- 1Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 2NUS Synthetic Biology for Clinical and Technological Innovation (SynCTI), Life Sciences Institute, National University of Singapore, Singapore, Singapore
- 3Agency for Science, Technology and Research (A∗STAR), Institute of Chemical and Engineering Sciences, Singapore, Singapore
Living organisms have evolved over millions of years to fine tune their metabolism to create efficient pathways for producing metabolites necessary for their survival. Advancement in the field of synthetic biology has enabled the exploitation of these metabolic pathways for the production of desired compounds by creating microbial cell factories through metabolic engineering, thus providing sustainable routes to obtain value-added chemicals. Following the past success in metabolic engineering, there is increasing interest in diversifying natural metabolic pathways to construct non-natural biosynthesis routes, thereby creating possibilities for producing novel valuable compounds that are non-natural or without elucidated biosynthesis pathways. Thus, the range of chemicals that can be produced by biological systems can be expanded to meet the demands of industries for compounds such as plastic precursors and new antibiotics, most of which can only be obtained through chemical synthesis currently. Herein, we review and discuss novel strategies that have been developed to rewrite natural metabolic blueprints in a bid to broaden the chemical repertoire achievable in microorganisms. This review aims to provide insights on recent approaches taken to open new avenues for achieving biochemical production that are beyond currently available inventions.
Introduction
Nature’s strength and beauty come from its diversity in biochemical systems, which not only generate but also degrade essential and non-essential chemical substances in living single cells or multicellular organisms through different biochemical reactions (i.e., metabolic pathways) that collectively form cellular metabolism. Consequently, a diverse range of biochemicals are present in nature. Many of these biochemicals are secondary metabolites to the native organisms but are of high biotechnological value to industries (Oksman-Caldentey and Inze, 2004; Harvey, 2008; Dhakal et al., 2017). Characterization of these secondary metabolites and exploration of the metabolic networks involved can potentially enable sustainable production of valuable and useful chemicals (Oksman-Caldentey and Inze, 2004). Although metabolic engineering has enabled the bioproduction of many valuable chemicals and has realized the aim of industrial-scale bio-based manufacturing of important compounds (e.g., 1,3-propanediol and artemisinin) (Zhu and Jackson, 2015), the range of compounds that can be generated are generally limited to those that occur naturally in living systems and with known biosynthesis pathways. Furthermore, the array of value-added chemicals required by industries is highly varied and many of these compounds can only be chemically synthesized as they are non-natural, and hence beyond the biosynthesis capabilities of existing biological systems. For example, different metabolic architectures have been identified from various organisms to produce carboxylic acids, which are central compounds in cellular metabolism (Lidén, 2017), thereby presenting promising sources for industrially important building blocks to manufacture bio-commodities, such as bioplastics. However, silicon-based materials can only be produced chemically because of the absence of organosilicon molecules in living organisms (Apeloig et al., 2001). Likewise, important secondary metabolites, such as polyketides and alkaloids, isolated from organisms (e.g., plants and marine microorganisms) can provide alternative therapeutics against health threats such as multidrug resistance bacteria and intractable cancers (Dhakal et al., 2016). Yet, while more potent analogs of these natural products may be discovered and obtained chemically (DeChristopher et al., 2012), novel biosynthesis routes to these superior non-natural drugs are elusive. Thus, it is imperative to diversify natural metabolic pathways to conceive novel ones that are capable of producing any desirable chemical through biological means. Herein, we will review novel strategies for rewriting natural metabolic blueprints and designing of biosynthetic pathways with computational tools (Figures 1, 2 and Table 1). We aim to provide current insights and future perspectives on how progress in the state-of-the-art approaches in metabolic pathway diversification for the production of novel value-added compounds will eventually facilitate the development of efficient designer microorganisms that can potentially meet most of the chemical needs of modern civilization.
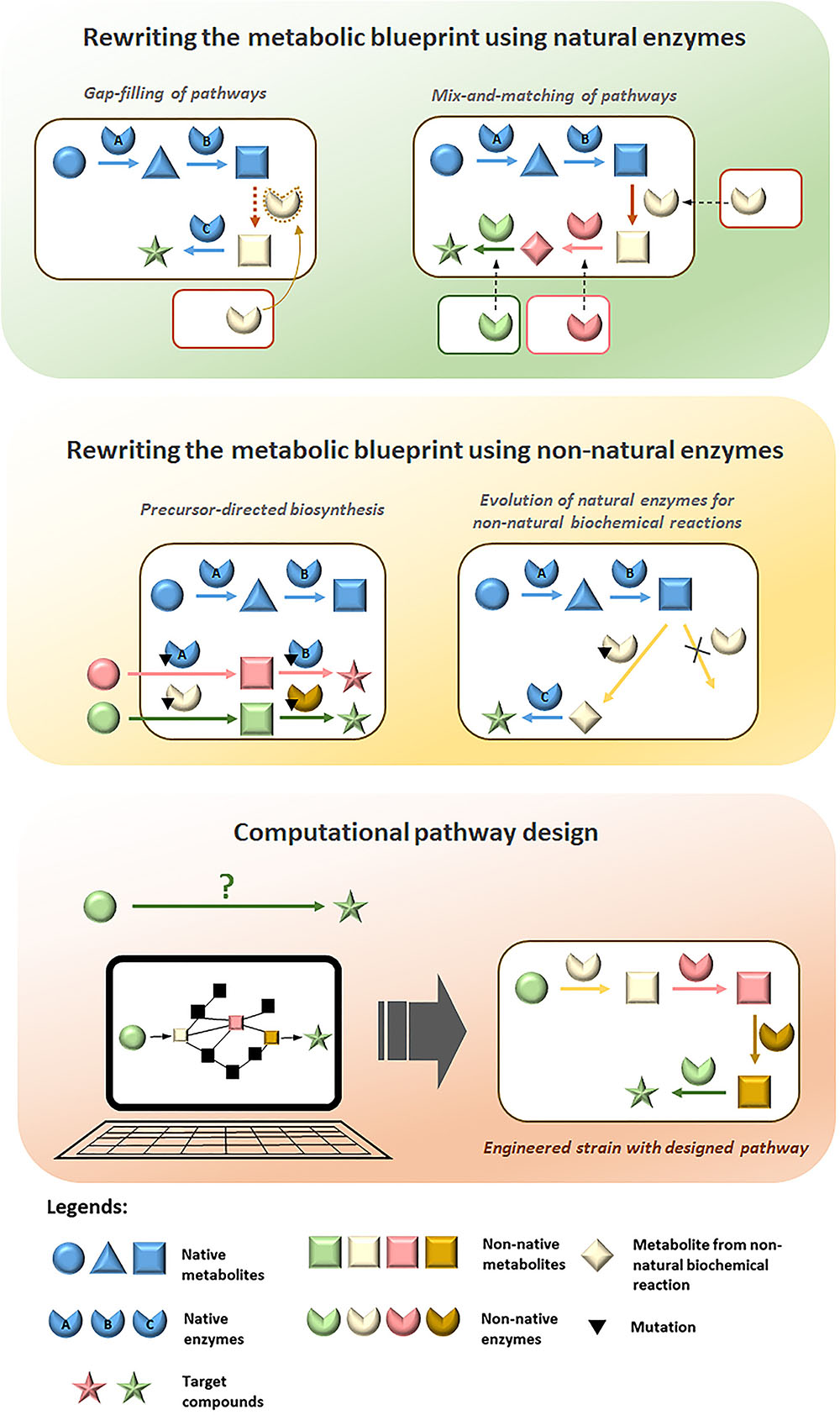
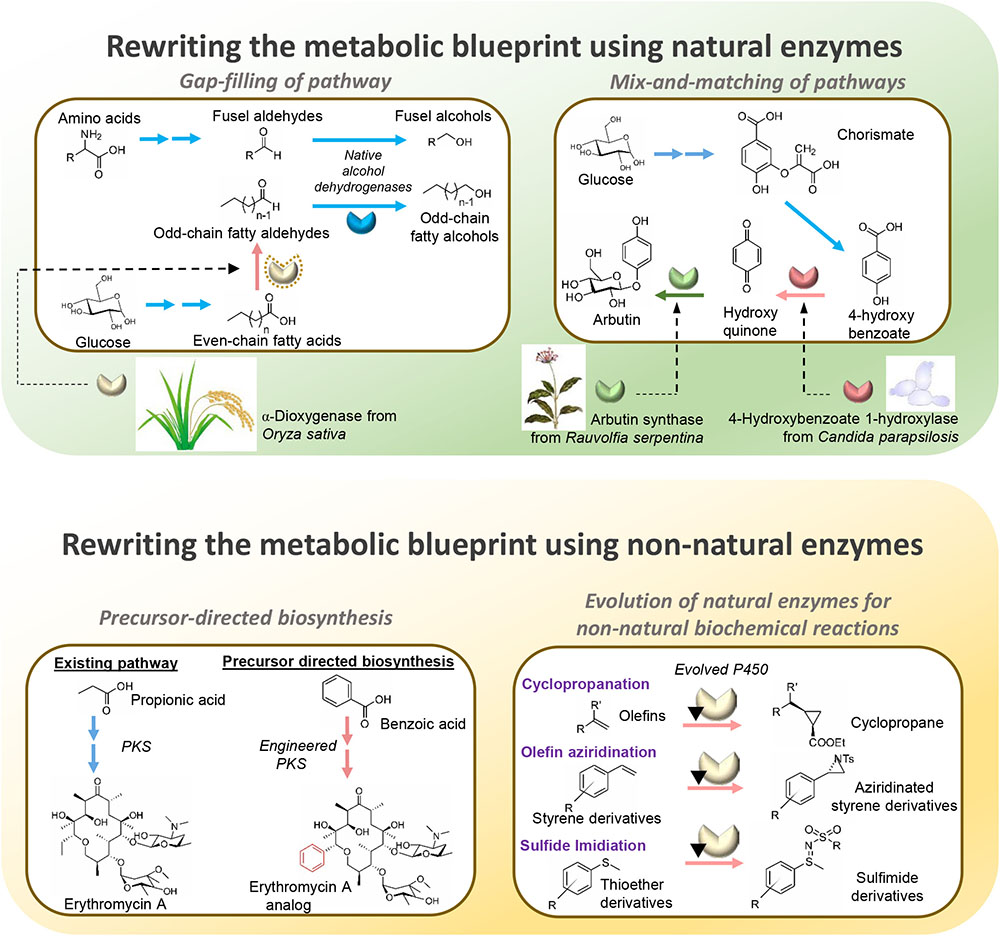
FIGURE 1. Overview of strategies employed for diversification of metabolic pathways to produce value-added compounds in microorganisms.
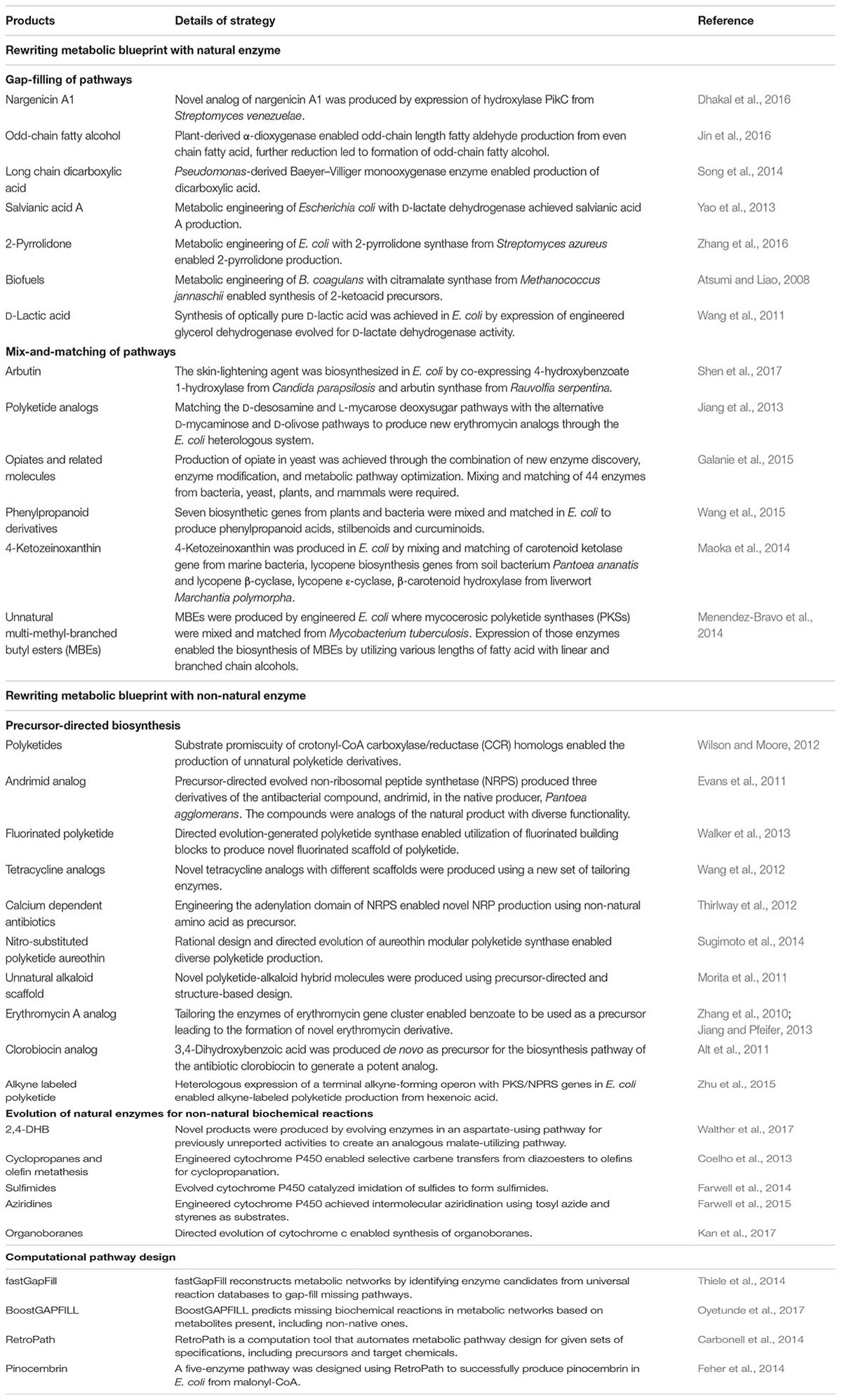
TABLE 1. Examples of notable novel valuable compounds produced in microbial cell factories through diversification of metabolic pathways and computational tools developed for pathway design.
Strategies to Diversify Metabolic Pathway for Synthesis of Value-Added Compounds
In order to produce novel compounds biologically, it is essential to rewire the native metabolic pathways in production hosts to form new ones that will lead to the desired compounds. Non-natural biosynthesis routes need to be designed, implemented and optimized through metabolic engineering for efficient production of the target compounds. In this section, we will review efforts in production of value-added compounds accomplished through rewriting of metabolic network with natural and non-natural enzymes, as well as computational design of non-natural metabolic pathways.
Rewriting the Metabolic Blueprint Using Natural Enzymes
Enzymes vary widely in the reactions they catalyze and in their substrate specificities. Homologs are present across different species thus there is a vast number of enzyme candidates that can be selected from nature for construction of novel pathways. By introducing non-native enzymes from different organisms into a production host, metabolic rewiring can be achieved to expand possibilities for biosynthesis of novel compounds. Two different approaches have been utilized to rewrite metabolic networks using natural enzymes: gap-filling and mix-and-matching of pathways. Here, we will review the employment of these strategies for diversifying metabolic pathways to produce value-added compounds.
Gap-Filling of Pathways
Native metabolic pathways in organisms are connected and insulated from one another to varying degrees (Hartwell et al., 1999). By gap-filling pathways with heterologous enzymes, shunts can be built between the pathways to create new and diverse biosynthesis routes toward desired metabolites (Shin et al., 2013; Lee and Kim, 2015). Essentially, by introducing a suitable non-native enzyme, a metabolite from a native pathway can be converted by the heterologous enzyme to a non-native intermediate to serve as a substrate of a previously disconnected native pathway, thus creating a novel biosynthesis pathway for producing target compounds. For example, the biosynthesis of odd-chain fatty alcohols, which is of industrial value, is made possible by the heterologous expression of α-dioxygenase from Oryza sativa in Saccharomyces cerevisiae (Jin et al., 2016). The α-dioxygenase can convert endogenous even-chain fatty acids to odd-chain fatty aldehydes which are subsequently reduced by native alcohol dehydrogenases to produce odd-chain fatty alcohols (Jin et al., 2016). Notably, although the biosynthesis pathway of salvianic acid A in the plant Salvia miltiorrhiza was unclear, the therapeutic antioxidant was remarkably produced in Escherichia coli by rerouting endogenously produced 4-hydroxyphenylpyruvate with D-lactate dehydrogenase derived from Lactobacillus pentosus to 4-hydroxyphenyllactate for conversion by a native hydroxylase complex to salvianic acid A (Yao et al., 2015). Similarly, novel nargenicin A1 derivatives were synthesized in Nocardia by introducing a hydroxylase (PikC) derived from the pikromycin gene cluster of Streptomyces venezuelae (Dhakal et al., 2016). The gap-filling strategy was also employed for the efficient synthesis of optically pure D-lactic acid in high titers by utilizing a glycerol dehydrogenase engineered for D-lactate dehydrogenase activity to gap-fill the pyruvate metabolism in Bacillus coagulans (Wang et al., 2011). Likewise, to produce biofuels such as propanol and butanol in high yield, a new pathway was engineered in E. coli for the production of 2-ketoacid precursors from pyruvate and acetyl-CoA by utilizing citramalate synthase from Methanococcus jannaschii (Atsumi and Liao, 2008). Other notable value-added chemicals produced by the gap-filling strategy, such as long-chain dicarboxylic acid (Song et al., 2014) and 2-pyrrolidone (Zhang et al., 2016), are summarized in Table 1. These examples illustrate that although gap-filling is a simple approach, relying on expression of a heterologous enzyme to diversify metabolic pathways, it demonstrates great potential in expanding the range of bio-based chemicals that can be produced. However, a major disadvantage of the gap-filling strategy is that it is applicable only when an intermediate metabolite can be found that links two native pathways to produce the target compound, thus limiting the possibilities of attainable pathway diversification. To allow greater diversification of biosynthetic pathways, rewiring of metabolic networks needs to be more extensive using other strategies such as mix-and-matching of pathways.
Mix-and-Matching of Pathways
The mix-and-match approach combinatorically expresses multiple genes or clusters from numerous organisms in a production host to produce metabolites of interest (Figure 1). Given the large number of annotated enzymes with known functions, artificial biosynthesis routes can therefore be built rationally by mix-and-matching suitable non-native enzymes to diversify native pathways toward target compounds. This strategy is particularly useful for reconstruction of pathways that are not fully elucidated and is exemplified by the production of the skin lightening agent arbutin in E. coli. Although the natural pathway for biosynthesizing arbutin is not fully understood, it was successfully produced in E. coli by diversifying the chorismic acid metabolic pathway with 4-hydroxybenzoate 1-hydroxylase from Candida parapsilosis to produce hydroquinone, which serves as the substrate for arbutin synthase from Rauvolfia serpentina to produce arbutin (Shen et al., 2017). The mix-and-match approach is also an effective strategy for producing variants of natural products, such as polyketides, carotenoids, phenyl propanoids and alkaloids, which involve gene clusters for biosynthesis. Genes in diverse sets of homologous gene clusters from different organisms can be viewed as “modules” for combinatorial assembly to produce novel derivatives. Using this strategy, novel pathways were constructed to enable biosynthesis of compounds with therapeutic and commercial value (Table 1). Taken together, the mix-and-match approach has demonstrated immense potential for diversifying metabolic pathways using combinations of natural enzymes for the production of novel value-added compounds. Nevertheless, mix-and-matching of pathways is often marred by low productivity due to substrate specificity constraints since the new metabolites are not natural substrates of the enzymes in the pathways assembled. Thus, the full potential of the mix-and-match strategy could only be realized by involving non-natural enzymes that have been evolved to suit the needs of the novel pathways.
Rewriting the Metabolic Blueprint with Non-natural Enzyme
Diversification of natural metabolic pathways to create novel biosynthesis routes will inevitably form intermediate metabolites that are beyond the substrate range which the natural enzymes involved can perform efficiently. Although nature has provided us with a gamut of enzymes to choose from, generation of non-natural enzymes is often essential for successful construction of efficient novel pathways to extensively rewrite metabolic blueprints (Foo et al., 2012). As most natural enzymes exhibit degrees of substrate promiscuity to analogs of their natural substrates (Gupta, 2016), promiscuous properties of natural enzymes can be exploited through protein engineering approaches to evolve their activities toward non-natural substrates. In addition to employment in gap-filling and mix-and-matching of pathways, non-natural enzymes have important applications for precursor-directed biosynthesis and catalysis of non-natural biochemical reactions, which are powerful strategies that we will review in this section for bio-based production of novel value-added chemicals.
Precursor-Directed Biosynthesis
Precursor-directed biosynthesis involves using evolved enzymes with altered substrate specificity to incorporate structurally diverse analogs of natural substrates into novel metabolic pathways in order to produce non-natural biochemicals. Commonly, substrate analogs are provided ex vivo to production hosts with non-natural enzymes with relaxed substrate specificity to form novel intermediates. While it is possible to rely on substrate promiscuity of the natural enzymes in the native pathways to convert the intermediates to the desired natural product derivatives, engineered enzymes are frequently required for the pathways to be efficient. For example, acyltransferase, the “gatekeeping” enzyme to polyketide synthesis, has been engineered to accept analogs of natural acyl-CoA precursors to initiate production of novel polyketide derivatives (Dunn and Khosla, 2013). Inclusion of enzymes modified by protein engineering in the downstream pathway can greatly facilitate the conversion of the resulting non-natural intermediates to a wide range of polyketide derivatives with potential therapeutic properties, as exemplified by the work of Lee et al. (2011) to create derivatives of the antibiotic erythromycin. Similar approaches were applied to enable biosynthesis of a wide range of novel derivatives of polyketides, non-ribosomal peptides (NRPs) and phenyl propanoids (Table 1). De novo precursor-directed biosynthesis can be achieved by co-expressing a pathway for generating substrate analog in vivo instead of ex vivo supplementation. This was demonstrated by Alt et al. for producing a novel derivative of the antibiotic clorobiocin by employing a 3,4-dihydroxybenzoic acid-producing pathway to provide the non-natural precursor to the clorobiocin biosynthesis pathway, resulting in the production of a potent DNA gyrase inhibitor (Alt et al., 2011).
With these examples, precursor-directed biosynthesis has proven to be an effective approach for generating a wide array of novel compounds rapidly with non-natural precursors. However, these precursors are often expensive and could only be synthesized chemically. Therefore, generating enzymes that can catalyze non-natural reactions are vital for progressing diversification of metabolic pathways.
Evolution of Natural Enzymes for Non-natural Biochemical Reactions
Metabolic engineering has advanced by leaps and bounds over the past decades facilitating the biosynthesis of many valuable compounds for pharmaceutical and industrial applications. However, chemical synthesis still has an edge in terms of the versatility of chemical structures that can be generated because biological synthesis is limited by the number of natural enzymes available (Wallace and Balskus, 2014). To circumvent this constraint, efforts have been directed toward creating enzymes that can perform reactions that have not been possible biologically for constructing novel metabolic pathways to produce compounds without known natural biosynthesis routes. For example, the versatile chemical, 2,4-dihydroxybutyric acid (2,4-DHB), was produced by exploiting a natural metabolic pathway involving aspartate to utilize malate, a structurally similar analog, as precursor (Walther et al., 2017). By engineering the enzymes in the natural pathway, novel enzymes with previously unreported activities in nature, namely malate semialdehyde reductase, malate kinase, and malate semialdehyde dehydrogenase, were generated to create an artificial 2,4-DHB-producing pathway. This illustrates the importance of non-natural enzymes for diversifying metabolic pathways. In recent years, numerous evolution and engineering strategies were applied to mechanistically diverse the superfamilies of biocatalysts to perform novel reactions (Gerlt and Babbitt, 2009). For instance, cytochrome P450s have been evolved to perform olefin aziridination (Farwell et al., 2015), carbene transfer to olefin (Coelho et al., 2013), and imidation of sulfides (Farwell et al., 2014), reactions which are unknown in the biological world. The promising trend of growing number of novel enzymes catalyzing non-natural biochemical reactions suggest the possibility of constructing fully artificial pathways to biosynthesize any compound that can be accomplished chemically.
Computational Pathway Design
As computing power grew exponentially in the past decade, computational tools became increasingly attractive as toolkits for metabolic engineering. While these tools initially were employed mainly to optimize metabolic pathways for achieving production of target compound with high yield (Copeland et al., 2012), they have been extended to facilitate designing of novel pathways. For example, to complement a rapid increase in the number of sequenced genomes, computational methods were created for data mining to identify and annotate genes (Blanco and Abril, 2009). However, it has reached a stage where the data generated exceeds the rate that they can be sifted and organized (Khosla, 2015). Automated annotation of these “big data” simply based on sequence homology has been unreliable or unsuccessful half of the time (Gerlt, 2016). To understand relationships between proteins in the databases, the “big data” generated could greatly benefit from tools developed by the Enzyme Function Initiative, such as EFI-EST and EFI-GNT (Gerlt, 2017), which use multidisciplinary approaches to accurately assign enzyme functions (Gerlt, 2016). Proper functional assignment is vital for identifying novel enzyme candidates from diverse superfamilies that can be deployed to construct non-natural biosynthesis routes. However, for diversification of metabolic pathways, creation of tools to predict and identify plausible routes are imperative to accelerate the design process for integrating heterologous genes to produce novel compounds in defined hosts. Therefore, computational tools such as BoostGAPFILL (Oyetunde et al., 2017) and fastGapFill (Thiele et al., 2014) have been developed to enable identification of candidate enzymes from a universal biochemical reaction database to fill network gaps. To enable the design of biologically feasible pathways to produce any target compound of interest, the retrosynthesis approach, which is a well-established method used in organic chemistry for identifying suitable precursors and synthesis routes, was combined with biological knowledge to create tools such as RetroPath (Carbonell et al., 2014) to automate the pathway design process. Several possible pathways consisting of various enzymes that can lead precursors to the target compound can be proposed and ranked based on selected criteria. Subsequently, RetroPath was successfully applied to design a pathway for biosynthesizing the antioxidant flavonoid pinocembrin, resulting in a five-enzyme pathway starting from malonyl-CoA as precursor (Feher et al., 2014). These promising computational tools and results suggest that computational pathway design will greatly facilitate the design of novel pathways and continued efforts in improving these computational suites will enable expansion of the chemical repertoire that can be produced biologically.
Conclusion and Future Perspectives
Metabolic engineering has progressed tremendously over decades with the aim to efficiently produce biochemicals from renewable resources to serve industrial needs. Indeed, there are many examples of engineered microbes that are able to produce industrially relevant levels of biochemical for commercialization (Zhu and Jackson, 2015). However, metabolic engineering is still largely limited to production of chemicals that exist naturally in biological systems. Therefore, the range of chemicals that metabolic engineering can generate is unable to meet that required by industries. Consequently, there is still heavy reliance on chemical synthesis from fossil resources for precursors needed by industries (Wallace and Balskus, 2014). Hence, diversification of metabolic pathways is crucial to achieve biosynthesis of non-natural compounds to rival the capabilities of chemical synthesis. Herein, we have reviewed several strategies that have been developed in the attempt to broaden the spectrum of compounds that can be created biologically. While the results have been promising, much still needs to be done before biological systems can be utilized to produce any target compound of choice.
Being a well-established field on its own, protein engineering is an area that can greatly benefit and complement efforts in metabolic pathway diversification (Foo et al., 2012). By increasing the number of available biocatalysts to catalyze non-natural reactions, protein engineering tools act as a driving force for designing new biosynthesis routes to create novel compounds. Further, these tailor-made enzymes catalyzing novel non-natural reactions uncover new arena to establish novel metabolic pathways. One remarkable example of the capability of protein engineering is the evolution of a natural enzyme to create carbon-silicon bonds, which are important moieties in many commercial products but do not exist in nature, essentially opening up possibilities of creating silicon-based biosynthetic pathways (Kan et al., 2016). Likewise, evolved Rhodothermus marinus cytochrome c have been employed to enable carbon–boron bond formation in E. coli to produce organoboranes, a class of compounds that are not found in biological systems but with applications in chemotherapeutics (Kan et al., 2017). With rapid advances in genome sequencing techniques, more enzymes will be identified to serve as candidates for engineering non-natural biochemical reactions. This may be complemented by the recent boom in microbiome studies, where metagenomics techniques have been developed and applied to sequence genomes of many microorganisms that were previously uncultivable. Computation-guided genome mining can thus be applied to discover novel natural products and their biosynthetic pathways (Medema and Fischbach, 2015).
Ultimately, biosynthesis pathways are a series of synchronized chemical reactions performed in biological chassis. Therefore, to create diverse non-natural metabolic pathways, it is imperative to incorporate chemical knowledge and techniques, particularly retrosynthesis, to complement our understanding of biological systems when designing biochemical routes required to produce novel target compounds. As discussed earlier, combining computational tools with retrosynthesis has already achieved some success in designing non-natural pathways for the biosynthesis of novel value-added compounds (Feher et al., 2014). By further developing user-friendly computational tools and databases that include all known natural and non-natural enzymes, automated platforms that integrate current strategies and knowledge pertinent to pathway diversification could be established to significantly accelerate designing and implementation of novel pathways.
Despite rapid advancement in the field of metabolic engineering and the potential tools available for interplaying with genetic material and metabolic networks of microbial workhorses, there are major bottlenecks that require resolution when forward engineering metabolic pathways. Alterations to metabolic pathways with heterologous or engineered enzymes in microbial hosts burden the microbial host and cause issues such as imbalance in metabolic pathways, poor growth, accumulation of toxic intermediates and other physiological stresses. Moreover, conventional model strains commonly used as production hosts often face issues under the harsh operating conditions of downstream industrial processes. These problems may be overcome by selecting suitable production hosts beyond the conventional ones, depending on the target compounds (Czajka et al., 2017). For example, the non-conventional oleaginous yeast Yarrowia lipolytica was exploited for production of fatty acid-derivatives (Zhu and Jackson, 2015). Furthermore, Pseudomonas strains possess metabolic, physiological and stress-tolerance characteristics that are favorable for metabolic engineering (Nikel et al., 2014) and Pseudomonas putida was demonstrated to be an ideal host for production of para-hydroxybenzoic acid due to its exceptional tolerance for aromatic compounds (Yu et al., 2016). In addition to strategic selection of production host, computation-guided pathway designing approaches, such as genome scale modeling and novel machine learning methods, have proven to be alternative solutions to solve metabolic imbalance issues by efficient strain engineering strategies (Wu G. et al., 2016; Wu S.G. et al., 2016). Concurrently, the lack of natural enzyme reaction cascade in engineered pathways brings about inefficient substrate conversion. To address this issue, artificial enzyme channels have been constructed to organize enzymes for efficient production of target compounds. This strategy has been reviewed extensively in literature (Proschel et al., 2015). These examples demonstrate that extensive efforts are ongoing for strain development to implement synthetic pathways efficiently, which is immensely beneficial for pathway diversification.
In conclusion, much progress in diversification of metabolic pathways has been made and the future for production of non-natural chemicals using biological systems is promising. There are fundamental concerns regarding the compatibility of non-natural pathways and products with living systems, such as substrate availability, toxicity of substrates and products, availability and intracellular balance of cofactors, and influx/efflux of substrates and products, that will impede the yield and productivity of the biosynthesis routes. Nevertheless, there are many tools available for strain engineering, such as targeted multi-site CRISPR/Cas9-based gene insertion or deletion (Jakociunas et al., 2015), riboswitch-based self-directed evolution of phenotype (Pham et al., 2017) and Synthetic Chromosome Recombination and Modification by LoxP-mediated Evolution (SCRaMbLE) in Synthetic Yeast 2.0 (Dymond and Boeke, 2012), that may be utilized to optimize host strains for tolerance towards non-natural pathways and products. Impressively, synthetic biology recently enabled the creation of a semi-synthetic organism to possess two additional letters in its codons that form an unnatural base pair. The organism can thus store increased genetic information and potentially create enzymes with novel activities, e.g., by incorporating non-canonical amino acids, thereby facilitate diversification of metabolic pathways (Zhang et al., 2017). In future, intensive research in pathway diversification might enable development of highly efficient microbial strains that can potentially carry out desired chemical reactions to produce any target compounds and eventually serve as a sustainable source for supplying valuable biochemicals to meet industrial needs.
Author Contributions
JLF, HL, WJC, and MWC conceptualized the content, figures, and table of the review. GSH, SPN, and LZ wrote the manuscript. JLF and HL revised the manuscript. T-KN and MWC proof corrected and reviewed the manuscript.
Funding
This work was funded by the Synthetic Biology Initiative of the National University of Singapore (DPRT/943/09/14), the Ministry of Education, Singapore (MOE/2014/T2/2/128), Global R&D Project Program, the Ministry of Knowledge Economy, the Republic of Korea (N0000677), and Synthetic Biology R&D Programme (SBP-P2) and Industry Alignment Fund-Industry Collaboration Project (ICP1600012) of the National Research Foundation, Singapore.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ping Han for her comments on the manuscript.
References
Alt, S., Burkard, N., Kulik, A., Grond, S., and Heide, L. (2011). An artificial pathway to 3,4-dihydroxybenzoic acid allows generation of new aminocoumarin antibiotic recognized by catechol transporters of E. coli. Chem. Biol. 18, 304–313. doi: 10.1016/j.chembiol.2010.12.016
Apeloig, Y., Rappoport, Z., and John Wiley and Sons (eds). (2001). “The chemistry of organic silicon compounds,” in The Chemistry of Functional Groups Supplement Si, Vol. 3, (New York, NY: Wiley).
Atsumi, S., and Liao, J. C. (2008). Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 74, 7802–7808. doi: 10.1128/AEM.02046-08
Blanco, E., and Abril, J. F. (2009). Computational gene annotation in new genome assemblies using GeneID. Methods Mol. Biol. 537, 243–261. doi: 10.1007/978-1-59745-251-9_12
Carbonell, P., Parutto, P., Baudier, C., Junot, C., and Faulon, J. L. (2014). Retropath: automated pipeline for embedded metabolic circuits. ACS Synth. Biol. 3, 565–577. doi: 10.1021/sb4001273
Coelho, P. S., Wang, Z. J., Ener, M. E., Baril, S. A., Kannan, A., Arnold, F. H., et al. (2013). A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo. Nat. Chem. Biol. 9, 485–487. doi: 10.1038/nchembio.1278
Copeland, W. B., Bartley, B. A., Chandran, D., Galdzicki, M., Kim, K. H., Sleight, S. C., et al. (2012). Computational tools for metabolic engineering. Metab. Eng. 14, 270–280. doi: 10.1016/j.ymben.2012.03.001
Czajka, J., Wang, Q., Wang, Y., and Tang, Y. J. (2017). Synthetic biology for manufacturing chemicals: constraints drive the use of non-conventional microbial platforms. Appl. Microbiol. Biotechnol. 101, 7427–7434. doi: 10.1007/s00253-017-8489-9
DeChristopher, B. A., Loy, B. A., Marsden, M. D., Schrier, A. J., Zack, J. A., and Wender, P. A. (2012). Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat. Chem. 4, 705–710. doi: 10.1038/nchem.1395
Dhakal, D., Chaudhary, A. K., Yi, J. S., Pokhrel, A. R., Shrestha, B., Parajuli, P., et al. (2016). Enhanced production of nargenicin A1 and creation of a novel derivative using a synthetic biology platform. Appl. Microbiol. Biotechnol. 100, 9917–9931. doi: 10.1007/s00253-016-7705-3
Dhakal, D., Pokhrel, A. R., Shrestha, B., and Sohng, J. K. (2017). Marine rare Actinobacteria: isolation, characterization, and strategies for harnessing bioactive compounds. Front. Microbiol. 8:1106. doi: 10.3389/fmicb.2017.01106
Dunn, B. J., and Khosla, C. (2013). Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J. R. Soc. Interface 10:20130297. doi: 10.1098/rsif.2013.0297
Dymond, J., and Boeke, J. (2012). The Saccharomyces cerevisiae SCRaMbLE system and genome minimization. Bioeng. Bugs 3, 168–171. doi: 10.4161/bbug.19543
Evans, B. S., Chen, Y., Metcalf, W. W., Zhao, H., and Kelleher, N. L. (2011). Directed evolution of the nonribosomal peptide synthetase AdmK generates new andrimid derivatives in vivo. Chem. Biol. 18, 601–607. doi: 10.1016/j.chembiol.2011.03.008
Farwell, C. C., McIntosh, J. A., Hyster, T. K., Wang, Z. J., and Arnold, F. H. (2014). Enantioselective imidation of sulfides via enzyme-catalyzed intermolecular nitrogen-atom transfer. J. Am. Chem. Soc. 136, 8766–8771. doi: 10.1021/ja503593n
Farwell, C. C., Zhang, R. K., McIntosh, J. A., Hyster, T. K., and Arnold, F. H. (2015). Enantioselective enzyme-catalyzed aziridination enabled by active-site evolution of a cytochrome P450. ACS Cent. Sci. 1, 89–93. doi: 10.1021/acscentsci.5b00056
Feher, T., Planson, A. G., Carbonell, P., Fernandez-Castane, A., Grigoras, I., Dariy, E., et al. (2014). Validation of RetroPath, a computer-aided design tool for metabolic pathway engineering. Biotechnol. J. 9, 1446–1457. doi: 10.1002/biot.201400055
Foo, J. L., Ching, C. B., Chang, M. W., and Leong, S. S. (2012). The imminent role of protein engineering in synthetic biology. Biotechnol. Adv. 30, 541–549. doi: 10.1016/j.biotechadv.2011.09.008
Galanie, S., Thodey, K., Trenchard, I. J., Filsinger Interrante, M., and Smolke, C. D. (2015). Complete biosynthesis of opioids in yeast. Science 349, 1095–1100. doi: 10.1126/science.aac9373
Gerlt, J. A. (2016). Tools and strategies for discovering novel enzymes and metabolic pathways. Perspect. Sci. 9(Suppl. C), 24–32. doi: 10.1016/j.pisc.2016.07.001
Gerlt, J. A. (2017). Genomic enzymology: web tools for leveraging protein family sequence-function space and genome context to discover novel functions. Biochemistry 56, 4293–4308. doi: 10.1021/acs.biochem.7b00614
Gerlt, J. A., and Babbitt, P. C. (2009). Enzyme (re)design: lessons from natural evolution and computation. Curr. Opin. Chem. Biol. 13, 10–18. doi: 10.1016/j.cbpa.2009.01.014
Gupta, R. D. (2016). Recent advances in enzyme promiscuity. Sustain. Chem. Process. 4:2. doi: 10.1186/s40508-016-0046-9
Hartwell, L. H., Hopfield, J. J., Leibler, S., and Murray, A. W. (1999). From molecular to modular cell biology. Nature 402(Suppl. 6761), C47–C52. doi: 10.1038/35011540
Harvey, A. L. (2008). Natural products in drug discovery. Drug Discov. Today 13, 894–901. doi: 10.1016/j.drudis.2008.07.004
Jakociunas, T., Bonde, I., Herrgard, M., Harrison, S. J., Kristensen, M., Pedersen, L. E., et al. (2015). Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 28, 213–222. doi: 10.1016/j.ymben.2015.01.008
Jiang, M., and Pfeifer, B. A. (2013). Metabolic and pathway engineering to influence native and altered erythromycin production through E. coli. Metab. Eng. 19, 42–49. doi: 10.1016/j.ymben.2013.05.005
Jiang, M., Zhang, H., Park, S. H., Li, Y., and Pfeifer, B. A. (2013). DeoxysugarI pathway interchange for erythromycin analogues heterologously produced through Escherichia coli. Metab. Eng. 20, 92–100. doi: 10.1016/j.ymben.2013.09.005
Jin, Z., Wong, A., Foo, J. L., Ng, J., Cao, Y. X., Chang, M. W., et al. (2016). Engineering Saccharomyces cerevisiae to produce odd chain-length fatty alcohols. Biotechnol. Bioeng. 113, 842–851. doi: 10.1002/bit.25856
Kan, S. B., Lewis, R. D., Chen, K., and Arnold, F. H. (2016). Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life. Science 354, 1048–1051. doi: 10.1126/science.aah6219
Kan, S. B. J., Huang, X., Gumulya, Y., Chen, K., and Arnold, F. H. (2017). Genetically programmed chiral organoborane synthesis. Nature 552, 132–136. doi: 10.1038/nature24996
Lee, H. Y., Harvey, C. J., Cane, D. E., and Khosla, C. (2011). Improved precursor-directed biosynthesis in E. coli via directed evolution. J. Antibiot. 64, 59–64. doi: 10.1038/ja.2010.129
Lee, S. Y., and Kim, H. U. (2015). Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 33, 1061–1072. doi: 10.1038/nbt.3365
Maoka, T., Takemura, M., Tokuda, H., Suzuki, N., and Misawa, N. (2014). 4-Ketozeinoxanthin, a novel carotenoid produced in Escherichia coli through metabolic engineering using carotenogenic genes of bacterium and liverwort. Tetrahedron Lett. 55, 6708–6710. doi: 10.1016/j.tetlet.2014.10.033
Medema, M. H., and Fischbach, M. A. (2015). Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648. doi: 10.1038/nchembio.1884
Menendez-Bravo, S., Comba, S., Sabatini, M., Arabolaza, A., and Gramajo, H. (2014). Expanding the chemical diversity of natural esters by engineering a polyketide-derived pathway into Escherichia coli. Metab. Eng. 24, 97–106. doi: 10.1016/j.ymben.2014.05.002
Morita, H., Yamashita, M., Shi, S. P., Wakimoto, T., Kondo, S., Kato, R., et al. (2011). Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase. Proc. Natl. Acad. Sci. U.S.A. 108, 13504–13509. doi: 10.1073/pnas.1107782108
Nikel, P. I., Martinez-Garcia, E., and de Lorenzo, V. (2014). Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12, 368–379. doi: 10.1038/nrmicro3253
Oksman-Caldentey, K. M., and Inze, D. (2004). Plant cell factories in the post-genomic era: new ways to produce designer secondary metabolites. Trends Plant Sci. 9, 433–440. doi: 10.1016/j.tplants.2004.07.006
Oyetunde, T., Zhang, M. H., Chen, Y. X., Tang, Y. J., and Lo, C. (2017). BoostGAPFILL: improving the fidelity of metabolic network reconstructions through integrated constraint and pattern-based methods. Bioinformatics 33, 608–611. doi: 10.1093/bioinformatics/btw684
Pham, H. L., Wong, A., Chua, N., Teo, W. S., Yew, W. S., and Chang, M. W. (2017). Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes. Nat. Commun. 8:411. doi: 10.1038/s41467-017-00511-w
Proschel, M., Detsch, R., Boccaccini, A. R., and Sonnewald, U. (2015). Engineering of metabolic pathways by artificial enzyme channels. Front. Bioeng. Biotechnol. 3:168. doi: 10.3389/fbioe.2015.00168
Shen, X., Wang, J., Wang, J., Chen, Z., Yuan, Q., and Yan, Y. (2017). High-level De novo biosynthesis of arbutin in engineered Escherichia coli. Metab. Eng. 42, 52–58. doi: 10.1016/j.ymben.2017.06.001
Shin, J. H., Kim, H. U., Kim, D. I., and Lee, S. Y. (2013). Production of bulk chemicals via novel metabolic pathways in microorganisms. Biotechnol. Adv. 31, 925–935. doi: 10.1016/j.biotechadv.2012.12.008
Song, J.-W., Lee, J.-H., Bornscheuer, U. T., and Park, J.-B. (2014). Microbial synthesis of medium-chain α,ω-dicarboxylic acids and ω-aminocarboxylic acids from renewable long-chain fatty acids. Adv. Synth. Catal. 356, 1782–1788. doi: 10.1002/adsc.201300784
Sugimoto, Y., Ding, L., Ishida, K., and Hertweck, C. (2014). Rational design of modular polyketide synthases: morphing the aureothin pathway into a luteoreticulin assembly line. Angew. Chem. Int. Ed. Engl. 53, 1560–1564. doi: 10.1002/anie.201308176
Thiele, I., Vlassis, N., and Fleming, R. M. (2014). fastGapFill: efficient gap filling in metabolic networks. Bioinformatics 30, 2529–2531. doi: 10.1093/bioinformatics/btu321
Thirlway, J., Lewis, R., Nunns, L., Al Nakeeb, M., Styles, M., Struck, A. W., et al. (2012). Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity. Angew. Chem. Int. Ed. Engl. 51, 7181–7184. doi: 10.1002/anie.201202043
Walker, M. C., Thuronyi, B. W., Charkoudian, L. K., Lowry, B., Khosla, C., and Chang, M. C. (2013). Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science 341, 1089–1094. doi: 10.1126/science.1242345
Wallace, S., and Balskus, E. P. (2014). Opportunities for merging chemical and biological synthesis. Curr. Opin. Biotechnol. 30, 1–8. doi: 10.1016/j.copbio.2014.03.006
Walther, T., Topham, C. M., Irague, R., Auriol, C., Baylac, A., Cordier, H., et al. (2017). Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2,4-dihydroxybutyric acid. Nat. Commun. 8:15828. doi: 10.1038/ncomms15828
Wang, P., Kim, W., Pickens, L. B., Gao, X., and Tang, Y. (2012). Heterologous expression and manipulation of three tetracycline biosynthetic pathways. Angew. Chem. Int. Ed. Engl. 51, 11136–11140. doi: 10.1002/anie.201205426
Wang, Q., Ingram, L. O., and Shanmugam, K. T. (2011). Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proc. Natl. Acad. Sci. U.S.A. 108, 18920–18925. doi: 10.1073/pnas.1111085108
Wang, S., Zhang, S., Xiao, A., Rasmussen, M., Skidmore, C., and Zhan, J. (2015). Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab. Eng. 29, 153–159. doi: 10.1016/j.ymben.2015.03.011
Wilson, M. C., and Moore, B. S. (2012). Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat. Prod. Rep. 29, 72–86. doi: 10.1039/c1np00082a
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., and Koffas, M. A. G. (2016). Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 34, 652–664. doi: 10.1016/j.tibtech.2016.02.010
Wu, S. G., Shimizu, K., Tang, J. K.-H., and Tang, Y. J. (2016). Facilitate collaborations among synthetic biology. Metabolic engineering and machine learning. ChemBioEng Rev. 3, 45–54. doi: 10.1002/cben.201500024
Yao, J., Guo, H., Chaiprasongsuk, M., Zhao, N., Chen, F., Yang, X., et al. (2015). Substrate-assisted catalysis in the reaction catalyzed by salicylic acid binding protein 2 (SABP2), a potential mechanism of substrate discrimination for some promiscuous enzymes. Biochemistry 54, 5366–5375. doi: 10.1021/acs.biochem.5b00638
Yao, Y. F., Wang, C. S., Qiao, J., and Zhao, G. R. (2013). Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab. Eng. 19, 79–87. doi: 10.1016/j.ymben.2013.06.001
Yu, S., Plan, M. R., Winter, G., and Kromer, J. O. (2016). Metabolic Engineering of Pseudomonas putida KT2440 for the Production of para-Hydroxy Benzoic Acid. Front. Bioeng. Biotechnol. 4:90. doi: 10.3389/fbioe.2016.00090
Zhang, H., Wang, Y., Wu, J., Skalina, K., and Pfeifer, B. A. (2010). Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 17, 1232–1240. doi: 10.1016/j.chembiol.2010.09.013
Zhang, J., Kao, E., Wang, G., Baidoo, E. E. K., Chen, M., and Keasling, J. D. (2016). Metabolic engineering of Escherichia coli for the biosynthesis of 2-pyrrolidone. Metab. Eng. Commun. 3(Suppl. C), 1–7. doi: 10.1016/j.meteno.2015.11.001
Zhang, Y., Ptacin, J. L., Fischer, E. C., Aerni, H. R., Caffaro, C. E., San Jose, K., et al. (2017). A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647. doi: 10.1038/nature24659
Zhu, Q., and Jackson, E. N. (2015). Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr. Opin. Biotechnol. 36, 65–72. doi: 10.1016/j.copbio.2015.08.010
Keywords: metabolic engineering, synthetic biology, pathway engineering, protein engineering, biochemical production
Citation: Hossain GS, Nadarajan SP, Zhang L, Ng T-K, Foo JL, Ling H, Choi WJ and Chang MW (2018) Rewriting the Metabolic Blueprint: Advances in Pathway Diversification in Microorganisms. Front. Microbiol. 9:155. doi: 10.3389/fmicb.2018.00155
Received: 14 November 2017; Accepted: 23 January 2018;
Published: 12 February 2018.
Edited by:
Ryan S. Senger, Virginia Tech, United StatesReviewed by:
Pablo Ivan Nikel, The Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), DenmarkYinjie Tang, Washington University in St. Louis, United States
Copyright © 2018 Hossain, Nadarajan, Zhang, Ng, Foo, Ling, Choi and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Wook Chang, matthew_chang@nuhs.edu.sg; bchcmw@nus.edu.sg
†These authors have contributed equally to this work.
 Gazi Sakir Hossain
Gazi Sakir Hossain Saravanan Prabhu Nadarajan
Saravanan Prabhu Nadarajan Lei Zhang
Lei Zhang Tee-Kheang Ng
Tee-Kheang Ng Jee Loon Foo
Jee Loon Foo Hua Ling
Hua Ling Won Jae Choi
Won Jae Choi Matthew Wook Chang
Matthew Wook Chang