- Institute of Microbiology, Charité – Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Department of Microbiology and Hygiene, Berlin, Germany
The World Health Organization has rated multi-drug resistant (MDR) Pseudomonas aeruginosa as serious threat for human health. It is, however, unclear, whether intestinal MDR P. aeruginosa carriage is associated with inflammatory responses in intestinal or even systemic compartments. In the present study, we generated with respect to their microbiota “humanized” mice by human fecal microbiota transplantation of secondary abiotic mice. Following peroral challenge with a clinical P. aeruginosa isolate on two consecutive days, mice harboring a human or murine microbiota were only partially protected from stable intestinal P. aeruginosa colonization given that up to 78% of mice were P. aeruginosa-positive at day 28 post-infection (p.i.). Irrespective of the host-specificity of the microbiota, P. aeruginosa colonized mice were clinically uncompromised. However, P. aeruginosa colonization resulted in increased intestinal epithelial apoptosis that was accompanied by pronounced proliferative/regenerative cell responses. Furthermore, at day 7 p.i. increased innate immune cell populations such as macrophages and monocytes could be observed in the colon of mice harboring either a human or murine microbiota, whereas this held true at day 28 p.i. for adaptive immune cells such as B lymphocytes in both the small and large intestines of mice with murine microbiota. At day 7 p.i., pro-inflammatory cytokine secretion was enhanced in the colon and mesenteric lymph nodes, whereas the anti-inflammatory cytokine IL-10 was down-regulated in the former at day 28 p.i. Strikingly, cytokine responses upon intestinal P. aeruginosa colonization were not restricted to the intestinal tract, but could also be observed systemically, given that TNF and IFN-γ concentrations were elevated in spleens as early as 7 days p.i., whereas splenic IL-10 levels were dampened at day 28 p.i. of mice with human microbiota. In conclusion, mere intestinal carriage of MDR P. aeruginosa by clinically unaffected mice results in pro-inflammatory sequelae not only in intestinal, but also systemic compartments.
Introduction
Pseudomonas aeruginosa constitute strictly aerobic Gram-negative bacteria that normally inhabit the soil and surfaces in aqueous environments. Their adaptive capabilities and intrinsic antibiotic resistance support survival in nature as well as in a plethora of settings including healthcare facilities (Gellatly and Hancock, 2013). Whereas a single flagellum and many cell surface pili facilitate bacterial motility and adherence to surfaces (Driscoll et al., 2007), alginate secretion, biofilm formation, quorum-sensing and an elaborate secretion system contribute to P. aeruginosa virulence (Driscoll et al., 2007; Gellatly and Hancock, 2013). In the hospital, typical environmental reservoirs of P. aeruginosa include sinks, water bottles and respiratory equipment, for instance (Morrison and Wenzel, 1984; Stratton, 1990). P. aeruginosa is a leading cause of nosocomial infections in ICUs such as ventilator-associated pneumonia, bloodstream infections, urinary tract infections or superinfections of burn wounds associated with mortality rates of more than 30% (Vincent, 2003). The emergence of MDR P. aeruginosa strains particularly due to extended-spectrum β-lactamases, carbapenemases and 16S rRNA methylases, has posed immuno-compromised individuals, patients suffering from cystic fibrosis and other chronic pulmonary morbidities or ICU patients at increased risk for prolonged hospital stay and mortality (Vincent, 2003; Oliver et al., 2015; Potron et al., 2015). In consequence, the WHO has rated MDR Gram-negative bacteria including P. aeruginosa as serious threat for human health and emphasized the urgent need for new treatment options (Tacconelli and Magrini, 2017). P. aeruginosa infection typically follows colonization that may happen either endogenously (hence arising from the patients’ inherent microbiota) or exogenously (if acquired from other patients or from the hospital environment by cross-contamination) (Cohen et al., 2017). Intestinal P. aeruginosa colonization before ICU admission has been shown to be associated with an almost 15 times increased risk of subsequent infection as compared to non-colonized individuals (Gomez-Zorrilla et al., 2015). Disruption of the complex and diverse commensal intestinal microbiota physiologically exerting colonization resistance to the host by antimicrobial compounds further facilitates establishment and invasion of opportunistic or obligate pathogens in mice and men (Hentges et al., 1985; Marshall et al., 1993; Bereswill et al., 2011; Fiebiger et al., 2016). We have recently shown that also the host-specificity of the complex intestinal microbiota is pivotal for effective colonization resistance given that mice harboring a murine, but not human gut microbiota were protected from intestinal colonization with enteropathogenic C. jejuni (Bereswill et al., 2011). Conversely, perorally with C. jejuni challenged “humanized” mice could be stably infected by the enteropathogen and displayed pro-inflammatory immune responses as seen in human campylobacteriosis (Bereswill et al., 2011; Masanta et al., 2013; Fiebiger et al., 2016).
To date, however, no data are available regarding host immune responses in otherwise healthy P. aeruginosa carriers with an intact intestinal microbiota composition. In the present study we therefore investigated, whether mice harboring a human vs. murine microbiota could be stably colonized by a clinical MDR P. aeruginosa strain and whether bacterial carriage was associated with intestinal or even systemic inflammatory host responses.
Materials and Methods
Ethics Statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin; registration numbers G0039/15). Animal welfare was monitored twice daily by assessment of clinical conditions and weight loss of mice.
Generation of Secondary Abiotic Mice
Female C57BL/6j mice were bred and maintained under SPF conditions in the Forschungsinstitute für Experimentelle Medizin (Charité – University Medicine, Berlin, Germany). Secondary abiotic mice with a virtually depleted microbiota were generated as described previously (Heimesaat et al., 2006). In brief, 8 weeks old mice were transferred into sterile cages and subjected to a broad-spectrum antibiotic treatment for 8 weeks by adding ampicillin plus sulbactam (1 g/L; Ratiopharm, Germany), vancomycin (500 mg/L; Cell Pharm, Germany), ciprofloxacin (200 mg/L; Bayer Vital, Germany), imipenem (250 mg/L; MSD, Germany) and metronidazole (1 g/L; Fresenius, Germany) to the drinking water (ad libitum). Cultural and culture-independent (i.e., 16S rRNA based molecular) quality control measures revealed virtual absence of bacteria in fecal samples as described earlier (Ekmekciu et al., 2017).
Human and Murine Fecal Microbiota Transplantation
Three days before association of secondary abiotic mice with a complex human or murine intestinal microbiota by FMT, the antibiotic cocktail was replaced by autoclaved tap water (ad libitum). Fresh fecal samples free of enteropathogenic bacteria (e.g., enteropathogenic Escherichia coli, Salmonella, Shigella, Yersinia, Campylobacter, Clostridium difficile), viruses (e.g., Noro-, Rota-, Enterovirus) and parasites (e.g., Giardia lamblia, Entamoeba histolytica, Blastocystis hominis and worms) were collected from five individual healthy human volunteers, dissolved in sterile PBS (Gibco, Life Technologies, United Kingdom), aliquoted and stored at -80°C as described earlier (Bereswill et al., 2011). Immediately before FMT, individual fecal aliquots were thawed and pooled (Bereswill et al., 2011). In addition, fresh murine fecal samples were collected from 10 age and sex matched SPF control mice, pooled, dissolved in 10 mL sterile PBS and the supernatant served as murine donor suspension. To generate human intestinal microbiota associated mice or reintroduce murine microbiota into mice, secondary abiotic animals were subjected to peroral FMT either with 0.3 mL of the human or murine donor suspension by gavage on two consecutive days (Bereswill et al., 2011). Of note, each donor suspension was tested negative for P. aeruginosa by both culture and culture-independent (i.e., 16S rRNA based) methods. Bacterial groups varied less than 0.5 logarithmic orders of magnitude between independent experiments. To assure proper establishment of the human or murine microbiota in the murine host, mice were kept for 3 weeks until P. aeruginosa infection. Immediately before peroral P. aeruginosa challenge individual fecal samples were collected for quantitative molecular analyses of main intestinal bacterial communities as described elsewhere (Bereswill et al., 2011).
Molecular Analysis of the Human and Murine Donor Suspensions and the Fecal Microbiota
DNA was extracted from fecal samples as described previously (Heimesaat et al., 2006). In brief, DNA was quantified by using Quant-iT PicoGreen reagent (Invitrogen, United Kingdom) and adjusted to 1 ng per μL. Then, TLs as well as the main bacterial groups abundant in the murine and human intestinal microbiota including EB, EC, LB, Bif, B/P, Clocc, Clept and MIB were assessed by qRT-PCR with species-, genera- or group-specific 16S rRNA gene primers (Tib MolBiol, Germany) as described previously (Heimesaat et al., 2010) and numbers of 16S rRNA gene copies per ng DNA of each sample determined.
MDR P. aeruginosa Infection and Quantitative Assessment of Fecal Loads
The MDR P. aeruginosa isolate was initially cultured from respiratory material of a patient suffering from nosocomial pneumonia and kindly provided by Prof. Dr. Bastian Opitz (Charité – University Medicine, Berlin, Germany). Notably, the clinical bacterial strain was tested resistant against piperacillin/tazobactam, ceftazidime, cefepim ± cefoxitin, imipenem, ciprofloxacin, gentamycin, tobramycin, amikacin, trimethoprim/sulfamethoxazole, and aztreonam (according to EUCAST interpretation guidelines) and displayed antimicrobial sensitivity to fosfomycin and colistin only (von Klitzing et al., 2017a). Prior infection, the P. aeruginosa strain was grown on cetrimide agar (Oxoid) for 48 h in an aerobic atmosphere at 37°C.
On days 0 and 1, mice were perorally challenged with 109 CFU of the MDR P. aeruginosa strain by gavage in a total volume of 0.3 mL PBS as reported earlier (von Klitzing et al., 2017a).
For quantitative assessment of fecal P. aeruginosa loads over time p.i., fecal samples were homogenized in sterile PBS, then serial dilutions streaked onto Columbia agar supplemented with 5% sheep blood (Oxoid, Germany) and cetrimide agar and incubated in an aerobic atmosphere at 37°C for 48 h as described previously (von Klitzing et al., 2017a). Fecal weights were determined by the difference of the sample weights before and after asservation. The detection limit of viable bacteria was 100 CFU per g.
Clinical Conditions
Macroscopic and/or microscopic abundance of fecal blood was assessed in individual mice on a daily basis by the Guajac method using Haemoccult (Beckman Coulter/ PCD, Germany) as described earlier (Haag et al., 2012a).
Sampling Procedures
Mice were sacrificed 28 days p.i. by isoflurane treatment (Abott, Germany). Cardiac blood (for serum) and tissue samples from spleen, MLN, ileum and colon were removed under sterile conditions. Intestinal samples were collected from each mouse in parallel for microbiological, immunological and immunohistochemical analyses.
Immunohistochemistry
Five μm thin paraffin sections of colonic and ileal ex vivo biopsies were used for in situ immunohistochemical analysis as reported previously (Heimesaat et al., 2010; Alutis et al., 2015a,b). In brief, primary antibodies against cleaved caspase-3 (Asp175, Cell Signaling, Beverly, MA, United States, 1:200), Ki67 (TEC3, Dako, Glostrup, Denmark, 1:100), F4/80 (#14-4801, clone BM8, eBioscience, 1:50) and B220 (eBioscience, 1:200) were used to assess apoptotic cells, proliferating cells, macrophages/monocytes, and B lymphocytes, respectively. The average numbers of positively stained cells within at least six HPFs (0.287 mm2; 400× magnification) were determined by an independent blinded investigator.
Cytokine Detection
Ex vivo biopsies (approximately 1 cm2) derived from colon and ileum (both cut longitudinally and washed in PBS), as well as MLN and spleen were placed into 24-flat-bottom well culture plates (Falcon, Germany) containing 500 mL serum-free RPMI 1640 medium (Gibco, Life Technologies) supplemented with penicillin (100 U/ml, Biochrom, Germany) and streptomycin (100 μg/ml; Biochrom). After 18 h at 37°C, culture supernatants and serum samples were tested for TNF, IFN-γ and IL-10 by the Mouse Inflammation Cytometric Bead Assay (CBA; BD Bioscience) on a BD FACSCanto II flow cytometer (BD Bioscience) as described earlier (Haag et al., 2012b).
Statistical Analysis
Medians and levels of significance were determined using appropriate tests (Mann–Whitney U test, one-way ANOVA and Kruskal–Wallis test) as indicated. Two-sided probability (p) values ≤ 0.05 were considered significant. Experiments were reproduced at least twice.
Results
Intestinal P. aeruginosa Colonization in Mice Harboring a Complex Human vs. Murine Gut Microbiota
In the present study we addressed whether a complex human or murine gut microbiota could sufficiently confer colonization resistance against MDR P. aeruginosa to the vertebrate host and if not, whether intestinal P. aeruginosa carriage resulted in intestinal or even systemic inflammatory sequelae. To accomplish this, secondary abiotic mice were associated with either a human or murine microbiota by peroral FMT. Culture-independent (i.e., molecular) analyses of the microbiota suspensions derived from human or murine donors revealed substantial host-specific differences in bacterial compositions. TLs and those of specific families/genera such as EB, Bif, Clocc, and Clept were higher in human as compared to murine bacterial suspensions (p < 0.01–0.001; Figure 1A), whereas LB or MIB were lower or even virtually absent in the former as compared to the latter (p < 0.001; Figure 1A). Three weeks following FMT and immediately before P. aeruginosa challenge (i.e., day 0), we further assessed respective microbiota compositions in fecal samples derived from individual mice. Molecular analyses revealed higher fecal gene numbers of B/P and Clept, but lower LB and MIB burdens as well as slightly lower total eubacterial gene numbers (less than one order of magnitude) in human microbiota transplanted mice as compared to murine counterparts (p < 0.001; Figure 1B). These results thus indicate stable intestinal establishment of distinct microbiota following respective FMT.
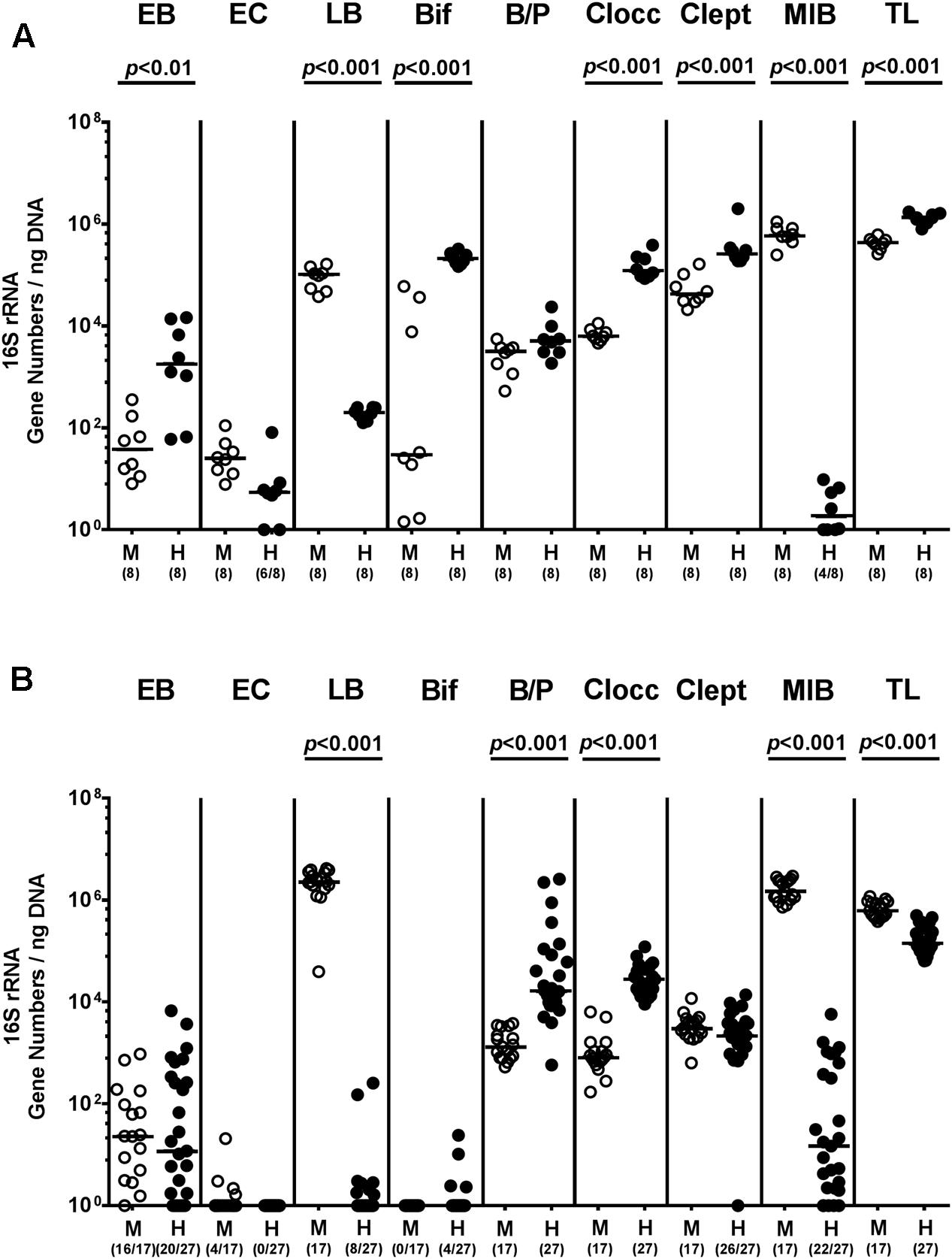
FIGURE 1. Intestinal bacterial colonization following human and murine FMT of secondary abiotic mice. Secondary abiotic mice were generated by broad-spectrum antibiotic treatment and associated with either a complex murine (M, open circles) or human (H, closed circles) intestinal microbiota by FMT on two consecutive days. (A) Respective donor suspensions and (B) individual fecal sample taken three weeks following FMT (and hence, immediately before MDR P. aeruginosa challenge) were subjected to a comprehensive fecal microbiota survey by quantitative Real-Time PCR amplifying variable regions of the bacterial 16S rRNA gene. The following main intestinal bacterial groups were determined (expressed as 16S rRNA gene numbers per ng DNA): Enterobacteria (EB), enterococci (EC), lactic acid bacteria (LB), bifidobacteria (Bif), Bacteroides/Prevotella spp. (B/P), Clostridium coccoides group (Clocc), Clostridium leptum group (Clept), Mouse Intestinal Bacteroides (MIB), and total eubacterial load (TL). Medians (black bars) and significance levels (p-values) determined by Mann–Whitney U-test are shown. Numbers of samples harboring the respective bacterial group out of the total number of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
On two consecutive days (i.e., days 0 and 1), we perorally challenged mice with 109 viable P. aeruginosa by gavage and quantitatively assessed fecal P. aeruginosa loads thereafter. Cultural analyses revealed that as early as 48 h following the latest bacterial challenge (i.e., day 3 p.i.), virtually all human microbiota transplanted mice harbored P. aeruginosa in their intestines with median loads of approximately 104 CFU per g, whereas bacterial counts were approximately two orders of magnitude lower in murine controls (p < 0.01; Figure 2 and Supplementary Figure S1). Of note, by day 3 p.i. P. aeruginosa could not be detected in 40% of mice with a murine gut microbiota. Until the end of the observation period, however, fecal P. aeruginosa loads were comparable in mice with either microbiota, whereas 64.7% of mice following human FMT and 77.8% of murine controls harbored the bacterial strain in their intestinal tract at day 28 p.i. (Figure 2 and Supplementary Figure S1). Hence, the complex intestinal microbiota, irrespective whether of human or murine origin, prevented only a subgroup of mice from long-term intestinal MDR P. aeruginosa colonization.
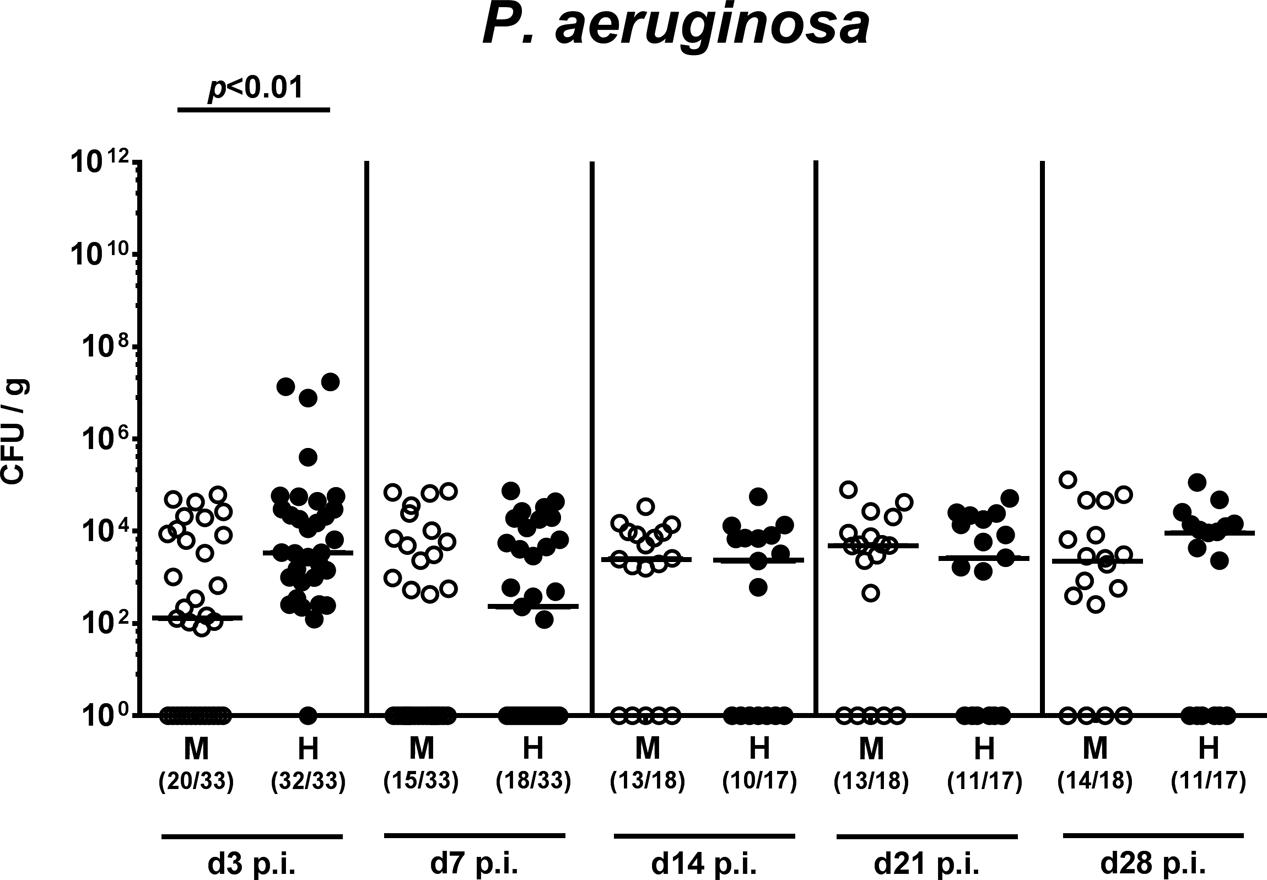
FIGURE 2. Intestinal colonization densities of MDR P. aeruginosa in mice harboring a human vs. murine microbiota. Mice with a murine (M, open circles) or human (H, closed circles) intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. Intestinal colonization densities were assessed in fecal samples at defined time points p.i. by culture. Medians (black bars) and significance levels (p-values) determined by Mann–Whitney U-test are shown. Numbers of samples harboring the respective bacterial group out of the total number of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
Intestinal Microbiota Changes upon P. aeruginosa Colonization of Mice Harboring a Complex Human vs. Murine Gut Microbiota
We next surveyed whether P. aeruginosa colonization was accompanied by distinct changes of the intestinal microbiota composition over time (Figures 3, 4). Quantitative culture-independent analysis revealed that four weeks following P. aeruginosa association, intestinal gene numbers of MIB had increased in mice that had been subjected to human FMT, but not conventionally colonized mice (p < 0.05; Figure 3). As early as one week following P. aeruginosa challenge, however, the respective gut microbiota did not differ from uninfected control groups, neither in mice harboring a human, nor murine microbiota (Figures 3, 4). Hence, no overt changes in intestinal microbiota composition could be observed relatively early (i.e., 1 week) upon P. aeruginosa colonization and were rather subtle thereafter.
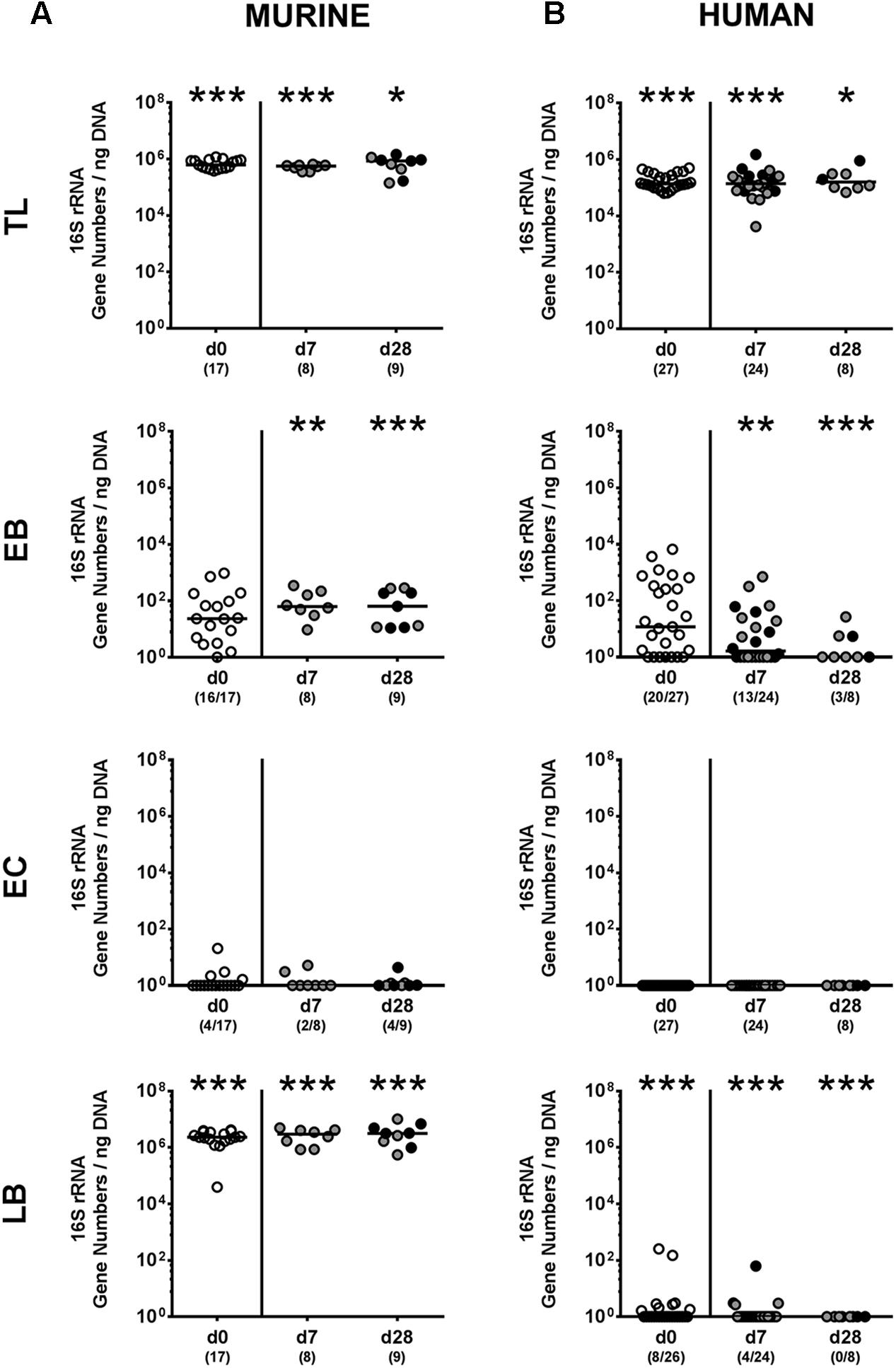
FIGURE 3. Intestinal total and aerobic bacterial changes following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with P. aeruginosa on day 0 and day 1. Immediately before (day 0, open circles) and at day 7 and day 28 post-challenge the intestinal microbiota composition of colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice was surveyed applying quantitative Real-Time PCR amplifying variable regions of the bacterial 16S rRNA gene. The following main intestinal bacterial groups were determined (expressed as 16S rRNA gene numbers per ng DNA): Total eubacterial load (TL), enterobacteria (EB), enterococci (EC), and lactic acid bacteria (LB). Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of samples harboring the respective bacterial group out of the total number of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
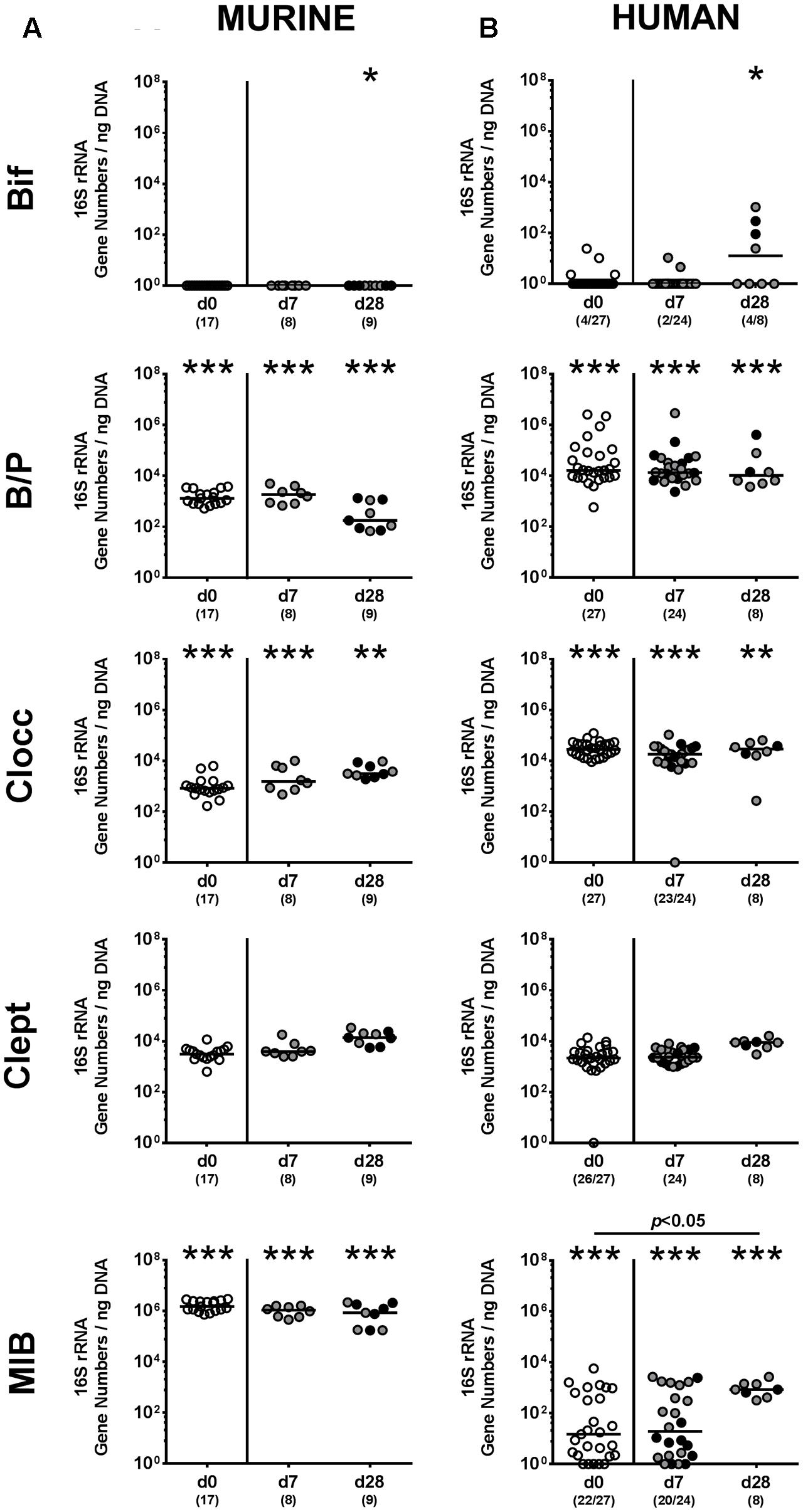
FIGURE 4. Intestinal anaerobic bacterial changes following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with P. aeruginosa on day 0 and day 1. Immediately before (day 0, open circles) and at day 7 and day 28 post-challenge the intestinal anaerobic microbiota composition of colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice was surveyed applying quantitative Real-Time PCR amplifying variable regions of the bacterial 16S rRNA gene. The following main intestinal bacterial groups were determined (expressed as 16S rRNA gene numbers per ng DNA): Bifidobacteria (Bif), Bacteroides/Prevotella spp. (B/P), Clostridium coccoides group (Clocc), Clostridium leptum group (Clept), and Mouse Intestinal Bacteroides (MIB). Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Data were pooled from four independent experiments.
Apoptotic and Proliferating Responses in Intestinal Epithelial Cells upon P. aeruginosa Colonization of Mice Harboring a Complex Human vs. Murine Gut Microbiota
Given that mice were not clinically compromised following P. aeruginosa challenge and did not display symptoms such as weight loss, diarrhea or fecal blood (data not shown), we next assessed potential microscopic sequelae of P. aeruginosa colonization. To address this, we quantitatively determined apoptotic epithelial cells in the large intestines applying in situ immunohistochemistry. Mice following human FMT displayed more than two-fold increased caspase-3 positive apoptotic cell numbers in their colonic epithelia at both day 7 and day 28 p.i. (p < 0.001; Figure 5B), whereas this was only the case later in the course of colonization of murine controls (p < 0.05; Figure 5A). Of note, at day 7 p.i. apoptotic epithelial cells were higher in the large intestines of mice that had been challenged with human gut microbiota as compared to murine controls (p < 0.001; Figures 5A,B). In the small intestines of mice with either microbiota, however, apoptotic cells were slightly higher at day 7 p.i. as compared to non-challenged controls (p < 0.05–0.01; Supplementary Figures S2A,B) and further increased until the end of the observation period (p < 0.05–0.001 vs. day 7 p.i.; Supplementary Figures S2A,B).
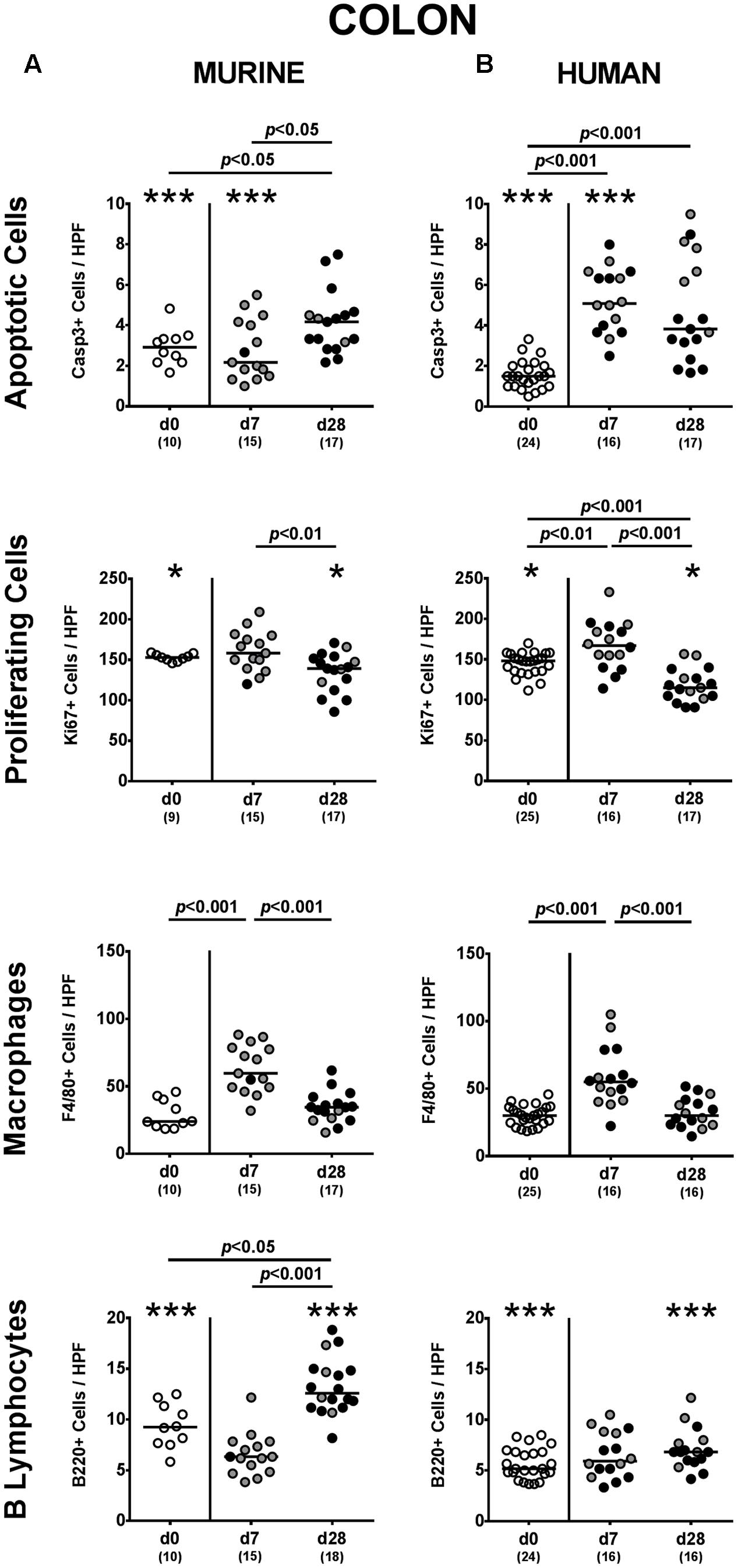
FIGURE 5. Apoptotic and proliferating epithelial cell as well as immune cell responses in the large intestines following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. At day 7 and day 28 post-challenge, the average number of apoptotic (Casp3+) and proliferating (Ki67+) cells as well as of macrophages/monocytes (F4/80+) and B lymphocytes (B220+) in at least six HPF were quantitatively assessed in colonic paraffin sections derived from colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice applying in situ immunohistochemistry. Unchallenged mice (day 0, open circles) harboring a respective murine or human gut microbiota served as negative controls. Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
We additionally assessed Ki67 positive cell numbers in intestinal epithelia indicative for proliferative/regenerative cell responses counteracting potential P. aeruginosa-induced intestinal inflammatory sequelae. Remarkably, only in mice following human FMT, Ki67 positive cells were elevated in large intestines as early as 7 day p.i. (p < 0.01 vs. none-challenged controls), but declined thereafter (p < 0.001; Figure 5B). In the small intestines, proliferative cell responses were highest early in the course of P. aeruginosa colonization, whereas Ki67 positive ileal epithelial cell numbers declined thereafter (Supplementary Figure S2). Hence, P. aeruginosa colonization of mice resulted in increased intestinal epithelial apoptosis that was accompanied by proliferative/regenerative epithelial cell responses.
Intestinal Immune Cell Responses upon P. aeruginosa Colonization of Mice Harboring a Complex Human vs. Murine Gut Microbiota
We next addressed whether colonization of human fecal microbiota transplanted mice with MDR P. aeruginosa was accompanied by an increased influx of distinct immune cell populations into the intestinal tract, again applying quantitative in situ immunohistochemistry. Irrespective of the host-specificity of the intestinal microbiota, numbers of innate immune cells such as F4/80 positive macrophages and monocytes were elevated in the large intestinal mucosa and lamina propria at day 7 p.i., but declined thereafter (p < 0.001; Figure 5). Only in mice with a murine microbiota, however, P. aeruginosa-induced adaptive immune cell responses were more pronounced later in the course of colonization as indicated by increased numbers of colonic B lymphocytes at day 28 p.i. (p < 0.05 vs. non-challenged controls; Figure 5A). In the small intestinal tract, increases in both numbers of macrophages/monocytes as well as of B lymphocytes could only be observed later in the course of P. aruginosa colonization in conventionally colonized, but not human fecal microbiota transplanted mice (p < 0.01–0.001; Supplementary Figure S2A). Hence, rather early in the course of P. aeruginosa colonization increased innate immune cell subsets could be observed in the large intestines of mice harboring either a human or murine gut microbiota, whereas this held true for adaptive immune cells such as B lymphocytes in the small and large intestinal tract of mice with a murine microbiota rather later in the course of P. aeruginosa colonization.
Intestinal Cytokine Responses upon P. aeruginosa Colonization of Mice Harboring a Complex Human vs. Murine Gut Microbiota
We next addressed whether changes in intestinal innate and adaptive immune cell numbers upon P. aeruginosa colonization of human vs. murine fecal microbiota transplanted mice was accompanied by pro- and/or anti-inflammatory cytokine responses within the intestinal tract. At day 7 p.i., increased concentrations of the pro-inflammatory cytokine TNF could be detected in colonic ex vivo biopsies derived from mice harboring a murine, but not human microbiota (p < 0.05; Figure 6A). Large intestinal secretion of the anti-inflammatory cytokine IL-10, however, was dampened at day 7 and day 28 p.i. of mice that had been subjected to human FMT (p < 0.001 and p < 0.05, respectively; Figure 6B) and at the later time point in mice with a murine microbiota (p < 0.01; Figure 6A). We further analyzed cytokine secretion in MLN. Alike the colon, TNF concentrations were elevated in MLN as early as 7 day p.i. of mice harboring a murine microbiota only (p < 0.001), but declined thereafter to basal levels (p < 0.01; Figure 7A). In addition, at day 7 following P. aeruginosa colonization increased IFN-γ secretion could be detected in MLN of both human and murine fecal microbiota transplanted mice (p < 0.001 and p < 0.01, respectively; Figure 7), whereas IL-10 levels remained unchanged (Figure 7). Hence, rather early in the course of murine P. aeruginosa colonization intestinal pro-inflammatory, but not anti-inflammatory cytokine responses were induced.
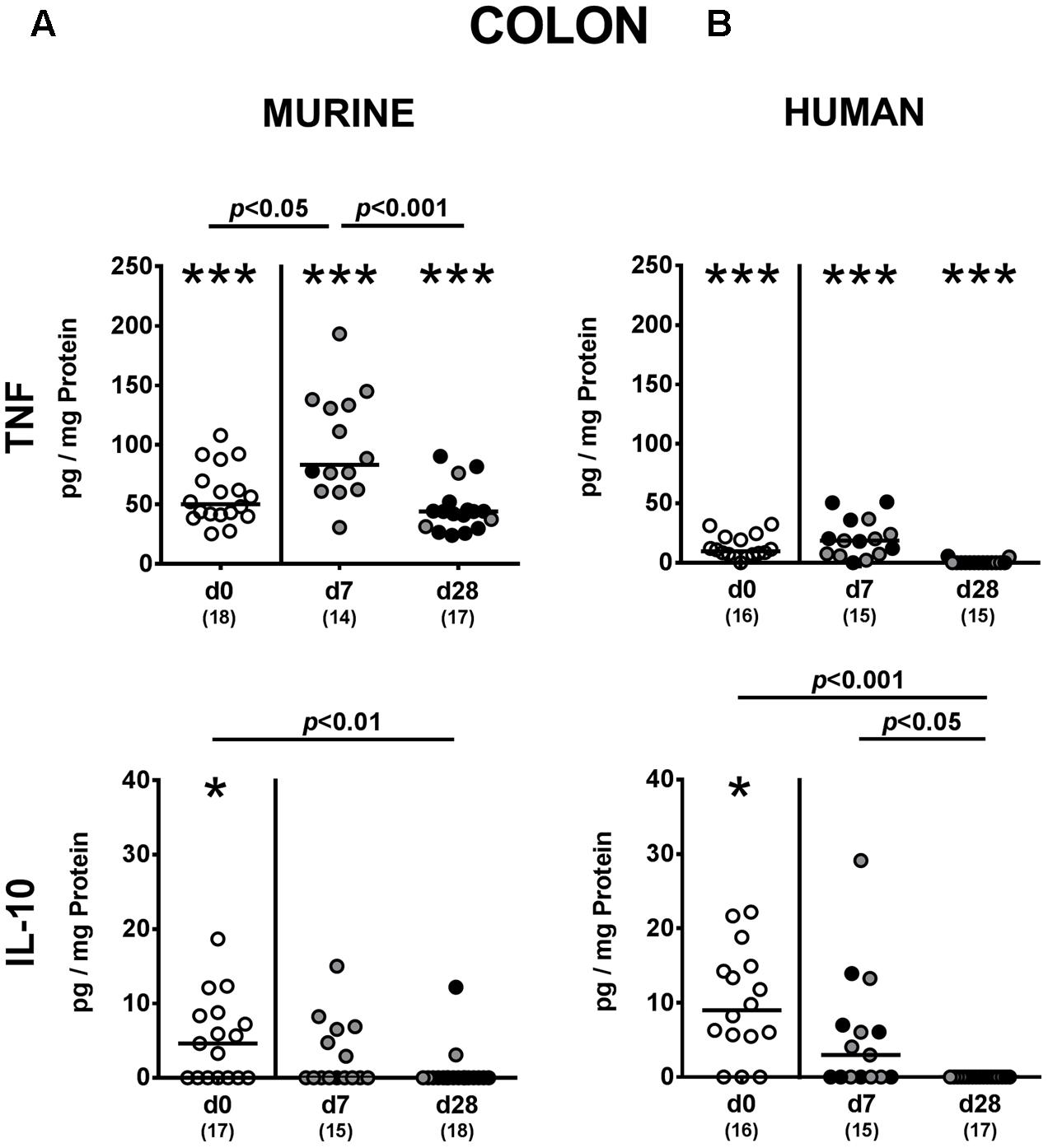
FIGURE 6. Colonic pro- and anti-inflammatory cytokine responses following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. At day 7 and day 28 post-challenge, TNF and IL-10 concentrations were determined in colonic ex vivo biopsies derived from colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice. Unchallenged mice (day 0, open circles) harboring a respective murine or human gut microbiota served as negative controls. Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
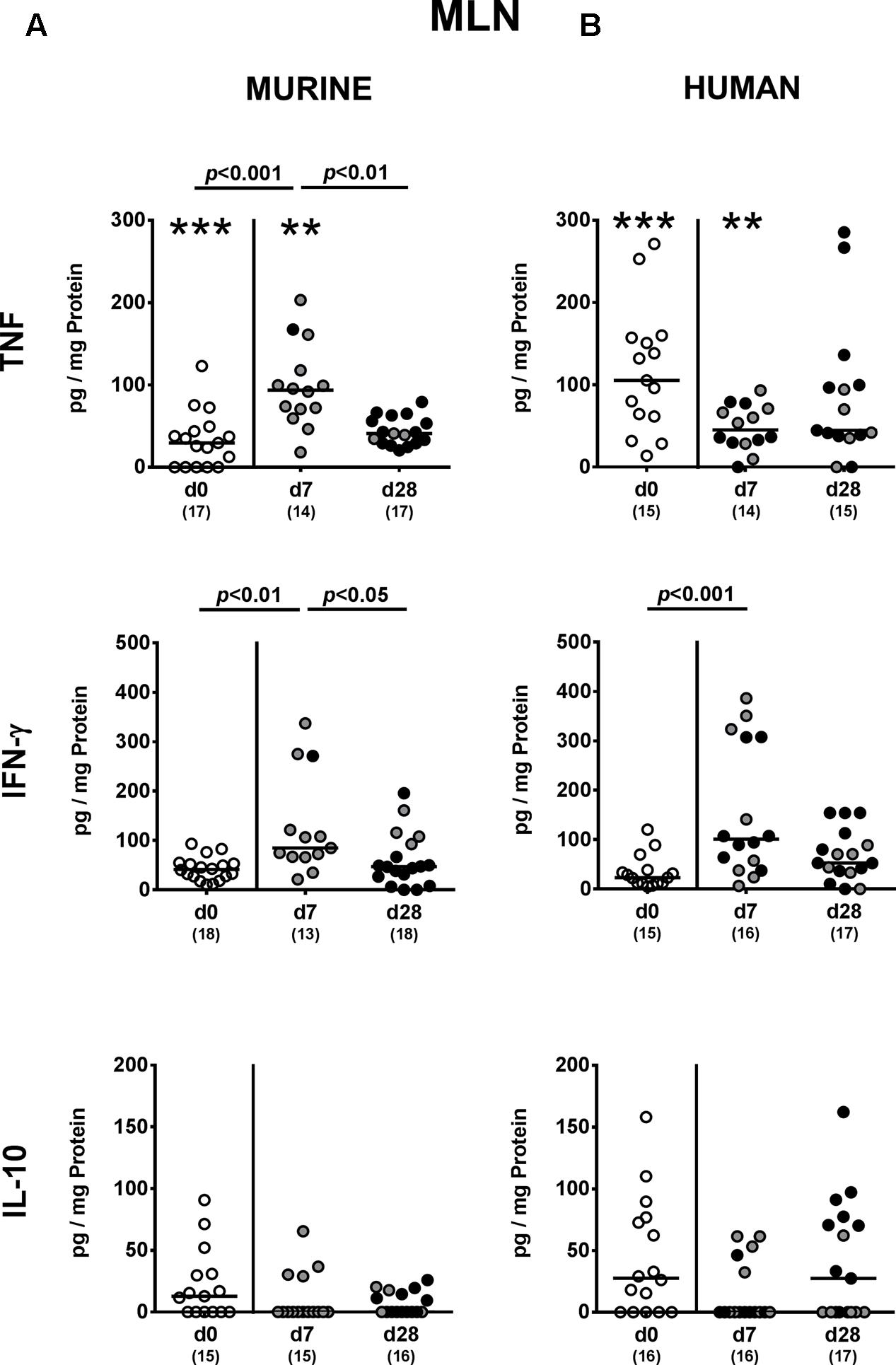
FIGURE 7. Pro- and anti-inflammatory cytokine responses in MLN following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. At day 7 and day 28 post-challenge, TNF, IFN-γ and IL-10 concentrations were determined in ex vivo biopsies derived from MLN of colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice. Unchallenged mice (day 0, open circles) harboring a respective murine or human gut microbiota served as negative controls. Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗∗p < 0.01; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
Systemic Cytokine Responses upon P. aeruginosa Colonization of Mice Harboring a Complex Human vs. Murine Gut Microbiota
We finally addressed whether MDR P. aeruginosa colonization of human and murine fecal microbiota transplanted mice not only induced local (i.e., intestinal), but also systemic cytokine responses and therefore measured pro- and anti-inflammatory cytokine secretion in splenic ex vivo biopsies. Seven days following P. aeruginosa challenge, increased TNF concentrations could be detected in spleens derived from mice harboring a murine gut microbiota, but decreased to naive levels until day 28 p.i. (p < 0.001; Figure 8). In mice that had been subjected to human FMT, a trend towards elevated splenic TNF concentrations could be observed at day 7 p.i. (n.s. due to high standard deviations; p < 0.05 vs. day 28 p.i.; Figure 8B). Whereas INF-γ concentrations were higher in spleens of mice with a murine microbiota at both day 7 and day 28 p.i. (p < 0.01 and p < 0.05, respectively; Figure 8A), splenic INF-γ levels increased in human fecal microbiota transplanted mice at day 7 p.i. only and reached basal concentrations thereafter (p < 0.05; Figure 8B). At day 28 p.i., IL-10 was down-regulated in the spleen of mice with human FMT (p < 0.01; Figure 8B). Hence, cytokine responses upon intestinal colonization with MDR P. aeruginosa were not restricted to the intestinal tract, but could also be observed systemically.
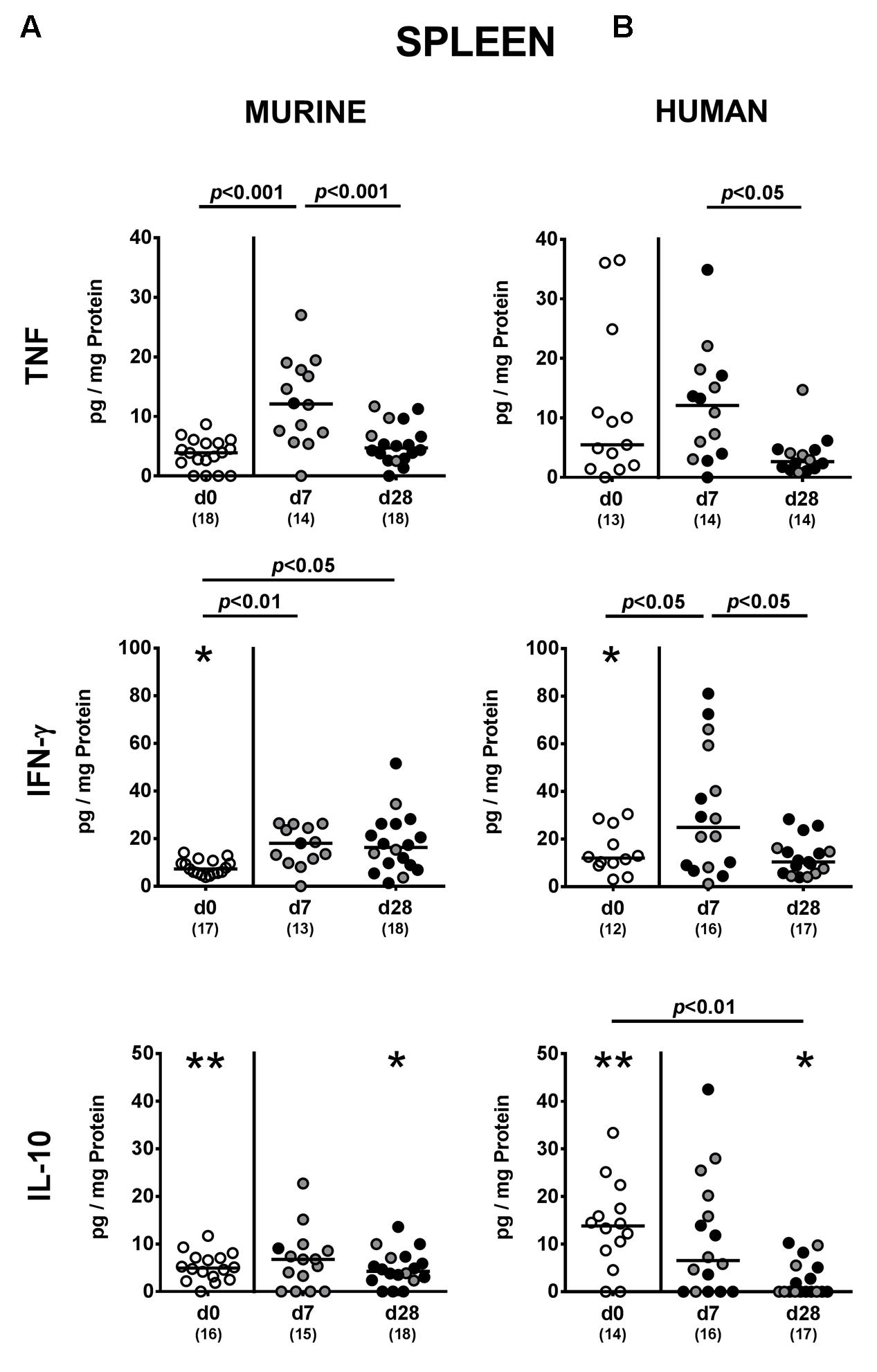
FIGURE 8. Systemic pro- and anti-inflammatory cytokine responses following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. At day 7 and day 28 post-challenge, TNF, IFN-γ and IL-10 concentrations were determined in splenic ex vivo biopsies derived from colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice. Unchallenged mice (day 0, open circles) harboring a respective murine or human gut microbiota served as negative controls. Medians (black bars) and significance levels (p-values) determined by one-way ANOVA or Kruskal–Wallis test (according to data distribution) are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗p < 0.01), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
Discussion
In recent years the rising incidences of infections with MDR Gram-negative bacterial including P. aeruginosa strains has gained increasing attention not only in the medical field, but also in the general public and global health politics (Livermore, 2009; Oliver et al., 2015). Infection and spread of P. aeruginosa have been mainly considered as hospital-associated issues so far, particularly in severely ill patients with antibiotic treatment. P. aeruginosa has not been considered part of the commensal microbiota and to date, only a few studies are available addressing the prevalence of P. aeruginosa carriage in the healthy population ranging from 0 to 24% (Darrell and Wahba, 1964; Shooter et al., 1966; Sutter and Hurst, 1966; Kessner and Lepper, 1967; Stoodley and Thom, 1970; Bleday et al., 1993; Speert et al., 1993; Bonten et al., 1999; Levy, 2000; Kerckhoffs et al., 2011). However, no data are available whether P. aeruginosa carriage, particularly of a MDR strain results in distinct host immune responses.
This prompted us to investigate, whether mice harboring a human vs. murine gut microbiota could be stably colonized by a clinical MDR P. aeruginosa strain and whether opportunistic pathogenic carriage was associated with intestinal or even systemic inflammatory host responses. Our present study revealed that the complex intestinal microbiota, irrespective whether of human or murine origin, only partially prevented mice from intestinal P. aeruginosa colonization. In fact, approximately 50% of mice harboring a human or murine microbiota were P. aeruginosa-negative one week upon supra-physiological challenge with 109 viable bacteria, whereas P. aeruginosa could be cultured at relatively low median loads of less than 104 CFU per g feces in up to 78% of mice at day 28 p.i. One might argue that the colonization resistance exerted by the complex intestinal microbiota composition was not fully restored upon broad-spectrum antibiotic pre-treatment and a 3-weeks period following complex bacterial reconstitution by FMT. Our previous infection studies performed at identical timelines, however, revealed that secondary abiotic mice reconstituted with a murine gut microbiota were protected from stable C. jejuni colonization and hence, from pathogen-induced pro-inflammatory sequelae (Bereswill et al., 2011). In an earlier study ingested P. aeruginosa could be cultured from feces derived from three healthy volunteers up to 6 days following peroral challenge without any clinical sequelae. In line with our results, fecal P. aeruginosa counts were lower as compared to those that had been ingested and subsequently declined over time post ingestion (Buck and Cooke, 1969).
One needs to take into consideration, however, that the applied mice harboring a human vs. murine gut microbiota following antibiotic microbiota depletion and subsequent FMT has its limitations – just like any experimental model – and does, hence, only to a certain degree mimic human (patho)physiological conditions. For instance, some members of the human microbiota, particularly obligate anaerobic species might be reduced or even lost upon freezing, thawing and further processing of the fecal donor samples (von Klitzing et al., 2017b) and/or might furthermore not have stably established within the intraluminal ecosystem in some cases as indicated by the observed differences in microbiota compositions of respective donor suspensions and fecal samples derived following FMT. Furthermore, behavioral and environmental factors including housing conditions, diet, genetic background, the distinct anatomic compartment with its inherent intraluminal milieu and respective immunological reportoire all together impact the composition of the complex host-specific gut microbiota (Turnbaugh et al., 2009; Neyrinck et al., 2011; Nguyen et al., 2015; von Klitzing et al., 2017b). Nevertheless, when taking both the strengths and the limitations of the applied experimental model into consideration, with respect to their gut microbiota “humanized” mice might constitute valuable tools in dissecting the interactions between gut commensals, (opportunistic) pathogens and host immunity in health and disease (von Klitzing et al., 2017b). For instance, mice with a human microbiota have been applied to unravel intestinal colonization properties of enteropathogens such as C. jejuni, Salmonella enterica, and C. difficile (Bereswill et al., 2011; Chung et al., 2012; Collins et al., 2015).
In the present study mice were also clinically uncompromised during the entire observation period following peroral P. aeruginosa challenge. On microscopic level, however, P. aeruginosa induced inflammatory sequelae could be assessed as indicated by increased apoptotic epithelial cell numbers in both small and large intestines as early as 7 days p.i., irrespective of the intestinal microbiota composition of mice. Of note, apoptosis was paralleled by increased proliferative/regenerative epithelial cell responses thereby counteracting potential cell damage. Furthermore, P. aeruginosa colonization was accompanied by a rather early innate immune response as indicated by increased numbers of macrophages and monocytes in the colonic mucosa and lamina propria of human fecal microbiota transplanted mice, whereas small and large intestinal numbers of adaptive immune cells such as B lymphocytes were elevated at the end of the observation period. Pro-inflammatory sequelae of P. aeruginosa colonization are further characterized by increased TNF secretion in both colon and MLN of mice at day 7 p.i., whereas anti-inflammatory IL-10 levels, however, remained unaffected in the intestinal tract. The P. aeruginosa induced immune responses are not surprising given the plethora of virulence factors of the opportunistic pathogen. For instance, Pseudomonas lipid A, a core component of bacterial lipopeptide, is capable of activating NFκB signaling via TLR-4 leading to pro-inflammatory cytokine secretion (Korneev et al., 2015). Subsequently, innate immune cells are recruited to the site of infection contributing to inflammatory host responses directed against P. aeruginosa (Gellatly and Hancock, 2013).
Strikingly, immune responses upon P. aeruginosa carriage were not restricted to the intestinal tract, but could also be observed systemically, given that TNF as well as IFN-γ concentrations were elevated in spleens as early as 7 days p.i., whereas splenic IL-10 levels were dampened in human fecal microbiota transplanted mice later-on. To the best of our knowledge, we here for the first time show that mere intestinal carriage of MDR P. aeruginosa by a clinically unaffected host results in pronounced intestinal as well as systemic sequelae. This is even more surprising, given that not all mice under investigation were stable P. aeruginosa carriers further underlining the pathogenic potential of MDR P. aeruginosa.
Despite several reports of diarrhea caused by P. aeruginosa in patients with prior antibiotic treatment, P. aeruginosa is not regarded as a common intestinal pathogen in the healthy host (Adlard et al., 1998; Kim et al., 2001). As early as 1918, however, “Shanghai fever”, a community acquired syndrome characterized by diarrhea, fever and P. aeruginosa sepsis with high mortality has been reported affecting neonates and children without pre-existing comorbidities primarily in Asian populations (Dold, 1918; Chusid and Hillmann, 1987; Martin-Ancel et al., 1993; Viola et al., 2006; Chuang et al., 2014).
Given the potential cross-talk between (opportunistic) pathogens and commensal intestinal bacteria, we addressed whether changes in respective gut microbiota compositions might occur during P. aeruginosa colonization. Interestingly, no overt gut microbiota shifts could be observed early (i.e., 1 week) upon P. aeruginosa colonization and were only minor in human fecal microbiota transplanted mice thereafter. Previous studies revealed that both acute and chronic small as well as large intestinal inflammation in mice were associated with increased luminal loads of commensal EB further perpetuating the inflammatory scenario via TLR-4 dependent signaling of lipopolysaccharide (LPS) as Gram-negative bacterial cell wall constituent (Heimesaat et al., 2006, 2007a,b, 2010; Erridge et al., 2010; Haag et al., 2012a,b; Otto et al., 2012). However, despite observed pro-inflammatory immune responses with pronounced intestinal epithelial apoptosis, sequelae of intestinal MDR P. aeruginosa carriage was not sufficient enough to drive gut microbiota shifts towards overgrowth with potentially pro-inflammatory commensals.
Conclusion
Our study demonstrates that in with a complex microbiota reconstituted secondary abiotic mice the established intestinal microbiota (irrespective whether of human or murine origin) does not sufficiently prevent the host from intestinal colonization with a MDR P. aeruginosa strain. Furthermore, mere intestinal carriage of MDR P. aeruginosa pose otherwise healthy hosts at risk of developing not only intestinal but even systemic pro-inflammatory sequelae. Future studies need to further unravel the molecular mechanisms underlying the interactions between P. aeruginosa, the commensal microbiota and host immunity in health and disease in order to develop strategies preventing from colonization with MDR Gram-negative strains including P. aeruginosa.
Author Contributions
EvK: designed and performed experiments, analyzed data, co-edited paper. IE: performed experiments, analyzed data, co-edited paper. SB: provided advice in design and performance of experiments, co-edited paper. MH: designed and performed experiments, analyzed data, wrote paper.
Funding
This work was supported by grants from the German Research Foundation (DFG) to SB (SFB633, TP A7), MH (SFB633, TP B6), and EvK and IE (SFB633, Immuco) and by the German Federal Ministeries of Education and Research (BMBF) to SB and MH (PAC-Campy 01KI1725D).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Michaela Wattrodt, Ursula Rüschendorf, Alexandra Bittroff-Leben, Ines Puschendorf, Ulrike Fiebiger, Gernot Reifenberger, and the staff of the animal research facility of the Charité – University Medicine Berlin for excellent technical assistance, and animal breeding.
Abbreviations
Bif, bifidobacteria; B/P, Bacteroides/Prevotella spp.; CFU, colony forming units; C. jejuni, Campylobacter jejuni; Clept, Clostridium leptum group; Clocc, Clostridium coccoides group; EB, enterobacteria; EC, enterococci; FMT, fecal microbiota transplantation; HPF, high power fields; ICU, intensive care unit; LB, lactic acid bacteria; MDR, multi-drug resistant; MIB, Mouse Intestinal Bacteroides; MLN, mesenteric lymph nodes; P, two-sided probability values; PBS, phosphate buffered saline; p.i., post-infection; qRT-PCR, quantitative real-time polymerase chain reaction; SPF, specific pathogen-free; TL, total eubacterial load; TLR, Toll-like receptor; WHO, World Health Organization.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02590/full#supplementary-material
FIGURE S1 | Fecal MDR P. aeruginosa loads in mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. Intestinal colonization densities were assessed in fecal samples at defined time points p.i. by culture. Medians (black bars) and significance levels (p-values) determined by Kruskal–Wallis test are shown. Numbers of samples harboring the respective bacterial group out of the total number of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
FIGURE S2 | Apoptotic and proliferating epithelial cell as well as immune cell responses in the small intestines following MDR P. aeruginosa colonization of mice harboring a human vs. murine microbiota. Mice with a (A) murine or (B) human intestinal microbiota were perorally challenged with MDR P. aeruginosa on day 0 and day 1. At day 7 and day 28 post-challenge, the average number of apoptotic (Casp3+) and proliferating (Ki67+) cells as well as of macrophages/monocytes (F4/80+) and B lymphocytes (B220+) in at least six HPF were quantitatively assessed in ileal paraffin sections derived from colonic P. aeruginosa carrying (black circles) and non-carrying (gray circles) mice applying in situ immunohistochemistry. Unchallenged mice (day 0, open circles) harboring a respective murine or human gut microbiota served as negative controls. Medians (black bars) and significance levels (p-values) determined by Mann–Whitney U-test are shown. Significant differences between mice with a murine vs. human microbiota at defined time points are indicated by asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001), whereas numbered p-values indicate differences between mice harboring the same microbiota. Numbers of analyzed samples are given in parentheses. Data were pooled from four independent experiments.
References
Adlard, P. A., Kirov, S. M., Sanderson, K., and Cox, G. E. (1998). Pseudomonas aeruginosa as a cause of infectious diarrhoea. Epidemiol. Infect. 121, 237–241. doi: 10.1017/S095026889800106X
Alutis, M. E., Grundmann, U., Fischer, A., Hagen, U., Kuhl, A. A., Gobel, U. B., et al. (2015a). The role of gelatinases in Campylobacter jejuni infection of gnotobiotic mice. Eur. J. Microbiol. Immunol. 5, 256–267. doi: 10.1556/1886.2015.00033
Alutis, M. E., Grundmann, U., Hagen, U., Fischer, A., Kuhl, A. A., Gobel, U. B., et al. (2015b). Matrix metalloproteinase-2 mediates intestinal immunopathogenesis in Campylobacter jejuni-infected infant mice. Eur. J. Microbiol. Immunol. 5, 188–198. doi: 10.1556/1886.2015.00020
Bereswill, S., Fischer, A., Plickert, R., Haag, L. M., Otto, B., Kuhl, A. A., et al. (2011). Novel murine infection models provide deep insights into the "menage a trois" of Campylobacter jejuni, microbiota and host innate immunity. PLOS ONE 6:e20953. doi: 10.1371/journal.pone.0020953
Bleday, R., Braidt, J., Ruoff, K., Shellito, P. C., and Ackroyd, F. W. (1993). Quantitative cultures of the mucosal-associated bacteria in the mechanically prepared colon and rectum. Dis. Colon Rectum 36, 844–849. doi: 10.1007/BF02047381
Bonten, M. J., Bergmans, D. C., Speijer, H., and Stobberingh, E. E. (1999). Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. Am. J. Respir. Crit. Care Med. 160, 1212–1219. doi: 10.1164/ajrccm.160.4.9809031
Buck, A. C., and Cooke, E. M. (1969). The fate of ingested Pseudomonas aeruginosa in normal persons. J. Med. Microbiol. 2, 521–525. doi: 10.1099/00222615-2-4-521
Chuang, C. H., Wang, Y. H., Chang, H. J., Chen, H. L., Huang, Y. C., Lin, T. Y., et al. (2014). Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut 63, 736–743. doi: 10.1136/gutjnl-2013-304786
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. doi: 10.1016/j.cell.2012.04.037
Chusid, M. J., and Hillmann, S. M. (1987). Community-acquired Pseudomonas sepsis in previously healthy infants. Pediatr. Infect. Dis. J. 6, 681–684. doi: 10.1097/00006454-198707000-00013
Cohen, R., Babushkin, F., Cohen, S., Afraimov, M., Shapiro, M., Uda, M., et al. (2017). A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob. Resist. Infect. Control 6, 7. doi: 10.1186/s13756-016-0167-7
Collins, J., Auchtung, J. M., Schaefer, L., Eaton, K. A., and Britton, R. A. (2015). Humanized microbiota mice as a model for recurrent Clostridium difficile disease. Microbiome 3:35. doi: 10.1186/s40168-015-0097-2
Darrell, J. H., and Wahba, A. H. (1964). Pyocine-typing of hospital strains of Pseudomonas pyocyanea. J. Clin. Pathol. 17, 236–242. doi: 10.1136/jcp.17.3.236
Dold, H. (1918). On pyocyaneus sepsis and intestinal infections in Shanghai due to Bacillus pyocyaneus. Chin. Med. J. 32:435.
Driscoll, J. A., Brody, S. L., and Kollef, M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368. doi: 10.2165/00003495-200767030-00003
Ekmekciu, I., von Klitzing, E., Fiebiger, U., Escher, U., Neumann, C., Bacher, P., et al. (2017). Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front. Immunol. 8:397. doi: 10.3389/fimmu.2017.00397
Erridge, C., Duncan, S. H., Bereswill, S., and Heimesaat, M. M. (2010). The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLOS ONE 5:e9125. doi: 10.1371/journal.pone.0009125
Fiebiger, U., Bereswill, S., and Heimesaat, M. M. (2016). Dissecting the interplay between intestinal microbiota and host immunity in health and disease: lessons learned from germfree and gnotobiotic animal models. Eur. J. Microbiol. Immunol. 6, 253–271. doi: 10.1556/1886.2016.00036
Gellatly, S. L., and Hancock, R. E. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632x.12033
Gomez-Zorrilla, S., Camoez, M., Tubau, F., Canizares, R., Periche, E., Dominguez, M. A., et al. (2015). Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob. Agents Chemother. 59, 5213–5219. doi: 10.1128/aac.04636-14
Haag, L. M., Fischer, A., Otto, B., Plickert, R., Kuhl, A. A., Gobel, U. B., et al. (2012a). Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10-/- mice via Toll-like-receptor-2 and -4 signaling. PLOS ONE 7:e40761. doi: 10.1371/journal.pone.0040761
Haag, L. M., Fischer, A., Otto, B., Plickert, R., Kuhl, A. A., Gobel, U. B., et al. (2012b). Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLOS ONE 7:e35988. doi: 10.1371/journal.pone.0035988
Heimesaat, M. M., Bereswill, S., Fischer, A., Fuchs, D., Struck, D., Niebergall, J., et al. (2006). Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 177, 8785–8795. doi: 10.4049/jimmunol.177.12.8785
Heimesaat, M. M., Fischer, A., Jahn, H. K., Niebergall, J., Freudenberg, M., Blaut, M., et al. (2007a). Exacerbation of murine ileitis by toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut 56, 941–948. doi: 10.1136/gut.2006.104497
Heimesaat, M. M., Fischer, A., Siegmund, B., Kupz, A., Niebergall, J., Fuchs, D., et al. (2007b). Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLOS ONE 2:e662. doi: 10.1371/journal.pone.0000662
Heimesaat, M. M., Nogai, A., Bereswill, S., Plickert, R., Fischer, A., Loddenkemper, C., et al. (2010). MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087. doi: 10.1136/gut.2009.197434
Hentges, D. J., Stein, A. J., Casey, S. W., and Que, J. U. (1985). Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect. Immun. 47, 118–122.
Kerckhoffs, A. P., Ben-Amor, K., Samsom, M., van der Rest, M. E., de Vogel, J., Knol, J., et al. (2011). Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J. Med. Microbiol. 60(Pt 2), 236–245. doi: 10.1099/jmm.0.022848-0
Kessner, D. M., and Lepper, M. H. (1967). Epidemiologic studies on gram-negative bacilli in the hospital and community. Am. J. Epidemiol. 85, 45–60. doi: 10.1093/oxfordjournals.aje.a120674
Kim, S. W., Peck, K. R., Jung, S. I., Kim, Y. S., Kim, S., Lee, N. Y., et al. (2001). Pseudomonas aeruginosa as a potential cause of antibiotic-associated diarrhea. J. Korean Med. Sci. 16, 742–744. doi: 10.3346/jkms.2001.16.6.742
Korneev, K. V., Arbatsky, N. P., Molinaro, A., Palmigiano, A., Shaikhutdinova, R. Z., Shneider, M. M., et al. (2015). Structural relationship of the lipid a acyl groups to activation of murine toll-like receptor 4 by lipopolysaccharides from pathogenic strains of Burkholderia mallei, Acinetobacter baumannii, and Pseudomonas aeruginosa. Front. Immunol. 6:595. doi: 10.3389/fimmu.2015.00595
Levy, J. (2000). The effects of antibiotic use on gastrointestinal function. Am. J. Gastroenterol. 95 1(Suppl.), S8–S10. doi: 10.1016/S0002-9270(99)00808-4
Livermore, D. M. (2009). Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl. 1), i29–i36. doi: 10.1093/jac/dkp255
Marshall, J. C., Christou, N. V., and Meakins, J. L. (1993). The gastrointestinal tract. The "undrained abscess" of multiple organ failure. Ann. Surg. 218, 111–119. doi: 10.1097/00000658-199308000-00001
Martin-Ancel, A., Borque, C., and del Castillo, F. (1993). Pseudomonas sepsis in children without previous medical problems. Pediatr. Infect. Dis. J. 12, 258–260. doi: 10.1097/00006454-199303000-00022
Masanta, W. O., Heimesaat, M. M., Bereswill, S., Tareen, A. M., Lugert, R., Gross, U., et al. (2013). Modification of intestinal microbiota and its consequences for innate immune response in the pathogenesis of campylobacteriosis. Clin. Dev. Immunol. 2013:526860. doi: 10.1155/2013/526860
Morrison, A. J. Jr., and Wenzel, R. P. (1984). Epidemiology of infections due to Pseudomonas aeruginosa. Rev. Infect. Dis. 6(Suppl. 3), S627–S642. doi: 10.1093/clinids/6.Supplement_3.S627
Neyrinck, A. M., Possemiers, S., Druart, C., Van de Wiele, T., De Backer, F., Cani, P. D., et al. (2011). Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLOS ONE 6:e20944. doi: 10.1371/journal.pone.0020944
Nguyen, T. L., Vieira-Silva, S., Liston, A., and Raes, J. (2015). How informative ist the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16. doi: 10.1242/dmm.017400
Oliver, A., Mulet, X., Lopez-Causape, C., and Juan, C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 22, 41–59. doi: 10.1016/j.drup.2015.08.002
Otto, B., Haag, L. M., Fischer, A., Plickert, R., Kuhl, A. A., Gobel, U. B., et al. (2012). Campylobacter jejuni induces extra-intestinal immune responses via toll-like-receptor-4 signaling in conventional IL-10 deficient mice with chronic colitis. Eur. J. Microbiol. Immunol. 2, 210–219. doi: 10.1556/EuJMI.2.2012.3.7
Potron, A., Poirel, L., and Nordmann, P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int. J. Antimicrob. Agents 45, 568–585. doi: 10.1016/j.ijantimicag.2015.03.001
Shooter, R. A., Walker, K. A., Williams, V. R., Horgan, G. M., Parker, M. T., Asheshov, E. H., et al. (1966). Faecal carriage of Pseudomonas aeruginosa in hospital patients. possible spread from patient to patient. Lancet 2, 1331–1334. doi: 10.1016/S0140-6736(66)92082-4
Speert, D. P., Campbell, M. E., Davidson, A. G., and Wong, L. T. (1993). Pseudomonas aeruginosa colonization of the gastrointestinal tract in patients with cystic fibrosis. J. Infect. Dis. 167, 226–229. doi: 10.1093/infdis/167.1.226
Stoodley, B. J., and Thom, B. T. (1970). Observations on the intestinal carriage of Pseudomonas aeruginosa. J. Med. Microbiol. 3, 367–375. doi: 10.1099/00222615-3-3-367
Stratton, C. W. (1990). Pseudomonas aeruginosa revisited. Infect. Control Hosp. Epidemiol. 11, 101–104. doi: 10.2307/30144269
Sutter, V. L., and Hurst, V. (1966). Sources of Pseudomonas aeruginosa infection in burns: study of wound and rectal cultures with phage typing. Ann. Surg. 163, 597–602. doi: 10.1097/00000658-196604000-00013
Tacconelli, E., and Magrini, N. (2017). Global Priority List of Antibiotic-Resistant Bacteria to Giude research Discovery, and Development of New Antibiotics. Geneva: WHO.
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Vincent, J. L. (2003). Nosocomial infections in adult intensive-care units. Lancet 361, 2068–2077. doi: 10.1016/s0140-6736(03)13644-6
Viola, L., Langer, A., Pulitano, S., Chiaretti, A., Piastra, M., and Polidori, G. (2006). Serious Pseudomonas aeruginosa infection in healthy children: case report and review of the literature. Pediatr. Int. 48, 330–333. doi: 10.1111/j.1442-200X.2006.02214.x
von Klitzing, E., Ekmekciu, I., Bereswill, S., and Heimesaat, M. M. (2017a). Acute ileitis facilitates infection with multidrug resistant Pseudomonas aeruginosa in human microbiota-associated mice. Gut Pathog. 9:4. doi: 10.1186/s13099-017-0154-4
Keywords: multidrug-resistant Gram-negative bacteria, intestinal Pseudomonas aeruginosa colonization, fecal microbiota transplantation, human microbiota associated mice, colonization resistance, host-pathogen-interaction, intestinal and systemic sequelae of colonization, broad-spectrum antibiotic treatment
Citation: von Klitzing E, Ekmekciu I, Bereswill S and Heimesaat MM (2017) Intestinal and Systemic Immune Responses upon Multi-drug Resistant Pseudomonas aeruginosa Colonization of Mice Harboring a Human Gut Microbiota. Front. Microbiol. 8:2590. doi: 10.3389/fmicb.2017.02590
Received: 29 August 2017; Accepted: 12 December 2017;
Published: 22 December 2017.
Edited by:
Bernd Kreikemeyer, University of Rostock, GermanyReviewed by:
Jan Buer, University of Duisburg-Essen, GermanyChristian U. Riedel, University of Ulm, Germany
Copyright © 2017 von Klitzing, Ekmekciu, Bereswill and Heimesaat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus M. Heimesaat, markus.heimesaat@charite.de
 Eliane von Klitzing
Eliane von Klitzing Ira Ekmekciu
Ira Ekmekciu Stefan Bereswill
Stefan Bereswill Markus M. Heimesaat
Markus M. Heimesaat