- Laboratories of General Pathology and Immunology “Giovanna Tosi”, Department of Medicine and Surgery, School of Medicine, University of Insubria, Varese, Italy
Human T cell leukemia virus type 1 (HTLV-1) is an oncogenic human retrovirus that has infected 10–15 million people worldwide. After a long latency, 3–5% of infected individuals will develop either a severe malignancy of CD4+ T cells, known as Adult T-cell Leukemia (ATL) or a chronic and progressive inflammatory disease of the nervous system designated Tropical Spastic Paraparesis/HTLV-1-Associated Myelopathy (HAM/TSP). The precise mechanism behind HTLV-1 pathogenesis still remains elusive. Two viral regulatory proteins, Tax-1 and HTLV-1 bZIP factor (HBZ) are thought to play a critical role in HTLV-1-associated diseases. Tax-1 is mainly involved in the onset of neoplastic transformation and in elicitation of the host’s inflammatory responses; its expression may be lost during cell clonal proliferation and oncogenesis. Conversely, HBZ remains constantly expressed in all patients with ATL, playing a role in the proliferation and maintenance of leukemic cells. Recent studies have shown that the subcellular distribution of HBZ protein differs in the two pathologies: it is nuclear with a speckled-like pattern in leukemic cells and is cytoplasmic in cells from HAM/TSP patients. Thus, HBZ expression and distribution could be critical in the progression of HTLV-1 infection versus the leukemic state or the inflammatory disease. Here, we reviewed recent findings on the role of HBZ in HTLV-1 related diseases, highlighting the new perspectives open by the possibility of studying the physiologic expression of endogenous protein in primary infected cells.
Tax-1 and HBZ: The Two Key Players in HTLV-1-Associated Pathologies
Human T-cell leukemia virus type 1 (HTLV-1) was the first oncogenic retrovirus identified in humans (Poiesz et al., 1980; Yoshida et al., 1982). It is currently estimated that HTLV-1 infects at least 10–15 million people worldwide. Large HTLV-1 endemic areas exist in Southern Japan, the Caribbean, Central and South America, the Middle East, Melanesia, and equatorial regions of Africa (Proietti et al., 2005; Gessain and Cassar, 2012). Although most HTLV-1 infected individuals remain asymptomatic carriers (AC) lifelong, about 3–5% of them develop, after many years of clinical latency, a severe malignancy of CD4+ T cells, known as Adult T-cell Leukemia (ATL).
Four clinical subtypes of HTLV-1 associated ATL are described: smoldering, chronic, lymphoma and acute (Shimoyama, 1991). Depending on the subtype and symptoms, the treatment is different and could include: ‘watchful waiting,’ chemotherapy, antiviral therapy, immunotherapy, allogeneic hematopoietic stem cell transplantation (alloHSCT) and targeted therapies (Utsunomiya et al., 2015). However, despite the available treatments, the prognosis is still extremely poor (Nasr et al., 2017). HTLV-1 is also the causative agent of a neurological disease called tropical spastic paraparesis/HTLV-1-associated myelopathy (HAM/TSP) (Gessain et al., 1985), characterized by different neurological features such as spasticity, muscle weakness and sensory deficits. In general, the central function and cranial nerves are strongly compromised and the clinical course is progressive and without remission (Martin et al., 2014). Due to the wide spectrum of symptoms and the inefficiency of the antiviral therapies, the symptomatic treatment represents the current standard of therapy of this disorder.
The pathogenetic mechanisms at the basis of HTLV-1 infection progressing toward ATL or HAM/TSP are not clearly understood although there are sedimented indications that interactions between the viral encoded proteins and a variety of cellular targets are crucial. Two viral proteins, Tax-1 and HTLV-1 bZIP factor (HBZ), encoded by the sense and antisense viral transcripts, respectively, are largely responsible for the malignant transformation and immortalization of the infected T cells. Tax-1 is a potent activator of viral transcription and is involved in neoplastic transformation through modulation of the expression of cellular genes and deregulation of diverse cell signaling pathways involved in cell proliferation, DNA damage repair, and apoptosis (Hall and Fujii, 2005; Matsuoka and Jeang, 2007; Chlichlia and Khazaie, 2010; Tosi et al., 2011; Forlani et al., 2013) The oncogenic properties of Tax-1 are strongly associated to its ability to constitutively activate the nuclear factor kappa B (NF-kB) pathway (Petropoulos et al., 1996; Forlani et al., 2016). It is of note that the alteration of NF-kB signaling pathway could also be involved in the inflammatory state observed in HAM/TSP (Peloponese et al., 2006). Indeed, it has been suggested that pro-inflammatory CD8+ T lymphocytes (CTL), specific for Tax-1-expressing CD4+ T cells, could infiltrate the central nervous system (CNS), kill the target cells and produce inflammatory cytokines, in particular NF-kB-inducible cytokines, that may participate to tissue damage (Asquith and Bangham, 2000). The high immunogenicity of Tax-1 renders it the major target of effectors CTL, and this event may be responsible for the loss of Tax-1 expression during the long clinically latent period leading to overt ATL (Asquith et al., 2005). Genetic and epigenetic modification in the proviral genome are also responsible of Tax-1 silencing (Takeda et al., 2004). Indeed only 40% of ATL patients can express Tax-1 mRNA suggesting that this viral factor is dispensable for the maintenance of leukemia (Matsuoka and Jeang, 2007). Similar data were reported for the inflammatory diseases, in which it was found that 50% of HAM/TSP patiens can express Tax-1 mRNA. (Usui et al., 2008; Saito et al., 2009; Andrade et al., 2013). Interestingly, our recent findings in a restricted sample of patients demonstrated that, at least at protein level, Tax-1 is found in 75% of HAM/TSP cases and in 100% of HTLV-1 AC, but not in ATL cases (Baratella et al., 2017).
At variance with Tax-1, HBZ mRNA (Matsuoka and Green, 2009) and HBZ protein (Raval et al., 2015) are constantly expressed all ATL cases and in HTLV-1 infected individuals, indicating that HBZ is essential not only for cellular transformation but also for the maintenance of leukemic state. HBZ expression increases the proliferation of HTLV-1 infected T cells in culture, and more importantly induces both T-cell lymphomas and systemic inflammation in mice (Satou et al., 2006, 2011). Several studies have suggested a crucial function of HBZ also in HTLV-1-associated inflammatory disorders as it is always found at both mRNA and protein levels in TSP/HAM patients (Andrade et al., 2013). Interestingly, and at variance with Tax-1, most AC do express HBZ mRNA (Saito et al., 2009; Andrade et al., 2013) but not the protein (Baratella et al., 2017). The fact that HBZ protein is scarcely produced in infected cells may explain why is not, or only marginally, discriminated by HBZ specific CTLs in HTLV-1-infected individuals and in HAM/TSP patients (Hilburn et al., 2011; Rowan et al., 2014). The lower immunogenicity of HBZ compared to Tax-1, and the ability of HBZ to inhibit most of Tax-1 activity, could favor the virus immune escape, thus promoting the spreading of infection and the persistence of viral latency. Several studies have indicated a key function of HBZ in supporting and/or maintaining the proliferation of HTLV-1 infected cells and by this, the initiation and persistence of ATL. It has been reported that HBZ promotes ATL cell proliferation by inhibiting apoptosis through different mechanisms: it impairs the binding of AFT3 to p53, thus affecting the activation of p53-mediated apoptosis signaling (Hagiya et al., 2011); it inhibits the transcriptional activation of pro-apoptotic genes as Bim and Fas Ligand, by interfering with FoxO3 (Tanaka-Nakanishi et al., 2014). It was also reported that HBZ impairs anti-viral immunity responses: it binds to NFAT and inhibits the production of Th1 cytokines (particularly IFN-γ); it induces the expression of TIGIT on the cell surface in ATL (Yasuma et al., 2016) and promotes cells migration and proliferation by enhancing CCR4 expression on T-cell surface (Sugata et al., 2016).
Interestingly, several studies have demonstrated that HBZ exerts opposite effects with respect to Tax-1 on signaling pathways. Tax-1 activates while HBZ selectively inhibits the classical NF-κB pathway by affecting the binding of p65-RelA to its consensus DNA sequence and promoting its degradation. This inhibition leads to the activation of the alternative NF-κB pathway (Zhao et al., 2009). It suppresses Tax-1-mediated viral transactivation by interacting with the KIX domain of p300/CBP and impairing the binding of these cellular factors to Tax-1 (Clerc et al., 2008). HBZ suppresses, while Tax-1 activates, Wnt pathway by interacting with the disheveled-associating protein with a high frequency of Leucine residues (DAPLE) (Ma et al., 2013).
Taken together, all these data emphasize the critical role of HBZ in promoting cellular proliferation and the persistence of the viral infection.
HBZ: Biochemical Aspects and Distinct Subcellular Distribution of Endogenous Protein in ATL and HAM/TSP
Since its discovery in 2002, HBZ has become a crucial hotspot in HTLV-1 research. HBZ gene is encoded by the minus strand of the HTLV-1 RNA genome (Gaudray et al., 2002) and transcribed by a functional promoter, contained in the U5 sequence of the 3′ Long Terminal Repeat (LTR) (Yoshida et al., 2008).
Three major HBZ transcriptional isoforms have been described: the unspliced (usHBZ) form and two alternative spliced form (SP1 and SP2) (Cavanagh et al., 2006; Murata et al., 2006). Regarding the two spliced HBZ isoforms, although both of them have been found in HTLV-1 infected cells, the SP2 variant occurred less frequently than SP1 (Cavanagh et al., 2006). The SP1 spliced and unspliced HBZ transcripts are translated into polypeptides of 206 and 209 amino acids, respectively, and they have almost identical sequences except for a stretch of seven amino acids at the N-terminus of the protein (MAAS for SP1 HBZ and MVNFVSA for usHBZ). However, the half-life of usHBZ protein is much shorter than that of SP1 HBZ (Yoshida et al., 2008) and the expression level of SP1 HBZ is four times higher than that of usHBZ in ATL cells (Usui et al., 2008). Nevertheless, it was reported that the two HBZ protein variants exhibit similar functions (Ma et al., 2016), as they were characterized by conserved functional domains: an N-terminal activation domain (AD), a central domain (CD) and a C-terminal basic ZIP domain (bZIP). HBZ contains three nuclear localization signals (NLS) responsible for its nuclear localization (Hivin et al., 2005; Zhao and Matsuoka, 2012) and two functional nuclear export signals (NES) within its N-terminal region (Mukai and Ohshima, 2014). Most of the reported sub-cellular localizations, biochemical interactions and functional aspects related to HBZ have been assessed in cells overexpressing tagged HBZ. Through its bZIP domain, HBZ was reported to interact with CREB/CREB-2, and this association was instrumental to suppress Tax-mediated HTLV-1 viral transcription (Gaudray et al., 2002). Similar experiments have shown that HBZ binds to different proteins of the JUN family via its bZIP domain. Upon binding to HBZ, JunB, and cJun were recruited in nuclear bodies and degraded, thus HBZ reduces the cJun/JunB-mediated transcriptional activation of a series of genes (Basbous et al., 2003; Thébault et al., 2004). Conversely, the association of HBZ to JunD did not inhibit the JunD-mediated transcriptional activation of target genes; indeed HBZ-JunD complex was reported to increase HBZ gene expression (Thébault et al., 2004; Gazon et al., 2012).
Most of the reported HBZ interactions, however, were assessed by artificial overexpressing systems (transfected cells) and thus it has been hard to extrapolate these results to the real situation encountered in infected cells or in leukemic cells from patients. Only recently, the availability of an anti-HBZ monoclonal antibody (mAb), 4D4-F3, isolated in our laboratory, has made it possible to assess endogenous HBZ expression, localization and interaction in vivo in HTLV-1 infected and in ATL patient (Raval et al., 2015). Indeed, endogenous HBZ interacts and co-localizes with p300 and JunD. Partial colocalization was observed also for CBP and CREB2 (Raval et al., 2015). By using the 4D4-F3 mAb we were able to quantify the HBZ protein demonstrating that the amount of HBZ in ATL patients is around 0.45 × 10-2 pg/cell corresponding to 17.461 molecules/cell, that is 20- to 50-fold lower than the amount expressed in HBZ transfected cells. Similar results were recently obtained by another group (Shiohama et al., 2016). Interestingly, our recent data generated by immunofluorescence with the 4D4-F3 mAb and careful confocal microscopy studies have demonstrated that HBZ protein is expressed in 80 to 100% of ATL cells, in 0.4 to 11% of PBMC of HAM/TSP cells, and very rarely, if any, in PBMC of asymptomatic HTLV-1 carriers (Raval et al., 2015; Baratella et al., 2017).
These studies have also demonstrated that endogenous HBZ protein is localized in the nucleus of ATL cells (Raval et al., 2015; Shiohama et al., 2016) with a similar speckle-like distribution as the one observed in cells transfected with tagged HBZ protein (Gaudray et al., 2002). However, the HBZ nuclear aggregates found in cells overexpressing tagged-HBZ, in particular GFP-HBZ, were shown to be artifacts of chimeric proteins. Although the composition of these nuclear structures containing HBZ is still unknown it has been demonstrated that HBZ-specific nuclear bodies did not overlap with Promyelocytic Leukemia (PML) nuclear bodies (Raval et al., 2015), indicating that HBZ did not co-localize with PML protein. Of note, PML bodies were shown to act as co-activators of Tax-1, without binding to or co-localizing with the viral transactivator (Ariumi et al., 2003)
One of the most important finding of our studies was the demonstration of the distinct subcellular localization of HBZ protein in HAM/TSP as compared to ATL cells. Until now, HBZ has been always assumed to have an exclusive nuclear localization. Thus, it was relatively unexpected to find that HBZ is exclusively localized in the cytoplasm of PBMC of HAM/TSP patients (Figure 1). This cytoplasmic localization was not affected by the presence of leptomycin B, a nuclear export inhibitor, indicating that the viral factor did not shuttle in and out of the nucleus (Baratella et al., 2017). Furthermore, cells expressing cytoplasmic HBZ were almost exclusively found in the CD4+ T cell compartment, very rarely (less than 1%) in CD8+ T cells, and never in B cells or NK cells. Interestingly, the CD4+ HBZ cells, did not expressed the CD25 T cell activation marker (Baratella et al., 2017), thus suggesting that they were either not in rapid proliferation or, if resting, not included in the classical regulatory T cell compartment. This goes in line with recent findings showing that the HBZ-specific humoral immune response correlated with reduced CD4+ T cell activation in HAM/TSP patients (Enose-Akahata et al., 2013). Within this context, the HBZ cytoplasmic localization in HAM/TSP patients may not be appropriate to the generation of peptides that can efficiently bind MHC class I molecules for presentation to, and scrutiny by CTLs. Of further interest was the finding that, at least in the patients’ sample analyzed in our studies, the expression of HBZ and Tax-1 was mutually exclusive (Baratella et al., 2017).
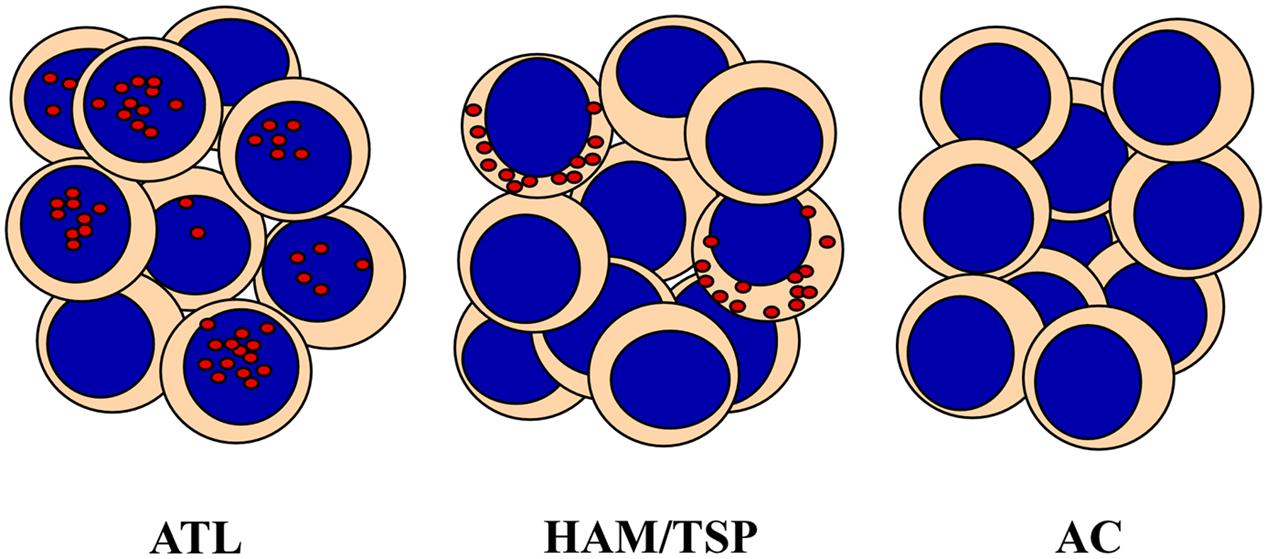
FIGURE 1. HTLV-1 bZIP factor (HBZ) distribution in PBMC from Adult T-cell Leukemia (ATL), HAM/TSP and asymptomatic carriers (AC). HBZ protein localizes in distinct subcellular compartments and with different percentage in leukemic cells of ATL patients and PBMC of HAM/TSP patients. HBZ is expressed in the nucleus of 90% and in the cytoplasm of 20% of cells isolated from ATL or HAM/TSP patients, respectively. AC do not express detectable levels of HBZ.
Taken together, these observations let us to propose the cytoplasmic localization of HBZ protein in PBMC of HAM/TSP patients as the first molecular marker of disease, since until now the only associative parameters with the neurological disease were either clinical correlates or the rather non-specific high proviral load (Nagai et al., 1998). Recently, it has been reported that HBZ-transfected cell lines, including the T cell line Jurkat, may partially segregate HBZ in the cytoplasm as result of interaction with THEMIS (Kinosada et al., 2017). With the limitations expressed above on the physiological correlates of HBZ in overexpressing systems, THEMIS may represent a potential HBZ interactor contributing to the cytoplasmic segregation of HBZ also in HAM/TSP patients’ cells. Future studies in patient settings should clarify this point.
Concluding Remarks and Future Perspectives: The Growing Relevance of HBZ in the Onset of HTLV-1 Associated Diseases
Although several studies have been focussed at clarifying the role of HBZ in HTLV-1-associated diseases, much research is still required to clearly define the molecular and cellular basis of the distinct outcome of viral infection, whether evolving versus the leukemia/lymphoma or versus the neuroinflammatory disorders. In this review, we have summarized the basic molecular aspects of HBZ expression and, importantly, the new findings related to the distinct subcellular localization of HBZ in HTLV-1 infected individuals, AC, affected by ATL or by HAM/TSP neurological disease.
A new paradigm is emerging that suggests a possible roadmap in the evolution of infection based on these new findings (Figure 2). We may think that primary HTLV-1 infection is characterized by the expression of Tax-1 (this stage may be observed in PBMC of still asymptomatic patients), followed by the possible transient coexpression of HBZ (yet to be demonstrated). As we have no specific demonstration of this intermediate step, we do not know whether HBZ protein can localize in the nucleus, cytoplasm or both cellular compartments at these early stages of infection. Immediately after, a compartmentalization of HBZ in the cytoplasm can take place in case of evolution of the infection toward HAM/TSP. At this stage, the evolving pathology can either present a concomitant Tax-1 expression in the same cells (rare), or a mutually exclusive expression of Tax-1 and HBZ in different cells, as we observed in our patients. Conversely, in case of evolution toward the leukemic stage, HBZ will be expressed always and only in the nucleus with or without the coexpression of Tax-1. This hypothetical scheme is useful to test several additional hypotheses and investigate previously unforeseen mechanisms. For example, can we pinpoint a stage in which HBZ is compartmentalized in the cytoplasm still without clinical signs of neurological disease and use this parameter also as a prognostic marker of future development to HAM/TSP? Moreover, what are the molecular mechanisms that drive the distinct nuclear or cytoplasmic localization of the HBZ protein? Are there specific interactors that retain HBZ in the two different cellular compartments? Are these mechanism related to the onset and/or the persistence of the distinct pathologies? These and other fascinating questions’ are now, we believe, experimentally amenable to scrutiny.
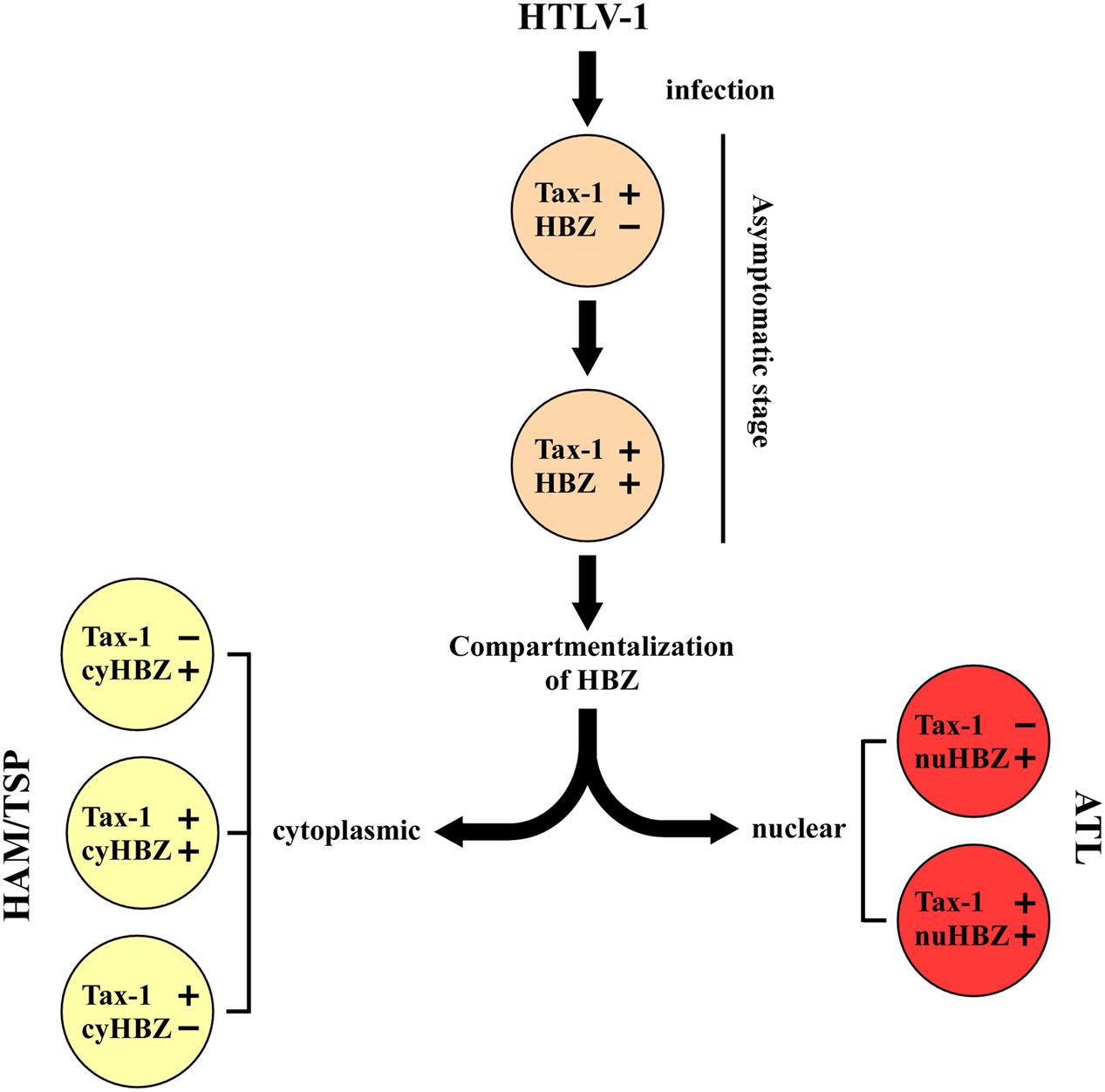
FIGURE 2. A hypothetical model of disease progression in HTLV-1 infected people. The teory is based on the expression and localization of Tax-1 and HBZ proteins at the single cell level. Primary HTLV-1 infection is associated to the expression of Tax-1. The progression of the disease leads to a different stage characterized by the concominant expression of Tax-1 and HBZ. The subsequent localization of HBZ in the nucleus (nuHBZ) or cytoplasm (cyHBZ) points the disease versus the leukemic or the inflammatory state, as more detailed in the text.
Author Contributions
MB, GF, and RA participated in the conception and design of the review and revised the manuscript. All the authors read, critiqued, and approved the final manuscript.
Funding
This work was supported by University of Insubria intramural grant FAR 2014-2016 to RA and GF; by The Institutional Grant 2017, University of Insubria, to RA; by European Community FP7 project HepaVAC Grant Number 602893 to RA (www.hepavac.eu).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Goutham Raval, Dr. Alessandra Tedeschi, and Dr. Farah Bou Nasser Eddine for their participation to several experiments on HBZ localization that were the ground for the results discussed in this review.
References
Andrade, R. G., Goncalves Pde, C., Ribeiro, M. A., Romanelli, L. C., Ribas, J. G., Torres, E. B., et al. (2013). Strong correlation between tax and HBZ mRNA expression in HAM/TSP patients: distinct markers for the neurologic disease. J. Clin. Virol. 56, 135–140. doi: 10.1016/j.jcv.2012.10.003
Ariumi, Y., Ego, T., Kaida, A., Matsumoto, M., Pandolfi, P. P., and Shimotohno, K. (2003). Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene 22, 1611–1619. doi: 10.1038/sj.onc.1206244
Asquith, B., and Bangham, C. R. M. (2000). The role of cytotoxic T lymphocytes in human T-cell lymphotropic virus Type 1 infection. J. Theor. Biol. 207, 65–79. doi: 10.1006/jtbi.2000.2156
Asquith, B., Mosley, A. J., Heaps, A., Tanaka, Y., Taylor, G. P., McLean, A. R., et al. (2005). Quantification of the virus-host interaction in human T lymphotropic virus I infection. Retrovirology 2:75. doi: 10.1186/1742-4690-2-75
Baratella, M., Forlani, G., Raval, G. U., Tedeschi, A., Gout, O., Gessain, A., et al. (2017). Cytoplasmic localization of HTLV-1 HBZ protein: a biomarker of HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). PLOS Negl. Trop. Dis. 11:e0005285. doi: 10.1371/journal.pntd.0005285
Basbous, J., Arpin, C., Gaudray, G., Piechaczyk, M., Devaux, C., and Mesnard, J. M. (2003). The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278, 43620–43627. doi: 10.1074/jbc.m307275200
Cavanagh, M. H., Landry, S., Audet, B., Arpin-André, C., Hivin, P., Paré, M. E., et al. (2006). HTLV-I antisense transcripts initiating in the 3’LTR are alternatively spliced and polyadenylated. Retrovirology 3:15. doi: 10.1186/1742-4690-3-15
Chlichlia, K., and Khazaie, K. (2010). HTLV-1 Tax: linking transformation, DNA damage and apoptotic T-cell death. Chem. Biol. Interact. 188, 359–365. doi: 10.1016/j.cbi.2010.06.005
Clerc, I., Polakowski, N., Andre-Arpin, C., Cook, P., Barbeau, B., Mesnard, J. M., et al. (2008). An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J. Biol. Chem. 283, 23903–23913. doi: 10.1074/jbc.M803116200
Enose-Akahata, Y., Abrams, A., Massoud, R., Bialuk, I., Johnson, K. R., Green, P. L., et al. (2013). Humoral immune response to HTLV-1 basic leucine zipper factor (HBZ) in HTLV-1-infected individuals. Retrovirology 10:19. doi: 10.1186/1742-4690-10-19
Forlani, G., Abdallah, R., Accolla, R. S., and Tosi, G. (2013). The MHC-II transactivator CIITA, a restriction factor against oncogenic HTLV-1 and HTLV-2 retroviruses: similarities and differences in the inhibition of Tax-1 and Tax-2 viral transactivators. Front. Microbiol. 4:234. doi: 10.3389/fmicb.2013.00234
Forlani, G., Abdallah, R., Accolla, R. S., and Tosi, G. (2016). The major histocompatibility complex class II transactivator CIITA inhibits the persistent activation of NF-kB by the human T cell lymphotropic virus Type 1 Tax-1 oncoprotein. J. Virol. 90, 3708–3721. doi: 10.1128/JVI.03000-15
Gaudray, G., Gachon, F., Basbous, J., Biard-Piechaczyk, M., Devaux, C., and Mesnard, J. M. (2002). The complementary strand of the human T-cell leukemia virus Type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76, 12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002
Gazon, H., Lemasson, I., Polakowski, N., Cesaire, R., Matsuoka, M., Barbeau, B., et al. (2012). Human T-cell leukemia virus type 1 (HTLV-1) bZIP factor requires cellular transcription factor JunD to upregulate HTLV-1 antisense transcription from the 3’ long terminal repeat. J. Virol. 86, 9070–9078. doi: 10.1128/JVI.00661-12
Gessain, A., and Cassar, O. (2012). Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 3:388. doi: 10.3389/fmicb.2012.00388
Gessain, A., Vernant, J. C., Maurs, L., Barin, F., Gout, O., Calender, A., et al. (1985). Antibodies to human t-lymphotropic virus type-i in patients with tropical spastic paraparesis. Lancet 326, 407–410. doi: 10.1016/S0140-6736(85)92734-5
Hagiya, K., Yasunaga, J., Satou, Y., Ohshima, K., and Matsuoka, M. (2011). ATF3, an HTLV-1 bZip factor binding protein, promotes proliferation of adult T-cell leukemia cells. Retrovirology 8:19. doi: 10.1186/1742-4690-8-19
Hall, W. W., and Fujii, M. (2005). Deregulation of cell-signaling pathways in HTLV-1 infection. Oncogene 24, 5965–5975. doi: 10.1038/sj.onc.1208975
Hilburn, S., Rowan, A., Demontis, M. A., MacNamara, A., Asquith, B., Bangham, C. R. M., et al. (2011). In vivo expression of human T-lymphotropic virus type 1 basic leucine-Zipper protein generates specific CD8+ and CD4+ T lymphocyte responses that correlate with clinical outcome. J. Infect. Dis. 203, 529–536. doi: 10.1093/infdis/jiq078
Hivin, P., Frédéric, M., Arpin-André, C., Basbous, J., Gay, B., Thébault, S., et al. (2005). Nuclear localization of HTLV-I bZIP factor (HBZ) is mediated by three distinct motifs. J. Cell Sci. 118(Pt 7), 1355–1362. doi: 10.1242/jcs.01727
Kinosada, H., Yasunaga, J.-I., Shimura, K., Miyazato, P., Onishi, C., Iyoda, T., et al. (2017). HTLV- enhances T-cell proliferation by impeding the suppressive signaling of co-inhibitory receptors. PLOS Pathog. 13:e1006120. doi: 10.1371/journal.ppat.1006120
Ma, G., Yasunaga, J., Fan, J., Yanagawa, S., and Matsuoka, M. (2013). HTLV-1 bZIP factor dysregulates the Wnt pathways to support proliferation and migration of adult T-cell leukemia cells. Oncogene 32, 4222–4230. doi: 10.1038/onc.2012
Ma, G., Yasunaga, J., and Matsuoka, M. (2016). Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology 13:16. doi: 10.1186/s12977-016-0249-x
Martin, F., Taylor, G. P., and Jacobson, S. (2014). Inflammatory manifestations of HTLV-1 and their therapeutic options. Expert Rev. Clin. Immunol. 10, 1531–1546. doi: 10.1586/1744666X.2014.966690
Matsuoka, M., and Green, P. L. (2009). The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology 6:71. doi: 10.1186/1742-4690-6-71
Matsuoka, M., and Jeang, K. T. (2007). Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280. doi: 10.1038/nrc2111
Mukai, R., and Ohshima, T. (2014). HTLV-1 HBZ positively regulates the mTOR signaling pathway via inhibition of GADD34 activity in the cytoplasm. Oncogene 33, 2317–2328. doi: 10.1038/onc.2013.181
Murata, K., Hayashibara, T., Sugahara, K., Uemura, A., Yamaguchi, T., Harasawa, H., et al. (2006). A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 80, 2495–2505. doi: 10.1128/jvi.80.5.2495-2505.2006
Nagai, M., Usuku, K., Matsumoto, W., Kodama, D., Takenouchi, N., Moritoyo, T., et al. (1998). Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 4, 586–593. doi: 10.3109/13550289809114225
Nasr, R., Marçais, A., Hermine, O., and Bazarbachi, A. (2017). Overview of targeted therapies for adult T-Cell leukemia/lymphoma. Methods Mol. Biol. 1582, 197–216. doi: 10.1007/978-1-4939-6872-5_15
Peloponese, J. M., Yeung, M. L., and Jeang, K. T. (2006). Modulation of nuclear factor-kappaB by human T cell leukemia virus type 1 tax protein: implications for oncogenesis and inflammation. Immunol. Res. 34, 1–12. doi: 10.1385/IR:34:1:1
Petropoulos, L., Lin, R., and Hiscott, J. (1996). Human T cell leukemia virus Type 1 tax protein increases NF-κB dimer formation and antagonizes the inhibitory activity of the IκBα regulatory protein. Virology 225, 52–64. doi: 10.1006/viro.1996.0574
Poiesz, B. J., Ruscetti, F. W., Gazdar, A. F., Bunn, P. A., Minna, J. D., and Gallo, R. C. (1980). Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 77, 7415–7419. doi: 10.1073/pnas.77.12.7415
Proietti, F. A., Carneiro-Proietti, A. B., Catalan-Soares, B. C., and Murphy, E. L. (2005). Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24, 6058–6068. doi: 10.1038/sj.onc.1208968
Raval, G. U., Bidoia, C., Forlani, G., Tosi, G., Gessain, A., and Accolla, R. S. (2015). Localization, quantification and interaction with host factors of endogenous HTLV-1 HBZ protein in infected cells and ATL. Retrovirology 12:59. doi: 10.1186/s12977-015-0186-0
Rowan, A. G., Suemori, K., Fujiwara, H., Yasukawa, M., Tanaka, Y., Taylor, G. P., et al. (2014). Cytotoxic T lymphocyte lysis of HTLV-1 infected cells is limited by weak HBZ protein expression, but non-specifically enhanced on induction of Tax expression. Retrovirology 11:116. doi: 10.1186/s12977-014-0116-6
Saito, M., Matsuzaki, T., Satou, Y., Yasunaga, J., Saito, K., Arimura, K., et al. (2009). In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 6:19. doi: 10.1186/1742-4690-6-19
Satou, Y., Yasunaga, J., Zhao, T., Yoshida, M., Miyazato, P., Takai, K., et al. (2011). HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLOS Pathog. 7:e1001274. doi: 10.1371/journal.ppat.1001274
Satou, Y., Yasunaga, J.-I., Yoshida, M., and Matsuoka, M. (2006). HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 103, 720–725. doi: 10.1073/pnas.0507631103
Shimoyama, M. (1991). Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the lymphoma study group (1984–87). Br. J. Haematol. 79, 428–437. doi: 10.1111/j.1365-2141.1991.tb08051.x
Shiohama, Y., Naito, T., Matsuzaki, T., Tanaka, R., Tomoyose, T., Takashima, H., et al. (2016). Absolute quantification of HTLV-1 basic leucine zipper factor (HBZ) protein and its plasma antibody in HTLV-1 infected individuals with different clinical status. Retrovirology 13:29. doi: 10.1186/s12977-016-0263-z
Sugata, K., Yasunaga, J., Kinosada, H., Mitobe, Y., Furuta, R., Mahgoub, M., et al. (2016). HTLV-1 viral factor HBZ induces CCR4 to promote T-cell migration and proliferation. Cancer Res. 76, 5068–5079. doi: 10.1158/0008-5472.CAN-16-0361
Takeda, S., Maeda, M., Morikawa, S., Taniguchi, Y., Yasunaga, J., Nosaka, K., et al. (2004). Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer 109, 559–567. doi: 10.1002/ijc.20007
Tanaka-Nakanishi, A., Yasunaga, J., Takai, K., and Matsuoka, M. (2014). HTLV-1 bZIP factor suppresses apoptosis by attenuating the function of FoxO3a and altering its localization. Cancer Res. 74, 188–200. doi: 10.1158/0008-5472.CAN-13-0436
Thébault, S., Basbous, J., Hivin, P., Devaux, C., and Mesnard, J. M. (2004). HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 562, 165–170. doi: 10.1016/s0014-5793(04)00225-x
Tosi, G., Forlani, G., Andresen, V., Turci, M., Bertazzoni, U., Franchini, G., et al. (2011). Major histocompatibility complex class II transactivator CIITA is a viral restriction factor that targets human T-cell lymphotropic virus type 1 Tax-1 function and inhibits viral replication. J. Virol. 85, 10719–10729. doi: 10.1128/JVI.00813-11
Usui, T., Yanagihara, K., Tsukasaki, K., Murata, K., Hasegawa, H., Yamada, Y., et al. (2008). Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells. Retrovirology 5:34. doi: 10.1186/1742-4690-5-34
Utsunomiya, A., Choi, I., Chihara, D., and Seto, M. (2015). Recent advances in the treatment of adult T-cell leukemia-lymphomas. Cancer Sci. 106, 344–351. doi: 10.1111/cas.12617
Yasuma, K., Yasunaga, J., Takemoto, K., Sugata, K., Mitobe, Y., Takenouchi, N., et al. (2016). HTLV-1 bZIP factor impairs anti-viral immunity by inducing co-inhibitory molecule, T cell immunoglobulin and ITIM domain (TIGIT). PLOS Pathog. 12:e1005372. doi: 10.1371/journal.ppat.1005372
Yoshida, M., Miyoshi, I., and Hinuma, Y. (1982). Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. U.S.A. 79, 2031–2035. doi: 10.1073/pnas.79.6.2031
Yoshida, M., Satou, Y., Yasunaga, J., Fujisawa, J., and Matsuoka, M. (2008). Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J. Virol. 82, 9359–9368. doi: 10.1128/JVI.00242-08
Zhao, T., and Matsuoka, M. (2012). HBZ and its roles in HTLV-1 oncogenesis. Front. Microbiol. 3:247. doi: 10.3389/fmicb.2012.00247
Keywords: HTLV-1, ATL, HAM/TSP, HBZ, Tax-1
Citation: Baratella M, Forlani G and Accolla RS (2017) HTLV-1 HBZ Viral Protein: A Key Player in HTLV-1 Mediated Diseases. Front. Microbiol. 8:2615. doi: 10.3389/fmicb.2017.02615
Received: 07 November 2017; Accepted: 14 December 2017;
Published: 22 December 2017.
Edited by:
Umberto Bertazzoni, University of Verona, ItalyReviewed by:
Jean-Marie Peloponese, UMR9004 Institut de Recherche en Infectiologie de Montpellier (IRIM), FranceMasao Matsuoka, Kyoto University, Japan
Copyright © 2017 Baratella, Forlani and Accolla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto S. Accolla, roberto.accolla@uninsubria.it
 Marco Baratella
Marco Baratella Greta Forlani
Greta Forlani Roberto S. Accolla
Roberto S. Accolla