- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, South Korea
- 2Centers for Disease Control and Prevention, National Institute of Health, Cheongju, South Korea
Between 2014 and 2015, the Klebsiella pneumoniae carbapenemase (KPC) was becoming endemic in South Korea. To assess this period of transition, we analyzed KPC producers in terms of molecular epidemiology. A total of 362 KPC-producing Enterobacteriaceae strains, including one from 2013, 13 from 2014, and 348 from 2015, were actively collected from 60 hospitals throughout the peninsula. Subtypes of KPC were determined by PCR and direct sequencing, and isotypes of Tn4401 (the transposon flanking the blaKPC gene) were specified by PCR using isotype-specific primers and direct sequencing. Sporadic occurrence of KPC-producing Enterobacteriaceae was initially observed around Seoul, which is the most crowded district of the country, and these strains rapidly disseminated in 2014, to the other parts of the country in 2015. The bacterial clones responsible for the extreme epidemiological transition were K. pneumoniae ST307 (46.2%) and ST11 (21.3%). Less frequently, E. coli (4.7%), Enterobacter spp. (1.4%), and other Enterobacteriaceae members (1.7%) producing the enzyme were identified. The blaKPC-2 gene bracketed by Tn4401a (72.1%) was the most prevalent mobile genetic element responsible for the dissemination, and the same gene carried either by Tn4401b (2.2%) or Tn4401c (6.6%) was identified at a lesser frequency. The genes blaKPC-3 (1.6%) and blaKPC-4 (6.4%), both flanked by Tn4401b, were occasionally identified. This study showed endemic dissemination of KPC producers in 2015 due to a clonal spread of two K. pneumoniae strains. Further systemic surveillance is needed to monitor dissemination of KPC-producers.
Introduction
Carbapenemases often jeopardize chemotherapy for infectious diseases caused by common nosocomial pathogens. Currently, Klebsiella pneumoniae carbapenemases (KPC) belonging to the Ambler class A carbapenemases are the most clinically worrisome enzymes because of their global spreads and a broad-spectrum of substrates including most of the β-lactam drugs, except cephamycins (Munoz-Price et al., 2013; Pitout et al., 2015). KPC producers were first identified for K. pneumoniae from the United States in 1996 (Yigit et al., 2001) and, so far, a total of 23 KPC variants have been identified (http://www.lahey.org/Studies). KPC producers, particularly among Enterobacteriaceae isolates, are now being extensively reported worldwide. In some regions of the world, such as North America (some regions of the USA), Latin America (Colombia, Brazil, and Argentina), Europe (Greece and Italy), the Middle East (Israel), and Asia (some regions of China), KPC dissemination has reached endemic levels (Albiger et al., 2015; Mehta et al., 2015).
The rapid dissemination of the blaKPC gene is known to be derived from the clonal spread of K. pneumoniae sequence type 258 (ST258) (Munoz-Price et al., 2013; Pitout et al., 2015). Besides, horizontal gene transfer drives KPC dissemination via a Tn3-type active transposon Tn4401 enclosing the blaKPC gene. The transposon is bracketed by 39-bp imperfect inverted repeat sequences capable of mobilizing the blaKPC gene at high frequency and it generates a 5-bp target duplication by insertion without target site specificity (Cuzon et al., 2011). At present, nine isotypes of Tn4401 (Tn4401a to Tn4401i) differing by the upstream sequences of the blaKPC gene were identified. The blaKPC gene promoters varied by the Tn4401 isotypes control the level of gene expression resulting in varied levels of carbapenem resistance (Naas et al., 2012; Cheruvanky et al., 2017).
The National Laboratory Surveillance System (NLSS) for CPEs started in November 2010 by the Korea Center for Disease Prevention and Control (KCDC) with 100 sentinel general hospitals (unpublished data). Through the surveillance study, an alarming transition of KPC epidemiological stages to inter-regional spread was observed between 2014 and 2015. The population of KPC producers and the inter-regional movement of core clones needed to be further investigated and, in this study, the molecular epidemiology was assessed with actively collected KPC producers located throughout the peninsula.
Materials and Methods
Strains
A total of 362 non-duplicate KPC-producing Enterobacteriaceae strains (one from 2013, 13 from 2014, and 348 from 2015) were collected voluntarily from 60 hospitals: 16 NLSS-non-participating hospitals including 12 general hospitals, two convalescent hospitals, and two clinics, and 44 NLSS-participating general hospitals. Further testing including the species identification by 16S rDNA sequencing was performed in a reference laboratory.
Antimicrobial Susceptibility and Resistance Analyses
Antimicrobial susceptibility to eight drugs (aztreonam, cefotaxime, ceftazidime, cefoxitin, gentamicin, amikacin, ciprofloxacin, and tigecycline) was evaluated by the disk diffusion method on Mueller-Hinton (MH) agar (Difco Laboratories, Detroit, MI), following CLSI guidelines (Clinical and Laboratory Standards Institute, 2015b). The MICs for colistin were assessed by the broth microdilution method using MH broth (Difco Laboratories), following the recommendations of the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group (The joint CLSI–EUCAST and Polymyxin Breakpoints Working Group, 2016). The MICs for imipenem and meropenem were determined by the agar dilution method, following CLSI guidelines (Clinical and Laboratory Standards Institute, 2015a). Both Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used for quality control. The blaKPC gene was detected by PCR using gene-specific primers (Supplementary Table), and direct sequencing was carried out for subtyping KPC. Additional resistance determinants were identified by PCR and direct sequencing: blaCTX-M-1-like, blaCTX-M-9-like, and blaCTX-M-25-like for CTX-M-type extended-spectrum β-lactamases (ESBLs); blaACT, blaACC, blaCMY, and blaDHA for plasmid-mediated AmpCs; and armA, rmtA, and rmtB for16S ribosomal methyltransferases (Perez-Perez and Hanson, 2002; Ryoo et al., 2005). Multiple-carbapenemase producers of positive amplification either of blaOXA-48-like, blaIMP, blaVIM, blaGES, and blaNDM genes were eliminated from the study.
Isotyping the Tn4401 Transposon
To determine the isotypes of Tn4401, isotype-specific forward primers for the five most common isotypes (a to e) (Naas et al., 2012) were newly designed (Supplementary Table) and PCR was carried out with a universal reverse primer targeting the blaKPC gene. The amplicons were sequenced for verification.
Multilocus Sequence Typing (MLST)
PCR and sequencing were carried out for seven housekeeping genes: gapA, infB, mdh, pgi, phoE, rpoB, and tonB for K. pneumoniae (Diancourt et al., 2005), and adk, fumC, gyrB, icd, mdh, purA, and recA for E. coli (Wirth et al., 2006). The nucleotide sequences obtained for both DNA strands were compared with sequences in the MLST database for each species (http://bigsdb.web.pasteur.fr/klebsiella for K. pneumoniae and http://mlst.warwick.ac.uk/mlst/dbs/Ecoli for E. coli), to determine allelic numbers and STs.
Results
The Epidemiology of KPC-Producing Enterobacteriaceae
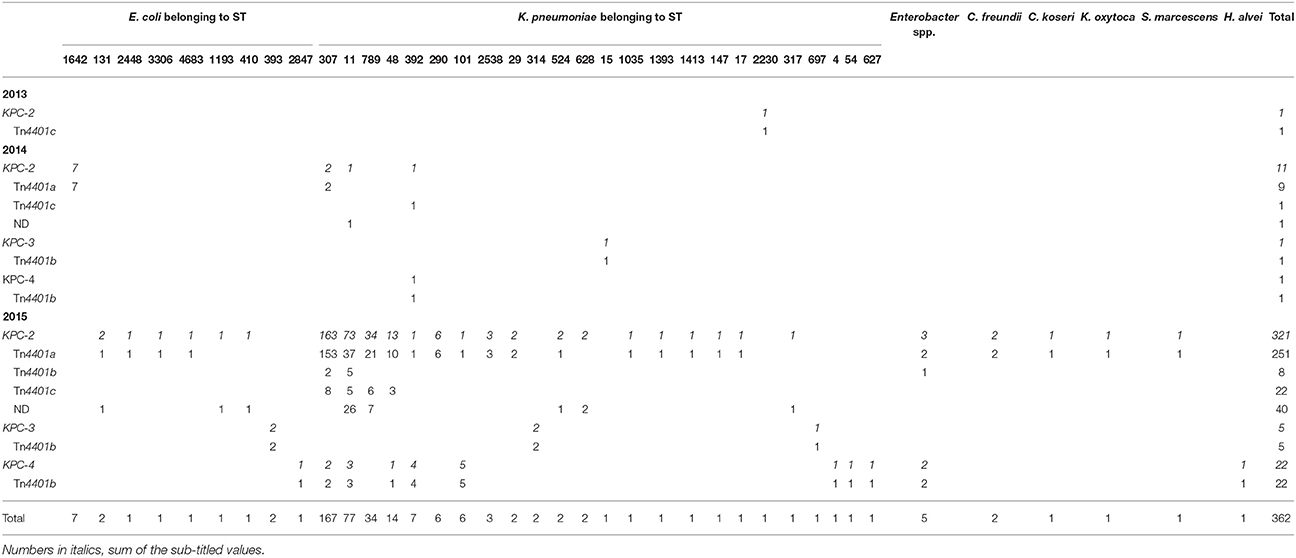
Among the 362 KPC-producing Enterobacteriaceae strains, K. pneumoniae (334/362, 92.2%) was the predominant bacterial species and E. coli (17/362, 4.7%) was followed by Enterobacter spp. (5/362, 1.4%), Citrobacter spp. (3/362, 0.8%), Klebsiella oxytoca (1/362, 0.3%), Serratia marcescens (1/362, 0.3%), and Hafnia alvei (1/362, 0.3%) were identified less frequently.
Three subtypes KPC-2 (334/362, 92.3%), KPC-3 (6/362, 1.7%), and KPC-4 (22/362, 6.1%) were identified. Major isotype of the Tn4401 bracketing the blaKPC gene was Tn4401a (261/362, 72.0%) and, less frequently, Tn4401b (36/362, 9.9%) and Tn4401c (24/362, 6.6%) were identified. Tn4401b was linked with all three subtypes of the blaKPC gene, while Tn4401a and Tn4401c carried only the blaKPC-2 gene. For the non-isotypable 41 transposons (11.3%) by the Tn4401-isotyping PCR, amplification between the 3′-end of the upstream ISKpn7 and the blaKPC gene was tried and only one of the 41 was identified having a truncated Tn3 at the isotype-determining region, while none produced any amplicons. The non-isotype-determined (ND) Tn4401s were coupled only with blaKPC-2 gene, whether in K. pneumoniae (n = 38) or in E. coli (n = 3).
The blaKPC-2 gene bracketed by Tn4401a (Tn4401a[blaKPC-2]) was harbored by K. pneumoniae (n = 243) of varied STs [mainly ST307 (n = 155), ST11 (n = 37), and ST789 (n = 21)], E. coli (n = 11) [mostly of ST1624 (n = 7)], and other Enterobacteriaceae [Enterobacter spp. (n = 2), C. freundii (n = 2), C. koseri (n = 1), K. oxytoca (n = 1), and S. marcescens (n = 1)] (Table 1). Tn4401b[blaKPC-2] was carried either by K. pneumoniae (n = 7), five ST11 and two ST307, or by Enterobacter spp. (n = 1). Tn4401c[blaKPC-2] was carried only by K. pneumoniae (n = 24) belonging to ST307 (n = 8), ST789 (n = 6), ST11 (n = 5), ST48 (n = 3), ST392 (n = 1), and ST2230 (n = 1). The Tn4401b[blaKPC-3] was identified in E. coli ST393 (n = 2) and in K. pneumoniae (n = 4, specifically two ST314 and one each of ST15 and ST697) and Tn4401b[blaKPC-4] was identified in K. pneumoniae (specifically five ST392, four ST101, three ST11, two ST307, and one of each ST4, ST48, ST54, and ST627), E. coli ST2847 (n = 1), Enterobacter spp. (n = 2), and Hafnia alvei (n = 1).
Dissemination of KPC Producers throughout the South Korean Peninsula from 2013 to 2015
One K. pneumoniae isolate ST2230/Tn4401c[blaKPC-2] was collected in Seoul in 2013. In 2014, 13 KPC producers were identified including six K. pneumoniae strains, i.e., two ST307/Tn4401a[blaKPC-2], one ST11/ND[blaKPC-2], one ST392/Tn4401c[blaKPC-2], one ST392/Tn4401b[blaKPC-4], and one ST15/Tn4401b[blaKPC-3], along with seven E. coli ST1642/Tn4401a[blaKPC-2] strains from a single-hospital outbreak in a suburb of Seoul (Incheon). In 2015, a total of 348 KPC producers were identified including 327 K. pneumoniae [mainly ST307/Tn4401a[blaKPC-2] (153/327, 46.8%), ST11/Tn4401a[blaKPC-2] (37/327, 11.3%), ST25/ND[blaKPC-2] (25/327, 7.6%), and ST789/Tn4401a[blaKPC-2] (21/327, 6.4%)] and 10 E. coli strains of nine subclones. Notably, five KPC-producing K. pneumoniae isolates were collected in 2015 from other healthcare facilities than general hospitals: ST307/Tn4401a[blaKPC-2], ST11/Tn4401a[blaKPC-2], and ST290/Tn4401a[blaKPC-2]) from two convalescent hospitals and ST48/Tn4401c[blaKPC-2] and ST307/Tn4401a[blaKPC-2] from two clinics.
The K. pneumoniae ST307/Tn4401a[blaKPC-2], ST11/ND[blaKPC-2], and ST392/Tn4401b[blaKPC-4] clones were identified over two consecutive years. In 2014, two K. pneumoniae ST307/Tn4401a[blaKPC-2] were first identified in Seoul. In 2015, the same clone was identified with 153 strains in Seoul (n = 49) and three more districts including a suburb of Seoul (Gyeonggi, n = 12), a southeastern peninsula (Busan, n = 91), and a southwestern peninsula (Jeonbuk, n = 1; Figure 1). Besides, the K. pneumoniae ST11/ND[blaKPC-2] clone was first identified in Seoul in 2014 and, in 2015, the same clone was identified in Seoul (n = 12), two suburbs of Seoul (Gyeonggi, n = 1; Incheon, n = 1), and in southeastern peninsula (Gyeongbuk, n = 2; Busan, n = 1). The K. pneumoniae ST392/Tn4401b[blaKPC-4] clone from Incheon in 2014 was subsequently identified with four isolates in 2015 from the same district.
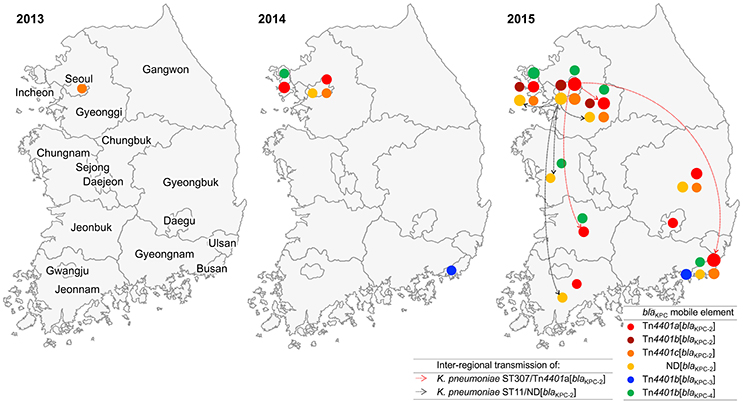
Figure 1. Appearance and inter-regional dissemination of KPC producers carrying varied Tn4401 isotypes between 2013 and 2015. Colored dots indicate the subtypes of the blaKPC gene and isotypes of Tn4401 that were located in each district. The size of each dot represents the number of isolates on a log scale. Inter-regional dissemination of the two KPC-producing clones is denoted with broken arrows.
Carbapenem Resistance Conferred by the blaKPC Gene
Overall MIC50s of imipenem and meropenem in KPC producers were 16 mg/L and 8 mg/L, respectively (Figure 2), and the values met the three predominant factors in the KPC populations: the host bacterial species (K. pneumoniae), the resistance gene (blaKPC-2), and the transposon encoding the gene (Tn4401a). The KPC-producing K. pneumoniae had three-dilution higher MIC50s of the two carbapenems than E. coli and KPC-2 producers had three- and four-dilution higher MIC50 values than to the KPC-3 and KPC-4 producers, respectively. The KPC producers carrying Tn4401a had one- or two-dilution higher MIC50 values of the carbapenemase than KPC producers carrying Tn4401b, whereas those carrying Tn4401c had the same MIC50 values as KPC producers carrying Tn4401a. The ND transposon encoding blaKPC-2 (n = 40) had the highest MIC50 of meropenem 32 mg/L that was two- to three-dilution higher than the three isotypes of Tn4401, whereas that of imipenem looked alike.
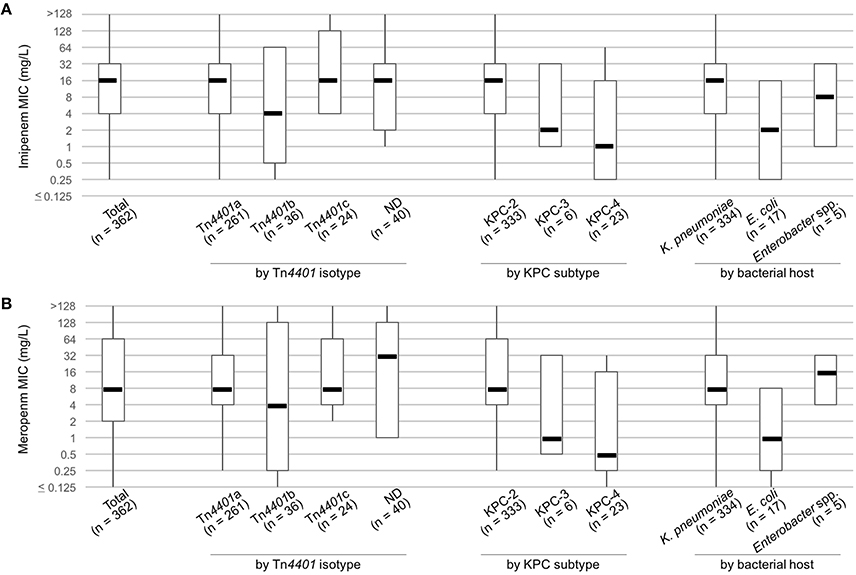
Figure 2. MICs of imipenem (A) and meropenem (B) in KPC producers. Boxplots present MIC50, MIC10, and MIC90 values, with whiskers showing the maximum and minimum MICs of the group.
Cross- and Co-resistance to Other Drugs
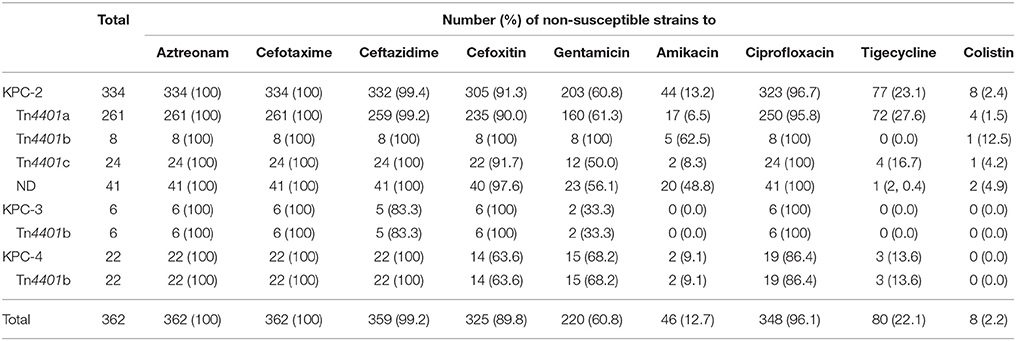
As a result of cross resistance from producing KPC, all or most of the 362 KPC producers were non-susceptible to aztreonam (100%), cefotaxime (100%), ceftazidime (n = 359, 99.2%), and cefoxitin (n = 325, 89.8%) (Table 2).
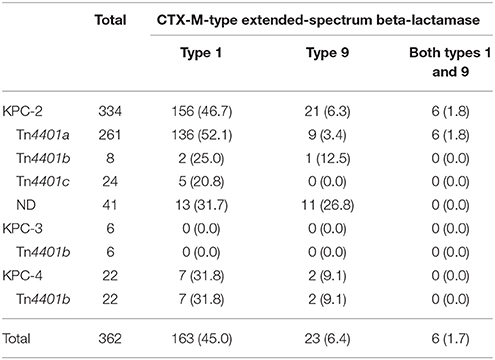
A total of 192 KPC producers (53.0%) produced at least one CTX-M-type ESBL (Table 3): 46.7% (169/362) CTX-M type 1 and 8.0% (29/362) CTX-M type 9. Of note, the blaCTX−M gene was often possessed by KPC-2 (183/334, 54.8%) and KPC-4 (9/22, 40.9%) producers, whereas none were identified in KPC-3 producers. Plasmid-mediated AmpC was rarely co-produced (13/362, 3.4%) by the KPC producers: DHA (n = 9), ACT (n = 1), or CMY (n = 1) production in KPC-2 producers and ACC (n = 2) production in KPC-4 producers.
Among the 362 KPC producers, 220 (60.8%) and 46 (12.7%) were resistant to gentamicin and amikacin, respectively, but only few KPC-2 producers possessed the 16S ribosomal methyltransferase armA (1/362, 0.3%) and rmtB (22/362, 6.1%) genes.
A total of 348 isolates (96.7%) were non-susceptible to ciprofloxacin and 80 (22.1%) and eight (2.2%) isolates were non-susceptible to tigecycline and colistin, respectively. None of the eight colistin-resistant strains possessed the mcr gene.
Discussion
KPC enzymes are able to hydrolyze penicillins, cephalosporins, monobactams, carbapenems, and even β-lactamase inhibitors. Therefore, the wide spread of the producers is one of the worrisome global issues encountering antimicrobial resistance. In accordance of occasional reports (Rhee et al., 2010; Hong et al., 2013; Yoo et al., 2013) and data from NLSS (unpublished data), only infrequent occurrence of the KPC producers were identified in South Korea until 2013. However, after a sudden expansion of KPC producers in 2014, rapid nation-wide spread supported by single-hospital outbreaks (Kim et al., 2017), frequently associated with inter-hospital or inter-regional spread (Jeong et al., 2016) led the endemic stage of KPC in 2015. Actually, the KPC producer in 2013 has never been identified again and the four K. pneumoniae subclones, ST307/Tn4401a[blaKPC-2], ST307/Tn4401b[blaKPC-4], ST392/Tn4401b[blaKPC-4], and ST11/ND[blaKPC-2] in 2014 triggered the inter-regional dissemination of KPC producers.
In agreement with the Healthcare Delivery System in South Korea, convalescent hospitals are in charge post-acute care and general hospitals are responsible for acute care, leading a habitual inter-hospital patient transfer. In general, the infection control system in convalescent hospitals is poorly operated compared to that in general hospitals and their role as a reservoir of CPE has been highly suspected. Indeed, the core KPC-producing subclone K. pneumoniae ST307/Tn4401a[blaKPC-2] was identified in all three types of healthcare facilities: 153 of the 362 KPC producers from general hospitals, one of the three isolates from the two convalescent hospitals and one of the two isolates from clinics. It suggested that the KPC has disseminated during the inter-hospital patient transfer and expanded up to the community level. Further study with more isolates from the two types of healthcare facility is needed.
The well-known KPC-producing clone in a global level is K. pneumoniae ST258 (Pitout et al., 2015; Sheppard et al., 2016), however, it was not the case in South Korea. The foremost South Korean KPC producer was K. pneumoniae ST307 clone and the ST11 was followed by presenting half the prevalence of ST307. Indeed, the ST258 (gapA-infB-mdh-pgi-phoE-rpoB-tonB, 3-3-1-1-1-1-79) is a deduced variant of ST11 (3-3-1-1-1-1-43) manufactured by multiple events of a genetic rearrangement (Breurec et al., 2013). K. pneumoniae ST307, which has gained attention as a potentially high-risk KPC producer, has been a notorious clone producing CTX-M-15 (Villa et al., 2017) and the clone co-producing KPC and CTX-M-15 was recently identified in Italy (Geraci et al., 2015). The South Korean ST307 clone are assumed to be close to the Italian clone in an evidence of 71.9% KPC-producing K. pneumoniae ST307 co-produced CTX-M enzyme. The second-most disseminated K. pneumoniae ST11 is one of the main KPC-producing clones in China, and it frequently co-harbors the rmtB genes (Cheng et al., 2016). The 22 RmtB-co-producers were all K. pneumoniae ST11 representing 28.7% (22/77) the clone.
KPC-2 and KPC-3 are the prevailing global subtypes of KPC, with regional variation (Munoz-Price et al., 2013). Among the South Korean KPC producers, KPC-4 was the second most prevalent subtype, although the number of KPC-4 producers was only one fifteenth that of the KPC-2 producers. Since the bacterial host species varied, the Tn4401b-associated plasmid (along with sporadic occurrence) was likely responsible for its observed dissemination. KPC-3, which was produced by fewer bacterial hosts, was limited to one district. KPC-2, KPC-3, and KPC-4 each confer a similar level of carbapenem resistance (Mehta et al., 2015). However, among the KPC producers studied, KPC-2 had the highest MIC50s for carbapenems, while KPC-3 and KPC-4 had 3- and 4-dilution-reduced MIC50s, respectively (Figure 2). It is likely that multiple factors (such as small relative group size, or low gene expression in the Tn4401b isotype) accounted for this unexpected result.
Although group size varied (334 vs. 17), the KPC-producing K. pneumoniae isolates had 3-dilution higher MIC50s for carbapenems compared to the KPC-producing E. coli isolates (Figure 2), pronouncing the importance of bacterial host background for revealing resistance. As well, the Tn4401 isotypes, differing the promoter of the blaKPC gene, is closely associated with the level of gene expression resulting in varied carbapenem susceptibility (Naas et al., 2012; Cheruvanky et al., 2017). For example, Tn4401a and Tn4401d possessing two promoters upstream from the gene confer high carbapenem resistance whereas Tn4401c and Tn4401e, with one promoter, confer low levels of resistance and the Tn4401b, with three promoters, confers moderate resistance (Naas et al., 2012; Cheruvanky et al., 2017). Three isotypes of the transposon that are Tn4401a, Tn4401b, and Tn4401c were identified in our study. Tn4401a had the highest MIC50s for carbapenems and the Tn4401b exhibited the lowest values. Tn4401c of similar MIC50 values to those of Tn4401a could be explained by the bacterial host, as K. pneumoniae likely, somehow, has the most efficient KPC production. The relatively high meropenem MIC50 by ND transposon encoding the blaKPC-2 compared to the other isotypes of Tn4401 needed to be further studied.
Though the global spread of KPC-producing Enterobacteriaceae is becoming more common, local epidemiology patterns need to be actively assessed. An awareness of the details of regional epidemiology is fundamental for understanding specific situations. Most endemic cases have occurred via the clonal dissemination of notorious KPC-producing clones, and the South Korean situation was no exception. The disseminated K. pneumoniae clones in South Korea have been linked to the Italian ST307 and Chinese ST11 strains, confirming the importance of international and intercontinental travel for clonal spread. There remains a paucity of antimicrobial options to treat pathogens capable of KPC production, which raises mortality rates. Only a few new drug candidates are in development at present. Combination therapy using both β-lactams and β-lactamase inhibitors that are stable in the presence of KPC enzymes (such as avibactam) is available in other parts of the world, but is not yet accessible in South Korea. Because South Korea is facing endemic dissemination, the situation is critical. Only systematic surveillance will empower researchers to control and prevent the spread of KPC producers.
Ethics Statement
The research, which has no involvement of human subject but the clinical isolates, does meet the exempt category without approval from Ethics Committee on Human Research of the Health Ministry in South Korea and the study design has not been reviewed by the committee.
Author Contributions
SJ and E-JY: contributed to idea conception, designed the study, conducted the experiments, interpreted the data, and wrote the manuscript; JK, DK, HL, JY, and KL: conducted the experiments and interpreted the data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by a fund (KCDC2017E4400100#) from the Research of Korea Centers for Disease Control and Prevention.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00056/full#supplementary-material
References
Albiger, B., Glasner, C., Struelens, M. J., Grundmann, H., Monnet, D. L., and European Survey of Carbapenemase-Producing Enterobacteriaceae Working Group (2015). Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill 20. doi: 10.2807/1560-7917.ES.2015.20.45.30062
Breurec, S., Guessennd, N., Timinouni, M., Le, T. A., Cao, V., Ngandjio, A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
Cheng, L., Cao, X. L., Zhang, Z. F., Ning, M. Z., Xu, X. J., Zhou, W., et al. (2016). Clonal dissemination of KPC-2 producing Klebsiella pneumoniae ST11 clone with high prevalence of oqxAB and rmtB in a tertiary hospital in China: results from a 3-year period. Ann. Clin. Microbiol. Antimicrob 15:1. doi: 10.1186/s12941-015-0109-x
Cheruvanky, A., Stoesser, N., Sheppard, A. E., Crook, D. W., Hoffman, P. S., Weddle, E., et al. (2017). Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob. Agents Chemother. 61:e00025–17. doi: 10.1128/AAC.00025-17
Clinical and Laboratory Standards Institute (2015a). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. (Wayne, PA: Clinical and Laboratory Standards Institute).
Clinical and Laboratory Standards Institute (2015b). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. (Wayne, PA: Clinical and Laboratory Standards Institute).
Cuzon, G., Naas, T., and Nordmann, P. (2011). Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob. Agents Chemother. 55, 5370–5373. doi: 10.1128/AAC.05202-11
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Geraci, D. M., Bonura, C., Giuffrè, M., Saporito, L., Graziano, G., Aleo, A., et al. (2015). Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin. Microbiol. Infect. 21, e15–e17. doi: 10.1016/j.cmi.2014.08.022
Hong, S. K., Yong, D., Kim, K., Hong, S. S., Hong, S. G., Khosbayar, T., et al. (2013). First outbreak of KPC-2-producing Klebsiella pneumoniae sequence type 258 in a hospital in South Korea. J. Clin. Microbiol. 51, 3877–3879. doi: 10.1128/JCM.01730-13
Jeong, S. H., Kim, H. S., Kim, J. S., Shin, D. H., Kim, H. S., Park, M. J., et al. (2016). Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann. Lab. Med. 36, 529–535. doi: 10.3343/alm.2016.36.6.529
Kim, J. O., Song, S. A., Yoon, E. J., Shin, J. H., Lee, H., Jeong, S. H., et al. (2017). Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn. Microbiol. Infect. Dis. 87, 343–348. doi: 10.1016/j.diagmicrobio.2016.12.012
Mehta, S. C., Rice, K., and Palzkill, T. (2015). Natural variants of the KPC-2 Carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog. 11:e1004949. doi: 10.1371/journal.ppat.1004949
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Naas, T., Cuzon, G., Truong, H. V., and Nordmann, P. (2012). Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob. Agents Chemother. 56, 4753–4759. doi: 10.1128/AAC.00334-12
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpCβ-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002
Pitout, J. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Rhee, J. Y., Park, Y. K., Shin, J. Y., Choi, J. Y., Lee, M. Y., Peck, K. R., et al. (2010). KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob. Agents Chemother. 54, 2278–2279. doi: 10.1128/AAC.00011-10
Ryoo, N. H., Kim, E. C., Hong, S. G., Park, Y. J., Lee, K., Bae, I. K., et al. (2005). Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J. Antimicrob. Chemother. 56, 698–702. doi: 10.1093/jac/dki324
Sheppard, A. E., Stoesser, N., Wilson, D. J., Sebra, R., Kasarskis, A., Anson, L. W., et al. (2016). Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778. doi: 10.1128/AAC.00464-16
The joint CLSI–EUCAST and Polymyxin Breakpoints Working Group (2016). Recommendations for MIC Determination of Colistin (polymyxin E) as Recommended by the Joint CLSI–EUCAST Polymyxin Breakpoints Working Group. Available online at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf (Accessed March 22, 2016).
Villa, L., Feudi, C., Fortini, D., Brisse, S., Passet, V., Bonura, C., et al. (2017). Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 3:e000110. doi: 10.1099/mgen.0.000110
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Keywords: KPC, Tn4401, carbapenem resistance, Klebsiella pneumoniae ST307, carbapenemase producing Enterobacteriaceae
Citation: Yoon E-J, Kim JO, Kim D, Lee H, Yang JW, Lee KJ and Jeong SH (2018) Klebsiella pneumoniae Carbapenemase Producers in South Korea between 2013 and 2015. Front. Microbiol. 9:56. doi: 10.3389/fmicb.2018.00056
Received: 16 October 2017; Accepted: 10 January 2018;
Published: 25 January 2018.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Mariagrazia Perilli, University of L'Aquila, ItalyAmy J. Mathers, University of Virginia, United States
Copyright © 2018 Yoon, Kim, Kim, Lee, Yang, Lee and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwang Jun Lee, kwangjun@korea.kr
Seok Hoon Jeong, kscpjsh@yuhs.ac
 Eun-Jeong Yoon1
Eun-Jeong Yoon1 Jung Ok Kim
Jung Ok Kim Seok Hoon Jeong
Seok Hoon Jeong