- Department of Microbiology and Immunology, University of Minnesota, Minneapolis, MN, United States
Many bacteria regulate gene expression in response to phosphate availability using a two-component signal transduction system, the activity of which is controlled by interaction with the Pst phosphate specific transporter and a cytoplasmic protein PhoU. Mycobacterium tuberculosis, the causative agent of tuberculosis, requires its phosphate sensing signal transduction system for virulence and antibiotic tolerance, but the molecular mechanisms of phosphate sensing remain poorly characterized. M. smegmatis serves as a model for studying mycobacterial pathogens including M. tuberculosis. M. smegmatis encodes two proteins with similarity to PhoU, but it was unknown if both proteins participated in signal transduction with the phosphate-responsive SenX3-RegX3 two-component system. We constructed phoU single and double deletion mutants and tested expression of genes in the RegX3 regulon. Only the ΔphoU1ΔphoU2 mutant exhibited constitutive activation of all the RegX3-regulated genes examined, suggesting that M. smegmatis PhoU1 and PhoU2 have overlapping functions in inhibiting activity of the SenX3-RegX3 two-component system when phosphate is readily available. The ΔphoU1ΔphoU2 mutant also exhibited decreased tolerance to several anti-tubercular drugs. However, a complex plasmid swapping strategy was required to generate the ΔphoU1ΔphoU2 mutant, suggesting that either phoU1 or phoU2 is essential for in vitro growth of M. smegmatis. Using whole-genome sequencing, we demonstrated that all five of the ΔphoU1ΔphoU2 mutants we isolated had independent suppressor mutations predicted to disrupt the function of the Pst phosphate transporter, suggesting that in the absence of the PhoU proteins phosphate uptake by the Pst system is toxic. Collectively, our data demonstrate that the two M. smegmatis PhoU orthologs have overlapping functions in both controlling SenX3-RegX3 activity in response to phosphate availability and regulating phosphate transport by the Pst system. Our results suggest that M. smegmatis can serve as a tractable model for further characterization of the molecular mechanism of phosphate sensing in mycobacteria and to screen for compounds that would interfere with signal transduction and thereby increase the efficacy of existing anti-tubercular antibiotics.
Introduction
In many bacteria, the transcriptional response to phosphate limitation is mediated by a two-component signal transduction system whose activity is controlled by a poorly understood interaction with the Pst (phosphate specific transport) inorganic phosphate (Pi) uptake system (Lamarche et al., 2008; Hsieh and Wanner, 2010). In the model organism Escherichia coli, the two-component system PhoR-PhoB is activated only when the bacteria are Pi starved; in Pi-replete conditions, the Pst Pi uptake system inhibits activation of PhoR-PhoB (Wanner, 1996). Although the precise molecular mechanism of inhibition is not known, null mutations in any component of the Pst system result in constitutive activation of PhoR-PhoB and constitutive expression of the Pi-starvation responsive Pho regulon (Wanner, 1996). Like E. coli, mycobacteria, including the human pathogen Mycobacterium tuberculosis, encode a two-component system SenX3-RegX3 that is activated by Pi limitation and that requires a functional Pst Pi uptake system for Pi sensing (Glover et al., 2007; Rifat et al., 2009; Tischler et al., 2013). In M. tuberculosis and other pathogens, both the Pst system and the two-component system are essential for virulence (Parish et al., 2003; Lamarche et al., 2008; Tischler et al., 2013). It is therefore of interest to understand at the molecular level how the Pst Pi uptake and two-component signal transduction systems interact, since small molecules targeting this interaction could have robust and broad spectrum antimicrobial activity.
In the E. coli model, an additional protein named PhoU is involved in Pi sensing. PhoU is not required for Pi uptake by the Pst system (Steed and Wanner, 1993), though it may regulate Pst system Pi transport activity (Rice et al., 2009). Mutation of phoU, like mutation of any component of the Pst system, results in constitutive activation of the PhoR-PhoB two-component system and expression of the Pho regulon in Pi-replete conditions (Li and Zhang, 2007). Recent evidence suggests that PhoU communicates signals from the Pst system to PhoR-PhoB via direct protein-protein interactions between PhoU and PstB, the Pst system ATPase, and between PhoU and the PhoR sensor kinase (Gardner et al., 2014). phoU mutants in E. coli and other bacterial species accumulate polyphosphate (PolyP), a linear polymer of Pi linked by high energy phosphoanhydride bonds, suggesting a possible role of PhoU in regulation of cellular Pi metabolism (Morohoshi et al., 2002; de Almeida et al., 2015; Lubin et al., 2016). Finally, E. coli phoU is necessary for the development of persisters, a sub-population that is phenotypically tolerant to antibiotics (Li and Zhang, 2007; Maissonneuve and Gerdes, 2014). However, the molecular mechanism by which PhoU promotes persister formation is unknown.
Mycobacteria are unusual in that they encode multiple copies of genes with similarity to E. coli phoU. In M. tuberculosis, the phoU orthologs were named phoY1 (rv3301c) and phoY2 (rv0821c). M. tuberculosis may have evolved two PhoU orthologs since it also encodes two complete Pst Pi uptake systems (Braibant et al., 2000), though only one of these systems functions in Pi sensing and signal transduction with SenX3-RegX3 (Tischler et al., 2013, 2016). In contrast, the fast-growing saprophytic species Mycobacterium smegmatis has a single Pst system, but encodes two phoU orthologs. One ortholog, which we have named phoU1 (Msmeg_5776), is encoded adjacent to the M. smegmatis pstSCAB operon. The second ortholog, which we have named phoU2 (Msmeg_1605), is encoded in a region of the genome distant from this locus. The PhoU1 and PhoU2 proteins exhibit 82% sequence similarity and are both encoded separately from the SenX3-RegX3 two-component system (Msmeg_0936 and Msmeg_0937). Previous reports have suggested that PhoY2 of M. tuberculosis is important for development of antibiotic tolerant persister variants (Shi and Zhang, 2010) and that PhoY2 in the related pathogen Mycobacterium marinum influences Pi homeostasis, energy and redox balance (Wang et al., 2013). However, we recently demonstrated that M. tuberculosis PhoY1 and PhoY2 function redundantly to control activity of the SenX3-RegX3 system and promote persister formation (Namugenyi et al., 2017). It is not known which of the M. smegmatis PhoU orthologs participates in Pi sensing and signal transduction or whether they contribute to antibiotic tolerance.
We hypothesized that the M. smegmatis PhoU orthologs would exhibit redundant function in transcriptional regulation and antibiotic tolerance, similar to the M. tuberculosis PhoY proteins. We generated deletions of phoU1 and phoU2 in M. smegmatis and evaluated expression of RegX3-regulated genes and antibiotic sensitivity. We found that it was necessary to delete both phoU1 and phoU2 to observe complete activation of the RegX3 regulon in Pi-rich conditions, suggesting partial functional redundancy between M. smegmatis PhoU1 and PhoU2. However, the ΔphoU1ΔphoU2 mutant was difficult to construct suggesting that PhoU1 or PhoU2 are jointly essential for M. smegmatis viability. Using whole-genome sequencing, we demonstrated that all five of the ΔphoU1ΔphoU2 mutants that we isolated have independent mutations predicted to disrupt the function of the Pst transporter. Collectively, our data suggest that the M. smegmatis PhoU proteins have overlapping functions in both controlling activation of SenX3-RegX3 and regulating Pi transport by the Pst system.
Materials and Methods
Bacterial Culture Conditions
M. smegmatis mc2155 and derivative strains were grown at 37°C in Middlebrook 7H9 (Difco) liquid culture medium supplemented with 10% albumin-dextrose-saline (ADS), 0.5% glycerol and 0.1% Tween-80 or on Luria Broth (LB) agar solid culture medium. Frozen stocks were prepared by growing cultures to mid-exponential phase (OD600 of 0.6–0.8), adding glycerol to 15% final concentration, and storing aliquots at -80°C. Antibiotics were used at the following concentrations: kanamycin (Kan) 15 μg/ml, hygromycin (Hyg) 50 μg/ml, rifampicin (RIF) 200 μg/ml, isoniazid (INH) 50 μg/ml, ethionamide (ETH) 200 μg/ml, ethambutol (ETB) 5 μg/ml.
Cloning
All plasmids used for cloning and strain construction are listed in Supplementary Table S1. Constructs for deletion of phoU1 (Msmeg_5776) or phoU2 (Msmeg_1605) in M. smegmatis were generated in the allelic exchange vector pJG1100 (Kirksey et al., 2011). Genomic regions 500–800 bp upstream and downstream of the genes to be deleted were PCR-amplified from M. smegmatis mc2155 genomic DNA using the oligonucleotides listed in Supplementary Table S2. Reverse primers for amplification of the upstream regions were designed with an AvrII restriction site in-frame with the translation start codon; corresponding forward primers for amplification of the downstream regions were designed with an AvrII restriction site in-frame with the stop codon. PCR products were cloned in pCR2.1-TOPO (Invitrogen) and sequenced. Upstream and downstream regions were removed from pCR2.1 by restriction with PacI/AvrII and AvrII/AscI, respectively and ligated together in pJG1100 between the PacI and AscI sites to generate the in-frame deletion constructs pBE101 (ΔphoU1), pBE102 (ΔphoU2).
Vectors for complementation of the phoU deletions were constructed in the episomal plasmid pMV261 under the control of the vector-encoded strong constitutive hsp60 promoter. The M. smegmatis phoU1 and phoU2 genes were PCR-amplified using the primers indicated in Supplementary Table S2, cloned in pCR2.1-TOPO and sequenced. The cloned genes were removed from pCR2.1 by restriction with EcoRI and HindIII and ligated in similarly digested pMV261 to generate pMVphoU1 and pMVphoU2.
For construction of Tet-inducible phoU1, the phoU1 gene was PCR-amplified using the primers indicated in Supplementary Table S2, cloned in pCR2.1-TOPO and sequenced. phoU1 was removed from pCR2.1 by restriction with HindIII and EcoRI and ligated in similarly digested pTIC10a or pJT6a to generate pTICphoU1 and pJTphoU1.
Strain Construction
M. smegmatis ΔphoU1 and ΔphoU2 deletion mutants were generated by a two-step homologous recombination method of allelic exchange, as previously described (Upton and McKinney, 2007) except that LB agar medium containing Kan and Hyg and LB agar medium containing 5% sucrose were used for selection of transformants and counter-selection of the pJG1100 vector, respectively. Integration of the vectors was confirmed with the following primer pairs, listed in Supplementary Table S2: ΔphoU1 upstream 5776F3/5776R4; ΔphoU1 downstream 5776F4/5776R3; ΔphoU2 upstream 1605F3/1605R4; ΔphoU2 downstream 1605F4/1605R3. Identification of deletion mutants was done with the following primer pairs: ΔphoU1 5776F3/5776R3; ΔphoU2 1605F3/1605R3. Complemented strains were constructed by electroporating the corresponding deletion mutant with the complementation plasmid pMVphoU1 or pMVphoU2, and selecting on LB containing Kan.
For construction of the ΔphoU1ΔphoU2 double deletion mutant, the ΔphoU2 mutant was electroporated with pTICphoU1 and transformants were selected on LB containing Kan. The resulting strain was then electroporated with the pBE101 (ΔphoU1) allelic exchange vector and transformants were selected on LB containing Kan and Hyg. Integration of the ΔphoU1 vector in the transformants was confirmed by PCR as described above. The resulting strain was then grown overnight in 7H9 medium containing 50 ng/ml anhydrotetracycline (ATc) to induce expression from the pTICphoU1 construct and subsequently plated on LB containing 5% sucrose and 100 ng/ml ATc to counterselect the pBE101 allelic exchange vector. Individual HygS colonies were tested for the ΔphoU1 deletion by PCR using the 5776F3/5776R3 primer pair. The resulting ΔphoU1ΔphoU2pTICphoU1 strain was then electroporated with pJT6a and transformants were selected on LB containing Hyg. Individual HygR transformants were patched to LB Kan and LB Hyg. Those that were HygR and KanS were selected for PCR analysis with primers pTfor and pTIC6a_R that amplify a 1.3 kbp fragment from pTICphoU1 and a 500 bp fragment from pJT6a. Isolates from which only the 500 bp product was amplified were tested for the deletions of phoU1 and phoU2 by PCR as described above.
Growth Curves
Cultures of M. smegmatis grown in complete 7H9 medium to mid-logarithmic phase were diluted to OD600 0.01 in 7H9 and 200 μl were inoculated in a 96 well plate (polystyrene rounded square well, Fisherbrand). Cultures were incubated at 37°C with shaking and OD600 was monitored hourly for 36 h using a Synergy H1 hybrid reader (BioTek).
Analysis of Clumping Phenotype
Cultures of M. smegmatis were grown at 37°C with shaking in either 14 ml polystyrene snap-cap tubes (Falcon) in 5 ml complete 7H9 medium to assess pellicle formation or a polystyrene flat-bottom 12-well plate (Falcon) in 4 ml complete 7H9 medium to test clumping. After 72 h of incubation, the tubes and plate were imaged.
Determination of Colony Sizes
Isolated single colonies of M. smegmatis were grown on LB agar for 4 days at 37°C. Plates were imaged with a FluorChem FC3 (Cell Biosciences) using the white light setting. Colony area for 50 representative colonies was measured on each of three plates grown from independent biological replicates using ImageJ.
Quantitative RT-PCR (qRT-PCR)
Bacteria were grown to mid-exponential phase (OD600 0.5) in 7H9 broth and RNA was extracted as described (Tischler et al., 2013). Equivalent amounts of total RNA were treated with Turbo DNase (Ambion) and reverse transcribed to cDNA with the Transcriptor First Strand cDNA Synthesis Kit (Roche) using random hexamers for priming and the following cycling conditions: 10 min at 25°C for annealing, 60 min at 50°C for extension, 5 min at 85°C for heat inactivation. cDNA was stored at -20°C until real-time PCR reactions were performed. Quantitative Real Time PCR reactions were prepared with 2× FastStart Sybr Green Master Mix (Roche), 2 μl cDNA, and 0.3 μM primers and were run in absolute quantification mode on a LightCycler 480 (Roche). PCR cycling conditions were: 95°C 10 min; 45 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 20 s with data collection once per cycle during the extension phase; one cycle of 95°C for 5 s, 60°C for 1 min, 97°C for 15 s with a 0.11°C/s ramp rate during the final denaturation to generate melting curves for confirmation of product specificity. Mock reactions (no RT) were performed on each sample to confirm the absence of genomic DNA contamination. Cp values were converted to copy numbers using standard curves for each gene. Target cDNA was internally normalized to sigA cDNA. Primers for real-time quantitative reverse transcriptase (RT) PCR used in this study are listed in Supplementary Table S3. Primers were designed using the Roche online Universal ProbeLibrary assay design center tool and were tested in standard PCR reactions using 100 M. smegmatis genome equivalents as template.
Persister Assay
Cultures of M. smegmatis grown in complete 7H9 medium overnight to late-logarithmic phase were diluted to OD600 0.05 in 20 ml complete 7H9 and antibiotics were added. Cultures were incubated at 37°C with aeration and viable CFU were determined at 0, 8, 24, 48, and 72 h by plating serially diluted cultures on LB agar medium. CFU were enumerated after 3–4 days of incubation at 37°C.
Minimal Inhibitory Concentration (MIC) Assay
Cultures of M. smegmatis grown in complete 7H9 medium to mid-logarithmic phase (OD600 of 0.5) were diluted to OD600 of 0.002 in fresh complete 7H9 medium. Antibiotics were added to 2 ml aliquots of the dilute culture in twofold decreasing concentrations with the highest concentration at 10x the MIC for wild-type M. smegmatis mc2155 (RIF 200 μg/ml, INH 50 μg/ml, EMB 5 μg/ml, ETH 200 μg/ml). Cultures were incubated for 48 h at 37°C with shaking and the OD600 was determined. The MIC90 was the concentration of antibiotic that inhibited 90% of the growth of a no drug control.
Alkaline Phosphatase Assay
Cultures of M. smegmatis were grown in complete 7H9 medium to mid-logarithmic phase (OD600 0.4–0.8) and centrifuged at low speed (150 × g for 5 min) to remove large clumps. The OD600 of the de-clumped culture was determined, and 0.5 ml of bacteria from this culture were pelleted by centrifugation (16000 × g for 10 min). The culture supernatant was removed and bacteria were resuspended in 0.1 ml 1M Tris buffer, pH 8.1 ml of 2 mM p-nitrophenyl phosphate was added and samples were incubated at 37°C in the dark for 10 min. Bacteria were removed by centrifugation and 1 ml of the supernatant was transferred to a cuvette. The OD420 of the sample was measured using 2 mM p-nitrophenyl phosphate as a blank. Alkaline phosphate units were calculated as (1000 × OD420)/(minutes of incubation × OD600 × 0.5 ml).
Polyphosphate Extraction and Quantification
Cultures of M. smegmatis were grown to mid-logarithmic phase (OD600 0.5–1.0) in complete 7H9. Cells from 20 ml of culture were pelleted (4700 × g for 15 min), then resuspended and lysed in 4 M guanidine isothiocyanate, 50 mM Tris-HCl (pH 7) at 95°C for 30 min. Total protein was quantified using 10 μl of the lysed sample (Pierce BCA Protein Concentration Assay, Thermo Scientific). PolyP was isolated from the remaining cell lysate using glassmilk (GeneClean) and quantified by binding to toluidine blue O (Sigma) dye solution (6 mg/L in 40 mM acetic acid) as previously described (Namugenyi et al., 2017).
Genomic DNA Extraction
M. smegmatis strains were grown to mid-logarithmic phase (OD600 0.5–0.8) and bacteria from 20 ml of culture were pelleted by centrifugation (2850 × g for 10 min). Genomic DNA was extracted by the cetyltrimethylammonium bromide (CTAB) – lysozyme method as described (Larsen et al., 2007) except that bacteria were lysed by incubation with lysozyme for 3 h at 37°C. DNA was resuspended in 100 μl TE buffer and stored at 4°C overnight. Concentration and purity of the genomic DNA were determined using a Nanodrop spectrophotometer (Thermo Scientific).
Whole-Genome Sequencing
Genomic DNA extracted from M. smegmatis mc2155 and five ΔphoU1ΔphoU2 isolates was diluted to 12 ng/μl in 25 μl DEPC-treated water and submitted to the University of Minnesota Genomics Center. Whole-genome sequencing libraries were created using the TruSeq Nano Library Preparation Kit (Illumina). Libraries were processed using the 350 bp shearing protocol according to the manufacturer’s instructions. Libraries were multiplexed and sequenced on 0.25 lane of a HiSeq 2500 (Illumina) in high-output mode using the cBot HiSeq PE Cluster Kit v4 (Illumina) to generate clusters, and HiSeq SBS Kit, v4 chemistry (Illumina) with paired-end 125 bp reads. Image analysis and base calling were done using the Illumina software package RTA v 1.18.62. De-multiplexing and fastq file creation were performed using bcl2fastq v2.17.1.14. Total sequence yields (in Gb) were: mc2155 = 13.6; ΔphoU1ΔphoU2 #504 = 5.1; ΔphoU1ΔphoU2 #518 = 9.1; ΔphoU1ΔphoU2 #521 = 4.4; ΔphoU1ΔphoU2 #625 = 8.4; ΔphoU1ΔphoU2 #664 = 4.8. Sequences from mc2155 were aligned to the reference M. smegmatis mc2155 sequence in Geneious version 10.0.9 software1 (Kearse et al., 2012) using the Geneious mapper to generate a Tischler lab WT mc2155 consensus sequence. Sequences from each of the five ΔphoU1ΔphoU2 mutants were then aligned to the WT mc2155 consensus using the Geneious mapper to identify variants. Single nucleotide polymorphisms (SNPs) and short duplications in Msmeg_1387, pstS, pstC, and pstB were confirmed by PCR amplification and sequencing using the primers listed in Supplementary Table S2.
Statistical Analysis
Student’s unpaired t-test (two-tailed) was used for pairwise comparisons between WT and mutant strains of M. tuberculosis. P values were calculated using GraphPad Prism 5.0 software (GraphPad Software, Inc.). P values < 0.05 were considered significant.
Results
M. smegmatis Encodes Two PhoU Orthologs That Function Redundantly in Signal Transduction
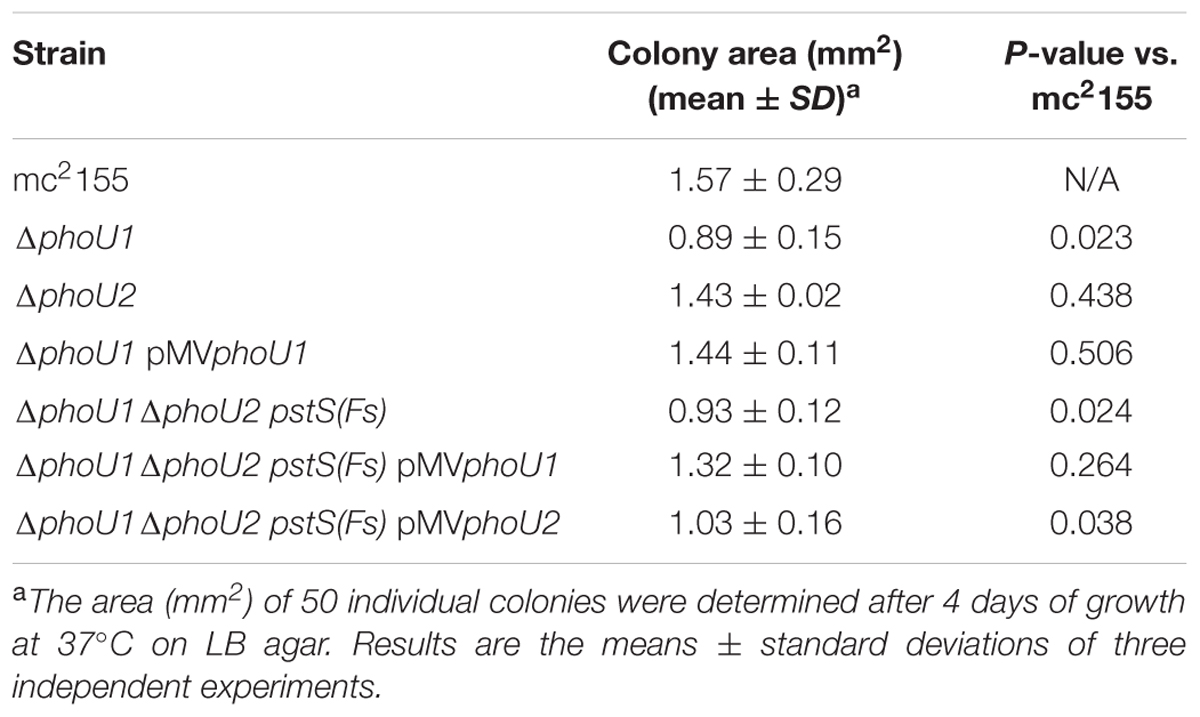
M. smegmatis encodes two putative orthologs of E. coli PhoU. To determine which of these proteins negatively regulates the M. smegmatis SenX3-RegX3 system when Pi is abundant, we constructed in-frame unmarked deletions of phoU1 (Msmeg_5776) and phoU2 (Msmeg_1605) by two-step allelic exchange (Upton and McKinney, 2007). The ΔphoU1 and ΔphoU2 deletions were confirmed by PCR (data not shown). Growth of each deletion mutant was monitored in complete 7H9 medium, which is Pi-rich. The ΔphoU2 mutant exhibited no change in replication rate or overall growth yield compared to wild-type (WT) M. smegmatis mc2155 in this medium (Figures 1A,B). Although the ΔphoU1 mutant grew at a similar rate as the WT control, it did not reach the same maximal optical density (Figure 1A). In addition, the ΔphoU1 mutant lost viability in stationary phase, with significantly fewer CFU recovered from cultures grown for 36 h as compared to the WT control (Figure 1B). The ΔphoU1 mutant also formed significantly smaller colonies than WT M. smegmatis on LB agar plates (Table 1). Colonies of the ΔphoU1 mutant had a sticky phenotype. The ΔphoU1 mutant also exhibited clumping phenotypes in liquid culture; when grown in tubes it formed a pellicle at the air-liquid interface (Figure 2A) and when grown in 12-well plates it formed large visible clumps with fewer dispersed cells compared to the WT control (Figure 2B) even in complete 7H9 medium, which contains Tween-80. Clumping may contribute to the reduced growth yield of the ΔphoU1 mutant. The growth yield, stationary phase viability, colony size and clumping phenotypes of the ΔphoU1 mutant were all complemented by providing the phoU1 gene in trans on pMVphoU1 (Figures 1A,B, 2 and Table 1), confirming that the phoU1 deletion is responsible for these phenotypes. These phenotypes were also unique to the ΔphoU1 mutant, as the ΔphoU2 deletion did not alter colony size (Table 1) or clumping in culture (Figure 2).
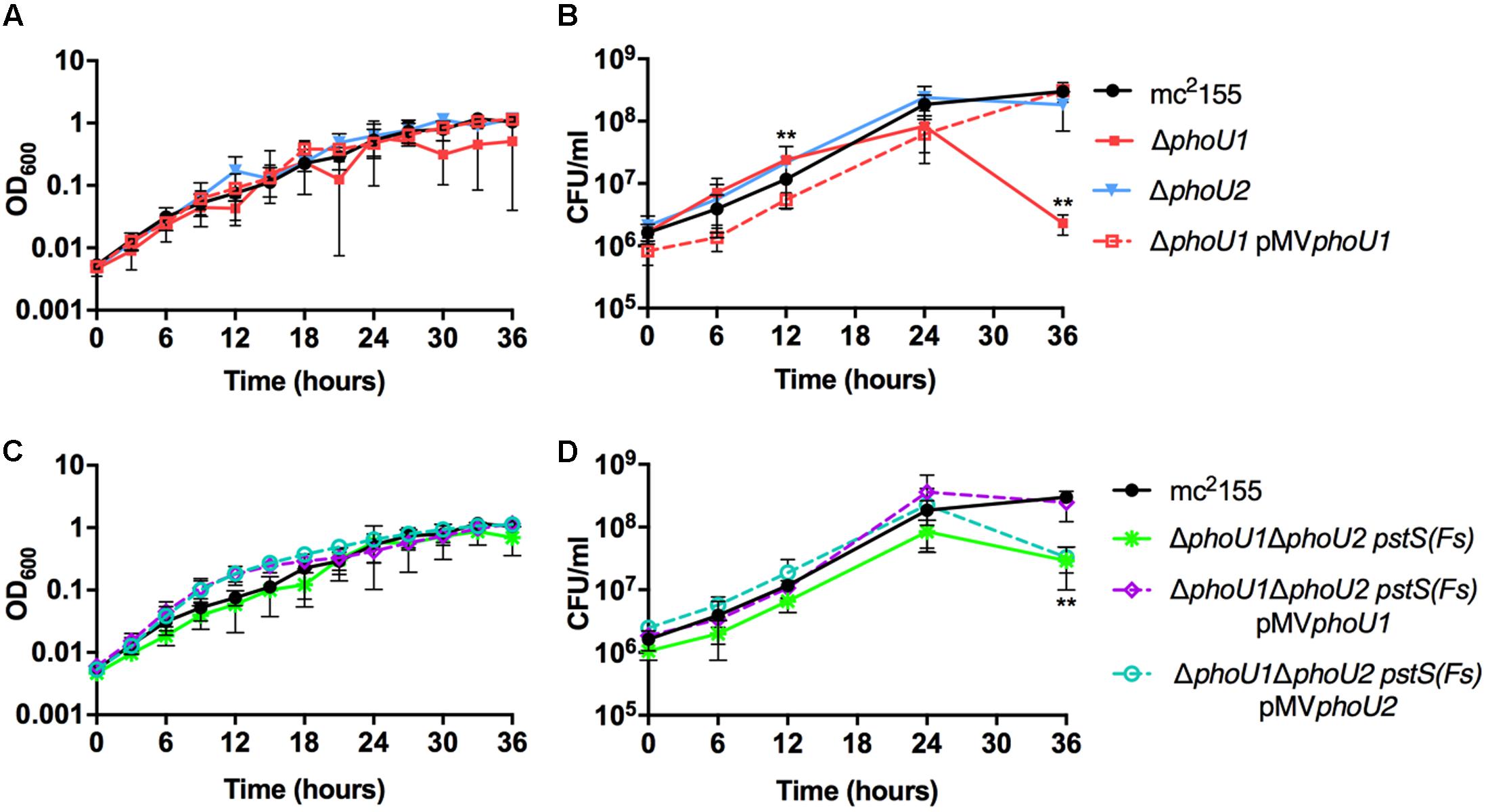
FIGURE 1. Growth curves of Mycobacterium smegmatis phoU deletion mutants. The indicated M. smegmatis strains were grown to mid-logarithmic phase and inoculated into complete 7H9 liquid medium at an OD600 of 0.01. Cultures were incubated at 37°C with shaking. (A,C) Cultures were incubated in a 96 well plate and the OD600 was measured hourly (OD600 values for every 3 h are shown). (B,D) Viable CFU were determined by plating serial dilutions of culture on LB agar. For all panels, results are the mean of three biological replicates ± standard deviations. Asterisks indicate statistically significant differences for ΔphoU1 (B) and ΔphoU1ΔphoU2 pstS(Fs) (D) compared to the mc2155 control: ∗∗P < 0.005.
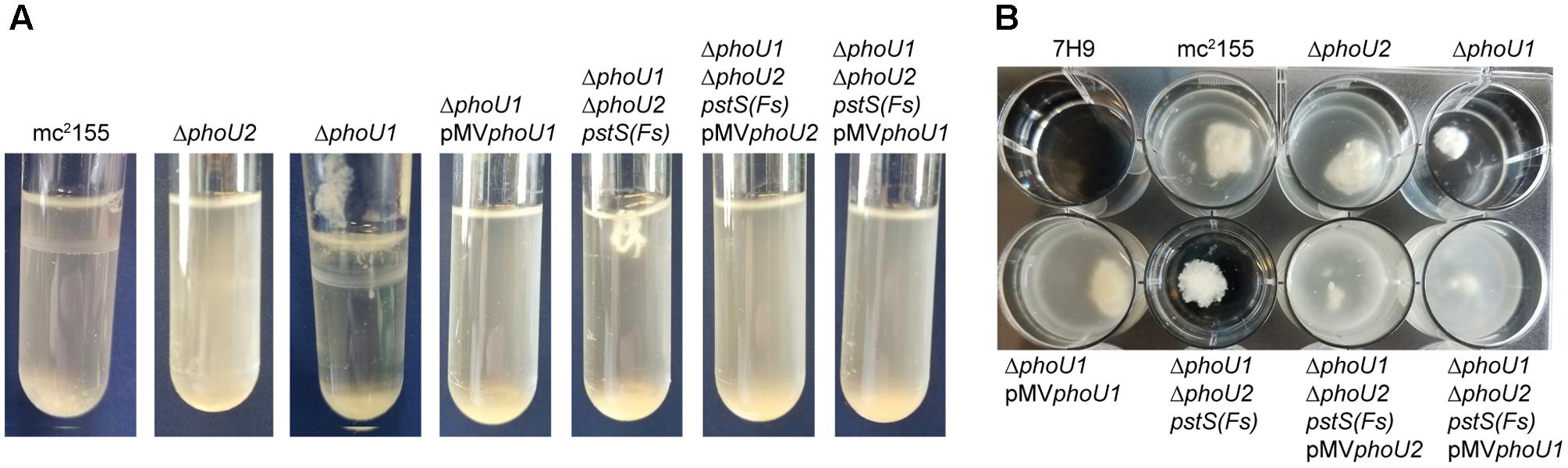
FIGURE 2. The ΔphoU1 and ΔphoU1ΔphoU2 pstS(Fs) mutants have clumping phenotypes in liquid culture. The indicated M. smegmatis strains were inoculated into complete 7H9 medium and grown at 37°C with shaking for 72 h in (A) tubes or (B) a 12-well plate and representative cultures were imaged.
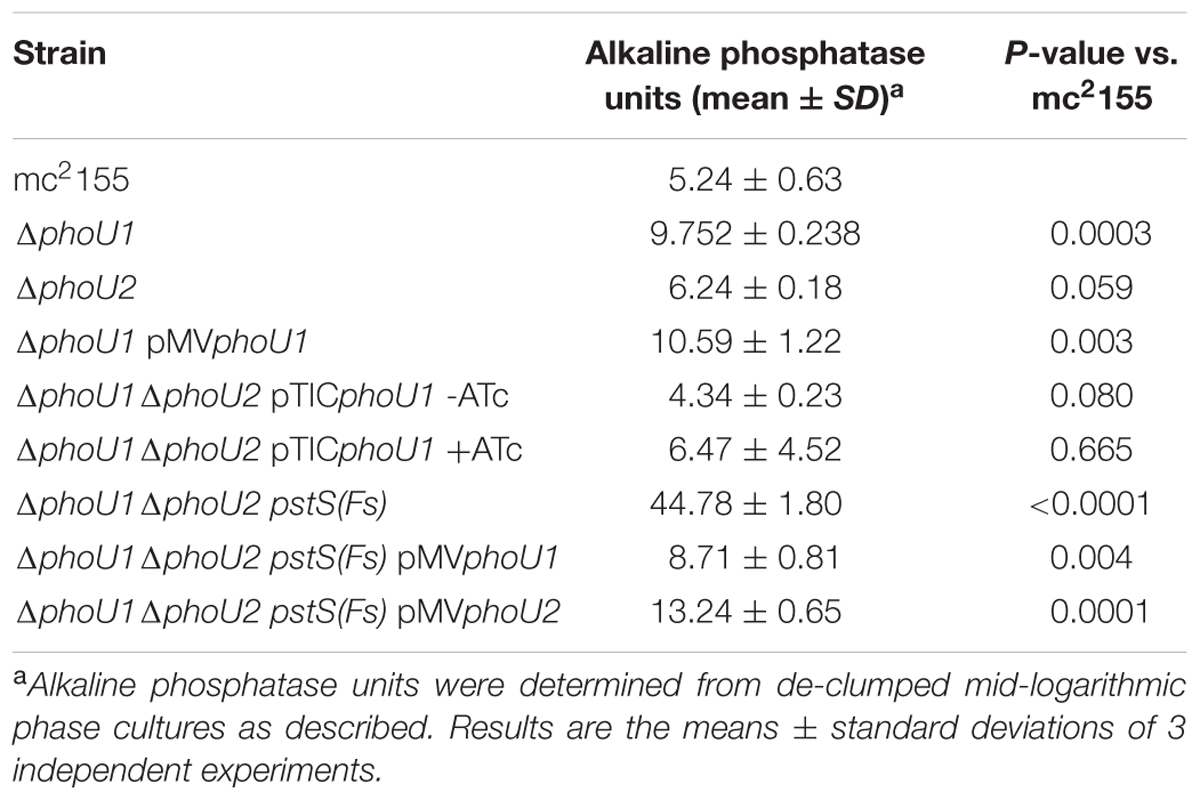
To determine if either phoU single deletion influences activity of SenX3-RegX3, we performed quantitative RT-PCR experiments on RNA extracted from M. smegmatis strains grown in Pi-rich 7H9 medium. We chose 3 known RegX3-dependent genes for analysis: regX3, pstS, which encodes the substrate-binding protein of the Pst system, and phoA, which encodes alkaline phosphatase (Glover et al., 2007). As controls, we tested expression of the phoU1 and phoU2 transcripts. The phoU1 and phoU2 transcripts were undetectable in the corresponding deletion mutant strains, verifying that the genes were deleted (Figure 3). Transcription of regX3, pstS, and phoA was not significantly altered by deletion of phoU2 (Figure 4). Similarly, the ΔphoU1 mutant showed no significant change in expression of either regX3 or phoA (Figures 4A,C). Consistent with these results, alkaline phosphatase activity was unchanged in the ΔphoU2 mutant and was increased less than twofold in the ΔphoU1 mutant (Table 2). Increased alkaline phosphatase activity of the ΔphoU1 mutant was not complemented by the pMVphoU1 plasmid (Table 2). In contrast, transcription of pstS was significantly increased in the ΔphoU1 mutant compared to the mc2155 control and this phenotype was complemented by pMVphoU1 (Figure 4B), suggesting that the phoU1 deletion is responsible for this phenotype. Because neither regX3 nor phoA transcription were altered in either the ΔphoU1 or ΔphoU2 mutants, our data suggest either that PhoU1 and PhoU2 have partially redundant function in regulating the activity of SenX3-RegX3, or that additional factors contribute to controlling SenX3-RegX3 activity in M. smegmatis.
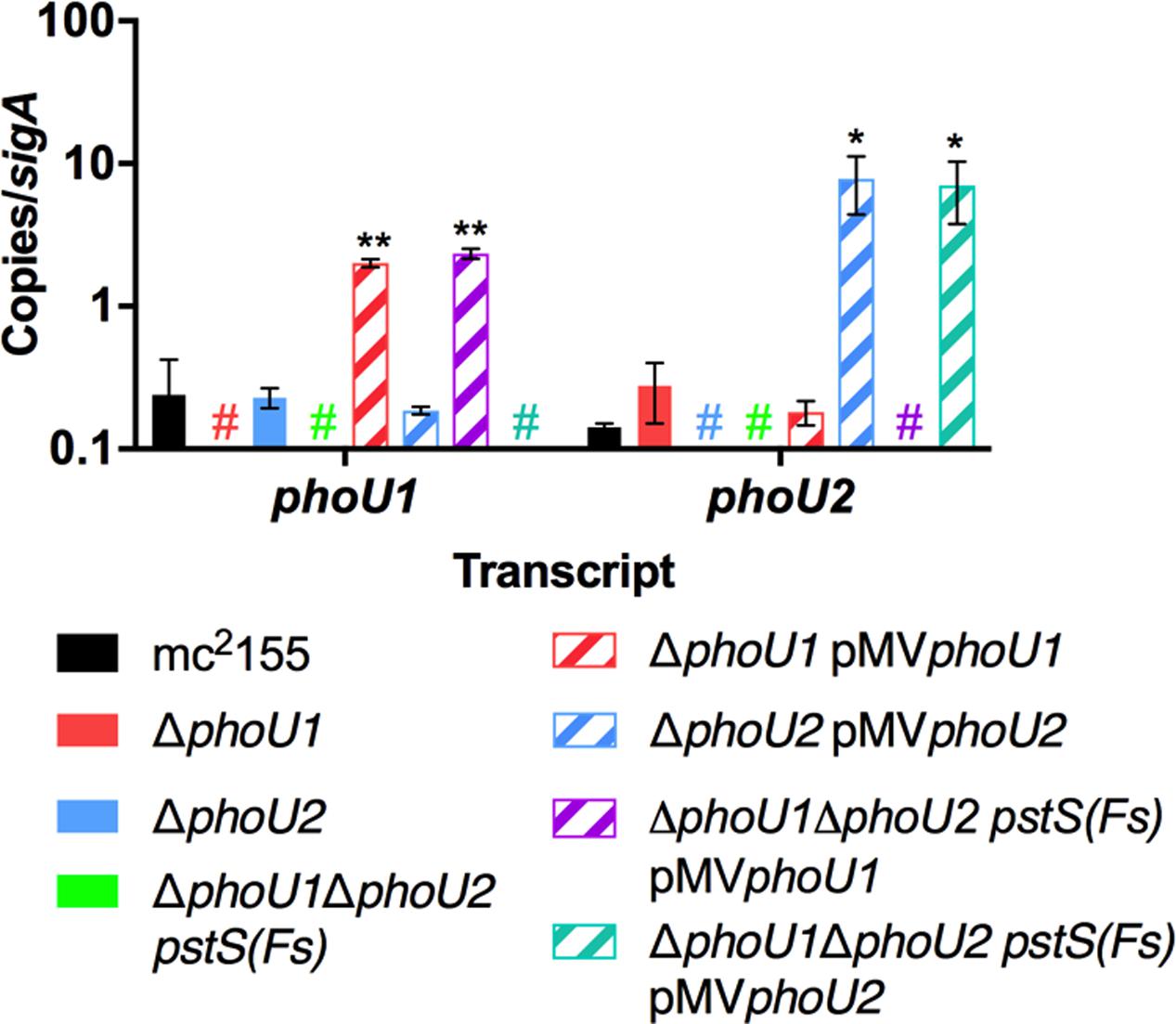
FIGURE 3. Verification of phoU deletion and complemented strains. RNA was extracted from the indicated strains grown in complete 7H9 medium to mid-logarithmic phase. Quantitative RT-PCR was performed to determine abundance of the phoU1 and phoU2 transcripts relative to the sigA housekeeping control. Results are the mean of three biological replicates ± standard deviations. Asterisks indicate statistically significant differences compared to the mc2155 control: ∗P < 0.05, ∗∗P < 0.005. Colored # indicates that no transcript was detected in the corresponding deletion mutant.
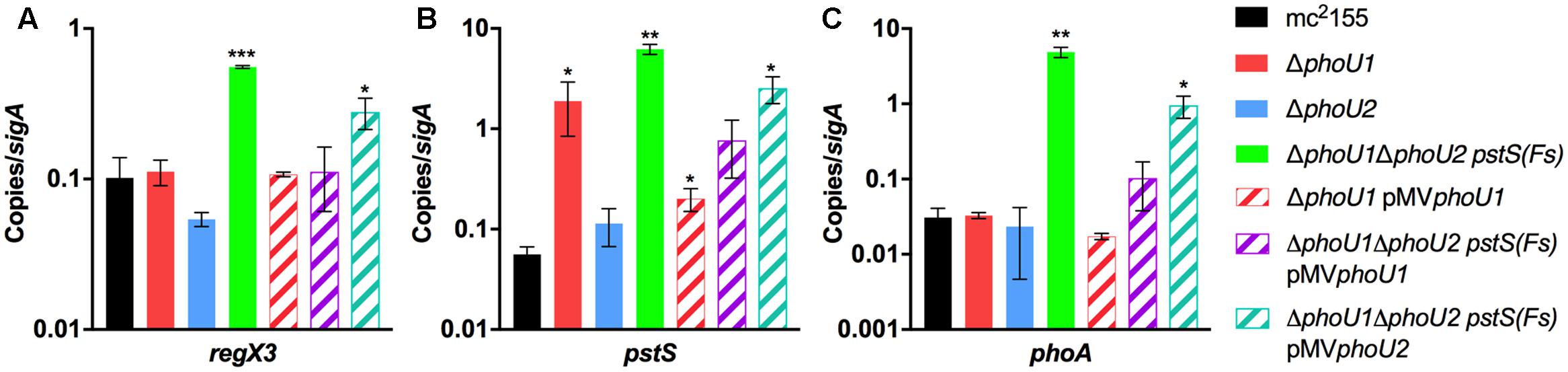
FIGURE 4. Deletion of both phoU orthologs causes aberrant expression of RegX3-regulated genes. RNA was extracted from the indicated M. smegmatis strains grown in complete 7H9 medium to mid-logarithmic phase. Quantitative RT-PCR was performed to determine abundance of the regX3 (A), pstS (B), and phoA (C) transcripts relative to the sigA housekeeping control. Results are the mean of three biological replicates ± standard deviations. Asterisks indicate statistically significant differences compared to the mc2155 control: ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.0001.
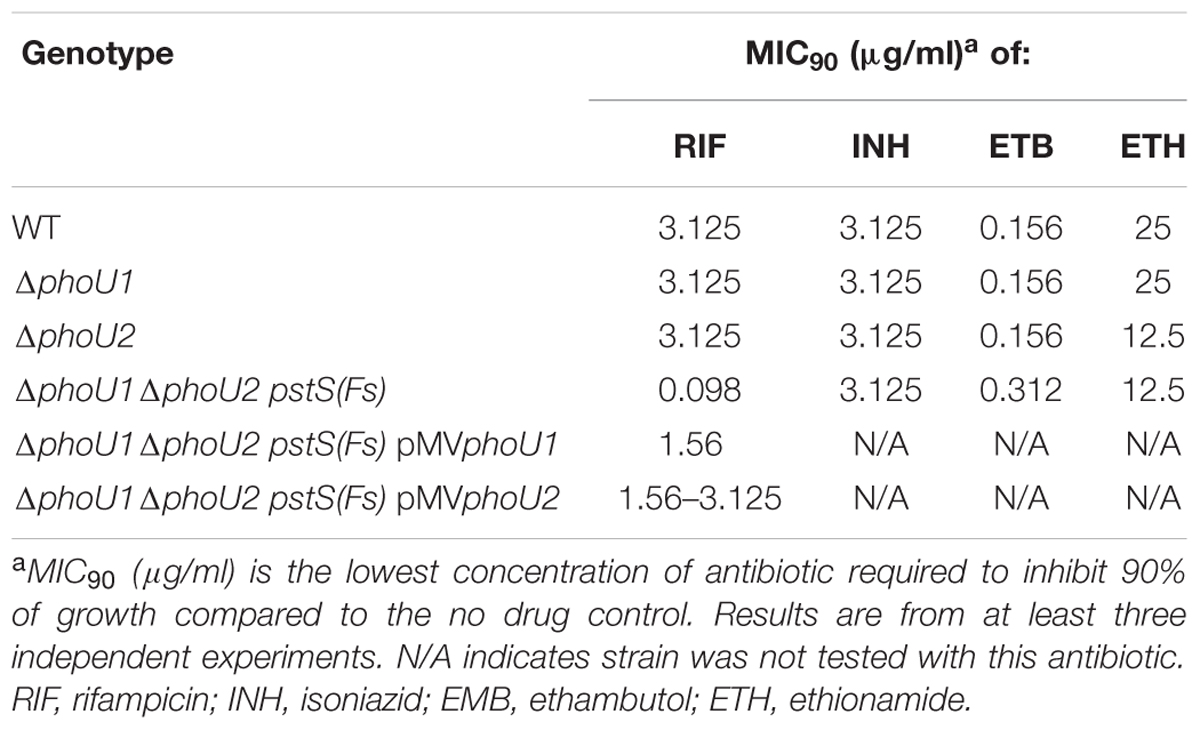
The ΔphoU1 and ΔphoU2 Mutants Exhibit Decreased Tolerance to Antibiotics Targeting the Cell Wall
In E. coli, PhoU has previously been implicated as a “persister switch” that is required for formation of antibiotic-tolerant persister variants (Li and Zhang, 2007). Similarly M. tuberculosis requires either PhoY1 or PhoY2 to promote persister formation (Namugenyi et al., 2017). To establish if M. smegmatis similarly requires PhoU1 or PhoU2 for antibiotic tolerance, we determined the minimal inhibitory concentrations (MICs) of the anti-tubercular drugs rifampicin (RIF), isoniazid (INH), ethambutol (ETB), or ethionamide (ETH) against these mutants. Neither the ΔphoU1 nor the ΔphoU2 mutant displayed a substantial change in susceptibility to these drugs (Table 3). We also analyzed the rates of death of these mutants following antibiotic treatment to assess persister formation. For the ΔphoU2 mutant, we observed little change in the kinetics of bacterial death compared to the WT mc2155 control during treatment with EMB or ETH (Figures 5C,D). The ΔphoU2 mutant exhibited modestly reduced tolerance to RIF at 24 h compared to the WT control, though this difference was not maintained at later time points (Figure 5A). INH killed the ΔphoU2 mutant more rapidly than mc2155, and this phenotype was partially complemented by the pMVphoU2 plasmid (Figure 5B). The ΔphoU1 mutant was equally susceptible as the mc2155 control to RIF (Figure 5A). The ΔphoU1 mutant showed slightly increased susceptibility to INH at 48 h (P = 0.035) that was partially complemented by pMVphoU1 (Figure 5B). We also observed modest increases in the death rate of the ΔphoU1 mutant exposed to either EMB or ETH, though the differences compared to the WT control were significant only for ETH at 24 h (Figures 5C,D). These phenotypes were both complemented by pMVphoU1 (Figures 5C,D). These data suggest that PhoU1 and PhoU2 may have independent effects on M. smegmatis physiology that influence tolerance to the cell wall targeting antibiotics INH and ETH and to the transcriptional inhibitor RIF.
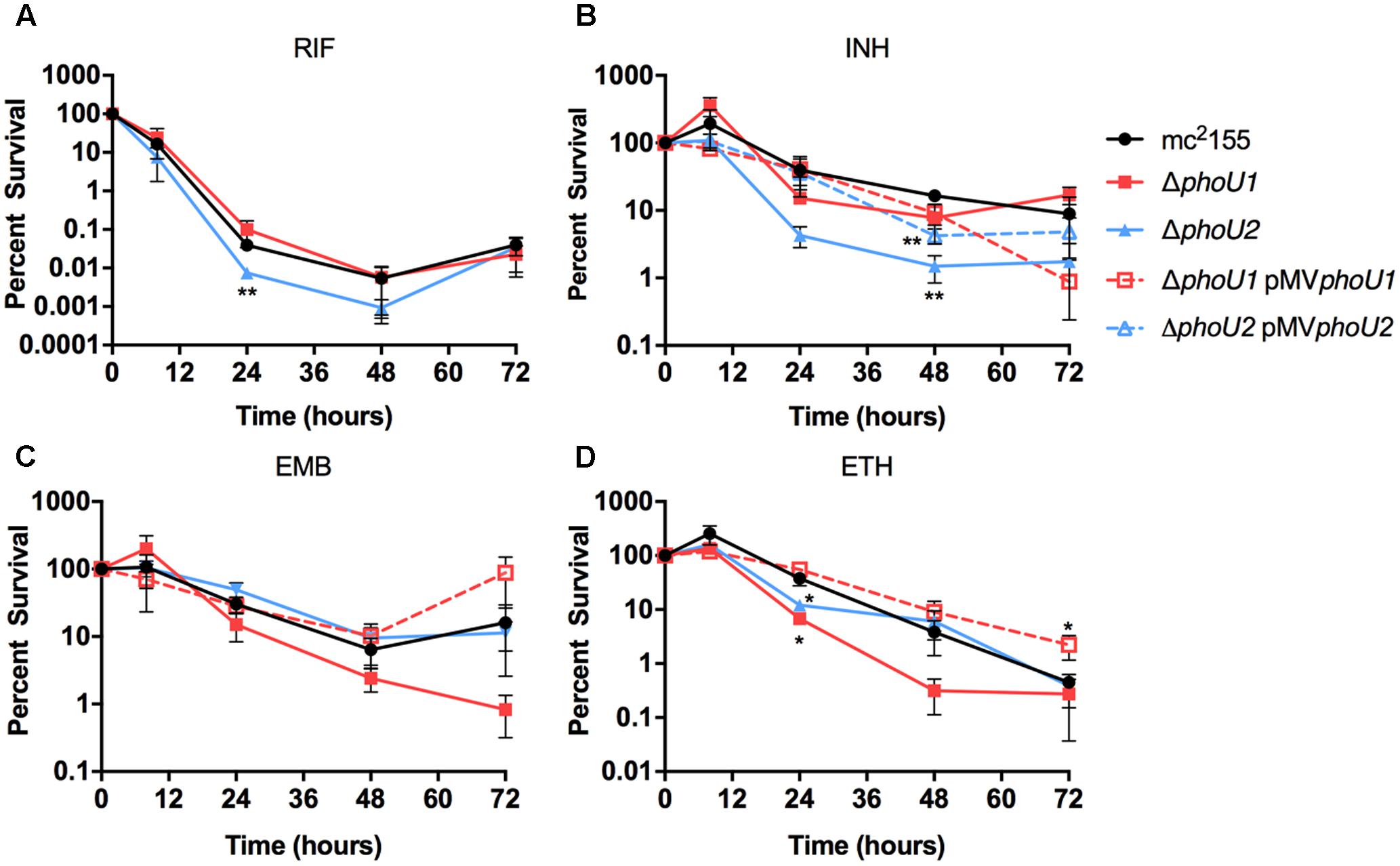
FIGURE 5. Formation of persister variants by M. smegmatis phoU single deletion mutant and complemented strains. The indicated strains were grown to late-logarithmic phase, diluted to OD600 of 0.05 in complete 7H9 liquid medium and exposed to the anti-tubercular drug (A) rifampicin (RIF, 200 μg/ml), (B) isoniazid (INH, 50 μg/ml), (C) ethambutol (EMB, 5 μg/ml), or (D) ethionamide (ETH, 200 μg/ml) for 72 h. Cultures were incubated at 37°C with aeration and the percent survival was calculated from viable CFU counts determined at 0, 8, 24, 48, and 72 h by plating serially diluted cultures on LB agar medium. Results are the mean of three biological replicates ± standard deviations. Asterisks indicate statistically significant differences compared to the mc2155 control: ∗P < 0.05, ∗∗P < 0.005.
Either PhoU1 or PhoU2 Is Required for M. smegmatis Replication in Vitro
Our results suggest that PhoU1 and PhoU2 have partially redundant function in negative regulation of SenX3-RegX3 activity in Pi-rich conditions. To test this idea, we attempted to construct a ΔphoU1ΔphoU2 double deletion mutant. We introduced the ΔphoU1 allelic exchange vector into the ΔphoU2 mutant and obtained 4 independent transformants that contained the vector integrated via the cloned regions of homology either upstream or downstream of the phoU1 gene. After counter-selection against the allelic exchange vector by plating on sucrose, we screened a total of 172 sucrose resistant, kanamycin sensitive (KanS), hygromycin sensitive (HygS) colonies for the ΔphoU1 deletion by PCR. None of these isolates harbored the ΔphoU1 deletion. These data suggested that M. smegmatis requires either PhoU1 or PhoU2 for viability under the growth conditions used.
To determine if phoU1 and phoU2 are jointly essential for M. smegmatis viability, we attempted to delete phoU1 in the ΔphoU2 background with a complementing copy of phoU1 provided in trans. We cloned phoU1 under the control of a tetracycline-inducible promoter on the integrating plasmid pTIC10a to enable conditional phoU1 expression. We created a ΔphoU2 pTICphoU1 strain and then electroporated this strain with the pJGΔphoU1 allelic exchange vector (Figure 6, steps 1–2). We obtained transformants in which the allelic exchange vector was integrated via either the upstream or downstream region of homology. After counter-selection on sucrose, we screened a total of 7 sucrose resistant, HygS colonies for the ΔphoU1 deletion by PCR. Two of these isolates had the ΔphoU1 deletion (Figure 6, step 3). The ease with which we constructed the ΔphoU1ΔphoU2 double mutant when a complementing copy of one gene was provided in trans suggests that M. smegmatis requires phoU1 or phoU2 for in vitro replication. The ΔphoU1ΔphoU2 pTICphoU1 strain exhibited alkaline phosphate activity similar to the WT control, even in the absence of induction with anhydrotetracycline (ATc) (Table 2), suggesting that leaky expression of phoU1 from the pTICphoU1 plasmid was sufficient for negative regulation of SenX3-RegX3. In fact, phoU1 was expressed at a significantly higher level from the pTICphoU1 plasmid even in the absence of ATc compared to the mc2155 control (Supplementary Figure S1).
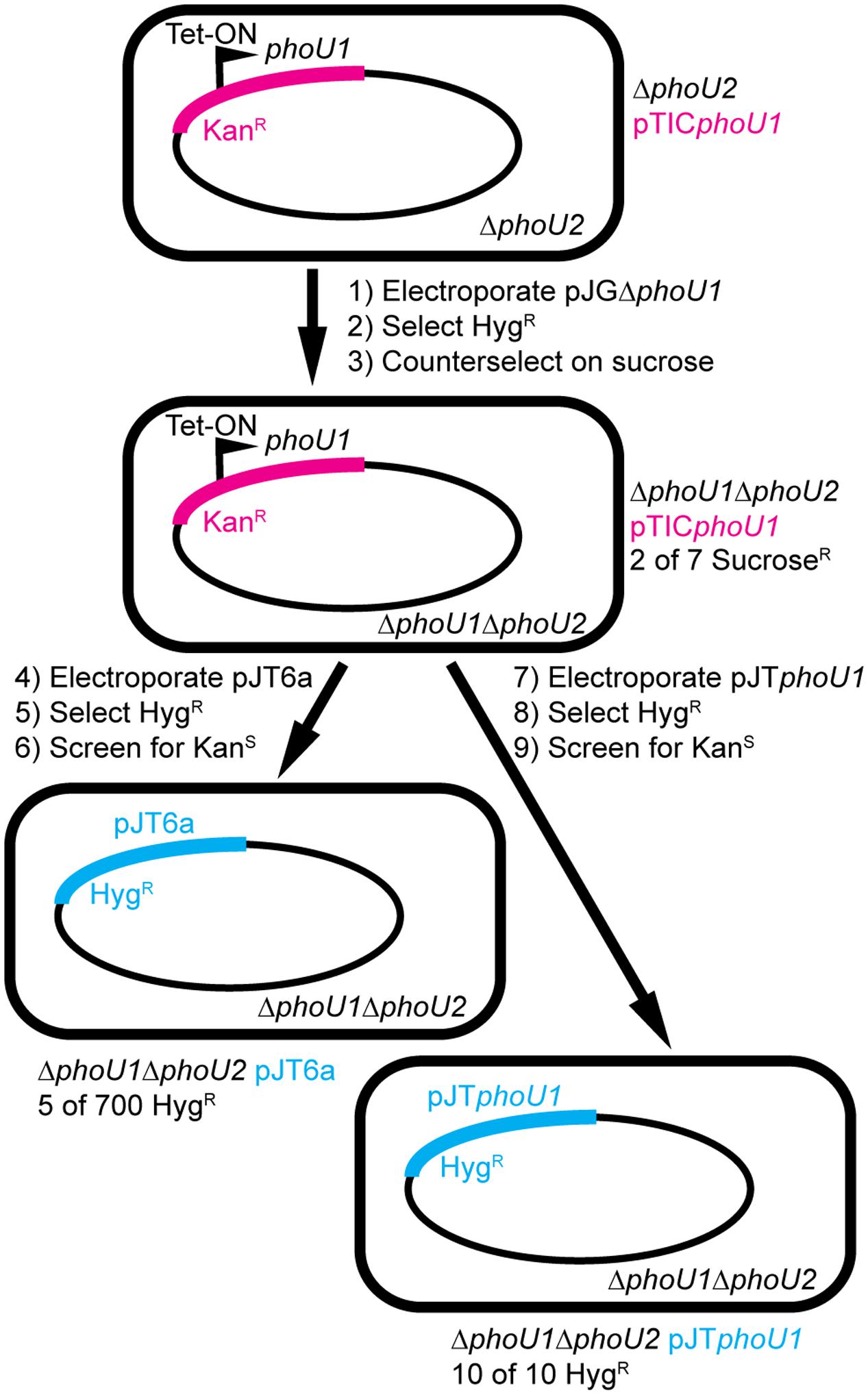
FIGURE 6. Construction of M. smegmatis ΔphoU1ΔphoU2 double deletion mutants by a plasmid swapping method. The phoU1 and phoU2 genes were deleted from the M. smegmatis chromosome by allelic exchange in a background containing a tetracycline-inducible copy of phoU1 provided in trans on the integrative plasmid pTICphoU1 that confers kanamycin (Kan) resistance (steps 1–3). The pTICphoU1 plasmid was then removed by swapping with either the empty non-compatible integrative plasmid pJT6a (steps 4–6) or pJTphoU1 (steps 7–9), which confer hygromycin (Hyg) resistance. The frequencies with which strains of the correct genotype were isolated are indicated.
To confirm that at least one of the phoU genes is essential for M. smegmatis viability, we attempted to swap the pTICphoU1 plasmid with a non-compatible plasmid containing a HygR marker, pJT6a (Rosen et al., 2017). The ΔphoU1ΔphoU2 pTICphoU1 strain was electroporated with pJT6a and transformants were selected on LB containing Hyg (Figure 6, steps 4–5). M. smegmatis mc2155 containing pTICphoU1 was similarly electroporated with pJT6a as a control. We obtained 4500 CFU/ml HygR transformants from the WT pTICphoU1 control, but only 150 CFU/ml HygR transformants from the ΔphoU1ΔphoU2 pTICphoU1 strain, suggesting that there is strong selective pressure against loss of the pTICphoU1 plasmid in the ΔphoU1ΔphoU2 background. The HygR transformants were then screened for loss of the KanR marker on pTICphoU1 (Figure 6, step 6). For the WT control, all 10 HygR colonies screened were sensitive to Kan, suggesting that they had lost the pTICphoU1 plasmid. PCR analysis on 8 of these HygR KanS isolates confirmed that they all contained the pJT6a plasmid and not pTICphoU1 (data not shown). In contrast, for the ΔphoU1ΔphoU2 pTICphoU1 strain, of the 700 HygR transformants that were screened, only 75 were sensitive to Kan, suggesting that the majority had not lost the pTICphoU1 plasmid. PCR analysis indicated that 63 of the HygR KanS isolates still harbored the pTICphoU1 plasmid and did not contain pJT6a, suggesting that some recombination event had occurred to swap the antibiotic resistance markers in these strains. Using PCR, we ultimately identified only five HygR KanS isolates that had successfully undergone the plasmid swap (Figure 6 and data not shown). All five of these isolates were also confirmed to have both the ΔphoU1 and ΔphoU2 deletions by PCR (data not shown).
To determine if the plasmid swap was inefficient in the ΔphoU1ΔphoU2 pTICphoU1 strain due to some deficiency in recombination as opposed to the loss of both phoU genes, we performed a similar plasmid swapping experiment using pJTphoU1 (Figure 6, steps 7–9). We observed a substantial increase in the number of transformants obtained with the pJTphoU1 plasmid compared to the empty pJT6a plasmid. We obtained 5800 CFU/ml HygR transformants from the WT mc2155 pTICphoU1 background and 4200 CFU/ml HygR transformants from the ΔphoU1ΔphoU2 pTICphoU1 strain. In addition, all 10 of the HygR transformants that we screened on each strain background were sensitive to Kan, suggesting that they had replaced pTICphoU1 with pJTphoU1 (Figure 6). Taken together, these data suggest that M. smegmatis requires either phoU1 or phoU2 for viability under the in vitro culture conditions that we used and that the five ΔphoU1ΔphoU2 pJT6a strains we generated may harbor secondary suppressor mutations that enable their growth.
Whole-Genome Sequencing Identifies Independent Suppressor Mutations in Genes Encoding the Pst Phosphate Transporter in the ΔphoU1ΔphoU2 Mutants
To determine if the ΔphoU1ΔphoU2 mutants that we isolated by the plasmid swap method contain suppressor mutations that enable their in vitro replication, we performed whole-genome sequencing on all five of the ΔphoU1ΔphoU2 pJT6a isolates. We compared the sequences of these strains to the sequence of our WT M. smegmatis mc2155 control, and identified two mutations in each strain. All five ΔphoU1ΔphoU2 pJT6a isolates harbored a G818T substitution in Msmeg_1387, which encodes a putative acyl-CoA dehydrogenase, resulting in a S273I transition in the Msmeg_1387 protein. We confirmed the G818T substitution in each ΔphoU1ΔphoU2 pJT6a isolate by PCR and sequencing, and determined that it was also present in the ΔphoU1ΔphoU2 pTICphoU1 parent strain, suggesting that this single nucleotide polymorphism (SNP) is not responsible for suppressing the essentiality of the phoU genes. Each ΔphoU1ΔphoU2 pJT6a strain also harbored an independent mutation in pstS, pstC or pstB, which encode components of the Pst Pi transporter (Table 4). These mutations were likewise confirmed by PCR and sequencing. Four of the strains (#504, 521, 625, and 664) contained SNPs that were predicted to severely disrupt the function of the encoded protein by causing a frame shift. The mutation in strain #518 was predicted to introduce a single glutamine residue into a region of PstB that contains primarily polar amino acids. This mutation may disrupt interaction of the PstB ATPase with other components of the Pst system. These data suggest that in the absence of PhoU1 and PhoU2, Pi import by the Pst system is toxic. All experiments described below used the ΔphoU1ΔphoU2 pJT6a #504 mutant, which harbors a frameshift mutation in pstS that is predicted to truncate more than 70% of the PstS protein (Table 4). Hereafter, we refer to this strain as the ΔphoU1ΔphoU2 pstS(Fs) triple mutant.
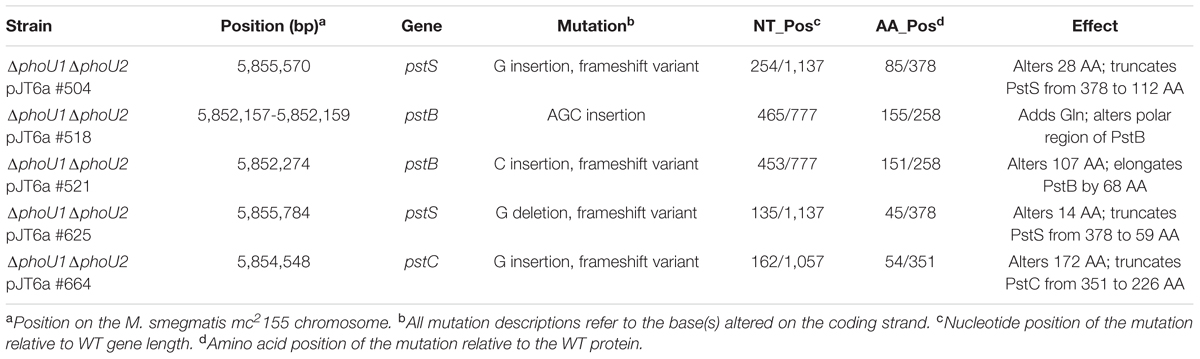
TABLE 4. Mutations in genes encoding Pst system components identified by whole-genome sequencing of five ΔphoU1ΔphoU2 pJT6a isolates.
The M. smegmatis ΔphoU1ΔphoU2 pstS(Fs) Mutant Has an in Vitro Growth Defect
To quantitatively assess the requirement of phoU1 and phoU2 for M. smegmatis growth in vitro, we performed growth curves. Like the ΔphoU1 mutant, the ΔphoU1 ΔphoU2 pstS(Fs) mutant grew at a similar rate as WT M. smegmatis in liquid medium in vitro (Figure 1C), but formed significantly smaller colonies on LB agar plates (Table 1). Colonies of the ΔphoU1ΔphoU2 pstS(Fs) mutant typically did not appear until after 3–4 days of incubation at 37°C while the WT and single deletion mutant colonies were visible after only 2 days. This growth defect on plates was complemented by pMVphoU1 (Table 1). The ΔphoU1ΔphoU2 pstS(Fs) mutant also lost viability in stationary phase, though this phenotype was less pronounced as compared to the ΔphoU1 single mutant (Figure 1D). Only phoU1 provided in trans could complement this stationary phase survival phenotype (Figure 1D). Finally, the ΔphoU1ΔphoU2 pstS(Fs) mutant produced a pellicle at the air-liquid interface of cultures grown in tubes (Figure 2A), and aggregated in complete 7H9 liquid medium similar to the ΔphoU1 mutant (Figure 2B). These clumping phenotypes were both complemented by providing either phoU1 or phoU2 in trans (Figure 2). These data suggest that M. smegmatis requires either phoU1 or phoU2 for normal replication in culture.
M. smegmatis PhoU1 and PhoU2 Have Overlapping Functions in Inhibition of SenX3-RegX3 Activation
To test if PhoU1 and PhoU2 function redundantly to regulate activity of SenX3-RegX3, we analyzed expression of known RegX3-regulated genes in the ΔphoU1ΔphoU2 pstS(Fs) mutant. By quantitative RT-PCR, the phoU1 and phoU2 transcripts were undetectable in this mutant (Figure 3), confirming that both phoU genes were deleted. Expression of regX3, pstS, and phoA was significantly increased 5-fold, 110-fold, and 158-fold, respectively, in the ΔphoU1ΔphoU2 pstS(Fs) triple mutant compared to the WT mc2155 control (Figure 4). Expression of pstS was also significantly increased in the ΔphoU1ΔphoU2 pstS(Fs) mutant relative to the ΔphoU1 single mutant (P = 0.004). We observed similar increases in the abundance of the regX3, pstS, and phoA transcripts in the #518 and #521 ΔphoU1ΔphoU2 pJT6a strains that each harbor an independent compensatory mutation in pstB (data not shown). We attempted to complement the gene expression phenotypes of the ΔphoU1ΔphoU2 pstS(Fs) mutant with either phoU1 or phoU2. The phoU1 and phoU2 transcripts were significantly elevated in the respective complemented strains compared to the WT mc2155 control (Figure 3). Complementation of the ΔphoU1ΔphoU2 pstS(Fs) mutant with pMVphoU1 restored expression of the regX3 and phoA transcripts to levels that were not significantly different from the WT mc2155 control (Figures 4A,C). Complementation with pMVphoU1 also reduced transcription of pstS to a level that was only 13-fold higher and not significantly different from the WT mc2155 control (Figure 4B). Complementation with pMVphoU2 only partially reversed the over-expression of the regX3, pstS, or phoA transcripts; levels of these transcripts were reduced compared to the ΔphoU1ΔphoU2 pstS(Fs) mutant but remained significantly elevated relative to the WT control (Figure 4). Consistent with these results, we observed a 8.5-fold increase in alkaline phosphatase activity in the ΔphoU1ΔphoU2 pstS(Fs) strain that was complemented by providing either phoU1 or phoU2 in trans, though full complementation was only observed with pMVphoU1 (Table 2). These data suggest that the PhoU1 and PhoU2 proteins have overlapping functions in negatively regulating SenX3-RegX3 activity in Pi-rich growth conditions. They further suggest that PhoU1 plays a more dominant role in fulfilling this Pi sensing signal transduction function. Since the gene expression phenotypes of the ΔphoU1ΔphoU2 pstS(Fs) mutant were complemented with pMVphoU1, our data also suggest that deletion of both phoU genes, not the pstS frameshift mutation in this strain, is responsible for activation of SenX3-RegX3.
The M. smegmatis ΔphoU1ΔphoU2 pstS(Fs) Mutant Is Hyper-Susceptible to RIF and Forms Persisters with Reduced Frequency
To determine if the M. smegmatis PhoU proteins are jointly required for antibiotic tolerance, similar to the M. tuberculosis PhoY proteins, we performed MIC and persister assays on the ΔphoU1ΔphoU2 pstS(Fs) mutant. MICs of the ΔphoU1ΔphoU2 pstS(Fs) mutant for INH, EMB and ETH, were unchanged relative to the WT control, but the MIC for RIF was reduced 32-fold (Table 3). Complementation with either phoU1 or phoU2 restored the RIF MIC to the WT level (Table 3). These data suggest that constitutive activation of SenX3-RegX3 contributes to increased susceptibility to RIF. The ΔphoU1ΔphoU2 pstS(Fs) mutant was also killed more rapidly by RIF, INH and ETH in liquid culture compared to the WT mc2155 control, suggesting that it forms fewer persister variants (Figures 7A,B,D). The ΔphoU1ΔphoU2 pstS(Fs) mutant also showed a trend toward decreased tolerance to EMB, though it was not statistically significant (Figure 7C). However, in most cases these phenotypes were only partially reversed by complementation with either phoU1 or phoU2. For RIF, complementation with pMVphoU1 restored tolerance at early time points, but failed to prevent the increased killing observed at later time points (Figure 7A). For INH, complementation with pMVphoU1 resulted in an intermediate antibiotic tolerance phenotype (Figure 7B), consistent with the fact that the ΔphoU2 single mutant exhibited a modest defect in survival of INH treatment (Figure 5B). Complementation with either pMVphoU1 or pMVphoU2 similarly led to intermediate tolerance to ETH (Figure 7D), consistent with the fact that the ΔphoU1 mutant was also more susceptible to this drug (Figure 5D). In contrast, both complemented strains exhibited further enhanced susceptibility to EMB (Figure 7C). It is possible that over-expression of phoU1 or phoU2 from the episomal plasmid is detrimental and prevents complete complementation of these antibiotic tolerance phenotypes. Alternatively, it is possible that the point mutation in pstS, which is predicted to disrupt Pi transport by the Pst system, is partially responsible for the decreased antibiotic tolerance.
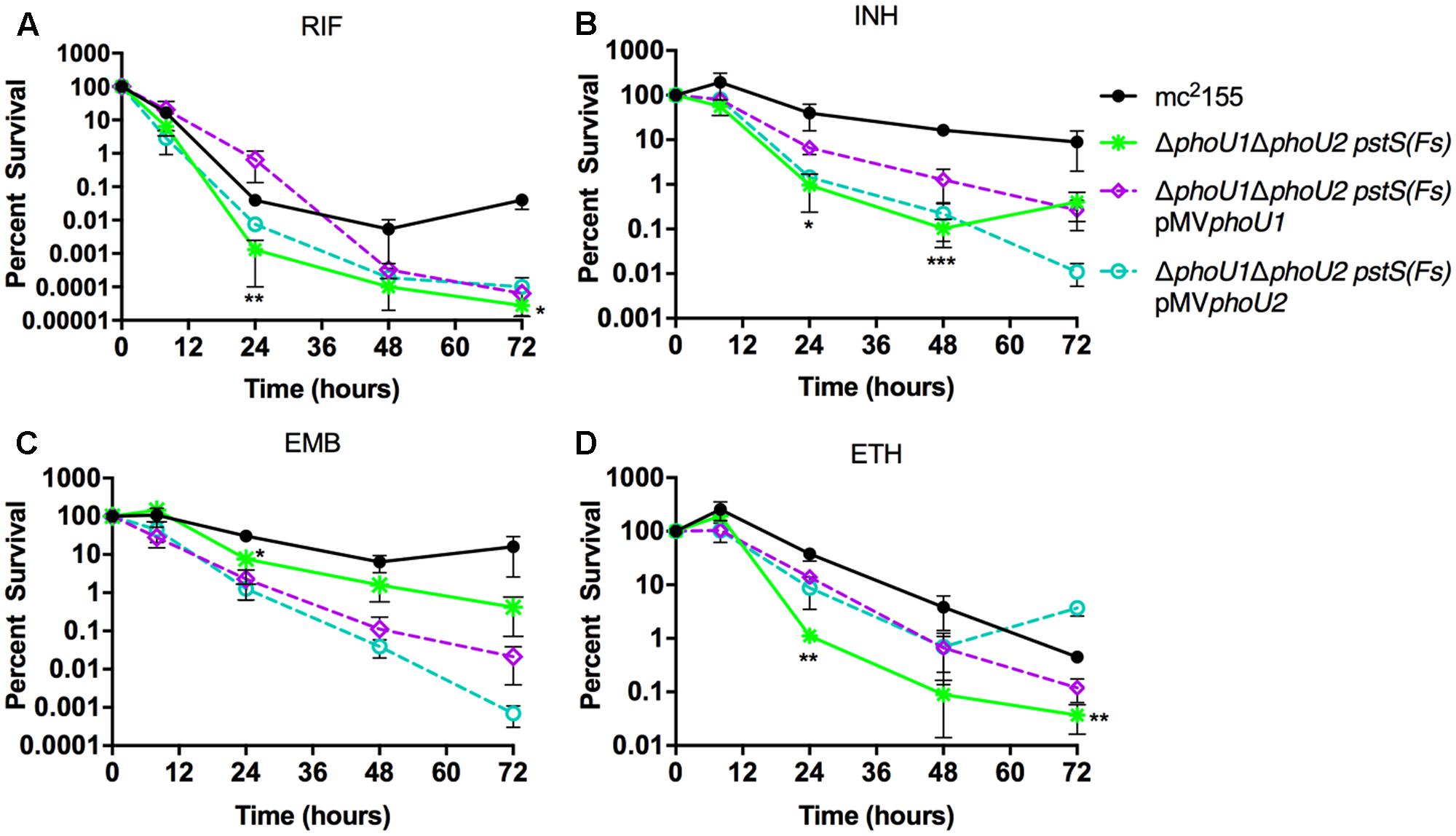
FIGURE 7. Formation of persister variants by M. smegmatis ΔphoU1ΔphoU2 pstS(Fs) mutant and complemented strains. The indicated strains were grown to late-logarithmic phase, diluted to OD600 of 0.05 in complete 7H9 liquid medium and exposed to the anti-tubercular drug (A) rifampicin (RIF, 200 μg/ml), (B) isoniazid (INH, 50 μg/ml), (C) ethambutol (EMB, 5 μg/ml), or (D) ethionamide (ETH, 200 μg/ml) for 72 h. Cultures were incubated at 37°C with aeration and the percent survival was calculated from viable CFU counts determined at 0, 8, 24, 48, and 72 h by plating serially diluted cultures on LB agar medium. Results are the mean of three biological replicates ± standard deviations. Asterisks indicate statistically significant differences compared to the mc2155 control: ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.0001.
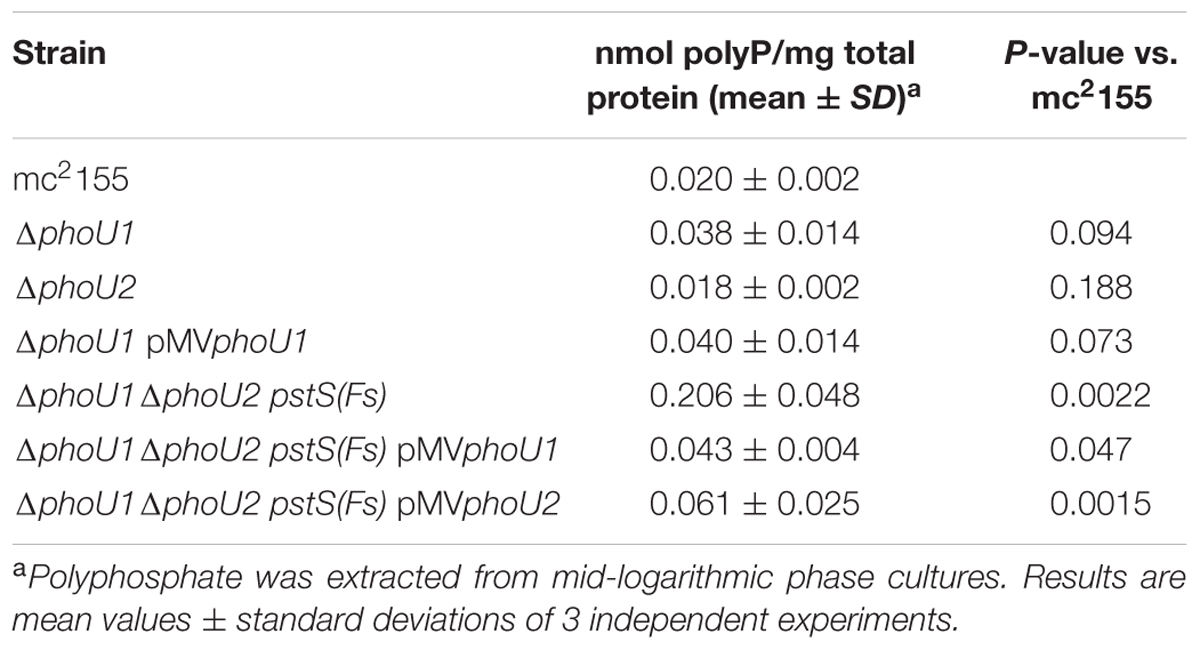
The ΔphoU1ΔphoU2 pstS(Fs) Mutant Accumulates Polyphosphate
Mutation of phoU has previously been reported to cause accumulation of polyphosphate (polyP) a linear polymer of phosphate residues linked by high-energy phosphoanhydride bonds (Kornberg et al., 1999), in a variety of bacterial species including M. marinum and M. tuberculosis (Morohoshi et al., 2002; Wang et al., 2013; Namugenyi et al., 2017). We therefore examined whether deletion of the M. smegmatis phoU genes altered polyP storage during growth in Pi-rich 7H9 medium. The ΔphoU2 mutant showed no significant alteration in polyP accumulation (Table 5). Intracellular polyP was increased approximately twofold in the ΔphoU1 mutant though this difference was not statistically significant and this phenotype was not complemented with the pMVphoU1 plasmid (Table 5). In contrast, the ΔphoU1ΔphoU2 pstS(Fs) triple mutant exhibited a statistically significant 10-fold increase in polyP storage relative to the mc2155 control (Table 5). Complementation with either phoU1 or phoU2 resulted in only a partial reduction in polyP accumulation, though pMVphoU1 complemented the phenotype more effectively than pMVphoU2 (Table 5). These data suggest that the M. smegmatis PhoU proteins regulate polyP storage. Since the ΔphoU1ΔphoU2 pstS(Fs) mutant accumulates polyP despite the mutation in pstS, our data further suggest that Pi uptake through the Pst system does not contribute to increased polyP storage.
Discussion
Although Pi limitation activates the M. smegmatis two-component signal transduction system SenX3-RegX3 (Glover et al., 2007), the molecular mechanisms mediating Pi sensing in this organism were not previously described. Here we demonstrate that the two M. smegmatis PhoU orthologs exhibit partially redundant functions in controlling activation of SenX3-RegX3 and that PhoU function is essential for M. smegmatis replication in vitro. Only deletion of both phoU1 and phoU2 led to constitutive expression of all three known RegX3-regulated genes. Each of the ΔphoU1ΔphoU2 double mutant strains that we isolated harbored independent compensatory mutations in genes encoding the Pst Pi transporter, which alleviated the essentiality of the two PhoU proteins. Our data are consistent with studies in other bacteria that implicated the Pst Pi transporter in mediating the growth defect or lethality associated with phoU inactivation (Hirota et al., 2013; Lubin et al., 2016; diCenzo et al., 2017). Mutations disrupting the M. smegmatis Pst transporter also constitutively activate SenX3-RegX3 (Kriakov et al., 2003; Glover et al., 2007). However, our data suggest that the deletion of both phoU genes causes constitutive expression of the RegX3 regulon in the ΔphoU1ΔphoU2 pstS(Fs) mutant independent of the pstS mutation because we could complement the gene expression phenotypes with phoU1 provided in trans. Our data therefore suggest that PhoU1 can directly regulate SenX3-RegX3 activity, independent of the pstS mutation, and that PhoU1 plays a more dominant role in Pi signaling than PhoU2. Furthermore, our data suggest that the M. smegmatis PhoU proteins function redundantly to regulate Pst Pi transport activity, since in their absence a functional Pst system is toxic.
Our results in M. smegmatis correspond to our previous report that the M. tuberculosis PhoU orthologs (PhoY1 and PhoY2) function redundantly to regulate SenX3-RegX3 activity (Namugenyi et al., 2017). However, we were able to generate the M. tuberculosis ΔphoY1ΔphoY2 double mutant using standard gene deletion methods, suggesting differences in the function of the PhoU orthologs between M. smegmatis and M. tuberculosis. It is possible that the M. tuberculosis PhoY proteins do not regulate Pi transport by the Pst system or that the M. tuberculosis Pst system transports Pi at a slower rate that does not cause toxic Pi accumulation. Further work will be required to conclusively establish the role of the M. smegmatis PhoU proteins or the M. tuberculosis PhoY proteins in regulating Pi uptake by the Pst system.
The M. smegmatis phoU mutants also exhibited reduced tolerance to several anti-tubercular drugs. In particular, the ΔphoU1ΔphoU2 pstS(Fs) mutant was markedly more susceptible to RIF, a phenotype we previously observed for the M. tuberculosis ΔphoY1ΔphoY2 mutant (Namugenyi et al., 2017). In M. tuberculosis, we demonstrated that constitutive activation of RegX3 caused RIF susceptibility (Namugenyi et al., 2017), so we predict that RegX3 activation is similarly responsible for RIF susceptibility in M. smegmatis. However, RegX3 is essential for growth of M. smegmatis in vitro (James et al., 2012), so we were unable to generate a regX3 deletion mutant to directly test this prediction. Our future studies will include performing transposon mutagenesis screens to identify factors that contribute to the enhanced RIF susceptibility of the ΔphoU1ΔphoU2 pstS(Fs) mutant.
The M. smegmatis phoU single and double deletion mutants also exhibited decreased phenotypic tolerance to the cell wall targeting antibiotics INH, EMB and ETH, revealed by examination of the death kinetics during antibiotic treatment. In general the ΔphoU1ΔphoU2 pstS(Fs) mutant was the most susceptible, but the ΔphoU1 mutant also exhibited reduced tolerance to ETH and EMB. Susceptibility to these drugs and the visible clumping phenotype of the ΔphoU1 mutant in liquid culture may both be caused by alterations to cell wall structure. In M. smegmatis, the Pst system has previously been implicated in regulating production of the alpha-glucan capsule (van de Weerd et al., 2016). However, we found no evidence for differential expression of genes involved in alpha-glucan synthesis in any of our ΔphoU mutants (data not shown) suggesting that other cell wall alterations are responsible for these phenotypes. Further study of changes to the cell wall in the ΔphoU1 mutant and determining whether these changes correlate with antibiotic susceptibility will be important as it could reveal new molecular targets that would enhance the activity of existing drugs.
The M. smegmatis ΔphoU1ΔphoU2 pstS(Fs) mutant also accumulated polyP, similar to the M. tuberculosis ΔphoY1ΔphoY2 mutant (Namugenyi et al., 2017), despite harboring a mutation predicted to render the PstS substrate binding domain of the Pst Pi transporter non-functional. Our data contrast with evidence that polyP accumulation by phoU mutants in E. coli, Pseudomonas aeruginosa, and Sinorhizobium meliloti requires a functional Pst system (Morohoshi et al., 2002; Hirota et al., 2013; diCenzo et al., 2017; Peng et al., 2017). It is possible that polyP would accumulate to an even higher level in a M. smegmatis phoU double mutant with a fully functional Pst system. Nevertheless, our data suggest either that Pi uptake via other transporters is dys-regulated in the M. smegmatis ΔphoU1ΔphoU2 pstS(Fs) mutant leading to increased polyP storage or that the M. smegmatis PhoU proteins directly modulate polyP accumulation by regulating expression or activity of enzymes that synthesize or degrade polyP. In addition to the Pst system, M. smegmatis has at least two other high-affinity Pi transporters, the Phn system and at least one other that remains to be identified (Gebhard et al., 2006). The PhoU proteins may regulate expression and/or activity of these alternative Pi transporters to influence polyP storage.
Although the M. smegmatis ΔphoU1 mutant did not constitutively express all genes in the RegX3 regulon, it exhibited several unique phenotypes including stickiness of colonies, clumping in liquid culture, reduced viability in stationary phase and over-expression of pstS. Each of these phenotypes could be complemented suggesting that they are caused by the phoU1 deletion. Furthermore, these phenotypes, particularly reduced viability in stationary phase, were more pronounced in the ΔphoU1 mutant compared to the ΔphoU1ΔphoU2 pstS(Fs) triple mutant. It is possible that the frameshift mutation in pstS partially alleviates the loss of phoU1 in the triple mutant strain. Our data suggest that M. smegmatis PhoU1 has one or more functions that are independent and distinct from PhoU2, which may include more robust regulation of Pst Pi transport activity. PhoU1 is encoded adjacent to the pstSCAB operon, suggesting there could be specificity conferred by co-evolution of the phoU1 and pstSCAB genes or a requirement for these genes to be close proximity to enable their co-regulation. Over-expression of pstS by the ΔphoU1 mutant suggests that PhoU1 influences activity of a transcriptional regulator that controls expression of the pst operon. RegX3 may be weakly activated in the ΔphoU1 mutant, leading to expression of genes with high affinity binding sites for the phosphorylated form of RegX3, which could include the pst genes. Alternatively, PhoU1 may control the activation of other transcriptional regulators. Gene expression profiling of the ΔphoU1 mutant could provide further insight into the basis for its in vitro growth phenotypes and clues to the transcriptional regulatory pathways that it influences.
Overall, our data suggest that both PhoU1 and PhoU2 participate in the Pi sensing system that controls activation of SenX3-RegX3 and expression of the Pi starvation RegX3 regulon in M. smegmatis. The PhoU1 and PhoU2 proteins exhibit 82% sequence similarity, so both proteins likely contain conserved domains that enable interactions with SenX3 and the Pst system. Further study of the interactions among these proteins could reveal the molecular basis for Pi sensing. Our work also suggests that M. smegmatis can serve as a model organism to discover compounds that inhibit PhoU1 and PhoU2 function to constitutively activate RegX3. Such compounds would be predicted to enhance susceptibility of mycobacteria to existing anti-tubercular drugs, including RIF.
Author Contributions
AT conceived the project and designed the strategy. AB, BE, MM, and JB carried out the experiments. AB and AT analyzed the data. AB and AT wrote the manuscript.
Funding
This work was supported by NIH Director’s New Innovator Award number 1DP2AI112245 (AT) and institutional start-up funds from the University of Minnesota (AT).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Anthony Baughn for providing plasmids pTIC10a and pJT6a, Hannah Cowan for technical assistance, the University of Minnesota Genomics Center for performing the whole-genome sequencing, and Joshua Thiede for assistance analyzing the whole-genome sequence data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02523/full#supplementary-material
Footnotes
References
Braibant, M., Gilot, P., and Content, J. (2000). The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24, 449–467. doi: 10.1111/j.1574-6976.2000.tb00550.x
de Almeida, L. G., Ortiz, J. H., Schneider, R. P., and Spira, B. (2015). phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl. Environ. Microbiol. 81, 3006–3015. doi: 10.1128/AEM.04168-14
diCenzo, G. C., Sharthiya, H., Nanda, A., Zamani, M., and Finan, T. M. (2017). PhoU allows rapid adaptation to high phosphate concentrations by modulating PstSCAB transport rate in Sinorhizobium meliloti. J. Bacteriol. 199:e00143-17. doi: 10.1128/JB.00143-17
Gardner, S. G., Johns, K. D., Tanner, R., and McCleary, W. R. (2014). The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 196, 1741–1752. doi: 10.1128/JB.00029-14
Gebhard, S., Tran, S. L., and Cook, G. M. (2006). The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 152, 3453–3465. doi: 10.1099/mic.0.29201-0
Glover, R. T., Kriakov, J., Garforth, S. J., Baughn, A. D., and Jacobs, W. R. Jr. (2007). The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503. doi: 10.1128/JB.00190-07
Hirota, R., Motomura, K., Nakai, S., Handa, T., Ikeda, T., and Kuroda, A. (2013). Stable polyphosphate accumulation by a pseudo-revertant of an Escherichia coli phoU mutant. Biotechnol. Lett. 35, 695–701. doi: 10.1007/s10529-012-1133-y
Hsieh, Y.-J., and Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203. doi: 10.1016/j.mib.2010.01.014
James, J. N., Hasan, Z., Ioerger, T. R., Brown, A. C., Personne, Y., Carroll, P., et al. (2012). Deletion of SenX3-RegX3, a key two-component regulatory system of Mycobacterium smegmatis, results in growth defects under phosphate-limiting conditions. Microbiology 158, 2724–2731. doi: 10.1099/mic.0.060319-0
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kirksey, M. A., Tischler, A. D., Siméone, R., Hisert, K. B., Uplekar, S., Guilhot, C., et al. (2011). Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect. Immun. 79, 2829–2838. doi: 10.1128/IAI.00097-11
Kornberg, A., Rao, N. N., and Ault-Riché, D. (1999). Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125. doi: 10.1146/annurev.biochem.68.1.89
Kriakov, J., Lee, S. H., and Jacobs, W. R. Jr. (2003). Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in Mycobacterium smegmatis. J. Bacteriol. 185, 4983–4991. doi: 10.1128/JB.185.16.4983-4991.2003
Lamarche, M. G., Wanner, B. L., Crépin, S., and Harel, J. (2008). The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473. doi: 10.1111/j.1574-6976.2008.00101.x
Larsen, M. H., Biermann, K., Tandberg, S., Hsu, T., and Jacobs, W. R. Jr. (2007). Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 6, 10A.2.1–10A.2.21. doi: 10.1002/9780471729259.mc10a02s6
Li, Y., and Zhang, Y. (2007). PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099. doi: 10.1128/AAC.00052-07
Lubin, E. A., Henry, J. T., Fiebig, A., Crosson, S., and Laub, M. T. (2016). Identification of the PhoB regulon and role of PhoU in the phosphate starvation response of Caulobacter crescentus. J. Bacteriol. 198, 187–200. doi: 10.1128/JB.00658-15
Maissonneuve, E., and Gerdes, K. (2014). Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548. doi: 10.1016/j.cell.2014.02.050
Morohoshi, T., Maruo, T., Shirai, Y., Kato, J., Ikeda, T., Takiguchi, N., et al. (2002). Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 68, 4107–4110. doi: 10.1128/AEM.68.8.4107-4110.2002
Namugenyi, S. B., Aagesen, A. M., Elliott, S. R., and Tischler, A. D. (2017). Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating Pst/SenX3-RegX3 phosphate sensing. mBio 8:e00494-17. doi: 10.1128/mBio.00494-17
Parish, T., Smith, D. A., Roberts, G., Betts, J., and Stoker, N. G. (2003). The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149, 1423–1435. doi: 10.1099/mic.0.26245-0
Peng, Y.-C., Lu, C. Y., Li, G., Eichenbaum, Z., and Lu, C.-D. (2017). Induction of the Pho regulon and polyphosphate synthesis against spermine stress in Pseudomonas aeruginosa. Mol. Microbiol. 104, 1037–1051. doi: 10.1111/mmi.13678
Rice, C. D., Pollard, J. E., Lewis, Z. T., and McCleary, W. R. (2009). Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75, 573–582. doi: 10.1128/AEM.01046-08
Rifat, D., Bishai, W. R., and Karakousis, P. C. (2009). Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 200, 1126–1135. doi: 10.1086/605700
Rosen, B. C., Dillon, N. A., Peterson, N. D., Minato, Y., and Baughn, A. D. (2017). Long-chain fatty acyl coenzyme a ligase FadD2 mediates intrinsic pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 61, e02130-16.
Shi, W., and Zhang, Y. (2010). PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1237–1242. doi: 10.1093/jac/dkq103
Steed, P. M., and Wanner, B. L. (1993). Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in phosphate regulation. J. Bacteriol. 175, 6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993
Tischler, A. D., Leistikow, R. L., Kirksey, M. A., Voskuil, M. I., and McKinney, J. D. (2013). Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect. Immun. 81, 317–328. doi: 10.1128/IAI.01136-12
Tischler, A. D., Leistikow, R. L., Ramakrishnan, P., Voskuil, M. I., and McKinney, J. D. (2016). Mycobacterium tuberculosis phosphate uptake system component PstA2 is not required for gene regulation or virulence. PLOS ONE 11:e0161467. doi: 10.1371/journal.pone.0161467
Upton, A. M., and McKinney, J. D. (2007). Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 153, 3973–3982. doi: 10.1099/mic.0.2007/011726-0
van de Weerd, R., Boot, M., Maaskant, J., Sparrius, M., Verboom, T., van Leeuwen, L. M., et al. (2016). Inorganic phosphate limitation modulates capsular polysaccharide composition in mycobacteria. J. Biol. Chem. 291, 11787–11799. doi: 10.1074/jbc.M116.722454
Wang, C., Mao, Y., Yu, J., Zhu, L., Li, M., Wang, D., et al. (2013). PhoY2 of mycobacteria is required for metabolic homeostasis and stress response. J. Bacteriol. 195, 243–252. doi: 10.1128/JB.01556-12
Keywords: Pst system, phosphate, PhoU, RegX3, tuberculosis, persister, antibiotic tolerance, polyphosphate
Citation: Brokaw AM, Eide BJ, Muradian M, Boster JM and Tischler AD (2017) Mycobacterium smegmatis PhoU Proteins Have Overlapping Functions in Phosphate Signaling and Are Essential. Front. Microbiol. 8:2523. doi: 10.3389/fmicb.2017.02523
Received: 09 October 2017; Accepted: 05 December 2017;
Published: 18 December 2017.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
George Colin DiCenzo, University of Florence, ItalyYasu S. Morita, University of Massachusetts Amherst, United States
Copyright © 2017 Brokaw, Eide, Muradian, Boster and Tischler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna D. Tischler, tischler@umn.edu
†Present address: Alyssa M. Brokaw, Department of Global Health, University of Washington, Seattle, WA, United States; Michael Muradian, Injury Research Center, Medical College of Wisconsin, Milwaukee, WI, United States; Joshua M. Boster, Internal Medicine, Brooke Army Medical Center, Fort Sam Houston, San Antonio, TX, United States
 Alyssa M. Brokaw
Alyssa M. Brokaw Benjamin J. Eide
Benjamin J. Eide Michael Muradian†
Michael Muradian† Joshua M. Boster
Joshua M. Boster Anna D. Tischler
Anna D. Tischler