. (1988). Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J. Biol. Chem. 263, 7124–7130.
Shimizu, M., Noguchi, Y., Sakiyama, Y., Kawakami, H., Katayama, T., and Takada, S. (2016). Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U.S.A. 113, E8021–E8030. doi: 10.1073/pnas.1609649113
Simmons, L. A., Felczak, M., and Kaguni, J. M. (2003). DnaA Protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 49, 849–858. doi: 10.1046/j.1365-2958.2003.03603.x
Skarstad, K., and Katayama, T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. doi: 10.1101/cshperspect.a012922
Smulczyk-Krawczyszyn, A., Jakimowicz, D., Ruban-Osmialowska, B., Zawilak-Pawlik, A., Majka, J., Chater, K., et al. (2006). Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: from in silico to in vivo studies. J. Bacteriol. 188, 6184–6194. doi: 10.1128/JB.00528-06
Speck, C., Weigel, C., and Messer, W. (1999). ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 18, 6169–6176. doi: 10.1093/emboj/18.21.6169
Stauffer, M. E., and Chazin, W. J. (2004). Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 279, 30915–30918. doi: 10.1074/jbc.R400015200
Su’etsugu, M., Harada, Y., Keyamura, K., Matsunaga, C., Kasho, K., Abe, Y., et al. (2013). The DnaA N-terminal domain interacts with Hda to facilitate replicase clamp-mediated inactivation of DnaA. Environ. Microbiol. 15, 3183–3195. doi: 10.1111/1462-2920.12147
Su’etsugu, M., Nakamura, K., Keyamura, K., Kudo, Y., and Katayama, T. (2008). Hda monomerization by ADP binding promotes replicase clamp-mediated DnaA-ATP hydrolysis. J. Biol. Chem. 283, 36118–36131. doi: 10.1074/jbc.M803158200
Sutton, M. D., Carr, K. M., Vicente, M., and Kaguni, J. M. (1998). Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262. doi: 10.1074/jbc.273.51.34255
Sutton, M. D., and Kaguni, J. M. (1997). Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 272, 23017–23024. doi: 10.1074/jbc.272.37.23017
Swinger, K. K., and Rice, P. A. (2004). IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14, 28–35. doi: 10.1016/j.sbi.2003.12.003
Tesfa-Selase, F., and Drabble, W. T. (1992). Regulation of the gua operon of Escherichia coli by the DnaA protein. Mol. Gen. Genet. 231, 256–264.
Travers, A., and Muskhelishvili, G. (2005). DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 3, 157–169.
von Freiesleben, U., Rasmussen, K. V., Atlung, T., and Hansen, F. G. (2000). Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 37, 1087–1093. doi: 10.1046/j.1365-2958.2000.02060.x
Waldminghaus, T., and Skarstad, K. (2009). The Escherichia coli SeqA protein. Plasmid 61, 141–150. doi: 10.1016/j.plasmid.2009.02.004
Weigel, C., Messer, W., Preiss, S., Welzeck, M., Morigen, and Boye, E. (2001). The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40, 498–507. doi: 10.1046/j.1365-2958.2001.02409.x
Wolański, M., Donczew, R., Zawilak-Pawlik, A., and Zakrzewska-Czerwińska, J. (2015). oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 5:735. doi: 10.3389/fmicb.2014.00735
Yoshida, Y., Obita, T., Kokusho, Y., Ohmura, T., Katayama, T., Ueda, T., et al. (2003). Identification of the region in Escherichia coli DnaA protein required for specific recognition of the DnaA box. Cell. Mol. Life Sci. 60, 1998–2008. doi: 10.1007/s00018-003-3176-7
Zhang, Q., Zhou, A., Li, S., Ni, J., Tao, J., Lu, J., et al. (2016). Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci. Rep. 6:30837. doi: 10.1038/srep30837
Keywords: DnaA, oriC, DARS, datA, chromosome replication
Citation: Katayama T, Kasho K and Kawakami H (2017) The DnaA Cycle in Escherichia coli: Activation, Function and Inactivation of the Initiator Protein. Front. Microbiol. 8:2496. doi: 10.3389/fmicb.2017.02496
Received: 18 October 2017; Accepted: 30 November 2017;
Published: 21 December 2017.
Edited by:
Ariel Amir, Harvard University, United StatesReviewed by:
James Aaron Kraemer, Massachusetts Institute of Technology, United StatesKirsten Skarstad, Oslo University Hospital, Norway
Copyright © 2017 Katayama, Kasho and Kawakami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsutomu Katayama, katayama@phar.kyushu-u.ac.jp
 Tsutomu Katayama
Tsutomu Katayama Kazutoshi Kasho
Kazutoshi Kasho Hironori Kawakami
Hironori Kawakami