- 1Institute for Epidemiology and Pathogen Diagnostics, Federal Research Centre for Cultivated Plants, Julius Kühn-Institut (JKI), Braunschweig, Germany
- 2Department Plant-Microbe Systems, Leibniz Institute of Vegetable and Ornamental Crops, Großbeeren, Germany
Biocontrol inoculants often show inconsistency in their efficacy at field scale and the reason for this remains often unclear. A high rhizosphere competence of inoculant strains is assumed to be a key factor for successful biocontrol effects as the biocontrol strain has to compete with the indigenous microbial community in the rhizosphere. It is known that many factors, among them plant species and soil type shape the rhizosphere microbial community composition. However, microbial community composition in the rhizosphere can also be influenced by the presence of a pathogen. We hypothesized that plant species, soil type, and a pathogen affect the rhizosphere competence of a biocontrol strain and its biocontrol effect against a soil-borne pathogen. To test the hypothesis, we used an experimental plot system with three soil types (diluvial sand, alluvial loam, loess loam) kept under similar agricultural management at the same field site for 12 years. We investigate the rhizosphere competence of Pseudomonas sp. RU47 in two plant species (potato and lettuce) and its biocontrol effect against Rhizoctonia diseases. The colonization density of a rifampicin resistant mutant of RU47 in the rhizosphere of both crops was evaluated by plate counts. Bacterial community compositions were analyzed by denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene fragments amplified from total community DNA. The inoculant RU47 was able to colonize the rhizosphere of both model crops in a sufficient density and to reduce disease severity of black scurf on potato and bottom rot on lettuce in all three soils. DGGE indicated that RU47 affected the bacterial community composition stronger in the rhizosphere of lettuce than in the potato rhizosphere. In contrast, the effect of the pathogen Rhizoctonia solani on the bacterial community was much stronger in the rhizosphere of potato than in the lettuce rhizosphere. A significant effect of RU47 on the Pseudomonas-specific gacA fingerprints of the rhizosphere was only observed in lettuce in alluvial soil. The soil type and plant species independent biocontrol effects of RU47 and its minor influence on the indigenous bacterial community composition might be important criteria for the registration and use of RU47 as biocontrol strain.
Introduction
Diseases caused by soil-borne pathogens such as Rhizoctonia solani (Kühn) are difficult to control, and the use of bacterial inoculants for disease suppression represents an environmentally friendly control method (Alabouvette et al., 2009). Still biocontrol inoculants often showed a lack of consistency in their biocontrol activity at the field scale (Barret et al., 2011).
Over decades, efforts have been made to unravel factors affecting biocontrol efficacy of bacterial inoculants (Bais et al., 2006; Barret et al., 2011; Ghirardi et al., 2012). Variation in the ability of bacterial inoculants to colonize the rhizosphere and survive at sufficiently high cell densities was thought to be a reason for inconsistency in disease suppression (Ghirardi et al., 2012). Therefore, a high rhizosphere competence was identified as a prerequisite for the expression of beneficial effects on plants (De Bellis and Ercolani, 2001; Barret et al., 2011; Ghirardi et al., 2012).
Research presently is just starting to unravel the complex interplay between plants and their microbiome including bacterial inoculants applied (Berendsen et al., 2012; van der Heijden and Hartmann, 2016; Berg et al., 2017). Studies done by 16S rRNA gene-based fingerprints provided a growing body of evidence that biotic factors (e.g., plant species/genotype, developmental stage, and plant pathogens) and abiotic factors (e.g., weather conditions, agricultural management) affect the bacterial community composition in the rhizosphere (Berg and Smalla, 2009; Weinert et al., 2009; Schreiter et al., 2014a).
Several studies have clearly shown that both plant and soil type shape the rhizosphere microbiome (Bulgarelli et al., 2012; Peiffer et al., 2013; Schreiter et al., 2014b). Different soil types varying in physicochemical properties were shown to contain distinct bacterial communities from which a subset of populations was enriched in the rhizosphere of the plant due to root exudates (Costa et al., 2006; Schreiter et al., 2014b) resulting in an increased relative abundance in the rhizosphere compared to the corresponding bulk soil.
Until recently, limited information was available on the effect of different soil types and the plant species on the rhizosphere competence and biocontrol activity of bacterial inoculants at field scale. Results of a previous study showed that although the soil types harbor a specific bacterial community no impact on the ability of the inoculant strains Serratia plymuthica 3Re4-18 and Pseudomonas sp. RU47 to colonize the rhizosphere of lettuce in a sufficient density was given at the field scale (Schreiter et al., 2014c).
The strain RU47, initially identified as Pseudomonas jessenii based on the 16S rRNA gene sequence, has been recently reclassified as its genome sequence is phylogenetically related to strains from P. koreensis group (Eltlbany et al., under revision). Based on the results of this study, we assume that the often-reported inconsistency of biocontrol effects in the field is more likely due to plant characteristics. The plant modulates via root exudates the rhizosphere microbiome by stimulating microorganisms with traits beneficial for plant growth and health (Dennis et al., 2010; Doornbos et al., 2012). About 30–50% of photosynthetically fixed carbon are released through the root system in the rhizosphere (Kumar et al., 2006). Hence, root exudates play an important role in provoking a substrate-driven competition between rhizosphere microorganisms including an applied biocontrol inoculant in colonization of roots (Bais et al., 2006). The root exudation pattern and the amounts of various exudate compounds synthesized and exuded by the roots are under the plant’s genetic control (Micallef et al., 2009). Therefore, the plant species and its unique root exudation pattern attract a specific rhizomicrobial community (Chapparo et al., 2014). A biocontrol inoculant introduced into the rhizosphere has to compete and interact with this plant specific microbial community. Moreover, the rhizosphere is a highly dynamic habitat due to the changing input by the plant. As a result, the rhizosphere microbial community composition and activity differs temporally and spatially (DeAngelis et al., 2009; Chowdhury et al., 2013).
The main objective of our study was to assess the rhizosphere competence and biocontrol activity of the bacterial inoculant RU47 against a soil-borne pathogen depending on the plant species in three soil types.
In 2011 the ability of RU47 to colonize the rhizosphere of lettuce and its biocontrol of R. solani was shown to be soil type independent but strikingly dependent on plant growth (Schreiter et al., 2014c). In the present study we compare the rhizosphere competence and biocontrol effects of the inoculant RU47 on lettuce (Lactuca sativa L.) with its rhizosphere competence and biocontrol effects on potato (Solanum tuberosum L.). There are great differences in systematic botany, phenotype and cultivation period between the model plants. The used model plant species potato and lettuce are members of different plant families (Solanaceae and Asteraceae, respectively) and the growing period for lettuce is around 5–6 weeks in the field, compared to potato with a cultivation period of around 4–5 months. We expect a pronounced different bacterial community structure in the rhizosphere of the used model plants. Both potato and lettuce are host plants of the soil-borne pathogen R. solani.
It is important to note that the fungal pathogen R. solani is a species complex of various genetic groups called anastomosis groups (AGs) with a distinct degree of host specificity. While R. solani AG1-IB is responsible for bottom rot on lettuce, R. solani AG3 causes black scurf disease on potato with striking differences in the genome (Wibberg et al., 2013) and in the interaction with the plant. Whereas R. solani AG3 colonizes the belowground parts of potato including the roots (Genzel et al., 2018), R. solani AG1-IB infects lettuce via the lower leaves in contact to the soil (Grosch et al., 2011). Hence, we expect a stronger impact of R. solani AG3 on the microbial community in the rhizosphere of potato than of R. solani AG1-IB in the rhizosphere of lettuce.
In view of the differences in plant-pathogen interaction of the two model plants we aimed to answer the question of whether RU47 is able to compete successfully with the bacterial community in the rhizosphere in both plant species independent from the soil type and to express biocontrol effects in the field on both crops. We used an experimental plot system with three soil types (diluvial sand, alluvial loam, loess loam) kept under similar agricultural management at the same field site. The effects of the inoculant strain RU47 as well as the pathogens on bacterial community composition in the rhizosphere were assessed by denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA gene fragments amplified from total community (TC)-DNA. The synthesis of secondary metabolites involved in antagonistic interactions and disease suppression is positively controlled by the global response regulator gene gacA (Haas and Defago, 2005). Being highly conserved among Pseudomonas species, gacA serves as a reliable phylogenetic marker for community fingerprinting (De Souza et al., 2003). Hence, we used gacA in addition to the 16S rRNA gene to analyze the effect of RU47 and the pathogens R. solani AG3 and AG1-IB on the bacterial and the Pseudomonas community in the rhizosphere of both model plants.
Materials and Methods
Field Experiments
A unique experimental plot system with independent experimental units for lettuce (unit 6) and potato (unit 7) was used for the experiments at the Leibniz-Institute of Vegetable and Ornamental Crops in Großbeeren (52° 33′ N, 13° 22′ E). Each unit consisted of three soil types characterized as Arenic-Luvisol (diluvial sand), Gleyic-Fluvisol (alluvial loam), and Luvic-Phaeozem (loess loam) that shared the comparable agricultural management at the same field site for more than 10 years (Rühlmann and Ruppel, 2005; Schreiter et al., 2014a). Each soil type was arranged in a separate block with 24 plots of 2 m × 2 m and a depth of 75 cm. From 2000 to 2011, following crops were cultivated in unit 6: pumpkin, nasturtium, pumpkin, amaranth, wheat, wheat, pumpkin, nasturtium, wheat, wheat, lettuce and lettuce; and in unit 7: broccoli, phacelia, broccoli, nasturtium, nasturtium, tomato, soybean, phacelia, wheat, phacelia, nasturtium, and phacelia.
On 5 May 2012, seed potato tubers (cv. Arkula, Norika GmbH, Groß Lüsewitz, Germany) were planted in unit 7 at a distance of 30 cm within a row, and with an intra-row distance of 65 cm. The number of tubers (diameter 35–60 mm) and the marketable tuber yield (MTY) were assessed at harvest 18 weeks after planting (on 5 September 2012). Additionally, the percentage of infestation of 20 randomly selected tubers per replicate with Rhizoctonia sclerotia was evaluated on a scale from 1 to 5 (1 – without sclerotia, 2 – <1% infestation, 3 – 5% infestation, 4 – 10% infestation, 5 – ≥15% infestation).
Seedling trays filled with the respective soil type were used to cultivate the lettuce seedlings (cv. Tizian, Syngenta, Bad Salzuflen, Germany) as described by Schreiter et al. (2014c). Lettuce seedlings were planted on 3 July 2012 at the three- to four-leaf stage 4 weeks after sowing with a within-row and intra-row distance of 30 cm to the experimental plot unit 6. The lettuce shoot dry mass (SDM) of each plant and the disease severity of bottom rot were assessed according to the following scale: (1) healthy plants without bottom rot symptoms; (2) symptoms on first lower leaves and small brown spots on the underside of leaf midribs; (3) brown spots on leaf midribs on lower and next upper leaf layers; and (4) severe disease symptoms on upper leaf layers and beginning of head rot to total head rot (Supplementary Figure S1) at harvest five weeks after planting (on August 8, 2012; typical size, form and firmness of head reached).
In both experimental units, plots of each soil type were adjusted to the same nitrogen amount of 157 kg ha-1 (Kalkamon, 27% N) before planting, based on nutrient analysis of the soil which was done according to the certified protocols of the Association of German Agricultural Analytic and Research Institutes (VDLUFA). The soils in unit 7 (potato) were also adjusted to the same amount of potassium of 210 kg ha-1 (Patentkali, K+S Kali GmbH, Kassel, Germany). Both crops were overhead irrigated based on the computer program ‘BEREST.’
The following treatments of potato and lettuce were studied: treatment without Pseudomonas sp. RU47 and without pathogen application (control), treatment with R. solani inoculation (+Rs), treatment with RU47 but without pathogen inoculation (RU47), and treatment with RU47 and R. solani inoculation (RU47+Rs). Each treatment included four replicates (plots) with 21 (potato) or 36 (lettuce) plants per replicate, randomly arranged per soil type.
Inoculation of the Pathogens
The R. solani isolate Ben3 was obtained from sclerotia of mature potato tuber and characterized by molecular tools as AG3 (Kuninaga et al., 2000). The isolate R. solani AG1-IB 7/3/14 (AJ868459) was originally isolated from lettuce plants with bottom rot symptoms and characterized by conventional and molecular tools (Grosch et al., 2007). The inocula of both pathogens were prepared on barley kernels as described by Schneider et al. (1997).
For pathogen inoculation of potato at planting, seed tubers were covered with a 3 cm thick soil layer upon which five barley kernels infected with the black scurf pathogen R. solani AG3 were placed in plots with pathogen inoculation. Non-infected kernels were placed on tubers in plots without pathogen inoculation.
To ensure a homogeneous pathogen pressure for bottom rot in unit 6, a total of 36 lettuce plants (nine or more true leaves unfolded) were shredded and evenly incorporated in the top soil (10 cm) together with 40 g of barley kernels infected with R. solani AG1-IB or of non-infected barley kernels (plots without pathogen inoculation) with a rotary hoe 2 weeks before initiating the experiment.
Preparation of RU47 Cell Suspension and Application Mode
A rifampicin resistant mutant of Pseudomonas sp.RU47 (strain collection of the Julius Kühn-Institut) was used to assess the colonization density in the field (Adesina et al., 2009). The inoculum of RU47 was prepared on King’s B agar plates (Merck KGaA, Darmstadt, Germany) for seed treatment, and in nutrient broth (NB II, SIFIN GmbH, Berlin, Germany) for treatment of young lettuce plants and seed potato tubers as described by Schreiter et al. (2014c). Both media were supplemented with rifampicin (75 μg mL-1). After a cultivation time of 16 h the culture was centrifuged at 13,000 g for 5 min, the supernatant discarded and the pellet was re-suspended in sterile 0.3% NaCl solution.
The potato seed tubers were inoculated with RU47 immediately before planting, spraying 20 mL suspension (108 colony forming units [CFU] mL-1) on 10 tubers, and the emerged potato plants were inoculated four weeks after planting (WAP) by watering each plant with 100 mL suspension (108 CFU mL-1).
A total of 100 lettuce seeds were coated on a Vortex Mixer (MSI, Minishaker, Staufen, Germany) with 250 μl of the bacterial cell suspension dropped on the seeds [108 CFU mL-1]. Each lettuce seedling at its three-leaf stage was treated with 20 mL cell suspension of RU47 (107 CFU mL-1) 1 week before transplanting them to the field plots and with 30 mL (108 CFU mL-1) 2 days after planting. Regarding respective control treatments for lettuce and potato, watering was carried out with 0.3% NaCl.
Sampling, Sample Processing and Determination of RU47 Counts
The rhizosphere competence of Pseudomonas sp. RU47 was analyzed on potato and on lettuce at two time points (7 and 12 WAP for potato, 2 and 5 WAP for lettuce) and the bacterial and Pseudomonas community composition (based on 16S rRNA or gacA gene, respectively) at one time point (7 WAP for potato, 2 WAP for lettuce) during the growth period of both plants.
Potato plants were sampled 7 WAP of seed potato tuber at growth stage 2 (formation of basal side shoots below and above soil surface) and 12 WAP at growth stage 7 (50% of berries in the first fructification have reached full size). The roots of two potato plants per replicate were combined at the first sampling time (7 WAP), and the roots of one plant per replicate were sampled at the second sampling time (12 WAP).
Lettuce plants were sampled 2 WAP when plants had nine or more true leaves unfolded, and 5 WAP when lettuce head reached typical size, form and firmness. For each treatment the roots of three lettuce plants per replicate were combined as a composite sample and considered as one replicate. In both crops the rhizosphere competence of RU47 and the rhizosphere bacteria of four replicates per treatment were analyzed.
Adhering soil was removed by a quick root wash step, and the root system was cut into 1 cm long pieces with sterile scissors and mixed before microorganisms were extracted as follows: 5 g of roots were placed in sterile Stomacher bags and treated by a Stomacher 400 Circulator (Seward Ltd., Worthing, United Kingdom) for 30 s at high speed after adding 15 ml of sterile 0.3% NaCl. The Stomacher blending step was repeated three times and one ml was taken from the combined 45 ml for plate counts while the microbial rhizosphere/rhizoplane fraction was harvested by centrifugation steps as described by Schreiter et al. (2014a).
Stomacher blending steps were immediately processed to determine the CFU of RU47 by plating serial dilutions on King’s B agar supplemented with rifampicin (75 μg mL-1) and cycloheximide (100 μg mL-1) after an incubation time of 48 h at 28°C. The CFU were calculated per gram root dry mass. For both plant species and all soil types, Stomacher supernatants obtained from the control plots were plated as well to determine the background of the indigenous rifampicin resistant bacteria.
Total DNA Extraction and Analysis of 16S rRNA and gacA Gene Fragments by DGGE Fingerprints
Total community DNA was extracted after a harsh lysis step by means of the FastDNA SPIN Kit for Soil® (MP Biomedicals, Heidelberg, Germany) from the same samples used for RU47 CFU determination (Schreiter et al., 2014a). PCR reactions were performed for amplification of 16S rRNA gene fragments with the bacterial primers F984-GC and R1378 using GoTaq® Flexi (Promega, Mannheim, Germany) (Nübel et al., 1996; Heuer et al., 1997). The PCR products were analyzed by DGGE as described by Weinert et al. (2009). For characterization of the effect of RU47 with (RU47+Rs) and without (RU47) R. solani inoculation on Pseudomonas community in the potato and lettuce rhizosphere, DGGE fingerprints of gacA gene amplicons were carried out according to Costa et al. (2007).
Data Analysis
All data obtained from the field were analyzed with the STATISTICA program (StatSoft Inc., Tulsa, OK, United States). The impact of the soil type, of the inoculant RU47 and of the pathogen R. solani on MTY of potato and SDM of lettuce was analyzed using three-way ANOVA combined with Tukey post hoc test (P ≤ 0.05). Effects of RU47 and the pathogen on MTY and SDM were analyzed for each soil type by two-way ANOVA. The data of disease severity was analyzed using the non-parametric Kruskal–Wallis test followed by Mann–Whitney U-test (P ≤ 0.05). The CFU per gram root dry mass (RDM) were calculated and logarithmically (Log10) converted. The effects of the soil type, the growth period/plant age (sampling time), and the pathogen on the plate counts of RU47 were evaluated using three-way ANOVA (P ≤ 0.05) combined with Tukey post hoc test (P ≤ 0.05). DGGE fingerprints were evaluated with GELCOMPAR II version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium) as described by Schreiter et al. (2014a). The Pearson correlation coefficient as a curve-based method was chosen to obtain pairwise similarities between fingerprints and similarities were clustered using unweighted pair group method using average linkages (UPGMA). These were used for statistical analysis by Permutation tests (P ≤ 0.05) where the d-value was calculated as average overall correlation coefficients within the groups minus the average overall correlation coefficients between samples from different groups as suggested by Kropf et al. (2004).
Results
Plant Species Effect on Rhizosphere Competence of RU47 in Three Soils
Our results demonstrated that Pseudomonas sp. RU47 was able to colonize the rhizosphere of potato and lettuce at a density of more than 5 Log10 CFU per gram root dry mass in all three soils at field scale at both sampling times. Three-way ANOVA revealed that neither the soil type (potato: P ≤ 0.19; lettuce: P ≤ 0.08) nor the presence of R. solani (AG3: P ≤ 0.10; AG1-IB: P ≤ 0.44) significantly affected the number of RU47 CFU counts in the rhizosphere of both crops. Two-way ANOVA showed for both crops significantly declined CFU counts of RU47 in the rhizosphere with increasing plant age in the treatments without (RU47) and with R. solani inoculation (RU47+Rs) in both loamy soils (alluvial loam: P ≤ 0.02; loess loam: P ≤ 0.002) but not in the diluvial soil (P ≥ 0.15). Interestingly, no plant age-dependent decline in the density of RU47 was observed in the rhizosphere of both crops in the treatments with R. solani inoculation (RU47+Rs) in all soils, except for lettuce grown in alluvial soil (Figure 1).
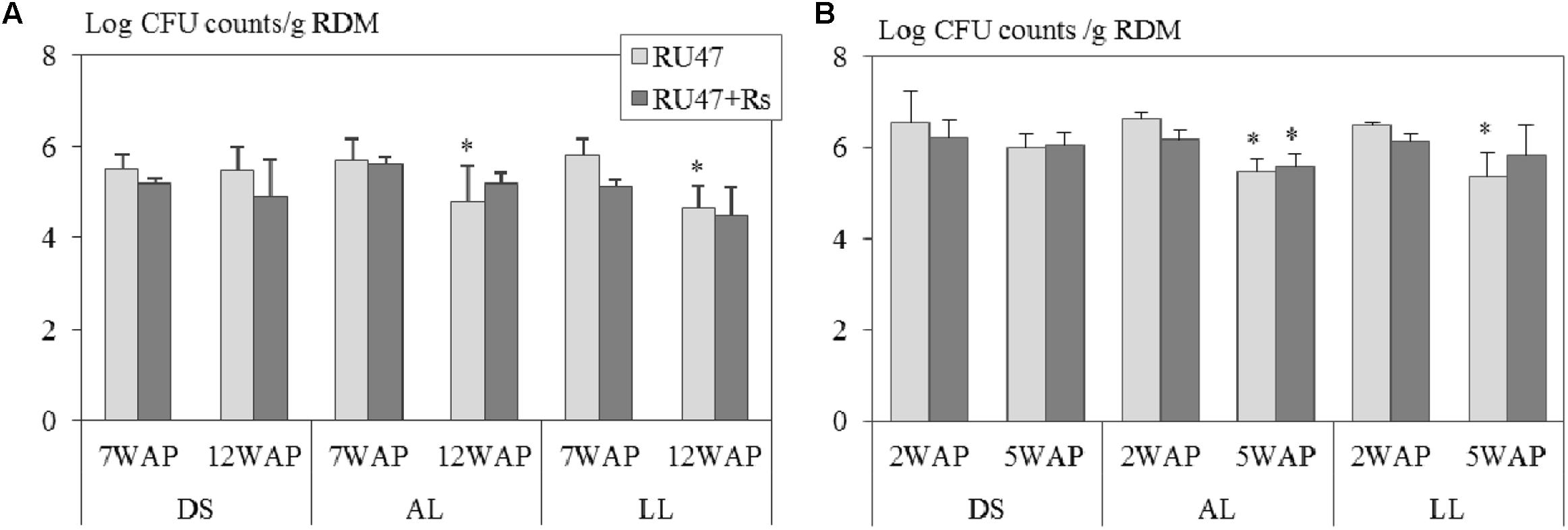
FIGURE 1. Number of colony forming units (CFU) per gram root dry mass (RDM) of Pseudomonas sp. RU47 estimated in the rhizosphere of (A) potato (cv. Arkula) and (B) lettuce (cv. Tizian) with RU47 and with RU47 + Rhizoctonia solani inoculation (RU47+Rs) 7 and 12 weeks after planting (7 WAP, 12 WAP) of potato and 2 and 5 weeks after planting (2 WAP, 5 WAP) of lettuce in the 2012 season. Plants were grown in three soils [diluvial sand (DS), alluvial loam (AL), loess loam (LL)] at the same field site. No significant effect of the soil type and the presence of R. solani on the number of CFU of RU47 was revealed (ANOVA, P ≥ 0.05). Asterisks indicate significant differences in CFU counts between the two sampling times within each soil type (Tukey post hoc test, P ≤ 0.05). The bars represent the standard deviation.
Soil Type, R. solani, and of RU47 Effects on Yield of Potato and Lettuce
A significant effect of the soil type (P ≤ 0.0001) and the pathogen R. solani AG3 (P ≤ 0.0001) on MTY was found but no effect of RU47 (P ≥ 0.15). Comparing the MTY of non-inoculated potato plants (control) in all soils, the lowest yield was recorded in diluvial sand (543.8 g plant-1) which differed significantly from MTY in both loamy soils (alluvial loam, 841.3 g plant-1; loess loam, 764.0 g plant-1; P ≤ 0.008) (Table 1). Two-way ANOVA indicated that the pathogen R. solani AG3 significantly reduced MTY of potato in both loamy soils (P ≤ 0.02) but not in diluvial sand (P ≤ 0.18) compared with control plants, respectively. In all soils, no effect by the inoculant RU47 on MTY of potato was observed in comparison to the non-inoculated control plants in absence (control vs. RU47) and in presence of R. solani (+Rs vs. RU47+Rs).
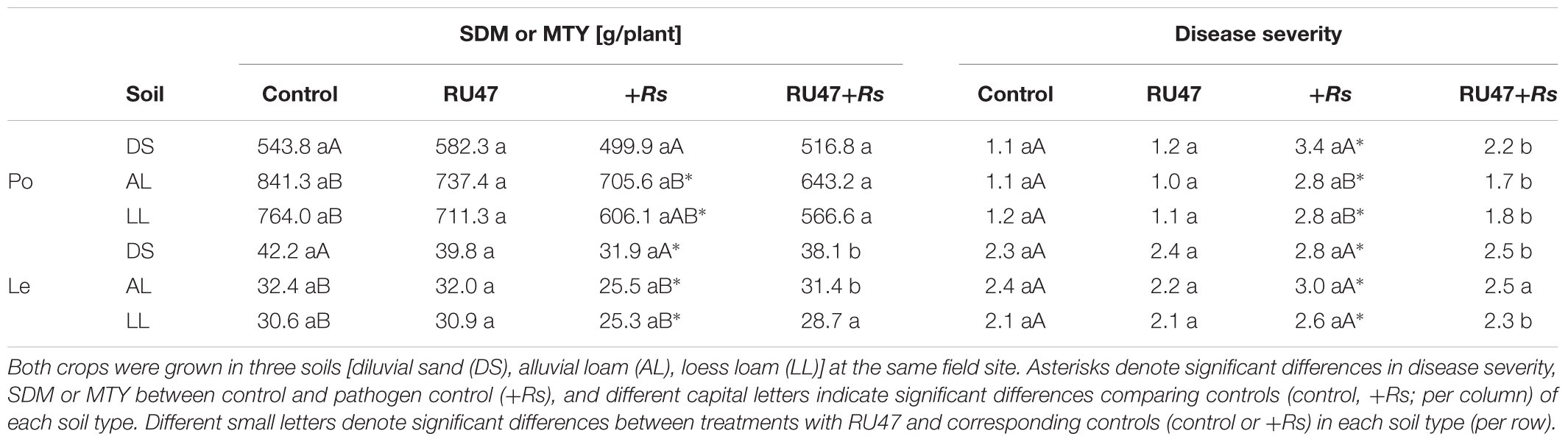
TABLE 1. Marketable tuber yield (MTY) of potato (Po, cv. Arkula) and shoot dry mass (SDM) of lettuce (Le, cv. Tizian) and disease severity of black scurf on potato and bottom rot on lettuce without (control) and with Pseudomonas sp. RU47 (RU47), and without and with Rhizoctonia solani inoculation (+Rs; RU47+Rs) in the 2012 season.
Three-way ANOVA indicated a significant effect of the soil type (P ≤ 0.0001) and the pathogen (P ≤ 0.0001) but also not of RU47 (P ≥ 0.07) on lettuce SDM. In contrast to potato, the highest SDM of lettuce was recorded in diluvial sand (42.2 g plant-1) which also differed significantly from SDM in both loamy soils (alluvial loam, 32.4 g plant-1; loess loam, 30.6 g plant-1; P ≤ 0.0003). The SDM was significantly reduced by R. solani AG1-IB in all soils compared to the control plants (Table 1). The inoculant RU47 did not improve the SDM of lettuce in any of the soils in the treatments without R. solani inoculation (control vs. RU47; P ≥ 0.45), but it significantly enhanced the lettuce growth in diluvial sand (P ≤ 0.01) and alluvial loam (P ≤ 0.01) in treatments with R. solani inoculation (+Rs vs. RU47+Rs).
Soil Type Effect on the RU47 Mediated Biocontrol Activity against R. solani
In all control treatments no, or only slight black scurf symptoms on potato tuber appeared at harvest. In contrast the inoculation of R. solani AG3 (+Rs) resulted in a significantly increased disease severity of black scurf on potato (P ≤ 0.05) in all soils (Table 1). Comparing disease severity of black scurf symptoms on potato in the pathogen controls (+Rs) of all three soils the highest and significant (P ≤ 0.03) disease severity was recorded on tubers harvested from diluvial sand. Comparable severity of black scurf (P ≤ 0.89) was observed on tubers yielded from plants grown in alluvial loam and loess loam (Table 1). RU47 was able to significantly suppress (P ≤ 0.03) the severity of black scurf on potato in all soils (+Rs vs. RU47+Rs).
Bottom rot symptoms on lettuce were revealed in the controls in each soil (Table 1). A significantly higher disease severity of bottom rot was revealed on plants in all soils after R. solani AG1-IB (+Rs) inoculation (P ≤ 0.05), while no significant differences (P ≥ 0.06) in disease severity were observed among the three soil types without (control) and with pathogen inoculation (+Rs; Table 1). The inoculant RU47 suppressed significantly (P ≤ 0.03) the severity of bottom rot on lettuce in diluvial sand and loess loam (+Rs vs. RU47+Rs). The disease severity of bottom root on plants grown in alluvial soil was reduced through RU47 application but not significantly (P ≤ 0.1).
R. solani and RU47 Effects on the Rhizosphere Bacterial Community Composition
Effects of the plant species in three soil types, the pathogens R. solani AG3 and AG1-1B, and inoculant RU47 on the bacterial communities were detected based on 16S rRNA gene DGGE fingerprints for samples taken 7 or 2 WAP of potato or lettuce plants, respectively.
DGGE fingerprints revealed distinct rhizosphere bacterial communities depending on the plant species (Supplementary Figures S2–S4). Significant effects of R. solani AG3 on rhizosphere bacterial communities of potato plants were observed with high d-values (Table 2), while the effects of AG1-1B on the bacterial community composition of the lettuce rhizosphere were negligible, indicated by low and not significant d-values (Table 2). The highest impact of both pathogens (AG3 and AG1-IB) on the rhizosphere bacterial community of potato and lettuce plants was revealed in alluvial loam (Table 2), indicating a soil type effect on the interaction of pathogen and indigenous bacterial community in the rhizosphere.
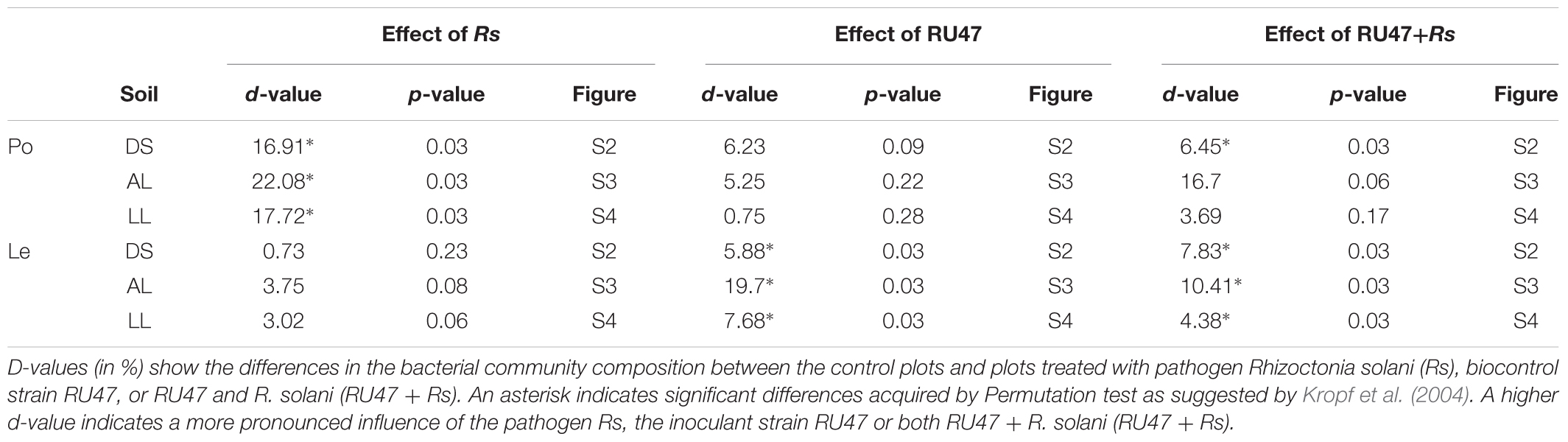
TABLE 2. D-values obtained by bacterial DGGE analysis of rhizosphere samples from potato (Po, cv. Arkula) and lettuce (Le, cv. Tizian) grown in three soils [diluvial sand (DS), alluvial loam (AL), loess loam (LL)] at the same field site.
The inoculant RU47 showed plant species dependent effects on the rhizosphere bacterial community composition as well. Based on the DGGE fingerprints and the d-values obtained by the comparison of the control and the treatment with RU47 fingerprints, the effects of RU47 were on average lower and not significant in the rhizosphere of potato in contrast to the rhizosphere of lettuce (Table 2).
An effect on the lettuce rhizosphere bacterial community composition was observed in the treatment with RU47 in presence of the pathogen R. solani for both crops. The highest effect was found consistently in alluvial loam albeit not significant for potato (Table 2).
RU47 and R. solani Effects on Pseudomonas Community in the Rhizosphere
The Pseudomonas community of the rhizosphere of both plant species was analyzed by DGGE of gacA genes for samples taken 7 or 2 WAP, respectively. A significant effect of the soil type was observed for lettuce in contrast to the potato rhizosphere which exhibited higher heterogeneity between replicates (control treatments in Supplementary Figures S5, S6). Inoculation with RU47 resulted only in small shifts of Pseudomonas community that were significant only for lettuce in alluvial loam soil (Table 3). The additional presence of R. solani AG3 (potato) or AG1-IB (lettuce) significantly changed the Pseudomonas community of potato grown in DS and LL and for lettuce grown in DS soil.
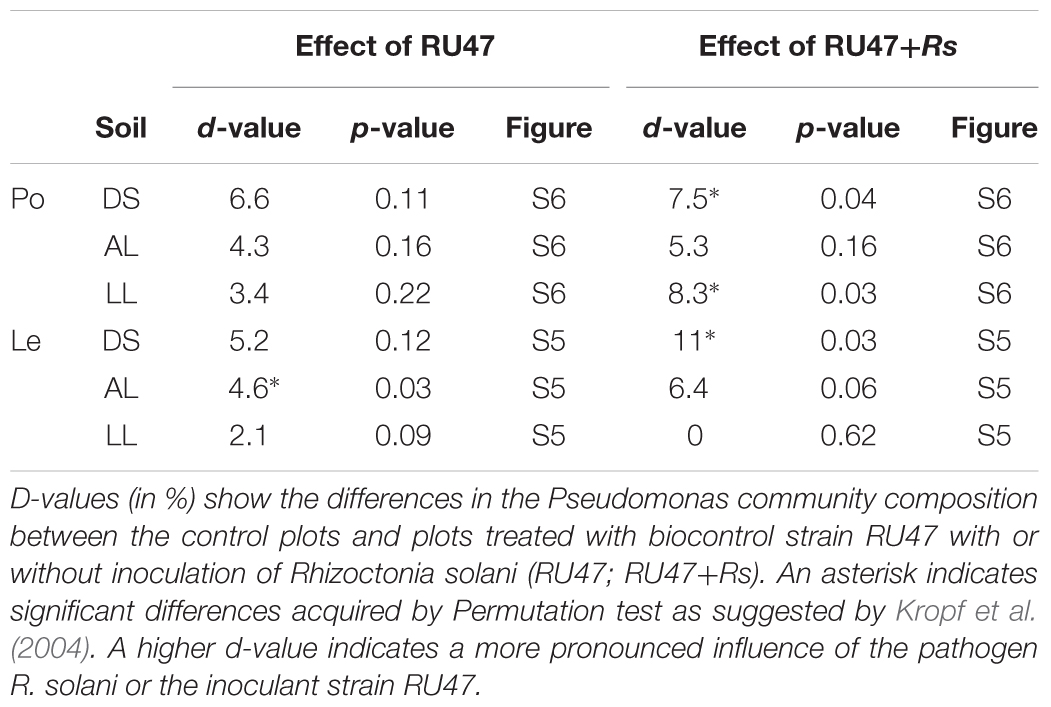
TABLE 3. D-values obtained by Pseudomonas-specific DGGE analysis of rhizosphere samples from potato (Po, cv. Arkula) and lettuce (Le, cv. Tizian) grown in three soils [diluvial sand (DS), alluvial loam (AL), loess loam (LL)] at the same field site.
Discussion
In this study, we compared the ability of RU47 to colonize the rhizosphere and to display biocontrol effects against Rhizoctonia diseases for two plant species grown in three different soil types. Hereby we confirm results of previous years obtained with lettuce in the same long term field trial and broaden our knowledge with a systematic, morphological different model plant (Schreiter et al., 2014a,b,c). Both, potato and lettuce are host plants of R. solani but the interaction of the model plants vary with the different anastomosis groups (AGs) of R. solani. The model plants were grown in the same three soil types at the same site. However, they were planted in two units of the experimental plot system which exhibited only a slight variation in cropping history. In contrast to other field studies the experimental set-up excluded site effects caused by different weather conditions and management practice on bacterial communities in the bulk soil. Potato plants have a much longer growth period than lettuce, and thus the analysis of bacterial community was performed 2 and 7 WAP (3 and 8 weeks after last application of RU47). We assumed that the bacterial community structure in the rhizosphere differed depending on the plant species in each soil and pyrosequencing (Schreiter et al., unpublished) and DGGE analysis underlined this assumption (Supplementary Figures S2–S4).
Contrasting effects of the soil types on MTY of potato and the SDM of lettuce in the untreated controls were assessed at harvest. The soil characteristics play an important role in the ability of a plant species to extract water and nutrients. Although the soil types were adjusted to the same amount of nitrogen it cannot be excluded that soil nitrogen, i.e., nitrate, leaches by irrigation water to a deeper, non-rooted soil level, especially in the diluvial sand. Availability of nitrogen was possibly limited in potato crop in diluvial sand within the growth period of 12 weeks compared to the loamy soils.
A negative effect of the pathogen R. solani AG1-IB on lettuce growth and of R. solani AG3 on the MTY of potato was observed in all soils. These results confirmed the observation for lettuce made in previous field experiments (Grosch et al., 2005; Schreiter et al., 2014c). The disease impact of the bottom rot pathogen was not revealed in the present study conducted in 2012 when plants were treated with RU47 confirming the biocontrol effects of RU47 observed in 2011 (Schreiter et al., 2014c). Our results are in accordance with findings for the inoculant strains Bacillus amyloliquefaciens FZB42 and Serratia plymuthica 3Re4-18 for lettuce (Grosch et al., 2005; Chowdhury et al., 2013). However, we found that the inoculant RU47 was not able to compensate the negative effect of the pathogen on MTY of potato. Compared to R. solani AG1-IB on lettuce which does not colonize the roots, R. solani AG3 was previously reported to affect all belowground parts of potato including the roots during the growth period (Atkinson et al., 2011; Genzel et al., 2018). Thus, it was not surprising that R. solani AG3 had a more pronounced effect on the composition of the bacterial community in the rhizosphere of potato as indicated by the higher d-values than R. solani AG1-IB on the rhizosphere bacterial community in lettuce. Windisch et al. (2017) showed that both the pathogen and the biocontrol strain induced changes in the root exudation pattern of lettuce, including higher amounts of antimicrobial compounds. An impact of pathogens such as Verticillium sp. and Pythium sp. on root exudation patterns of cotton and basil plants was reported by Bais et al. (2006) and Zhang et al. (2011). It is assumed that R. solani AG3 may also cause changes in root exudation patterns of potato and thus affect the interaction of RU47 with the plant. Although RU47 was found in a sufficient density in the rhizosphere of potato and was able to reduce severity of black scurf symptoms in all soils the potato yield losses were not compensated.
Our results revealed that despite the plant species dependent rhizosphere bacterial community composition observed (Supplementary Figures S2–S4), the inoculant RU47 was able to successfully colonize the rhizosphere of potato and lettuce. The rhizosphere competence of RU47 in potato and lettuce was not influenced by the soil type, as already revealed in a previous experiment (Schreiter et al., 2014c). Higher CFU counts of RU47 were found in the rhizosphere of lettuce in comparison to the potato rhizosphere. The finding might be due to the differences in sampling time after RU47 application (3 and 8 weeks after the last application for potato; 2 and 5 weeks after the last application for lettuce). Furthermore, the CFU counts were calculated per gram of root dry mass and not per cm2 root surface. In contrast to potato, the fine root system of lettuce, which is likely to provide a higher root surface than the potato root system, might be a reason for the observed higher CFU counts in lettuce. A decline in the population density of RU47 within the growth period was found for both crops and confirmed previous observations of RU47 and the inoculant strains 3Re4-18, and FZB42 (Chowdhury et al., 2013; Schreiter et al., 2014c). It is assumed that the decrease in CFU counts is likely linked to changes in root morphology as the number of fine roots decreased with the growing plant. In both crops no impact of R. solani AG3 or AG1-IB on the rhizosphere competence of RU47 was revealed. These results were consistent with the study of Chowdhury et al. (2013) where the rhizosphere competence of FZB42 was also not affected by the bottom rot pathogen R. solani AG1-IB in lettuce. Our results underline that the ability of the inoculant RU47 to colonize the rhizosphere of potato and lettuce plants in a sufficient density correlated with effective biocontrol activity in both crops and in all three soils.
Black scurf symptoms were observed only on potato tubers originating from treatments inoculated with R. solani AG3. In contrast, the bottom rot symptoms on lettuce were also detected in all control treatments and confirmed previous reports on the occurrence of R. solani AG1-IB in arable soils (Grosch et al., 2011). In accordance with the results of the 2011 conducted field experiment (Schreiter et al., 2014c), the lowest disease severity of bottom rot on lettuce (control, +Rs) was assessed in loess loam (+Rs). But the differences in disease severity between the soil types were not significant in the treatment with R. solani AG1-IB as determined previously. An impact of the soil type on bean hypocotyl rot severity caused by R. solani AG4 was also revealed by Nerey et al. (2010). In addition, Tamm et al. (2010) showed that the soil type is a major determinant for suppressiveness against soil-borne diseases. Several studies suggested that physicochemical properties of different soils have a strong effect on microbial community structure and function (reviewed in Berg and Smalla, 2009). In accordance, we found distinct rhizosphere bacterial communities in the different soil types with the best soil disease suppression in loess loam (Schreiter et al., 2014c). The disease severity of bottom rot in the growing period 2011 was much higher in all soils compared to the growing period 2012. Pathogen inoculum density affects the disease severity and additional inoculation increased the density in all soils. Moreover, results of previous experiments over different growing periods showed that also weather conditions have a pronounced impact on disease development (Grosch et al., 2011). In the growing period 2011 the conditions were more favorable for disease development of bottom rot compared to 2012.
Although DGGE fingerprints do not provide taxonomic information on community composition and responders to the treatments, they provide rapid insights into dynamic changes and the differences between treatments. Recently, we could show that the pyrosequencing data for lettuce rhizosphere and bulk soil from the experimental plot units confirmed the DGGE findings (Schreiter et al., 2014a,b).
The effect of RU47 on rhizosphere bacterial communities differed depending on plant species and soil type as revealed by bacterial and Pseudomonas-specific DGGE fingerprints. A significant effect of RU47 was found in all three soils in lettuce in the treatments without and with additional pathogen inoculation. This is in accordance to previous findings on lettuce where the impact of RU47 was assessed over three consecutive years. In the previous study an increasing effect of RU47 on rhizosphere bacterial communities could be observed over the years 2010 until 2012. Nevertheless, the influence of RU47 was negligible compared to community changes caused by the soil type (Schreiter et al., 2014b). The comparison of this and the previous study conducted in 2011 confirmed the results obtained for lettuce which therefore shows the importance of repeating field experiments for validation and reliable assessment of biocontrol agents. Furthermore, also a comparison of the impact of a biocontrol agent between economically important model plants is needed. Therefore, we used TC-DNA extracted from the same lettuce root samples for 2012 as used in Schreiter et al. (2014b), performed a separated amplification, DGGE run and analysis to compare to potato. In contrast to lettuce, on average a minor influence of RU47 on the total bacterial as well as Pseudomonas communities was revealed in the potato rhizosphere and might be explained by differences in the response of the plant-specific microbial community to RU47 inoculation. Recent results of Schreiter et al. (2014b) on lettuce in the 2011 field experiment indicated that different taxonomic groups responded to RU47 application depending on the soil type. An increased relative abundance of bacteria belonging to the genera Bacillus and Paenibacillus was observed in the rhizosphere of lettuce grown in alluvial loam. Thus, mutualistic effects of the inoculant with distinct enriched populations or indirect effects involving other bacterial rhizosphere populations cannot be excluded. Numerous strains of these genera were previously reported to display antagonistic activity and may support the observed biocontrol effect of RU47 (Kröber et al., 2014). The observed effect on rhizosphere bacterial communities in potato of the treatment RU47 in presence of the pathogen (RU47+Rs) seems to be caused by R. solani AG3. The strong effect of the pathogen on bacterial and Pseudomonas communities could not be compensated by the inoculant RU47.
Conclusion
The results underline that the bacterial community structure in the rhizosphere differed depending on the plant species in all soils analyzed. Despite the influence of the plant, the soil type, and the pathogen, the inoculant RU47 was capable of colonizing the rhizosphere of both crops in a density sufficient manner to reduce black scurf disease severity on potato and bottom rot on lettuce in all three soils. In both crops the rhizosphere competence of RU47 was neither affected by the soil type nor by the presence of the respective pathogen R. solani. In contrast to lettuce, the colonization of the belowground part of potato including the roots by the pathogen R. solani AG3 has had a pronounced effect on the bacterial community composition in the potato rhizosphere. Interestingly, the pathogen R. solani affected the Pseudomonas community in both crops whereas the inoculant RU47 influenced especially the bacterial as well as the Pseudomonas community in the rhizosphere of lettuce. The plant species and soil type independent rhizosphere competence of RU47 and biocontrol of Rhizoctonia diseases observed under field conditions underlines the biocontrol potential of RU47.
Author Contributions
KS and RG preparing the project proposal for third-party funds. KS, RG, and SS project planning, design and performance of experiments. SS and RG determination of rhizosphere competence and plant characteristics, disease assessment, analysis of these data. SS and DB molecular work and analysis of microbial community composition in the rhizosphere. SS, RG, KS, and DB preparation of the manuscript.
Funding
The authors acknowledge that DFG SM59/11-1/GR568121 funded the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Petra Zocher, Ute Zimmerling, Sabine Breitkopf, and Angelika Fandrey for their skilled technical assistance and Ilse-Marie Jungkurth for her helpful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00097/full#supplementary-material
References
Adesina, M. F., Grosch, R., Lembke, A., Vatchev, T. D., and Smalla, K. (2009). In vitro antagonists of Rhizoctonia solani tested on lettuce: rhizosphere competence, biocontrol efficiency and rhizosphere microbial community response. FEMS Microbiol. Ecol. 69, 67–74. doi: 10.1111/j.1574-6941.2009.00685.x
Alabouvette, C., Olivain, C., Migheli, Q., and Steinberg, C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184, 529–544. doi: 10.1111/j.1469-8137.2009.03014.x
Atkinson, D., Thornton, M. K., and Miller, J. S. (2011). Development of Rhizoctonia solani on stems, stolons and tubers of potato II. Efficacy of chemical applications. Am. J. Potato Res. 88, 96–103. doi: 10.1007/s12230-010-9172-1
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Barret, M., Morrissey, J. P., and O’Gara, F. (2011). Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol. Fertil. Soils 47, 729–743. doi: 10.1007/s00374-011-0605-x
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bulgarelli, D., Rott, M., Schläppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Chapparo, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chowdhury, S. P., Dietel, K., Rändler, M., Schmid, M., Junge, H., Borriss, R., et al. (2013). Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLOS ONE 8:e68818. doi: 10.1371/journal.pone.0068818
Costa, R., Gomes, N. C. M., Krögerrecklenfort, E., Opelt, K., Berg, G., and Smalla, K. (2007). Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ. Microbiol. 9, 2260–2273. doi: 10.1111/j.1462-2920.2007.01340.x
Costa, R., Götz, M., Mrotzek, N., Lottmann, J., Berg, G., and Smalla, K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56, 236–249. doi: 10.1111/j.1574-6941.2006.00026.x
De Bellis, P., and Ercolani, G. L. (2001). Growth interactions during bacterial colonization of seedling rootlets. Appl. Environ. Microbiol. 67, 1945–1948. doi: 10.1128/AEM.67.4.1945-1948.2001
De Souza, J. T., Mazzola, M., and Raaijmakers, J. M. (2003). Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 5, 1328–1340. doi: 10.1111/j.1462-2920.2003.00438.x
DeAngelis, K. M., Brodie, E. L., DeSantis, T. Z., Andersen, G. L., Lindow, S. E., and Firestone, M. K. (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178. doi: 10.1038/ismej.2008.103
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. doi: 10.1111/j.1574-6941.2010.00860.x
Doornbos, R., van Loon, C. L., and Bakker, P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32, 227–243. doi: 10.1007/s13593-011-0028-y
Genzel, F., Franken, P., Witzel, K., and Grosch, R. (2018). Systemic induction of salicylic acid-related plant defences in potato in response to Rhizoctonia solani AG3PT. Plant Pathol. 67, 337–348. doi: 10.1111/ppa.12746
Ghirardi, S., Dessaint, F., Mazurier, S., Corberand, T., Raaijmakers, J. M., Meyer, J. M., et al. (2012). Identification of traits shared by rhizosphere-competent strains of fluorescent pseudomonads. Microb. Ecol. 64, 725–737. doi: 10.1007/s00248-012-0065-3
Grosch, R., Faltin, F., Lottmann, J., Kofoet, A., and Berg, G. (2005). Effectiveness of 3 antagonistic bacterial isolates to control Rhizoctonia solani Kühn on lettuce and potato. Can. J. Microbiol. 51, 345–353. doi: 10.1139/w05-002
Grosch, R., Schneider, J. H. M., Kofoet, A., and Feller, C. (2011). Impact of continuous cropping of lettuce on the disease dynamics of bottom rot and genotypic diversity of Rhizoctonia solani AG 1-IB. J. Phytopathol. 159, 35–44. doi: 10.1111/j.1439-0434.2010.01708.x
Grosch, R., Schneider, J. H. M., Peth, A., Waschke, A., Kofoet, A., Franken, P., et al. (2007). Development of a specific PCR assay for the detection of Rhizoctonia solani AG 1-IB using SCAR primers. J. Appl. Microbiol. 102, 806–819. doi: 10.1111/j.1365-2672.2006.03125.x
Haas, D., and Defago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Heuer, H., Krsek, M., Baker, P., Smalla, K., and Wellington, E. M. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63, 3233–3241.
Kröber, M., Wibberg, D., Grosch, R., Eikmeyer, F., Verwaaijen, B., Chowdhury, S. P., et al. (2014). Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol. 5:252. doi: 10.3389/fmicb.2014.00252
Kropf, S., Heuer, H., Grüning, M., and Smalla, K. (2004). Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57, 187–195. doi: 10.1016/j.mimet.2004.01.002
Kumar, R., Pandey, S., and Pandey, A. (2006). Plant root and carbon sequestration. Curr. Sci. 91, 885–890.
Kuninaga, S., Carling, D. E., Takeuchi, T., and Yokosawa, R. (2000). Comparison of rDNA-ITS sequences between potato and tobacco strains in Rhizoctonia solani AG3. J. Gen. Plant Pathol. 66, 2–11. doi: 10.1007/PL00012917
Micallef, S. A., Channer, S., Shiaris, M. P., and Colón-Carmona, A. (2009). Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 4, 777–780. doi: 10.1093/jxb/erp053
Nerey, Y., Van Beneden, S., Franca, S. C., Jimenez, A., Cupull, R., Herrera, L., et al. (2010). Influence of soil type and indigenous pathogenic fungi on bean hypocotyl rot caused by Rhizoctonia solani AG4 HGI in Cuba. Soil Biol. Biochem. 42, 797–803. doi: 10.1016/j.soilbio.2010.01.015
Nübel, U., Engelen, B., Felske, A., Snaidr, J., Wieshuber, A., Amann, R. I., et al. (1996). Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178, 5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Rühlmann, J., and Ruppel, S. (2005). Effects of organic amendments on soil carbon content and microbial biomass - results of the long-term box plot experiment in Grossbeeren. Arch. Agron. Soil Sci. 51, 163–170. doi: 10.1080/03650340400026651
Schneider, J. H. M., Schilder, M. T., and Dijst, G. (1997). Characterization of Rhizoctonia solani AG 2 isolates causing bare patch in field grown tulips in the Netherlands. Eur. J. Plant Pathol. 103, 265–279. doi: 10.1023/A:1008643311984
Schreiter, S., Ding, G.-C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014a). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Schreiter, S., Ding, G.-C., Grosch, R., Kropf, S., Antweiler, K., and Smalla, K. (2014b). Soil type dependent effects of a potential biocontrol inoculant on indigenous bacterial communities in the rhizosphere of field-grown lettuce. FEMS Microbiol. Ecol. 90, 718–730. doi: 10.1111/1574-6941.12430
Schreiter, S., Sandmann, M., Smalla, K., and Grosch, R. (2014c). Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLOS ONE 9:e103726. doi: 10.1371/journal.pone.0103726
Tamm, L., Thürig, B., Bruns, C., Fuchs, J. G., Köpke, U., Laustela, M., et al. (2010). Soil type, management history, and soil amendments influence the development of soil-borne (Rhizoctonia solani, Pythium ultimum) and air-borne (Phytophthora infestans, Hyaloperonospora parasitica) diseases. Eur. J. Plant Pathol. 127, 465–481. doi: 10.1007/s10658-010-9612-2
van der Heijden, M. G. A., and Hartmann, M. (2016). Networking in the Plant Microbiome. PLOS Biol. 14:e1002378. doi: 10.1371/journal.pbio.1002378
Weinert, N., Meincke, R., Gottwald, C., Heuer, H., Gomes, N. C. M., Schloter, M., et al. (2009). Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl. Environ. Microbiol. 75, 3859–3865. doi: 10.1128/AEM.00414-09
Wibberg, D., Jelonek, L., Rupp, O., Hennig, M., Eikmeyer, F., Goesmann, A., et al. (2013). Establishment and interpretation of the genome sequence of the phytopathogenic fungus Rhizoctonia solani AG1-IB isolate 7/3/14. J. Biotechnol. 167, 142–155. doi: 10.1016/j.jbiotec.2012.12.010
Windisch, S., Bott, S., Ohler, M. A., Mock, H.-P., Lippmann, R., Grosch, R., et al. (2017). Rhizoctonia solani and bacterial inoculants stimulate root exudation of antifungal compounds in lettuce in a soil-type specific manner. Agronomy 7:44. doi: 10.3390/agronomy7020044
Keywords: Rhizoctonia solani, plant health, biocontrol, plant disease, bacterial community, DGGE
Citation: Schreiter S, Babin D, Smalla K and Grosch R (2018) Rhizosphere Competence and Biocontrol Effect of Pseudomonas sp. RU47 Independent from Plant Species and Soil Type at the Field Scale. Front. Microbiol. 9:97. doi: 10.3389/fmicb.2018.00097
Received: 24 October 2017; Accepted: 16 January 2018;
Published: 01 February 2018.
Edited by:
Corné M. J. Pieterse, Utrecht University, NetherlandsReviewed by:
Monika Maurhofer, ETH Zurich, SwitzerlandDavid Dowling, Institute of Technology Carlow, Ireland
Copyright © 2018 Schreiter, Babin, Smalla and Grosch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rita Grosch, grosch@igzev.de
† Present address: Susanne Schreiter, Department of Sustainable Agriculture Science, Rothamsted Research, Harpenden, United Kingdom
 Susanne Schreiter
Susanne Schreiter Doreen Babin
Doreen Babin Kornelia Smalla
Kornelia Smalla Rita Grosch
Rita Grosch