- 1Yanchi Research Station, School of Soil and Water Conservation, Beijing Forestry University, Beijing, China
- 2Key Laboratory of State Forestry Administration on Soil and Water Conservation, Beijing Forestry University, Beijing, China
- 3Engineering Research Center of Forestry Ecological Engineering, Ministry of Education, Beijing Forestry University, Beijing, China
Desert microbes are expected to be substantially sensitive to global environmental changes, such as precipitation changes and elevated nitrogen deposition. However, the effects of precipitation changes and nitrogen enrichment on their diversity and community composition remain poorly understood. We conducted a field experiment over 2 years with multi-level precipitation and nitrogen addition in a desert shrubland of northern China, to examine the responses of soil bacteria and fungi in terms of diversity and community composition and to explore the roles of plant and soil factors in structuring microbial communities. Water addition significantly increased soil bacterial diversity and altered the community composition by increasing the relative abundances of stress-tolerant (dormant) taxa (e.g., Acidobacteria and Planctomycetes); however, nitrogen addition had no substantial effects. Increased precipitation and nitrogen did not impact soil fungal diversity, but significantly shifted the fungal community composition. Specifically, water addition reduced the relative abundances of drought-tolerant taxa (e.g., the orders Pezizales, Verrucariales, and Agaricales), whereas nitrogen enrichment decreased those of oligotrophic taxa (e.g., the orders Agaricales and Sordariales). Shifts in microbial community composition under water and nitrogen addition occurred primarily through changing resource availability rather than plant community. Our results suggest that water and nitrogen addition affected desert microbes in different ways, with watering shifting stress-tolerant traits and fertilization altering copiotrophic/oligotrophic traits of the microbial communities. These findings highlight the importance of resource availability in driving the desert microbial responses to short-term environmental changes.
Introduction
Deserts occupy approximately one-third of the Earth’s land surface (Laity, 2009) and are currently experiencing widespread global environmental changes, including precipitation changes and elevated nitrogen deposition (Galloway et al., 2008; Maestre et al., 2012). As desert ecosystems are usually characterized by stressful conditions of low water and nutrient availability (Noy-Meir, 1973; Makhalanyane et al., 2015), they are expected to be substantially more sensitive to global environmental changes than other ecosystems (Bobbink et al., 2010; Maestre et al., 2016). Soil microbial diversity and community composition in desert biomes have been shown to differ remarkably from those in non-desert biomes (Fierer et al., 2012), suggesting that their responses to environmental changes might also be distinct. However, the impacts of environmental changes on desert microbial communities are not well-understood.
Soil microbes play a crucial role in biogeochemical cycles, and thus their responses to environmental changes can provide feedback to influence plant communities and climate systems (Nie et al., 2013; Wei et al., 2013). Recent high-throughput sequencing data have shown that both precipitation changes and nitrogen deposition can alter the microbial community composition and diversity (Evans and Wallenstein, 2014; Leff et al., 2015; Li et al., 2016; Zeng et al., 2016). In general, nitrogen addition tends to decrease microbial diversity, increase the relative abundance of copiotrophic (i.e., fast-growing, low carbon-use efficiency) taxa (e.g., Proteobacteria and Bacteroidetes), and reduce that of oligotrophic (i.e., slow-growing, high carbon-use efficiency) taxa (e.g., Acidobacteria and Basidiomycota) (Fierer et al., 2007; Ramirez et al., 2012; Leff et al., 2015; Ho et al., 2017). It is generally hypothesized that precipitation enrichment, similarly to nitrogen addition, could favor copiotrophic taxa over oligotrophic taxa due to water-induced increases in nitrogen availability (Li et al., 2017b); however, results of the impacts of precipitation changes on microbial communities are inconsistent. Data from many field experiments have suggested that background precipitation variability (e.g., seasonal or interannual variability) more strongly shapes the microbial community composition than does the direct effect of experimental precipitation treatments (Cregger et al., 2012; Gutknecht et al., 2012; Curiel Yuste et al., 2014). Data from other studies have indicated that indirect environmental factors (e.g., the plant community) that shift under precipitation changes might play a larger role in determining microbial community composition than direct changes to soil moisture (Evans et al., 2014; Li et al., 2016). Water-induced indirect effects on microbial communities might be ecosystem-specific, which may contribute to previous inconsistent results among different precipitation-manipulation experiments. Collectively, these previous findings suggest that nitrogen addition consistently impacts microbial communities, whereas the inconsistent responses of microbes to precipitation changes may result from ecosystem-specific background/history precipitation regimes and/or water-induced indirect environmental factors.
Precipitation changes and nitrogen addition not only directly affect microbial physiology but also indirectly influence microbial abundances by changing plant and soil properties (Chen et al., 2015; Li et al., 2017b). Water and nitrogen availability can change the quantity and quality of plant residua (e.g., root exudates and litter), which represent major resource inputs to soil (Ren et al., 2015; Liu et al., 2016). These changes can impact microbial structure and function (Wardle et al., 2006; Bardgett and van der Putten, 2014). Several lines of empirical evidence have shown that nitrogen-induced soil acidification exerts native effects on microbes (Chen et al., 2015; Zeng et al., 2016), while precipitation increment can dampen these effects by increasing the soil pH (Zhang et al., 2014; Li et al., 2016). Similarly, precipitation and nitrogen enrichment have counteractive effects on plant species richness, with water-induced increases and nitrogen-induced declines in plant diversity (Xu Z. et al., 2015). Plant diversity has been documented as one of the major drivers in structuring microbial communities (Prober et al., 2015; Chen et al., 2017; Li et al., 2017a), suggesting that increased precipitation might alleviate the effects of nitrogen-induced loss of plant richness on microbial communities through increasing plant diversity. Collectively, previous findings have shown the very complex nature of the interactive influences of precipitation changes and nitrogen addition on microbial communities, which occur via the alteration of plant communities and soil properties.
To examine the effects of precipitation and nitrogen addition on soil microbial communities in desert ecosystems, we conducted a field experiment by varying precipitation and nitrogen addition to a desert shrubland in the Mu Us Desert of northern China. Our main goals were (i) to test how soil bacterial and fungal communities respond to increased precipitation and nitrogen and (ii) to identify the relative roles of the direct and indirect effects of water and nitrogen addition on microbial communities, thereby advancing a potential mechanistic understanding of the responses of desert microbial communities to global environmental changes.
Materials and Methods
Study Site and Experimental Design
This study was conducted at the Yanchi Research Station (37°04′–38°10′ N, 106°30′–107°41′ E, elevation 1,530 m above sea level), which is located on the southwestern edge of the Mu Us Desert in Ningxia, China. This region is characterized by a semiarid continental monsoon climate with an average annual temperature of 8.1°C and a mean annual precipitation of 284.8 mm (1955–2013). Approximately 80% of the precipitation occurs from May to September. The mean annual potential evapotranspiration is 2,024 mm (Jia et al., 2016). The soil type is quartisamment based on the US Soil Taxonomy (Gao et al., 2014). The dominant shrub species in this region is Artemisia ordosica, although sparse shrubs (Hedysarum mongolicum, Salix psammophila, and Caragana korshinskii) and the grass Agropyron cristatum are also indigenous to the region (She et al., 2015). The study was conducted in a fenced area, in which grazing has been prohibited since the late 1990s and vegetation was allowed to recover for over a decade (Sun et al., 2016).
A two-factor field experiment performed with constant precipitation and nitrogen addition was established in October 2014. There were three precipitation levels (W0: ambient; W20: ambient + 20%; and W40: ambient + 40%) and two nitrogen levels (N0: ambient, 0 kg N ha-1 yr-1 and N60: fertilization, 60 kg N ha-1 yr-1), resulting in a total of six treatments (including all permutations of both factors), each with four replications. Twenty-four 5 m × 5 m plots were laid out in a randomized block design (four blocks with six treatment plots within each block). Plots were separated by a 1 m wide buffer zone.
The amount of added precipitation was determined by referring to the long-term mean annual precipitation in this area (1955–2013: 284.8 mm). Specifically, the W20 and W40 treatment plots received 20% (∼56 mm) and 40% (∼112 mm) of supplementary precipitation, respectively. According to the magnitude and distribution of long-term mean monthly precipitation in this area (see details in She et al., 2016), water was applied with a sprinkler irrigation system as nine equal applications during the growing season (May–September), three times in July and August and once in May, June, and September. Water was added following natural rainfall events to avoid altering the precipitation regime. In this study, we defined annual precipitation as the water-year precipitation received between October 1 and September 30 in the following year. Ambient water-year precipitation was 288 and 369 mm in 2015 and 2016, respectively.
Fertilization treatments involved five equal applications of NH4NO3 solution at the beginning of each month during the growing season, corresponding to a total fertilization rate of 60 kg N ha-1 yr-1 for the N60 treatment. For each fertilization event, NH4NO3 (analytical grade) was weighed and dissolved in 10 L tap water. The same amount of water, equivalent to a 2 mm precipitation, was applied to the nitrogen control plots. The rate of nitrogen addition in the N60 treatment was nearly fivefold greater than the background deposition rate in the study site (12 kg N ha-1 yr-1; She, unpublished data), but was well within the range of current deposition rates in northern China (an average value of 56.2 kg N ha-1 yr-1; Xu W. et al., 2015).
Plant and Soil Measurements
In mid-September 2016, herbaceous vegetation structure (i.e., plant density, height, and coverage) was investigated using two permanent 1 m × 1 m quadrats in each plot. One of the quadrats was placed under a shrub canopy, and another was placed in the surrounding interspace between shrubs. The number, height, and coverage of shrub species were estimated in each 5 m × 5 m plot. Using these measurements, the plant Shannon–Wiener (SW) index was calculated as the relative abundance of each plant species. Herbaceous aboveground biomass was measured by randomly clipping a 1 m × 1 m quadrat within each plot. The harvested plants were sorted by species, oven-dried at 75°C for 48 h, and weighed. We used herbaceous aboveground biomass measurements to estimate the herbaceous aboveground net primary productivity (ANPP) and classified herbs into two functional groups according to their life forms, including perennial herbs (PH) and annuals (AS). The shrub ANPP was estimated by using an improved method (see details in She et al., 2016) based on the length and number of current-year plant twigs.
After harvesting plant biomass, soil samples were collected to depths of 0-20 cm from the interspaces between shrubs. For each plot, three sample cores were randomly collected using a 3.8 cm diameter soil auger and then composited into a single sample. All collected samples were transported immediately to our laboratory. Fresh samples were sieved through 2 mm screens and stored at 4°C before chemical analysis and at -80°C before soil DNA extraction. All soil samples were separated into two portions. One portion was maintained fresh for measurements of soil moisture and dissolvable inorganic nitrogen (nitrate and ammonium). The other portion was air-dried for determinations of soil pH, soil organic carbon (SOC), soil total nitrogen (STN), and soil total phosphorous (STP). Soil moisture was determined after oven-drying at 105°C for 24 h. Soil nitrate and ammonium were extracted with 2 M KCl and analyzed by dual-wavelength ultraviolet spectrophotometry and indophenol blue colorimetry, respectively. Soil pH was measured in a soil/water (1:2.5) suspension. SOC was measured using the potassium dichromate oxidation method. STN was analyzed by the micro-Kjeldahl method. STP was determined by the Mo–Sb anti-spectrophotometric method.
Soil DNA Extraction, Amplification, and Sequencing
Genomic DNA was isolated from 0.25 g of each soil sample using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, Inc., United States) following the manufacturer’s instructions. DNA concentrations and purities were assessed by 1% agarose gel electrophoresis. DNA samples were diluted to 1 ng μL-1 in sterile water.
The V4 hypervariable region of bacterial 16S rRNA was amplified from bacterial DNA samples with a barcoded primer set, including primers 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′). The V4 hypervariable region of fungal 18S rRNA was amplified using a barcoded primer set, including primers 528F (5′-GCG GTA ATT CCA GCT CCA A-3′) and 706R (5′-AAT CCR AGA ATT TCA CCT CT-3′). All PCR procedures were performed in 30 μL reaction mixtures containing 15 μL of Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England BioLabs, Inc., United States), 0.2 μM of each primer, and approximately 10 ng template DNA. Thermal cycling involved an initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 60 s, with a final extension at 72°C for 5 min.
Polymerase chain reaction products were assessed by 2% agarose gel electrophoresis, and those with a bright main band between 400 and 450 bp were chosen for further experiments. Equal amounts of the PCR product from each sample were pooled and then purified using the GeneJET Gel Extraction Kit (Thermo Scientific, Inc., United States). A sequencing library was generated using the NEB Next® UltraTM DNA Library Prep Kit for Illumina (New England BioLabs, Inc., United States) following the manufacturer’s instructions, and then sequencing adapters were added to the 5′ ends of the amplicons. The library quality was assessed on the Qubit® 2.0 Fluorometer (Thermo Scientific, Inc., United States) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Inc., United States). Finally, the qualified library was sequenced using an Illumina HiSeq 2500 platform (Illumina, Inc., United States) at Novogene (Beijing, China), producing 250-bp paired-end reads.
Bioinformatics Processing
Paired-end reads were assigned to samples based on their unique barcodes and were truncated by filtering out the barcode and primer sequences. Paired-end reads from the original amplicon were merged using FLASH software (Magoc and Salzberg, 2011) which is a very fast and accurate analysis tool used to merge paired-end reads when the original DNA fragments are shorter than twice the length of reads. The obtained splicing sequences were referred to raw tags. Quality filtering of the raw tags was performed under specific filtering conditions to obtain high-quality clean tags (Bokulich et al., 2013) using QIIME software (Caporaso et al., 2010) as a quality-control process. The clean tags were compared with the reference database using the UCHIME algorithm (Edgar et al., 2011) to detect chimeric sequences (Haas et al., 2011). After removing the chimeric sequences, we obtained effective tags.
Effective tags were clustered into operational taxonomic units (OTUs) at ≥97% sequence similarity with the UPARSE program (Edgar, 2013), and singleton OTUs (with only one read) were removed. A representative sequence from each OTU was selected and annotated for taxonomic information using the Ribosomal Database Project classifier (Wang et al., 2007) against the SILVA Database (Quast et al., 2013). Phylogenetic relationships of different OTUs were conducted using MUSCLE software (Edgar, 2004). OTU abundance tables were constructed using USEARCH software. The relative abundances of species at different taxonomic levels were calculated using the OTU abundance tables. To rarify all data sets to the same level of sampling effort, OTU abundance tables were rarefied to 45,469 bacterial 16S rRNA sequences and 3,138 fungal 18S rRNA sequences (the minimum number of sequences for a sample) for each soil sample. Further analysis of alpha diversity and beta diversity were performed based on these rarefied OTU tables. Finally, microbial diversity was assessed based on the observed species (species richness) and SW index, and Bray–Curtis dissimilarities were used to assess differences in microbial community composition among treatment groups.
All raw sequence reads generated in this study were archived in the Sequence Read Archive database of the National Center for Biotechnology Information under accession number SRP126812.
Statistical Analyses
The Shapiro–Wilk test was conducted to examine the normality of data that were used for analysis of variance (ANOVA). Data that did not meet the assumption of normality were log/sqrt-transformed prior to analyses to normalize their distributions. Two-way ANOVA was performed to test the impacts of water, nitrogen addition, and their interactions on plant ANPP and diversity, soil chemical properties, microbial diversity, and the relative abundances of dominant microbial taxa. One-way ANOVA with Duncan’s multiple-range tests was performed to compare the effects of watering on each response variable at each nitrogen-addition rate and to compare the effects of nitrogen at each water-treatment level. To estimate the effect sizes of water or nitrogen treatment on the relative abundances of the dominant microbial taxa, the response ratio was calculated as ln(Xij / Xic), where Xij is the observed value for variable i in each experimental plot j and Xic is the mean value of the variable i in the control treatment (Byrne et al., 2017). One-sample t-test was applied to determine whether each response ratio was significantly different from zero. Differences in microbial community composition (Bray–Curtis dissimilarities) among the water- and nitrogen-treatment groups were assessed by permutational multi-variate ANOVA (PERMANOVA) and visualized using principal coordinate analysis (PCoA). Spearman’s rank correlation analysis (Mantel test) was used to test the relationships between plant/soil variables and the microbial community composition. Stepwise regression analysis was conducted to explore the multi-variate effects of plant and soil factors on the microbial diversity and relative abundances of dominant microbial phyla. Prior to performing stepwise regression, environmental factors that showed a high collinearity with other factors (r > 0.6) were removed, and other factors (plant SW, shrub ANPP, PH ANPP, AS ANPP, soil moisture, dissolvable inorganic nitrogen (DIN), and STN) were retained.
We conducted structural equation modeling (SEM) to specifically test the direct and indirect effects of water and nitrogen addition on the composition of soil microbial communities (as assessed by PCo1 of the Bray–Curtis dissimilarity matrix). Prior to performing SEM analysis, we hypothesized that increased precipitation and nitrogen would impact the bacterial/fungal communities, directly by increasing water and nitrogen availability, or indirectly through changing plant ANPP, based on our ANOVA, Mantel test, and stepwise regression results. Subsequently, a priori model of the above hypothetical relationships was constructed (Supplementary Figure S1). Thus, the pairwise correlations among these environmental factors, including the water- and nitrogen-addition rates, soil moisture, DIN, shrub ANPP, PH ANPP, AS ANPP, and bacterial/fungal PCo1 values, were examined by Spearman’s rank correlation analysis. The matrix of R values derived from the Spearman’s correlation analysis was retained to conduct SEM. The data matrix was fitted to the model using the maximum likelihood estimation method (Grace, 2006). The overall goodness-of-fit of our model was characterized by a non-significant chi-square test (P > 0.05), low Akaike information criteria (AIC), low root-mean-square error of approximation (RMSEA < 0.05 and P > 0.1), and a comparative fit index (CFI) > 0.95. The final model was improved by removing relationships between observed variables from a priori models, according to these indices.
All analyses and figures were performed with R software, version 3.3.1 (R Core Team, 2016). Statistical significance was determined at a level of P ≤ 0.05. We used the “vegdist,” “adonis,” and “capscale” functions in the vegan package (Oksanen et al., 2013) for computing the Bray–Curtis dissimilarity, conducting PERMANOVA and Mantel test, and implementing PCoA, respectively. We employed the “stepAIC” function in the MASS package (Venables and Ripley, 2002) to conduct stepwise regression analysis, the psych package (Revelle, 2016) to construct a Spearman’s rank correlation matrix, the lavaan package (Yves, 2012) for SEM, and the ggplot2 package (Wickham, 2009) for generating graphs.
Results
Plant and Soil Properties
Water addition significantly increased plant SW, but did not affect plant ANPP (Supplementary Table S1). Plant SW increased from 1.14 ± 0.05 (W0 treatment) to 1.23 ± 0.04 (W20) and 1.40 ± 0.05 (W40), amounting to average increment rates of 7.89 and 22.81%, respectively. Nitrogen addition significantly enhanced PH ANPP, but did not alter plant SW, shrub ANPP, and AS ANPP (Supplementary Table S1). Nitrogen-induced increase in PH ANPP under N60 treatment (58.13 ± 14.66 g m-2) was 178.13% higher than that under ambient treatment (N0, 20.90 ± 6.15 g m-2). No significant interactive effects of water and nitrogen addition were found among any plant properties.
Water and nitrogen enrichment significantly influenced soil moisture and nitrogen availability, but did not affect soil pH, SOC, STN, or STP (Supplementary Table S1). Soil moisture significantly increased only in plots with added water, from 4.18 ± 0.27 (W0) to 6.04 ± 0.38 (W20) and 7.53 ± 0.36% (W40), an average increase of 44.50 and 80.14%, respectively. Nitrogen-induced increase in soil nitrogen availability in the plots with fertilization (N60, 2.40 ± 0.17 mg N kg-1) was 144.90% higher than that in unfertilized plots (N0, 0.98 ± 0.08 mg N kg-1). No significant interactive effects were detected among any soil properties.
Diversity and Composition of Microbial Communities
Soil bacterial species richness and SW index showed positive responses to water addition, but were unresponsive to nitrogen addition and water–nitrogen interactions (Figure 1A and Supplementary Table S1). Relative to ambient treatment, water addition increased bacterial species richness and SW index by 4.28-7.91% and 1.35-2.50%, respectively. Shifts in soil bacterial community composition were detected under different water treatments (PERMANOVA, F = 1.800, P = 0.005), but not under nitrogen treatments (PERMANOVA, F = 0.826, P = 0.675) and water–nitrogen interaction treatments (PERMANOVA, F = 0.952, P = 0.533) (Table 1 and Figure 2A).
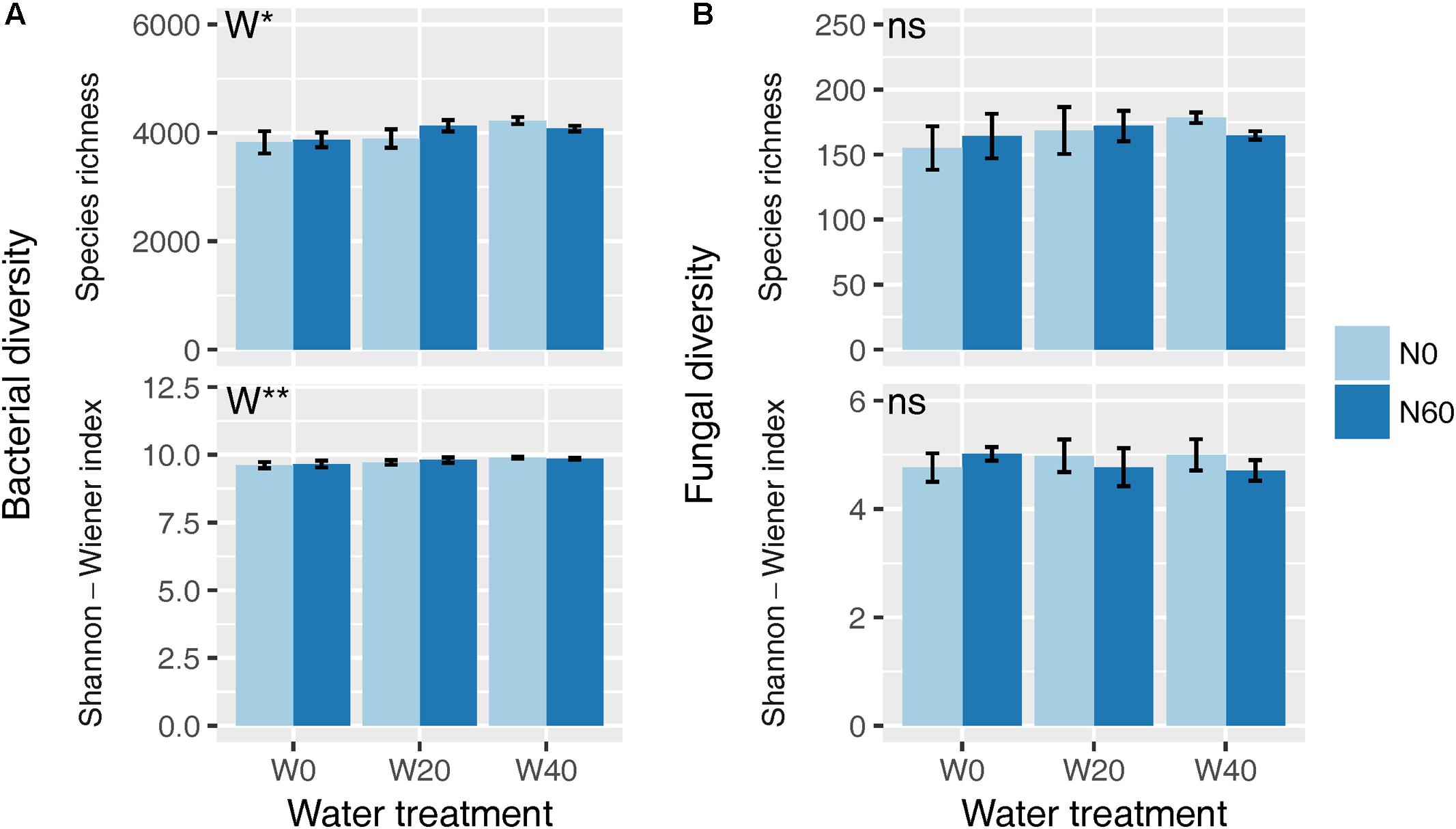
FIGURE 1. Effects of water and nitrogen addition on soil bacterial (A) and fungal (B) diversity. ∗P < 0.05, ∗∗P < 0.01; ns, not significant.
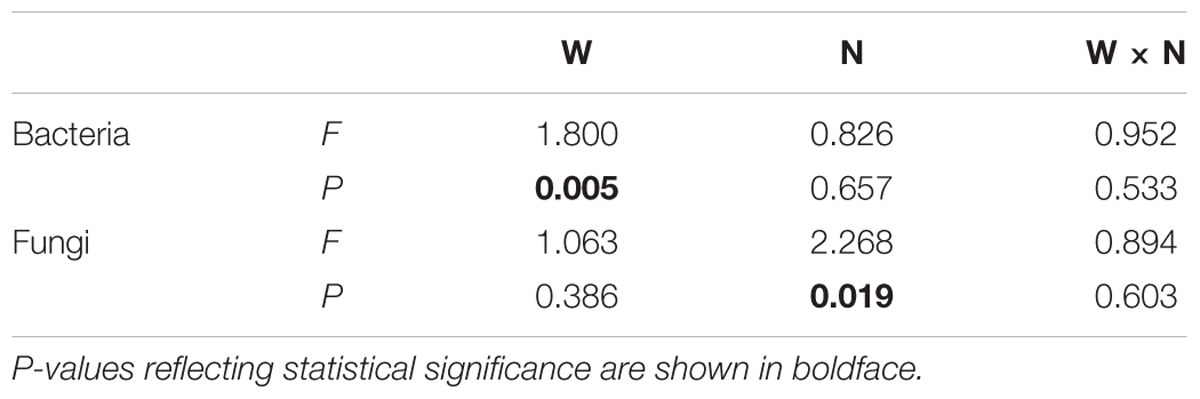
TABLE 1. Effects of water addition (W), nitrogen addition (N), and their interaction (W × N) on the compositions of soil microbial communities, as determined by PERMANOVA.
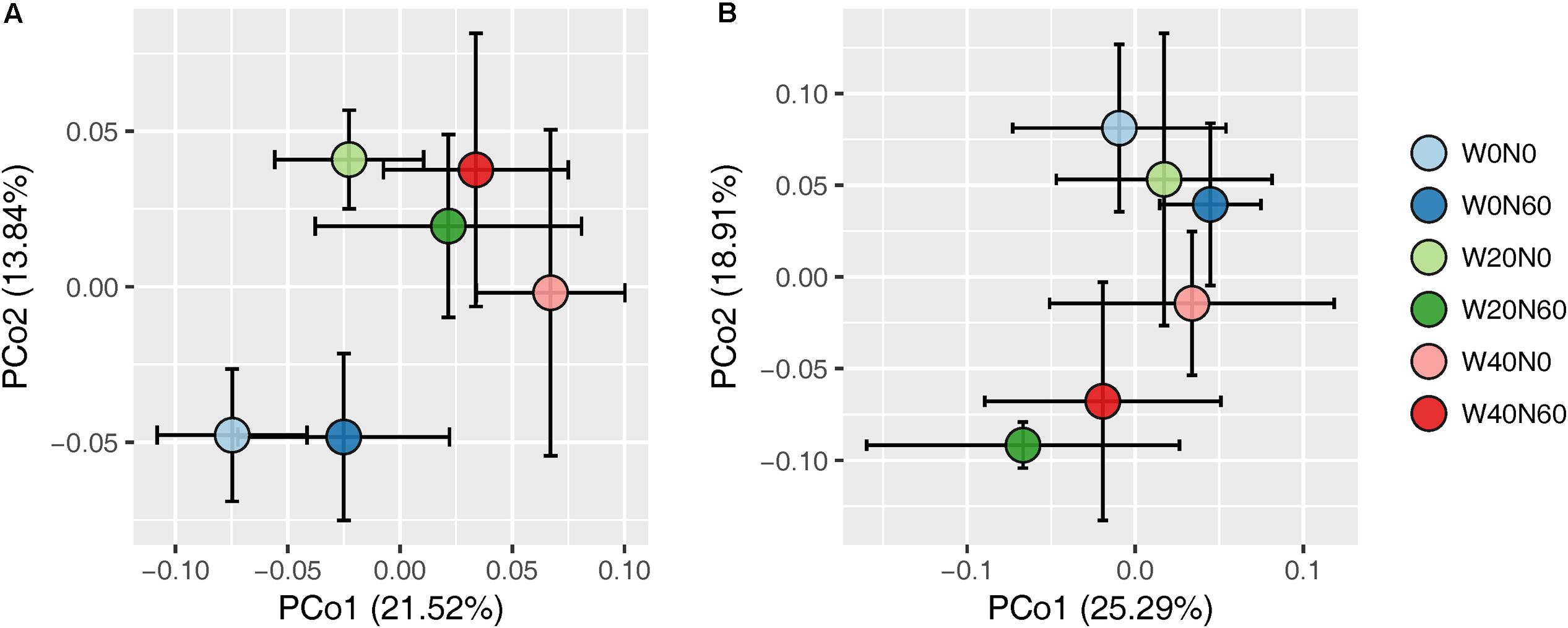
FIGURE 2. Principal coordinate analysis of soil bacterial (A) and fungal (B) community differences (Bray–Curtis dissimilarities) following different water and nitrogen treatments.
Both water and nitrogen addition had no influences on fungal species richness and SW index (Figure 1B and Supplementary Table S1). Soil fungal community composition was significantly altered under nitrogen treatments (PERMANOVA, F = 2.268, P = 0.019), but not under water treatments (PERMANOVA, F = 1.063, P = 0.386) and water–nitrogen interaction treatments (PERMANOVA, F = 0.894, P = 0.603) (Table 1 and Figure 2B).
Relative Abundance of Dominant Microbial Taxa
Soil bacterial community was dominated by Proteobacteria (38.11%), Actinobacteria (31.09%), Acidobacteria (5.46%), Bacteroidetes (4.79%), Gemmatimonadetes (4.52%), Planctomycetes (4.49%), Chloroflexi (4.07%), Cyanobacteria (2.40%), and Firmicutes (1.04%), based on the analysis of all soil samples (Supplementary Figure S2). Water addition had significant impacts on the relative abundance of Bacteroidetes, whereas nitrogen addition, alone or in combination with water, had no effects on the dominant bacterial phyla (Supplementary Table S2). Watering reduced the relative abundance of Bacteroidetes from 5.72 ± 0.59 (W0) to 4.61 ± 0.48 (W20) and 4.06 ± 0.24% (W40). Response ratio analysis showed that the relative abundances of Proteobacteria, Actinobacteria, and Bacteroidetes decreased with water addition, while Acidobacteria and Planctomycetes exhibited the opposite trend (Figure 3A). Nitrogen enrichment increased the response ratio of the Cyanobacteria relative abundance, but had no effects on that of other dominant bacterial phyla (Figure 3B).
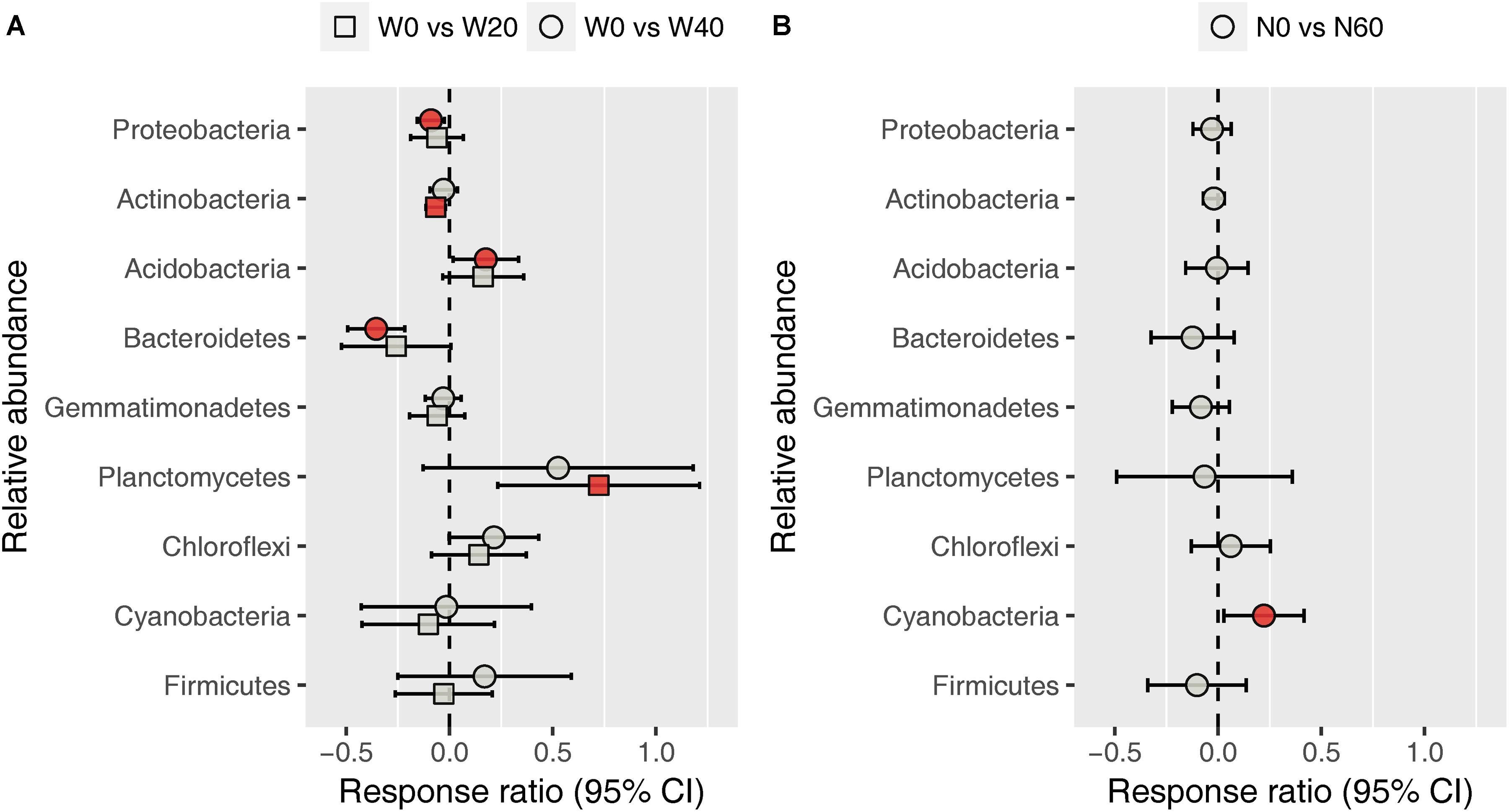
FIGURE 3. Response ratio analysis of changes in the relative abundance of dominant bacterial phyla in response to water treatment (A) and nitrogen treatment (B) compared to the control treatment, at the 95% confidence interval. Red points indicate significant changes compared with the control treatment.
The fungal community was dominated by Ascomycota (88.03%) and Basidiomycota (8.32%), while Zygomycota (1.59%) and Chytridiomycota (1.03%) were minor phyla under all treatments (Supplementary Figure S3). At the order level, we identified nine dominant orders (Pleosporales, 38.21%; Chaetothyriales, 15.63%; Hypocreales, 6.68%; Sordariales, 4.13%; Pezizales, 2.86%; Capnodiales, 2.72%; Verrucariales, 1.88%; Eurotiales, 1.80%; and Lichinales, 1.18%) in the Ascomycota division, and one order each in the Basidiomycota (Agaricales, 5.03%) and Zygomycota (Mortierellales, 1.57%) division among all soil samples studied (Supplementary Figure S4). ANOVA results showed that nitrogen enrichment significantly influenced the relative abundances of the Ascomycota and Basidiomycota phyla and the Pleosporales, Sordariales, and Agaricales orders, whereas water addition and water–nitrogen interactions showed no effects on the relative abundances of any dominant fungal taxa (Supplementary Table S3). Relative to the unfertilized plots, nitrogen addition increased the relative abundances of Ascomycota and Pleosporales by 6.38 and 23.53%, respectively, but reduced those of Basidiomycota, Sordariales, and Agaricales by 41.12, 37.53, and 67.73%, respectively. Response ratio analysis showed similar results to ANOVA, in terms of the phylum- and order-level responses of fungi to nitrogen addition (Figure 4B). In addition, response ratio analysis also demonstrated that water addition had remarkable impacts on dominant fungal taxa, with Ascomycota increasing but Basidiomycota, Agaricales, Pezizales, and Verrucariales decreasing in their relative abundances (Figure 4A).
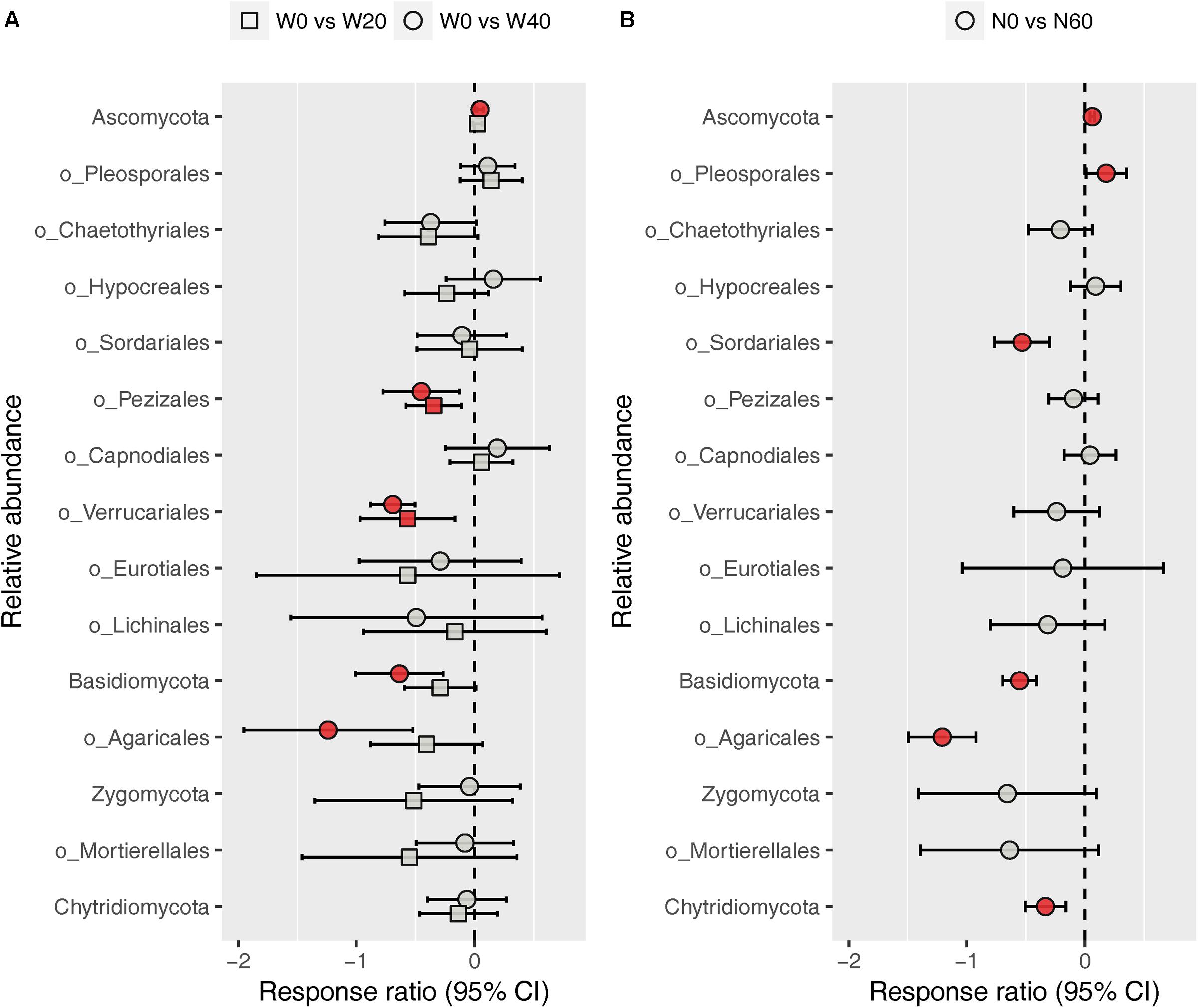
FIGURE 4. Response ratio analysis of changes in the relative abundance of dominant fungal phyla/orders in response to water treatment (A) and nitrogen treatment (B) compared to the control treatment, at the 95% confidence interval. Red points indicate significant changes compared with the control treatment.
Relationships between Microbial Communities and Plant/Soil Properties
Soil bacterial community composition strongly correlated with PH ANPP, STN, and STP, but weakly correlated with soil moisture (Table 2). Stepwise regression analysis demonstrated that bacterial diversity and the relative abundances of Bacteroidetes and Planctomycetes, whose value being altered by water addition, were mainly affected by soil moisture and total nitrogen content; those of Proteobacteria and Acidobacteria were shifted by watering, but were unrelated to any plant and soil properties measured in our study; and that of Cyanobacteria, whose abundance being increased by nitrogen addition, showed positive correlations with the ANPP of perennial shrub and herbs (Table 3). SEM results showed that water addition directly affected soil bacterial community by increasing soil moisture, while nitrogen enrichment indirectly influenced the bacterial community by changing PH ANPP (Figure 5A). The total net effect of water addition on the bacterial community was marginally significant [standardized coefficient = 0.28, Zvalue (dividing the regression weight estimate by its standard error) = 1.79, P = 0.073], whereas that of nitrogen addition was non-significant (standardized coefficient = 0.11, Zvalue = 1.35, P = 0.176).
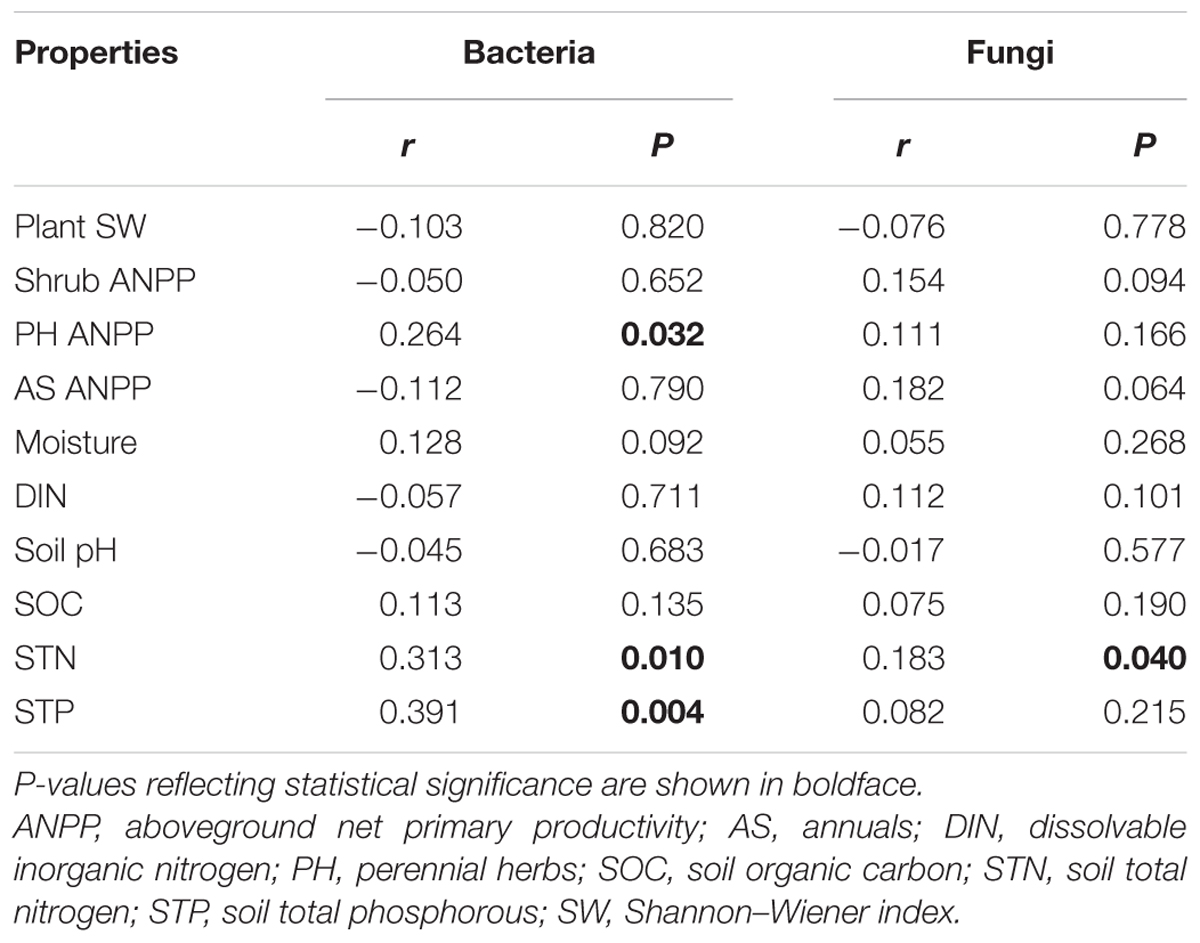
TABLE 2. Correlations between plant/soil properties and soil microbial communities (Bray–Curtis dissimilarities), as determined by Mantel test.
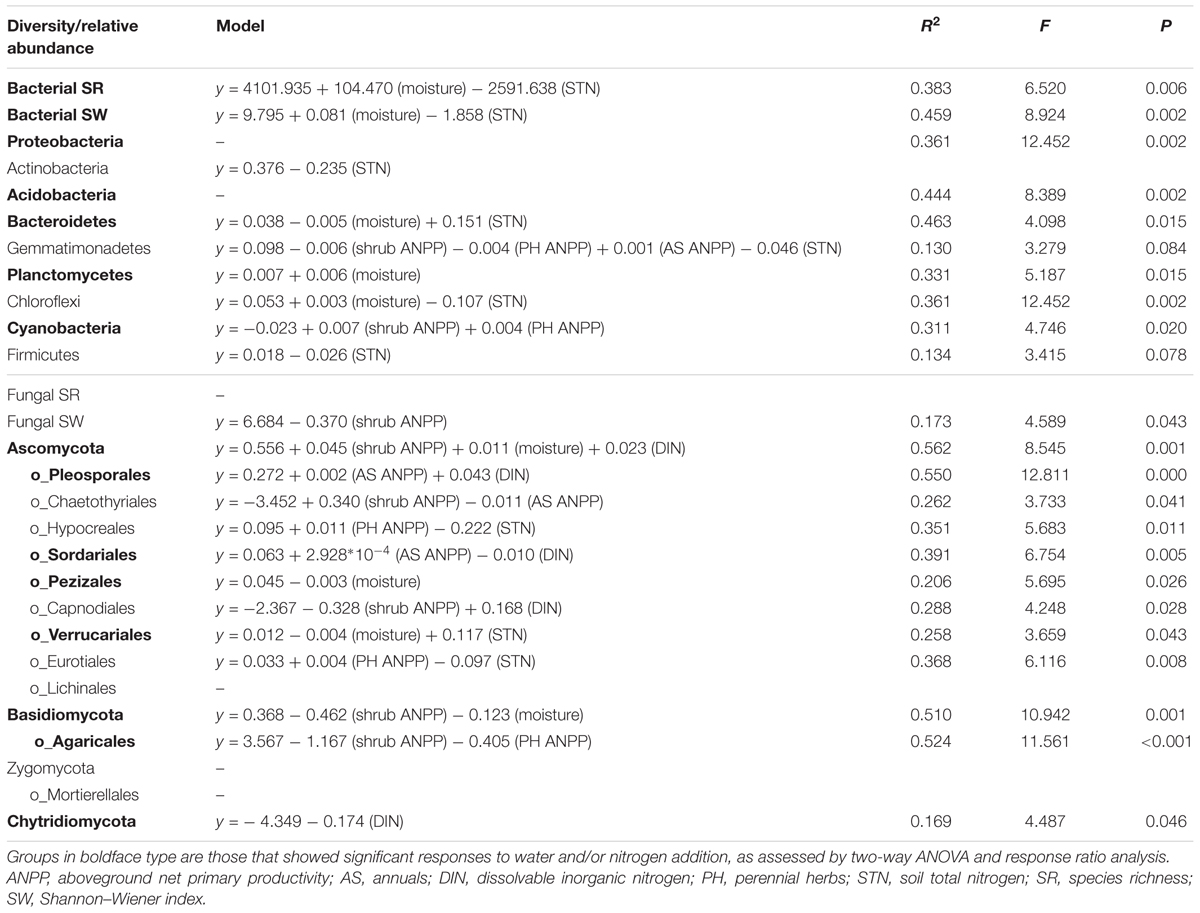
TABLE 3. Results of stepwise regression analysis of the relationships between plant/soil properties and the diversity and relative abundance of various microbial groups.
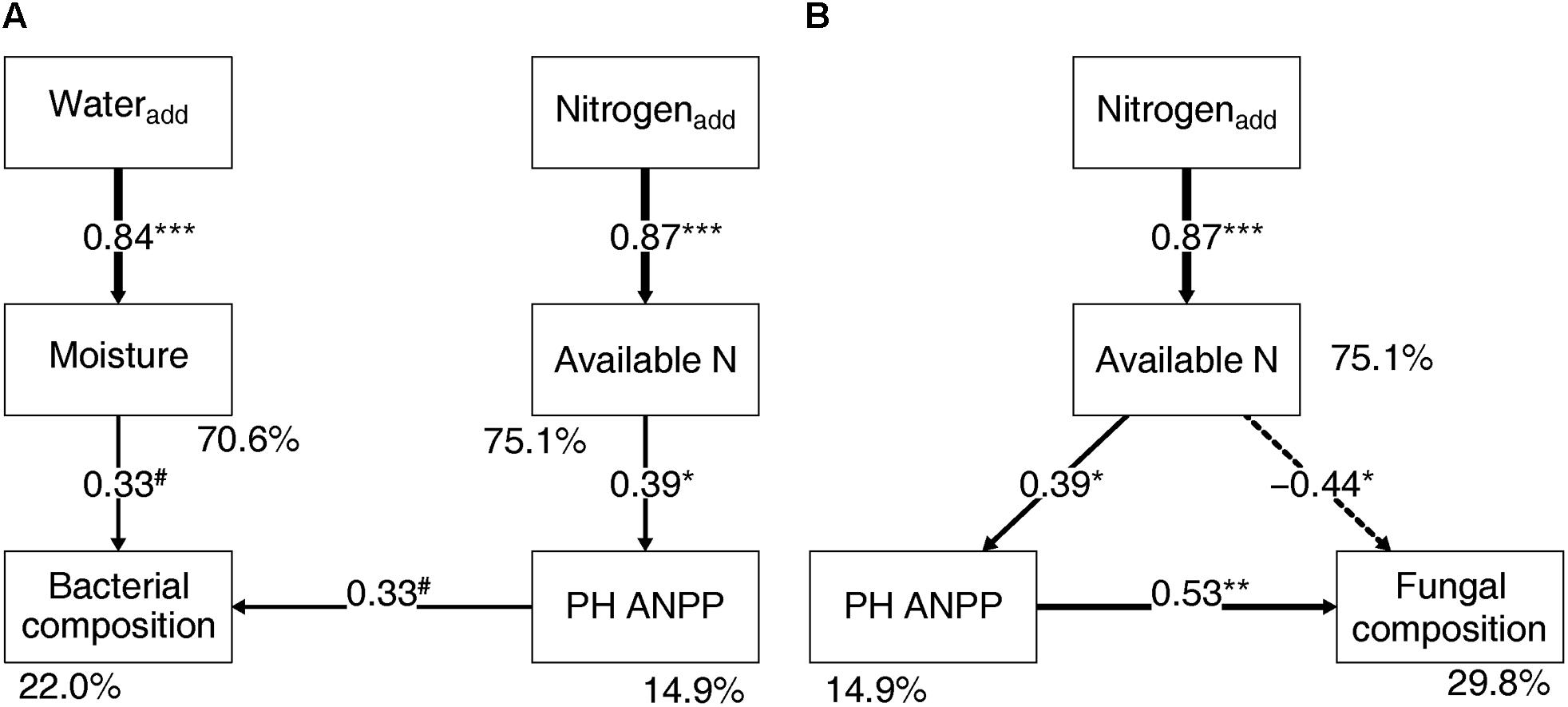
FIGURE 5. Structural equation modeling showing the relationships between plant/soil properties and the bacterial (A) and fungal (B) community compositions. Solid arrows indicate positive effects, and the dashed arrow indicates a negative correlation. The standardized path coefficients are adjacent to the arrows and indicate the effect size of the relationship. Arrow widths are proportional to the strength of each relationship. Percentages beside the response variables refer to the proportion of variance explained by the model (R2). Results of model fitting: (A) bacteria: χ2 = 10.096, df = 9, P = 0.343; CFI = 0.984; AIC = 353.187; RMSEA = 0.071, P = 0.391; (B) fungi: χ2 = 2.052, df = 2, P = 0.358; CFI = 0.999; AIC = 238.585; RMSEA = 0.033, P = 0.380. PH ANPP, the aboveground net primary productivity of perennial herbs. #P < 0.07, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
The fungal community composition strongly correlated with STN and weakly correlated with the ANPP of shrub and annual plants (Table 2). Stepwise regression analysis revealed that fungal diversity and the relative abundances of Ascomycota and Basidiomycota strongly correlated with shrub ANPP (Table 3). The dominant fungal phyla Ascomycota, Basidiomycota, and Chytridiomycota were shifted by water and nitrogen addition, and also showed strong correlations with soil moisture and/or available nitrogen (Table 3). In the order level, the relative abundances of Pezizales and Verrucariales, whose abundance being reduced by water addition, were negatively correlated with soil moisture and total nitrogen content; that of Pleosporales increased with nitrogen enrichment and showed positive correlations with AS ANPP and soil available nitrogen; that of Sordariales, whose abundance being reduced by nitrogen fertilization, showed a positive correlation with AS ANPP, but a negative correlation with soil available nitrogen; that of Agaricales, whose abundance being decreased in both water and nitrogen treatments, showed negative correlations with the ANPP of perennial shrub and herbs (Table 3). SEM analysis demonstrated that nitrogen enrichment could directly affect the fungal community by increasing nitrogen availability and indirectly via changing PH ANPP (Figure 5B). The total net direct effect of nitrogen enrichment was significant (standardized coefficient = -0.38, Zvalue = -2.30, P = 0.022), whereas the indirect effect was non-significant (standardized coefficient = 0.18, Zvalue = 1.64, P = 0.101).
Discussion
Precipitation Increment But Not Nitrogen Enrichment Significantly Influenced Desert Bacterial Community
Our results showed that water addition significantly increased soil bacterial diversity, and altered the bacterial community composition, with Acidobacteria and Planctomycetes increasing but Proteobacteria and Bacteroidetes decreasing in terms of their relative abundances (Figures 1–3 and Table 1). These results suggest that water addition tended to promote flourishing of oligotrophic taxa and depress copiotrophic taxa. Many species of oligotrophic taxa (e.g., Acidobacteria) have been shown to act as stress tolerators, which can enter a dormant state to evade stressful conditions (Fierer, 2017; Tecon and Or, 2017). As desert environments are water-stressful, alleviating that stress following water addition would revive dormant microorganisms and increase their abundances (Tecon and Or, 2017). However, this shifting trend in bacterial taxa in our study contrasted with the results from studies performed in a temperate grassland, in which water addition tends to increase the relative abundance of Proteobacteria and decrease that of Acidobacteria (Zhang et al., 2013, 2014). One possible mechanism underlying the responses of bacterial phyla to water addition in grasslands was that watering could increase nitrogen availability by stimulating the mineralization of soil organic matter (Li et al., 2016, 2017b). However, in our study, soil nitrogen availability was not affected by water addition, probably due to the quite low amount of soil organic matter (Supplementary Table S1), which is roughly equivalent to one-seventh of that in grassland soils (Zeng et al., 2016; Li et al., 2017b). Alternatively, this contrary response was likely due to the high sensitivity of oligotrophic taxa to water addition in the more stressful desert environment. Evidence from aridity-gradient studies in northern China indicates that the relative abundances of Acidobacteria and Planctomycetes increase with an increasing aridity index (AI, estimated by the ratio of precipitation to potential evapotranspiration) in more arid areas (AI < 0.2), but exhibit no further variation at higher AI values (Wang X. et al., 2015; Wang et al., 2017); whereas the relative abundances of Bacteroidetes and most Proteobacteria subphyla show non-linear relationships with AI, with the lowest value being present at AI ≈ 0.2 (Wang X. et al., 2015). The AI in our study site is 0.14; thus, oligotrophic taxa were expected to increase and copiotrophic taxa were expected to decrease with water addition. The dormancy of oligotrophic taxa might be more prevalent in more stressful environments, resulting in a larger population recovery when conditions improve and further depress copiotrophic taxa.
In contrast to previous findings, neither the bacterial diversity nor the community composition was substantially affected by nitrogen addition in our study (Figures 1, 2 and Table 1). Among all dominant bacterial phyla identified, only Cyanobacteria were slightly increased with nitrogen input in terms of the relative abundance (Figure 3B). Previously, the relative abundance of Cyanobacteria has been shown a non-linear response to nitrogen enrichment, with an increase under moderate nitrogen input (35-70 kg N ha-1 yr-1) and a decrease under excess nitrogen addition (140 kg N ha-1 yr-1) (Wang J. et al., 2015). The overall unresponsiveness of the bacterial community following nitrogen enrichment was also reported in fertilization experiments conducted in semiarid grasslands (Carey et al., 2015; McHugh et al., 2017). There are several potential explanations regarding these insensitive responses. The low nitrogen-retention capacity of desert soil is likely to weaken the effects of nitrogen addition (Jin et al., 2015; McHugh et al., 2017). In addition, data from numerous fertilization studies have shown that nitrogen-induced soil acidification is an important mechanism in shifting the bacterial community composition (Chen et al., 2015; Zeng et al., 2016; Zhang et al., 2017b). However, in our study, nitrogen addition did not influence the soil pH (Supplementary Table S1), which was likely due to the high buffering capacity of the soil (Liu et al., 2015). Although direct effects of nitrogen addition on the bacterial community were not detected, our results suggest that nitrogen addition could indirectly influence the bacterial community by changing PH ANPP (Figure 5A). In grasslands, many field studies have shown that nitrogen-induced changes in aboveground plant biomass are important drivers of shifting bacterial communities (Chen et al., 2015; Yuan et al., 2016). We also found that nitrogen addition increased the biomass of PH (Supplementary Table S1), although this indirect effect was weak (Figure 5A). It is possible that the effects of nitrogen deposition, as seen in the fertilization experiments, take longer to emerge than the 2-year observation period of this study.
Increased Precipitation and Nitrogen Altered the Drought-Tolerant and Trophic Traits of Desert Fungal Community
In contrast to the bacterial community, both water and nitrogen addition significantly impacted the fungal community composition, but not the fungal diversity (Figures 1, 2 and Table 1). Nitrogen addition mainly increased the relative abundance of Ascomycota and decreased that of Basidiomycota (Figure 4B and Supplementary Table S3). It was suggested that the phyla Ascomycota and Basidiomycota could roughly grouped into copiotrophic and oligotrophic taxa, respectively (Ho et al., 2017; Yao et al., 2017), suggesting that they would respond oppositely to nitrogen enrichment. Moreover, many field experiments conducted in diverse ecosystems have also revealed that Basidiomycota decreased in relative abundance following nitrogen input (Allison et al., 2007; Entwistle et al., 2013; Chen et al., 2018). We also found that the relative abundance of Chytridiomycota decreased under nitrogen treatment; however, its relative proportion was quite small (∼1%), and thus its variation can be negligible. The increased relative abundance of Ascomycota following nitrogen addition was primarily due to an increased relative abundance of Pleosporales, which was the predominant fungal order in our study. Members in the order Pleosporales have been shown to live as plant endophytes in arid grass species and were shown to transfer nutrients between plants and nearby soil (Green et al., 2008; Porras-Alfaro et al., 2011). Our stepwise regression analysis also indicated that the relative abundance of Pleosporales was positively correlated with annual plants and soil inorganic nitrogen (Table 3). Previous data also demonstrated that the relative abundance of Pleosporales increases with nitrogen enrichment (Lowell and Klein, 2001; Wang J. et al., 2015), which supports our findings. In contrast to Pleosporales, members of the orders Sordariales (Ascomycota) and Agaricales (Basidiomycota) are considered as the potent degraders of lignin and predominantly show oligotrophic features (Poggeler, 2011; Entwistle et al., 2013; Ho et al., 2017). Results from stepwise regression showed that the relative abundance of Sordariales was negatively correlated with soil available nitrogen, while that of Agaricales showed a negative correlation with PH ANPP, suggesting that nitrogen enrichment was likely to directly affect Sordariales relative abundance via increasing nitrogen availability, whereas indirectly influence Agaricales by changing PH ANPP (Table 3). Taken together, our results indicated that nitrogen enrichment tended to depress oligotrophic fungal taxa, but to facilitate taxa regarding nutrient transfer.
Our response ratio analysis indicated that water addition tended to favor Ascomycota over Basidiomycota (Figure 4A). The ANOVA and PERMANOVA results showed that the fungal community composition was unresponsive to water addition (Table 1, Figure 2, and Supplementary Table S3), probably due to the large variation in our fungal data. Data from many field experiments have demonstrated that drought treatment decreases the relative abundance of Ascomycota and increases that of Basidiomycota (McHugh and Schwartz, 2014; He et al., 2016; Bastida et al., 2017), suggesting that Basidiomycota adapt better to drought conditions. Our stepwise regression analysis also revealed that the relative abundance of Ascomycota was positively correlated with soil moisture, while that of Basidiomycota exhibited an opposite pattern (Table 3). Analyzing the order-level responses of fungi to water addition demonstrated that the relative abundances of Pezizales (Ascomycota), Verrucariales (Ascomycota), and Agaricales (Basidiomycota) decreased with water input, indicating that the change of Basidiomycota was mainly caused by Agaricales and the increase in Ascomycota relative abundance was likely driven by many low-abundance taxa. Results from stepwise regression showed that the relative abundances of Pezizales and Verrucariales were negatively correlated with soil moisture (Table 3), suggesting that these fungal orders might prefer to live in drought conditions. Previous studies have also revealed that species of the order Pezizales are important members of ectomycorrhizal fungi (Tedersoo et al., 2006; Healy et al., 2013) and adapt well in water-stressed environments (Smith et al., 2006; Gordon and Gehring, 2011). In contrast to Pezizales, most species of the order Verrucariales are characterized as lichen-forming fungi (Gueidan et al., 2007; Wang et al., 2014). Previous physiological data indicated that, in arid areas, these fungi are extremely drought-tolerant and, thus, are expected to adapt well in desert environments (Wang Y. et al., 2015; Zhang et al., 2017a). These studies indirectly supported our findings that water addition reduced the relative abundances of drought-adapted fungi.
Precipitation and Nitrogen Addition Primarily Directly Affected Desert Microbial Communities via Changing Resource Availability
The present study indicated that water and nitrogen addition resulted in stronger direct effects on soil microbial communities through changing resource availability rather than indirect influences via changes of plant community (Table 3 and Figure 5). Specially, shifts in the fungal community composition following water and nitrogen enrichment were primarily caused by the changes of water and nitrogen availability, whereas shifts in the bacterial community composition were mainly driven by changes in soil moisture. Fungal and bacterial communities responded in different ways to resource availability, probably due to their distinctive adaptive strategies to desert environments. Fungi are typically more drought-tolerant and nutrient-sensitive than bacteria, attributed to their ability to access soil water and nutrients better through hyphal networks (Boer et al., 2005; Yuste et al., 2011; Manzoni et al., 2012). Consequently, when water and nitrogen availability are improved, drought- and oligotrophic-adapted fungal taxa are expected to be suppressed (Crowther et al., 2014). However, desert bacteria probably show limited responses to a moderate rate of nitrogen fertilization due to their relatively low ability for nutrient acquisition and the low nutrient-retention capacity of desert soils. In contrast to nitrogen availability, desert bacteria are likely more sensitive to the improvement of soil moisture, probably due to the revival of dormant taxa when water stress is alleviated (Lennon and Jones, 2011; Tecon and Or, 2017).
The SEM results suggest that nitrogen enrichment could indirectly influence soil microbial communities via altering PH ANPP, while the indirect effects of water addition were not detectable (Figure 5). Although the indirect nitrogen effects were found in our study, their strength was relatively weaker than the direct effects. Our findings were not in line with the results reported in grassland ecosystems, where nitrogen-induced shifts in microbial community composition are mainly indirectly mediated by changes of soil pH and/or plant community rather than directly via changing resource availability (Chen et al., 2015; Yuan et al., 2016; Zeng et al., 2016). It is likely that desert ecosystems are more resource limited than grasslands, suggesting that desert microbial communities are likely more sensitive to resource availability. The high buffering capacity of our soils might weaken the effects of nitrogen-induced soil acidification. It is also possible that a stronger indirect effect of nitrogen enrichment can be seen in a long-term experimental treatment. Owing to limitations of our short-term experiment, further filed studies with longer observation period will be necessary to disentangle the direct and indirect influences of global environmental changes on desert microbial communities.
Conclusions
In summary, our results indicated that soil microbial communities responded differently to increased precipitation and nitrogen in this desert ecosystem. Watering increased soil bacterial diversity and shifted the community composition by promoting the flourishing of stress-tolerant (dormant) taxa, whereas nitrogen enrichment had no substantial effects. Water and nitrogen addition did not influence soil fungal diversity, but significantly altered the community composition with drought-adapted taxa being suppressed following water addition and with oligotrophic-adapted taxa being suppressed under nitrogen enrichment. Water- and nitrogen-induced changes in soil microbial communities arose mainly through altering resource availability rather than plant community. Our results suggest that water addition affected desert microbial communities by altering their stress-tolerant traits, while nitrogen enrichment shifted their copiotrophic/oligotrophic traits. Although, in a short-term experiment, our findings highlight the importance of resource availability in driving the desert microbial responses to altered environmental conditions, further long-term study is needed to help in better understanding the responses of desert microbial communities to global environmental changes.
Author Contributions
WS, YZ, and SQ conceived of the study and designed the methodology. WS, YB, and JZ collected the data. WS analyzed the data. WS led the writing of the manuscript. YB, YZ, SQ, WF, YS, and BW assisted with revising the draft manuscript. All authors approved of the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31470711) and the National Key Research and Development Program of China (Grant No. 2016YFC0500905).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shijun Liu, Jie Fu, Yangui Qiao, and the staff of the Yanchi Research Station for their assistance in the field and laboratory. We are grateful to the reviewers for their valuable comments that helped improve this paper.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00186/full#supplementary-material
References
Allison, S. D., Hanson, C. A., and Treseder, K. K. (2007). Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 39, 1878–1887. doi: 10.1016/j.soilbio.2007.02.001
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Bastida, F., Torres, I. F., Andres-Abellan, M., Baldrian, P., Lopez-Mondejar, R., Vetrovsky, T., et al. (2017). Differential sensitivity of total and active soil a microbial communities to drought and forest management. Glob. Change Biol. 23, 4185–4203. doi: 10.1111/gcb.13790
Bobbink, R., Hicks, K., Galloway, J., Spranger, T., Alkemade, R., Ashmore, M., et al. (2010). Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl. 20, 30–59. doi: 10.1890/08-1140.1
Boer, W., Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Byrne, K. M., Adler, P. B., and Lauenroth, W. K. (2017). Contrasting effects of precipitation manipulations in two Great Plains plant communities. J. Veg. Sci. 28, 238–249. doi: 10.1111/jvs.12486
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carey, C. J., Beman, J. M., Eviner, V. T., Malmstrom, C. M., and Hart, S. C. (2015). Soil microbial community structure is unaltered by plant invasion, vegetation clipping, and nitrogen fertilization in experimental semi-arid grasslands. Front. Microbiol. 6:466. doi: 10.3389/fmicb.2015.00466
Chen, D. M., Lan, Z. C., Hu, S. J., and Bai, Y. F. (2015). Effects of nitrogen enrichment on belowground communities in grassland: relative role of soil nitrogen availability vs. soil acidification. Soil Biol. Biochem. 89, 99–108. doi: 10.1016/j.soilbio.2015.06.028
Chen, W., Xu, R., Wu, Y., Chen, J., Zhang, Y., Hu, T., et al. (2018). Plant diversity is coupled with beta not alpha diversity of soil fungal communities following N enrichment in a semi-arid grassland. Soil Biol. Biochem. 116, 388–398. doi: 10.1016/j.soilbio.2017.10.039
Chen, Y. L., Xu, T. L., Veresoglou, S. D., Hu, H. W., Hao, Z. P., Hu, Y. J., et al. (2017). Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biol. Biochem. 110, 12–21. doi: 10.1016/j.soilbio.2017.02.015
Cregger, M. A., Schadt, C. W., McDowell, N. G., Pockman, W. T., and Classen, A. T. (2012). Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl. Environ. Microbiol. 78, 8587–8594. doi: 10.1128/AEM.02050-2012
Crowther, T. W., Maynard, D. S., Crowther, T. R., Peccia, J., Smith, J. R., and Bradford, M. A. (2014). Untangling the fungal niche: the trait-based approach. Front. Microbiol. 5:579. doi: 10.3389/fmicb.2014.00579
Curiel Yuste, J., Fernandez-Gonzalez, A. J., Fernandez-Lopez, M., Ogaya, R., Penuelas, J., Sardans, J., et al. (2014). Strong functional stability of soil microbial communities under semiarid Mediterranean conditions and subjected to long-term shifts in baseline precipitation. Soil Biol. Biochem. 69, 223–233. doi: 10.1016/j.soilbio.2013.10.045
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Entwistle, E. M., Zak, D. R., and Edwards, I. P. (2013). Long-term experimental nitrogen deposition alters the composition of the active fungal community in the forest floor. Soil Sci. Soc. Am. J. 77, 1648–1658. doi: 10.2136/sssaj2013.05.0179
Evans, S. E., and Wallenstein, M. D. (2014). Climate change alters ecological strategies of soil bacteria. Ecol. Lett. 17, 155–164. doi: 10.1111/ele.12206
Evans, S. E., Wallenstein, M. D., and Burke, I. C. (2014). Is bacterial moisture niche a good predictor of shifts in community composition under long-term drought? Ecology 95, 110–122. doi: 10.1890/13-0500.1
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 109, 21390–21395. doi: 10.1073/pnas.1215210110
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Gao, G. L., Ding, G. D., Zhao, Y. Y., Wu, B., Zhang, Y. Q., Qin, S. G., et al. (2014). Fractal approach to estimating changes in soil properties following the establishment of Caragana korshinskii shelterbelts in Ningxia, NW China. Ecol. Indic. 43, 236–243. doi: 10.1016/j.ecolind.2014.03.001
Gordon, G. J., and Gehring, C. A. (2011). Molecular characterization of pezizalean ectomycorrhizas associated with pinyon pine during drought. Mycorrhiza 21, 431–441. doi: 10.1007/s00572-010-0349-8
Grace, J. (2006). Structural Equation Modeling and Natural Systems. New York, NY: Cambridge University Press. doi: 10.1017/CBO9780511617799
Green, L. E., Porras-Alfaro, A., and Sinsabaugh, R. L. (2008). Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J. Ecol. 96, 1076–1085. doi: 10.1111/j.1365-2745.2008.01388.x
Gueidan, C., Roux, C., and Lutzoni, F. (2007). Using a multigene phylogenetic analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycol. Res. 111, 1145–1168. doi: 10.1016/j.mycres.2007.08.010
Gutknecht, J. L. M., Field, C. B., and Balser, T. C. (2012). Microbial communities and their responses to simulated global change fluctuate greatly over multiple years. Glob. Change Biol. 18, 2256–2269. doi: 10.1111/j.1365-2486.2012.02686.x
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
He, D., Xiang, X. J., He, J. S., Wang, C., Cao, G. M., Adams, J., et al. (2016). Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan Plateau. Biol. Fertil. Soils 52, 1059–1072. doi: 10.1007/s00374-016-1142-4
Healy, R. A., Smith, M. E., Bonito, G. M., Pfister, D. H., Ge, Z. W., Guevara, G. G., et al. (2013). High diversity and widespread occurrence of mitotic spore mats in ectomycorrhizal Pezizales. Mol. Ecol. 22, 1717–1732. doi: 10.1111/mec.12135
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93;fix006. doi: 10.1093/femsec/fix006
Jia, X., Zha, T. S., Gong, J. N., Wang, B., Zhang, Y. Q., Wu, B., et al. (2016). Carbon and water exchange over a temperate semi-arid shrubland during three years of contrasting precipitation and soil moisture patterns. Agric. For. Meteorol. 228, 120–129. doi: 10.1016/j.agrformet.2016.07.007
Jin, Z., Zhu, Y., Li, X., Dong, Y., and An, Z. (2015). Soil N retention and nitrate leaching in three types of dunes in the Mu Us Desert of China. Sci. Rep. 5:14222. doi: 10.1038/srep14222
Leff, J. W., Jones, S. E., Prober, S. M., Barberan, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130. doi: 10.1038/nrmicro2504
Li, H., Xu, Z., Yan, Q., Yang, S., Van Nostrand, J. D., Wang, Z., et al. (2017a). Soil microbial beta-diversity is linked with compositional variation in aboveground plant biomass in a semi-arid grassland. Plant Soil doi: 10.1007/s11104-017-3524-2
Li, H., Yang, S., Xu, Z. W., Yan, Q. Y., Li, X. B., van Nostrand, J. D., et al. (2017b). Responses of soil microbial functional genes to global changes are indirectly influenced by aboveground plant biomass variation. Soil Biol. Biochem. 104, 18–29. doi: 10.1016/j.soilbio.2016.10.009
Li, H., Xu, Z., Yang, S., Li, X., Top, E. M., Wang, R., et al. (2016). Responses of soil bacterial communities to nitrogen deposition and precipitation increment are closely linked with aboveground community variation. Microb. Ecol. 71, 974–989. doi: 10.1007/s00248-016-0730-z
Liu, J., Wu, N., Wang, H., Sun, J., Peng, B., Jiang, P., et al. (2016). Nitrogen addition affects chemical compositions of plant tissues, litter and soil organic matter. Ecology 97, 1796–1806. doi: 10.1890/15-1683.1
Liu, J. B., Fa, K. Y., Zhang, Y. Q., Wu, B., Qin, S. G., and Jia, X. (2015). Abiotic CO2 uptake from the atmosphere by semiarid desert soil and its partitioning into soil phases. Geophys. Res. Lett. 42, 5779–5785. doi: 10.1002/2015gl064689
Lowell, J. L., and Klein, D. A. (2001). Comparative single-strand conformation polymorphism (SSCP) and microscopy-based analysis of nitrogen cultivation interactive effects on the fungal community of a semiarid steppe soil. FEMS Microbiol. Ecol. 36, 85–92. doi: 10.1111/j.1574-6941.2001.tb00828.x
Maestre, F. T., Eldridge, D. J., Soliveres, S., Kefi, S., Delgado-Baquerizo, M., Bowker, M. A., et al. (2016). Structure and functioning of dryland ecosystems in a changing world. Annu. Rev. Ecol. Evol. Syst. 47, 215–237. doi: 10.1146/annurev-ecolsys-121415-032311
Maestre, F. T., Salguero-Gomez, R., and Quero, J. L. (2012). It is getting hotter in here: determining and projecting the impacts of global environmental change on drylands. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 3062–3075. doi: 10.1098/rstb.2011.0323
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Makhalanyane, T. P., Valverde, A., Gunnigle, E., Frossard, A., Ramond, J. B., and Cowan, D. A. (2015). Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 39, 203–221. doi: 10.1093/femsre/fuu011
Manzoni, S., Schimel, J. P., and Porporato, A. (2012). Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93, 930–938. doi: 10.1890/11-0026.1
McHugh, T. A., Morrissey, E. M., Mueller, R. C., Gallegos-Graves, V., Kuske, C. R., and Reed, S. C. (2017). Bacterial, fungal, and plant communities exhibit no biomass or compositional response to two years of simulated nitrogen deposition in a semiarid grassland. Environ. Microbiol. 19, 1600–1611. doi: 10.1111/1462-2920.13678
McHugh, T. A., and Schwartz, E. (2014). Changes in plant community composition and reduced precipitation have limited effects on the structure of soil bacterial and fungal communities present in a semiarid grassland. Plant Soil 388, 175–186. doi: 10.1007/s11104-014-2269-4
Nie, M., Pendall, E., Bell, C., Gasch, C. K., Raut, S., Tamang, S., et al. (2013). Positive climate feedbacks of soil microbial communities in a semi-arid grassland. Ecol. Lett. 16, 234–241. doi: 10.1111/ele.12034
Noy-Meir, I. (1973). Desert ecosystems: environment and producers. Annu. Rev. Ecol. Syst. 4, 25–51. doi: 10.1146/annurev.es.04.110173.000325
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Package ‘vegan’. Community ecology package, version 2.9. Available at: http://CRAN.R-project.org/package=vegan
Poggeler, S. (2011). Evolution of multicopper oxidase genes in coprophilous and non-coprophilous members of the order sordariales. Curr. Genomics 12, 95–103. doi: 10.2174/138920211795564368
Porras-Alfaro, A., Herrera, J., Natvig, D. O., Lipinski, K., and Sinsabaugh, R. L. (2011). Diversity and distribution of soil fungal communities in a semiarid grassland. Mycologia 103, 10–21. doi: 10.3852/09-297
Prober, S. M., Leff, J. W., Bates, S. T., Borer, E. T., Firn, J., Harpole, W. S., et al. (2015). Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95. doi: 10.1111/ele.12381
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramirez, K. S., Craine, J. M., and Fierer, N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 18, 1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x
Ren, H. Y., Xu, Z. W., Huang, J. H., Lu, X. T., Zeng, D. H., Yuan, Z. Y., et al. (2015). Increased precipitation induces a positive plant-soil feedback in a semi-arid grassland. Plant Soil 389, 211–223. doi: 10.1007/s11104-014-2349-5
Revelle, W. (2016). psych: Procedures for Personality and Psychological Research. Evanston: Northwestern University.
She, W., Zhang, Y., Qin, S., Wu, B., and Bai, Y. (2016). Increased precipitation and nitrogen alter shrub architecture in a desert shrubland: implications for primary production. Front. Plant Sci. 7:1908. doi: 10.3389/fpls.2016.01908
She, W. W., Zhang, Y. Q., Qin, S. G., Wu, B., Liu, Z., Liu, J., et al. (2015). Habitat effect on allometry of a xeric shrub (Artemisia ordosica Krasch) in the Mu Us Desert of northern China. Forests 6, 4529–4539. doi: 10.3390/f6124385
Smith, M. E., Trappe, J. M., and Rizzo, D. M. (2006). Genea, Genabea and Gilkeya gen. nov.: ascomata and ectomycorrhiza formation in a Quercus woodland. Mycologia 98, 699–716. doi: 10.1080/15572536.2006.11832642
Sun, Y., Zhang, Y., Feng, W., Qin, S., Liu, Z., Bai, Y., et al. (2016). Effects of xeric shrubs on soil microbial communities in a desert in northern China. Plant Soil 414, 281–294. doi: 10.1007/s11104-016-3111-y
Tecon, R., and Or, D. (2017). Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 41, 599–623. doi: 10.1093/femsre/fux039
Tedersoo, L., Hansen, K., Perry, B. A., and Kjoller, R. (2006). Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol. 170, 581–596. doi: 10.1111/j.1469-8137.2006.01678.x
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S, 4th Edn. New York, NY: Springer. doi: 10.1007/978-0-387-21706-2
Wang, J., Bao, J. T., Su, J. Q., Li, X. R., Chen, G. X., and Ma, X. F. (2015). Impact of inorganic nitrogen additions on microbes in biological soil crusts. Soil Biol. Biochem. 88, 303–313. doi: 10.1016/j.soilbio.2015.06.004
Wang, J., Zhang, T., Li, L., Li, J., Feng, Y., and Lu, Q. (2017). The patterns and drivers of bacterial and fungal beta-diversity in a typical dryland ecosystem of northwest China. Front. Microbiol. 8:2126. doi: 10.3389/fmicb.2017.02126
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Van Nostrand, J. D., Deng, Y., Lu, X., Wang, C., Zhou, J., et al. (2015). Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. FEMS Microbiol. Ecol. 91:fiv133. doi: 10.1093/femsec/fiv133
Wang, Y., Zhang, X., Zhou, Q., Zhang, X., and Wei, J. (2015). Comparative transcriptome analysis of the lichen-forming fungus Endocarpon pusillum elucidates its drought adaptation mechanisms. Sci. China Life Sci. 58, 89–100. doi: 10.1007/s11427-014-4760-9
Wang, Y. Y., Liu, B., Zhang, X. Y., Zhou, Q. M., Zhang, T., Li, H., et al. (2014). Genome characteristics reveal the impact of lichenization on lichen-forming fungus Endocarpon pusillum Hedwig (Verrucariales, Ascomycota). BMC Genomics 15:34. doi: 10.1186/1471-2164-15-34
Wardle, D. A., Yeates, G. W., Barker, G. M., and Bonner, K. I. (2006). The influence of plant litter diversity on decomposer abundance and diversity. Soil Biol. Biochem. 38, 1052–1062. doi: 10.1016/j.soilbio.2005.09.003
Wei, C., Yu, Q., Bai, E., Lu, X., Li, Q., Xia, J., et al. (2013). Nitrogen deposition weakens plant-microbe interactions in grassland ecosystems. Glob. Change Biol. 19, 3688–3697. doi: 10.1111/gcb.12348
Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis, 2nd Edn. New York, NY: Springer. doi: 10.1007/978-0-387-98141-3
Xu, W., Luo, X. S., Pan, Y. P., Zhang, L., Tang, A. H., Shen, J. L., et al. (2015). Quantifying atmospheric nitrogen deposition through a nationwide monitoring network across China. Atmos. Chem. Phys. 15, 12345–12360. doi: 10.5194/acp-15-12345-2015
Xu, Z., Ren, H. Y., Li, M. H., van Ruijven, J., Han, X. G., Wan, S. Q., et al. (2015). Environmental changes drive the temporal stability of semi-arid natural grasslands through altering species asynchrony. J. Ecol. 103, 1308–1316. doi: 10.1111/1365-2745.12441
Yao, F., Yang, S., Wang, Z., Wang, X., Ye, J., Wang, X., et al. (2017). Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front. Microbiol. 8:2071. doi: 10.3389/fmicb.2017.02071
Yuan, X., Knelman, J. E., Gasarch, E., Wang, D. L., Nemergut, D. R., and Seastedt, T. R. (2016). Plant community and soil chemistry responses to long-term nitrogen inputs drive changes in alpine bacterial communities. Ecology 97, 1543–1554. doi: 10.1890/15-1160.1
Yuste, J. C., Penuelas, J., Estiarte, M., Garcia-Mas, J., Mattana, S., Ogaya, R., et al. (2011). Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Glob. Change Biol. 17, 1475–1486. doi: 10.1111/j.1365-2486.2010.02300.x
Yves, R. (2012). lavaan: an R package for structural equation modeling. J. Stat. Softw. 48, 1–36. doi: 10.18637/jss.v048.i02
Zeng, J., Liu, X. J., Song, L., Lin, X. G., Zhang, H. Y., Shen, C. C., et al. (2016). Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 92, 41–49. doi: 10.1016/j.soilbio.2015.09.018
Zhang, X., Zhang, G., Chen, Q., and Han, X. (2013). Soil bacterial communities respond to climate changes in a temperate steppe. PLOS ONE 8:e78616. doi: 10.1371/journal.pone.0078616
Zhang, X. M., Wei, H. W., Chen, Q. S., and Han, X. G. (2014). The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem. Soil Biol. Biochem. 72, 26–34. doi: 10.1016/j.soilbio.2014.01.034
Zhang, Y., Li, H., Wang, Y., and Wei, J. (2017a). The calcium-binding protein EpANN from the lichenized fungus Endocarpon pusillum enhances stress tolerance in yeast and plants. Fungal Genet. Biol. 108, 36–43. doi: 10.1016/j.fgb.2017.09.003
Keywords: copiotrophic/oligotrophic, global environmental changes, microbial diversity and community composition, nitrogen deposition, precipitation changes, soil bacteria and fungi, stressful environment, stress-tolerant
Citation: She W, Bai Y, Zhang Y, Qin S, Feng W, Sun Y, Zheng J and Wu B (2018) Resource Availability Drives Responses of Soil Microbial Communities to Short-term Precipitation and Nitrogen Addition in a Desert Shrubland. Front. Microbiol. 9:186. doi: 10.3389/fmicb.2018.00186
Received: 10 October 2017; Accepted: 26 January 2018;
Published: 09 February 2018.
Edited by:
Hongchen Jiang, China University of Geosciences Wuhan, ChinaReviewed by:
Hui Li, Institute of Applied Ecology, Chinese Academy of Sciences, ChinaKristen M. DeAngelis, University of Massachusetts Amherst, United States
Copyright © 2018 She, Bai, Zhang, Qin, Feng, Sun, Zheng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing Zhang, zhangyqbjfu@gmail.com
 Weiwei She
Weiwei She Yuxuan Bai
Yuxuan Bai Yuqing Zhang
Yuqing Zhang Shugao Qin1,3
Shugao Qin1,3