- 1Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
- 2Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Wuhan, China
- 3Oil Crops and Lipids Process Technology National and Local Joint Engineering Laboratory, Wuhan, China
- 4Hubei Key Laboratory of Lipid Chemistry and Nutrition, Wuhan, China
- 5Agriculture and Food, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Canberra, ACT, Australia
Medium chain hydroxy fatty acids (HFAs) at ω-1, 2, or 3 positions (ω-1/2/3) are rare in nature but are attractive due to their potential applications in industry. They can be metabolically engineered in Escherichia coli, however, the current yield is low. In this study, metabolic engineering with P450BM3 monooxygenase was applied to regulate both the chain length and sub-terminal position of HFA products in E. coli, leading to increased yield. Five acyl-acyl carrier protein thioesterases from plants and bacteria were first evaluated for regulating the chain length of fatty acids. Co-expression of the selected thioesterase gene CcFatB1 with a fatty acid metabolism regulator fadR and monooxygenase P450BM3 boosted the production of HFAs especially ω-3-OH-C14:1, in both the wild type and fadD deficient strain. Supplementing renewable glycerol to reduce the usage of glucose as a carbon source further increased the HFAs production to 144 mg/L, representing the highest titer of such HFAs obtained in E. coli under the comparable conditions. This study illustrated an improved metabolic strategy for medium chain ω-1/2/3 HFAs production in E. coli. In addition, the produced HFAs were mostly secreted into culture media, which eased its recovery.
Introduction
Hydroxy fatty acids (HFAs) are widely found in plants, animals, and microorganisms, although rare in medium-chain length (C8-C14). Their physiological roles, including anti-biotic, anti-inflammatory, and anti-diabetic effects, have been uncovered recently. In particular, the position of hydroxyl groups in the fatty acyl chain shows a key effect on the activity against certain plant pathogenic fungi. Branched chain HFAs generated by gut microbes are demonstrated to affect human health (Hou, 2009; Kishino et al., 2013; Sakurama et al., 2014; Yore et al., 2014). Compared to non-hydroxylated fatty acids, HFAs have higher reactivity, solvent miscibility, stability, and viscosity. Therefore, HFAs have been widely used in food, chemical, pharmaceutical, cosmetic industries, and as synthetic precursors (Yang et al., 2014; Meesapyodsuk et al., 2015; Bowen et al., 2016). Omega HFAs (ω-HFAs) are fatty acids with a hydroxyl group near the methyl end, or omega end. The hydroxylation at different positions of the fatty acid chain is needed for different chemical industries. For example, ω-HFAs can be used as renewable starting materials for the synthesis of polymers, with high resistance to heat or chemicals, flexibility, biocompatibility, and non-toxicity (Seo et al., 2015; Bowen et al., 2016). Meanwhile, the geometry of sub-terminal ω-1/2/3 HFA combined with different chain length provides diversity of characteristics for polymer synthesis from HFA, generating potential for wide range of commercial use.
Despite the wide applications of HFAs, direct synthesis of desired specific hydroxylation is limited and requires harsh reaction conditions. Enzymatic hydroxylation is an alternative approach due to its specificity for the positions to be hydroxylated. In general, four types of enzymes are involved in the production of diverse HFAs in nature, including cytochrome P450 monooxygenase, hydratase, hydroxylase, and lipoxygenase. P450 monooxygenases are classified into α-hydroxylase (carboxyl-terminal hydroxylase) and ω-hydroxylase (terminal or sub-terminal hydroxylase). Several P450 monooxygenases have been recently characterized with distinct substrate specificities from plant, yeast, and bacterium. Hydratases catalyze the hydration of a specific double bond. They are only found in microbes including Stenotrophnomonas, Lactobacillus, Pediococcus, Selenomonas, and Enterococcus (Hirata et al., 2015; Takeuchi et al., 2015, 2016). Hydroxylases introduce a hydroxyl group at single bond of fatty acid. Δ12-hydroxylases, for example, have been identified from plants Ricinus communis, Lesquerella fendleri, Lesquerella lindheimeri, or Hiptage benghalensis, and fungus Claviceps purpurea (Mavraganis et al., 2010; Zhou et al., 2013; Kim and Chen, 2015; Lee et al., 2015), which introduce a hydroxyl group at Δ12 position of oleic acid. Lipoxygenases use polyunsaturated fatty acids as substrates to form corresponding HFAs (Kim and Oh, 2013). Among those P450 monooxygenases, P450BM3 from Bacillus megaterium is the first one that has been characterized as a self-sufficient enzyme. It transfers electrons from NADPH during its oxidation as it contains a flavin reductase domain as a redox-partner. Such self-sufficient P450 enzymes are suitable as efficient and yet simple catalytic system due to their independence of an additional reductase enzyme and high turnover numbers (Narhi and Fulco, 1986). P450BM3 has broad substrate specificity, in addition to fatty acids, hydroxylating alkanols and alkylamides at sub-terminal positions as well. Purified P450BM3 hydroxylates fatty acids with chain length ranging from C12 to C18, and exhibits the highest specificity for C14:0, followed by C15:0 and C12:0, whereas, C16:0, C17:0, and C18:0 are used with the least efficiency (Schneider et al., 1998; Hilker et al., 2008). Three amino acids in P450BM3, R47, Y51, and F87, are crucial for substrate binding (Li and Poulos, 1999). Unlike fungal fatty acid hydroxylases, only sub-terminal positions are hydroxylated by the action of P450BM3.
Although considerable effort has been made in engineering of oil crops or microbes for producing HFAs, there has been limited success in the production of medium chain fatty acid (MCFA) sub-terminal HFAs (Meesapyodsuk et al., 2015; Bowen et al., 2016). Different strategies have been applied to increase MCFA sub-terminal or terminal HFAs production. Examples include overexpression of acyl-acyl carrier protein (acyl-ACP) thioesterase (or TE) to provide more MCFAs as substrates in E. coli, reducing fatty acid turnover, overexpression of NAD+ dependent formate dehydrogenase to produce cofactor NADH as well as optimization of fermentation process (Wang et al., 2012; Sung et al., 2015; Cao et al., 2016).
Since E. coli uses a Type II fatty acid biosynthesis, targeted chemicals of various chain lengths including MCFAs can be synthesized. In general, chain length is determined by 3-ketoacyl-ACP synthases and a variety of acyltransferases in E. coli. However, several plant TEs are able to regulate the chain length of E. coli fatty acid biosynthesis end products by hydrolyzing the acyl-ACP intermediates and releasing FFA. Of them, a few plant TEs (designated as FatBs) show preference for medium chain acyl-ACPs (Reynolds et al., 2015). Previous studies have demonstrated that the overexpression of MCFA specific TE such as bacterial TesA' or plant Umbellularia californica FatB2 (UcFatB2) alleviates the feedback inhibition caused by acyl-ACPs and yields MCFAs (Wang et al., 2012; Cao et al., 2016). Compared to CnFatB1, CnFatB2 from Cocos nucifera and UcFatB2, the expression of CnFatB3 in Nicotiana benthamiana results in the highest MCFA (Yuan et al., 1995; Reynolds et al., 2015). However, CnFatB3 was not evaluated in E. coli in those studies. CcFatB1 from Cinnamomum camphora shows high specificity mainly for C14:0 in E. coli and N. benthamiana leaves (Yuan et al., 1995; Reynolds et al., 2015). CcFatB1 is also used for production of MCFA ethyl esters (Fan et al., 2013). CpFatB1 from Cuphea palustris with preference for C8 has been expressed in yeast for synthesis of short chain fatty acids, while CpFatB2 exhibits the highest specificity for C14:0 (24 mol%) when expressed in Camelina seeds (Kim and Chen, 2015). Bacterial LpFat from Lachnoclostridium phytofermentans shows preference for both C12 and C14 substrates, while CtFat from Clostridium tetani utilizes mainly C14 as substrate (Nawabi et al., 2011). Therefore, application of MCFA-specific TEs is crucial for the production of desired MCFAs and their derived HFAs.
In this study, three plant TE genes, CnFatB3, CcFatB1, and CpFatB2 from C. nucifera C. camphora, and C. palustris, respectively, as well as the two bacterial TE genes, CtFat and LpFat, were first assessed in E. coli. The selected TE-encoding gene was then coexpressed with P450BM3 to produce medium chain HFAs in E. coli. Next, the effect of a fatty acid metabolism regulator fadR and an acyl-CoA synthase encoding gene fadD on HFA production was investigated. In addition, we also tested if supplementation of renewable glycerol could further improve the biomass as well as the yield of medium chain HFAs.
Materials and Methods
Materials
Restriction enzymes, polymerase, and ligase were purchased from New England Biolabs (Beverly, USA). The plasmid miniprep kit, total DNA extraction kit, PCR purification kit, and gel extraction kit were purchased from TianGen (Beijing, China). Primers and genes were synthesized by TsingKe (Beijing, China). Fatty acid standards were purchased from Larodan (Stockholm, Sweden). Chemical reagents were purchased from Sigma-Aldrich (St. Louis, USA) or Sinopharm (Shanghai, China). His-tag and S-tag antibodies were purchased from Proteintech (Wuhan, China).
Microbial Strains and Plasmids
Bacterial strains and plasmids were listed in Table S1. E. coli DH5α was used for plasmid construction and propagation. E. coli BL21(DE3), E. coli Rossetta(DE3), or E. coli BL21ΔfadD were used for FFA and HFA production. Primers used for plasmid construction are summarized in Table S2.
Plant TE genes CnFatB3, CcFatB1, CpFatB2, and bacterial TE genes CtFat, LpFat, based on GenBank accession numbers JF338905, U31813, U38189, CTC_RS00430, and ABX40638 respectively, were chemically synthesized with codon optimized for E. coli. The gene P450BM3 (GenBank accession AAA87602) from B. megaterium ATCC 14581 responsible for the ω-HFA synthesis was chemically synthesized.
For overexpression of individual TE genes, each gene was inserted into the EcoRI/XhoI double-digested pET28a(+). For co-expression of TE gene and P450BM3, P450BM3 was inserted into EcoRI/NotI sites and the TE gene was inserted into BglII/XhoI sites of the pCDFDuet-1 or pACYCDuet-1, respectively.
Fatty acid metabolism regulator FadR (NP_415705) from E. coli MG 1655, and the putative fatty acid metabolism regulators predicted based on Rhodococcus opacus PD630 genome sequence namely RoTetR1 (AHK36109), RoTetR2 (AHK34097), and RoTetR3 (AHK34039), were used in this study. The gene fadR was amplified with primers fadR-F and fadR-R (Table S2). The PCR product was inserted into the BamHI and HindIII sites of pET28a(+), resulting in plasmid pEF. R. opacus RoTetR1/2/3 were chemically synthesized, and inserted into the BamHI and HindIII sites of pET28a(+), resulting in plasmid pER1, pER2, or pER3, respectively.
Detection of Recombinant Proteins by SDS-PAGE and Western Blot Analysis
E. coli cells were cultivated at 37°C in Luria-Bertani medium (LB medium, 5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl) supplemented with corresponding antibiotics until their OD600 reached 0.6. Then, cells were cultured at 30°C for 4 h after addition of 1 mM isopropyl ß-D-thiogalactoside (IPTG). Cells were subsequently collected from 1.5 mL of bacterial cultures by centrifugation and re-suspended in sodium dodecyl sulfate (SDS) sample buffer. Proteins were heated in boiling water for 5 min. The resulting supernatant was analyzed by SDS-polyacrylamide gel electrophoresis and Western blot.
For Western blot analysis, protein samples were separated on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane. After blocking non-specific binding using 5% skim milk for 1 h, the blots were incubated with anti His-tag monoclonal antibody or anti S-tag antibody at 4°C overnight. Membranes were washed three times with TTBS buffer (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.6) and incubated with an appropriate secondary antibodies conjugated to horseradish peroxidase for 1 h at room temperature. After being washed three times with TTBS, direct chemiluminescence imaging of the blots was performed using the ChemiDox XRS (BioRad) imaging system.
Culture Conditions for Fatty Acid Production
For HFA production, cells were cultivated in flasks in M9 medium (15.13 g/L Na2HPO4·12H2O, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 2 mM MgSO4) containing 2% (w/w) glucose at 37°C with shaking at 200 rpm. Corresponding antibiotics were added when necessary. When the optical density of the cells reached 0.6 at 600 nm, IPTG was added to a final concentration of 1 mM. The cultures were incubated at 30°C with shaking at 150 rpm for further 16 h. Glycerol at final concentrations of 0, 0.5, 1, 2, 3, 4, and 5% were added into M9 medium with 2% glucose before culturing when tested. Further optimization of glucose concentration was also performed at final concentrations of 0, 0.5, 1, 1.5, and 2% in M9 medium containing 2% glycerol.
Extraction of Fatty Acid from Cell Lysate and Supernatant
The cell pellet and the supernatant of the cultures were collected separately. The cells were freeze-dried overnight while the supernatants were used to extract fatty acids directly. Before extraction, C21:0-FFA (25 μg) and 3-OH-C16:0-FFA (25 μg) were added to the supernatant as internal standards, respectively. Four milliliters of supernatants were first acidified with acetic acid. The total lipids were extracted with 2 mL of ethyl acetate by vigorous vortexing for 30 min. The solvent in the collected ethyl acetate layer was blown off with nitrogen and the lipids were methylated with 1 M HCl/CH3OH at 80°C for 3 h to generate fatty acid methyl esters (FAMEs). The FAMEs were then extracted with hexane. The dry cell pellets were methylated using the same methods to generate FAMEs. For detection of HFAs, the FAMEs were further converted into their trimethylsilyl (TMS) derivatives by incubating at 90°C for 1 h, with an excess of N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA).
Fatty Acid Analysis
The FAMEs were analyzed using a 7890A gas chromatography (GC, Agilent technologies, USA) equipped with a flame ionization detector (FID) and an HP-FFAP column (30 m × 250 μm i.d. × 0.25 μm thickness). The oven temperature was initially set at 150°C for 1 min, then increased by 5°C/min to 230°C, holding at that temperature for 5 min. The temperature of the inlet and detector were 260 and 280°C, respectively. Structure of ω-HFAs was further confirmed by GC-mass spectrometry (GC-MS) analysis using Agilent 5975C with Triple-Axis detector. Two microliters of the sample were injected by split mode (split ratio 20:1) and analyzed using a nonpolar capillary HP-5 column (5% phenyl methyl siloxane capillary 30 m × 320 μm i.d. × 0.25 μm thickness). The oven temperature was set at 50°C for 1 min, and then increased by 15°C/min to 250°C, holding at this temperature for 10 min. The temperature of the inlet was 250°C, while the temperature of the detector was 280°C.
Lipid Analysis
Total lipids were extracted from dry cell pellets (20 mg) with chloroform-methanol (2:1, v/v) (Rottig et al., 2015). Prior to lipid extraction, 50 μg of internal standard C21:0-FFA was added into each sample. The lower organic phase was extracted, evaporated, and resolved. For thin layer chromatography (TLC), a certain amount of each sample was loaded on silica gel plates (silica gel 60, 20 × 20 cm, EMD Chemicals, Germany) with a solvent system containing hexane-diethyl ether-acetic acid (70:30:1, v/v). Lipid classes were visualized by spraying with 0.01% primuline (Sigma-Aldrich) in 80% acetone. The spots corresponding to FFA standard were scraped into glass vials and methylated as above. The resulting FAMEs were analyzed by GC as above.
Results and Discussion
Assessment of Plant and Bacterial TEs for MCFA Production in E. coli
For selective production of medium chain ω-1/2/3 HFAs, several steps involved in the biosynthesis pathway were tested in this study (Figure 1). The first step is to release MCFA as substrate for P450BM3 monooxygenase. Three plant TE genes, CnFatB3, CcFatB1, and CpFatB2, and two bacterial TE genes, CtFat and LpFat, were chosen to be expressed in E. coli BL21(DE3) individually. The neutral lipid fraction of total lipids from both cell pellet and supernatant were first separated by TLC. Only FFA and a small amount of diacylglycerol (DAG) were observed from cell neutral lipids after staining by primuline (Figure 2A), while in the supernatant there was only a FFA band observed (data not shown). Expressing TEs increased the cellular FFA amount, although there was no significant effect on the biomass and total fatty acid (TFA) amounts (Table 1). The amounts of MCFA including C12 and C14 had a slight increase in cellular FFA, but significantly increased in extracellular FFA, by expressing these TEs, compared to BL21(DE3) with empty vector BE (Table 1). The cellular content of C16:0 decreased while C14:0 and C14:1 increased, indicating that the expression of these TEs enhanced the release of MCFAs (Figures 2B,C, left panel, detailed profile shown in Tables S3, S4). Furthermore, CcFatB1 and CpFatB2 mainly released C14 FFA, while CnFatB3 released both C12 and C14 FFAs.
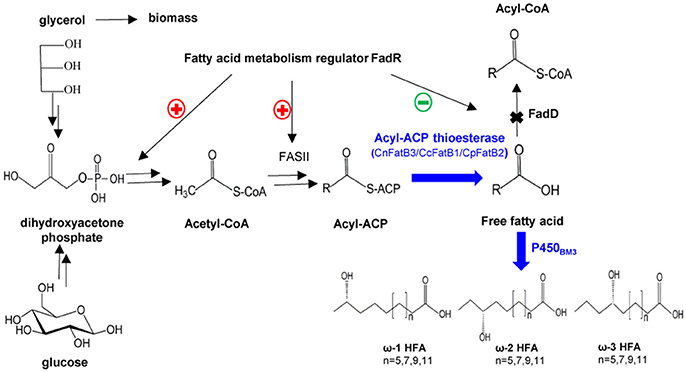
Figure 1. A schematic representation of the engineered pathway for production of medium chain ω-1/2/3 HFAs in E. coli. CnFatB3, CcFatB1, and CpFatB2 represent acyl-ACP thioesterases from Cocos nucifera, Cinnamomum camphora, and Cuphea palustris, respectively. FadD, acyl-CoA synthase from E. coli. P450BM3, P450 monooxygenase from Bacillus megaterium.
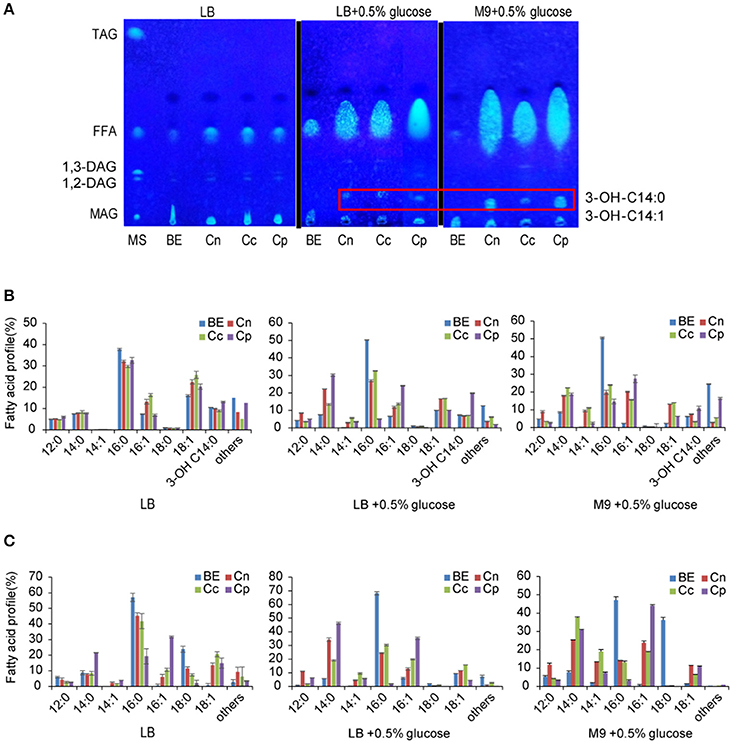
Figure 2. Overexpression of individual acyl-ACP thioesterase gene in E. coli. (A) Separation of neutral lipid from recombinant E. coli cell lysates by thin layer chromatography (TLC). (B) Fatty acid profile of cellular TFA of recombinant E. coli, trace amount of 3-OH-C14:1 was included in others. (C) Fatty acid profile of cellular FFA of recombinant E. coli. Trace amount of 3-OH-C14:0 and 3-OH-C14:1 were included in others. Cells were cultured in LB medium, LB medium plus 0.5% glucose, or M9 medium plus 0.5% glucose by IPTG induction for 24 h. MS, mixed standards for TLC containing 18:1/18:1/18:1-TAG, 18:1-FFA, 18:1/18:1-DAG, and 18:1-MAG; BE, BL21(DE3) with empty vector; Cn, BL21(DE3) harboring CnFatB3 in pECn; Cc, BL21(DE3) haroboring CcFatB1 in pECc; Cp, BL21(DE3) harboring CpFatB2 in pECp. All data are the mean ± standard deviation from triplicate cultures.
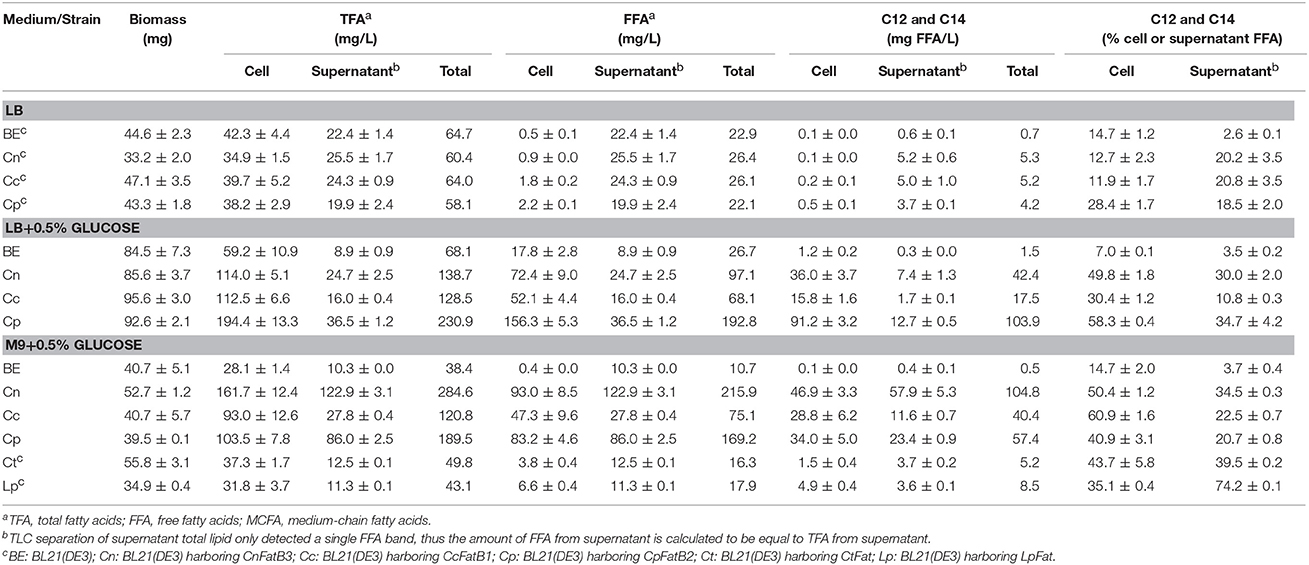
Table 1. A survey of acyl-ACP thioesterase activity on the production of TFA, FFA, and MCFA in recombinant E. coli.
Next, cell culture was supplemented with glucose in media. Supplementing 0.5% of glucose in LB medium significantly enhanced both biomass and TFA production compared to without glucose supplementation (Table 1). Importantly, all the recombinant cells expressing TEs had significantly higher TFA or FFA compared to BL21(DE3) expressing empty vector BE (Table 1), especially a huge increase in FFA (Figure 2A). Of them, strain Cp (E. coli harboring CpFatB2) yielded the highest TFA (194.4 mg/L), FFA (156.3 mg/L), and MCFA (91.2 mg/L) in the cell. Supplementing the same amount of glucose in M9 medium also increased the TFA but did not affect the biomass (Table 1). The TFA in the supernatant (appearing as FFA) were also increased in the presence of glucose except for the bacterial TEs. The ratio of FFA to TFA was about 60, 50, and 80% by Cn (E. coli harboring CnFatB3), Cc (E. coli harboring CcFatB1), and Cp, respectively, in both LB and M9 media with glucose (Table 1). Glucose could be converted into acetyl-CoA in E. coli, thus used for fatty acid synthesis. Indeed, TFA was enhanced in both LB and M9 media with supplementation of glucose. The increased biomass in LB medium but not M9 medium in the presence of glucose was probably due to the insufficient nitrogen in M9 medium.
HFAs in the form of FFA from cell lysate were also detected and later confirmed as the mixture of 3-OH-C14:0 and a trace amount of 3-OH-C14:1 by GC-MS (Figure S1). The 3-OH-C14:0 and 3-OH-C14:1 in FFA were visible on TLC plate (Figure 2A). It has been reported that the expression of TEs from Clostridium acetobutylicum and C. phytofermentans leads to a significant production of 3-OH-FFA with chain length ranging from C10 to C14 (Nawabi et al., 2011). However, the authors did not observe the same phenomenon when Arabidopsis thaliana TE (AtFatA) was overexpressed in E. coli, and thus it was concluded that plant TE could not lead to the production of 3-OH-FFA (Nawabi et al., 2011). In this study, the TLC result clearly demonstrated that plant TEs, CnFatB3, CcFatB1, and CpFatB2, led to the formation of 3-OH-FFA in E. coli as well. The native enzyme responsible for the formation of such HFA in E. coli has not been resolved yet.
Reduced C16:0 and increased C14:0, C14:1, or C16:1 caused by expressing plant TE was further enhanced by supplementation of glucose. C16:0 proportion in cellular TFA was sharply decreased from about 50% in the cells carrying empty vector (BE) to less than 20% in all the recombinant cells when supplied with glucose in either LB or M9 medium (Figure 2B). A similar effect was also observed in cellular FFA (Figure 2C). The most dramatic change was found with C16:0 in FFA, decreased from 68% in BE to 2% in the cells expressing Cp, while C14:0 and C14:1 increased from about 6% in BE to 50% in Cp in LB media with glucose (Figure 2C). Glucose could be converted to acetyl-CoA via glycolysis in E. coli, and subsequently condensed to yield acyl-ACP through the FASII system, leading to the enhanced FA synthesis.
CnFatB3 produced the highest TFA (284.6 mg/L), FFA (215.9 mg/L), and MCFA (104.8 mg/L) when cultured in M9 plus 0.5% of glucose, whereas CpFatB2 yielded the highest TFA (230.9 mg/L), FFA (192.8 mg/L), and MCFA (103.9 mg/L) in LB plus 0.5% of glucose. Unfortunately, two bacterial TEs generated much less FFA and MCFA contents than those released by plant TEs, although the proportion of C12 and C14 to the FFA was elevated as well (Table 1). Thus three plant TEs were selected for subsequent experiments.
Sequence Comparison of MCFA-specific TEs
The primary amino acid sequences and tertiary structures of these TEs were also compared. Amino acid sequence alignment showed that CnFatB3, CcFatB1, and CpFatB2 shared 74.2, 74.4, and 42.0% identities to UcFatB2, respectively. All these four TE proteins had the conserved residues NXHX34C as indicated in Figure S2A. Only CnFatB3 and CcFatB1 had the putative plastid-targeting peptide cleavage site (LPDW) which was located at the N-terminus (Figure S2A), as predicted for many other plant TEs. N-terminal domain is responsible for the enzyme specificity of plant TEs. Although the sequences of two bacterial TEs were much shorter than those of plant TEs, they also had conserved NXH structure. This might partly explain the much lower TFA, FFA, and MCFA contents obtained from bacterial TEs compared with those from plant TEs. No transmembrane structure was predicted among all these enzymes by the TMHMN tool (http://www.cbs.dtu.dk/services/TMHMM/). 3D models of CnFatB3, CcFatB1, and CpFatB2 were achieved through online software (http://zhanglab.ccmb.med.umich.edu/I-TASSER/, Figure S2B). The C-score of modeled CnFatB3, CcFatB1, and CpFatB2 were −2.75, −2.04, and −2.52, respectively, which were relatively low. They were all modeled based on the structure of oleoyl TE from Lactobacillus plantarum (PDB file: 2OWN), which shows preference for C6 and C8 acyl-ACP substrates. This is partly due to the lack of tertiary structural information on other TEs, especially medium chain acyl-ACP TEs. Nevertheless, a hot-dog fold pattern was found in CnFatB3, CcFatB1, and CpFatB2, which was described as the active site of the structure-resolved acyl-ACP TEs (PDB file: 2OWN and 2ESS). In addition, a few catalytic residues, for example Thr124 and Val183 in CcFatB1, Gly150, and Tyr234 in CpFatB2, were predicted. The above analysis of tested TEs might guide further improvement or application of those enzymes although the existing information on tertiary structures was limited.
Increased ω-1/2/3 Medium Chain HFA Content in E. coli by Co-expression of P450BM3 and Plant Acyl-ACP Thioesterase Gene
The sub-terminal carbons of released FFAs were then hydroxylated by a self-sufficient and region-selective P450BM3 to produce ω-1/2/3 HFAs. Such HFAs are valuable and more stable than terminal ω-HFAs. We first constructed the strains CPCn (E. coli harboring P450BM3 and CnFatB3), CPCc (E. coli harboring P450BM3 and CcFatB1), and CPCp (E. coli harboring P450BM3 and CpFatB2), to co-express P450BM3 and a single plant TE gene. Western blot analysis confirmed the expression of both genes in each recombinant strain (Figure S3). E. coli cells expressing P450BM3 alone (EP) did not produce detectable ω-1/2/3 HFAs (Figure 3). When coupled P450BM3 with different plant TE genes, the recombinant strains produced significant amounts of HFAs as shown in Figure 3. HFAs were comprised of 5.9, 68.1, and 79.4% of total FFA in the supernatant of CPCn, CPCc, and CPCp, respectively (Figure 4A). Among those combinations, strain CPCc yielded the highest amount of HFAs, which was up to 38.1 mg/L in the medium (Figures 3B,C). Unlike the previous results of single TE expression without P450BM3, only a small increase of MCFAs from the cell lysate was detected in CPCn (6.3 mg/L), CPCc (7.2 mg/L), and CPCp (7.4 mg/L), compared to 5.7 mg/L in control EP (Figure 3). This suggested that the MCFAs could be further converted into corresponding HFAs by P450BM3 monooxygenase. To our surprise, CPCn could only produce up to 1 mg/L HFAs. Neither BL21(DE3) nor BL21 Rosseta(DE3) harboring pCPCn produced HFA higher than 1 mg/L in the medium (Figure 3C and data not shown). In addition, FFA content from the CPCn cell lysate dramatically decreased, even to the level of the control cells. However, Western blot results confirmed the proteins were expressed although the protein levels were low (Figure S3). The combination of CnFatB3 and P450BM3 might negatively affect the fatty acid metabolism in a mechanism yet to be understood.
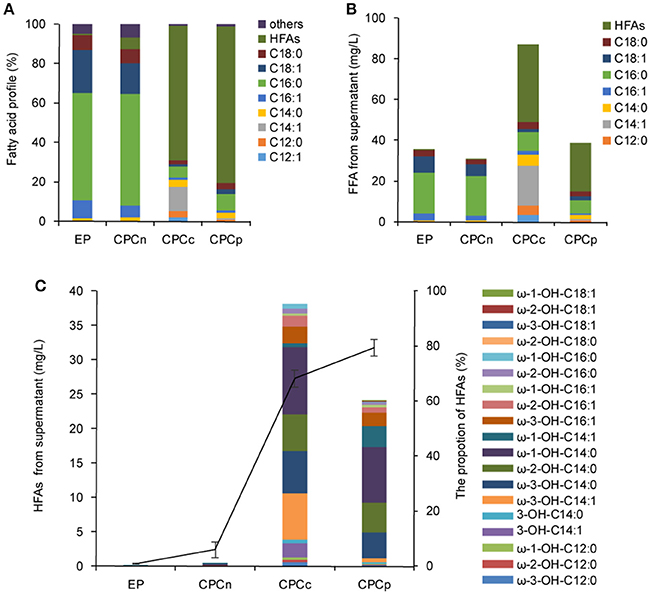
Figure 3. Coexpression of P450BM3 and plant thioesterase gene in E. coli. (A) Fatty acid profile of the supernatant FFA from the recombinant cells. (B) Fatty acid concentration of the supernatant FFA. (C) The concentration and the profile of the supernatant HFA. Black line represents the percentage of HFAs in the supernatant FFA. EP, BL21 harboring P450BM3 alone; CPCn, BL21 harboring P450BM3 and CnFatB3; CPCc, BL21 harboring P450BM3 and CcFatB1; CPCp, BL21 harboring P450BM3 and CpFatB2. All data are the mean ± standard deviation from triplicate cultures.
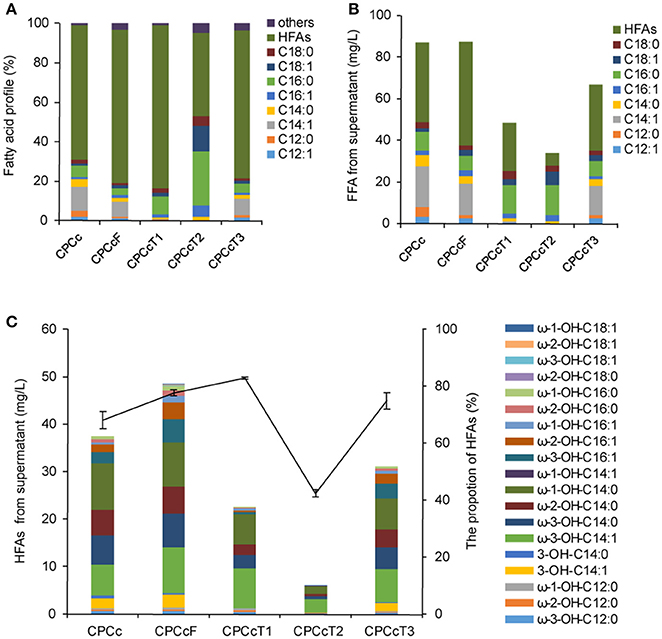
Figure 4. Coexpression of P450BM3, CcFatB1 with additional fatty acid metabolism regulators in strain CPCc. (A) Fatty acid profile of the supernatant FFA. (B) The concentration of the supernatant FFA. (C) The concentration of HFAs from supernatant FFA. CPCc, BL21 harboring P450BM3 and CcFatB1; CPCcF, BL21 harboring P450BM3, CcFatB1, and fadR from E. coli; CPCcT1~CPCcT3, BL21 harboring P450BM3, CcFatB1 and RoTetR1, or RoTetR2, or RoTetR3 from R. opacus PD630. The vector pCDFDuet-1 is used for coexpression of P450BM3 and CcFatB1. pET28a is used for expression of individual fadR, RoTetR1, RoTetR2, or RoTetR3. All data are the mean ± standard deviation from triplicate cultures.
Based on these results, there was a positive correlation between FFA production obtained in the TE-only strains and HFA production in the derived strain containing both the respective TE and P450BM3. However, when FFA reached 90 mg/L or above, the production of HFA was reduced (Table 1 and Figure 3). Thus, precise modular control of the expression ratio of P450BM3 and MCFA-specific TE may be important to the production of high HFA levels.
Although dominated by C14 HFA, as many as 19 HFAs including ω-1-OH-C12/C14/C16/C18, ω-2-OH-C12/C14/C16/C18, ω-3-OH-C12/C14/C16/C18, 3-OH-C14:0, and 3-OH-C14:1 were detected from the supernatant FFA in stains CPCc and CPCp and confirmed by GC-MS (Figure 3C). The mass spectra of the main HFAs are shown in Figure S4. The HFA profile discovered here was more complicated than that of previous reports (Wang et al., 2012; Cao et al., 2016), where only four types of HFAs including 9-OH-C10, 11-OH-C12, 10-OH-C16, and 12-OH-C18, were detected in recombinant E. coli harboring P450BM3 and tesA' (Cao et al., 2016). Although the production of multiple HFAs might complicate the downstream purification process, variable ratios of different HFAs in the mixture from different gene combinations might also provide the unique polymer property from these HFAs.
For medium chain HFAs, CPCc exhibited strong preference for producing ω-1/2/3-OH-C14, which was up to 31.2 mg/L, corresponding to 81.8% of total HFA. Of them, ω-3-OH-C14:1, ω-3-OH-C14:0, ω-2-OH-C14:0, and ω-1-OH-C14:0 were the most abundant (Figure 3C and Figure S5). This result was consistent with the previous in vitro result that P450BM3 oxidized C14:0 very efficiently (Schneider et al., 1998), thus yielded abundant ω-1/2/3-OH-C14:0 in this study. Besides, a large amount of monounsaturated ω-3-OH-C14:1 was also produced. Only a small amount of ω-1/2/3-OH-C12:0 was detected which might be partly due to the insufficient supply of free C12:0 in strains APCc or CPCc. Nevertheless, our results showed that co-expressing CcFatB1 or CpFatB2 with P450BM3 greatly increased the production of medium chain ω-1/2/3-HFAs.
Co-expression of Transcriptional Factor Further Boosted the Production of Medium Chain HFA in BL21(DE3) and BL21ΔfadD
Fatty acid metabolism regulator FadR from E. coli plays dual roles in both repression of fatty acid degradation and activation of fatty acid biosynthesis (Zhang et al., 2012). Therefore, we were interested to test if FadR would enhance HFA production. In addition, we were also interested in screening more similar regulators from oleaginous bacteria. Three putative fatty acid metabolism regulators of oleaginous bacterium R. opacus PD630 were predicted in this work and designated as RoTetR1, RoTetR2, and RoTetR3, based on the published genome sequence of this strain. RoTetR1/2/3 showed high homology to other TetR family proteins, although phylogeny analysis indicated that E. coli FadR was substantially different from the TetR family (Figure S6). FadR overexpression in E. coli has resulted in increased TFA (Zhang et al., 2012). In this study, three putative R. opacus fatty acid metabolism regulators RoTetRs were individually overexpressed in E. coli BL21(DE3). However, no obvious increase of TFA or change of fatty acid profile was observed with RoTetRs (Figure S7). Then each fadR or RoTetR was expressed in HFA-producing strain CPCc. Expression of FadR resulted in the increase of HFAs from 38.1 to 49.8 mg/L (Figure 4). In contrast, RoTetRs reduced the content of total FFA (Figure 4B), leading to the lower amount of HFAs. However, the ratio of HFA in total extracellular FFA was increased by RoTetR1 or RoTetR3 (Figure 4A). It has been reported that the overexpression of fadR enhances TFA titer by 7.5-fold, and the increased proportion of unsaturated fatty acids, which has been explained by FadR-mediated activation of fabA and fabB (Zhang et al., 2012). It could partly explain why unsaturated ω-3-OH-C14:1 was up-regulated (Figure 4C and Figure S8). Unlike previous reports, TFA yield was barely elevated in this study which might be due to the subsequent conversion of FFA by P450BM3. Our results showed that RoTetR alone had no obvious effect on the amount of TFA, but reduced the supernatant FFA when expressed in the HFA producing strain, with an increased HFA ratio in the supernatant FFA by RoTetR1 or RoTetR3.
Using a low copy number vector for the expression of TE could alleviate the unintended consequences caused by modulating of acyl-ACP pools (Lennen and Pfleger, 2012). Thus pACYCDuet-1 with a copy number of about 10–12, which was much lower than that of pCDFDuet-1, was used in this study as well. Results showed that the production of HFAs by CcFatB1 from the low copy number vector in strain APCc was 41.8 mg/L, higher than 38.1 mg/L by CcFatB1 from the high copy number vector in strain CPCc (Figures 4C, 5C). Again, CnFatB3 from the low copy number vector in APCn only yielded 1.6 mg/L of HFAs (Figure 5C).
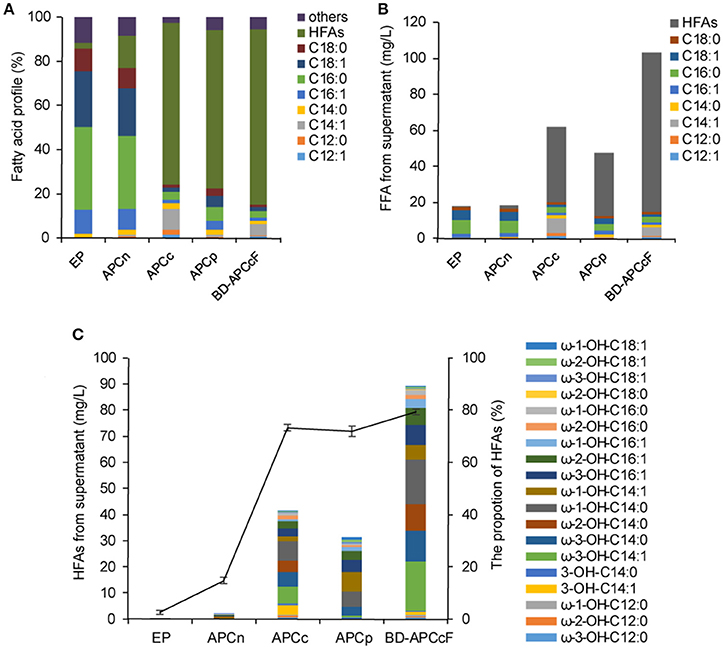
Figure 5. Coexpression of P450BM3, plant thioesterase gene and fadR in BL21 or BL21ΔfadD. (A) The proportion of the supernatant FFA. (B) The titer of each category of FFA. (C) The concentration of HFAs from the supernatant FFA. EP, BL21 harboring P450BM3 alone; APCn, BL21 harboring P450BM3 and CnFatB3; APCc, BL21 harboring P450BM3 and CcFatB1; APCp, BL21 harboring P450BM3 and CpFatB2, BD-APCcF: BL21ΔfadD harboring P450BM3, CcFatB1, and fadR. The vector used for P450BM3 and plant TE was pACYCDuet-1 instead of pCDFDuet-1 in this test. The cultures were incubated at 30°C with shaking at 150 rpm for 16 h after IPTG induction. All data are the mean ± standard deviation from triplicate cultures.
Inactivation of fadD, the only gene coding for acyl-CoA synthetase in E. coli, would substantially inhibit the degradation of fatty acids by preventing acyl-CoA synthesis (Fujita et al., 2007). Low copy number plasmid, pAPCcF, and high copy number plasmid, pCPCcF, were transformed in to BL21ΔfadD mutant respectively, for construction of BD-APCcF and BD-CPCcF. Indeed, HFAs concentration was doubled and reached up to 86.1 mg/L (Figures 5A,B). Notably, the levels of medium chain HFAs including ω-3-OH-C14:1, ω-3-OH-C14:0, ω-2-OH-C14:0, ω-1-OH-C14:0 were all doubled or tripled in BD-APCcF, up to 18.4, 11.5, 9.8, and 16.7 mg/L, respectively (Figure 5C and Figure S9). The concentration of ω-1/2/3-OH-C16:1 was enhanced as well, calculated as 17.2 mg/L, mainly due to the existence of naturally high amounts of C16:1 in E. coli (Figure S9). BD-APCcF produced higher amounts of HFA than BD-CPCcF, thus BD-APCcF was used in the following study. The utilization of pCDFDuet-1 led to a significantly lower amount of HFAs, when compared to pACYCDuet-1 in this study. They differed in the resistance marker (chloramphenicol vs. streptomycin), the type of replicon (CloDF13 vs. P15A) and the copy number (20–40 vs. 10–12). We thus proposed that a lower copy number vector might be beneficial for the high yield of HFAs in E. coli.
Since the translation efficiency decreased with the increasing distance from the gene to the promoter in expression plasmid, CcFatB1 was then arranged after P450BM3 under the same T7 promoter in pCDFDuet-1 or pACYCDuet-1 (see Table S1, strains BD-APCc-1 and BD-CPCc-1). However, none of these constructs exceeded the HFAs production of BD-APCcF. The highest titer of HFAs produced by strain BD-APCc-1 achieved only 17.6 mg/L, while strain BD-CPCc-1 achieved 14.0 mg/L. Therefore, more precise fine-tuning of P450BM3 and CcFatB1 might be needed considering the balance of metabolism burden and product. Besides, β-oxidation reversal has been successfully applied to produce carboxylic acids and MCFAs (Clomburg et al., 2015; Wu et al., 2017). Theoretically, overexpression of genes responsible for β-oxidation reversal along with P450BM3 might further improve the desired HFA production by using acyl-CoA pool directly.
Addition of Glycerol Led to Higher HFA Accumulation
HFAs are intracellularly synthesized in E. coli, thus the biomass would be a key factor that determines the amount of produced HFAs. Glycerol as an inexpensive carbon source has been applied in cultivation of E. coli for enhanced biomass (Chen and Liu, 2016). In this study, 0.5–5% (w/v) of glycerol was added into the M9 medium containing 2% (w/v) glucose at the beginning of culturing. The biomass of BD-APCcF was indeed increased after incubating with different concentrations of glycerol, rising from 28 to 37 mg per 50 mL medium, although the biomass was still slightly lower than that of control EP, which was 39 mg. As expected, the HFA concentration of strain BD-APCcF was increased from 86.1 to 107.9 mg/L when supplemented with 2% of glycerol, although the HFA profile was not changed (Figure 6A).
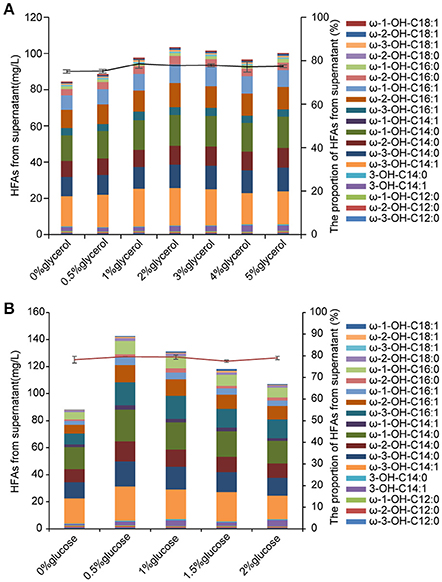
Figure 6. The effect of glycerol and glucose on HFAs production. (A) The concentration and the profile of HFAs from supernatant in M9 medium plus 2% glucose along with 0–5% glycerol. (B) The concentration and the profile of HFAs from supernatant in M9 medium plus 2% glycerol along with 0–2% glucose. All data are the mean ± standard deviation from triplicate cultures.
In general, 2% of glucose was used for production of HFA in previously published work (Wang et al., 2012; Kim et al., 2015; Cao et al., 2016). Here, we found that the reduced amount of glucose to as low as 0.5% along with 2% glycerol in the medium led to 144 mg/L HFAs and 50.6 mg of cell dry weight. In addition, the HFA titer from the culture without supplementation of glucose was much lower than that of culture with 0.5–2% glucose (Figure 6B). The presence of glucose in medium could hinder E. coli to utilize other carbon resources such as lactose and glycerol due to the glucose effect. After consumption of glucose by cells for growth, glycerol could then be channeled into glycolysis via production of dihydroxyacetone phosphate, glyceraldehyde phosphate, and pyruvate. Pyruvate would subsequently be converted into acetyl-CoA, the precursor for fatty acid synthesis. Glycerol is cheaper than glucose, and more importantly, it has a higher degree of reduction than glucose, thus it is proposed that the theoretical yield of fatty acid, oils, or other reduced chemicals on glycerol is higher than that of glucose (Chen and Liu, 2016). With the combinational effect of glucose and glycerol, the HFA titer was further elevated. The cost of feed-stock accounts for a heavy proportion of the total cost of HFA production. Previous work that feeding glucose instead of FFA in culture is an important step forwards commercial application (Wang et al., 2012). Our results demonstrated that the addition of abundant, inexpensive and renewable glycerol with the reduction of glucose usage elevated the desired HFA production as well.
Conclusion
This study demonstrated that the overexpression of plant acyl-ACP TEs from C. nucifera, C. camphora, or C. plaustris in E. coli substantially increased the production of MCFA both in cell and in supernatant. Co-expression of P450BM3, plant acyl-ACP TE, and fatty acid metabolism regulator FadR yielded substantially increased amount of ω-1/2/3 position HFAs, especially ω-1/2/3-OH-C14. Reducing usage of glucose and supplementing with cheaper glycerol further boosted the production of HFAs. Importantly, such HFAs were secreted into the medium which was convenient for downstream purification of HFAs. In summary, E. coli had been metabolic engineered to selectively produce medium chain omega HFA by utilization of renewable glucose and glycerol.
Author Contributions
KX, X-HY, LW, F-HH, and XW: conceived the study; KX, X-HY, and LX: conducted experiments; XW, KX, W-CC, and X-RZ: analyzed the data and wrote the manuscript; All authors read and approved the final manuscript.
Funding
Agricultural Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI), National Key R & D Program (2016YFD0501209) and Earmarked Fund for China Agriculture Research System (CARS-13).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Huizhou Liu from Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences for kindly providing the strain BL21ΔfadD.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00139/full#supplementary-material
Figure S1. Mass spectra of 3-OH-C14:0 (A) and 3-OH-C14:1 (B) TMS derivatives. *Indicates the characteristic mass ions.
Figure S2. Amino acid sequence comparison and structure prediction of Acyl-ACP TEs. (A) Alignment of amino acid sequences of CnFatB3 (JF338905) from Cocos nucifera, CcFatB1 (U31813) from Cinnamomum camphora, CpFatB2 (U38189) from Cuphea plaustris, UcFatB2 (U17097) from Umbellularia californica, CtFat (CTC_RS00430) from Clostridium tetani and LpFat (ABX40638) from Lachnoclostridium phytofermentans. The secondary structure elements of each protein are shown above the alignment. Identical amino acids are shown in white on a red background, while similar residues are shown in red. Three conserved residues that constitute the catalytic triad are indicated by black arrows. The start of mature proteins is indicated by blue arrow. (B) Structural prediction analysis of CnFatB3 (left), CcFatB1 (middle), and CpFatB2 (right).
Figure S3. Western blot analysis of recombinant cells. The proteins from cells harboring P450BM3 along with CnFatB3 (left), CcFatB1 (middle), or CpFatB2 (right) were blotted with anti His-tag monoclonal antibody (A) for P450BM3 fusion protein or anti S-tag antibody (B) for TE fusion proteins.
Figure S4. The chemical structure and mass spectra of HFA TMS derivatives from strain CPCc. Mass spectra of 3-OH-C14:0 and 3-OH-C14:1 TMS derivatives were same as Figure S1. *Indicates the characteristic mass ions.
Figure S5. Hydroxyl fatty acid profile for recombinant strain EP, CPCn, CPCc, and CPCp, respectively.
Figure S6. Phylogeny tree of three potential fatty acid metabolism regulators RoTetRs and FadR from E. coli.
Figure S7. Lipid profile of recombinant E. coli hells expressing individual RoTetRs. (A) TLC showing the neutral lipids; (B) fatty acid profile.
Figure S8. Hydroxyl fatty acid profile for recombinant strain CPCc, CPCcF, CPCcT1, CPCcT2, and CPCcT3, respectively.
Figure S9. Hydroxyl fatty acid profile for recombinant strain EP, APCn, APCc, APCp, and BD-APCcF, respectively.
Table S1. Strains and plasmids used in this study.
Table S2. Details of primer sequences used in this test.
Table S3. Fatty acid profile of cellular total fatty acid (TFA) produced by BL21(DE3) harboring individual plant acyl-ACP thioesterase gene.
Table S4. Fatty acid profile of cellular cell free fatty acid (FFA) a produced by BL21(DE3) harboring individual plant acyl-ACP thioesterase gene.
Abbreviations
ACP, Acyl-carrier protein; TAG, triacylglycerol; DAG, Diacylglycerol; MAG, monoacylglycerol; FAME, Fatty acid methyl ester; FatB, Fatty acyl thioesterase B; FFA, Free fatty acid; HFA, hydroxyl fatty acid; IPTG, Isopropyl ß-D-thiogalactoside; MCFA, Medium chain fatty acid; SDS, Sodium dodecyl sulfate; TE, Acyl-ACP thioesterase; TFA, Total fatty acid; TLC, Thin layer chromatography; TMS, Trimethylsilyl.
References
Bowen, C. H., Bonin, J., Kogler, A., Barba-Ostria, C., and Zhang, F. (2016). Engineering Escherichia coli for conversion of glucose to medium-chain ω-hydroxy fatty acids and α,ω-dicarboxylic acids. ACS Synth. Biol. 5, 200–206. doi: 10.1021/acssynbio.5b00201
Cao, Y., Cheng, T., Zhao, G., Niu, W., Guo, J., Xian, M., et al. (2016). Metabolic engineering of Escherichia coli for the production of hydroxy fatty acids from glucose. BMC Biotechnol. 16:26. doi: 10.1186/s12896-016-0257-x
Chen, Z., and Liu, D. (2016). Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol. Biofuels 9:205. doi: 10.1186/s13068-016-0625-8
Clomburg, J. M., Blankschien, M. D., Vick, J. E., Chou, A., Kim, S., and Gonzalez, R. (2015). Integrated engineering of β-oxidation reversal and omega-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids. Metab. Eng. 28, 202–212. doi: 10.1016/j.ymben.2015.01.007
Fan, L., Liu, J., Nie, K., Liu, L., Wang, F., Tan, T., et al. (2013). Synthesis of medium chain length fatty acid ethyl esters in engineered Escherichia coli using endogenously produced medium chain fatty acids. Enzyme Microb. Technol. 53, 128–133. doi: 10.1016/j.enzmictec.2013.03.012
Fujita, Y., Matsuoka, H., and Hirooka, K. (2007). Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66, 829–839. doi: 10.1111/j.1365-2958.2007.05947.x
Hilker, B. L., Fukushige, H., Hou, C., and Hildebrand, D. (2008). Comparison of Bacillus monooxygenase genes for unique fatty acid production. Prog. Lipid Res. 47, 1–14. doi: 10.1016/j.plipres.2007.09.003
Hirata, A., Kishino, S., Park, S. B., Takeuchi, M., Kitamura, N., and Ogawa, J. (2015). A novel unsaturated fatty acid hydratase toward C16 to C22 fatty acids from Lactobacillus acidophilus. J. Lipid Res. 56, 1340–1350. doi: 10.1194/jlr.M059444
Hou, C. T. (2009). Biotechnology for fats and oils: new oxygenated fatty acids. N. Biotechnol. 26, 2–10. doi: 10.1016/j.nbt.2009.05.001
Kim, H. J., Silva, J. E., Vu, H. S., Mockaitis, K., Nam, J. W., and Cahoon, E. B. (2015). Toward production of jet fuel functionality in oilseeds: identification of FatB acyl-acyl carrier protein thioesterases and evaluation of combinatorial expression strategies in Camelina seeds. J. Exp. Bot. 66, 4251–4265. doi: 10.1093/jxb/erv225
Kim, H. U., and Chen, G. Q. (2015). Identification of hydroxy fatty acid and triacylglycerol metabolism-related genes in lesquerella through seed transcriptome analysis. BMC Genomics 16:230. doi: 10.1186/s12864-015-1413-8
Kim, K. R., and Oh, D. K. (2013). Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 31, 1473–1485. doi: 10.1016/j.biotechadv.2013.07.004
Kishino, S., Takeuchi, M., Park, S. B., Hirata, A., Kitamura, N., Kunisawa, J., et al. (2013). Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. U.S.A. 110, 17808–17813. doi: 10.1073/pnas.1312937110
Lee, K. R., Chen, G. Q., and Kim, H. U. (2015). Current progress towards the metabolic engineering of plant seed oil for hydroxy fatty acids production. Plant Cell Rep. 34, 603–615. doi: 10.1007/s00299-015-1736-6
Lennen, R. M., and Pfleger, B. F. (2012). Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 30, 659–667. doi: 10.1016/j.tibtech.2012.09.006
Li, H., and Poulos, T. L. (1999). Fatty acid metabolism, conformational change, and electron transfer in cytochrome P-450(BM-3). Biochim. Biophys. Acta 1441, 141–149. doi: 10.1016/S1388-1981(99)00161-4
Mavraganis, I., Meesapyodsuk, D., Vrinten, P., Smith, M., and Qiu, X. (2010). Type II diacylglycerol acyltransferase from Claviceps purpurea with ricinoleic acid, a hydroxyl fatty acid of industrial importance, as preferred substrate. Appl. Environ. Microbiol. 76, 1135–1142. doi: 10.1128/AEM.02297-09
Meesapyodsuk, D., Chen, Y., Ng, S. H., Chen, J., and Qiu, X. (2015). Metabolic engineering of Pichia pastoris to produce ricinoleic acid, a hydroxy fatty acid of industrial importance. J. Lipid Res. 56, 2102–2109. doi: 10.1194/jlr.M060954
Narhi, L. O., and Fulco, A. J. (1986). Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 261, 7160–7169.
Nawabi, P., Bauer, S., Kyrpides, N., and Lykidis, A. (2011). Engineering Escherichia coli for biodiesel production utilizing a bacterial fatty acid methyltransferase. Appl. Environ. Microbiol. 77, 8052–8061. doi: 10.1128/AEM.05046-11
Reynolds, K. B., Taylor, M. C., Zhou, X. R., Vanhercke, T., Wood, C. C., Blanchard, C. L., et al. (2015). Metabolic engineering of medium-chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front. Plant Sci. 6:164. doi: 10.3389/fpls.2015.00164
Rottig, A., Zurek, P. J., and Steinbuchel, A. (2015). Assessment of bacterial acyltransferases for an efficient lipid production in metabolically engineered strains of E. coli. Metab. Eng. 32, 195–206. doi: 10.1016/j.ymben.2015.09.016
Sakurama, H., Kishino, S., Mihara, K., Ando, A., Kita, K., Takahashi, S., et al. (2014). Biohydrogenation of C20 polyunsaturated fatty acids by anaerobic bacteria. J. Lipid Res. 55, 1855–1863. doi: 10.1194/jlr.M045450
Schneider, S., Wubbolts, M. G., Sanglard, D., and Witholt, B. (1998). Biocatalyst engineering by assembly of fatty acid transport and oxidation activities for in vivo application of cytochrome P-450BM-3 monooxygenase. Appl. Environ. Microbiol. 64, 3784–3790.
Seo, J. H., Lee, S. M., Lee, J., and Park, J. B. (2015). Adding value to plant oils and fatty acids: biological transformation of fatty acids into omega-hydroxycarboxylic, alpha,omega-dicarboxylic, and omega-aminocarboxylic acids. J. Biotechnol. 216, 158–166. doi: 10.1016/j.jbiotec.2015.10.024
Sung, C., Jung, E., Choi, K. Y., Bae, J. H., Kim, M., Kim, J., et al. (2015). The production of omega-hydroxy palmitic acid using fatty acid metabolism and cofactor optimization in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 6667–6676. doi: 10.1007/s00253-015-6630-1
Takeuchi, M., Kishino, S., Hirata, A., Park, S. B., Kitamura, N., and Ogawa, J. (2015). Characterization of the linoleic acid Δ9 hydratase catalyzing the first step of polyunsaturated fatty acid saturation metabolism in Lactobacillus plantarum AKU 1009a. J. Biosci. Bioeng. 119, 636–641. doi: 10.1016/j.jbiosc.2014.10.022
Takeuchi, M., Kishino, S., Park, S. B., Hirata, A., Kitamura, N., Saika, A., et al. (2016). Efficient enzymatic production of hydroxy fatty acids by linoleic acid Δ9 hydratase from Lactobacillus plantarum AKU 1009a. J. Appl. Microbiol. 120, 1282–1288. doi: 10.1111/jam.13088
Wang, X., Li, L., Zheng, Y., Zou, H., Cao, Y., Liu, H., et al. (2012). Biosynthesis of long chain hydroxyfatty acids from glucose by engineered Escherichia coli. Bioresour. Technol. 114, 561–566. doi: 10.1016/j.biortech.2012.02.119
Wu, J., Zhang, X., Xia, X., and Dong, M. (2017). A systematic optimization of medium chain fatty acid biosynthesis via the reverse beta-oxidation cycle in Escherichia coli. Metab. Eng. 41, 115–124. doi: 10.1016/j.ymben.2017.03.012
Yang, B., Chen, H., Gu, Z., Tian, F., Ross, R. P., Stanton, C., et al. (2014). Synthesis of conjugated linoleic acid by the linoleate isomerase complex in food-derived lactobacilli. J. Appl. Microbiol. 117, 430–439. doi: 10.1111/jam.12524
Yore, M. M., Syed, I., Moraes-Vieira, P. M., Zhang, T., Herman, M. A., Homan, E. A., et al. (2014). Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332. doi: 10.1016/j.cell.2014.09.035
Yuan, L., Voelker, T. A., and Hawkins, D. J. (1995). Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proc. Natl. Acad. Sci. U.S.A. 92, 10639–10643. doi: 10.1073/pnas.92.23.10639
Zhang, F., Ouellet, M., Batth, T. S., Adams, P. D., Petzold, C. J., Mukhopadhyay, A., et al. (2012). Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab. Eng. 14, 653–660. doi: 10.1016/j.ymben.2012.08.009
Keywords: Escherichia coli, medium chain fatty acid, hydroxy fatty acid, acyl-ACP thioesterase, fatty acid metabolism regulator
Citation: Xiao K, Yue X-H, Chen W-C, Zhou X-R, Wang L, Xu L, Huang F-H and Wan X (2018) Metabolic Engineering for Enhanced Medium Chain Omega Hydroxy Fatty Acid Production in Escherichia coli. Front. Microbiol. 9:139. doi: 10.3389/fmicb.2018.00139
Received: 30 August 2017; Accepted: 22 January 2018;
Published: 07 February 2018.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
M. Kalim Akhtar, United Arab Emirates University, United Arab EmiratesAlvaro R. Lara, Universidad Autónoma Metropolitana, Mexico
Copyright © 2018 Xiao, Yue, Chen, Zhou, Wang, Xu, Huang and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Wan, wanxia@oilcrops.cn
 Kang Xiao1
Kang Xiao1 Xue-Rong Zhou
Xue-Rong Zhou Xia Wan
Xia Wan