- 1Plant Protection Research Division, Agroscope, Zurich, Switzerland
- 2Competence Division Methods Development, Analytics and SIB Swiss Institute of Bioinformatics, Agroscope, Zurich, Switzerland
- 3School of Biological Sciences, University of Canterbury, Christchurch, New Zealand
- 4Biomolecular Interaction Centre, University of Canterbury, Christchurch, New Zealand
In light of public concerns over the use of pesticides and antibiotics in plant protection and the subsequent selection for spread of resistant bacteria in the environment, it is inevitable to broaden our knowledge about viable alternatives, such as natural antagonists and their mode of action. The genus Pseudomonas is known for its metabolic versatility and genetic plasticity, encompassing pathogens as well as antagonists. We characterized strain Pseudomonas orientalis F9, an isolate from apple flowers in a Swiss orchard, and determined its antagonistic activity against several phytopathogenic bacteria, in particular Erwinia amylovora, the causal agent of fire blight. P. orientalis F9 displayed antagonistic activity against a broad suite of phytopathogenic bacteria in the in vitro tests. The promising results from this analysis led to an ex vivo assay with E. amylovora CFBP1430Rif and P. orientalis F9 infected detached apple flowers. F9 diminished the fire blight pathogen in the flowers but also revealed phytotoxic traits. The experimental results were discussed in light of the complete genome sequence of F9, which revealed the strain to carry phenazine genes. Phenazines are known to contribute to antagonistic activity of bacterial strains against soil pathogens. When tested in the cress assay with Pythium ultimum as pathogen, F9 showed results comparable to the known antagonist P. protegens CHA0.
Introduction
Pseudomonas is a diverse genus, including pathogens, tritagonists (microorganisms of unknown ecological function) and mutualists that are impacting on larger hosts (Silby et al., 2011; Freimoser et al., 2015). Members of the genus are known to synthesize an extensive number of metabolites including those that directly stimulate plant growth or that inhibit the growth of harmful microorganisms. Strains of the species Pseudomonas fluorescens are biocontrol agents and include P. fluorescens A506, and P. fluorescens EPS62e that are known for their antagonistic activity against Erwinia amylovora, the causal agent of bacterial fire blight in Rosaceae (Wilson and Lindow, 1993; Haas and Défago, 2005; Pujol et al., 2005). As Rosaceae include apple and pear, fire blight is a major threat to pome fruit production worldwide causing substantial crop and economic loss. To date, the most effective control against fire blight is the application of antibiotics, e.g., streptomycin, during the flowering period. The application of antibiotics is limited due to regulatory restrictions and the rise and spread of multiple resistant pathogens (Chiou and Jones, 1995; Manulis et al., 1998; McGhee et al., 2011). In Switzerland, the application of streptomycin was banned from field applications since early 2016 and only a limited range of pest management strategies remain to hold E. amylovora at bay. In light of the restricted regulations and public concerns over the application of antibiotics and pesticides, the intensified use of beneficial and antagonistic organisms in agriculture is a much favorable alternative (Johnson and Stockwell, 2000; Seibold et al., 2004; Born et al., 2017).
The prerequisite for intensified biological pest management is a versatile selection of potential antagonists and an understanding of their mode of action. Some of the secondary metabolites of pseudomonads counteracting plant pathogens have already been identified. These include antibiotics, e.g., phenazines, pyrrolnitrin, pyoluteorin, and siderophores, e.g., pyoverdin, pyochelin, pseudomonin (Cornelis and Matthijs, 2002; Haas and Défago, 2005). E. amylovora antagonistic pseudomonads were shown to compete for space, to be more efficient in the use of nutrients or to act by cell-to-cell interference (Cabrefiga et al., 2007; Paternoster et al., 2010).
During a screen for novel antagonists against E. amylovora, we isolated a P. orientalis strain (designated F9) from apple flowers in a Swiss orchard. P. orientalis was first described in 1999 and isolated from Lebanese spring waters (Dabboussi et al., 1999). There is only scarce information available concerning the metabolic potential of this species, but some strains of the species that produced phenazines have been isolated from roots of dryland wheat. These isolates were associated with antagonistic activity against the Rhizoctonia root rot of wheat (Parejko et al., 2013; Jaaffar et al., 2017).
We evaluated the antagonistic activity of P. orientalis F9 against several plant pathogens in vitro and against the fire blight pathogen, E. amylovora, in vitro and ex vivo. The strain revealed antagonistic but also phytotoxic traits in the performed assays. In order to combine the experimental data with a data mining effort for genes potentially responsible for the observed traits, we sequenced P. orientalis F9 and de novo assembled the complete genome. The presence of phenazine genes prompted us to test F9 for its antagonistic activity against Pythium ultimum in the cress assay where F9 revealed an antagonistic activity comparable to P. protegens CHA0.
Materials and Methods
Cultivation of Bacteria and P. ultimum
Bacterial overnight cultures were grown at 26°C in Tryptic Soy broth (TSB, Oxoid, Karlsruhe, Germany) or King’s B medium (King et al., 1954). PSTB medium was prepared according to Pusey et al. (2008b). P. ultimum was cultivated on malt agar for 2–3 days (Amresco) at room temperature. Microorganisms used in this study are listed in Table 1.
MALDI Biotyping
Matrix-assisted laser desorption/ionization (MALDI) biotyping was performed according to Gekenidis et al. (2014) using a MicroFlex biotyper, and the MALDI Biotyper software (Database Version 4.0.0.1, Bruker Daltonics Germany). Briefly, bacterial cell material from a colony on agar plate was smeared onto a MALDI target using a toothpick. The smear was then covered with 1 μl matrix solution (10 mg ml-1 HCCA (α-cyano-4-hydroxycinnamic acid) dissolved in acetonitrile-water-trifluoroacetic acid (TFA) (50:47.5:2.5 [vol/vol/vol]) (Sigma–Aldrich). After drying of the matrix solution, the target was placed in the MALDI biotyper and processed using the instrument’s standard settings for bacteriological classification.
Bacterial Growth Rate Analysis
For growth rate analysis the Bioscreen C (Oy Growth Curves Ab Ltd, Helsinki) automatic microbiology growth curve analysis system was used and bacteria grown on KB plates overnight. Fresh colonies were resuspended in 1 × PBS (K2HPO4 2.5 g l-1, KH2PO4 1.2 g l-1) and adjusted to OD600 nm = 1. Nine hundred μl medium were pipetted into a reaction, 200 μl of which were not inoculated but used as a negative control for the corresponding growth curves. The remaining 700 μl were inoculated with 3 μl of the bacterial suspension. Three replicates of the inoculated medium, each 200 μl, were loaded in wells of a Bioscreen C honeycomb plate. The plates were incubated at 26°C and shaken for 10 s before absorbance at OD600 nm. Absorbance was determined every 30 min for 24 h. Growth experiments were performed in two independent trials.
Growth Inhibition Test Using the Double Layer Assay
For the double layer assay P. orientalis F9 and the selected plant pathogenic strains were cultivated on KB plates overnight. Fresh colonies were resuspended in 1 × PBS and adjusted to OD600 nm = 1. Approximately 5 × 108 bacteria were added to 10 ml of 0.75% KB top agar. Four ml of the top agar were poured onto a standard KB plate, respectively. Ten μl of the P. orientalis F9 suspension were spotted onto the solidified top layer surfaces. Growth halos were detected after 1–2 days of incubation at 26°C and diameter of the inhibition zones measured. The experiment was performed in duplicates.
Siderophore Indicator Test
Siderophore production was tested on CAS (Chrome azurol S) agar, by applying 10 μl of overnight cultures of the bacterial strain to be tested. The CAS assay relies on the color change from a green-blue CAS-iron complex to orange desferrated CAS around siderophore-producing colonies (producing a CAS halo, or CAS positive) (Schwyn and Neilands, 1987).
Biochemical Profiling of P. orientalis F9
For biochemical characterization of P. orientalis F9, Biolog GN2 and AN plates (Biolog Inc., United States) were used. The strain was cultivated overnight in 25 ml MM2 medium (4 g l-1 L-asparagine, 2 g l-1 K2HPO4, 0.2 g l-1 MgSO4, 3 g l-1 NaCl, 10 g l-1 sorbitol) at 28°C and 240 rpm. Bacteria were harvested by centrifugation at 3500 × g for 10 min, the cell pellet washed thrice in 1 × PBS buffer, and bacteria resuspended in 1 × PBS to an optical density of OD600 nm = 0.1. This suspension was used for inoculation of the Biolog plates which were incubated for several days at 28°C and analyzed for changes of their optical density at 590 nm using a microtiter plate reader (Infinite M200, Tecan, Switzerland).
Detached Flower Assay
For the detached flower assay (Pusey, 1997) freshly opened flowers of 2-year-old potted Malus domestica ‘Golden Delicious’ were used. Freshly grown colonies of E. amylovora CFBP1430Rif, P. vagans C9-1 and P. orientalis F9 grown on KB plates were resuspended in 1 × PBS buffer to an OD600 nm = 1. Twenty μl E. amylovora CFBP1430Rif and 20 μl P. vagans C9-1 respectively P. orientalis F9 were add to 960 μl 1 × PBS. For the control inoculum, 20 μl E. amylovora CFBP1430Rif was added to 980 μl of 1 × PBS. Twenty μl of the bacterial mixtures were directly pipetted onto the hypanthium of individual flowers. Mock treatments were performed with 1 × PBS. For each combination, 32–48 flowers were tested in two independent replicates. After inoculation, flowers were incubated at 26°C for 4–5 days. After incubation flowers were visually graded based on the following scale: grade 1: calyx green; grade 2: calyx necrotic (brownish), pedicel green; grade 3: calyx and pedicel necrotic. The severity grade was defined according to Llop et al. (2011).
Cell Count Estimation
To estimate bacterial colony forming unit (CFU), present in single flowers, petals, pedestals, stamps, and stigmas were removed. The remains were shaken in 1 ml 1 × PBS buffer for 30 min at 1400 rpm and afterward vortexed for 30 s. A serial dilution of the bacterial suspension was performed up to 10-7 and 3 μl of each dilution were spotted onto TSB plates supplemented with Rifampicin (100 μg/ml) in duplicates.
Cress Assay
For the cress assay (Rosendahl and Olson, 1992), two 1 cm diameter punched-out agar plugs of P. ultimum were laid on the bottom of a 9 cm diameter petri dish counterpart and carefully overlaid with 14 g twice autoclaved soil. P. protegens CHA0 and P. orientalis F9 grown on KB plates overnight, were resuspended in 1 × PBS buffer to an OD600 nm = 1 followed by a 1:10 dilution. Ten ml of the diluted bacterial suspensions or the mock treatment (1 × PBS), were evenly spread over the soil surface. Subsequently, 0.4 g cress seeds (Lepidium sativum) were evenly distributed onto the soil. Four replicas were performed for each treatment. Plates were incubated at 22°C and 65% humidity. After 2, 4, and 6 days, 20 ml of autoclaved tap water was added. After 7 days, cress seedlings were harvested by cutting them off on ground-level and their biomass was determined by weighing. Statistical analysis was performed using the software Prism 7.0 (Graphpad Software).
Chromosomal DNA Preparation
For chromosomal DNA isolation, cells were grown overnight in TSB and total DNA was isolated using the method of Davis (Davis et al., 1990). The quality and quantity of the extracted high molecular weight DNA was evaluated on a 0.8% (w/v) agarose gel followed by measuring absorption ratios 260 nm/280 nm and 260 nm/230 nm on a Nanodrop 1000 (Thermo Scientific), and by performing a Qubit dsDNA GR assay (Life Technologies, United States).
Genome Sequencing and Assembly
Genomic DNA of P. orientalis F9 was sequenced on the Pacific Biosciences RS II platform and on Illumina MiSeq at the Functional Genomics Center Zurich. PacBio RS II sequencing resulted in a total of 1.03 Gbp of raw data (using two SMRT cells and P6-C4 chemistry). After quality filtering, 133,068 subreads with a mean length of 6,748 bp were obtained, which were de novo assembled with SMRT Portal 2.3.0 protocol RS_HGAP_Assembly.3 (Chin et al., 2013). Terminal repeats were removed, the genome was circularized and its start position aligned with dnaA using Circlator 1.1.2 (Hunt et al., 2015). Remaining assembly errors were corrected by performing two rounds of resequencing (SMRT Portal 2.3.0, protocol RS_Resequencing.1). This resulted in one 5,986,236-bp contig of the chromosome with an average coverage depth of 130-fold. The PacBio assembly did not contain any plasmids. MiSeq sequencing was performed with a 2 × 300 bp paired end library resulting in 2.6 million read pairs. MiSeq reads were mapped to our de novo assembly (average coverage of 222-fold) and allowed us to detect and correct 17 single nucleotide indel errors. To capture small plasmids, which may potentially have been missed by PacBio RS II sequencing due to the size selection step during library preparation, we assembled the Illumina reads which did not map to the chromosome assembly (about 0.5% of the Illumina reads) with SPAdes 3.9 (Bankevich et al., 2012). However, no evidence for small plasmids was found. The high quality complete genome was deposited at Genbank with accession number CP018049 (Bioproject accession: PRJNA353169).
Genome Annotation and Properties
Genome annotation was done by NCBI [Prokaryotic Genome Annotation Pipeline version 3.3 (Tatusova et al., 2016)]. COG classification was performed by searching the predicted CDSs against the eggNOG 4.5 database (Gammaproteobacteria specific dataset, “gproNOG”) (Huerta-Cepas et al., 2016) and subsequent extraction of COG categories. Hits with e-value higher than 0.001 were discarded. On top of the NCBI annotation, CRISPR repeats were searched with CRISPRFinder (Grissa et al., 2007) and PILER-CR (Edgar, 2007). A search for putative prophages was performed with PHASTER (Arndt et al., 2016). Genomic islands and antibiotic resistance genes within genomic islands were predicted using IslandViewer 3 (Dhillon et al., 2015). Ori-Finder (Gao and Zhang, 2008) and oriloc (Frank and Lobry, 2000) were used to extract putative positions of origin and terminus of replication. The RAST Server (Meyer et al., 2008) was used to mine specific genome properties.
Phylogenetic Placement of P. orientalis F9 Based on Multilocus Sequence Analysis
For the construction of a multilocus sequence analysis-based (MLSA) phylogenetic tree (Glaeser and Kämpfer, 2015), chromosomal sequences for 12 representative Pseudomonas species and Pantoea vagans C9-1, serving as outgroup, were obtained from NCBI GenBank/RefSeq (Supplementary Table 3). Nine species of the P. fluorescens group including P. orientalis F9, two partial P. orientalis genome sequences, and the known E. amylovora antagonist P. fluorescens A506, were chosen. Additionally, the chromosomal sequences of less related strains P. syringae pv. persicae NCPPB 2254, P. syringae pv. actinidiae ICMP 9617, P. syringae pv. syringae ACW (representing plant pathogenic Pseudomonas species) but also P. graminis UASWS1507, and P. citronellolis P3B5 were added to cover a broader phylogenetic spectrum (Takikawa et al., 1989; Remus-Emsermann et al., 2016). The nucleotide sequences of the housekeeping genes gyrB, recA, rpoB and rpoD were extracted from all genome sequences, respectively. Multiple sequence alignment (MSA) using MUSCLE 3.8 (Edgar, 2004) was performed on the individual genes to ensure that the extracted sequences were suitable for phylogenetic analysis. The nucleotide sequences of the four genes were concatenated for every strain and a final MUSCLE MSA was performed. With the resulting alignment of 9,520 nucleotides length, a maximum likelihood based phylogenetic inference was performed using RAxML 8.1 (Stamatakis, 2014).
Results
P. orientalis F9 Is a Member of the Fluorescent Pseudomonas Group
Pseudomonas orientalis F9 was isolated in spring 2014 from Boskoop apple flowers in a Swiss orchard during a screen for novel bacterial biocontrol agents of E. amylovora. F9 was classified as P. orientalis using MALDI biotyping with a moderate score. P. orientalis is a member of the P. fluorescens group within the genus (Dabboussi et al., 1999). The fluorescence is due to the synthesis of siderophores, iron chelators that are secreted under low iron conditions (Neilands, 1995). Indeed, isolate F9 exhibited a halo on CAS agar, which is indicative for siderophore production, and showed fluorescence when cultivated under low iron conditions (data not shown).
Genome Properties of P. orientalis F9
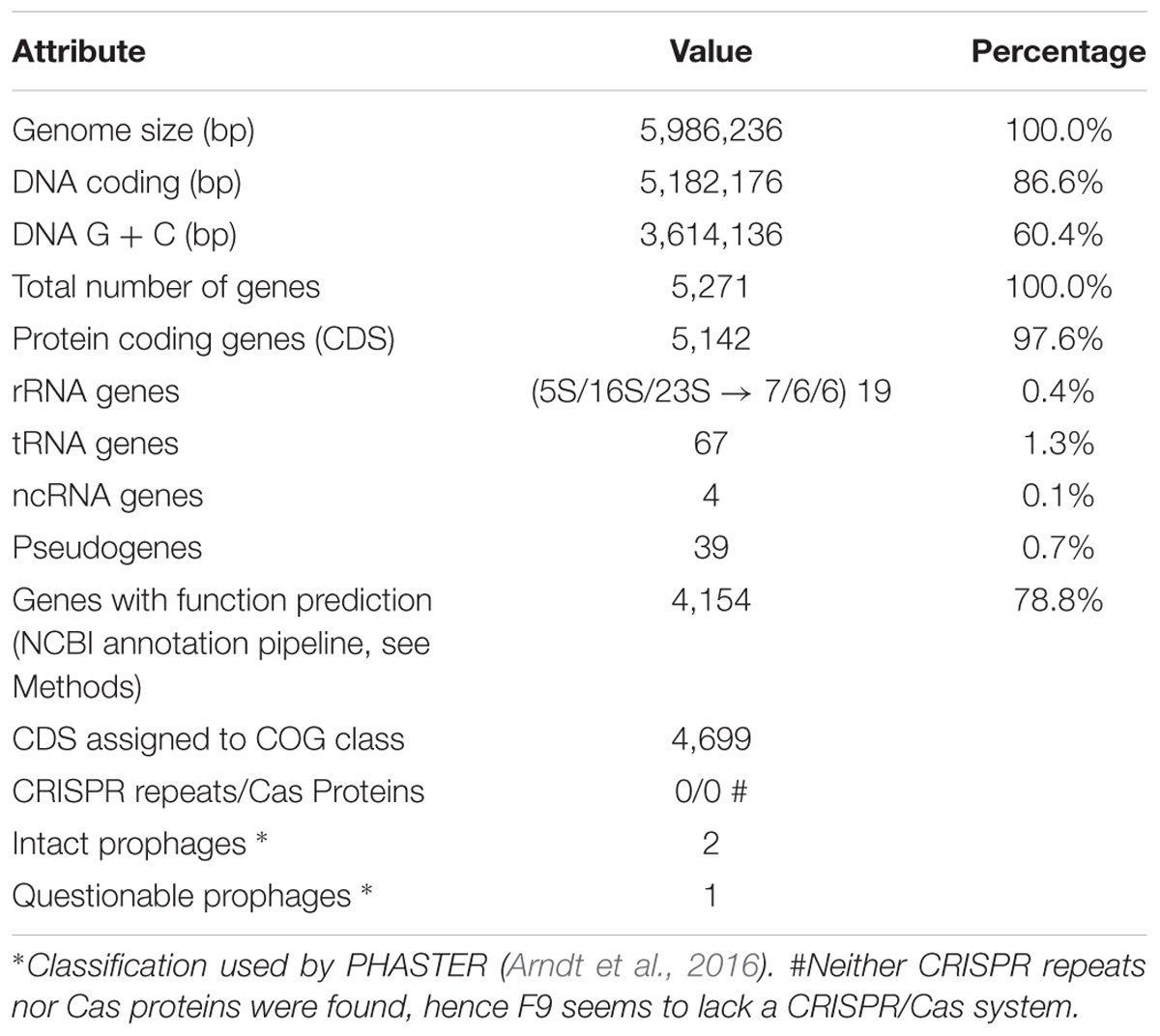
To verify the identification as a P. orientalis isolate and to further analyze its genome properties, strain F9 was sequenced and de novo assembled into a closed, high quality complete genome using a hybrid strategy (Koren et al., 2012), i.e., relying on PacBio long reads and Illumina MiSeq short reads (see Methods). No plasmids were detected. The 5.99 Mbp genome has an average GC content of 60.4%. Of the 5,271 predicted genes, 5,142 (97.6%) were predicted as protein coding sequences (CDS), 19 as rRNAs (0.4%), 67 as tRNAs (1.3%), 4 as ncRNAs (0.1%) and 39 as pseudogenes (0.7%) (Table 2). Figure 1 shows a circular view of the genome. The GC content deviates from the mean value in regions where prophages, genomic islands or rDNA clusters are located. The GC skew splits up at the putative origin of replication [confirmed using Ori-Finder (Gao and Zhang, 2008) and oriloc (Frank and Lobry, 2000)] and the putative terminus of replication (confirmed by oriloc). This provided additional evidence for the correctness of the assembly. PHASTER (Arndt et al., 2016) detected two intact prophages (Locus tags BOP93_05630 to BOP93_05745 and BOP93_13270 to BOP93_13665) and one questionable prophage (Locus tags BOP93_15105 to BOP93_15245) (Table 2), while IslandViewer 3 (Dhillon et al., 2015) detected a large number of genomic islands (53, positions on the genome provided in Supplementary Table 4). Information about the COG classification of all CDSs can be found in Supplementary Table 1. A brief comparison of the two already available P. orientalis genome assemblies (strains BS2775 and DSM 17489) versus our complete de novo assembly can be found in Supplementary Table 2.
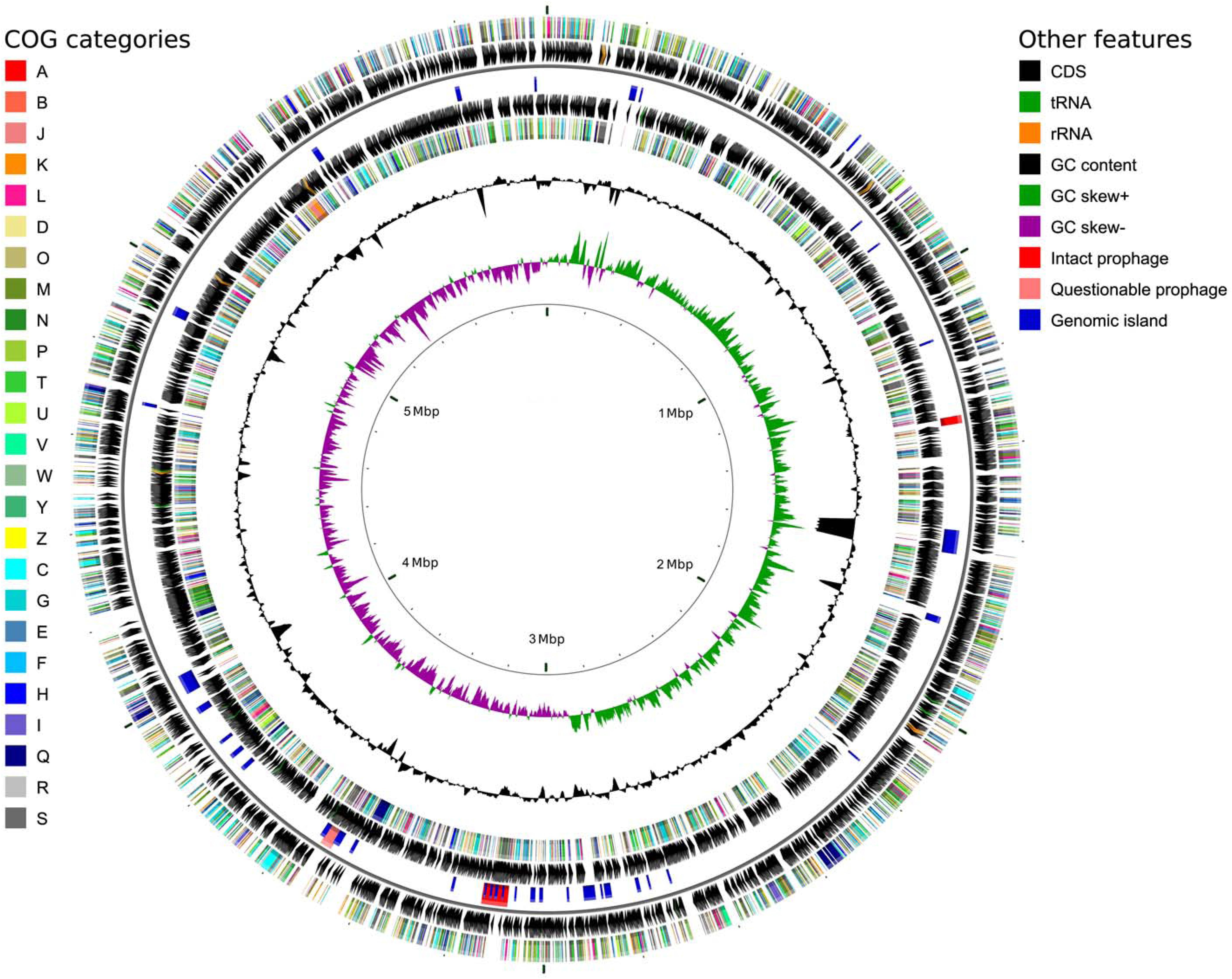
FIGURE 1. Circular genome map for P. orientalis F9, generated using CGView (Stothard and Wishart, 2005). The following features are shown (moving from the outermost track inward, origin of replication at 0 Mbp): (1) CDS on forward strand colored according to COG category, (2) CDS (black), tRNA (green) and rRNA (orange) on forward strand, (3) Intact prophages (red), questionable prophages (light red) and genomic islands (blue) for both strands, (4) CDS (black), tRNA (green) and rRNA (orange) on reverse strand, (5) CDS on reverse strand colored according to COG category, (6) GC content (black), (7) positive and negative GC skew (green and purple, respectively) and (8) genome position in Mbp.
Phylogenetic Placement of P. orientalis F9
A MLSA phylogenetic tree based on the housekeeping genes gyrB, recA, rpoB, and rpoD was calculated to compare the placement of P. orientalis F9 with twelve representative Pseudomonas species and Pantoea vagans C9-1 (outgroup) phylogenetically. In accordance with MALDI biotyping, the resulting tree places P. orientalis F9 closely to two other P. orientalis strains (Figure 2) within the group of the fluorescent pseudomonads. The phylogeny presented in the phylogenetic tree is in accordance with a recent phylogenomics and systematics study by Gomila et al. (2015) although a detailed comparison is difficult since the taxonomy of many Pseudomonas type strains is outdated.
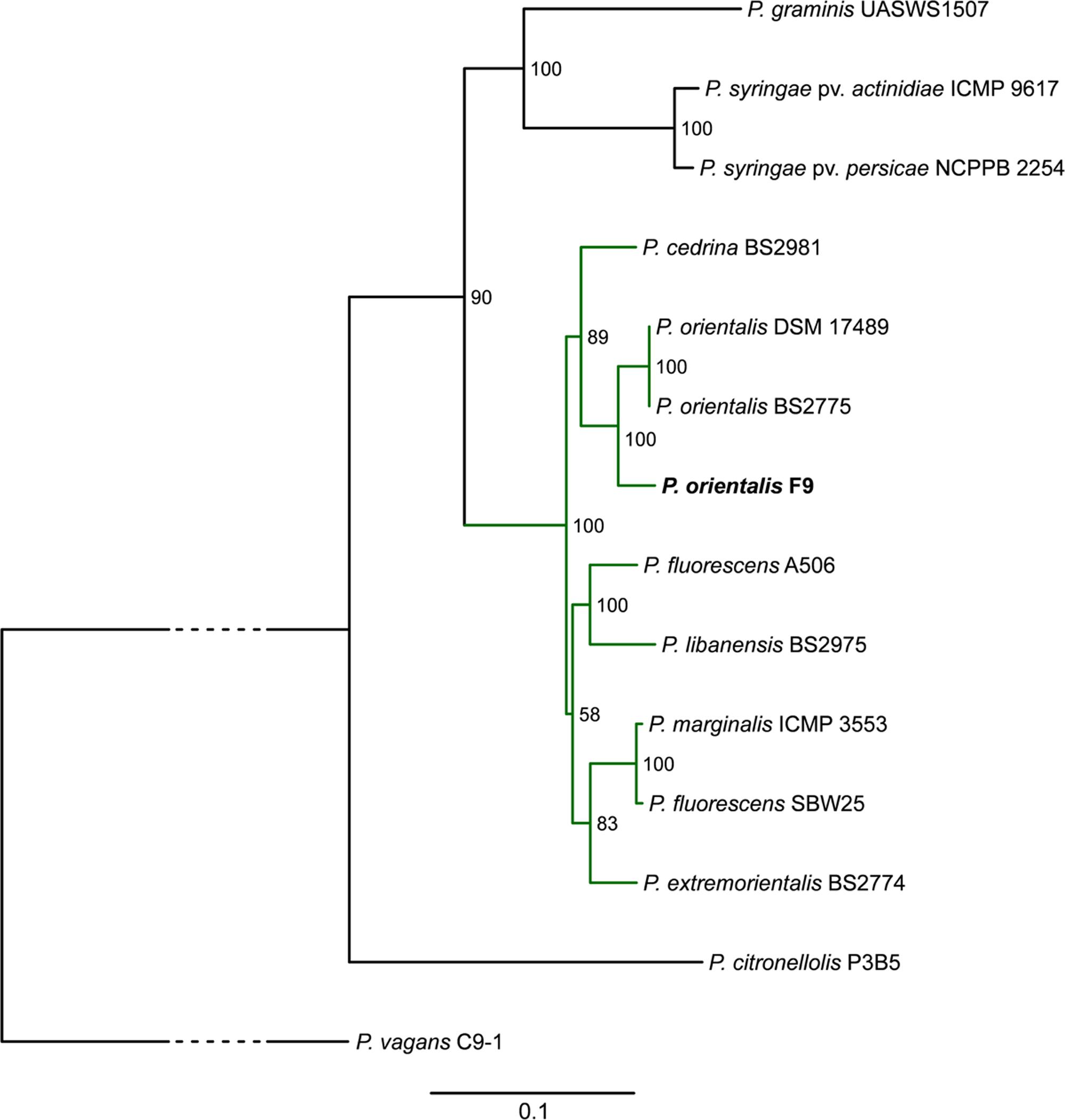
FIGURE 2. Phylogenetic tree highlighting the position of P. orientalis F9 (bold) relative to other representative Pseudomonas species (green subtree is representing the P. fluorescens group). The tree is based on MLSA of the four housekeeping genes gyrB, recA, rpoB, and rpoD. Maximum likelihood based phylogenetic inference was performed with RAxML. Numbers at the branches indicate the percentage of replicate trees in which associated taxa clustered in the bootstrap test (100 replicates). P. vagans C9-1 served as outgroup; the bottom bar reflects the estimated number of nucleotide changes per site between two nodes in the tree. The dashed lines represent regions of the phylogenetic tree where the branches were collapsed. Accession numbers of the strains are given in Supplementary Table 3.
Biochemical Profile and Growth of P. orientalis F9 in PSTB and KB Medium
The metabolic versatility of P. orientalis F9 with regard to the ability to metabolize major components present on the stigma was assessed using Biolog GN2, AN2 and GenIII plates. The strain was able to utilize L-asparagine, L-aspartic acid, L-proline, D-sorbitol but not D-fructose. As P. orientalis F9 was isolated from apple flowers, we evaluated its growth in PSTB medium that mimics the nutrient composition on stigma (Pusey et al., 2008a). P. orientalis F9’s growth in PSTB was compared to that of E. amylovora CFBP1430Rif and P. vagans C9-1 (Figure 3). F9 was growing in the sugar-rich PSTB medium (max growth rate μ = 0.33) but slower than both the fire blight pathogen (μ = 0.39) and P. vagans C9-1 (μ = 0.41). In contrast, P. orientalis F9 grew faster (μ = 0.49) than both strains (CFBP1430Rif: μ = 0.31, C9-1: μ = 0.41) in iron limited KB medium. Due to the ability of F9 to efficiently grow in iron deficient medium, we tested its potential as biocontrol agent under iron limited conditions.
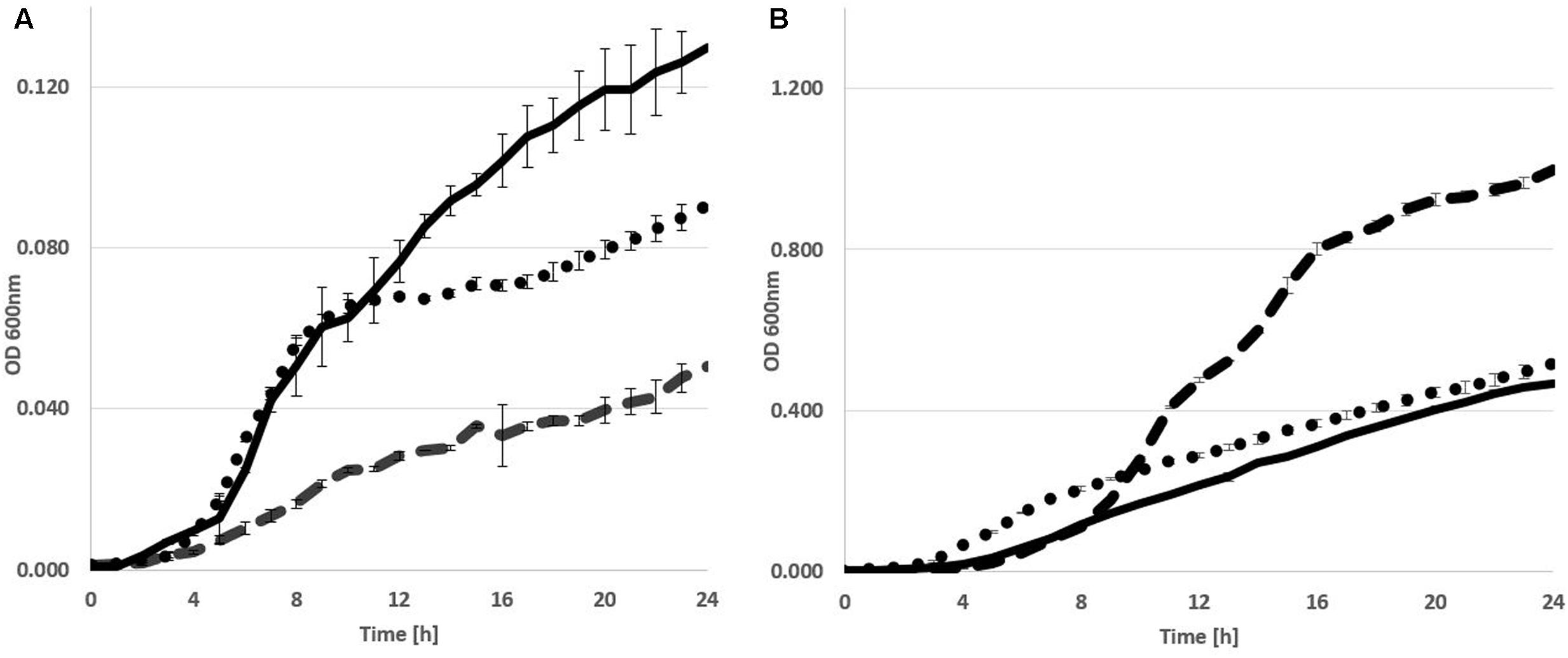
FIGURE 3. Growth curve of E. amylovora CFBP1430 Rif (Ea CFBP1430, black line), P. vagans C9-1 (Pv C9-1, dotted line), and P. orientalis F9 (Po F9, broken line), in PSTB (A) or KB medium (B). Measurement of OD (A600 nm) was performed every 30 min for 24 h. Error bars represent standard deviations. Area under curve in PSTB: Ea CFBP1430 = 1.68 (±0.02); Pv C9-1 = 1.3 (±0.005); Po F9 = 0.59 (±0.007) in KB: Ea CFBP1430 = 5.19 (±0.015); Pv C9-1 = 6.5 (±0.033); Po F9 = 11.2 (±0.033).
P. orientalis F9 Induces Growth Deficiency of E. amylovora CFBP1430Rif and P. syringae Pathovars in Vitro
When analyzing the P. orientalis F9 genome for potential siderophore synthesis genes, genes related to pyoverdine production were detected (CDSs BOP93_10400 to BOP93_10440). Pyoverdine is the generic name given to a vast family of fluorescent green-yellowish pigments produced by Pseudomonas species and represents their primary siderophore (Cornelis and Matthijs, 2002; Cornelis, 2010).
The impact of pyoverdine on the growth of E. amylovora CFBP1430 in vitro was analyzed in a double layer assay. The assay was performed with iron limited KB medium and E. amylovora CFBP1430Rif seeded in the top layer. Application of P. orientalis F9 onto the surface of the top layer led to a clear growth halo of E. amylovora CFBP1430Rif, that could not be abrogated by addition of FeCl3 (Figure 4). To test if P. orientalis F9 affects the growth of additional bacteria, plant pathogenic strains P. syringae pv. syringae ACW (causing bacterial canker of pome and stonefruit, P. syringae pv. actinidiae ICMP 9617 [bacterial canker of kiwifruit (Takikawa et al., 1989)], and P. syringae pv. persicae NCPPB 2254 (bacterial die-back in peach, nectarine, Japanese plum) and also P. vagans C9-1, were added to the assay (Paulin and Samson, 1973; Ishimaru, 1988; Young, 1988; Takikawa et al., 1989).
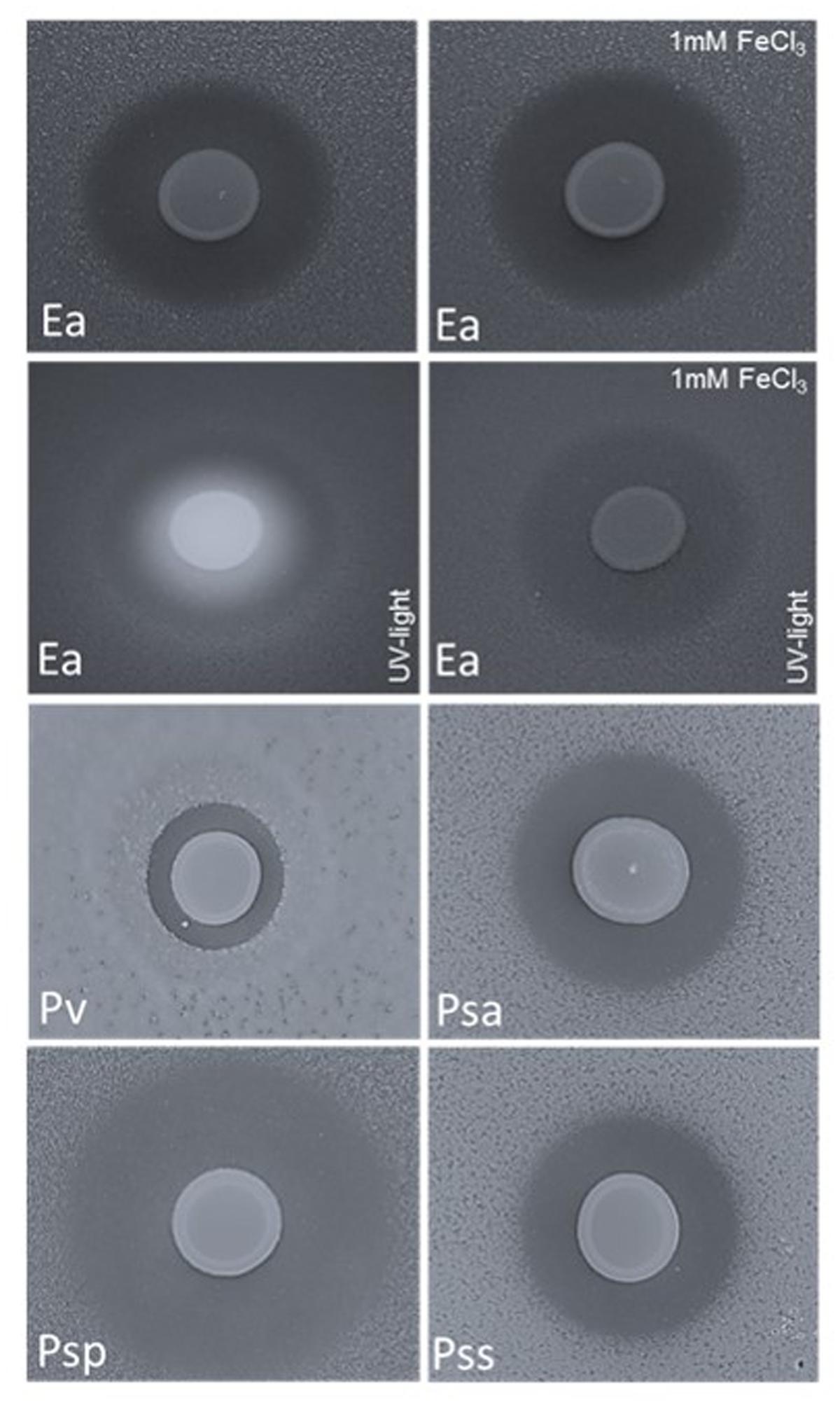
FIGURE 4. Double layer assay with E. amylovora CFBP1430Rif (Ea); P. vagans C9-1 (Pv, halo size 0.3 cm); P. syringae pv. actinidiae ICMP 9617 (Psa, halo size 0.7 cm); P. syringae pv. persicae NCPPB 2254 (Psp, halo size 1.2 cm) and P. syringae pv. syringae ACW (Pss, halo size 0.6) seeded in the top layer and P. orientalis F9 applied onto the surface (10 μl of a P. orientalis F9 suspension of OD600 nm = 1). Application of Fe3+ (1 mM FeCl3) to the medium had no impact on the growth halo formed by the fire blight pathogen but abolished the fluorescence that appears concomitantly with siderophore production.
P. orientalis F9 induced halos in all tested strains after 1–2 days of incubation (Figure 4). Again, addition of iron to the medium had no visible impact on halo formation or halo size (data not shown) implying that iron deprivation caused by pyoverdine synthesis of P. orientalis F9 is not the basic cause for growth deficiency of the tested strains.
The Genome of P. orientalis F9 Harbors the Safracin and Phenazine Clusters
We mined the P. orientalis F9 genome to address the question of potential mode of actions for the demonstrated antagonistic activity and could identify several potential candidate genes: (i) P. orientalis F9 carries a safracin cluster (Locus tags BOP93_17395 to BOP93_17440) that is almost 100% identical to the cluster present in P. fluorescens A2-2 (Accession: AY061859.1). Safracin is a compound with potent broad-spectrum antibacterial activities (Velasco et al., 2005). (ii) F9 harbors, the phzABCDEFG operon (Locus tags BOP93_12290 to BOP93_12320). Phenazine production of P. orientalis strains could also be observed in case of isolates from dryland wheat (Parejko et al., 2013). All phenazine-producing bacteria contain orthologs of this core biosynthesis operon which is required for the conversion of chorismic acid into the broad-spectrum antibiotic phenazine-1-carboxylate. Phenazines are a diverse group of secondary metabolites with broad-spectrum antibiotic activity against bacteria, fungi, and eukaryotes. They are actively involved in the suppression of plant pathogens and phenazines produced by P. agglomerans on apple flowers contribute to the suppression of E. amylovora, (Thomashow et al., 1990; Giddens et al., 2003). Furthermore, genes phzI (BOP93_12330) and phzR (BOP93_12325) that encode a quorum-sensing circuit which regulates the phenazine production (Pierson et al., 1994) are located upstream of the phz-operon.
P. orientalis F9 Proliferates in Apple Flowers
P. orientalis F9 was isolated from apple flowers. To confirm its ability to successfully proliferate in apple flowers the strain was inoculated at low densities (20–70 CFU) onto the hypanthium. It reached population densities of 107 CFU per flower after 5 days of incubation, a value comparable to that reported for other flower colonizing bacteria (Cabrefiga et al., 2007).
P. orientalis F9 Shows Ambiguous Results in the “Detached Flower Assay”
As P. orientalis F9 has the ability to proliferate in apple flowers and induces growth deficiency of E. amylovora CFBP1430Rif in vitro, its antagonistic activity was tested ex vivo using the “detached flower assay” (Pusey, 1997). In two independent experiments detached apple flowers (24 each) were inoculated with binary strain mixtures, combining E. amylovora CFBP1430Rif with either the well-described antagonist P. vagans C9-1 or P. orientalis F9, respectively. As a control, the fire blight pathogen was inoculated as single culture. After 4–5 days of incubation the flowers were visually rated (Figure 5). In both trials, less than 10% of the flowers infected with the pathogen only were evaluated with grade 1 (indicative for a phenotypically healthy flower). The severity of infection for the control was 69 (Ea_a) in the first - and 66 (Ea_b) in the second trial. When co-inoculated with P. vagans C9-1, a significant increase of healthy flowers was observed (severity Ea + Pv_a = 38, Ea + Pv_b = 40). Flowers inoculated with P. orientalis F9 and E. amylovora CFBP1430Rif yielded ambiguous results between the trials. Even though an increase of healthy flowers (ca. 60%) could be detected in the first trial (severity Ea + Po_a = 46), there was no difference to the E. amylovora CFBP1430Rif control infection in the second (severity Ea + Po_b = 65).
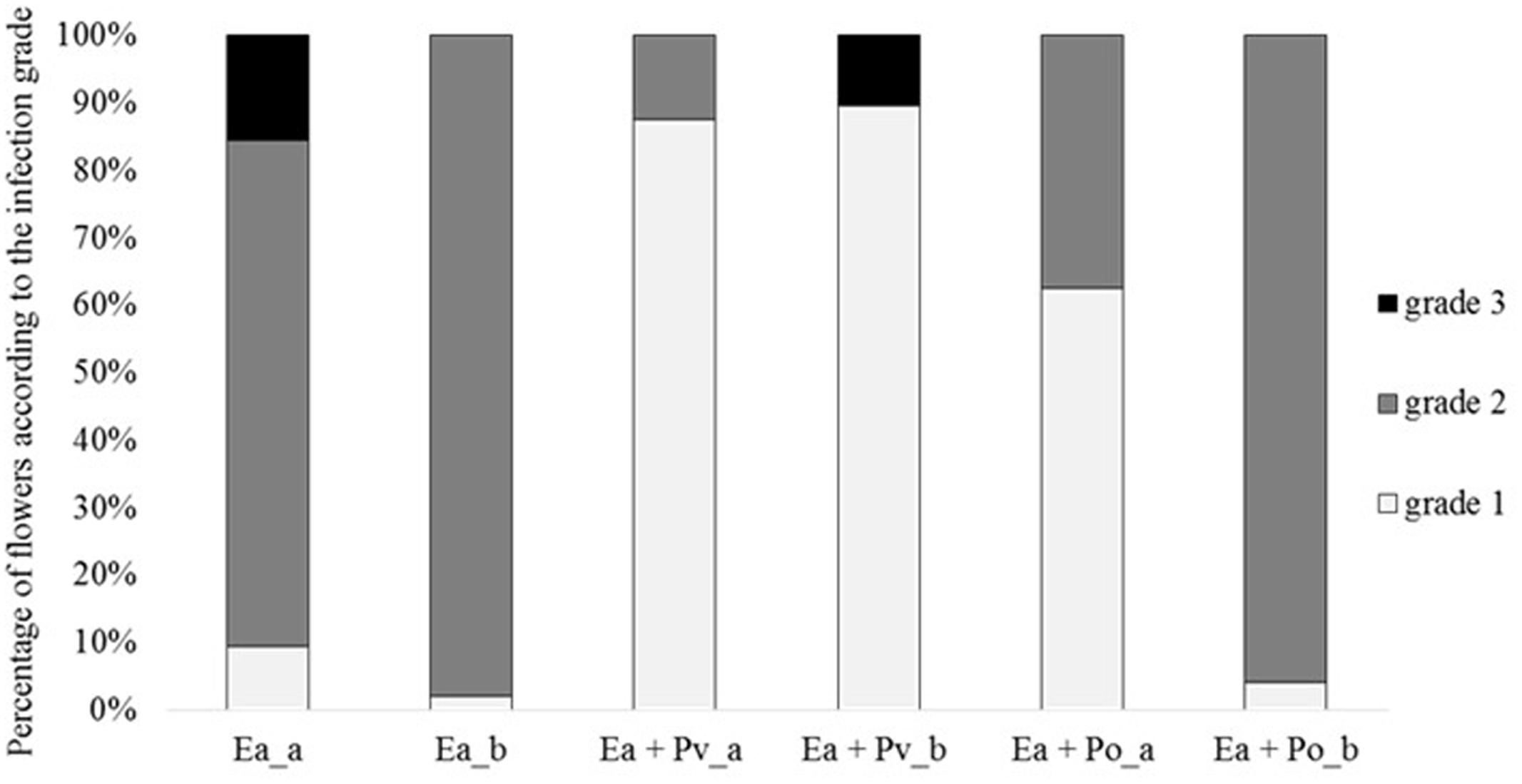
FIGURE 5. Percentage of flowers according to the infection grade of two independent detached flower assay experiments (a and b). Each column represents 32 (a) or 48 (b) flowers. Flowers were inoculated with E. amylovora CFBP1430Rif (Ea), E. amylovora CFBP1430Rif co-inoculated with P. vagans C9-1 (Ea + Pv) or E. amylovora CFBP1430Rif co-inoculated with P. orientalis F9 (Ea + Po). Evaluation of the flowers were performed according to the following scale: grade 1: calyx green; grade 2: calyx necrotic; grade 3: calyx and pedicel necrotic.
Co-inoculation with P. orientalis F9 Diminishes E. amylovora CFBP1430Rif CFU in Apple Flowers
Results of the second trial prompted us to test infected flowers for the presence of E. amylovora. Bacteria were re-isolated from flowers and plated onto rifampicin containing TSB agar to select for E. amylovora CFBP1430Rif. Flowers infected with only E. amylovora, consistently carried > 107 CFU. Co-inoculation with P. vagans C9-1 decreased the number of E. amylovora CFBP1430Rif recovered from flowers, 70% of the flowers carried 105 to 107 E. amylovora. Co-inoculation of E. amylovora CFBP1430Rif with P. orientalis F9 resulted in the lowest numbers of the fire blight pathogen (Figure 6). In 13 apple flowers (27%) no pathogen could be detected. Whilst having the lowest estimated CFU of the pathogen, the visual rating of the E. amylovora CFBP1430Rif/P. orientalis F9 infection was comparable to that of the infection control. This discrepancy points to a phytotoxic potential of P. orientalis F9 in apple flowers.
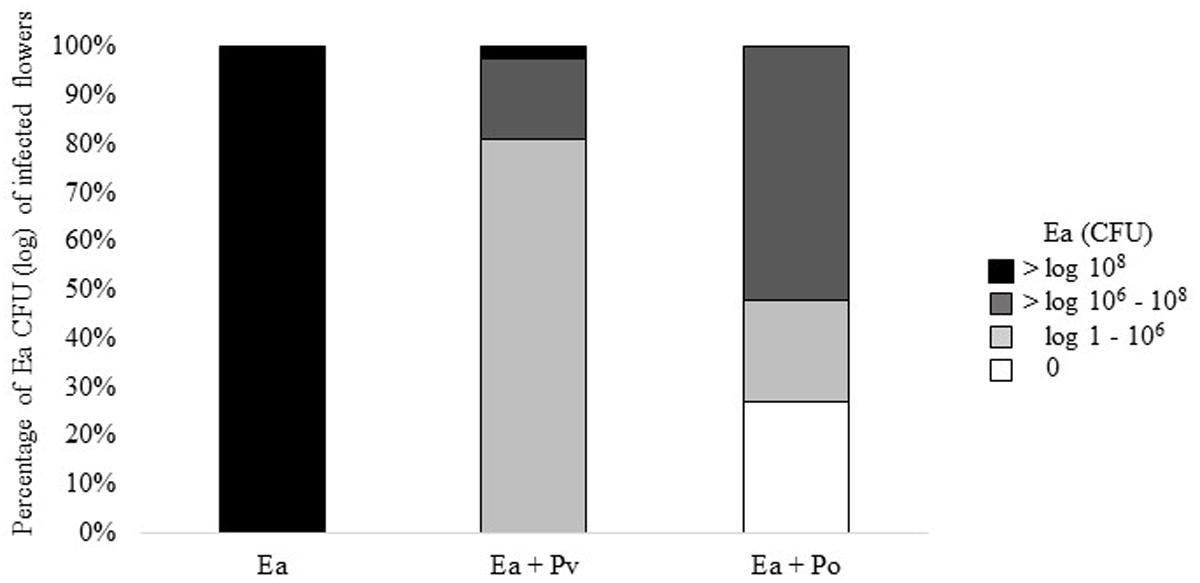
FIGURE 6. Percentage of log CFU and 0 CFU of E. amylovora CFBP1430Rif recovered from infected apple flowers. Forty eight flowers (=100%) were inoculated with E. amylovora CFBP1430Rif (Ea), E. amylovora CFBP1430Rif co-inoculated with P. vagans C9-1 (Ea + Pv) or E. amylovora CFBP1430Rif co-inoculated with P. orientalis F9 (Ea + Po).
P. orientalis F9 Reveals Phytotoxic Traits in Detached Apple Flowers
To evaluate its phytotoxic traits, P. orientalis F9 was inoculated (ca. 5 × 105) onto the hypanthium of apple flowers. Two, four, six, and eight days post infection (dpi), flowers were visually evaluated and the CFU of P. orientalis F9 determined. After day six, half of the flowers showed a necrotic phenotype (Figure 7). The increase of flowers necrosis and CFU correlated, implying the degradation of flowers to be an additional nutrient source for the strain. When mining the F9 genome for P. syringae related phytotoxins (syringomycin, phaseolotoxin, syringopeptin, tabtoxin) as source for the observed phenotype, their presence could not be confirmed.
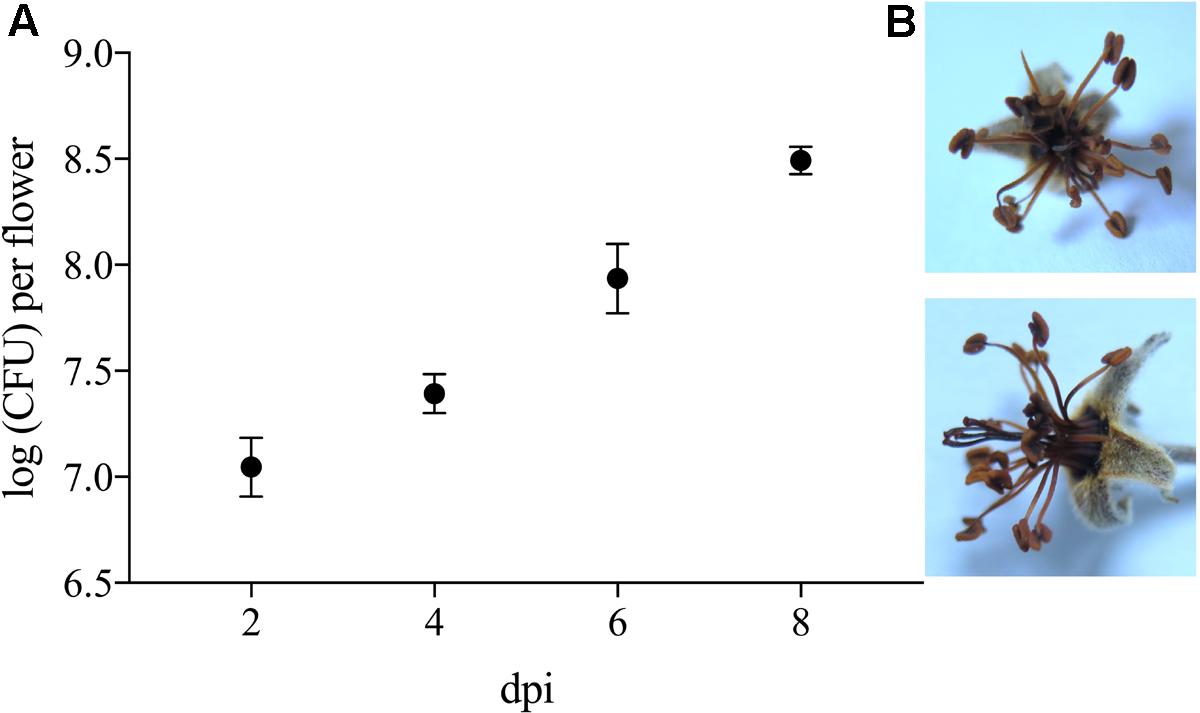
FIGURE 7. (A) Log CFU of P. orientalis F9 recovered from detached apple blossom on day two, four, six and eight post infection. Error bars represent the standard error of the mean. (B) Necrotic flower 6 days post infection.
P. orientalis F9 Exhibits a Protective Phenotype in the Cress Assay
Genome mining revealed that P. orientalis F9 is possessing the phenazine cluster, which is also present in pseudomonas strains that antagonize wheat pathogens (Jaaffar et al., 2017). To evaluate the potential of F9 as antagonist of soil-borne pathogens we used the cress assay (Rosendahl and Olson, 1992). The oomycete P. ultimum was used as a model soil pathogen and P. protegens CHA0 served as a reference strain for antagonistic activity. CHA0 is a biocontrol agent that protects cucumber from several fungal pathogens, including Pythium spp. (Keel et al., 1992) and is often used as model strain in biocontrol experiments. Results of the cress assay showed that after infection with P. ultimum the average cress biomass was 0.35 g per petri dish. In presence of P. ultimum and P. protegens CHA0 the biomass increased to 1.13 g on average. When treated with P. ultimum and P. orientalis F9, the biomass accumulated to 1.32 g (Figure 8). Both pseudomonads have an effect on cress weight when compared to mock treated controls cress only. There is no evidence that this is due to the phytotoxic effect that F9 exhibits on blossoms, as both strains have the effect (Figure 8).
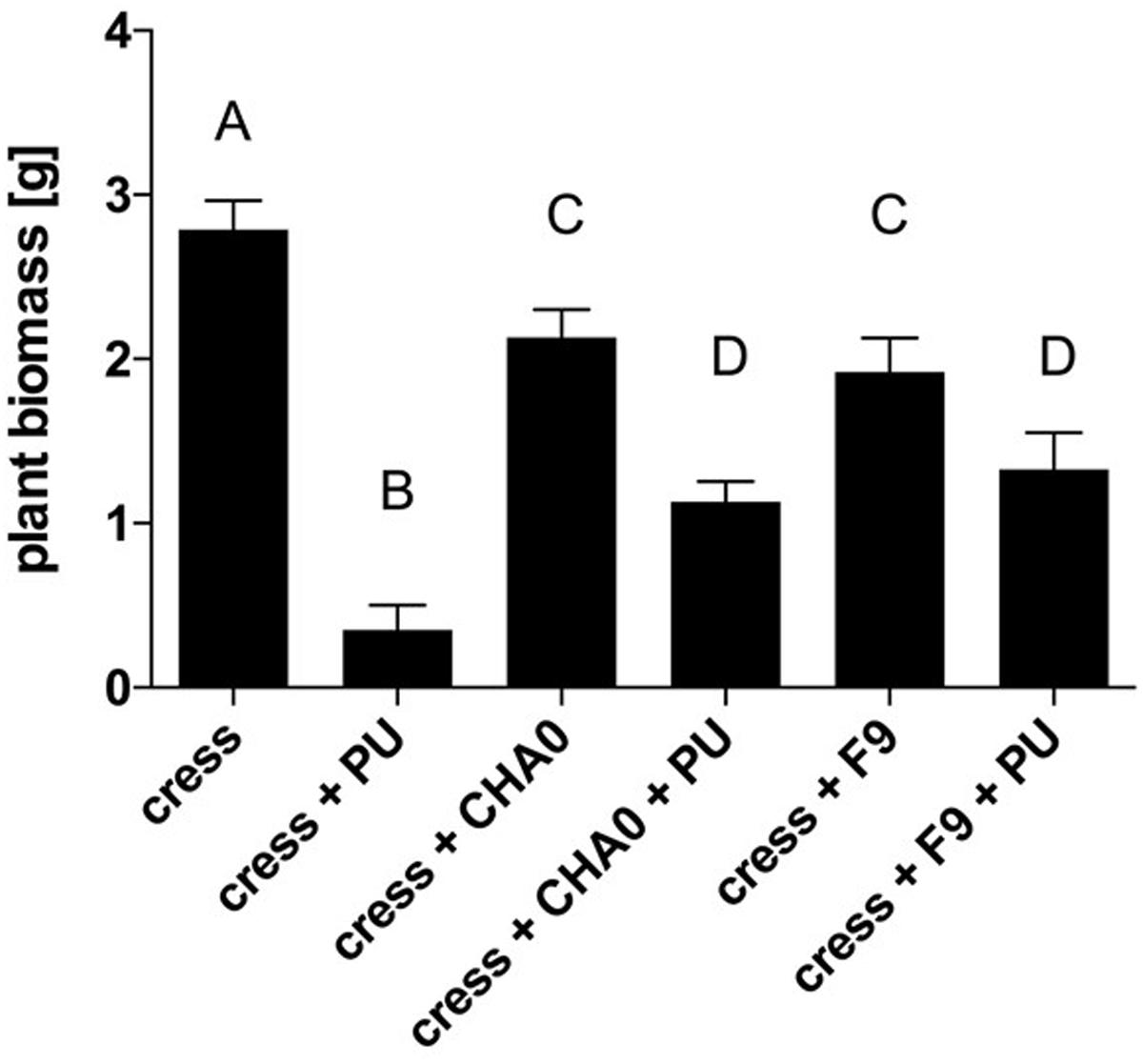
FIGURE 8. Cress biomass after treatment with P. ultimum (PU), P. protegens CHA0 (CHA0), and/or P. orientalis F9 (Po). Cress biomass was harvested 7 days post inoculation. Error bars represent standard error of the means. Bars with the same letter represent results that are not significantly different (P > 0.05, one-way ANOVA with Tukey’s multiple comparisons test).
Discussion
In light of public concerns over the use of antibiotics and also pesticides in plant protection, effective biological control in form of antagonistic bacteria should be considered as an alternative in fire blight management. A prerequisite for the successful application of antagonists with high biocontrol efficacy and stability is a broad set of well-defined microorganisms with antagonistic activity and an understanding of their modes of action. This would allow to choose optimal antagonists for application under a wide range of given environmental conditions and diseases. Pseudomonads, known for their metabolic versatility and genetic plasticity, encompass plant pathogens but also antagonists that are being applied against plant pathogens including E. amylovora, the cause of fire blight (Wilson and Lindow, 1993; Galasso et al., 2002; Paternoster et al., 2010; Mikiciński et al., 2016).
We isolated bacterial strain F9, from apple flowers in a Swiss orchard. F9 was classified by MALDI-TOF biotyping as P. orientalis. We sequenced and de novo assembled the complete genome of P. orientalis F9 using a hybrid approach of PacBio long reads and Illumina MiSeq data, providing the basis for accurate and comprehensive genome annotation (Omasits et al., 2017). The assembly of the reads revealed a 5.99 Mbp genome with an average GC content of 60.4% and 5,142 predicted protein coding genes. A MLSA with four housekeeping genes of both strain F9 and twelve related Pseudomonas species strains confirmed F9 to be a P. orientalis strain. To the best of our knowledge, the here presented F9 genome represents the first complete and circularized full genome sequence of a P. orientalis strain (Supplementary Table 2).
Pseudomonas orientalis was first described in Dabboussi et al. (1999); but even to date an in-depth characterization of the species is lacking. We tested P. orientalis F9 for its antagonistic activity against E. amylovora CFBP 1430. The pathogen grows epiphytically on stigmata before it enters through the nectaries from where it spreads into the tissue, resulting in flower and shoot blight symptoms (Thomson, 2000). Current control efforts focus on the limitation of the pathogen in the flower. The application of antibiotics (e.g., streptomycin) is thus far the most successful control measure against fire blight, however, it is also controversially discussed, due to concerns about possible antibiotic resistance spread (Jones and Schnabel, 2000; McManus et al., 2002). In vitro the strain successfully antagonized the fire blight pathogen in the KB double layer assay. In addition, F9 induced growth deficiency not only in E. amylovora CFBP1430, but also in several tested P. syringae pathovar strains and in P. vagans C9-1, an E. amylovora antagonist (Figure 4). We tested if F9‘s mode of action in the assay is based on siderophore production as KB is an iron deficient medium. Under iron limiting conditions bacteria secrete siderophores, high affinity iron chelators that bind Fe3+ and thereby make it available for the cell (Braun and Winkelmann, 1987; Neilands, 1995). E. amylovora harbors one siderophore system; the hydroxamate siderophore desferrioxamine E (DFO E). The importance of this system for the pathogenicity of E. amylovora has been demonstrated by siderophore synthesis or uptake mutants that exhibit decreased pathogenicity on apple flowers (Dellagi et al., 1998).
Pseudomonads are known for their large repertoire of siderophores and pyoverdines are the main siderophores produced by fluorescent pseudomonads. F9 exhibits strong fluorescence when cultivated on KB medium and indeed, the strain harbors pyoverdine synthesis genes in the genome (Cornelis and Matthijs, 2002). However, as the induced growth halos neither disappeared nor were reduced in size after addition of iron to the medium, siderophore production of F9 and subsequent iron deprivation of the tested strains are unlikely to be the cause for growth inhibition. An alternative explanation for the ability of F9 to serious growth deficiencies in strains belonging to different genera is given by the biosynthesis genes of phenazine and safracin that are located on the chromosome. Both components have anti-bacterial activity (Velasco et al., 2005). Phenazines are known to antagonize plant pathogens especially in the rhizosphere but also in the apple flower (Giddens et al., 2003). For Pantoea strains such as P. agglomerans Eh325, P. agglomerans Eh318 or P. vagans C9-1, the antagonistic effect against the fire blight pathogen is mainly attributed to the production of antibiotics (Wright et al., 2001; Stockwell et al., 2002; Pusey et al., 2011; Kamber et al., 2012). In case of P. agglomerans Eh325 the antibiotic is even specific for E. amylovora (Pusey et al., 2008b) which is not true for the F9 antibiotics that exhibited a broad host range and effected all bacteria tested. P. synxantha 2-79 is known for its antagonistic activity against the fungus Gaeumannomyces graminis var. tritici, a major root disease of wheat. P. synxantha 2–79 produces phenazine and pyoverdin. Pyoverdin mutants of P. synxantha 2–79 controlled the pathogen as effectively as the fluorescent parental strains, revealing the phenazine antibiotic to be the dominant factor in the antagonistic activity of the strain (Hamdan et al., 1991).
F9’s promising antagonistic traits in the in vitro double layer assay prompted us to test for its antagonistic activity against the fire blight pathogen ex vivo. Before performing the “detached flower assay” the growth of P. orientalis F9 was tested in detached apple flowers. The strain not only persisted but successfully proliferated in apple flowers and reached up to 107 CFU. In the following two independent trials P. orientalis F9 was co-inoculated with E. amylovora CFBP1430Rif in detached apple flowers. While an increase of healthy flowers (ca. 60%) could be detected in the first trial, there was no difference to the E. amylovora CFBP1430Rif control infection in the second (Figure 5). In contrast to the visual rating, the CFU of E. amylovora CFBP1430Rif in apple flowers co-inoculated with P. orientalis F9 flowers was lower than in the control with 13 apple flowers (27%) free of the pathogen. The discrepancy between visual rating and CFU of E. amylovora in the detached flower assay can be explained by phytotoxicity of P. orientalis F9 in apple flowers that could be confirmed by inoculation of flowers with F9 only. Flower necrosis and increase of F9 CFU correlated. Necrosis was only detectable when the initial inoculate of the strain was approximately 106 CFU. Flowers inoculated with less than 102 bacteria showed no necrotic symptoms after 5 dpi and presence of P. syringae related phytotoxins (syringomycin, phaseolotoxin, syringopeptin, tabtoxin) on the F9 chromosome could not be confirmed. A recent publication showed that P. orientalis strains may be pathogenic to citrus species after leaf infiltration (Beiki et al., 2016). Here, we provide further evidence that P. orientalis may act as a plant pathogen, however, we propose that on plant it is an opportunistic pathogen rather than a bona fide disease agent. Data mining of the P. orientalis F9 genome, implied F9 to be capable of producing phenazines. Phenazines producing P. orientalis strains successfully antagonize wheat pathogens (Jaaffar et al., 2017). Thus, the ability of F9 as protective agent against soil-born pathogens was tested in the cress assay. Indeed, when compared to the extensively studied antagonist P. protegens CHA0, P. orientalis F9 protected cress to a comparable degree (Figure 8). The antagonistic activity of P. protegens CHA0 is not attributed to phenazines but mainly to the production of the antimicrobial polyketides 2,4-diacetylphloroglucinol and pyoluteorin. P. orientalis F9 is as efficient as CHA0 in the cress assay. It has to be elucidated, if these antagonistic traits are effective in longer term protection experiments and which genes are responsible for the exhibited traits. In the presented paper genome mining was the tool that led to the shift in antagonistic studies of a bacterial isolate from phyllosphere to rhizosphere with promising results. Genome comparison of F9 to additional P. orientalis and Pseudomonas strains will be an important tool to understand and characterize pseudomonads and their mode of actions. In the future, it might reveal additional factors that specify the environment in which F9 evolve its optimal potential as an antagonist.
MALDI-TOF biotyping and genome sequencing are modern techniques that facilitate the selection of antagonists and may ultimately provide the basis to unravel their mode of action. MALDI-TOF is a fast tool to eliminate phytopathogenic or restricted bacteria from the antagonist screening process. Annotated complete genome sequences of potential and tested antagonists can provide a first indication of underlying mechanisms and of the genes involved by e.g., applying principles of gene trait matching. Further validations, e.g., constructing mutants of the in silico predicted genes can confirm their significance in the antagonistic activity of bacteria. Future approaches in the evaluation process of potential antagonists will involve genome mining complemented with gene expression analysis under different conditions (Omasits et al., 2013). This will not only provide important information to decipher genes and pathways active under the given conditions but also potential mechanisms underlying the antagonistic effects, criteria that can be used for an improved strain selection.
Author Contributions
VZ isolated the strain, performed detached flower assays, inhibition assays, MALDI, and determined the CFU. MS planned and performed the bioinformatic analysis, assembled and annotated the genome, performed phylogenetic analysis and contributed to the writing of the manuscript. MB performed studies on the proliferation of F9 in the apple flowers. DM performed the cress assay and critically read the manuscript. MR-E performed and analyzed the biochemical characterization, analyzed data and wrote the manuscript. CA planned and supervised the bioinformatic analysis, and contributed to the writing of the manuscript. CP conceived the study, isolated DNA, performed the CAS assay and wrote the manuscript. All authors critically read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Elisabeth Eugster Meier and Barbara Guggenbühl for launching the Agroscope research program, Microbial Diversity (MicBioDiv), their colleagues for constructive discussions, Monika Maurhofer and Matthias Lutz for providing P. protegens CHA0 and P. ultimum, and acknowledge the support of the Functional Genomics Center Zurich. This work was financially supported by the Agroscope research program MicBioDiv.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00145/full#supplementary-material
References
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beiki, F., Busquets, A., Gomila, M., Rahimian, H., Lalucat, J., and García-Valdés, E. (2016). New Pseudomonas spp. are pathogenic to citrus. PLOS ONE 11:e0148796. doi: 10.1371/journal.pone.0148796
Born, Y., Fieseler, L., Thöny, V., Leimer, N., Duffy, B., and Loessner, M. J. (2017). Engineering of bacteriophages Y2::dpoL1-C and Y2::luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 83, e00341-17. doi: 10.1128/AEM.00341-17
Braun, V., and Winkelmann, G. (1987). Microbial iron transport structure and function of siderophores. Prog. Clin. Biochem. Med. 5, 67–99. doi: 10.1007/978-3-642-72902-7_4
Cabrefiga, J., Bonaterra, A., and Montesinos, E. (2007). Mechanisms of antagonism of Pseudomonas fluorescens EPS62e against Erwinia amylovora, the causal agent of fire blight. Int. Microbiol. 10, 123–132.
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Chiou, C. S., and Jones, A. L. (1995). Expression and identification of the strA-strB gene pair from streptomycin-resistant Erwinia amylovora. Gene 152, 47–51. doi: 10.1016/0378-1119(94)00721-4
Cornelis, P. (2010). Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645. doi: 10.1007/s00253-010-2550-2
Cornelis, P., and Matthijs, S. (2002). Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4, 787–798. doi: 10.1046/j.1462-2920.2002.00369.x
Dabboussi, F., Hamze, M., Elomari, M., Verhille, S., Baida, N., Izard, D., et al. (1999). Taxonomic study of bacteria isolated from Lebanese spring waters: proposal for Pseudomonas cedrella sp.nov. and P. orientalis sp. nov. Res. Microbiol. 150, 303–316. doi: 10.1016/S0923-2508(99)80056-4
Davis, L. G., Dibner, M. D., and Battley, J. F. (1990). Basic Methods in Molecular Biology. New York, NY: Elsevier.
Dellagi, A., Brisset, M. N., Paulin, J. P., and Expert, D. (1998). Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 11, 734–742. doi: 10.1094/MPMI.1998.11.8.734
Dhillon, B. K., Laird, M. R., Shay, J. A., Winsor, G. L., Lo, R., Nizam, F., et al. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43, W104–W108. doi: 10.1093/nar/gkv401
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2007). PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics 8:18. doi: 10.1186/1471-2105-8-18
Frank, A. C., and Lobry, J. R. (2000). Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 16, 560–561. doi: 10.1093/bioinformatics/16.6.560
Freimoser, F. M., Pelludat, C., and Remus-Emsermann, M. N. P. (2015). Tritagonist as a new term for uncharacterised microorganisms in environmental systems. ISME J. 10, 1–3. doi: 10.1038/ismej.2015.92
Galasso, O., Sponza, G., Bazzi, C., and Vanneste, J. L. (2002). Characterization of two fluorescent strains of Pseudomonas as biocontrol agents against fire blight. Acta Hortic. 590, 299–307. doi: 10.17660/ActaHortic.2002.590.44
Gao, F., and Zhang, C.-T. (2008). Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 9:79. doi: 10.1186/1471-2105-9-79
Gekenidis, M.-T., Studer, P., Wüthrich, S., Brunisholz, R., and Drissner, D. (2014). Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: in search of microorganism-specific tryptic peptides enabling discrimination of subspecies. Appl. Environ. Microbiol. 80, 4234–4241. doi: 10.1128/AEM.00740-14
Giddens, S. R., Houliston, G. J., and Mahanty, H. K. (2003). The influence of antibiotic production and pre-emptive colonization on the population dynamics of Pantoea agglomerans (Erwinia herbicola) Eh1087 and Erwinia amylovora in planta. Environ. Microbiol. 5, 1016–1021. doi: 10.1046/j.1462-2920.2003.00506.x
Glaeser, S. P., and Kämpfer, P. (2015). Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 38, 237–245. doi: 10.1016/j.syapm.2015.03.007
Gomila, M., Peña, A., Mulet, M., Lalucat, J., and García-Valdés, E. (2015). Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 6:214. doi: 10.3389/fmicb.2015.00214
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hamdan, H., Weller, D. M., and Thomashow, L. S. (1991). Relative importance of fluorescent siderophores and other factors in biological control of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2-79 and M4-80R. Appl. Environ. Microbiol. 57, 3270–3277.
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. doi: 10.1186/s13059-015-0849-0
Imperiali, N., Dennert, F., Schneider, J., Laessle, T., Velatta, C., Fesselet, M., et al. (2017). Relationships between root pathogen resistance, abundance and expression of pseudomonas antimicrobial genes, and soil properties in representative swiss agricultural soils. Front. Plant Sci. 8:427. doi: 10.3389/fpls.2017.00427
Ishimaru, C. A. (1988). Multiple antibiotic production by Erwinia herbicola. Phytopathology 78, 746–750. doi: 10.1094/Phyto-78-746
Jaaffar, A. K. M., Parejko, J. A., Paulitz, T. C., Weller, D. M., and Thomashow, L. S. (2017). Sensitivity of Rhizoctonia isolates to phenazine-1-carboxylic acid and biological control by phenazine-producing Pseudomonas spp. Phytopathology 107, 692–703. doi: 10.1094/PHYTO-07-16-0257-R
Johnson, K. B., and Stockwell, V. O. (2000). “Biological control of fire blight,” in Fire Blight: the Disease and its Causative Agent, Erwinia amylovora, ed. J. L. Vanneste (Wallingford: CABI Publishing), 319–334. doi: 10.1079/9780851992945.0319
Jones, A. L., and Schnabel, E. L. (2000). “The development of streptomycin-resistant strains of Erwinia amylovora,” in Fire blight: the disease and its causative agent, Erwinia amylovora, ed. J. L. Vanneste (Wallingford: CABI Publishing), 235–251. doi: 10.1079/9780851992945.0235
Kamber, T., Lansdell, T. A., Stockwell, V. O., Ishimaru, C. A., Smits, T. H. M., and Duffy, B. (2012). Characterization of the biosynthetic operon for the antibacterial peptide herbicolin in Pantoea vagans biocontrol strain C9-1 and incidence in Pantoea Species. Appl. Environ. Microbiol. 78, 4412–4419. doi: 10.1128/AEM.07351-11
Keel, C., Schnider, U., Maurhofer, M., Voisard, C., Laville, J., Burger, U., et al. (1992). Suppression of root diseases by Pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5, 4–13. doi: 10.1094/MPMI-5-004
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307.
Koren, S., Schatz, M. C., Walenz, B. P., Martin, J., Howard, J. T., Ganapathy, G., et al. (2012). Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30, 693–700. doi: 10.1038/nbt.2280
Llop, P., Cabrefiga, J., Smits, T. H. M., Dreo, T., Barbé, S., Pulawska, J., et al. (2011). Erwinia amylovora novel plasmid pEl70: complete sequence, biogeography, and role in aggressiveness in the fire blight phytopathogen. PLOS ONE 16:e28651. doi: 10.1371/journal.pone.0028651
Manulis, S., Zutra, D., Kleitman, F., Dror, O., David, I., Zilberstaine, M., et al. (1998). Distribution of streptomycin-resistant strains of Erwinia amylovora in Israel and occurrence of blossom blight in the autumn. Phytoparasitica 26, 223–230. doi: 10.1007/BF02981437
McGhee, G. C., Guasco, J., Bellomo, L. M., Blumer-Schuette, S. E., Shane, W. W., Irish-Brown, A., et al. (2011). Genetic analysis of streptomycin-resistant (SmR) strains of Erwinia amylovora suggests that dissemination of two genotypes Is responsible for the current distribution of SmR E. amylovora in Michigan. Phytopathology 101, 182–191. doi: 10.1094/PHYTO-04-10-0127
McManus, P. S., Stockwell, V. O., Sundin, G. W., and Jones, A. L. (2002). Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465. doi: 10.1146/annurev.phyto.40.120301.093927
Meyer, F., Paarmann, D., D’Souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386
Mikiciński, A., Sobiczewski, P., Puławska, J., and Malusa, E. (2016). Antagonistic potential of Pseudomonas graminis 49M against Erwinia amylovora, the causal agent of fire blight. Arch. Microbiol. 198, 531–539. doi: 10.1007/s00203-016-1207-7
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Omasits, U., Quebatte, M., Stekhoven, D. J., Fortes, C., Roschitzki, B., Robinson, M. D., et al. (2013). Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome. Genome Res. 23, 1916–1927. doi: 10.1101/gr.151035.112
Omasits, U., Varadarajan, A. R., Schmid, M., Goetze, S., Melidis, D., Bourqui, M., et al. (2017). An integrative strategy to identify the entire protein coding potential of prokaryotic genomes by proteogenomics. Genome Res. 27, 2083–2095. doi: 10.1101/153213
Parejko, J. A., Mavrodi, D. V., Mavrodi, O. V., Weller, D. M., and Thomashow, L. S. (2013). Taxonomy and distribution of phenazine-1-carboxylic acid-producing Pseudomonas spp. in the dryland agroecosystem of the Inland Pacific Northwest. Appl. Environ. Microbiol. 79, 3887–3891. doi: 10.1128/AEM.03945-12
Paternoster, T., Défago, G., Duffy, B., Gessler, C., and Pertot, I. (2010). Selection of a biocontrol agent based on a potential mechanism of action: degradation of nicotinic acid, a growth factor essential for Erwinia amylovora. Int. Microbiol. 13, 195–206.
Paulin, J. P., and Samson, R. (1973). Le feu bactérien en France. II.caractères des souches d’Erwinia amylovora (Burril) Winslow et al. 1920, isolées du foyer franco-belge. Annal. Phytopathol. 5, 389–397.
Pierson, L. S. III, Keppenne, V. D., and Wood, D. W. (1994). Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176, 3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994
Pujol, M., Badosa, E., Cabrefiga, J., and Montesinos, E. (2005). Development of a strain-specific quantitative method for monitoring Pseudomonas fluorescens EPS62e, a novel biocontrol agent of fire blight. FEMS Microbiol. 249, 343–352. doi: 10.1016/j.femsle.2005.06.029
Pusey, P. L. (1997). Crab apple blossoms as a model for research on biological control of fire blight. Phytopathology 87, 1096–1102. doi: 10.1094/PHYTO.1997.87.11.1096
Pusey, P. L., Rudell, D. R., Curry, E. A., and Mattheis, J. P. (2008a). Characterization of stigma exudates in aqueous extracts from apple and pear flowers. HortScience 43, 1471–1478.
Pusey, P. L., Stockwell, V. O., Reardon, C. L., Smits, T. H. M., and Duffy, B. (2011). Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 101, 1234–1241. doi: 10.1094/PHYTO-09-10-0253
Pusey, P. L., Stockwell, V. O., and Rudell, D. R. (2008b). Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora. Phytopathology 98, 1136–1143. doi: 10.1094/PHYTO-98-10-1136
Remus-Emsermann, M. N. P., Schmid, M., Gekenidis, M.-T., Pelludat, C., Frey, J. E., Ahrens, C. H., et al. (2016). Complete genome sequence of Pseudomonas citronellolis P3B5, a candidate for microbial phyllo-remediation of hydrocarbon-contaminated sites. Stand. Genomic Sci. 11:75. doi: 10.1186/s40793-016-0190-6
Rosendahl, C. N., and Olson, L. W. (1992). An in vivo screening method for antifungal activity against the plant pathogen Pythium ultimum Trow. J. Phytopathol. 134, 324–328. doi: 10.1111/j.1439-0434.1992.tb01240.x
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Seibold, A., Fried, A., Kunz, S., Moltmann, E., Lange, E., and Jelkmann, W. (2004). Yeasts as antagonists against fireblight. Eppo Bull. 34, 389–390. doi: 10.1111/j.1365-2338.2004.00766.x
Silby, M. W., Winstanley, C., Godfrey, S. A. C., Levy, S. B., and Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stockwell, V. O., Johnson, K. B., Sugar, D., and Loper, J. E. (2002). Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 92, 1202–1209. doi: 10.1094/PHYTO.2002.92.11.1202
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Stutz, E. W., Defago, G., and Kern, H. (1986). Naturally occurring fluorescent pseudomonads involved in suppression of black root-rot of tobacco. Phytopathology 76, 181–185. doi: 10.1094/Phyto-76-181
Takikawa, Y., Serizawa, S., Ichikawa, T., Tsuyumu, S., and Goto, M. (1989). Pseudomonas syringae pv. actinidiae pv. nov.: the causal bacterium of canker of kiwifruit in Japan. Jpn. J. Phytopathol. 55, 437–444. doi: 10.1094/PHYTO-03-12-0064-R
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thomashow, L. S., Weller, D. M., Bonsall, R. F., and Pierson, L. S. (1990). Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56, 908–912.
Thomson, S. V. (2000). “Epidemiology of fire blight,” in Fire Blight: the Disease and its Causative Agent, Erwinia amylovora, ed. J. L. Vanneste (Wallingford: CABI Publishing), 9–36. doi: 10.1079/9780851992945.0009
Velasco, A., Acebo, P., Gomezyu, A., Schleissner, C., Rodríguez, P., Aparicio, T., et al. (2005). Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2-2: designing new cytotoxic compounds. Mol. Microbiol. 56, 144–154. doi: 10.1111/j.1365-2958.2004.04433.x
Wilson, M., and Lindow, S. E. (1993). Interaction between the biological control agent Pseudomonas fluorescens A506 and Erwinia amylovora in pear blossoms. Phytopathology 83, 117–123. doi: 10.1094/Phyto-83-117
Wright, S. A. I., Zumoff, C. H., Schneider, L., and Beer, S. V. (2001). Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 67, 284–292. doi: 10.1128/AEM.67.1.284-292.2001
Keywords: Pseudomonas orientalis, Erwinia amylovora, antagonistic traits, phytotoxicity, whole genome sequencing
Citation: Zengerer V, Schmid M, Bieri M, Müller DC, Remus-Emsermann MNP, Ahrens CH and Pelludat C (2018) Pseudomonas orientalis F9: A Potent Antagonist against Phytopathogens with Phytotoxic Effect in the Apple Flower. Front. Microbiol. 9:145. doi: 10.3389/fmicb.2018.00145
Received: 28 September 2017; Accepted: 23 January 2018;
Published: 09 February 2018.
Edited by:
Jesús Mercado-Blanco, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Gustavo Santoyo, Universidad Michoacana de San Nicolás de Hidalgo, MexicoJordi Cabrefiga Olamendi, University of Girona, Spain
Copyright © 2018 Zengerer, Schmid, Bieri, Müller, Remus-Emsermann, Ahrens and Pelludat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cosima Pelludat, cosima.pelludat@agroscope.admin.ch
†These authors have contributed equally to this work.
 Veronika Zengerer
Veronika Zengerer Michael Schmid
Michael Schmid Marco Bieri
Marco Bieri Denise C. Müller
Denise C. Müller Mitja N. P. Remus-Emsermann
Mitja N. P. Remus-Emsermann Christian H. Ahrens
Christian H. Ahrens Cosima Pelludat
Cosima Pelludat