- School of Energy and Environment, City University of Hong Kong, Kowloon, Hong Kong
Nitrification plays a crucial role in global nitrogen cycling and treatment processes. However, the relationships between the nitrifier guilds of ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB) are still poorly understood, especially in freshwater habitats. This study examined the physiological interactions between the AOB and NOB present in a freshwater aquarium biofilter by culturing them, either together or separately, in a synthetic medium. Metagenomic and 16S rRNA gene sequencing revealed the presence and the draft genomes of Nitrosomonas-like AOB as well as Nitrobacter-like NOB in the cultures, including the first draft genome of Nitrobacter vulgaris. The nitrifiers exhibited different growth rates with different ammonium (NH4+) or nitrite concentrations (50–1,500 μM) and the growth rates were elevated under a high bicarbonate (HCO3-) concentration. The half-saturation constant (Ks for NH4+), the maximum growth rate (μmax), and the lag duration indicated a strong dependence on the synergistic relationships between the two guilds. Overall, the ecophysiological and metagenomic results in this study provided insights into the phylogeny of the key nitrifying players in a freshwater biofilter and showed that interactions between the two nitrifying guilds in a microbial community enhanced nitrification.
Introduction
Nitrification, the biological oxidation of ammonia (NH3) to nitrate (NO3-) via nitrite (NO2-), is a vital oxidative process that links reduced and oxidized inorganic nitrogen to sustain the global nitrogen cycle in soils (Wang et al., 2015) and aquatic systems (Hou et al., 2013). This process is mainly carried out by two different but interdependent guilds of lithoautotrophic microorganisms, namely the ammonia-oxidizing bacteria (AOB) (e.g., Nitrosomonas and Nitrosospira) and the ammonia-oxidizing archaea (AOA) as one guild that convert NH3 to NO2- using the key enzymes ammonia monooxygenase (encoded by the amoCAB genes) and hydroxylamine dehydrogenase (encoded by the haoAB genes) (Sedlacek et al., 2016), and the nitrite-oxidizing bacteria (NOB) (e.g., Nitrospira and Nitrobacter) as another that convert NO2- to NO3- with the enzyme nitrite oxidoreductase (encoded by the nxrAB genes) (Dionisi et al., 2002). Recent reports have also identified single organisms that are capable of performing the comammox reaction, which leads to complete nitrification from NH3 to NO3- (Daims et al., 2015; van Kessel et al., 2015).
The coexistence of AOB and NOB has been demonstrated in a variety of environments (Graham et al., 2007; Keuter et al., 2011; Wang et al., 2015) and microscopic examinations of samples from nitrifying reactors have often showed AOB and NOB in physical contact with each other (Okabe et al., 1999). Previous studies have shown that nitrification rates are closely related to environmental parameters such as temperature, NH3 and/or bicarbonate (HCO3-) concentrations (French et al., 2012; Jiang et al., 2015). Even though AOB and NOB can function independently (Koops et al., 1991; Lücker et al., 2010), their synergistic relationships (Schink, 2002) benefit both species; for example, the growth of AOB is elevated in the presence of NOB, as the latter helps prevent the accumulation of NO2- that could inhibit AOB (Kim et al., 2006), while oxidation of NH3 by AOB benefits NOB by providing a consistent energy supply as NO2- and by maintaining NH3 concentrations below the toxicity threshold (Laanbroek and Gerards, 1993). In some cases, AOB benefit more than NOB (Perez et al., 2015). However, the tight coupling between AOB and NOB means that even minor perturbations in the abundance of ammonia oxidizers can lead to large changes in the abundance of nitrite oxidizers (Knapp and Graham, 2007), and subsequently to erratic nitrification activities (Graham et al., 2007).
Our current understanding of the synergistic relationships between nitrifiers arises mainly from studies of artificially constructed co-cultures of isolates (e.g., pairing Nitrosomonas europaea and Nitrobacter winogradskyi) (Laanbroek and Gerards, 1993; Perez et al., 2015). These conditions may not be representative of some of the real-life environmental conditions in a microbial community in an ecosystem. We explored the relationships between ammonia-oxidizing and nitrite-oxidizing populations found in freshwater systems by instead culturing one or both of these nitrifying guilds from a microbial community inhabiting a freshwater biofilter in a synthetic medium. The community composition was determined by 16S rRNA gene and metagenomic sequencing, with the recovery of draft genomes of the nitrifiers, including the first draft genome of Nitrobacter vulgaris. The ecophysiological characteristics of the cultures were studied in greater depth by testing different concentrations of NH4+ and NO2- (50–1,500 μM), and HCO3- (1,000 and 3,000 μM). New insights were obtained into the interactions and physiology of the freshwater AOB and NOB and clues (e.g., kinetic parameters) were provided for future manipulation of microbial interactions to enhance the efficiency of microbial nitrification processes.
Materials and Methods
Cultivation of Nitrifiers
The inoculum used to establish the nitrifying cultures was obtained from the biofilter of a household-size freshwater fish tank. The tank housed five small tropical fish and nitrification occurred (i.e., NO3- was detected). The biofilter was selected for use as inoculum since the microbial community had adapted to a relatively low NH4+ condition (∼500 μM was typically measured in the influent to the biofilter) and the biofilter had been in stable operation for more than two years. The synthetic sponge filter (36 cm2) was inoculated (March 2014) into 60-ml serum bottles sealed with butyl rubber stoppers with 30 ml of synthetic medium containing 4 μM KH2PO4, 10 ml/L mineral salts, 0.1 ml/L selenite/tungstate solution, and 1 ml/L trace metals (Biebl and Pfennig, 1978). After autoclaving, 3 μl of filter-sterilized vitamin solution (Balch et al., 1979) were added, together with NH4Cl and NaHCO3 at final concentrations of 500 and 1,000 μM, respectively. The NOB were eliminated by amending a separate set of cultures with sodium chlorate (10 mM) (Belser and Mays, 1980) until no NO3- was detected in subsequent transfers. The cultures with NH4+ without chlorate are referred to as ‘Culture01,’ the ones with NH4+ and chlorate are ‘Culture02,’ while the Culture01 fed with NO2- instead of NH4+ for experimental purposes are ‘Culture03.’ Cultures were grown at 25°C (akin to the temperature of the aquarium) in the dark without shaking (Martens-Habbena et al., 2009). Late exponential-phase cultures (∼10% v/v) were routinely transferred to fresh media after an incubation period of about 5 to 7 days for 15 months (until the cultures became stable) before characterizations of the cultures were performed.
Characterizations of the Cultures
All experiments were conducted in triplicate in 30 ml medium as described in Section “Cultivation of Nitrifiers” with 3 ml of late exponential-phase culture as inoculum. A range of NH4+ concentrations (50, 200, 500, 1,000, and 1,500 μM) was used to test the growth rates of the nitrifiers in Culture01 and Culture02. The growth of NOB was tested using a range of NO2- concentrations (50, 200, 500, 1,000, and 1,500 μM) in Culture03. Different HCO3- concentrations (1,000 and 3,000 μM) were tested for all cultures. The potential inhibition effects of NO2- on the AOB in Culture02 when grown with 500 μM of NH4+ were studied by adding a range of NO2- concentrations (500, 1,500, or 10,000 μM). Liquid samples were withdrawn at regular intervals during the exponential phase (5–24 h) to determine the NH4+, NO2- and/or NO3- concentrations, and the collected samples were filtered (0.2 μm) prior to storage at -80°C. The ultimate dissolved oxygen concentration and pH of the cultures were measured using a portable oximeter (SevenGo Duo Pro-SG68, Mettler Toledo, Switzerland). The dissolved oxygen concentration was more than 5.6 mg/L and pH ranged from 6.8 to 7.0 in all cultures.
16S rRNA Gene Amplicon Sequencing and Analysis
The composition of the microbial communities was determined by collecting a section of the biofilter (36 cm2) and 20 ml of Culture01 and Culture02 for DNA extraction using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, United States), as described previously (Lu et al., 2013). The 515F/806R universal primer pair (Caporaso et al., 2011) was used to amplify the V4 region of the 16S rRNA gene of the genomic DNA. The PCR conditions, amplicon purification, and library preparation were as described previously (Leung et al., 2014). The samples were sequenced on an Illumina MiSeq platform (Genentech Corporation, Taipei, Taiwan), which generated paired-end 250-bp reads with ∼60,000 paired-end raw reads per sample.
Reads obtained from the sequencing platform were first processed by removing the barcodes and primers, followed by alignment of the reads using FLASH (V1.2.7) (Magoč and Salzberg, 2011). The forward and reverse reads gave similar results, so the forward reads were used for analysis. The aligned sequences were filtered using the QIIME pipeline (v.1.8.0) (Caporaso et al., 2010b) with the script “split_library_fastq.py.” Chimera sequences were identified and removed with UCHIME (Edgar et al., 2011) against the GOLD database (Bernal et al., 2001). OTU formation was performed following the UPARSE pipeline (Edgar, 2013) and the dereplicated reads were clustered into OTUs at a 97% sequence similarity threshold. Singleton OTUs were removed and the remaining high-quality sequence reads were aligned with PyNAST (Caporaso et al., 2010a) against the Ribosomal Database Project (RDP, release 11.3).
Metagenomic Sequencing and Assembly
Based on the growth profiles of the cultures, three time series samples (days 1, 3, and 5) were collected from each culture (Culture01 and Culture02) for metagenomic sequencing and to facilitate the subsequent genome binning. The samples were named according to the day in which they were collected (e.g., the sample collected on day 1 is referred to as ‘Culture01_1’ and ‘Culture02_1’). Genomic DNA was extracted as indicated in Section “16S rRNA Gene Amplicon Sequencing and Analysis” from 120 ml of each culture at each time point. DNA concentrations and quality were determined using a spectrophotometer (NanoDrop 2000; Thermo Scientific, United States).
Paired-end sequencing libraries were prepared using the Illumina HiSeq 3000/4000 PE Cluster Kit with 2 μg of DNA according to the manufacturer’s instructions. The libraries were sequenced on an Illumina HiSeq 4000 sequencer, which generated 150 bp paired-end reads at the sequencing core facility of UC Berkeley. Raw sequencing reads were trimmed and filtered with a minimum quality score of 32 using Trimmomatic (version 0.35) (Bolger et al., 2014). Read pairs with either end shorter than 80 bp were discarded. De novo assembly on the filtered read pairs was performed using the IDBA-UD assembler (Peng et al., 2012) with a maximum k-mer size of 100 and a minimum contig length of 1.2 kb. The six metagenomes were assembled independently. The targeted nitrifiers were more abundant in the mid-point samples (day 3) so the contigs reconstructed from the mid-point metagenome (Culure01_3 and Culture02_3) served as sequence templates for coverage estimation. Paired-end information was extracted from the SAM files generated by mapping filtered read pairs to sequence templates using bwa 0.7.1 (Li and Durbin, 2010). The taxonomic compositions of the metagenomes were determined using both the reads and assembled contigs via the MG-RAST platform as described previously (Cai et al., 2016).
Genomes Binning of the Metagenomic Sequencing Data
We distinguished individual genomes in the metagenomes using a differential coverage binning approach similar to that reported in a previous study (Albertsen et al., 2013), with modifications to include multiple time-point samples. Coverage of the individual contig from each time point was determined by mapping the filtered reads to the respective sequence templates (Culure01_3 for Culture01 and Culture02_3 for Culture02). The contigs were binned into genome bins by plotting contig coverage estimates of any two time points or, to obtain a better resolving power, by plotting the contig coverage estimates for multiple time points with multidimensional scaling (MDS). The inclusion of three metagenomes gave the best resolution. The draft genome bins were refined based on sequence compositions (GC content and tetra-nucleotide frequencies) and taxonomic compositions. In addition, contigs not included in the previous steps or wrongly assigned were recruited to, or removed from, the genome bins according to paired-end information. Reconstructed draft genomes were compared with the results generated by the expected-maximization based method MaxBin 2.0 (Wu et al., 2016). The quality of the draft genomes (e.g., completeness and contamination) was evaluated using CheckM (Version 1.0.4) (Parks et al., 2015). Genome-wide average nucleotide identity (ANI) and average amino acid identity (AAI) analyses were calculated using the online ANI and AAI calculators (Rodriguez-R and Konstantinidis, 2016).
Protein coding genes were inferred from the assembled contigs using Prodigal (v2.60) (Hyatt et al., 2010) with metagenome mode enabled. The genes were functionally annotated by searching the gene sequence against the NCBI non-redundant database using DIAMOND (v0.8.17.79) (Buchfink et al., 2015) and submitting to the KEGG Automatic Annotation Server (Moriya et al., 2007). The hallmark gene amoC was partially assembled, so the region-specific Sanger sequencing primer pairs were designed for amplifying the gene. Genes related to nitrogen metabolism and carbon fixation (K numbers) were compared to those from the reference genomes Nitrosomonas sp. AL212 (Yuichi et al., 2011) and N. winogradskyi Nb-255 (Starkenburg et al., 2006). The comparative results were visualized using Circos (Krzywinski et al., 2009). The metabolic pathways of the nitrifier bins were manually curated and reconstructed using EC numbers as described previously (Cai et al., 2016).
Phylogenetic Analyses
The phylogeny of the nitrifiers in the cultures was determined using both the 16S rRNA genes and nitrification-related functional genes (i.e., amoA and nxrA) by the neighbor-joining algorithm in MEGA (v 5.2) (Tamura et al., 2011). Reference 16S rRNA gene sequences of the nitrifiers were recruited by searching the keywords ‘Nitrosomonas 16S’ and ‘Nitrobacter 16S’ in the NCBI database, while the amino acid sequences of the hallmark genes were obtained by searching the keywords ‘ammonia, methane, amo, pmo or monooxygenase’ and ‘nxr, nitrite reductase or nitrite oxidoreductase.’ The recruited sequences that showed a high similarity to the genes in our samples were retained for phylogenetic calculations. For the genome tree, all complete and draft genomes of AOB and NOB were obtained from NCBI. The phylogenetic analysis was performed using PhyloPhlAn (Segata et al., 2013) with default parameters.
Inorganic Nitrogen Measurement and Nitrification Kinetics Calculation
The concentrations of NH4+ were determined using a colorimetric assay (Bollmann et al., 2011) with a spectrophotometer (SpectraMax M2, Molecular Devices, United States), while NO2- and NO3- were determined using ion chromatography (Dionex-ICS-1100) with an Ion Pac AS18 4 mm × 250 mm analytical column. Growth rates of the nitrifiers were calculated based on the slope of the log-transformed concentrations of NO2- + NO3- for AOB or NO3- for NOB plotted against time (h) during the exponential growth phase as described previously (Bollmann et al., 2011; French et al., 2012). The coefficients of determination (R2) of all the slopes were ≥0.99. The Monod kinetics model μ = μmax(S / (Ks + S)) was used to determine the characteristics (μmax and Ks) of the cultures. Here, μ is the growth rate at different substrate concentration, μmax is the maximum growth rate, Ks is the half-saturation constant, and S is the rate-limiting substrate concentration. The lag duration of a culture in an experiment was calculated from the time point immediately after inoculation to the first time point used for the growth rate calculation.
Cell Concentration Analysis
The cell concentrations of different cultures were determined by collecting 12–15 ml of the inoculum and samples at the end of incubation and extracting the genomic DNA, as indicated above. The absence/presence of specific nitrifying populations (i.e., AOA, AOB, and NOB) was verified using PCR with the primers listed in Supplementary Table S1, according to the thermocycling protocols of the respective references. The abundance of the bacterial amoA gene was quantified using qPCR with the primers amoA-1F and amoA-2R (Rotthauwe et al., 1997), while the primers F1norA and R2norA (Attard et al., 2010) were used to quantify the nxrA gene. The qPCR was performed on a StepOnePlus Real-Time PCR Systems (Applied Biosystems) with the SYBR Green master mix, according to previous studies (Attard et al., 2010; Limpiyakorn et al., 2011). Serially diluted DNA standards were prepared for absolute gene copy quantification using purified amoA and nxrA genes PCR products according to a previous study (Ritalahti et al., 2006). Melt curves were performed with each assay to confirm the specificity of the primers. For all experimental samples, the average of biological triplicates was calculated.
Nucleotide Sequence Accession Number
The amplicon sequences, metagenomic sequences, and the AOB and NOB draft genomes generated in this study have been deposited in the NCBI BioProject Database under accession number PRJNA343684.
Results
Descriptions of the Nitrifying Cultures
Two cultures of nitrifiers (Culture01 and Culture02) were established from a freshwater aquarium biofilter (Figure 1). The concentrations of NH4+ in the influent to the biofilter were in the range of 500 μM; therefore, a synthetic medium containing 500 μM NH4+ was used to culture the nitrifiers to simulate the in situ process. After 15 months, a stable culture was established that converted NH4+ to NO3- via transient production of NO2- within 120 h (Culture01) (Figure 1A). Selection only for ammonia oxidizers was made by amending with chlorate, which resulted in a culture that stoichiometrically converted NH4+ to NO2- within 180 h (including 75 h of lag time) with no NO3- production (Culture02) (Figure 1B). We tested the activities of the nitrite oxidizers by transferring cells of Culture01 to a medium containing only NO2- without NH4+, in which gave a stoichiometric conversion of NO2- to NO3- but required a much longer time (375 h) (Culture03) (Figure 1C).
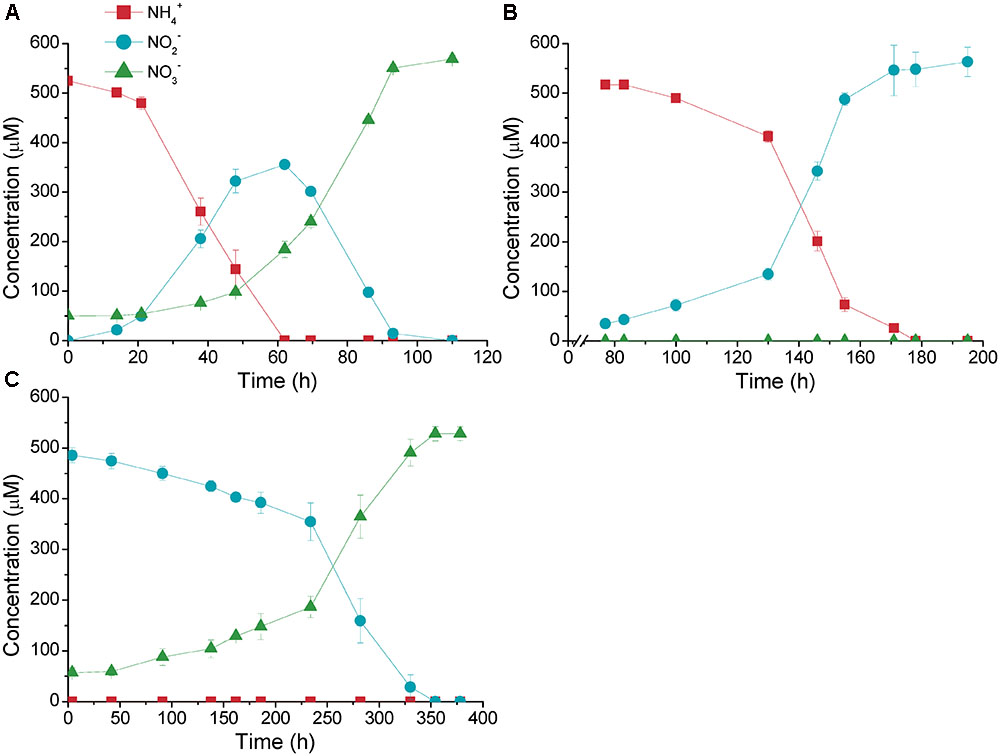
FIGURE 1. Nitrification activities of (A) Culture01, (B) Culture02, and (C) Culture03. The amendment was 500 μM of NH4+ (for Culture01 and Culture02) or NO2- (for Culture03). Error bars represent one standard deviation from biological triplicate experiments.
Composition of the Microbial Communities in the Biofilter and Cultures via 16S rRNA Gene Sequencing
The microbial community structures of the biofilter, Culture01, and Culture02 were evaluated using 16S rRNA gene amplicon sequencing. A total of 8,728 OTUs were recovered from all the samples. After 15 months of culturing, the relative abundances of nitrifying bacteria in Culture01 and Culture02 were 19.1 and 12.4%, respectively, compared to 4.4% for the biofilter (Supplementary Figure S1). Of the OTUs pertinent to the AOB guild, three (i.e., OTU01, OTU02, and OTU03) were closely related to the Nitrosomonas genus. As shown in Figure 2 and Supplementary Table S2, Culture01 and Culture02 shared the same Nitrosomonas-like AOB (OTU01) and this most abundant AOB clustered closely (98% sequence identity) to the 16S rRNA gene of Nitrosomonas oligotropha Nm45. The eight OTUs classified as nitrite oxidizers were distributed within the Nitrospira (n = 5; OTU04-08) and Nitrobacter clusters (n = 3; OTU09-11). Of the eight NOB-like OTUs present in the biofilter, only two increased in relative abundance in Culture01 (OTU05 and OTU09, Figure 2A).
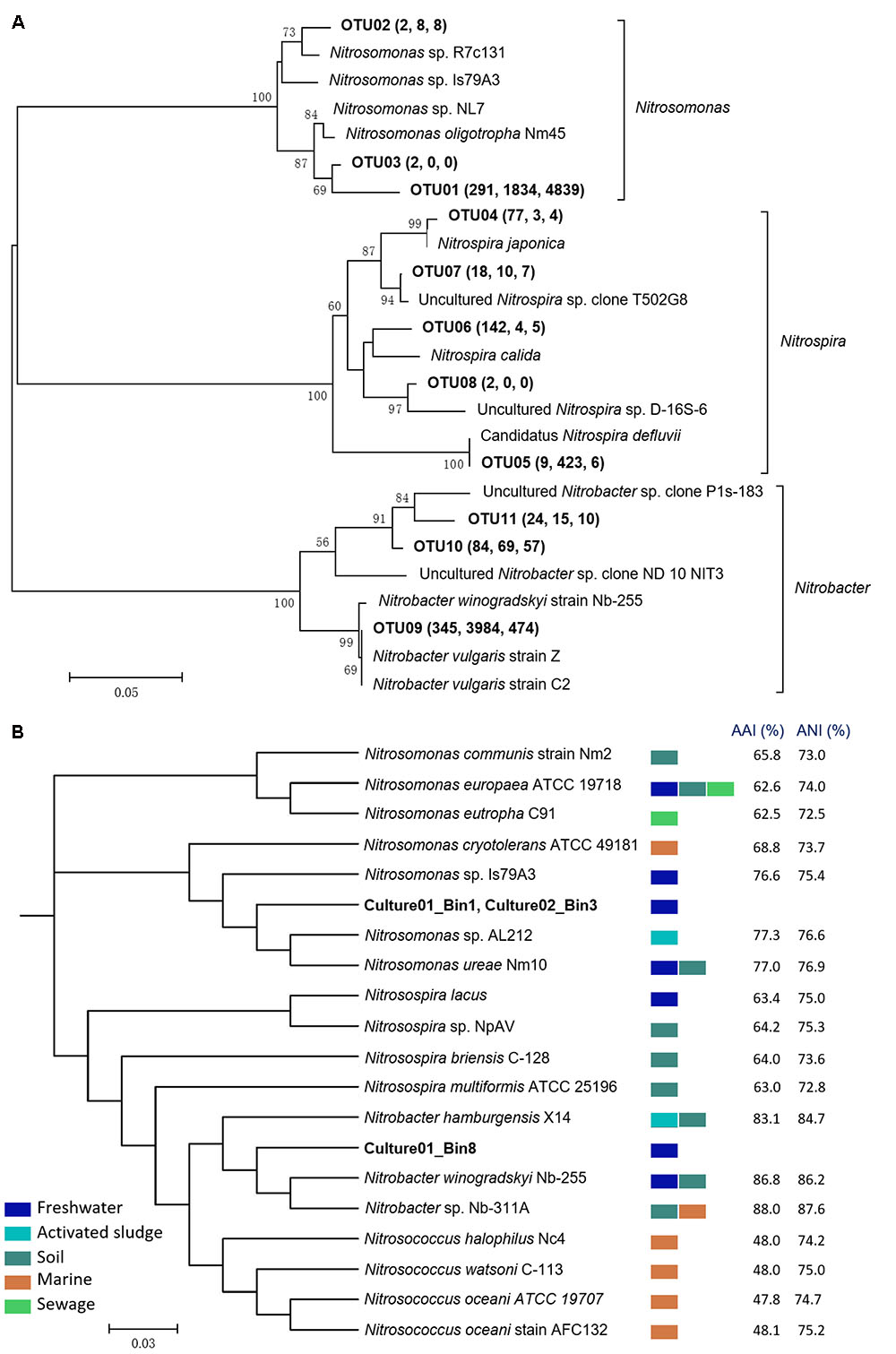
FIGURE 2. (A) Phylogenetic tree of the nitrifiers in the biofilter and cultures based on 16S rRNA genes. The tree was built using the neighbor-joining algorithm in MEGA 5.2. OTUs (bolded) were defined based on a similarity threshold of 97% and bootstrap values (1,000 iterations) greater than 50% are shown. The values in the brackets indicate the numbers of reads found in the biofilter, Culture01, and Culture02, respectively. (B) Phylogenetic tree based on the complete and draft genomes using PhyloPhlAn. AAI and ANI for Nitrosomonas and Nitrosospira were calculated against Culture01_Bin1, while Nitrobacter and Nitrosococcus were calculated against Culture01_Bin8. All internal nodes have a high bootstrap value (65–100%). Colored rectangles represent the typical habitats of the nitrifiers.
An OTU of the Nitrososphaera family (3.6%) belonging to the AOA was found in the biofilter, but no sequence belonging to the AOA was subsequently detected in either culture. PCR with primers targeting the archaea amoA gene failed to detect the presence of AOA in Culture01 or Culture02 (Supplementary Table S1). Genera belonging to Bosea (increased by 1.5% in Culture01 and 6.1% in Culture02), Aminobacter (increased by 1.9% in Culture01 and 4.0% in Culture02), Sediminibacterium (increased by 2.5% in Culture01 and 2.6% in Culture02), and Acidovorax (increased by 10.0% in Culture01 and 18.4% in Culture02) were higher in both cultures relative to the biofilter (Supplementary Figure S1), while Azospirillum decreased (by 8.8% in Culture01 and 8.9% in Culture02). Interestingly, Desulfitobacterium was abundant in the biofilter (29.1%) and remained at a relatively high abundance after cultivation (20.0% in Culture01 and 16.7% in Culture02). Overall, the organisms found in the cultures were representative of the in situ key participants in the aquarium biofilter.
Metagenomic Sequencing of the Cultures and Draft Genomes of the Nitrifiers
The 16S rRNA gene sequencing was supplemented with metagenomic sequencing, applied to query the genomic contents of the cultures. The variation in the community composition during growth was taken into account by collecting samples of Culture01 and Culture02 on days 1, 3, and 5, based on the nitrification profiles (Figure 1). The samples contained 1.0–2.8 Gb of paired-end sequences after quality control (Supplementary Table S3). The qualified reads were assembled into contigs with lengths ranging from 1.2–1,066 kb, generating a total of 45.7–59.2 Mb per sample (Supplementary Table S3). The resulting contigs were resolved and assigned into genomic bins using differential coverage binning (Supplementary Figure S2). A total of eight high-quality bins could be identified in the metagenomes of Culture01, while nine were found in Culture02 (Supplementary Table S4), with a high similarity shared between some of the bins of the two cultures (Supplementary Table S2). The high completeness (87.8–100%, except for Culture02_Bin9) and the low number of contaminating sequences found in the genomes (0–2.4%, Supplementary Table S4) suggest a high quality for the resulting bins. Furthermore, the high similarity of the bins (72.7–100%) obtained using two different binning approaches (MaxBin and differential coverage binning, Supplementary Table S5) indicates the reliability of the binning results.
Phylogenetic analysis using PhyloPhlAn based on whole genomes shows that the two AOB draft genomes (with about 97% completeness) from Culture01 (Culture01_Bin1) and Culture02 (Culture02_Bin3) are closely related to the complete genome of Nitrosomonas ureae Nm10 [found in freshwater and soils (Koops et al., 1991)] and Nitrosomonas sp. AL212 [found in activated sludge (Prosser et al., 2014)], with AAI and ANI similarities around 77% (Figure 2B). Pair-wise comparison of the two Nitrosomonas-like draft genomes in the two cultures shows an identity of more than 98% (Supplementary Table S2) and an AAI and ANI similarity of 100%. The two draft genomes of the Nitrosomonas-like AOB (Supplementary Table S6) contained genes encoding ammonia monooxygenase (i.e., amoCAB sequences). Likewise, genes encoding hydroxylamine dehydrogenase (haoAB) and its electron carriers, cytochromes C554 and Cm552, were identified (Supplementary Figure S3). As shown in Supplementary Figures S3, S4, the identified hallmark genes and gene loci of the Nitrosomonas-like AOB (amoCAB and haoA) in both cultures were highly similar to the genes of the Nitrosomonas cluster, consistent with the whole genome-based phylogenetic tree (Figure 2B), whereas they were much lower in similarity with the genes of the comammox Nitrospira species.
The whole genome-based phylogenetic tree also shows that a draft genome binned from Culture01 (Culture01_Bin8) was most closely related to N. winogradskyi Nb-255 [found in freshwater and soils (Koops and Pommerening Röser, 2001; Wertz et al., 2008)] and Nitrobacter sp. Nb-311A [found in marine and soils (Moran et al., 2007; Wertz et al., 2008)], with AAI and ANI similarities around 87% (Figure 2B). The draft genome of the Nitrobacter-like NOB (Supplementary Table S7) contained genes encoding nitrite oxidoreductase (i.e., nxrA and partial nxrB sequences) and its cognate electron carriers (i.e., cytochrome c) (Supplementary Figure S3). The sequences of the nxrA and nxrB genes and their loci were highly similar to N. winogradskyi Nb-255 and Nitrobacter sp. Nb-311A, in agreement with the phylogenetic tree of the whole genomes (Figure 2B), but very different from the comammox Nitrospira species (Supplementary Figure S3). Consideration of unsequenced species clustered the nxr genes of the cultured Nitrobacter-like NOB closer to N. vulgaris (Supplementary Figure S4).
Reconstruction of the key carbon metabolic pathways in the draft genomes of both the Nitrosomonas-like AOB (Culture01_Bin1 and Culture02_Bin3) and Nitrobacter-like NOB (Culture01_Bin8) revealed the presence of a complete set of genes in the metabolic pathways of both the carbon fixation and citric acid cycles, as well as acetate metabolism, which is consistent with the reference genomes from the same genus (e.g., Nitrosomonas sp. AL212 and N. winogradskyi Nb-255, Supplementary Figures S5, S6). The pathways for nitrogen metabolism in the draft genomes are also consistent with the reference genomes (Supplementary Figure S6).
Physiological Activities of the Nitrifiers in the Cultures
The physiological activities of the nitrifying populations in the cultures were tested using five different concentrations (50–1,500 μM) of NH4+ or NO2-. Increasing the substrate concentrations increased the duration of the lag phase for Culture01 (23–45 h), but shortened it for Culture02 (250–120 h) (data not shown). The lag time for Culture03 increased from 88–146 h in response to increasing NO2- concentration; this duration was longer than that observed for Culture01 with NH4+ (data not shown).
The growth rates of Culture01 and Culture02 increased with NH4+ amendment up to 500 μM, but showed no substantial changes with higher concentrations (Figure 3A). Culture03 showed only incremental changes in the growth rates with increasing NO2- concentration (Figure 3A). In general, the growth rates were higher for Culture01 (0.019–0.028 h-1) than for either Culture02 (0.0091–0.021 h-1) or Culture03 (0.0029–0.0051 h-1) (Figure 3A). The NO2- inhibition test indicated that the cultured AOB in Culture02 could completely transform 500 μM NH4+ to NO2- even following spiking with a relatively high concentration of NO2- (10 mM). The Ks values for NH4+ uptake for Culture01 and Culture02 were 25.9 and 71.8 μM, respectively. The variations in NO2- concentration in Culture01 during cultivation precluded an accurate determination of the activities of NOB in Culture01, while the data of Culture03 fitted the Monod kinetics model poorly.
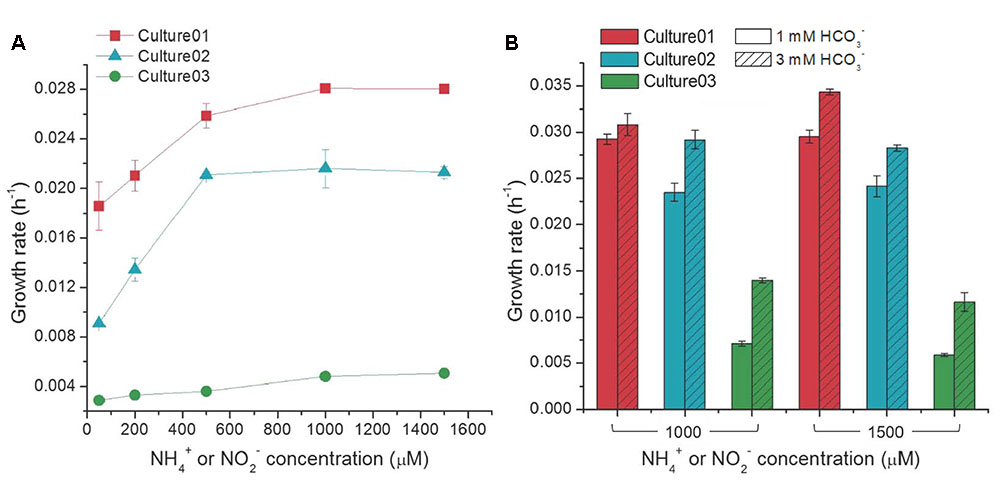
FIGURE 3. Influence of different concentrations of (A) NH4+ or NO2- (with 1,000 μM HCO3-), and (B) HCO3- on the growth rates of the nitrifiers. Culture01 and Culture02 were fed NH4+, while Culture03 was fed NO2-. Error bars represent one standard deviation from biological triplicate experiments.
High concentrations of NH4+ or NO2- (1,000 or 1,500 μM) resulted in incomplete oxidation in the three cultures. The possibility of insufficient carbons and buffering capacity in the cultures was tested by amending with a higher HCO3- concentration (3,000 μM versus 1,000 μM routinely used). The growth rates of the nitrifiers increased significantly with increasing HCO3- concentration [Student’s t-test, p < 0.01, except Culture01 (1,000 μM NH4+)] (Figure 3B) and complete oxidation of the input nitrogen occurred. When compared to Culture01 and Culture02, the growth rate of Culture03 was more responsive to the increase in HCO3- concentrations [i.e., the growth rate almost doubled in response to increasing HCO3- concentration] (Figure 3B). The possible presence of carbon dioxide (CO2) arising from ambient air in the headspace was eliminated by purging the serum bottles with a pure gas mixture of N2:O2 (79:21%) without CO2 and no difference was noted with or without purging.
Growth of Nitrifying Populations
The influences of different nitrogen and carbon concentrations on the growth of nitrifiers in the cultures were assessed by qPCR of the amoA and nxrA genes, representing the ammonia and nitrite oxidizers, respectively. Culture01 showed no increase in the extent of growth for the Nitrosomonas-like AOB with increasing NH4+ concentrations and no substantial growth for the Nitrobacter-like NOB under NH4+ concentrations of 1,000 and 1,500 μM when 1,000 μM HCO3- was supplied (Figure 4). A HCO3- concentration of 1,000 μM increased the growth of AOB in Culture02 at NH4+ concentration ≤500 μM and NOB in Culture03 at NO2- concentrations ≤200 μM, but this growth became steady when nitrogen concentration increased (Figure 4). By contrast, an excess of HCO3- (3,000 μM) promoted higher growth of AOB and NOB in all cultures (Student’s t-test, p < 0.01) when compared to cultures with 1,000 μM HCO3- (Figure 4).
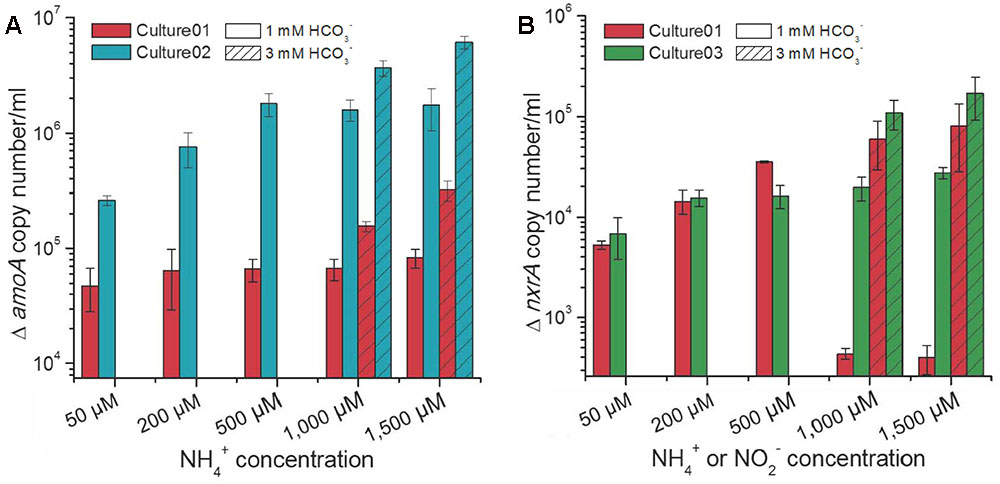
FIGURE 4. Increases in the (A) amoA and (B) nxrA gene copy numbers at different NH4+ or NO2- concentrations. Culture01 and Culture02 were fed NH4+, while Culture03 was fed NO2-. The amoA gene copy numbers at time zero for Culture01 and Culture02 were 2.2 × 104 ± 7.9 × 102 and 4.0 × 104 ± 1.3 × 103 per ml, respectively. The nxrA gene copy number at time zero for Culture01 and Culture03 was 2.0 × 103 ± 2.5 × 102 per ml. Error bars represent one standard deviation from biological triplicate experiments.
Under the same substrate (HCO3- and NH4+) concentrations, the growth of AOB was significantly higher (Student’s t-test, p < 0.01) in Culture02 than in Culture01, with the amoA gene showing increases from 4.8 × 104 ± 1.9 × 104 to 3.2 × 105 ± 6.3 × 104 per ml for Culture01 and increases from 2.6 × 105 ± 2.5 × 104 to 6.1 × 106 ± 7.5 × 105 per ml for Culture02 (Figure 4A). Growth of the Nitrobacter-like NOB was similar between Culture01 and Culture03 when the HCO3- was in excess, with increases in the nxrA gene ranging from 5.3 × 103 ± 5.0 × 102 to 8.0 × 104 ± 3.8 × 104 per ml culture for Culture01 and from 6.8 × 103 ± 3.0 × 103 to 1.6 × 105 ± 7.7 × 104 per ml for Culture03 (Figure 4B).
Discussion
In this study, a biofilter from an aquarium was used to culture AOB and NOB, which have functional roles in removing NH3 and NO2- that are toxic to aquatic life (Keuter et al., 2011; French et al., 2012; Wu et al., 2013). A culture-dependent approach was adopted to allow testing of the bacterial physiology. In particular, nitrification activities were examined with and without the synergistic interactions between the AOB and NOB. A 15-month cultivation period (over 60 transfers) with NH4+ as the sole energy source for the nitrifying community successfully cultured a number of AOB and NOB, as confirmed by both 16S rRNA gene amplicon and metagenomic sequencing. Culture02, cultured using chlorate, contained a few Nitrobacter 16S rRNA gene sequences (Supplementary Figure S1), but the NOB were unlikely to be functional as stoichiometric conversion of NH4+ to NO2- was obtained and the nxrA gene was not amplified via PCR (Supplementary Table S1). The AOA belonging to Nitrososphaera were initially found in the biofilter, but no archaeal amoA gene or 16S rRNA gene was subsequently detected in the cultures. The relatively low culturing temperature (25°C as opposed to the 37°C preferred by AOA) (Wu et al., 2013), short retention time (Xia et al., 2011), high NH4+ concentration (Martens-Habbena et al., 2009; Ke et al., 2013) and high dissolved oxygen levels (Yan et al., 2012) likely favored AOB over AOA. We attempted to use ampicillin to isolate AOA with 500 μM NH4+ in synthetic medium (Mosier and Francis, 2008), but we detected no nitrification activity after incubating for more than one month.
Similar to the biofilter, the other major bacteria present in the cultures were from the Acidovorax lineage (Supplementary Figures S1, S7), which could reduce NO3- under aerobic conditions at 20°C in the presence of polycyclic aromatic hydrocarbons (Eriksson et al., 2003), and different classes of Proteobacteria (Supplementary Figure S1). Despite the high oxygen concentration in the medium (>5.6 mg/L), anaerobic bacteria from the Desulfitobacterium lineage (Supplementary Figures S1, S7) remained in the cultures, similar to the biofilter [some Desulfitobacterium strains can exhibit high tolerance toward oxygen stress (De Wildeman et al., 2004; Kim et al., 2012)]. No nitrification-related genes were found in populations other than the AOB and NOB.
Physiology of the Cultured AOB
The AOB in the cultures were assigned to the Nitrosomonas lineage (Figure 2), corroborating previous studies confirming that the bacteria in this lineage can typically function in environments with a low NH4+ concentration, such as soils (Wang et al., 2015), lake sediments (French et al., 2012), biofilms (Stehr et al., 1995), and freshwater aquarium biofilters (Burrell et al., 2001). The OTU with the most reads assigned to the AOB in the cultures (OTU01) showed the closest match (98% sequence identity) to the 16S rRNA gene of N. oligotropha Nm45, which grows at a relatively low NH4+ concentrations (1,000–5,000 μM) (Prosser et al., 2014) and has one of the highest affinities for NH4+ of all AOB studied (Koops et al., 1991; Martens-Habbena et al., 2009; Kits et al., 2017). Similarly, the hallmark genes (amoCAB and haoA) and the gene loci in the draft genomes of the AOB show high similarity to the two known species (i.e., N. ureae Nm10 and Nitrosomonas sp. AL212, Supplementary Figure S3) in the N. oligotropha lineage normally found in low NH4+ habitats, such as freshwater rivers and lakes (Koops et al., 1991; Prosser et al., 2014).
The growth rates (0.009–0.028 h-1) of the Nitrosomonas-like AOB in the cultures are within the range of those reported for other AOB (0.002–0.088 h-1) (French et al., 2012; Prosser and Nicol, 2012) and are most similar to the strains within N. oligotropha (0.02–0.06 h-1); this included the growth rate characteristic of no further increases at the NH4+ concentration of ∼500 μM (French et al., 2012). Growth rates of nitrifiers are closely related to inorganic carbon concentration as it has been shown that the change in HCO3- concentrations can lead to greater changes in growth rate than NH3 and oxygen (Wett and Rauch, 2003; Jiang et al., 2015), indicating that the availability of inorganic carbons is a major factor that influences nitrification in freshwater environments. The higher growth rates observed at the NH4+ concentrations of 1,000 and 1,500 μM with 3 mM HCO3- are likely the results of availability of additional inorganic carbons to the nitrifiers and a stronger capacity to buffer the protons released during NH3 oxidation (Ahn, 2006).
Previous studies have shown that the Ks of NH4+ for species within the Nitrosomonas lineage ranges from 30–3,900 μM, with strains of N. oligotropha having the lowest values (30–118 μM) among the reported AOB (Suwa et al., 1994; Martens-Habbena et al., 2009). Although the present case was not a pure culture, the Ks of NH4+ for Culture01 (25.9 μM) is lower than the previously reported values and the Ks for Culture02 is within the range reported for N. oligotropha (Supplementary Figure S8), suggesting that the AOB in the cultures are competitive in an oligotrophic environment. However, the affinity for NH4+ is definitely far weaker than the recently reported comammox bacterium Nitrospira inopinata (0.84 μM) (Kits et al., 2017).
Similar to the closely related Nitrosomonas sp. AL212 and N. ureae Nm10 (Yuichi et al., 2011; Kozlowski et al., 2016), the cultured Nitrosomonas-like AOB may not be obligately lithoautotrophic, as inference from the genome suggests they may be able to potentially use both organic and inorganic carbon (Supplementary Figure S5), which has to be further verified experimentally.
Physiology of the Cultured NOB
For the nitrite oxidizers, five OTUs related to Nitrobacter and three OTUs to Nitrospira were detected in the biofilter. After cultivation, one of the more dominant nitrite oxidizers was OTU05, which shared 100% sequence identity to the 16S rRNA gene of Candidatus Nitrospira Defluvii [found in activated sludge (Spieck et al., 2006) and can survive under low NO2- conditions (Lücker et al., 2010)]. Nitrospira are speculated to represent the most prevalent nitrite oxidizers in low-nitrogen environments, such as soils (Xia et al., 2011; Wang et al., 2015) and freshwater systems (Hovanec et al., 1998; Regan et al., 2002). We were unable to extract the Nitrospira genome in the cultures from the metagenomes, possibly due to its low abundance [Supplementary Table S8; confirmed by weak or no band in PCR (Supplementary Table S1)]. The other dominant NOB are related to OTU09, which clusters closely (100% 16S rRNA gene sequence identity) to the species N. vulgaris Z, one of the slowest lithoautotrophically growing nitrite oxidizers (doubling time 140 h) widely present in soils and freshwater (Bock et al., 1990). The nxrAB genes of the Nitrobacter-like NOB in the cultures are also highly similar to those found in N. vulgaris (Supplementary Figure S4); however, the genome of N. vulgaris is not available for whole genome-based phylogenetic analysis. The draft genome of Culture01_Bin8 represents the first sequenced genome of the species N. vulgaris. The nitrite reductase gene (nirK) was present in the draft genomes of the Nitrosomonas-like AOB and Nitrobacter-like NOB and confirmed via PCR (Supplementary Table S1); however, this gene is unlikely to be functional in our study, since we found a stoichiometric conversion of NH4+ to NO2- in our cultures (Figure 1). In addition, our cultivation was performed in an oxygen-rich condition (>5.6 mg/L), contrary to previous studies that showed functioning of the nirK gene in a low oxygen environment (0–4%) (Starkenburg et al., 2008; Attard et al., 2010).
Nitrobacter NOB are usually of high abundance in environments with a high nitrogen content, such as wastewater (Wagner and Loy, 2002) and high-nutrient soils (Attard et al., 2010). The accumulated NO2- concentration (maximum 355 μM) in Culture01 is not considered high; nevertheless, the Nitrobacter-like NOB was much more abundant than Nitrospira (Figure 2 and Supplementary Figure S1). This contrasts with other aquarium biofilter studies that found Nitrospira-like NOB to be the most common nitrite oxidizers (Hovanec et al., 1998; Burrell et al., 2001). This difference could be due to the likelihood of a faster growth rate of the Nitrobacter-like NOB in our cultures, as N. vulgaris has a short generation time (13 h) compared with other species of NOB (∼26–43 h) (Nowka et al., 2015) and the Ks of NO2- is lower for N. vulgaris (49 μM) than for other Nitrobacter-like NOB and is similar to Nitrospira (Nowka et al., 2015), suggesting that N. vulgaris can be as competitive as Nitrospira in oligotrophic environments.
Synergistic Interactions between the AOB and NOB in the Cultures
AOB and NOB coexist in both natural (e.g., rice paddy soils) (Ke et al., 2013; Wang et al., 2015) and engineered systems (e.g., wastewater treatment plants) (Dionisi et al., 2002), but previous studies investigating their relationships were usually carried out using artificially constructed co-cultures of AOB and NOB isolates (Laanbroek and Gerards, 1993; Perez et al., 2015). These approaches may not reflect the nitrification processes in a real-life ecosystem, and most studies also tended to focus on nitrifiers in activated sludge (Sliekers et al., 2005; Winkler et al., 2012), with fewer co-culture studies on members of freshwater biofilters (van Kessel et al., 2010).
Metagenomic and 16S rRNA gene sequencing, together with physiological experiments, indicated that the nitrifying partnership in the cultures involves a Nitrosomonas-like AOB and a Nitrobacter-like NOB that are adapted to a relatively low NH4+ condition. The combination of AOB and NOB in Culture01 reduced the lag time compared to that in Culture02 and Culture03. For example, incubation at the lowest NH4+ or NO2- concentration tested (50 μM) gave longer lag phases for Culture02 and Culture03 than for Culture01 of 10- and 3.5-fold, respectively. Although nitrifiers are reportedly sensitive to the initial incubation conditions (e.g., substrate concentrations) (Graham et al., 2007), the duration of the lag phase also varies significantly when incubated with the same NH4+ concentration, depending on whether the nitrifying partner is present. Furthermore, similar to a previous study (Sedlacek et al., 2016), the growth rate also highly depends on the presence of the nitrifying partner, with Culture01 showing the highest rate compared to the two other cultures, and especially Culture03 (Figure 3A). The extent of cell growth for the AOB was lower in Culture01 than in Culture02 and the NOB in Culture 01 had similar growth to that seen in Culture03 (Figure 4). The major difference between Culture01 and Culture02 was the composition of the nitrifiers; therefore, the decoupling between the growth of AOB and nitrification activities might reflect how the cells utilize the energy derived from NH4+ in the presence NOB. In line with the previous finding that AOB had a higher Ks value for NH4+ in the absence of NOB (Sedlacek et al., 2016), the Ks of NH4+ for Culture02 without NOB was threefold higher than for Culture01 with NOB, suggesting that the synergistic interactions between AOB and NOB enable the robust oxidation of NH4+ at a low concentration. The presence of heterotrophic bacteria with AOB had also been shown to increase the expression of proteins related to the ammonia oxidation pathway of AOB and promote nitrification (Sedlacek et al., 2016). The kinetic parameters and the synergistic relationships can potentially be used to enhance nutrient removal in freshwater treatment systems, which could benefit aquatic life and prevent eutrophication (Hou et al., 2013).
The physical contact between AOB and NOB allows the efficient transfer of compounds between them (Okabe et al., 1999). Reciprocal feeding between AOB and NOB has been experimentally demonstrated by co-culturing AOB and NOB, whereby Nitrospira moscoviensis breaks down urea or cyanate to NH4+ to provide an energy source for N. europaea or Nitrosomonas nitrosa, which in turn produce NO2- for the NOB (Palatinszky et al., 2015). This source of NH4+ is important for the functioning of AOB in oligotrophic environments (Prosser et al., 2014). Interestingly, the gene encoding cyanate hydratase (Starkenburg et al., 2006), which converts cyanate to NH3 and CO2 in the presence of HCO3-, is found in the genome of the Nitrobacter-like NOB (Culture01_Bin8) (also present in N. winogradskyi Nb-255), and this could aid in supporting AOB when urea or cyanate is available.
Conclusion
We used a culture-dependent approach to simulate the in situ nitrification process in a freshwater biofilter to study the ecophysiology of the ammonia-oxidizing and nitrite-oxidizing guilds at a relatively low nitrogen concentration and the synergistic relationships between these guilds. The cultured N. vulgaris may be as competitive as Nitrospira-like NOB in oligotrophic environments. The nitrification kinetics of the cultures are influenced by NH4+ and/or NO2-, and HCO3- concentrations. Metagenomic sequencing indicated the draft genomes of the nitrifying partners (Nitrosomonas-like AOB and Nitrobacter-like NOB), and that their growth rate, substrates affinity, and lag duration strongly depended on the presence of each partner. Although the AOB and NOB could function independently, when both were present together, robust nitrification occurred. Overall, the observations in this study indicate the competitiveness of the cultured Nitrosomonas-like AOB and Nitrobacter-like NOB in an oligotrophic environment and a strong dependence of their activities on the synergistic relationships between the two guilds. These results provide insights for possible manipulations of multi-species interactions to optimize nitrification treatment processes.
Author Contributions
MC performed the research, analyzed the data, and wrote the manuscript. S-KN, CKL, HL, and YJ performed the research and analyzed the data. PKHL designed the experiments and wrote the manuscript.
Funding
This research was supported by the Research Grants Council of Hong Kong through project 116413.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley for providing sequencing services. We also express our gratitude to Marcus Leung and David Wilkins for their assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00280/full#supplementary-material
References
Ahn, Y.-H. (2006). Sustainable nitrogen elimination biotechnologies: a review. Process Biochem. 41, 1709–1721. doi: 10.1016/j.procbio.2006.03.033
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K. L., Tyson, G. W., and Nielsen, P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Attard, E., Poly, F., Commeaux, C., Laurent, F., Terada, A., Smets, B. F., et al. (2010). Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326. doi: 10.1111/j.1462-2920.2009.02070.x
Balch, W., Fox, G., Magrum, L., Woese, C., and Wolfe, R. (1979). Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43, 260–296.
Belser, L. W., and Mays, E. L. (1980). Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Appl. Environ. Microbiol. 39, 505–510.
Bernal, A., Ear, U., and Kyrpides, N. (2001). Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 29, 126–127. doi: 10.1093/Nar/29.1.126
Biebl, H., and Pfennig, N. (1978). Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch. Microbiol. 117, 9–16. doi: 10.1007/Bf00689344
Bock, E., Koops, H. P., Moller, U. C., and Rudert, M. (1990). A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch. Microbiol. 153, 105–110. doi: 10.1007/Bf00247805
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bollmann, A., French, E., and Laanbroek, H. J. (2011). Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. Method Enzymol. 486, 55–88. doi: 10.1016/S0076-6879(11)86003-9
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Burrell, P. C., Phalen, C. M., and Hovanec, T. A. (2001). Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl. Environ. Microbiol. 67, 5791–5800. doi: 10.1128/AEM.67.12.5791-5800.2001
Cai, M. W., Wilkins, D., Chen, J. P., Ng, S. K., Lu, H. Y., Jia, Y. Y., et al. (2016). Metagenomic reconstruction of key anaerobic digestion pathways in municipal sludge and industrial wastewater biogas-producing systems. Front. Microbiol. 7:778. doi: 10.3389/Fmicb.2016.00778
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
De Wildeman, S., Linthout, G., Van Langenhove, H., and Verstraete, W. (2004). Complete lab-scale detoxification of groundwater containing 1, 2-dichloroethane. Appl. Microbiol. Biot. 63, 609–612. doi: 10.1007/s00253-003-1363-y
Dionisi, H. M., Layton, A. C., Harms, G., Gregory, I. R., Robinson, K. G., and Sayler, G. S. (2002). Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68, 245–253. doi: 10.1128/Aem.68.1.245-253.2002
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/Nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eriksson, M., Sodersten, E., Yu, Z., Dalhammar, G., and Mohn, W. W. (2003). Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from northern soils. Appl. Environ. Microbiol. 69, 275–284. doi: 10.1128/AEM.69.1.275-284.2003
French, E., Kozlowski, J. A., Mukherjee, M., Bullerjahn, G., and Bollmann, A. (2012). Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl. Environ. Microbiol. 78, 5773–5780. doi: 10.1128/Aem.00432-12
Graham, D. W., Knapp, C. W., Van Vleck, E. S., Bloor, K., Lane, T. B., and Graham, C. E. (2007). Experimental demonstration of chaotic instability in biological nitrification. ISME J. 1, 385–393. doi: 10.1038/ismej.2007.45
Hou, J., Song, C., Cao, X., and Zhou, Y. (2013). Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res. 47, 2285–2296. doi: 10.1016/j.watres.2013.01.042
Hovanec, T. A., Taylor, L. T., Blakis, A., and Delong, E. F. (1998). Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64, 258–264.
Hyatt, D., Chen, G. -L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jiang, D., Khunjar, W., Wett, B., Murthy, S., and Chandran, K. (2015). Characterizing the metabolic trade-off in Nitrosomonas europaea in response to changes in inorganic carbon supply. Environ. Sci. Technol. 49, 2523–2531. doi: 10.1021/es5043222
Ke, X. B., Angel, R., Lu, Y. H., and Conrad, R. (2013). Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ. Microbiol. 15, 2275–2292. doi: 10.1111/1462-2920.12098
Keuter, S., Kruse, M., Lipski, A., and Spieck, E. (2011). Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ. Microbiol. 13, 2536–2547. doi: 10.1111/j.1462-2920.2011.02525.x
Kim, D. J., Lee, D. I., and Keller, J. (2006). Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour. Technol. 97, 459–468. doi: 10.1016/j.biortech.2005.03.032
Kim, S., Harzman, C., Davis, J. K., Hutcheson, R., Broderick, J. B., Marsh, T. L., et al. (2012). Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 12:21. doi: 10.1186/1471-2180-12-21
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., Bulaev, A., Pjevac, P., et al. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Knapp, C. W., and Graham, D. W. (2007). Nitrite-oxidizing bacteria guild ecology associated with nitrification failure in a continuous-flow reactor. FEMS Microbiol. Ecol. 62, 195–201. doi: 10.1111/j.1574-6941.2007.00380.x
Koops, H., Böttcher, B., Möller, U., Pommerening-Röser, A., and Stehr, G. (1991). Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. Microbiology 137, 1689–1699. doi: 10.1099/00221287-137-7-1689
Koops, H., and Pommerening Röser, A. (2001). Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37, 1–9. doi: 10.1111/j.1574-6941.2001.tb00847.x
Kozlowski, J. A., Kits, K. D., and Stein, L. Y. (2016). Complete genome sequence of Nitrosomonas ureae strain Nm10, an oligotrophic group 6a Nitrosomonad. Genome Announc. 4:e00094-16. doi: 10.1128/genomeA.00094-16
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Laanbroek, H. J., and Gerards, S. (1993). Competition for limiting amounts of oxygen between Nitrosomonas europaea and Nitrobacter winogradskyi grown in mixed continuous cultures. Arch. Microbiol. 159, 453–459. doi: 10.1007/Bf00288593
Leung, M. H. Y., Wilkins, D., Li, E. K. T., Kong, F. K. F., and Lee, P. K. H. (2014). Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl. Environ. Microbiol. 80, 6760–6770. doi: 10.1128/Aem.02244-14
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Limpiyakorn, T., Sonthiphand, P., Rongsayamanont, C., and Polprasert, C. (2011). Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour. Technol. 102, 3694–3701. doi: 10.1016/j.biortech.2010.11.085
Lu, X. Y., Rao, S., Shen, Z. Y., and Lee, P. K. H. (2013). Substrate induced emergence of different active bacterial and archaeal assemblages during biomethane production. Bioresour. Technol. 148, 517–524. doi: 10.1016/j.biortech.2013.09.017
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Moran, M., Belas, R., Schell, M., Gonzalez, J., Sun, F., Sun, S., et al. (2007). Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73, 4559–4569. doi: 10.1128/AEM.02580-06
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Mosier, A. C., and Francis, C. A. (2008). Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10, 3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x
Nowka, B., Daims, H., and Spieck, E. (2015). Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753. doi: 10.1128/Aem.02734-14
Okabe, S., Satoh, H., Watanabe, Y. (1999). In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65, 3182–3191.
Palatinszky, M., Herbold, C., Jehmlich, N., Pogoda, M., Han, P., von Bergen, M., et al. (2015). Cyanate as an energy source for nitrifiers. Nature 524, 105–108. doi: 10.1038/nature14856
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Perez, J., Buchanan, A., Mellbye, B., Ferrell, R., Chang, J., Chaplen, F., et al. (2015). Interactions of Nitrosomonas europaea and Nitrobacter winogradskyi grown in co-culture. Arch. Microbiol. 197, 79–89. doi: 10.1007/s00203-014-1056-1
Prosser, J. I., Head, I. M., Stein, L. Y. (2014). “The family Nitrosomonadaceae,” in The Analysis of Gene Expression Data, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson (Berlin: Springer), 901–918.
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Regan, J. M., Harrington, G. W., and Noguera, D. R. (2002). Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68, 73–81. doi: 10.1128/Aem.68.1.73-81.2002
Ritalahti, K. M., Amos, B. K., Sung, Y., Wu, Q. Z., Koenigsberg, S. S., and Loffler, F. E. (2006). Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72, 2765–2774. doi: 10.1128/Aem.72.4.2765-27774.2006
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712.
Schink, B. (2002). Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81, 257–261. doi: 10.1023/A:1020579004534
Sedlacek, C. J., Nielsen, S., Greis, K. D., Haffey, W. D., Revsbech, N. P., Ticak, T., et al. (2016). Effects of bacterial community members on the proteome of the ammonia-oxidizing bacterium Nitrosomonas sp. strain Is79. Appl. Environ. Microbiol. 82, 4776–4788. doi: 10.1128/AEM.01171-16
Segata, N., Bornigen, D., Morgan, X. C., and Huttenhower, C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/Ncomms3304
Sliekers, A. O., Haaijer, S. C. M., Stafsnes, M. H., Kuenen, J. G., and Jetten, M. S. M. (2005). Competition and coexistence of aerobic ammonium- and nitrite-oxidizing bacteria at low oxygen concentrations. Appl. Microbiol. Biot. 68, 808–817. doi: 10.1007/s00253-005-1974-6
Spieck, E., Hartwig, C., McCormack, I., Maixner, F., Wagner, M., Lipski, A., et al. (2006). Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ. Microbiol. 8, 405–415. doi: 10.1111/j.1462-2920.2005.00905.x
Starkenburg, S. R., Arp, D. J., and Bottomley, P. J. (2008). Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ. Microbiol. 10, 3036–3042. doi: 10.1111/j.1462-2920.2008.01763.x
Starkenburg, S. R., Chain, P. S., Sayavedra-Soto, L. A., Hauser, L., Land, M. L., Larimer, F. W., et al. (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72, 2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006
Stehr, G., Bottcher, B., Dittberner, P., Rath, G., and Koops, H. P. (1995). The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17, 177–186. doi: 10.1111/j.1574-6941.1995.tb00141.x
Suwa, Y., Imamura, Y., Suzuki, T., Tashiro, T., and Urushigawa, Y. (1994). Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 28, 1523–1532. doi: 10.1016/0043-1354(94)90218-6
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
van Kessel, M. A., Harhangi, H. R., van de Pas-Schoonen, K., van de Vossenberg, J., Flik, G., Jetten, M. S., et al. (2010). Biodiversity of N-cycle bacteria in nitrogen removing moving bed biofilters for freshwater recirculating aquaculture systems. Aquaculture 306, 177–184. doi: 10.1016/j.aquaculture.2010.05.019
van Kessel, M. A. H. J., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J. M., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Wagner, M., and Loy, A. (2002). Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotech. 13, 218–227. doi: 10.1016/S0958-1669(02)00315-4
Wang, B. Z., Zhao, J., Guo, Z. Y., Ma, J., Xu, H., and Jia, Z. J. (2015). Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils. ISME J. 9, 1062–1075. doi: 10.1038/ismej.2014.194
Wertz, S., Poly, F., Le Roux, X., and Degrange, V. (2008). Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol. Ecol. 63, 261–271. doi: 10.1111/j.1574-6941.2007.00416.x
Wett, B., and Rauch, W. (2003). The role of inorganic carbon limitation in biological nitrogen removal of extremely ammonia concentrated wastewater. Water Res. 37, 1100–1110. doi: 10.1016/S0043-1354(02)00440-2
Winkler, M. K. H., Bassin, J. P., Kleerebezem, R., Sorokin, D. Y., and van Loosdrecht, M. C. M. (2012). Unravelling the reasons for disproportion in the ratio of AOB and NOB in aerobic granular sludge. Appl. Microbiol. Biot. 94, 1657–1666. doi: 10.1007/s00253-012-4126-9
Wu, Y. C., Ke, X. B., Hernandez, M., Wang, B. Z., Dumont, M. G., Jia, Z. J., et al. (2013). Autotrophic growth of bacterial and archaeal ammonia oxidizers in freshwater sediment microcosms incubated at different temperatures. Appl. Environ. Microbiol. 79, 3076–3084. doi: 10.1128/Aem.00061-13
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Xia, W. W., Zhang, C. X., Zeng, X. W., Feng, Y. Z., Weng, J. H., Lin, X. G., et al. (2011). Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5, 1226–1236. doi: 10.1038/ismej.2011.5
Yan, J., Haaijer, S. C. M., den Camp, H. J. M. O., van Niftrik, L., Stahl, D. A., Konneke, M., et al. (2012). Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ. Microbiol. 14, 3146–3158. doi: 10.1111/j.1462-2920.2012.02894.x
Keywords: freshwater, biofilter, ammonia-oxidizing bacteria, nitrite-oxidizing bacteria, synergistic relationships, metagenomics
Citation: Cai M, Ng S-K, Lim CK, Lu H, Jia Y and Lee PKH (2018) Physiological and Metagenomic Characterizations of the Synergistic Relationships between Ammonia- and Nitrite-Oxidizing Bacteria in Freshwater Nitrification. Front. Microbiol. 9:280. doi: 10.3389/fmicb.2018.00280
Received: 14 November 2017; Accepted: 07 February 2018;
Published: 27 February 2018.
Edited by:
Chris Francis, Stanford University, United StatesReviewed by:
Annette Bollmann, Miami University, United StatesLisa Y. Stein, University of Alberta, Canada
Copyright © 2018 Cai, Ng, Lim, Lu, Jia and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick K. H. Lee, patrick.kh.lee@cityu.edu.hk
 Mingwei Cai
Mingwei Cai Siu-Kin Ng
Siu-Kin Ng Chee Kent Lim
Chee Kent Lim Yangyang Jia
Yangyang Jia Patrick K. H. Lee
Patrick K. H. Lee