- 1Department of Clinical Laboratory, The 306th Hospital of the People’s Liberation Army, Beijing, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 3The 302nd Hospital of the People’s Liberation Army, Beijing, China
- 4Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
Forty-five KPC-producing Enterobacteriaceae strains were isolated from multiple departments in a Chinese public hospital from 2014 to 2015. Genome sequencing of four representative strains, namely Proteus mirabilis GN2, Serratia marcescens GN26, Morganella morganii GN28, and Klebsiella aerogenes E20, indicated the presence of blaKPC-2-carrying IncX6 plasmids pGN2-KPC, pGN26-KPC, pGN28-KPC, and pE20-KPC in the four strains, respectively. These plasmids were genetically closely related to one another and to the only previously sequenced IncX6 plasmid, pKPC3_SZ. Each of the plasmids carried a single accessory module containing the blaKPC-2/3-carrying ΔTn6296 derivatives. The ΔTn6292 element from pGN26-KPC also contained qnrS, which was absent from all other plasmids. Overall, pKPC3_SZ-like blaKPC-carrying IncX6 plasmids were detected by PCR in 44.4% of the KPC-producing isolates, which included K. aerogenes, P. mirabilis, S. marcescens, M. morganii, Escherichia coli, and Klebsiella pneumoniae, and were obtained from six different departments of the hospital. Data presented herein provided insights into the genomic diversity and evolution of IncX6 plasmids, as well as the dissemination and epidemiology of blaKPC-carrying IncX6 plasmids among Enterobacteriaceae in a hospital setting.
Introduction
Klebsiella pneumoniae carbapenamase (KPC), a class A β-lactamase, can hydrolyze almost all β-lactams, including carbapenems (Bush and Fisher, 2011). At least 31 variants (KPC-2 to KPC-32; KPC-1 is essentially identical to KPC-2) of the KPC enzyme have been identified to date1. Two Tn3-family unit transposons, Tn4401 and Tn6296, which are genetically divergent from each other, represent the two major prototype genetic platforms carrying blaKPC genes (Wang et al., 2015). Tn4401 and its derivatives are frequently identified in KPC-encoding plasmids of different incompatibility groups in bacterial isolates from European and American countries, but are rarely found in isolates from China (Feng et al., 2017). The blaKPC genetic environment in isolates from China is predominantly associated with Tn6296 and its derivatives (Wang et al., 2015).
Plasmids belonging to incompatibility group X (IncX) are 30–80 kb in size and were initially discovered in the pre-antibiotic era (Datta and Hughes, 1983). IncX plasmids have a narrow host range and are mainly circulated among Enterobacteriaceae species (Norman et al., 2008). The backbones of all IncX plasmids have a pir–parA–hns–hha–topB–pilX (tivB)–actX–taxC (rlx)–taxA (dtr) organization, but are quite divergent with respect to nucleotide and amino acid sequences similarity (Johnson et al., 2012). Comparative genomic analysis has shown that IncX plasmids can be phylogenetically grouped into seven major IncX subgroups, IncX1 to IncX6 (Du et al., 2016), along with another IncX6 subgroup (Bustamante and Iredell, 2017) that is re-designated herein IncX7.
blaKPC-2-harboring plasmids have been identified among the IncX3, IncX5, and IncX6 subgroups, including over a dozen plasmids belonging to subgroup IncX3 [e.g., pKpS90 (GenBank accession number JX461340) (Kassis-Chikhani et al., 2013) and pMNCRE44_5 (GenBank accession number CP010881) (Hargreaves et al., 2015)], two IncX5 plasmids [pKPC_CAV1492 (GenBank accession number CP011639) and pBK31567 (GenBank accession number JX193302) (Chen et al., 2013)], and a single IncX6 plasmid [pKPC3_SZ (GenBank accession number KU302800) (Du et al., 2016)]. Interestingly, blaKPC-2-harboring plasmids have not been found among the other IncX subgroups.
This study provides evidence for the dissemination of genetically highly similar KPC-2-encoding IncX6 plasmids among at least six Enterobacteriaceae species collected in a Chinese public hospital from 2014 to 2015. The complete nucleotide sequences of plasmids pGN2-KPC, pGN26-KPC, pGN28-KPC, and pE20-KPC, extracted from strains belonging to four representative species, were determined to be genetically closely related to the IncX6 reference plasmid pKPC3_SZ. In addition, all five plasmids carried a single accessory region that harbored the blaKPC-2/3 gene.
Materials and Methods
Bacterial Identification
Bacterial species identification was performed on the basis of 16S rRNA gene sequencing (Frank et al., 2008). The major plasmid-borne carbapenemase and extended-spectrum β-lactamase genes were screened for by PCR (Chen et al., 2014). All PCR amplicons were sequenced using an ABI 3730 Sequencer (Life Technologies, Carlsbad, CA, United States) using the primers used for PCR.
Plasmid Transfer
Plasmid conjugal transfer experiments were carried out using rifampin-resistant Escherichia coli strain EC600 (LacZ-, NalR, RifR) as the recipient and each of Proteus mirabilis GN2, Serratia marcescens GN26, Morganella morganii GN28, and Klebsiella aerogenes E20 as the donor (Feng et al., 2016). Aliquots (3 ml) of overnight cultures of each of the donor and recipient strains were mixed together, harvested, and resuspended in 80 μl of brain heart infusion broth (BD Biosciences, Franklin Lakes, NJ, United States). The mixture was spotted onto a 1-cm2 hydrophilic nylon membrane filter with a 0.45-μm pore size (Millipore, Billerica, MA, United States) placed onto the surface of a brain heart infusion agar (BD Biosciences, Franklin Lakes, NJ, United States) plate. Plates were incubated for mating at 37°C for 12–18 h. Bacteria were washed from the filter membrane and spotted on Mueller-Hinton agar (BD Biosciences, Franklin Lakes, NJ, United States) plates containing 1 mg/ml rifampin and 2 μg/ml imipenem to select the transconjugants containing the blaKPC marker.
Phenotypic Assays
Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test (Feng et al., 2016). Bacterial antimicrobial susceptibility was examined using the broth dilution method, and interpreted as per the Clinical and Laboratory Standards Institute guidelines (CLSI, 2015).
Genomic DNA Sequencing and Sequence Assembly
Genomic DNA was isolated from Enterobacteriaceae isolates GN2, GN26, GN28, and E20 using a Blood and Cell Culture DNA Maxi Kit (Qiagen, Hilden, Germany). Genome sequencing was performed for isolate GN2 using a sheared DNA library with an average size of 15 kb (ranging from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA, United States), as well as with a paired-end library with an average insert size of 400 bp (ranging from 150 to 600 kb) on a HiSeq sequencer (Illumina, San Diego, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads using proovread (Hackl et al., 2014), then the corrected PacBio reads were assembled de novo using SMARTdenovo2.
Genomic DNA from isolates GN26, GN28, and E20 was sequenced from a mate-pair libraries with an average insert size of 5 kb (ranging from 2 to 10 kb) using a MiSeq sequencer (Illumina, San Diego, CA, United States). DNA contigs that were not matched with the reference chromosome sequences of S. marcescens (GenBank accession number HG738868), M. morganii (GenBank accession number CP023505) or K. aerogenes (GenBank accession number FO203355) were assembled based on their contig coverage values using Newbler 2.6 (Nederbragt, 2014). Gaps between contigs were filled using a combination of PCR and Sanger sequencing using an ABI 3730 Sequencer.
Sequence Annotation and Genome Comparison
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Resistance genes, mobile elements, and other features were annotated using online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), and the Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.13.
Nucleotide Sequence Accession Numbers
The complete sequences of plasmids pE20-KPC, pGN2-KPC, pGN26-KPC, and pGN28-KPC and the draft sequences of the E20, GN2, GN26, and GN28 chromosomes were submitted to GenBank under accession numbers MF156709 to MF156712, CP026722, CP026581, CP026650, and CP026651, respectively.
Results
blaKPC-Carrying Isolates
From 2014 to 2015, a total of 143 carbapenem-resistant Alcaligenes xylosoxidans (n = 6), Acinetobacter baumannii (n = 24), Pseudomonas aeruginosa (n = 47), Pseudomonas putida (n = 5), K. aerogenes (n = 6), E. coli (n = 2), Enterobacter cloacae (n = 2), K. pneumoniae (n = 35), P. mirabilis (n = 8), S. marcescens (n = 5), and M. morganii (n = 3) isolates were obtained from the 143 different patients with various infections at a Chinese public hospital. Of these carbapenem-resistant isolates, 45 (31.5%) demonstrated class A carbapenemase activity and contained blaKPC genes, while three isolates (2.1%) had class B carbapenemase activity and carried blaNDM genes. Carbapenemase activity and major plasmid-borne carbapenemase genes were not detected in the remaining strains (66.4%). All of the carbapenemase-positive isolates were identified as Enterobacteriaceae.
These 45 blaKPC-carrying isolates consisted of K. pneumonia (n = 31), K. aerogenes (n = 6), S. marcescens (n = 5), and one isolate each of M. morganii, E. coli, and P. mirabilis. In total, 36 of the isolates were recovered from sputum specimens, while the remaining isolates were obtained from urine specimens. The 45 isolates came from 10 different hospital departments: 25 from the Intensive Care Unit, 6 from the Department of Gerontology, 5 from the Department of Respiratory Medicine, 2 from the Department of Neurology, 2 from the Department of Urology, and 1 each from the Department of General Surgery, the Department of Neurosurgery, the Emergency Department, the Department of Endocrinology, and the Department of Traditional Chinese Medicine (Supplementary Table S1). Thirty-six (80.0%) of the 45 isolates carried one or more β-lactamase genes [blaTEM, blaSHV, blaCTX-M-1G (Group), blaCTX-M-9G, blaOXA-1 and blaOXA-2] in addition to blaKPC.
pKPC3_SZ-Like IncX6 Plasmids From blaKPC-2-Carrying Isolates
Four blaKPC-positive isolates, P. mirabilis GN2, S. marcescens GN26, M. morganii GN28, and K. aerogenes E20, were arbitrarily selected for genome sequencing. GN2 was isolated from the urine specimens of an elderly female with urinary tract infection in 2014, while GN26 and GN28 (in 2015) and E20 (in 2014) were isolated from sputum specimens from three different elderly males suffering from pulmonary infections. These four patients were admitted to the hospital because of primary diseases consisting of myocardial infarction, cerebral infarction sequelae, cerebral contusion and pneumonia, respectively, and developed the above hospital-acquired infections during hospitalization.
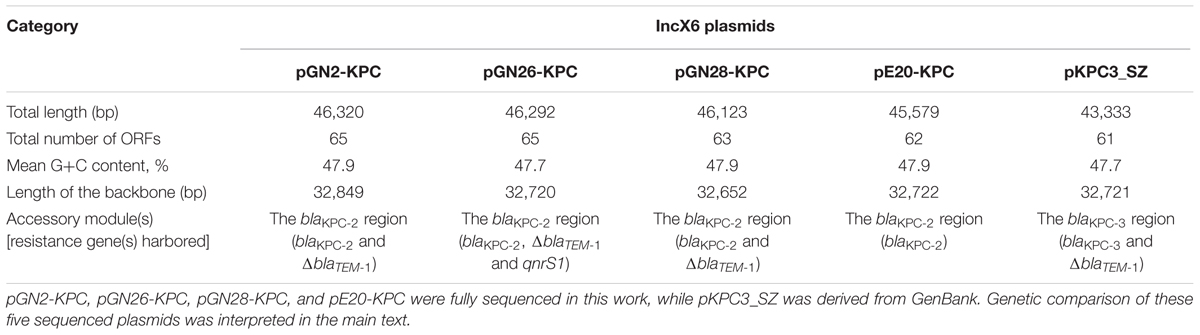
GN2, GN26, GN28, and E20 each contained an IncX6 plasmid, designated pGN2-KPC, pGN26-KPC, pGN28-KPC, and pE20-KPC, respectively. These plasmids were 45.6–46.3 kb in length, with 62–65 predicted ORFs (Table 1). The modular structure of each plasmid was divided into the backbone regions along with a single accessory module, which was defined as an acquired DNA region associated with mobile elements, and was inserted into the backbone (Figure 1 and Supplementary Figure S1). A total of three resistance genes were identified: blaKPC-2 was located in all four plasmids, while ΔblaTEM-1 was identified in pGN2-KPC, pGN26-KPC, and pGN28-KPC, and qnrS1 was found in pGN26-KPC. All these resistance genes were located in the accessory modules.
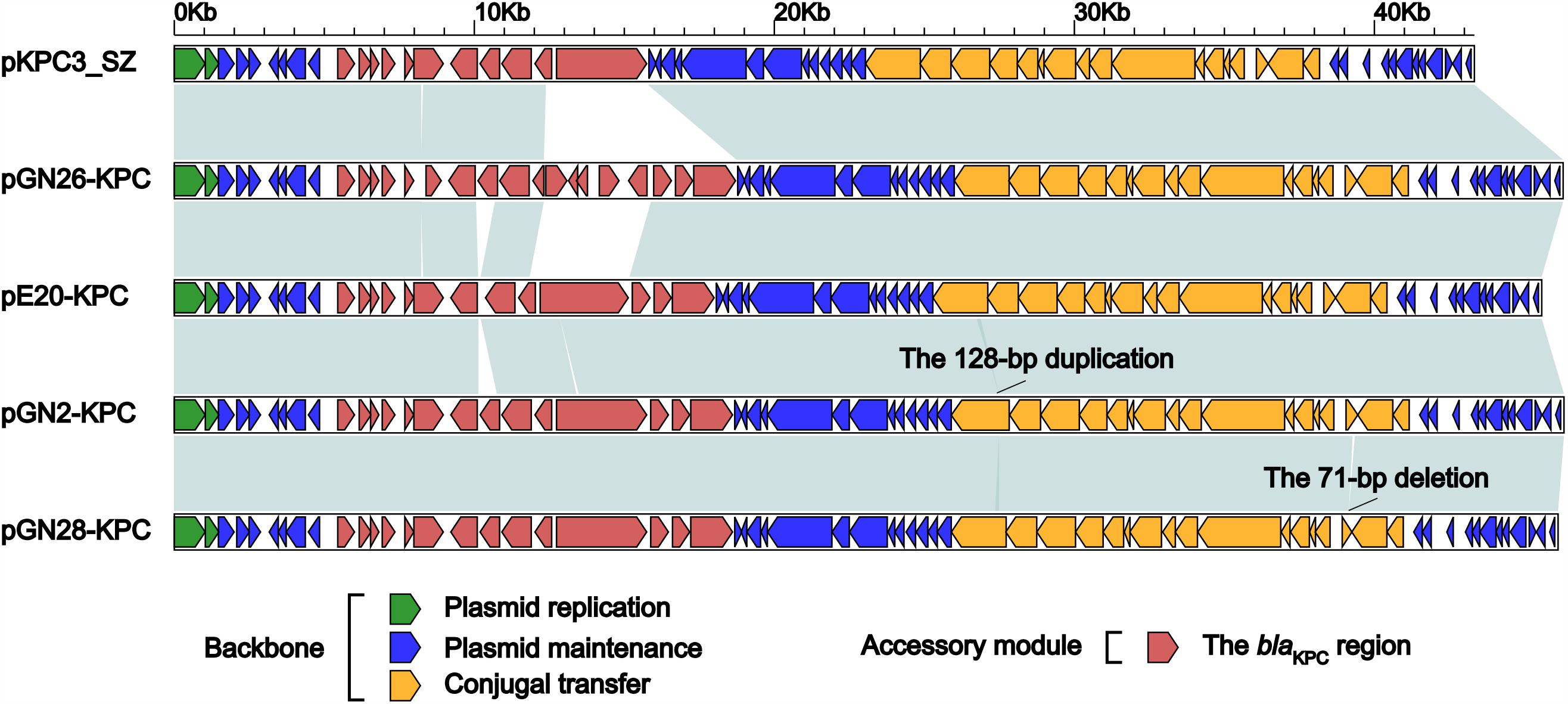
FIGURE 1. Linear comparison of IncX6 plasmids. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
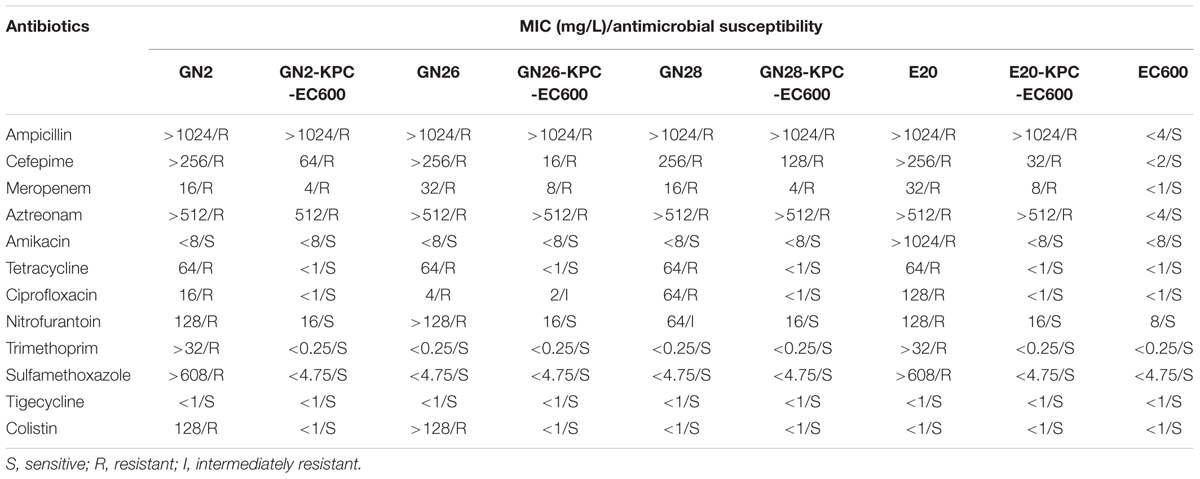
All four blaKPC-2-carrying plasmids could be transferred to E. coli EC600 via conjugation, generating the corresponding blaKPC-positive E. coli transconjugants GN2-KPC-EC600, GN26-KPC-EC600, GN28-KPC-EC600, and E20-KPC-EC600. Class A carbapenemase activity was detected for all transconjugants, and resulted from the production of the KPC-2 enzyme. Both the wild-type and transconjugant strains were resistant to ampicillin, cefepime, meropenem, and aztreonam. Moreover, GN26 was resistant to ciprofloxacin, but its transconjugant was intermediately resistant to this drug due to the presence of qnrS1 known to mediate the low-level resistance to fluoroquinolones (Table 2). In conclusion, each of GN2, GN26, GN28, and E20 harbored a conjugative blaKPC-carrying IncX6 plasmid, which accounted for the carbapenem resistance phenotype.
Based on the complete sequences of the five IncX6 plasmids (pGN2-KPC, pGN26-KPC, pGN28-KPC, pE20-KPC, and pKPC3_SZ), a total of nine genes were arbitrarily selected to screen for the prevalence of pKPC3_SZ-like IncX6 plasmids among the 45 blaKPC-positive isolates. Of these nine selected genes, eight [replication initiation: repA (replication initiation protein); maintenance: parA (partitioning ATPase), topB (type III topoisomerase), dnaJ (molecular chaperone), and ftsH (cell division protein); conjugal transfer: tivB3-4 (P-type type IV secretion, inner-membrane component of translocation channel and ATPase), tivB6 (P-type type IV secretion, inner-membrane component of translocation channel), and tivB10 (P-type type IV secretion, outer-membrane component of translocation channel)] were from backbone regions, while the remaining one was quinolone-resistance gene qnrS1. PCR analysis and amplicon sequencing showed that all eight backbone genes were present in 24 isolates, including 11 K. pneumoniae isolates, all 6 K. aerogenes isolates, 4 S. marcescens isolates, and 1 isolate each of M. morganii, E. coli, and P. mirabilis, indicating that these isolates harbored IncX6 plasmids. All five replication and maintenance genes, but none of the three conjugal transfer genes, were detected in another S. marcescens isolate, probably indicating that this isolate contained an IncX6 plasmid that had lost the conjugal transfer genes. None of the eight selected genes were detected in the remaining 20 isolates, signifying that these isolates did not carry IncX6 plasmids. qnrS1 was detected in 26 K. pneumoniae isolates, 6 K. aerogenes isolates, and 4 S. marcescens isolates, denoting coexistence of blaKPC and qnrS1 in these isolates.
Genomic Comparison of IncX6 Plasmids
pGN2-KPC, pGN26-KPC, pGN28-KPC, and pE20-KPC showed the highest sequence identity to the IncX6 reference plasmid pKPC3_SZ (Du et al., 2016), with >92% query coverage and >99% nucleotide identity. The major backbone genes or gene loci included repA and its iterons (replication initiation), parA and topB–hha–hns (maintenance), and rlx, dtr, tivB, cpl, and eex (conjugal transfer). repA coded for the IncX6-specific replication initiation protein and was not identified in any other available sequences. A 253-bp region containing seven imperfect GGTTTTTAAATCCCGata direct repeats was located 73-bp upstream of repA, and may function as iterons that bind the RepA protein. ParA was the partitioning ATPase responsible for plasmid segregation and stability (Schumacher, 2008), however, centromere-binding protein ParB and its binding sites parC could be not located. The gene expression modulation (gem) region, composed of topB, hha (transcriptional regulator), and hns (histone-like DNA-binding protein), was involved in plasmid maintenance (Norman et al., 2008). The conjugal transfer region was composed of a complete set of P-type conjugative DNA transfer genes, including rlx and dtr (DNA transfer; encoding relaxase Rlx and an auxiliary protein, Dtr), tivB1–tivB11 (encoding P-type type IV secretion system elaborating the pilus for mating pair formation), cpl (encoding a coupling protein that links DNA transfer and mating pair formation), and eex (entry exclusion preventing nucleoprotein transport between donors) (De La Cruz et al., 2010; Chen et al., 2013; Thomas et al., 2017).
The backbones of these five plasmids displayed only two major modular differences (Figure 1): (i) a 128-bp duplication in cpl of pGN2-KPC resulted in frameshift mutation, turning cpl into a pseudogene but retaining the conjugal transfer ability of pGN2-KPC, and (ii) a 71-bp deletion within orf393 (coding for an XRE-family Helix-turn-helix protein) was identified in pGN28-KPC, again causing the hypothetical gene orf393 to become a pseudogene.
The accessory modules of pGN2-KPC, pGN26-KPC, pGN28-KPC, pE20-KPC, and pKPC3_SZ were named the blaKPC regions (Figure 1), and were highly similar to one another (Figure 2). The blaKPC regions from pGN2-KPC, pGN28-KPC, and pE20-KPC comprised a ΔTn6296 derivative and an ISKpn19 element, while that from pGN26-KPC consisted of a ΔTn6296 derivative and an ISKpn19-containing ΔTn6292 derivative. The blaKPC region from pKPC3_SZ contained only a ΔTn6296 derivative (Figure 2).
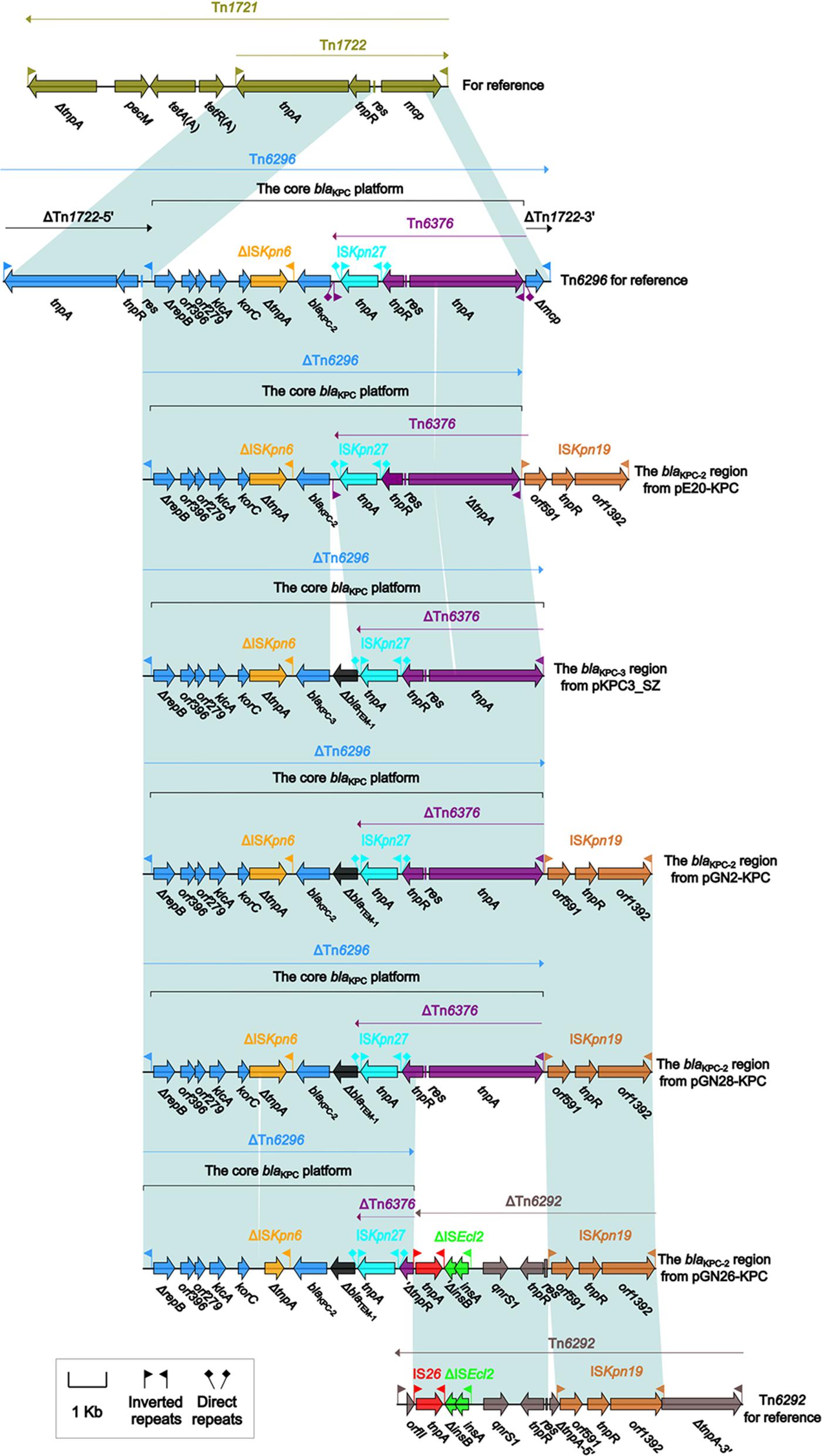
FIGURE 2. The blaKPC regions from IncX6 plasmids, and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
Tn6296 was originally identified in plasmid pKP048 from K. pneumoniae (Jiang et al., 2010). It was generated by the insertion of the core blaKPC-2 genetic platform (Tn6376–blaKPC-2–ΔISKpn6–korC–orf6–klcA–ΔrepB) into the mcp (methyl-accepting chemotaxis protein) gene of the cryptic transposon Tn1722, truncating mcp and splitting Tn1722 into ΔTn1722-5′ [IRL (inverted repeat left)–tnpAR–res] and ΔTn1722-3′ [Δmcp–IRR (inverted repeat right)]. The Tn3-family unit transposon Tn6292, as observed in pIMP-HZ1 from K. pneumoniae, carried a core quinolone resistance genetic platform, qnrS1-ΔISEcl2, and contained an ISKpn19 insertion within tnpA (Feng et al., 2016; Liang et al., 2017).
The ΔTn6296 derivatives from these five plasmids were slightly different from each other, with deletions and insertions relative to the prototype Tn6296 (Figure 2 and Supplementary Table S2). First, ΔTn1722-5′ was lost from all five ΔTn6296 elements. Second, a 70-bp deletion within tnpA of Tn6376 and a 70-bp deletion within ΔtnpA of ΔISKpn6 were found in pE20-KPC and pGN26-KPC, respectively, leading to frameshift mutations of these two coding regions. Third, the insertion of a 624-bp ΔblaTEM-1-containing region between ISKpn27 and blaKPC-2/3 was identified in pGN26-KPC, pGN2-KPC, pGN28-KPC, and pKPC3_SZ. Two promoters, consisting of the intrinsic P1 promoter and an upstream Tn6376-provided P2 promoter, were found to govern the blaKPC-2 expression of Tn6296 (Wang et al., 2015). The insertion of the ΔblaTEM-1-containing region resulted in the loss of IRLTn6376 and P1 (Supplementary Figure S2), leaving P2 as the only promoter for blaKPC expression. Finally, the 3′-terminal regions of these five ΔTn6296 derivatives were truncated in different formats: (i) ΔTn1722-3′ was absent in each of pE20-KPC, pGN2-KPC, and pGN28-KPC due to connection of ISKpn19; (ii) in pGN26-KPC, the qnrS1-containing ΔTn6292 element (6.4 kb in length) was connected with a 1.6-kb ΔTn6376 remnant with deletion of IRL–tnpA–res and truncation of tnpR, and the introduction of ΔTn6292 into pGN26-KPC probably resulted from homologous recombination between Tn6292 and a pre-existing ISKpn19 element (as observed in pE20-KPC, pGN2-KPC, and pGN28-KPC), with ISKpn19 acting as the common region necessary for recombination; and (iii) ΔTn1722-3’ was also lost in pKPC3_SZ, although none of the ISKpn19-related elements or any other regions were found to be adjacent to the 3′-end of ΔTn6296.
In summary, complex transposition and homologous recombination events, particularly those involving the three prototype mobile elements Tn6296, Tn6292, and ISKpn19, occurred to promote the assembly and mobilization of the blaKPC regions in these plasmids.
Discussion
IncX6 plasmid backbones have very limited sequence identity (<18% BLAST coverage and <84% nucleotide sequence identity) to those of other subgroups. Indeed, dramatic genetic diversities are presented among different IncX subgroups. Nevertheless, IncX6 plasmids contain the core IncX backbone makers responsible for plasmid replication initiation (repA and bis), maintenance (parA, hns–hha–topB, relEB, and dnaJ), and conjugal transfer (rlx, dtr, tivB, cpl, eex, and actX).
Previously sequenced IncX plasmids mostly belong to the IncX1–IncX4 subgroups, with very few representatives of IncX5–IncX7 plasmids. Currently, only five IncX6 plasmids have been fully sequenced, including pKPC3_SZ (Du et al., 2016) and the pGN2-KPC, pGN26-KPC, pGN28-KPC, and pE20-KPC plasmids sequenced in the current study. The five plasmids all originate from clinical isolates belonging to different Enterobacteriaceae species, namely E. cloacae, P. mirabilis, S. marcescens, M. morganii, and K. aerogenes, respectively, all of which come from China. Each of these five IncX6 plasmids contains a single accessory module containing two or three resistance genes, with all five carrying blaKPC-2/3, pKPC3_SZ, pGN2-KPC, and pGN28-KPC harboring ΔblaTEM-1, and pGN26-KPC containing both ΔblaTEM-1 and qnrS1. None of these IncX6 plasmids encodes multi-drug resistance. IncX6 plasmids appear to be an important vehicle for blaKPC genes in China, and the core genetic environments of blaKPC genes are close derivatives of Tn6296.
pGN2-KPC, pGN26-KPC, and pGN28-KPC were the only plasmids detected in their corresponding host strains. pKPC3_SZ coexists with pNDM1_SZ1, a multidrug resistance IncC plasmid that carries genes contributing to resistance to carbapenems (blaNDM-1), macrolides [mph(E)], aminoglycosides (strAB and addA2), and sulphonamides (sul1 and sul2) (Du et al., 2016). pE20-KPC coexists with four additional plasmids, including pE20-FIIA (GenBank accession number MG288681; IncFII), pE20-HI3 (GenBank accession number MG288682; IncHI3), pE20-NR (GenBank accession number MG288683), and pE20-qnrS (GenBank accession number MG288684) belonging to two unknown incompatibility groups [GenBank accessions not yet released]. Other than pE20-NR, the plasmids that co-exist with the IncX6 plasmid described in the current study all confer resistance to one or more antimicrobial agents, and mediate resistance to at least seven classes of antibiotics (β-lactams including carbapenems, quinolones, macrolides, aminoglycosides, amphenicols, sulphonamides, and trimethoprims). This severely limits the choice of antibiotics for treatment of infections caused by these bacterial strains.
Genomic and epidemiological analyses herein show that blaKPC-carrying IncX6 plasmids are present in 44.4% of the analyzed blaKPC-positive isolates from a single hospital and have disseminated among at least six different Enterobacteriaceae species from six distinct departments of this hospital, indicating wide spread of these plasmids in this hospital. Further studies are needed to determine the prevalence of IncX6 plasmids among various geographic areas to understand the contribution of IncX6 plasmids to blaKPC epidemiology among Enterobacteriaceae isolates.
Author Contributions
DZ and BoL: conception and design of the study. BiL, JF, DZ, BoL, ZZ, ZY, QJ, PW, XC, BG, JH, PM, WW, WC, YT and JW: acquisition of data. BiL, JF, DZ, and BoL: analysis and interpretation of data. BiL, JF, DZ, and BoL: drafting the article. BiL, JF, DZ, BoL, ZZ, ZY, QJ, PW, XC, BG, JH, PM, WW, WC, YT, and JW: critical revision. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program (2017YFC1200800) of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00478/full#supplementary-material
FIGURE S1 | Schematic maps of IncX6 plasmids. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and gray, respectively. The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
FIGURE S2 | Alignment of promoter-proximal regions of blaKPC-2. The 898–354 bp upstream sequences together with the start codon of the blaKPC-2 genes from IncX6 plasmids and Tn6296 are aligned by MUSCLE. Shown are core promoter regions, -35 and -10 elements, transcription starts, Shine-Dalgarno (SD) sequences for ribosome recognition and translation starts.
TABLE S1 | KPC-producing Enterobacteriaceae strains.
TABLE S2 | Major features of ΔTn6296 derivatives compared to Tn6296.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/pathogens/beta-lactamase-data-resources/
- ^ https://github.com/ruanjue/smartdenovo
- ^ https://inkscape.org/en/
References
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Bustamante, P., and Iredell, J. R. (2017). Carriage of type II toxin-antitoxin systems by the growing group of IncX plasmids. Plasmid 91, 19–27. doi: 10.1016/j.plasmid.2017.02.006
Chen, L., Chavda, K. D., Fraimow, H. S., Mediavilla, J. R., Melano, R. G., Jacobs, M. R., et al. (2013). Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57, 269–276. doi: 10.1128/AAC.01648-12
Chen, Z., Fang, H., Wang, L., Sun, F., Wang, Y., Yin, Z., et al. (2014). IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci. Rep. 4:7212. doi: 10.1038/srep07212
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. Wayne, PA: CLSI.
Datta, N., and Hughes, V. M. (1983). Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature 306, 616–617.
De La Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Du, H., Chen, L., Chavda, K. D., Pandey, R., Zhang, H., Xie, X., et al. (2016). Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 Carbapenemases. Antimicrob. Agents Chemother. 60, 2519–2523. doi: 10.1128/AAC.03053-15
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, J., Yin, Z., Zhao, Q., Zhao, Y., Zhang, D., Jiang, X., et al. (2017). Genomic characterization of novel IncFII-type multidrug resistant plasmids p0716-KPC and p12181-KPC from Klebsiella pneumoniae. Sci. Rep. 7:5830. doi: 10.1038/s41598-017-06283-z
Feng, W., Zhou, D., Wang, Q., Luo, W., Zhang, D., Sun, Q., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Hackl, T., Hedrich, R., Schultz, J., and Forster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Hargreaves, M. L., Shaw, K. M., Dobbins, G., Snippes Vagnone, P. M., Harper, J. E., Boxrud, D., et al. (2015). Clonal dissemination of Enterobacter cloacae harboring blaKPC-3 in the Upper Midwestern United States. Antimicrob. Agents Chemother. 59, 7723–7734. doi: 10.1128/AAC.01291-15
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, Y., Yu, D., Wei, Z., Ping, S., Zhou, Z., and Yu, Y. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54, 3967–3969. doi: 10.1128/AAC.00137-10
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kassis-Chikhani, N., Frangeul, L., Drieux, L., Sengelin, C., Jarlier, V., Brisse, S., et al. (2013). Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57, 618–620. doi: 10.1128/AAC.01712-12
Liang, Q., Yin, Z., Zhao, Y., Liang, L., Feng, J., Zhan, Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113.
Norman, A., Hansen, L. H., She, Q., and Sorensen, S. J. (2008). Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60, 59–74. doi: 10.1016/j.plasmid.2008.03.003
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Schumacher, M. A. (2008). Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412, 1–18. doi: 10.1042/BJ20080359
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Thomas, C. M., Thomson, N. R., Cerdeno-Tarraga, A. M., Brown, C. J., Top, E. M., and Frost, L. S. (2017). Annotation of plasmid genes. Plasmid 91, 61–67. doi: 10.1016/j.plasmid.2017.03.006
Wang, L., Fang, H., Feng, J., Yin, Z., Xie, X., Zhu, X., et al. (2015). Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 6:838. doi: 10.3389/fmicb.2015.00838
Keywords: plasmid, IncX6, genomics, epidemiology, blaKPC
Citation: Li B, Feng J, Zhan Z, Yin Z, Jiang Q, Wei P, Chen X, Gao B, Hou J, Mao P, Wu W, Chen W, Tong Y, Wang J, Li B and Zhou D (2018) Dissemination of KPC-2-Encoding IncX6 Plasmids Among Multiple Enterobacteriaceae Species in a Single Chinese Hospital. Front. Microbiol. 9:478. doi: 10.3389/fmicb.2018.00478
Received: 22 November 2017; Accepted: 28 February 2018;
Published: 19 March 2018.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Zhiyong Zong, West China Hospital, ChinaJason Sahl, Northern Arizona University, United States
Roberta Migliavacca, University of Pavia, Italy
Copyright © 2018 Li, Feng, Zhan, Yin, Jiang, Wei, Chen, Gao, Hou, Mao, Wu, Chen, Tong, Wang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhou, dongshengzhou1977@gmail.com Boan Li, lba@263.net
†These authors have contributed equally to this work.
 Bing Li1†
Bing Li1† Zhe Yin
Zhe Yin Yigang Tong
Yigang Tong Dongsheng Zhou
Dongsheng Zhou