- 1Department of Food Safety and Quality, Veterinary Academy, Lithuanian University of Health Sciences, Kaunas, Lithuania
- 2Department of Veterinary and Animal Sciences, University of Copenhagen, Frederiksberg, Denmark
Recently, the number of reports on isolation of ciprofloxacin resistant Campylobacter jejuni has increased worldwide. The aim of this study was to determine the prevalence of resistance to ciprofloxacin and its genetic determinants among C. jejuni isolated from humans (n = 100), poultry products (n = 96) and wild birds (n = 96) in Lithuania. 91.4% of the C. jejuni isolates were phenotypically resistant to ciprofloxacin. DNA sequence analyses of the gyrA gene from 292 isolates revealed that a change in amino acid sequence, Thr86Ile, was the main substition conferring resistance to ciprofloxacin. This change was significantly associated with isolates from poultry products (P < 0.05) and humans (P < 0.05). A total of 26.7% of C. jejuni isolates from human (n = 47), poultry products (n = 30) and wild bird (n = 1), had a mutation from Ser at position 22, and six had an additional mutation from Ala at position 39. Eight isolates from poultry and two isolates from human, corresponding to 67.0% of isolates with MICs ≥128 μg/ml, showed missense mutations Thr86Ile (ACA → ATA) and Ser22Gly (AGT → GGT) together, whereas isolates without these mutations showed lower MIC values (from 4 to 64 μg/ml). Two hundred forty-five C. jejuni isolates showed one or more silent mutations, and 32.4% of examined isolates possessed six silent mutations. In addition to the ciprofloxacin resistant isolates harboring only Thr86Ile point mutation (110 isolates), the current study identified resistant isolates (n = 101) harboring additional point mutations (Ser22Gly, Ala39Ser, Arg48Lys, Thr85Ala Ala122Ser, Glu136Asp, Vall49Ile), and strains (n = 57) having only Glu136Asp point mutation. The study highlight the potential public health problem with elevated ciprofloxacin resistance in Campylobacters from poultry meat, wild birds and humans, and the need for extensive surveillance enabling to follow changes of antimicrobial resistance development in this species.
Introduction
Campylobacter jejuni is a leading foodborne pathogen in many countries (Engberg et al., 2001). It colonizes the gastrointestinal tracts of a wide range of wild, domestic, and livestock animals, and foods of animal origin are a significant source of campylobacteriosis in humans (Colles et al., 2008; Silva et al., 2011). Transmission of C. jejuni to humans most often occurs via to consumption of contaminated food, especially chicken meat, or by direct contact with feces (Miller et al., 2010). Although most infections are mild and self-limiting and do not require antimicrobial therapy, treatment is indicated in immunocompromised patients, or if the infection is extraintestinal (Goodman et al., 1984). Fluoroquinolones, such as ciprofloxacin, are the antimicrobial agents of choice for treatment of campylobacteriosis (Guerrant et al., 2001; Blaser and Engberg, 2008). Other antimicrobials, however, such as erythromycin, azithromycin and gentamicin may be viable alternatives (Wieczorek and Osek, 2013; Wimalarathna et al., 2013). Moreover, fluoroquinolones have been used as first-line antibiotics against bacterial gastroenteritis in the absence of microbiological diagnosis (Wieczorek, 2011). The quinolones target two large, bacterial enzymes, DNA gyrase and topoisomerase IV, and binding to these enzymes inhibit the synthesis of bacterial DNA, causing cell death. Resistance to fluoroquinolones is mainly due to amino acids substitutions in the quinolone resistance-determining region (QRDR) of the topoisomerase (Wieczorek and Osek, 2013). Due to common use of flouroquinolones in livestock in some countries, and for treatment of campylobacteriosis in humans, resistance to this antimicrobial group has emerged worldwide in C. jejuni and has become a public health issue (Abay et al., 2014; Sahin et al., 2015). Resistance to quinolones in Campylobacter is usually mediated by a single point mutation in the quinolone resistance-determining region (QRDR) of the gyrA gene at codon 86 (ACA → ATA), leading to isoleucine substitution for threonine (Wang et al., 1993; Ruiz et al., 1998; Zirnstein et al., 1999). This alteration is most often associated with high MIC values for fluoroquinolones (Wang et al., 1993). Other substitutions in the QRDR are also known and described, but these changes are less common (Wang et al., 1993; Hakanen et al., 2003).
The prevalence of resistance to ciprofloxacin in Campylobacter species vary considerably between countries, with high prevalence reported in Poland (Wieczorek and Osek, 2013), Italy (Di Giannatale et al., 2014), Switzerland (Kittl et al., 2013) and some other EU countries (EFSA, 2014a,b). The rapid emergence of antimicrobial resistance is a cause for global concern (Engberg et al., 2001; Hein et al., 2003). To our knowledge, there are no published data on the genetic background for flouroquinolone resistance in C. jejuni isolated from different sources in Lithuania. Therefore, the aim of this study was to determine the prevalence of resistance to ciprofloxacin among C. jejuni isolated from humans, poultry products and wild birds in Lithuania, and to assess the gyrA mutations responsible for the resistance among the C. jejuni isolates.
Materials and Methods
Bacterial Strains and Culture Conditions
A total of 292 C. jejuni isolates from infected children (n = 100), raw (n = 77), and marinated (n = 19) broiler products and wild birds: pigeons (n = 39) and crows (n = 57) were selected. Food and wild bird isolates were from the Campylobacter collection at the Department of Food Safety and Quality, Veterinary Academy, Lithuanian University of Health Sciences. The human C. jejuni isolates were received from the Microbiological laboratory of Kaunas Clinical Hospital and were isolated in the period from 2011 to 2012. Identification of Campylobacter isolates was performed with multiplex PCR as described by Wang et al. (2002) with the minor modifications described previously by Ramonaite et al. (2015).
Campylobacter isolates were stored as frozen stocks at −80°C in in brain heart infusion broth (BHI) (Oxoid Ltd., Basingstoke, UK) with 30% glycerol (Stanlab, Poland). They were recovered from frozen stocks on Blood agar base No. 2 (Oxoid, Basingstoke, Hampshire, England) supplemented with 5% defibrinated horse blood (E&O Laboratories, Burnhouse, Bonnybridge, Scotland) and incubated under microaerophilic conditions (5% oxygen, 10% carbon dioxide and 85% nitrogen) at 37°C for 48 h.
Antimicrobial Resistance Testing
Minimum inhibitory concentrations (MIC) were determined according to the recommendations of the Clinical and Laboratory Standarts Institute (CLSI) (CLSI, 2006). Antimicrobial susceptibility was evaluated using the quality control strain C. jejuni ATCC 33560. Briefly, suspension of C. jejuni isolates adjusted to an OD600 = 0.1 were prepared in phosphate buffered saline and inoculated onto Mueller-Hinton agar (LiofilChem, Milan Italy) supplemented with 5% lysed sheep blood (E&O Laboratories, Burnhouse, Bonnybridge, Scotland) and ciprofloxacin (Sigma-Aldrich, Saint-Louis, USA) in concentrations ranging from 0.25 to 256 μg/ml and incubated under microaerophilic conditions at 42°C for 24 h. The MIC interpretive criterion for resistance to ciprofloxacin was ≥4 μg/ml.
DNA Extraction
Bacterial isolates were grown at 37°C on blood agar plates for 48 h under microaerophilic conditions. After sufficient growth was obtained, one 1 μl loopful of bacteria was suspended in Eppendorf tubes containing 200 μl of PrepMan Ultra Sample Preparation Reagent (PrepMan™ Ultra, Applied Biosystems, USA). DNA extraction was carried out following the instructions of the supplier including heating of bacterial suspension at 100°C for 10 min., centrifugation at 16,000 g for 3 min. and transferring the supernatants into new tubes before storage in the freezer at −20°C until use.
PCR Detection of Antibiotic Resistance Determinants
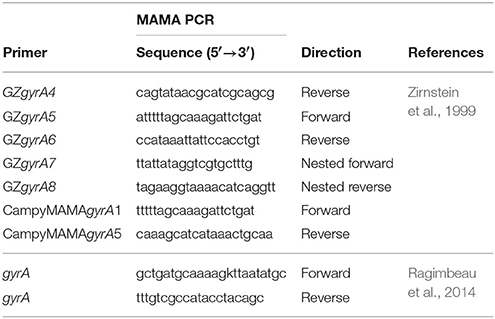
The QRDR of the gyrA gene of the C. jejuni isolates was amplified by MAMA PCR as described by Zirstein (Zirnstein et al., 1999) using GzgyrA5 and GzgyrA6 primers for amplification of a 673 bp product (Table 1). Forward primer CampyMAMAgyrA1 and a reverse primer CampyMAMAgyrA5 (Table 1) were used to generate a 256 bp PCR product that was a positive indication of the presence of the Thr-86-Ile (ACA → ATA) mutation in the C. jejuni gyrA gene. Primer GZgyrA4, a conserved reverse primer, was used in conjugation with primer CampyMAMAgyrA1 to produce a positive PCR control product of 368 bp with any C. jejuni gyrA gene. PCR cycling conditions were as follow: 3 min initial denaturation at 94°C, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min and extension at 72°C for 1 min, with a final step at 72°C for 5 min.
DNA Sequencing and Sequence Analysis
Nested primers GZgyrA7 and GZgyrA8 (Table 1), which are internal to the 673 bp gyrA PCR products produced above, were used for sequencing. The partial gene sequence of gyrA, generated by use of CampyMAMAgyrA1 and a reverse primer CampyMAMAgyrA5 targeting the quinolone resistance determining region (QRDR), was sequenced with the forward and reverse primers described by Ragimbeau et al. (Table 1). The amplification protocol consisted of 95°C for 10 min, followed by 35 cycles of 95°C 30 s, 55°C 30 s, and 72°C 50 s. The reaction was completed by a final extension of 5 min at 72°C.
The PCR amplicons were purified using the GeneJet PCR purification system (Thermo Scientific, EU). Sequencing reactions were carried out using the BigDye Terminator 3.1 cycle Sequencing Kit (Applied Biosystems, USA) according to instructions from the manufacturer. Duplicate forward and reverse sequencing reactions were run with forward and reverse primers, respectively, for each sample. The samples were analyzed with a 3500 Genetic Analyzer (Applied Biosystems). The BioNumerics program 7.0 (Applied Maths NV, USA, EU) was used to evaluate the specific genomic mutations associated with resistance to ciprofloxacin, including comparison to the reference C. jejuni strain ATCC 700819 genome (NCTC11168).
Statistical Analysis
The statistical package SPSS (Statistics 20, IBM) was used. Comparison of association between phenotypic resistance and resistance genes in C. jejuni from humans, poultry products and wild birds isolates and distributions of resistant isolates were evaluated using the Chi-square test and Fisher's exact test. A p-value of < 0.05 was used to indicate statistically significant results.
Results
Phenotypic Antimicrobial Resistance
Among the 292 isolates tested, 267 (91.4%) were phenotypically resistant to ciprofloxacin (Table 2). In particular, all C. jejuni isolates from poultry products were resistant to ciprofloxacin with MIC ranging from 4 μg/ml up to 256 μg/ml.
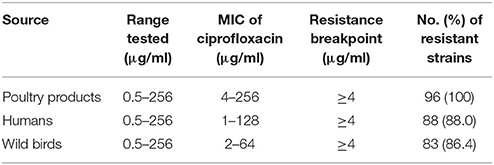
Table 2. Antimicrobial resistance of C. jejuni from poultry products, humans, and wild-bird samples.
PCR Typing of Resistant Isolates
Phenotypic resistance to ciprofloxacin matched genotypic resistance of all isolates from broiler products and the vast majority (93%) of isolates from humans. Out of the 267 ciprofloxacin-resistant isolates, the majority (76.4%; n = 204) were positive for the Thr86Ile substitution of the gyrA gene as demonstrated by PCR. The Thr86Ile substitution was not detected in 62 wild bird isolates, which showed phenotypic resistance to ciprofloxacin.
Amino Acid Sequences of the gyrA Gene in Ciprofloxacin Resistant C. jejuni
We identified eight different missense amino acid substitutions in the gyrA gene of the ciprofloxacin resistant C. jejuni isolates (Table S1 in Supplementary Materials). One hundred and ten C. jejuni isolates harbored a single Thr86Ile substitution in gyrA without any other amino acid changes in this region. One of the other substitutions was Ser22Gly, which is known to confer fluoroquinolones resistance, often along with the Thr86Ile substitution (Jesse et al., 2006). The combined Thr86Ile (ACA → ATA) and Ser22Gly (AGT → GGT) changes were detected in 10 out of the 15 ciprofloxacin-resistant C. jejuni isolates with MICs ≥128 μg/ml (67.0%). Seven isolates carried the Ala39Ser substitution, and these isolates also had the Thr86Ile amino acid change. Six isolates carried the Ser22Gly, Ala39Ser and the Thr86Ile changes in combination, whereas only one isolate carried four substitutions: Ser22Gly, Arg48Lys, Thr86Ile, and Glu136Asp. Interestingly, four ciprofloxacin resistant C. jejuni isolates had no point mutations in QRDR. Seventy-four ciprofloxacin resistant C. jejuni isolates had a Glu136 point mutation. This mutation was significantly more common in isolates from wild birds (P < 0.05) than in isolates from poultry product and humans. Table S1 in Supplementary Materials lists the observed combinations of mutations in the gyrA QRDR.
The study revealed at least one silent mutation in 245 C. jejuni isolates (Table 3), and 6 C. jejuni isolates harbored more than six silent mutations. Details on the observed silent mutations in relation to clonal lines can be seen from Table S1 in the Supplementary Material.
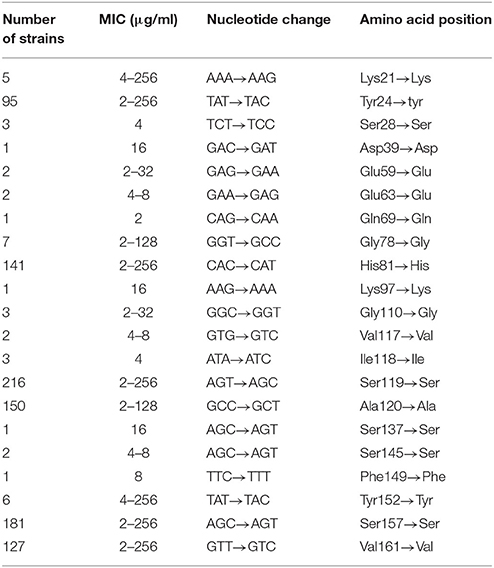
Table 3. Silent mutations in the gyrA QRDR of C. jejuni isolates and associated ranges of MICs of ciprofloxacin.
Association between MLST Genotypes and Phenotypic Antimicrobial Resistance
MLST types of the isolates investigated were published by Ramonaite et al. (2015, 2017). In the current investigation, we related QRDR sequence types to MLST type. The gyrA gene point-mutations A64G, G118T and C257T were observed in four isolates with ST-257 (CC257), in one isolate with ST-51 (CC443) and one isolate with ST-6413 (CC353). In total 40 isolates having the Thr-86-Ile substitution were assigned to CC353 and 31 of 33 isolates belonging to CC21. More than half of the isolates (57.7%) belonging to an undefined clonal complex is isolated from wild birds and had the Glu136Asp novel mutation (Table S1 in Supplementary Materials).
Discussion
Ciprofloxacin resistant Campylobacter isolates were observed already from the late 1980s, indicating animals as the main source of resistant bacteria. Currently, flouroquinolone resistance in C. jejuni is emerging globally and is considered a problem of public health importance (Wardak et al., 2007; Luangtongkum et al., 2009; Wieczorek, 2011). In this study, C. jejuni isolates originating from humans, poultry products and wild birds in Lithuania were examined for resistance to ciprofloxacin.
A high prevalence of C. jejuni resistance to ciprofloxacin was observed irrespective of the source of isolation (91.4%). This is much higher resistance prevalence than has been reported in other parts of the Baltic region. For example, Estonia reported only 16.7% prevalence of this resistance (Mäesaar et al., 2016). On the other hand, it is very similar to what has previously been reported from isolates obtained from broiler meat in Latvia and Lithuania (87.5 and 84.8%, respectively) (Mäesaar et al., 2016). The Baltic region is not exceptional, as elevated levels of ciprofloxacin resistance in C. jejuni have also been reported from Poland (Wieczorek and Osek, 2013), Italy (Di Giannatale et al., 2014), Switzerland (Kittl et al., 2013) and some other EU countries (EFSA, 2014a,b). However, it should be mentioned, that prevalence of resistance can change dramatically over time.
Resistance to fluoroquinolones in Campylobacter is mainly due to amino acids substitutions in the quinolone resistance-determining region (QRDR) of gyrA (Wieczorek and Osek, 2013). The most frequently observed mutation is the C257T mutation in the gyrA gene, which leads to the Thr86Ile substitution in the gyrase, and confers high-level resistance to this class of antimicrobials (Wang et al., 1993; Payot et al., 2006). In confirmation of this and other studies (Kinana et al., 2006; Tang et al., 2017), the Thr86Ile substitution was the most frequently observed amino acid change in the current study. Previous studies (Sonnevend et al., 2006; Boonmar et al., 2007; Wieczorek, 2011; Duarte et al., 2014) have reported that all ciprofloxacin resistant C. jejuni carried the Thr86Ile amino acid substitution in the QRDR of gyrA. However, while our study showed this mutation in 100% of the broiler products isolates and 98.9% of the human isolates, only 25.3% of the ciprofloxacin-resistant wild bird isolates carried the corresponding mutation. Other mechanisms of resistance, including decreased outer membrane permeability and efflux systems, have been described (Charvalos et al., 1996), and these may contribute to the phenotypic resistance observed in the strains, where no amino acid changes in gyrA were observed.
The Ser22Gly substitution alone may not confer ciprofloxacin resistance, since it has been identified in susceptible strains (Jesse et al., 2006; Tang et al., 2017). However, the double substitution, Thr86Ile/Ser22Gly, is known to confer high-level ciprofloxacin resistance in Campylobacter (Oishi et al., 2015). We also identified the mutation leading to Ser22Gly substitution in combination with the mutation leading to Thr86Ile substitution, and the presence of both mutations was associated with high MIC values. Interestingly, one of the isolates with this combination carried an additional double novel mutation, from Arg at position 48 and from Glu at position 136. More research is needed to clarified whether these novel mutations contribute to phenotypic ciprofloxacin resistance in C. jejuni.
Silent mutation are often described in ciprofloxacin resistant and sensitive strains and a high number of combinations of transitions and mutations may exist (Frasao et al., 2015). We confirmed this observation, and some of the most frequently observed silent mutations at His-81 → His, Ala-119 → Ala, and Ser-120 → Ser corresponds to mutations described in Campylobacter isolated in Finland and Brazil (Hakanen et al., 2002; Frasao et al., 2015). Other transitions without amino acid changes were frequently found in the QRDR, generating many different gyrA alleles, TCT → TCC at the codon 84 described by Carattoli et al. (2002) and Gly-110 → Gly (GGC → GGT) described by Beckmann et al. (2004). However, we did not observe silent mutation at Ser-75 → Ser (GCT → GAT) and Ser-79 → Ser (CGT → AGT), as described by Frasao et al. (2015).
Conclusions
In conclusion, the phenotypic and genotypic antimicrobial resistance of C. jejuni from human, poultry products and wild birds to ciprofloxacin have been investigated and reported for the first time in Lithuania. Our study demonstrated a high prevalence of ciprofloxacin resistance in the isolates. The data presented here confirmed previous findings that mutations in the gyrA gene at position 257 are mainly responsible for ciprofloxacin resistance of C. jejuni strains from poultry products and humans. In addition, our results indicated that Glu136Asp mutation is associated with high-level ciprofloxacin resistance among wild bird isolates.
Author Contributions
JA, SR, and MM: conceived of the presented idea; JA and SR: performed laboratory work and analysis; JA: performed statistical analysis, and drafted the manuscript; JO and MM: contributed to the interpretation of the results. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Funding
This study was funded by the Research Council of Lithuania (MIP-041/2015).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00203/full#supplementary-material
References
Abay, S., Kayman, T., Otlu, B., Hizlisoy, H., Aydin, F., and Ertas, N. (2014). Genetic diversity and antibiotic resistance profiles of Campylobacter jejuni isolates from poultry and humans in Turkey. Int. J. Food Microbiol. 178, 29–38. doi: 10.1016/j.ijfoodmicro.2014.03.003
Beckmann, L., Müller, M., Luber, P., Schrader, C., Bartelt, E., and Klein, G. (2004). Analysis of gyrA mutations in quinolone-resistant and -susceptible Campylobacter jejuni isolates from retail poultry and human clinical isolates by non-radioactive single-strand conformation polymorphism analysis and DNA sequencing. J. Appl. Microbiol. 96, 1040–1047. doi: 10.1111/j.1365-2672.2004.02242.x
Blaser, M. J., and Engberg, J. (2008). “Clinical aspects of Campylobacter jejuni and Campylobacter coli infections” in Campylobacter, 3rd Edn., eds I. Nachamkin, C. Szymanski, and M. Blaser (Washington, DC: ASM Press), 99–121. doi: 10.1128/9781555815554.ch6
Boonmar, S., Morita, Y., Fujita, M., Sangsuk, L., Suthivarakom, K., Padungtod, P., et al. (2007). Serotypes, antimicrobial susceptibility, and gyr a gene mutation of Campylobacter jejuni isolates from humans and chickens in Thailand. Microbiol. Immunol. 51, 531–537. doi: 10.1111/j.1348-0421.2007.tb03941.x
Carattoli, A., Dionisi, A. M., and Luzzi, I. (2002). Use of a lightcycler gyrA mutation assay for identification of ciprofloxacin-resistant Campylobacter coli. FEMS Microbiol. Lett. 214, 87–93. doi: 10.1111/j.1574-6968.2002.tb11329.x
Charvalos, E., Peteinaki, E., Spyridaki, I., Manetas, S., and Tselentis, Y. (1996). Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand conformation polymorphism and direct DNA sequencing. J. Clin. Lab. Anal. 10, 129–133. doi: 10.1002/(SICI)1098-2825(1996)10:3<129::AID-JCLA3>3.0.CO;2-6
CLSI. (2006). M100-S16, Performance standards for Antimicrobial Susceptibility Testing; 16th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Colles, F. M., Jones, T. A., McCarthy, N. D., Sheppard, S. K., Cody, A. J., Dingle, K. E., et al. (2008). Campylobacter infection of broiler chickens in a free-range environment. Environ. Microbiol. 10, 2042–2050. doi: 10.1111/j.1462-2920.2008.01623.x
Di Giannatale, E., Di Serafino, G., Zilli, K., Alessiani, A., Sacchini, L., Garofolo, G., et al. (2014). Characterization of antimicrobial resistance patterns and detection of virulence genes in campylobacter isolates in Italy. Sensors 14, 3308–3322. doi: 10.3390/s140203308
Duarte, A., Santos, A., Manageiro, V., Martins, A., Fraqueza, M. J., Caniça, M., et al. (2014). Human, food and animal Campylobacter spp. isolated in Portugal: high genetic diversity and antibiotic resistance rates. Int. J. Antimicrob. Agents 44, 306–313. doi: 10.1016/j.ijantimicag.2014.06.012
EFSA. (2014a). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2012 – 2014 - EFSA Journal - Wiley Online Library [WWW Document]. Available online at: http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2014.3547/full (Accessed September 23, 2017).
EFSA. (2014b). The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2012 – 2014 - EFSA Journal - Wiley Online Library [WWW Document]. Available online at: http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2014.3590/full (Accessed September 23, 2017).
Engberg, J., Aarestrup, F. M., Taylor, D. E., Gerner-Smidt, P., and Nachamkin, I. (2001). Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7, 24–34. doi: 10.3201/eid0701.700024
Frasao, B., Medeiros, V., Barbosa, A. V., Aguiar, D., Silva, W., Cosendey, M. H., et al. (2015). Detection of fluoroquinolone resistance by mutation in gyrA gene of Campylobacter spp. isolates from broiler and laying (Gallus gallus domesticus) hens, from Rio de Janeiro State, Brazil. Ciênc. Rural 45, 2013–2018. doi: 10.1590/0103-8478cr20141712
Goodman, L. J., Fliegelman, R. M., Trenholme, G. M., and Kaplan, R. L. (1984). Comparative in vitro activity of ciprofloxacin against Campylobacter spp. and other bacterial enteric pathogens. Antimicrob. Agents Chemother. 25, 504–506.
Guerrant, R. L., Van Gilder, T., Steiner, T. S., Thielman, N. M., Slutsker, L., Tauxe, R. V., et al. (2001). Infectious diseases society of America, 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32, 331–351. doi: 10.1086/318514
Hakanen, A., Jalava, J., Kotilainen, P., Jousimies-Somer, H., Siitonen, A., and Huovinen, P. (2002). gyrA polymorphism in Campylobacter jejuni: detection of gyrA mutations in 162 C. jejuni isolates by single-strand conformation polymorphism and DNA Sequencing. Antimicrob. Agents Chemother. 46, 2644–2647. doi: 10.1128/AAC.46.8.2644-2647.2002
Hakanen, A. J., Lehtopolku, M., Siitonen, A., Huovinen, P., and Kotilainen, P. (2003). Multidrug resistance in Campylobacter jejuni strains collected from finnish patients during 1995-2000. J. Antimicrob. Chemother. 52, 1035–1039. doi: 10.1093/jac/dkg489
Hein, I., Schneck, C., Knögler, M., Feierl, G., Plesss, P., Köfer, J., et al. (2003). Campylobacter jejuni isolated from poultry and humans in Styria, Austria: epidemiology and ciprofloxacin resistance. Epidemiol. Infect. 130, 377–386. doi: 10.1017/S0950268803008380
Jesse, T. W., Englen, M. D., Pittenger-Alley, L. G., and Fedorka-Cray, P. J. (2006). Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle*. J. Appl. Microbiol. 100, 682–688. doi: 10.1111/j.1365-2672.2005.02796.x
Kinana, A. D., Cardinale, E., Tall, F., Bahsoun, I., Sire, J. M., Garin, B., et al. (2006). Genetic diversity and quinolone resistance in Campylobacter jejuni isolates from poultry in Senegal. Appl. Environ. Microbiol. 72, 3309–3313. doi: 10.1128/AEM.72.5.3309-3313.2006
Kittl, S., Korczak, B. M., Niederer, L., Baumgartner, A., Buettner, S., Overesch, G., et al. (2013). Comparison of genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli on chicken retail meat and at slaughter. Appl. Environ. Microbiol. 79, 3875–3878. doi: 10.1128/AEM.00493-13
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Mäesaar, M., Kramarenko, T., Meremäe, K., Sõgel, J., Lillenberg, M., Häkkinen, L., et al. (2016). Antimicrobial resistance profiles of campylobacter spp. isolated from broiler chicken meat of estonian, latvian and lithuanian origin at estonian retail level and from patients with severe enteric infections in Estonia. Zoonoses Public Health 63, 89–96. doi: 10.1111/zph.12208
Miller, R. S., Miller, W. G., Behringer, M., Hariharan, H., Matthew, V., and Oyarzabal, O. A. (2010). DNA identification and characterization of Campylobacter jejuni and Campylobacter coli isolated from caecal samples of chickens in Grenada. J. Appl. Microbiol. 108, 1041–1049. doi: 10.1111/j.1365-2672.2009.04507.x
Oishi, A., Murakami, K., Etoh, Y., Sera, N., and Horikawa, K. (2015). Antimicrobial susceptibility and resistance mutations in Campylobacter jejuni and C. coli isolates from human and meat sources. Kansenshogaku Zasshi 89, 244–253. doi: 10.11150/kansenshogakuzasshi.89.244
Payot, S., Bolla, J. M., Corcoran, D., Fanning, S., Mégraud, F., and Zhang, Q. (2006). Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes. Infect. 8, 1967–1971. doi: 10.1016/j.micinf.2005.12.032
Ragimbeau, C., Colin, S., Devaux, A., Decruyenaere, F., Cauchie, H. M., Losch, S., et al. (2014). Investigating the host specificity of Campylobacter jejuni and Campylobacter coli by sequencing gyrase subunit A. BMC Microbiol. 14:205. doi: 10.1186/s12866-014-0205-7
Ramonaite, S., Novoslavskij, A., Zakariene, G., Aksomaitiene, J., and Malakauskas, M. (2015). High prevalence and genetic diversity of Campylobacter jejuni in wild crows and pigeons. Curr. Microbiol. 71, 559–565. doi: 10.1007/s00284-015-0881-z
Ramonaite, S., Tamuleviciene, E., Alter, T., Kasnauskyte, N., and Malakauskas, M. (2017). MLST genotypes of Campylobacter jejuni isolated from broiler products, dairy cattle and human campylobacteriosis cases in Lithuania. BMC Infect. Dis. 17:430. doi: 10.1186/s12879-017-2535-1
Ruiz, J., Goñi, P., Marco, F., Gallardo, F., Mirelis, B., Jimenez De Anta, T., et al. (1998). Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol. Immunol. 42, 223–226.
Sahin, O., Kassem, I. I., Shen, Z., Lin, J., Rajashekara, G., and Zhang, Q. (2015). Campylobacter in poultry: ecology and potential interventions. Avian Dis. 59, 185–200. doi: 10.1637/11072-032315-Review
Silva, J., Leite, D., Fernandes, M., Mena, C., Gibbs, P. A., and Teixeira, P. (2011). Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2:200. doi: 10.3389/fmicb.2011.00200
Sonnevend, A., Rotimi, V. O., Kolodziejek, J., Usmani, A., Nowotny, N., and Pál, T. (2006). High level of ciprofloxacin resistance and its molecular background among Campylobacter jejuni strains isolated in the United Arab Emirates. J. Med. Microbiol. 55, 1533–1538. doi: 10.1099/jmm.0.46744-0
Tang, Y., Sahin, O., Pavlovic, N., LeJeune, J., Carlson, J., Wu, Z., et al. (2017). Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci. Rep. 7:494. doi: 10.1038/s41598-017-00584-z
Wang, G., Clark, C. G., Taylor, T. M., Pucknell, C., Barton, C., Price, L., et al. (2002). Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40, 4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002
Wang, Y., Huang, W. M., and Taylor, D. E. (1993). Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37, 457–463.
Wardak, S., Szych, J., Zasada, A. A., and Gierczynski, R. (2007). Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob. Agents Chemother. 51, 1123–1125. doi: 10.1128/AAC.01187-06
Wieczorek, K. (2011). Resistance to quinolones and tetracycline and its molecular background among Campylobacter strains isolated in Poland. Bull. Vet. Inst. Puławy 55, 613–618.
Wieczorek, K., and Osek, J. (2013). Antimicrobial resistance mechanisms among campylobacter. Biomed Res. Int. 2013:e340605. doi: 10.1155/2013/340605
Wimalarathna, H. M. L., Richardson, J. F., Lawson, A. J., Elson, R., Meldrum, R., Little, C. L., et al. (2013). Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol. 13:160. doi: 10.1186/1471-2180-13-160
Keywords: Campylobacter jejuni, antimicrobial resistance, sequencing identification, ciprofloxacin, QRDR
Citation: Aksomaitiene J, Ramonaite S, Olsen JE and Malakauskas M (2018) Prevalence of Genetic Determinants and Phenotypic Resistance to Ciprofloxacin in Campylobacter jejuni from Lithuania. Front. Microbiol. 9:203. doi: 10.3389/fmicb.2018.00203
Received: 16 October 2017; Accepted: 29 January 2018;
Published: 14 February 2018.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Issmat Kassem, American University of Beirut, LebanonXimin Zeng, University of Tennessee, Knoxville, United States
Copyright © 2018 Aksomaitiene, Ramonaite, Olsen and Malakauskas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jurgita Aksomaitiene, jurgita.aksomaitiene@lsmuni.lt
 Jurgita Aksomaitiene
Jurgita Aksomaitiene Sigita Ramonaite
Sigita Ramonaite John E. Olsen
John E. Olsen Mindaugas Malakauskas1
Mindaugas Malakauskas1