- 1Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China, Ministry of Education, Southwest Forestry University, Kunming, China
- 2Key Laboratory of Biodiversity Conservation in Southwest China, State Forest Administration, Southwest Forestry University, Kunming, China
- 3College of Life Science, Jinggangshan University, Ji'an, China
- 4Central Laboratory of Xiangyang No.1 Hospital, College of Basic Medical Sciences, Hubei Key Laboratory of Wudang Local Chinese Medicine Research, Hubei University of Medicine, Shiyan, China
Being a sister species of Saccharomyces cerevisiae, Saccharomyces uvarum shows great potential regarding the future of the wine industry. The sulfite tolerance of most S. uvarum strains is poor, however. This is a major flaw that limits its utility in the wine industry. In S. cerevisiae, FZF1 plays a positive role in the transcription of SSU1, which encodes a sulfite efflux transport protein that is critical for sulfite tolerance. Although FZF1 has previously been shown to play a role in sulfite tolerance in S. uvarum, there is little information about its action mechanism. To assess the function of FZF1, two over-expression vectors that contained different FZF1 genes, and one FZF1 silencing vector, were constructed and introduced into a sulfite-tolerant S. uvarum strain using electroporation. In addition, an FZF1-deletion strain was constructed. Both of the FZF1-over-expressing strains showed an elevated tolerance to sulfite, and the FZF1-deletion strain showed the opposite effect. Repression of FZF1 transcription failed, however, presumably due to the lack of alleles of DCR1 and AGO. The qRT-PCR analysis was used to examine changes in transcription in the strains. Surprisingly, neither over-expressing strain promoted SSU1 transcription, although MET4 and HAL4 transcripts significantly increased in both sulfite-tolerance increased strains. We conclude that FZF1 plays a different role in the sulfite tolerance of S. uvarum compared to its role in S. cerevisiae.
Introduction
Saccharomyces uvarum is a sister species of Saccharomyces cerevisiae that was initially considered a synonym of Saccharomyces bayanus, but is now considered a species in its own right. Subsequently, S. uvarum is an object of interest for scientists working on applied and fundamental research (Naumov et al., 2011; Masneuf-Pomarede et al., 2016). It hybridizes with its sister species, including S. pastorianus, S. eubayanus, and S. cerevisiae, to form hybrids that are important in the beer industry (Nguyen et al., 2011). As S. uvarum and S. cerevisiae both belong to Saccharomyces sensu stricto, these two species have similar characteristics, but in the later stage of the Sauvignon Blanc fermentation process, S. uvarum plays a more important role in producing alcohol from anaerobic respiration (Sipiczki and Ciani, 2002; Naumov et al., 2011). It has already been used in wine, but it is mainly used in cider (González Flores et al., 2017). It may have great potential in the wine industry in the future, however, as it can ferment at low temperatures (Zhang et al., 2015).
Yeast resistance to sulfite is an important technological characteristic for wine making (Nadai et al., 2016). In wine production, a variety of antioxidants are added, both to prevent oxidation and to inhibit the growth of unwanted yeast and bacteria. Sulfite, added as metabisulfite or as sulfur dioxide, is widely used in wine fermentation (Taylor et al., 1986), and usually at a dosage of 50 mg/L free sulfite in grape juice (Doneche, 1993). High concentrations of sulfite are also toxic to Saccharomyces cells, however (Nardi et al., 2010; Divol et al., 2012; Nadai et al., 2016). Hence, sulfite resistance has become one of the most important indexes for the screening of wine-making yeast strains.
In S. cerevisiae, the SSU1 gene is the key regulator of sulfite tolerance. It acts as a “sulfite efflux pump” in cells, removing toxic sulfite from them (Donalies and Stahl, 2002). Meanwhile, sulfite also forms special flavors of wine, i.e., thiol esters, in the process, thereby improving the quality of the wine (Avram and Bakalinsky, 1997; Park and Bakalinsky, 2000; Donalies and Stahl, 2002; Nardi et al., 2010). Among wine yeast strains, two translocations have been identified that provide up-regulation of SSU1 and increased sulfite tolerance (Pérez-Ortín et al., 2002; Yuasa et al., 2005; Zimmer et al., 2014). A second locus, FZF1, also provides sulfite resistance in S. cerevisiae (Breitwieser et al., 1993). S. cerevisiae FZF1 is a transcription factor that has 203 targets and plays a positive role in the transcription of SSU1 according to the yeast search for transcriptional regulators and consensus tracking (YEASTRACT) database (Teixeira et al., 2014). Efflux of sulfite via active transport appears to be the predominant method utilized by S. cerevisiae to improve the sulfite tolerance of the cell (Zimmer et al., 2014). Genome-wide microarray analysis of ribonucleic acid (RNA) expression identified a second possible transcriptional target for the FZF1 gene besides SSU1 in S. cerevisiae: GCV1 (Hu et al., 2007), which encodes a T subunit of the mitochondrial glycine decarboxylase complex (McNeil et al., 1997; Piper et al., 2000).
In S. uvarum, relatively little is known about sulfite tolerance. Some strains show tolerance, however, and in one case, tolerance was shown to be linked to an allele of FZF1 that had been introgressed from S. eubayanus, but was not linked to the SSU1 locus (Zhang et al., 2015).
Studying the function of the FZF1 gene in sulfite tolerance is vital for interpreting the metabolism mechanism of sulfite in S. uvarum. Here, we describe the construction of two different FZF1-over-expressing strains, one FZF1-silencing strain, and one FZF1-deletion strain. The function of the FZF1 gene of S. uvarum was analyzed via sulfite-tolerant phenotype screening, polymerase chain reaction (PCR) analysis, growth curves, quantitative reverse transcription polymerase chain reaction (qRT-PCR), and transcriptome analysis.
Materials and Methods
Materials
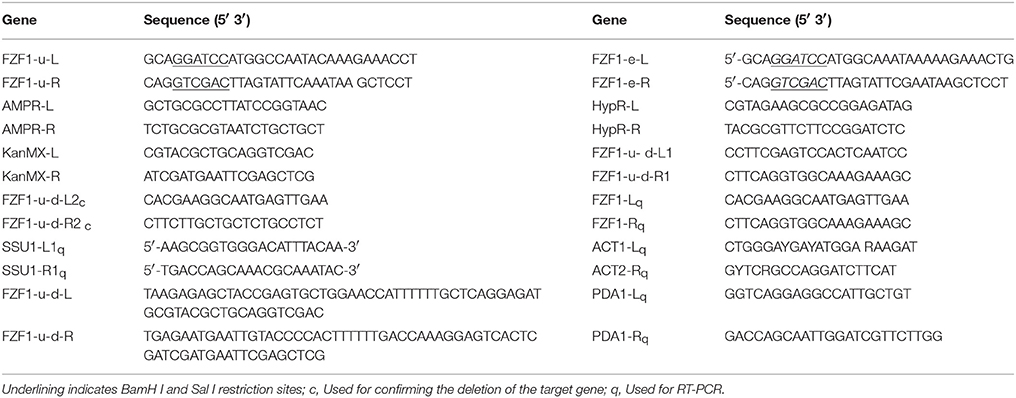
S. uvarum strain A9 (Zhang et al., 2015), pSilent, pYIP5, and Escherichia coli DH5α, were obtained from collections at Southwest Forestry University. Two RNA interference (RNAi) fragments, TTCATAAGACCATGTCATTTAA and TTAAATGACATGGTCTTATGAA, were designed using the website http://www.broadinstitute.org/rnai/public/seq/search, and were synthesized and constructed in a pSilent vector by Zoonbio Biotechnology Co. Ltd., China. The sequence of FZF1-u (Genbank accession number KY905342) was identical to that in the sulfite-tolerant S. uvarum strain A9, which originally came from S. eubayanus (Zhang et al., 2015), and the sequence of FZF1-e (Genbank accession number KY905343) was derived from S. uvarum strain ACY338. Fifteen percent of nucleotide sequences were different between FZF1-u and FZF1-e. FZF1-u and FZF1-e genes were synthesized with extra BamH I and Sal I sites at their termini, and constructed in the pGEM–T easy vector by Nextomics Biosciences Co. Ltd. The genotypes of all the strains used or created are listed in Table 2.
Construction of the FZF1 Gene Expression Vector and Transformation of S. uvarum
Procedures for the manipulation of plasmid deoxyribonucleic acid (DNA), and transformation and sequence validation, were performed as previously described (Sambrook et al., 1989). Using BamH I and Sal I, the PCR products of the FZF1-u and FZF1-e genes from the vectors and expression plasmid pYIP5 were digested. The digested fragments of the expression vector and PCR products were then ligated, and shuttle vectors were constructed. The vectors were again subjected to restriction analysis using BamH I and Sal I to confirm that the target gene had been correctly inserted into the vector. The vectors were also sent to Sangon (Shanghai Sangon Co., Shanghai, China) for sequencing to test for putative mutations. pYIP5-FZF1-u, pYIP5-FZF1-e, and pSilent-FZF1-u-s were transferred into Escherichia coli DH5α and the sulfite-tolerant S. uvarum strain A9 via electrotransformation as described as Kozak et al. (2014). DNA was extracted using the cetyltrimethyl ammonium bromide (CTAB) method described by Zhang et al. (2010). The URA3 gene was used as a selectable marker for pYIP5-FZF1-u and pYIP5-FZF1-e transformants, and the hygromycin resistance gene was utilized for pSilent-FZF1-u-s transformants.
Candidate transformant colonies were subjected to PCR analysis to confirm. The primers used in the PCR are listed in Table 2. For the FZF1 over-expression candidates, 10 transformant colonies of both FZF1-u and FZF1-e that could grow well on the medium containing 5-fluoroorotic acid (5-FOA) were selected for PCR analysis. The colonies of transformations could produce a 312-bp band specific to the AmpR gene. For the FZF1 repression expression candidates, 10 transformed candidates with hygromycin resistance were selected for the PCR analysis. The transformant colonies were expected to generate a 760-bp specific band from the hygromycin resistance gene, and a 312-bp band specific to the AmpR gene.
Construction of the FZF1-Deletion Strain
The methods used were the same as on the website (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). Candidate FZF1-deletion colonies used the KanMX gene as a selectable marker to screen. The deletion of the target gene was confirmed by PCR using the primer pairs FZF1-u-d-L1 c/FZF1-u-d-R1 c and FZF1-u- d-L2 c/FZF1-u- d-R2 c (see Table 2).
Sulfite Tolerance Genotyping
After culturing for 24 h in liquid yeast extract peptone dextrose (YPD), 100 μL of the yeast colonies were diluted with liquid YPD (1:10, 1:100, 1:1000, and 1:10000). After dilution, 3 μL of the yeast colonies after dilution were inoculated onto fresh YPD plates containing 5, 10, 20, 40, 60, 80, 100, and 120 mM of sodium sulfite, and 80 mM of succinic acid (pH 3.5). After growing on the medium for 3 days at 30°C, the sulfite tolerance levels of the colonies were recorded by visual analysis of growth.
RNA Extraction and Complementary Deoxyribonucleic Acid (cDNA) Synthesis
Yeast colonies were collected after culturing for 24 h in liquid YPD without adding sulfite, then placed for 20 mins in liquid YPD with 20 mM of sodium sulfite and 80 mM of succinic acid, for RNA extraction. Total RNA was extracted using a Qiagen kit. The RNA was then reverse-transcribed into cDNA using a Fermentas kit.
qRT-PCR Analysis
qRT-PCR was performed according to the method described by Chen et al. (2008) with the ABI7500 fluorescence quantitative PCR instrument. Primers were FZF1-Lq and FZF1-Rq (see Table 1), and the 2−ΔΔCT method was used to assay the data (Livak and Schmittgen, 2001), with PDA1 and ACT1 as housekeeping genes for normalizing (Divol et al., 2006). Three parallel experiments were conducted for each gene.
Determination of Growth Curves of Recombinant Strains
Cultures of individual clones were grown for 64 h in liquid YPD, then diluted 100-fold into 0.05 L of fresh liquid YPD. The strains were the starting A9 strain, and transformed derivatives A9-FZF1-u, A9-FZF1-e, and A9-FZF1-u-s. Cultures were incubated at 30 degrees C, with shaking at 220 rpm, and sampled every 4 h, with growth measured using light absorption at 600 nm. Using the YPD liquid culture medium without inoculation as a control, the growth curves of the strains were plotted as optical density (OD600) against time. Different stages of the growth of the strains were used to calculate the maximum growth rates: For A9, A9-FZF1-u-s, and A9-FZF1-u-d we used the stage of 28 h to 32 h after inoculation; while for A9-FZF1-u and A9-FZF1-e, we used the stage of 20 h to 24 h. These stages were used as these were the points when the strains grew the fastest.
Transcriptome Analysis
For comparison of differential transcription genes between strains, next generation sequencing was employed to assess the RNA samples that were extracted from colonies cultured in YPD for 24 h, and then in 20 mM of sodium sulfite and 80 mM of succinic acid for 10 min. The RNA was prepared using Illumina TruSeq RNA-Seq kits, and RNA-Seq was performed by Nextomics Biosciences Co. Ltd using Illumina HiSeq™. The RNA-Seq data were submitted to the Genome Sequence Archive of Beijing Institute of Genomics (BIG) Data Center (publicly accessible at http://gsa.big.ac.cn, Accession No. PRJCA000414). Transcriptome analysis was performed using DEGseq (Wang et al., 2010) and DESeq (Anders and Huber, 2010).
Statistical Analysis
Data were analyzed by the graphing of histograms in Excel 2007 (Microsoft, Redmond, WA), mean ± standard errors (SE) were calculated, and significant differences between different strains were calculated using a one-way analysis of variance (ANOVA). Differences were considered statistically significant if the p-values were <0.05. For the transcriptome analysis, the Q-values were calculated based on p values to identify differentially expressed genes (Wang et al., 2010). If the Q-values were <0.01, differences were considered statistically significant.
Results
Sulfite Resistance of Transformations
Forty transformants (10 transformants for each strain) with the right genotypes (see Table 2) and PCR products were used for the sulfite resistant test. These 40 transformants, together with the starting strain A9, were inoculated and grown on YPD plates with different sulfite concentrations to screen the sulfite tolerant genotype. The results showed that A9-FZF1-u and A9-FZF1-e were the most tolerant colonies, and that they could grow on media containing 100 mM sulfite (see Figure 1). The sulfite tolerance of FZF1-u-s transformations, however, was not changed at all compared to the parent A9 strain as both could grow on 20 mM (see Table 1). These data demonstrate that over-expression of the FZF1 genes from both S. eubayanus and S. uvarum could increase the sulfite resistance of S. uvarum, while the silencing of the FZF1 gene via RNAi had no effect. The deletion strain could not grow on YPD with 10 mM sodium sulfite, but could grow on 5 mM. This indicated that the deletion of the FZF1 gene in the S. uvarum genome may havedecreased the sulfite resistance.

Table 2. The genotype and sulfite resistance ability of different S. uvarum stains or transformations.
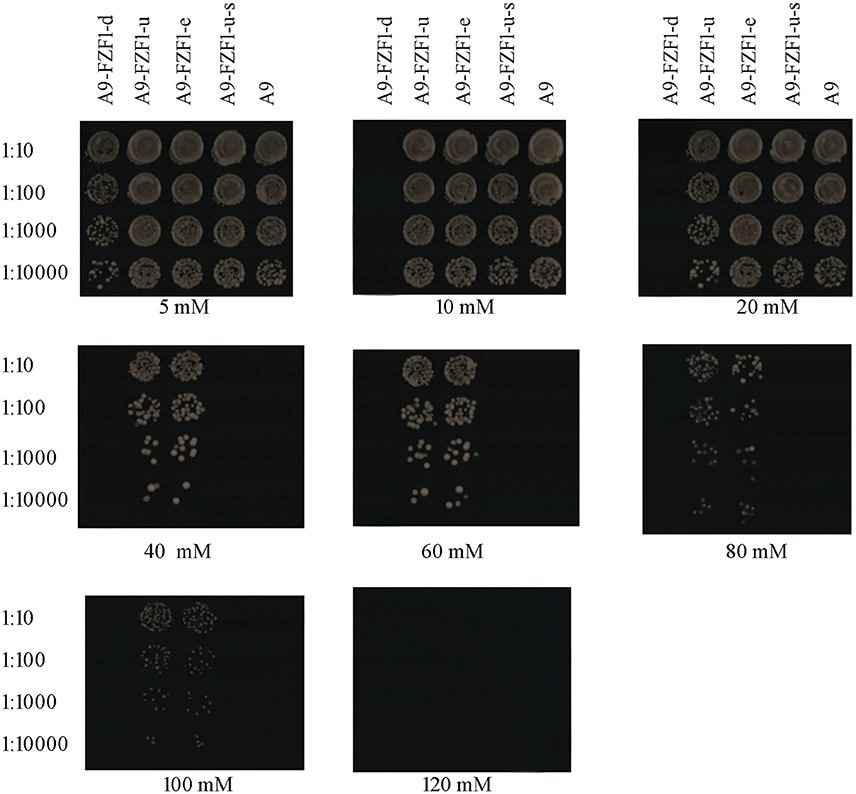
Figure 1. Drop off test experiment: yeast colonies were cultured for 24 h in liquid YPD without adding sulfite, then 100 uL of them were diluted with liquid YPD into 1:10, 1:100, 1:1000, and 1:10000 time. Three microliter droplets of yeast dilutions were inoculated onto fresh YPD plates containing 5, 10, 20, 40, 60, 80, 100, and 120 mM of sodium sulfite, and 80 mM of succinic acid (pH 3.5). After growing on the medium for 3 days, the sulfite tolerance levels of the colonies were recorded by visual analysis of growth.
qRT-PCR Analysis
One transformant colony of each vector with the right genotypes and the expected PCR amplified bands was selected as representing that strain. qRT-PCR was used to quantify the average transcript concentration of the FZF1 gene in these strains together with the starting strain A9. Figure 2A shows that A9-FZF1-u and A9-FZF1-e demonstrated increased transcription relative quantification (RQ) of FZF1 by 4.87- and 4.72-fold, respectively, over that of A9, while transcription RQ in A9-FZF1-u-s was 0.96 times that of the starting strain (as shown in Figure 2). The increases in the two over-expressing strains were significant (p < 0.01), but there was no significant difference between A9 and A9-FZF1-u-s. These data show that both FZF1 over-expressions were successful.
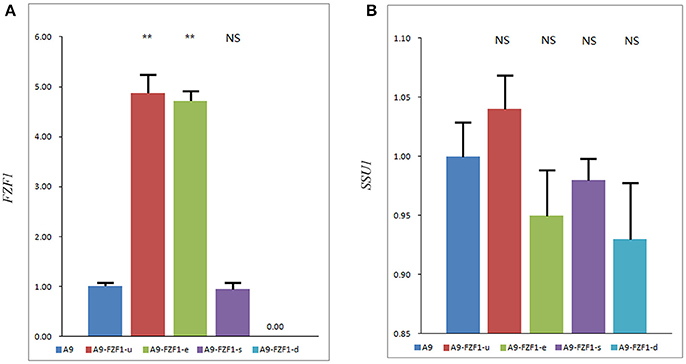
Figure 2. The expression level of the FZF1 (A) and SSU1 (B) genes in different strains. The expression levels of genes were assessed using a 2-ΔΔCT method to determine the relative gene expression from qPCR data with ACT1 as a housekeeping gene. Values are 2-ΔΔCT Mean ± SE (n = 3). NS: P > 0.05, not significant, **P < 0.01, with one-way ANOVA. Note that different scales are used in the two figures. Total RNA failed to be extracted from A9-FZF1-u-d after treatment with sulfite, so the data of A9-FZF1-u-d were absent.
The expression levels of the SSU1 gene's three transformed strains were also assessed using qRT-PCR (as shown in Figure 2B). All three strains showed similar levels of SSU1 transcription, with no significant differences from the starting strain. These results show that over-expression of FZF1 does not affect the expression level of the SSU1 gene in S. uvarum, which is in direct contrast to the situation in S. cerevisiae.
Determination of the Growth Curves of the Recombinant Strains
The three representative strains, and the parental A9 strain, were cultured in YPD liquid medium in shake flasks. The lag phases (absorbance at 600 nm <0.10) of A9-FZF1-u-s and A9 were almost the same: around 20 h. Meanwhile, the lag phases of the FZF1-over-expressing strains were also similar to each other at around 12 h, but were significantly shorter than the other two strains (p < 0.00001). The average maximum growth rates for the triplicate values of A9, A9-FZF1-u, A9-FZF1-e, A9-FZF1-u-s, and A9-FZF1-u-d were 0.201 (±0.014), 0.220 (±0.013), 0.331 (±0.023), 0.209 (±0.027), and 0.196 (±0.007) h−1, respectively. There was no significant difference (p > 0.05) between the maximum growth rates of A9, A9-FZF1-u-s, and A9-FZF1-u-d. All of them were significantly smaller than that of A9-FZF1-u and A9-FZF1-e (p < 0.001), however. The average final titers for the triplicate values of these strains were all similar (1.975 (±0.009), 1.995 (±0.015), 1.969 (±0.028), 1.969 (±0.022), and 1.965 (±0.012) × 107 cells/ml, respectively). The experimental results showed that the FZF1-over-expressing strains grew faster than the starting strain A9, and that A9-FZF1-u-s did almost the same as A9 (see Figure 3).
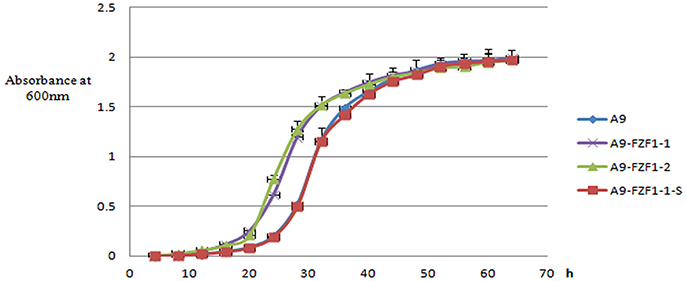
Figure 3. The growth curve of different strains. The data are plotted as the means and standard deviations of the triplicate values. The culture medium was fresh liquid YPD, and the data were collected every 4 h, with growth measured using light absorption at 600 nm.
Transcriptome Analysis
RNA sequencing was used to quantify the transcripts in each of the four strains (A9, A9-FZF1-u, A9-FZF1-e, and A9-FZF1-u-s) in which RNA samples were extracted from colonies after treatment with 20 mM of sodium sulfite and 80 mM of succinic acid for 10 min. Total sequence reads varied from 27 to 40 million per strain, and all of the total clean reads ratio of these strains were very high (from 99.64% in A9-FZF1-u-s, to 99.94% in A9, see Table S1).
All 8490 unigene sequences were clustered using the clusters of orthologous groups (COG) function classification. There are 25 functional categories, such as RNA processing and modification, chromatin structure and dynamics, and energy production and conversion. The unigene sequences numbers within each cluster ranged from two to 1241, with an average number of 339.6 unigene sequences per group. There were 726 unigene sequences up-regulated in both A9-FZF1-e and A9-FZF1-u after subtracting vector-derived genes out of this total. Of these, 514 unigene sequences were also found to be up-regulated in A9-FZF1-u-s. Therefore only 212 unigene sequences were found to be up-regulated in FZF1-over-expressing strains. These unigene sequences represent candidates for altered regulation directed by the over-expression of FZF1 in the two strains (see Table S2). There were 364 unigene sequences up-regulated in A9-FZF1-u-s, and 219 of them were also found to be up-regulated in A9-FZF1-e and A9-FZF1-u.
A total of 112 transcription factors (see Table S3) appeared in all four strains. The Log2 FPKM (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) values of the transcription factors ranged from -9.9658 in all strains to 11.5954 in A9, 9.8497 in A9-FZF1-u-s, 10.0773 in A9-FZF1-u, and 10.05140 in A9-FZF1-e. Results showed that the two over-expressing FZF1 gene transformants have the most similar overall expression pattern of their transcription factors.
Analysis of the differentially expressed genes (DEGs) by function showed that the most enriched pathways differed between strains over-expressing different FZF1 genes and the FZF1-RNAi fragment (see Figure 4). Among them, all the genes concerning a sulfur relay system were down-regulated in the A9-FZF1-u-s, but there were up-regulated and down-regulated genes in the transformed strains of the different FZF1 genes. The gene numbers of categories, such as membrane transport and translation in over-expression strains, were increased compared to those in the silenced one, while the gene numbers of categories such as replication and repair, energy metabolism, lipid metabolism, amino acid metabolism, carbohydrate metabolism, and cell growth and death in over-expression strains decreased.
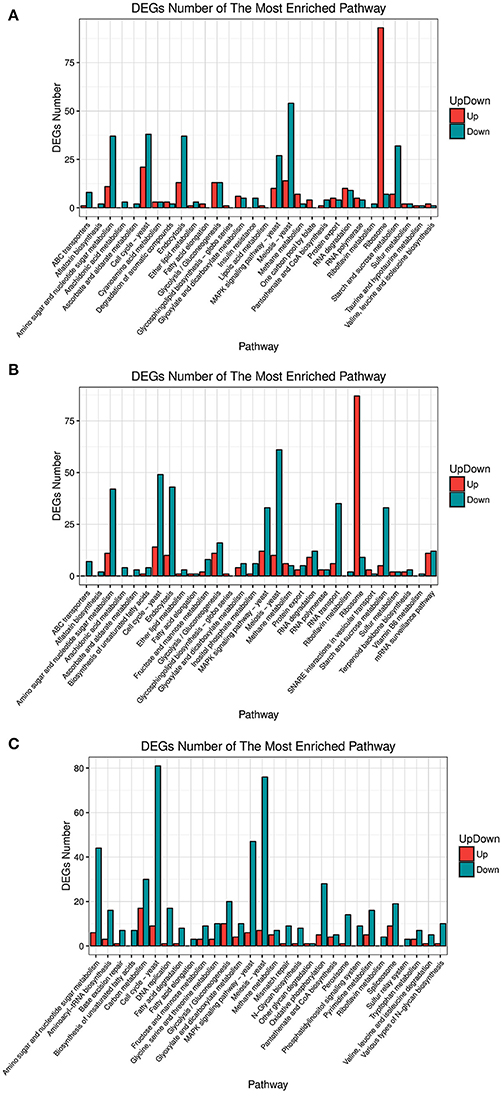
Figure 4. Pathway enrichment of different expression genes between FZF1 transformed strains and A9 with Gene Ontology (GO) interpretation. (A) A9-FZF1-u vs. A9; (B) A9-FZF1-e vs. A9; (C) A9-FZF1-u-s vs. A9.
Table 3 shows the expression data for a selection of sulfur-related genes assessed for the ratio of expression (comparison of the average of the two overexpressed strains to the average of the A9 and silenced strain). Among them, MET4 and HAL4 were up-regulated in both FZF1-over-expressing strains, PRPD and CYS3 were up-regulated only in A9-FZF1-u, and ILV3, ELP3, NFS1, and CFD1 were significantly up-regulated only in A9-FZF1-e (see Table 3). Thus, MET4 and HAL4 are the leading candidates for involvement in sulfite tolerance via up-regulation of FZF1.
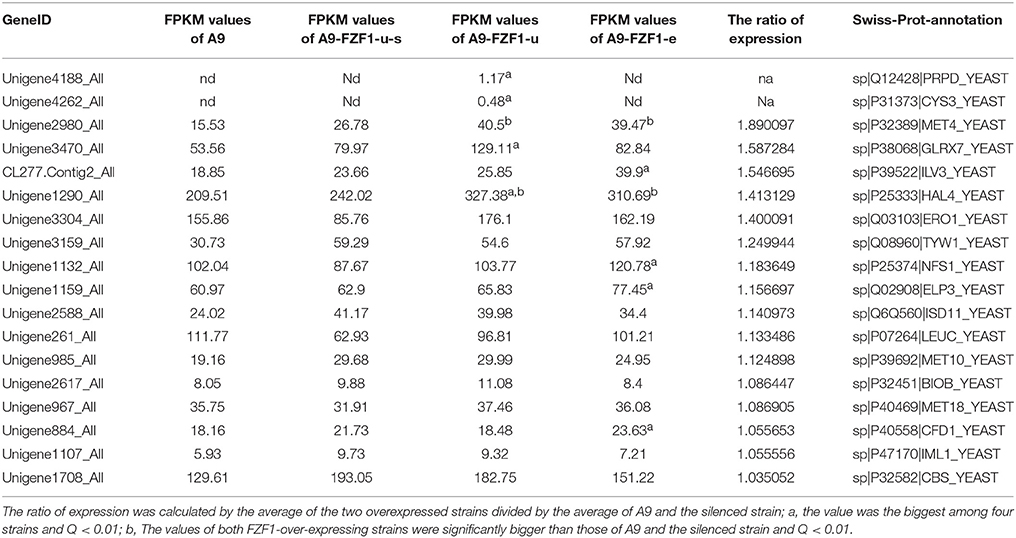
Table 3. List of up-regulated genes related to sulfite or sulfur metabolism in over-expressed strains compared with A9 and the silenced strain.
Discussion
In S. cerevisiae, the FZF1 gene was found to be a positive regulator of SSU1 and to be involved in sulfite tolerance. It was found that multi-copy FZF1 genes could result in producing more efflux acting through Ssu1p and Met20p (Avram and Bakalinsky, 1996). Here, we have confirmed that over-expression of FZF1 also confers sulfite tolerance in S. uvarum. We also found, however, that over-expressing of the FZF1 gene in S. uvarum did not lead to the expression change of the SSU1 gene, but that over-expressing and deletion of it led to a change of sulfite tolerance. It confirmed the previous findings that sulfite resistance in the S. uvarum isolates is linked to FZF1, but not to SSU1.
Although GCV1, as well as SSU1, was found to be the target gene of FZF1 besides SSU1 in S. cerevisiae in a previous study (Hu et al., 2007), there was no evidence to infer that GCV1 was influenced by the over-expressing of FZF1 in this study. Compared to the 203 target genes of S. cerevisiae in the YEASTRACT database (Teixeira et al., 2014), however, 212 unigene sequences were found to be up-regulated in both FZF1-over-expressing strains, but not in A9-FZF1-u-s, and this group of genes should include the candidates for the FZF1 targets. The sulfur-related genes among these included MET4 and HAL4, which showed a 50-80% increase in transcription in both FZF1-over-expressing strains compared to A9 and the silenced strain.
Previously, studies have revealed that MET4 and HAL4 are involved in the sulfur metabolism in S. cerevisiae (Fauchon et al., 2002; Lee et al., 2010; Gey et al., 2014). In this study, MET4 and HAL4 were significantly up-regulated in both FZF1-over-expressing strains, meaning that they could be involved in the sulfite metabolism in S. uvarum. The protein encoded by MET4 is a leucine zipper, and once it assembles onto the promoters, Met4p can recruit other transcriptional coactivator complexes, including Mediator and Spt-Ada-Gcn5-Acetyltransferase (SAGA) (Leroy et al., 2006; Su et al., 2008). This might be one of the reasons why there were up to 112 transcription factors (see Table S3) that changed their expression levels in this study. Here, MET4 and HAL4 were first found to be positively regulated by FZF1 in S. uvarum.
Around 15% of the nucleotide sequences are different between FZF1-u and FZF1-e, which leads to 21% of the coded protein sequences being different. The changes in protein sequences may lead to a change of protein structure and function, and in the expression of the other genes involved. Two genes, namely PRPD and CYS3, were up-regulated only in A9-FZF1-u, and ILV3, ELP3, NFS1, and CFD1 were significantly up-regulated in A9-FZF1-e. These differences might be caused by the expression of different sequences of the FZF1 genes. Regarding sulfur metabolism, however, these differential genes concerning sulfur metabolism could not cause a sulfite tolerance difference as there was no significant difference between A9-FZF1-u and A9-FZF1-e.
RNAi is an ancient mechanism present in plants, animals, and most fungi, and is considered to be a type of genetic immune system (Ghildiyal and Zamore, 2009; Malone and Hannon, 2009). In this study, however, the results showed that the RNAi of the FZF1 gene had failed. As the alleles of DCR1 and AGO play vital roles in the mechanism of RNAi (Moazed, 2009), we checked the whole genome of A9, which has been previously sequenced (Zhang et al., 2015), but no alleles of DCR1 and AGO were found. The idea that RNAi had been lost during the evolution of budding yeasts (Moazed, 2009) also seems to have been confirmed in this study. Although the depression of FZF1 with RNAi was not working, it was still good to use A9-FZF1-u-s as another control besides A9 for comparing with the FZF1-overexpression strains. Among those changed genes, the SLM3, NCS6, and TCD2 genes, which might concern the sulfite metabolism, were all up-regulated in A9-FZF1-u-s, but the mechanism was unknown and should be studied in future research.
Several of the mechanisms and strain-dependent strategies used to obtain sulfite resistance can deeply influence wine quality (Nadai et al., 2016). Zinc finger proteins represent some superfamily of nucleic acid binding proteins in eukaryotes cell that take part in a variety of cellular activities, such as differentiation, development, and cell cycle. Here, we have provided direct evidence that the FZF1 gene plays an important role in regulating the sulfite tolerance in S. uvarum, and suggest the involvement of MET4 rather than SSU1 in this process. Again, the results confirm the conclusion of previous studies that FZF1 is required for sulfite tolerance in S. uvarum, but that SSU1 is not linked to this trait. This is the first observation that the different genes of the sulfur metabolism network can be up-regulated or down-regulated when an FZF1 gene of different origin is over-expressed in the cells of S. uvarum.
Conclusion
In conclusion, our results confirmed that the FZF1 gene is important in the sulfite resistance mechanism of S. uvarum. Over-expression of FZF1 derived from the S. uvarum strain A9 did not increase sulfite tolerance compared to the over-expression of FZF1 derived from S. uvarum strain ACY338. Meanwhile, the deletion of the FZF1 gene led to a decrease in sulfite tolerance. Instead of promoting the expression of SSU1, the expression of MET4 and HAL4 was increased. The FZF1 gene, therefore, plays a different role in S. uvarum compared to its role in S. cerevisiae.
Author Contributions
XZL analyzed the experimental data and drafted the manuscript. XZL, XPL, and ZZ conducted the study. MS and XS helped analyze the experimental data. CH and PX participated in the coordination of the study. HZ conceived of the study and contributed to writing the manuscript. All authors approved the final version of the manuscript.
Funding
The study was supported by grants from National Natural Science Foundation of China (31760450, 31360404), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (212209), and China Scholarship Council Fund (20155103).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Richard C. Gardner of the University of Auckland for critical review and revising the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://journal.frontiersin.org/articles/10.3389/fmicb.2018.00096/full#supplementary-material
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Gen. Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Avram, D., and Bakalinsky, A. T. (1996). Multicopy FZF1 (SUL1) suppresses the sulfite sensitivity but not the glucose derepression or aberrant cell morphology of a grr1 mutant of Saccharomyces cerevisiae. Genetics 144, 511–521.
Avram, D., and Bakalinsky, A. T. (1997). SSU1 encodes a putative transporter with a central role in a network of proteins conferring sulfite tolerance in Saccharomyces cerevisiae. J. Bacteriol. 179, 5971–5974. doi: 10.1128/jb.179.18.5971-5974.1997
Breitwieser, W., Price, C., and Schuster, T. (1993). Identification of a gene encoding a novel zinc finger protein in Saccharomyces cerevisiae. Yeast 9, 551–556. doi: 10.1002/yea.320090512
Chen, Y., Shen, S., Wang, Y., and Xiao, D. (2008). Effect of SSU1 multi-copy expression on Saccharomyces cerevisiae sulphite production. Acta Microbiol. Sin. 48, 1609–1615.
Divol, B., Miot-Sertier, C., and Lonvaud-Funel, A. (2006). Genetic characterization of strains of Saccharomyces cerevisiae responsible for 'refermentation' in Botrytis-affected wines. J. Appl. Microbiol. 100, 516–526. doi: 10.1111/j.1365-2672.2005.02818.x
Divol, B., du Toit, M., and Duckitt, E. (2012). Surviving in presence of sulphur dioxide: strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 95, 601–613. doi: 10.1007/s00253-012-4186-x
Donalies, U. E., and Stahl, U. (2002). Increasing sulphite formation in Saccharomyces cerevisiae by overexpression of MET14 and SSU1. Yeast 19, 475–484. doi: 10.1002/yea.849
Doneche, B. (1993). “Botrytized wines,” in Wine Microbiology and Biotechnology, ed G. H. Fleet (Chur: Harwood Academic Publishers), 327–351.
Fauchon, M., Lagniel, G., Aude, J. C., Lombardia, L., Soularue, P., Petat, C., et al. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9, 713–723. doi: 10.1016/S1097-2765(02)00500-2
Gey, U., Czupalla, C., Hoflack, B., Krause, U., and Rödel, G. (2014). Proteomic analysis reveals a novel function of the kinase Sat4p in Saccharomyces cerevisiae mitochondria. PLoS ONE 9:e103956. doi: 10.1371/journal.pone.0103956
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
González Flores, M., Rodríguez, M. E., Oteiza, J. M., Barbagelata, R. J., and Lopes, C. A. (2017). Physiological characterization of Saccharomyces uvarum and Saccharomyces eubayanus from Patagonia and their potential for cidermaking. Int. J. Food Microbiol. 249, 9–17. doi: 10.1016/j.ijfoodmicro.2017.02.018
Hu, Z., Killion, P. J., and Iyer, V. R. (2007). Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 39, 683–687. doi: 10.1038/ng2012
Kozak, B. U., van Rossum, H. M., Luttik, M. A., Akeroyd, M., Benjamin, K. R., Wu, L., et al. (2014). Engineering acetyl coenzyme a supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. mBio 5, e01696–e01614. doi: 10.1128/mBio.01696-14
Lee, T. A., Jorgensen, P., Bognar, A. L., Peyraud, C., Thomas, D., and Tyers, M. (2010). Dissection of combinatorial control by the met4 transcriptional complex. Mol. Biol. Cell 21, 456–469. doi: 10.1091/mbc.E09-05-0420
Leroy, C., Cormier, L., and Kuras, L. (2006). Independent recruitment of mediator and SAGA by the activator Met4. Mol. Cell Biol. 26, 3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Malone, C. D., and Hannon, G. J. (2009). Small RNAs as guardians of the genome. Cell 136, 656–668. doi: 10.1016/j.cell.2009.01.045
Masneuf-Pomarede, I., Salin, F., Börlin, M., Coton, E., Coton, M., Jeune, C. L., et al. (2016). Microsatellite analysis of Saccharomyces uvarum diversity. FEMS Yeast Res. 16:fow002. doi: 10.1093/femsyr/fow002
McNeil, J. B., Zhang, F., Taylor, B. V., Sinclair, D. A., Pearlman, R. E., and Bognar, A. L. (1997). Cloning, and molecular characterization of the GCV1 gene encoding the glycine cleavage T-protein from Saccharomyces cerevisiae. Gene 186, 13–20. doi: 10.1016/S0378-1119(96)00670-1
Nadai, C., Treu, L., Campanaro, S., Giacomini, A., and Corich, V. (2016). Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. Appl. Microbiol. Biotechnol. 100, 797–813. doi: 10.1007/s00253-015-7169-x
Nardi, T., Corich, V., Giacomini, A., and Blondin, B. (2010). A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiol. 156, 1686–1696. doi: 10.1099/mic.0.036723-0
Naumov, G. I., Naumova, E. S., Martynenko, N. N., and Masneuf-Pomaréde, I. (2011). Taxonomy, ecology, and genetics of the yeast Saccharomyces bayanus: a new object for science and practice. Microbiology 80, 723–730. doi: 10.1134/S0026261711060154
Nguyen, H. V., Legras, J. L., Neuvéglise, C., and Gaillardin, C. (2011). Deciphering the hybridisation history leading to the Lager strainage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380. PLoS ONE 6:e25821. doi: 10.1371/journal.pone.0025821
Park, H., and Bakalinsky, A. T. (2000). SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16, 881–888. doi: 10.1002/1097-0061(200007)16:10<881::AID-YEA576>3.0.CO2-3
Pérez-Ortín, J. E., García-Martínez, J., and Alberola, T. M. (2002). DNA chips for yeast biotechnology. The case of wine yeasts. J. Biotechnol. 98, 227–241. doi: 10.1016/S0168-1656(02)00134-7
Piper, M. D., Hong, S. P., Ball, G. E., and Dawes, I. W. (2000). Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 275, 30987–30995. doi: 10.1074/jbc.M004248200
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press
Sipiczki, M., and Ciani, M. (2002). “Taxonomic and physiological diversity of Saccharomyces bayanus,” in Biodiversity and Biotechnology of Wine Yeasts (Kerala: Research Signpost), 53–69.
Su, N. Y., Ouni, I., Papagiannis, C. V., and Kaiser, P. (2008). A dominant suppressor mutation of the met30 cell cycle defect suggests regulation of the Saccharomyces cerevisiae Met4-Cbf1 transcription complex by Met32. J. Biol. Chem. 283, 11615–11624. doi: 10.1074/jbc.M708230200
Taylor, S. L., Higley, N. A., and Bush, R. (1986). Sulfites in food: uses, analytical methods, residues, fate, exposure assessment, metabolism, toxicity, and hypersensitivity. Adv. Food Res. 30, 1–75. doi: 10.1016/S0065-2628(08)60347-X
Teixeira, M. C., Monteiro, P. T., Guerreiro, J. F., Gonçalves, J. P., Mira, N. P., dos Santos, S. C., et al. (2014). The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucl. Acids Res. 42, D161–D166. doi: 10.1093/nar/gkt1015
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Yuasa, N., Nakagawa, Y., Hayakawa, M., and Iimura, Y. (2005). Two alleles of the sulfite resistance genes are differentially regulated in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 69, 1584–1588. doi: 10.1271/bbb.69.1584
Zhang, H., Richards, K. D., Wilson, S., Lee, S. A., Sheehan, H., Roncoroni, M., et al. (2015). Genetic characterization of strains of Saccharomyces uvarum from New Zealand wineries. Food Microbial. 46, 92–99. doi: 10.1016/j.fm.2014.07.016
Zhang, H. Y., Skelton, A., Gardner, R. C., and Goddard, M. R. (2010). Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for global migration from Europe and hybrids between the species. FEMS Yeast Res. 10, 941–947. doi: 10.1111/j.1567-1364.2010.00681.x
Keywords: S. uvarum, FZF1 gene, sulfite tolerance, functional analysis, qRT-PCR, transcriptome analysis
Citation: Liu X, Liu X, Zhang Z, Sang M, Sun X, He C, Xin P and Zhang H (2018) Functional Analysis of the FZF1 Genes of Saccharomyces uvarum. Front. Microbiol. 9:96. doi: 10.3389/fmicb.2018.00096
Received: 31 July 2017; Accepted: 16 January 2018;
Published: 06 February 2018.
Edited by:
Haitao Lu, Shanghai Jiao Tong University, ChinaReviewed by:
Zhao Chen, University of California, Davis, United StatesGiovanna Suzzi, Università di Teramo, Italy
Copyright © 2018 Liu, Liu, Zhang, Sang, Sun, He, Xin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanyao Zhang, zhanghanyao@hotmail.com
†These authors are equal contributors to this work.
 Xiaozhen Liu1,2†
Xiaozhen Liu1,2† Hanyao Zhang
Hanyao Zhang