- 1University of Chinese Academy of Sciences, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2State Key Laboratory of Applied Microbiology, Southern China and Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangdong Institute of Microbiology, Guangzhou, China
- 3Department of Food Science and Technology, Jinan University, Guangzhou, China
- 4College of Food Science, South China Agricultural University, Guangzhou, China
Bacillus cereus is a common and important food-borne pathogen that can be found in various food products. Due to low-temperature sterilization for a short period of time, pasteurization is not sufficient for complete elimination of B. cereus in milk, thereby cause severe economic loss and food safety problems. It is therefore of paramount importance to perform risk assessment of B. cereus in pasteurized milk. In this study, we isolated B. cereus from pasteurized milk samples in different regions of China, and evaluated the contamination situation, existence of virulence genes, antibiotic resistance profile and genetic polymorphism of B. cereus isolates. Intriguingly, 70 samples (27%) were found to be contaminated by B. cereus and the average contamination level was 111 MPN/g. The distribution of virulence genes was assessed toward 10 enterotoxigenic genes (hblA, hblC, hblD, nheA, nheB, nheC, cytK, entFM, bceT, and hlyII) and one emetic gene (cesB). Forty five percent strains harbored enterotoxigenic genes hblACD and 93% isolates contained nheABC gene cluster. The positive rate of cytK, entFM, bceT, hlyII, and cesB genes were 73, 96, 75, 54, and 5%, respectively. Antibiotic susceptibility assessment showed that most of the isolates were resistant to β-lactam antibiotics and rifampicin, but susceptible to other antibiotics such as ciprofloxacin, gentamicin and chloramphenicol. Total multidrug-resistant population was about 34%. In addition, B. cereus isolates in pasteurized milk showed a high genetic diversity. In conclusion, our findings provide the first reference on the prevalence, contamination level and characteristics of B. cereus isolated from pasteurized milk in China, suggesting a potential high risk of B. cereus to public health and dairy industry.
Introduction
The opportunistic pathogen Bacillus cereus is known to cause food-borne outbreaks in humans. B. cereus generally causes two types of gastrointestinal illness including emesis and diarrhea after consumption of a contaminated food, which contains more than 104–105 spores or vegetative cells of B. cereus per gram (Jensen et al., 2003; Bamnia and Kaul, 2015). The investigation during 1960 to 1992 had shown that food-borne outbreaks associated with B. cereus ranged from 1 to 22% in Europe, Japan, and North America (Griffiths and Schraft, 2002). B. cereus has emerged as the second food-borne pathogen in France after Staphylococcus aureus (Santé Publique France, 2015; Glasset et al., 2016), and has ranked third in China (Mao et al., 2010).
The pathogenicity of B. cereus is caused by different toxins produced by this bacterium. Diarrhea is associated with a series of enterotoxins including hemolysin BL (Hbl), non-hemolytic enterotoxin (Nhe), cytotoxin K (CytK) and enterotoxin FM (Fagerlund et al., 2004; Ehling-Schulz et al., 2006), as well as potential enterotoxins hemolysin II (HlyII) and enterotoxin T (BceT) (Agata et al., 1995a). Emetic syndrome is caused by the toxin cereulide which is synthesized by non-ribosomal peptide synthetases encoded by ces gene cluster (Ehling-Schulz et al., 2005, 2015). Unlike enterotoxins, cereulide is a thermo- and acidic-stable depsipeptide (Rajkovic et al., 2008) that is preformed in contaminated foods. Moreover, B. cereus involves in many serious and potentially fatal non-gastrointestinal-tract infections such as severe eye infections, osteomyelitis, hepatitis and inflammatory responses (Bottone, 2010; Rishi et al., 2013), and even death (Lund et al., 2000; Posfay-Barbe et al., 2008).
Antibiotic therapy is still the primary treatment method for the infections of B. cereus. However, emergence of antibiotic resistant B. cereus strains, mainly due to antibiotic misusage (Barbosa and Levy, 2000) or acquisition of resistance genes through horizontal gene transfer (Bogdanova et al., 1998; Agerso et al., 2002; Brown et al., 2003), results in the failure of antibiotic treatment. Thus, obtaining the B. cereus antibiotics resistance profile is highly relevant to public health.
Bacillus cereus has been found in milk with a high contamination rate (Rather et al., 2011; Gundogan and Avci, 2014; Chaves et al., 2017; Owusu-Kwarteng et al., 2017; Saleh-Lakha et al., 2017; Lan et al., 2018), especially pasteurized milk using low-temperature sterilizing process that cannot fully eliminate B. cereus spores (Zwietering et al., 1996). Consequently, it could lead to a variety of milk defects and foodborne diseases (Stenfors Arnesen et al., 2008). Besides, B. cereus in milk is probably associated with a higher risk of hazard. For example, thermophilic strains of B. cereus from milk had higher toxicity (Miller et al., 2016) and milk-originated B. cereus sensu lato strains harbored Bacillus anthracis-like plasmids, suggesting that B. cereus could gain more pathogenic genes through the interflow among Bacillus groups in milk (Bartoszewicz and Marjańska, 2017).
B. cereus contamination in dairy products have been reported (Gundogan and Avci, 2014; Yobouet et al., 2014; Drean et al., 2015; Chaves et al., 2017; Owusu-Kwarteng et al., 2017; Saleh-Lakha et al., 2017; Lan et al., 2018); however, the overall scale, especially in pasteurized milk, is quite small. Since pasteurization has a low inactivation rate of B. cereus spores and detection of B. cereus is not required for the dairy microorganism test of Chinese food security standard (The Hygiene Ministry of China, 2010b), it may increase the risk of B. cereus in dairy products. Thus, it is necessary to assess the prevalence and microbiological traits of B. cereus in pasteurized milk products. Here we used enterobacterial repetitive intergenic consensus sequences polymerase chain reaction (ERIC-PCR) to analyze genotypic diversity of B. cereus isolated from pasteurized milk products in China and combined with the pathogenic potential, antimicrobial resistance characters, aiming to provide an overview of the risk assessment for B. cereus isolated from pasteurized milk in China.
Materials and Methods
Sample Collection
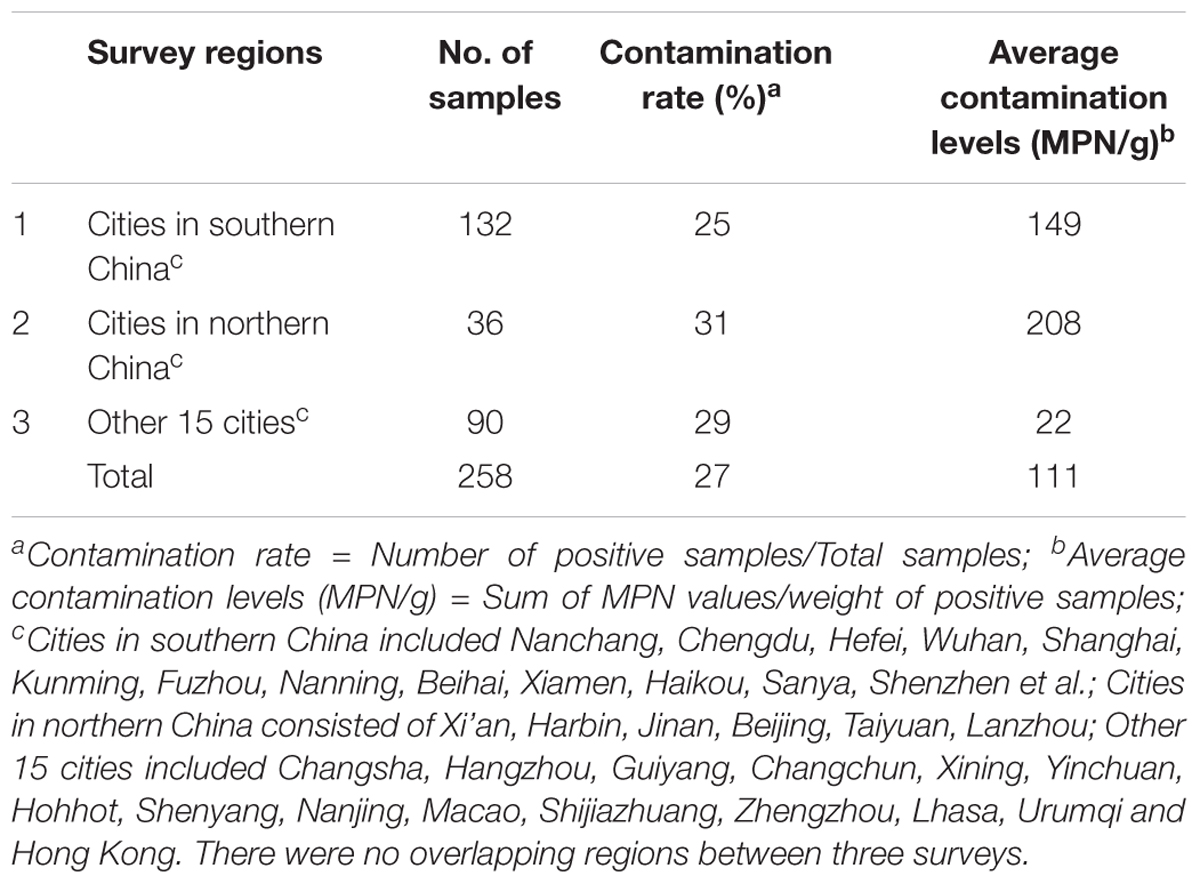
From July 2011 to May 2016, two hundred and seventy six pasteurized milk samples were collected from major cities in China (Supplementary Figure S1). The investigation process was divided into three stages. The first-stage investigation mainly focused on cites in southern china, and the second stage included six cities in northern China. The third one was a national wide survey that included other 15 cities. Details of sample distribution were shown in Table 1.
Qualitative and Quantitative Detection of B. cereus
The qualitative and quantitative detection of B. cereus were performed according to National Food Safety Standard (The Hygiene Ministry of China, 2010a) with minor modification. In brief, 25 ml of sample was mixed and homogenized with 225 ml Trypticase-soy-polymyxin (TSB) broth (Huankai, China) at 30°C for 48 h. Then cultures were streaked on the Mannitol-egg yolk-polymyxin (MYP) agar plate (Selective media; Huankai, China) and Chromogenic plate (Huankai, China) and incubated at 30°C for 24 h. Colonies with pink sparkle in blue or blue-green precipitation on Chromogenic plate were picked for further biochemical identification using the B. cereus biochemistry assessor (Huankai, China). The quantitative detection assay was conducted by B. cereus most probable number (MPN) counting method in Food Safety Standards (The Hygiene Ministry of China, 2010a).
Detection of Virulence Genes
The genomic DNA of various strains isolated from pasteurized milk samples was extracted using the HiPure Bacterial DNA Kit (Magene, United States) in accordance with the manufacturer’s instruction.
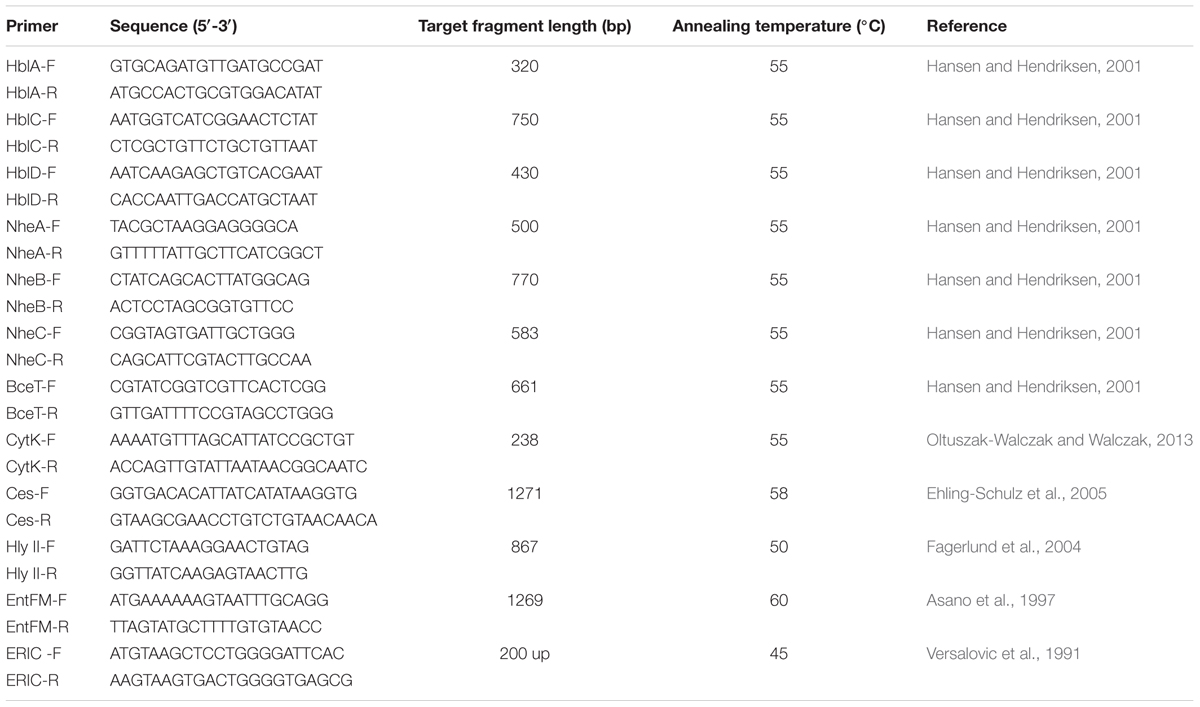
PCR screening was employed to detect the presence of seven enterotoxigenic genes (hblA, hblC, hblD, nheA, nheB, nheC, cytK), three potential virulence genes (bceT, entFM, hlyII) and one cereulide synthetase gene (cesB). The PCR reaction mixture (25 μl) consisted of 50 ng genomic DNA, 2 μM primers, and 12.5 μl PCR Premix TaqTM (Takara, China). The amplification was performed as described previously (Asano et al., 1997; Hansen and Hendriksen, 2001; Fagerlund et al., 2004; Ehling-Schulz et al., 2005; Oltuszak-Walczak and Walczak, 2013). The detailed sequence information and annealing temperature of these primer sets were shown in Table 2.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of all B. cereus isolates was evaluated by the Kirbye–Bauer disk diffusion method according to performance standards for antimicrobial susceptibility testing of the Clinical and Laboratory Standards Institute (The Clinical, and Laboratory Standards Institute [CLSI], 2010/2015) for Staphylococcus aureus. Twenty one antibiotics (Oxoid, United Kingdom) were tested, including ampicillin (AMP, 10 μg), amoxicillin-clavulanic acid (AMC, 20 μg/10 μg), penicillin (P, 10 U), trimethoprim-sulfamethoxazole (SXT, 1.25 μg/23.75 μg), cephalothin (KF, 30 μg), cefoxitin (FOX, 30 μg), cefotetan (CTT, 30 μg), imipenem (IPM, 10 μg), gentamicin (CN, 10 μg), kanamycin (K, 30 μg), erythromycin (E, 15 μg), telithromycin (TEL, 15 μg), vancomycin (VA, 30 μg), teicoplanin (TEC, 30 μg), ciprofloxacin (CIP, 5 μg), chloramphenicol (C, 30 μg), tetracycline (TE, 30 μg), clindamycin (DA, 2 μg), rifampin (RD, 5 μg), quinupristin (QD, 15 μg), and nitrofurantoin (FD, 300 μg). After incubating for 24 h at 37°C, the inhibition zones were measured and interpreted referring to the zone diameter interpretive criteria of S. aureus in Supplementary Table S1.
Genetic Biodiversity Assay
The ERIC-PCR method was performed for typing and comparing the pasteurized milk isolates. ERIC–PCR was carried out using the primers of ERIC-F and ERIC-R (Table 1) as described by Versalovic et al. (1991). The PCR reaction mixture (25 μl) contained 12.5 μl of ExTaq Mix (Takara, China), 1.0 μM of each primer, 50–100 ng genomic DNA. Amplification was performed as follows: an initial denaturation at 94°C for 3 min; 35 cycles each consists of 30 s at 94°C, 40 s at 45°C and 3 min at 72°C; and a final extension at 72°C for 10 min. Thereafter the amplicons (10 μl) were electrophoresed on 1.5% agarose gel. The DNA fingerprint was analyzed by BioNumerics 7.1 software (Applied Maths, Belgium). ERIC-PCR cluster analysis was assessed by Simpson’s diversity index (Hunter and Gaston, 1988).
Results
Prevalence Analysis of Bacillus cereus in Pasteurized Milk
Bacillus cereus contamination was found to occur in pasteurized milk samples from 32 out of 39 cities in China (Supplementary Figure S1). Of 258 milk samples evaluated, 70 (27%) samples contained B. cereus. Intriguingly, the overall contamination level of B. cereus is significantly higher (111 MPN/g), indicating that B. cereus risk in pasteurized milk is serious. According to the sample collection sites, the contamination of B. cereus was 31% (11/36) in northern China and 25% (33/132) in southern China (Table 1). Among different sample collection cities, contamination is quite serious in Guangzhou, Nanjing, Xining and Shenzhen. In contrast, no contamination was detected in Shantou, Sanya, Nanning, Shanghai, Nanchang, Xi’an, and Lhasa (Supplementary Figure S1).
Distribution of Virulence Genes Among B. cereus Isolates
The distribution of virulence genes was evaluated and summarized in Figure 1 and Supplementary Table S2. According to the pathogenic characteristics of B. cereus, the virulence genes are divided into two categories, namely enterotoxin genes (nheABC, hblACD, cytK, bceT, hly II, entFM) and cereulide synthetase genes (cesB). In enterotoxin genes, the gene cluster nheABC encoding the non-hemolytic enterotoxin (Nhe) complexes present in most of the strains (93%) with only a small portion of strains missing nheC or nheA. Forty five percent strains harbored enterotoxigenic genes hblACD. But the detection rate of hblA (46%) was significantly lower than hblC (66%) and hblD (67%). CytK, entFM and bceT were detected in more than 70% of the strains. In contrast, the detection rate of cereulide synthetase gene cesB was found to be only 5%, demonstrating that diarrheal strains in pasteurized milk samples are more common than emetic strains.
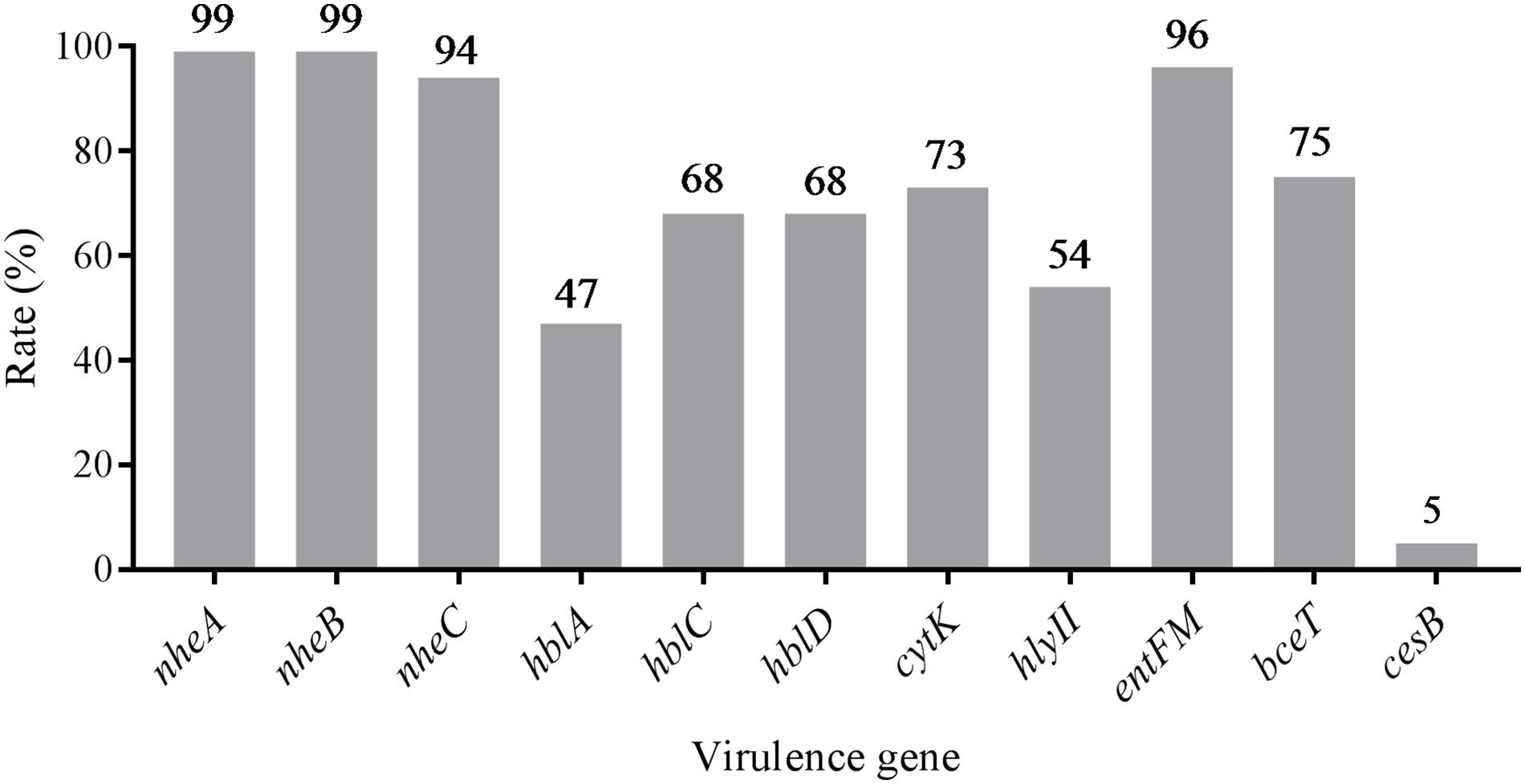
FIGURE 1. Distribution of virulence genes in Bacillus cereus isolated from pasteurized milk in China.
Based on the distribution of virulence genes, all isolates were divided into 32 virulence genes profiles. As shown in Figure 2, only two isolates (2833-2A-Bc in G2 and 3732-Bc in G5) contained all 11 virulence genes, and one isolate harbored the least virulence gene profile (2583-Bc in G3, nheA-nheB-entFM). The main gene profile was hblA-hblC-hblD-nheA-nheB-nheC-cytK-hlyII-entFM-bceT (28%), revealing that a large number of potentially diarrheal strains exist in the collected samples.
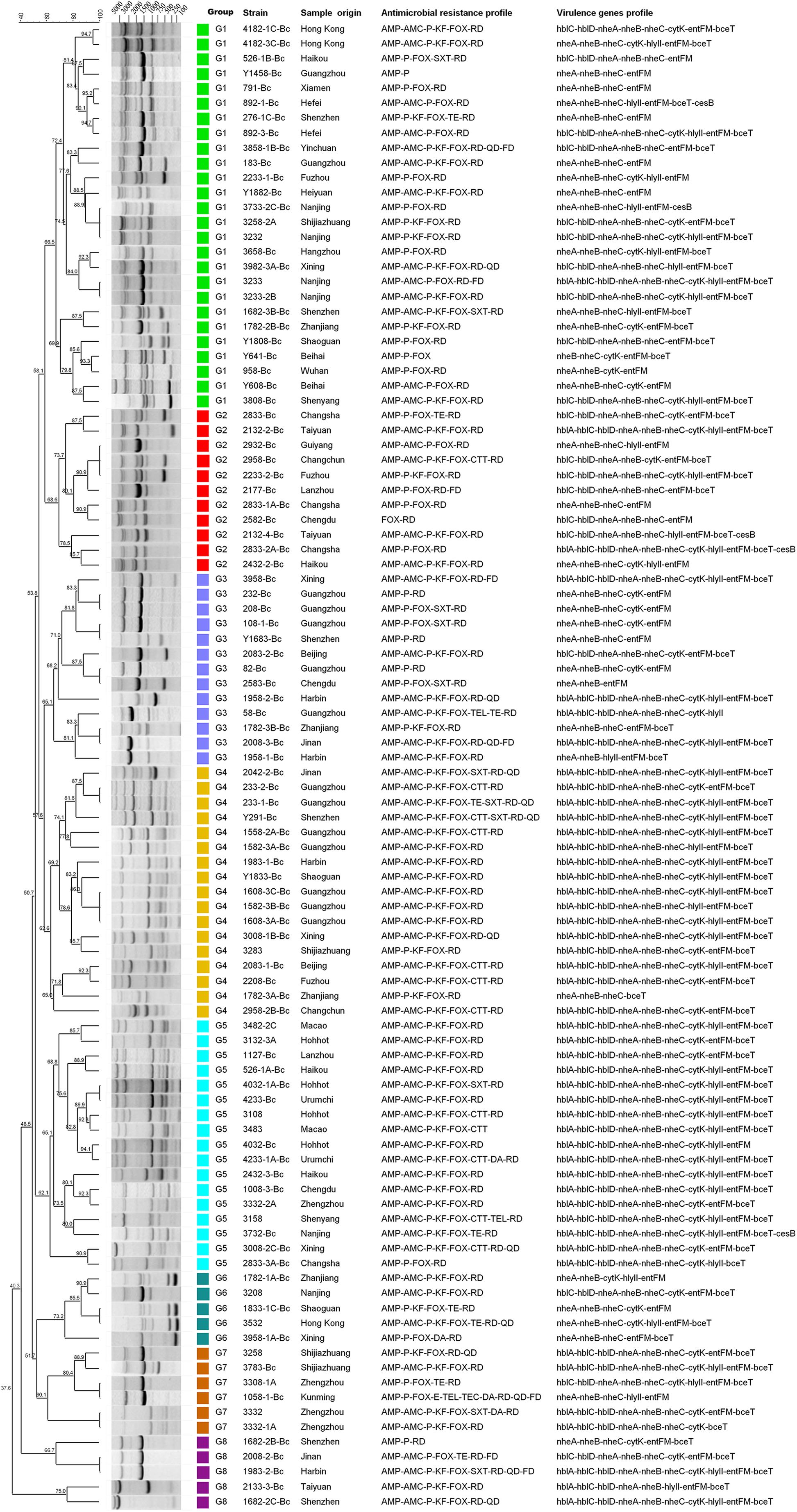
FIGURE 2. Dendrogram of Bacillus cereus isolated from pasteurized milk in China. Similarity (%) between fingerprints generated by ERIC-PCR was calculated by using the Dice index. The data were sorted by the UPGMA method. Different colors represent different groups, and the virulence genes profile and antimicrobial resistance characteristics of all isolates are listed in this dendrogram.
Antimicrobial Susceptibility Tests of B. cereus Isolates
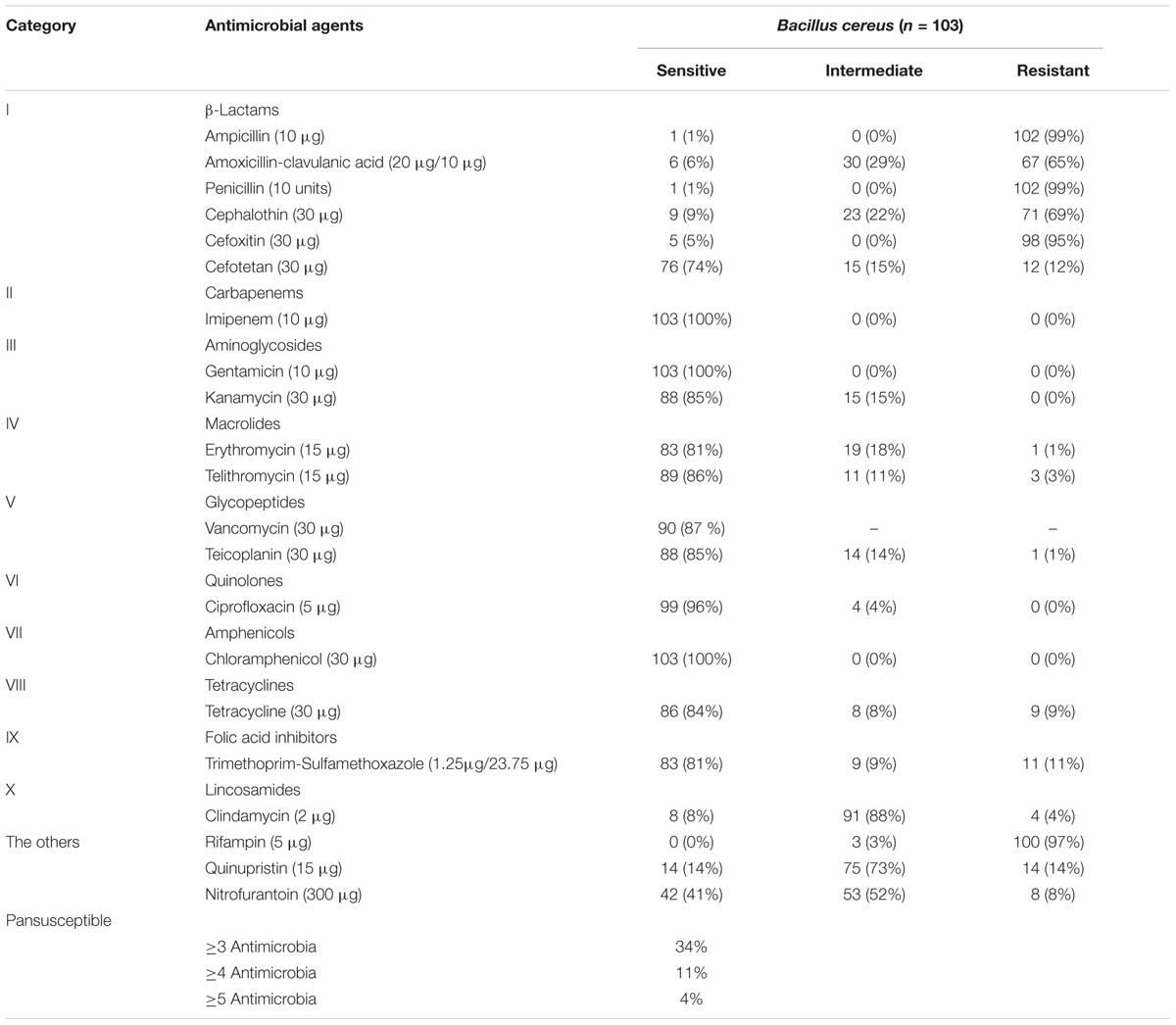
All B. cereus isolates were tested for antimicrobial susceptibilities to 21 selected antibiotics. As shown in Table 3 and Figure 3, most of the isolates were resistant to ampicillin (AMP; 99%), penicillin (P; 99%), cefoxitin (FOX; 95%), amoxicillin-clavulanic acid (AMC; 65%) and cephalothin (KF; 69%), which belong to β-lactams. Besides, rifampin (RD) had no effect on most strains (97%). Nearly all isolates were sensitive to the remaining antibiotics, such as imipenem (IPM; 100%), gentamicin (CN; 100%), kanamycin (K; 85%), telithromycin (TEL; 86%), teicoplanin (TEC; 85%), ciprofloxacin (CIP; 96%), chloramphenicol (C; 100%) et al. In addition, most of the isolates were moderately resistant to clindamycin (DA; 88%), quinupristin (QD; 73%) and nitrofurantoin (FD; 52%). On the other hand, we also found part of isolates (13%) was not sensitive to vancomycin (VA) according to the standard of The Clinical, and Laboratory Standards Institute [CLSI] (2010/2015) for Kirbye–Bauer disk diffusion method.
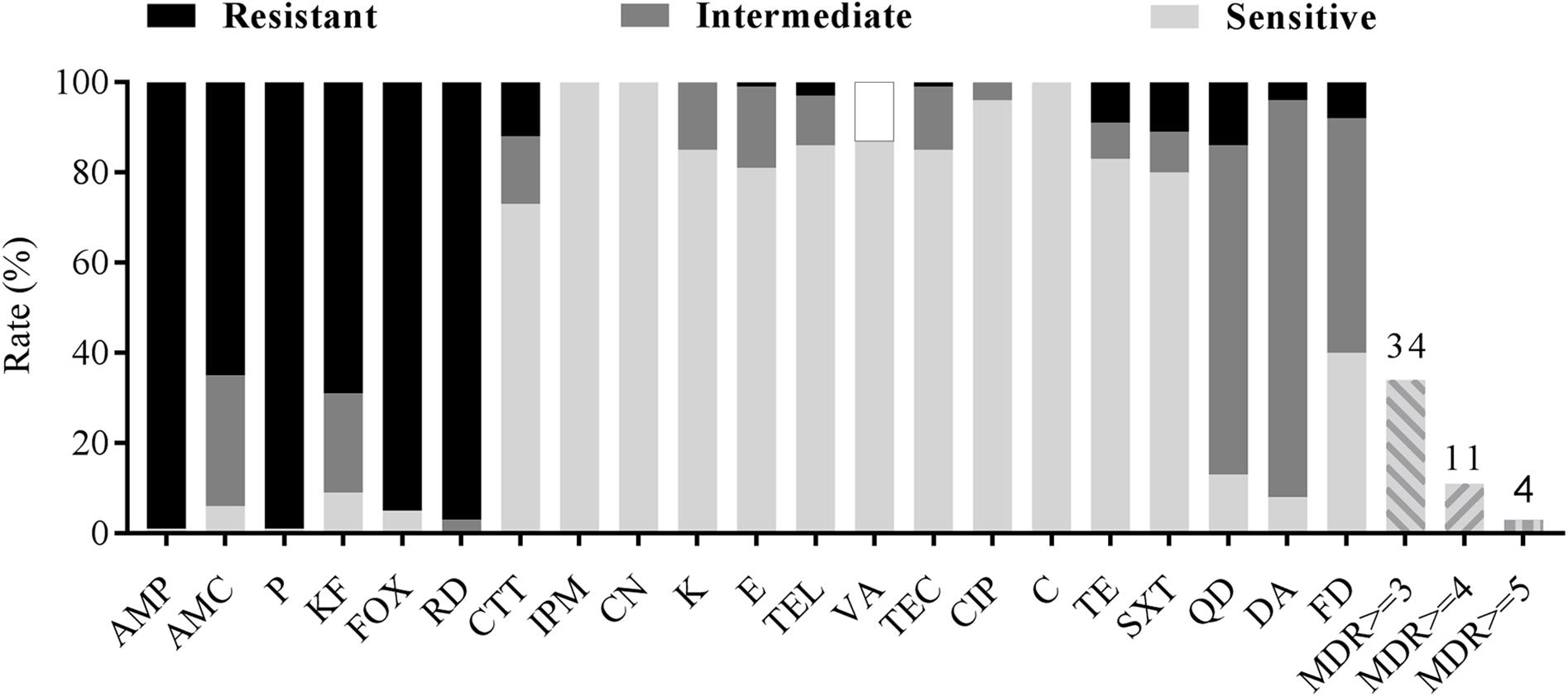
FIGURE 3. Antibiotic susceptibility of Bacillus cereus isolated from pasteurized milk in China. Blank bar, dark gray bar and light gray bar represent the proportion of resistant strains, moderately resistant strains and sensitive strains respectively. Gray stripe bars represent the proportion of multidrug resistance (MDR) strains. Vancomycin (VA) tested by Kirbye–Bauer disk diffusion method could only identify sensitive strains, so the black circle represents the percentage of remaining resistant and intermediate ones.
There were 34 antimicrobial resistance profiles for all isolates. As shown in Figure 2, the most susceptible strain resisted to only two antibiotics, while the most resistant strain resisted to ten antibiotics. AMP-AMC-P-KF-FOX-RD was the most common antimicrobial resistance profile. Multidrug resistance (MDR) profiles, defined as simultaneously resistant to more than three types of antibiotics (Magiorakos et al., 2012), were also evaluated. Thirty four percent of all isolates displayed resistance to three or more antibiotics, and 4% of the isolates displayed resistance to over five types of antibiotics (Table 3 and Figure 3).
ERIC-PCR Typing and Cluster Analysis
Molecular typing is a good way for tracing the sources and understanding the epidemiology of food-borne pathogens, such as random amplification of polymorphic DNA (RAPD, Nilsson et al., 1998), multi-locus sequence typing (MLST, Helgason et al., 2004), amplified fragment length polymorphism (AFLP, Hill et al., 2004), pulse-field gel electrophoresis (PFGE, Merzougui et al., 2014) and so on. Among these methods, ERIC-PCR (Shangkuan et al., 2000) is a simple and efficient approach to study the genetic diversity among strains that can explain the association of phenotypic and genotypic characters quite well. For our samples, we explored ERIC-PCR for genetic diversity analysis. The fingerprints gained by ERIC-PCR consisted of 2–11 distinct bands ranging in size from 250 base pairs to 5000 base pairs. Together with the distribution of virulence genes and antimicrobial susceptibility, the dendrogram was generated by using the software BioNumerics 7.0 (Figure 2).
All isolates showed 83 ERIC-PCR patterns. As shown in the Figure 2, when the relative similarity coefficient is 65%, 103 isolates could be divided into 8 clusters (G1 to G8). The G1 strains were dominant in our national isolates of which the fingerprints consisted of 6 to 8 bands and shared the common bands in sizes of about 1500 base pairs, 3000 base pairs and 4000 base pairs. As a discriminatory index, Simpson’s diversity is used to evaluate different typing methods (de la Puente-Redondo et al., 2000; Almeida et al., 2016; Wasfi et al., 2016), which produces values in the ranges of 0.0–1.0. The value 1.0 indicates that a typing method is able to distinguish each member of a population. Conversely, 0.0 indicates that all members of a population are of an identical type (Hunter and Gaston, 1988). If Simpson’s index (DI value) of a typing method is greater than 0.90, it is suggested that the typing method has generated a good result to distinguish all isolates. In our study, the DI value was 0.996, indicating the ERIC-PCR typing method could discriminate all the isolates well. Moreover, G1 (25.2%), G4 (16.5%), and G5 (16.5%) were the dominant groups. The fingerprints of G4 and G5 were quite similar, as well as their drug resistance spectra and virulence gene profiles. These two groups accounted for 41% of all strains, with hblA/C/D-nheA/B/C-cytK-hlyII-entFM-bceT as the main virulence gene type and AMP-AMC-P-KF-FOX-RD as the main drug resistance spectrum, indicating that the genetic diversity of these isolates were somehow conservative.
Discussion
The Prevalence and Genetic Diversity of B. cereus Isolates
Till now, the prevalence studies of pathogenic B. cereus in pasteurized milk in China are scant. In this study, B. cereus was detected in 27% of pasteurized milk samples collected from major cities in China, suggesting a potential risk of B. cereus. Compared to previous surveys in other countries, it is a medium contamination level compared with 27% in Abidjan (Yobouet et al., 2014), 27.37% in Brazil (Reis et al., 2013), 14% in Slovakia (Acai et al., 2014), 47% in Ghana (Owusu-Kwarteng et al., 2017) and 37% in India (Rather et al., 2011). Traced to the source of B. cereus contamination in pasteurized milk, heat stable B. cereus spores in raw milk and the post-pasteurization contamination along the milk processing lines were major sources (Lin et al., 1998; Saleh-Lakha et al., 2017). Besides, the environments for milk production, handling and processing could introduce B. cereus into dairy products (Cui et al., 2016a; Kumari and Sarkar, 2016). Some studies reported the storage temperature of dairy products also affected the number and toxicity of B. cereus (Fermanian et al., 1997; Notermans et al., 1997) as toxic strains could produce toxin even at 8°C. Together, these analyses imply the high prevalence of B. cereus and existence of potential hazards in consuming of the contaminated pasteurized milk.
The epidemiological typing is considered a crucial tool for studying the prevalence of food-borne bacteria. ERIC-PCR is based on the targeting of repeated DNA sequences with oligonucleotide primers, which has been broadly employed to perform the epidemiological typing of micro-organisms and widely applied into the risk survey of pathogenic bacteria such as Salmonella Typhimurium (Almeida et al., 2016), Staphylococcus aureus (Miao et al., 2016), Klebsiella pneumonia (Wasfi et al., 2016) and Arcobacter spp. (Vicente-Martins et al., 2018) as well as B. cereus strains (Hsueh et al., 1999; Shangkuan et al., 2000; Lopez and Alippi, 2007; Freitas et al., 2008; Avsar et al., 2017). In this study, we used ERIC-PCR to analyze the genetic and biological diversity of B. cereus isolates from pasteurized milk in China. The results showed that ERIC typing was suitable for studying the relationship between genetic and biological characteristics (Figure 2). The dominant genotype defined by ERIC fingerprints had the main virulence gene profile hblA/C/D-nheA/B/C-cytK-hlyII-entFM-bceT and drug resistance spectrum AMP-AMC-P-KF-FOX-RD, indicating the correlation between these different traits. Previous studies showed that fingerprinting patterns were used to distinguish different species such as AFLP typing in grouping B. anthracis, B. cereus, and B. thuringiensis isolates (Hill et al., 2004) and RAPD typing for discrimination of the B. cereus groups (Kuwana et al., 2012). However, there are very few studies available on fingerprint typing to correlate the genetic characteristics with biological traits of different strains. Here, in this study, we found that ERIC-PCR had great advantages in distinguishing and classifying strains with different biological characteristics.
Virulence Genes of B. cereus Isolates
Unlike emesis, diarrhea is associated with a series of enterotoxins produced by B. cereus in the small intestine, and the predominant protein responsible for this has not been yet determined other than enterotoxins (Jessberger et al., 2015). Nevertheless, pore-forming cytotoxins haemolysin BL (Hbl), non-haemolytic enterotoxin (Nhe) and cytotoxin K (CytK) have been identified as etiological agents of the diarrheal disease (Lund and Granum, 1996; Lund and Granum, 1999; Lund et al., 2000). In this study, 10 enterotoxigenic genes were detected and the proportion of nheABC, hblACD and cytK in B. cereus were found to be 93, 45, and 73%, respectively, which is quite similar to previous studies in France (nhe 96%, hbl 40%, cytk 42%, Glasset et al., 2016) or in China (nhe 100%, hbl 78.3%, Cui et al., 2016b), but were higher than an investigation of dairy products in Turkey (hbl 13%, nhe 60%, cytk 75%, Yibar et al., 2017), indicating that diarrheal strains have a wider distribution and a higher risk of B. cereus infections exists in consuming pasteurized milk in China.
Emetic symptom is triggered by the heat-stable dodecadepsipeptide cereulide, and a minimal emesis-causing dose was reported to be 8–10 μg/kg body weight in animal experiments (Agata et al., 1995b; Shinagawa et al., 1995). The emetic disease has often been connected with the consumption of fried and cooked rice (Agata et al., 2002; Kim et al., 2017), or pasta, pastry and noodles (Schoeni and Wong, 2005). In dairy products, strains with emetic toxin encoding genes were rare (Svensson et al., 2006; Saleh-Lakha et al., 2017). In our samples, the emetic strain was identified to be about 5%, which is not comparable to recent studies (10.2%, Biesta-Peters et al., 2016; 9%, Owusu-Kwarteng et al., 2017), but is much higher than others (1.0–3.8%, Svensson et al., 2006; 2%, Chaabouni et al., 2015; 1.1%, Cui et al., 2016a). According to previous study, strains isolated from dairy products present strong toxicity (7–15.3 folds higher than the reference emetic strain; Cui et al., 2016b). Congruently, earlier report showed that emetic strains could produce cereulide even at low temperature (Thorsen et al., 2006). So the emetic B. cereus is also a potential risk in pasteurized milk.
Antimicrobial Susceptibility of B. cereus
Bacillus cereus may cause severe diseases and infections that even lead to death (Lund et al., 2000; Dierick et al., 2005). Effective antibiotic therapy is considered as a predominant treatment to eliminate B. cereus infections, which has necessitated the investigation of antimicrobial susceptibility of B. cereus. In our study, B. cereus isolates were resistant to β-lactam antibiotics and rifampicin, but were susceptible to quinolones, aminoglycosides and macrolides. Consistent with previous studies, B. cereus were resistant to β-lactam antibiotics (Luna et al., 2007; Fernandes et al., 2014; Kim et al., 2015; Yibar et al., 2017) owing to the β-lactamase production (Bottone, 2010). It is worth mentioning that B. cereus was resistant to cephalosporin (cefoxitin, cephalothin) except for the third generation cephalosporin (cefotetan). Since broad-spectrum cephalosporin is the main antibiotic used in the treatment of gastrointestinal diseases caused by bacterial infections, it should be avoided as clinical treatment to gastroenteritis caused by B. cereus (Savic et al., 2016). Nowadays, vancomycin is considered as one of the most proper choice for B. cereus infections (Tatara et al., 2013; Torkar et al., 2016). However, a portion of our isolates (about 13%) detected were not sensitive to this antibiotic, suggesting the existence of potential risk for B. cereus infections. For multiple drug resistance, B. cereus isolates displayed resistance to three or more antibiotics were 34% which should be paid more attention to.
Conclusion
Despite the potential health risks associated with B. cereus, the prevalence and molecular analysis of genotypes of them have not been fully explored in pasteurized milk in China. In this study, we showed the high prevalence of B. cereus and its antibiotic resistance characteristics in pasteurized milk all over China. ERIC-PCR analysis demonstrated high genetic diversity of B. cereus in pasteurized milk samples. To the best of our knowledge, this is the first comprehensive investigation about the prevalence of B. cereus virulence factors, antibiotic resistance phenotypes and genotypes by ERIC typing in pasteurized milk from diverse regions of China that revealed a potential high risk of B. cereus to public health and dairy industry.
Author Contributions
QW, YD, JW, JZ, and TG conceived the project and designed the experiments. TG, SY, PY, CL, LK, ZF, MC, SW, HZ, and HW performed the experiments. QW and YD supervised the project. TG and YD analyzed the data and wrote the article. QW, JW, and YD complemented the writing.
Funding
The authors would like to acknowledge the financial support of National Natural Science Foundation of China (Nos. 31730070 and 31701195); Science and Technology Program of Guangdong Province (2015B020230006); Science and Technology Program of Guangzhou, China (201504010036 and 201604016068); State Key Laboratory of Applied Microbiology Southern China (SKLAM004-2016 and SKLAM006-2016); GDAS’ Special Project of Science and Technology Development (2017GDASCX-0201).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
YD is awarded the 1000-Youth Elite Program (The Recruitment Program of Global Experts in China). We thank Dr. Srinivasan Balamurugan for his comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00533/full#supplementary-material
FIGURE S1 | Map of China showing the cities where the Bacillus cereus isolates were collected.
TABLE S1 | Zone diameter interpretive criteria in this study.
TABLE S2 | Prevalence of virulence genes in Bacillus cereus isolated from pasteurized milk in China.
References
Acai, P., Valik, L., and Liptakova, D. (2014). Quantitative risk assessment of Bacillus cereus in pasteurised milk produced in the Slovak Republic. Czech J. Food Sci. 32, 122–131. doi: 10.17221/106/2013-CJFS
Agata, N., Ohta, M., Arakawa, Y., and Mori, M. (1995a). The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology 141, 983–988. doi: 10.1099/13500872-141-4-983
Agata, N., Ohta, M., Mori, M., and Isobe, M. (1995b). A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol. Lett. 129, 17–20. doi: 10.1016/0378-1097(95)00119-p
Agata, N., Ohta, M., and Yokoyama, K. (2002). Production of Bacillus cereus emetic toxin (cereulide) in various foods. Int. J. Food Microbiol. 73, 23–27. doi: 10.1016/S0168-1605(01)00692-4
Agerso, Y., Jensen, L. B., Givskov, M., and Roberts, M. C. (2002). The identification of a tetracycline resistance gene tet (M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 214, 251–256. doi: 10.1111/j.1574-6968.2002.tb11355.x
Almeida, F., Medeiros, M. I., Kich, J. D., and Falcao, J. P. (2016). Virulence-associated genes, antimicrobial resistance and molecular typing of Salmonella typhimurium strains isolated from swine from 2000 to 2012 in Brazil. J. Appl. Microbiol. 120, 1677–1690. doi: 10.1111/jam.13110
Asano, S. I., Nukumizu, Y., Bando, H., Iizuka, T., and Yamamoto, T. (1997). Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 63, 1054–1057.
Avsar, C., Civek, S., and Aras, E. S. (2017). Phenotypic and genotypic characterization of foodborne bacteria isolated from Sinop Province. Turkey. Food Biotechnol. 31, 141–161. doi: 10.1080/08905436.2017.1331450
Bamnia, M., and Kaul, G. (2015). Cereulide and diarrheal toxin contamination in milk and milk products: a systematic review. Toxin Rev. 34, 119–124. doi: 10.3109/15569543.2015.1063070
Barbosa, T. M., and Levy, S. B. (2000). The impact of antibiotic use on resistance development and persistence. Drug Resist. Updat. 3, 303–311. doi: 10.1054/drup.2000.0167
Bartoszewicz, M., and Marjańska, P. S. (2017). Milk-originated Bacillus cereus sensu lato strains harbouring Bacillus anthracis-like plasmids are genetically and phenotypically diverse. Food Microbiol. 67, 23–30. doi: 10.1016/j.fm.2017.05.009
Biesta-Peters, E. G., Dissel, S., Reij, M. W., Zwietering, M. H., and in’t Veld, P. H. (2016). Characterization and exposure assessment of emetic Bacillus cereus and cereulide production in food products on the Dutch market. J. Food Prot. 79, 230–238. doi: 10.4315/0362-028x.jfp-15-217
Bogdanova, E. S., Bass, I. A., Minakhin, L. S., Petrova, M. A., Mindlin, S. Z., Volodin, A. A., et al. (1998). Horizontal spread of mer operons among Gram-positive bacteria in natural environments. Microbiology 144, 609–620. doi: 10.1099/00221287-144-3-609
Bottone, E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. doi: 10.1128/cmr.00073-09
Brown, J. R., Gentry, D., Becker, J. A., Ingraham, K., Holmes, D. J., and Stanhope, M. J. (2003). Horizontal transfer of drug-resistant aminoacyl-transfer-RNA synthetases of anthrax and Gram-positive pathogens. EMBO Rep. 4, 692–698. doi: 10.1038/sj.embor.embor881
Chaabouni, I., Barkallah, I., Hamdi, C., Jouini, A., Saidi, M., Mahillon, J., et al. (2015). Metabolic capacities and toxigenic potential as key drivers of Bacillus cereus ubiquity and adaptation. Ann. Microbiol. 65, 975–983. doi: 10.1007/s13213-014-0941-9
Chaves, J. Q., de Papa, E. P., Rabinovitch, L., and Vivoni, A. M. (2017). Molecular characterization and risk assessment of Bacillus cereus sensu lato isolated from ultrahigh-temperature and pasteurized milk marketed in Rio de Janeiro. Brazil. J. Food Prot. 80, 1060–1065. doi: 10.4315/0362-028x.jfp-16-448
Cui, Y., Liu, X., Dietrich, R., Märtlbauer, E., Cao, J., Ding, S., et al. (2016a). Characterization of Bacillus cereus isolates from local dairy farms in China. FEMS Microbiol. Lett. 363:fnw096. doi: 10.1093/femsle/fnw096
Cui, Y., Liu, Y., Liu, X., Xia, X., Ding, S., and Zhu, K. (2016b). Evaluation of the toxicity and toxicokinetics of cereulide from an emetic Bacillus cereus strain of milk origin. Toxins 8, 156–166. doi: 10.3390/toxins8060156
de la Puente-Redondo, V. A., del Blanco, N. G., Gutierrez-Martin, C. B., Garcia-Pena, F. J., and Rodriguez Ferri, E. F. (2000). Comparison of different PCR approaches for typing of Francisella tularensis strains. J. Clin. Microbiol. 38, 1016–1022.
Dierick, K., Coillie, E., Van Swiecicka, I., Meyfroidt, G., Devlieger, H., Meulemans, A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43, 4277–4279. doi: 10.1128/jcm.43.8.4277-4279.2005
Drean, P., McAuley, C. M., Moore, S. C., Fegan, N., and Fox, E. M. (2015). Characterization of the spore-forming Bacillus cereus sensu lato group and Clostridium perfringens bacteria isolated from the Australian dairy farm environment. BMC Microbiol. 15:38. doi: 10.1186/s12866-015-0377-9
Ehling-Schulz, M., Frenzel, E., and Gohar, M. (2015). Food-bacteria interplay: pathometabolism of emetic Bacillus cereus. Front. Microbiol. 6:704. doi: 10.3389/fmicb.2015.00704
Ehling-Schulz, M., Guinebretiere, M. H., Monthan, A., Berge, O., Fricker, M., and Svensson, B. (2006). Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol. Lett. 260, 232–240. doi: 10.1111/j.1574-6968.2006.00320.x
Ehling-Schulz, M., Vukov, N., Schulz, A., Shaheen, R., Andersson, M., Martlbauer, E., et al. (2005). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113. doi: 10.1128/aem.71.1.105-113.2005
Fagerlund, A., Ween, O., Lund, T., Hardy, S. P., and Granum, P. E. (2004). Genetic and functional analysis of the cytK family of genes in Bacillus cereus. Microbiology 150, 2689–2697. doi: 10.1099/mic.0.26975-0
Fermanian, C., Lapeyre, C., Fremy, J. M., and Claisse, M. (1997). Diarrhoeal toxin production at low temperature by selected strains of Bacillus cereus. J. Dairy Res. 64, 551–559. doi: 10.1017/S0022029997002379
Fernandes, M. D. S., Fujimoto, G., Schneid, I., Kabuki, D. Y., and Kuaye, A. Y. (2014). Enterotoxigenic profile, antimicrobial susceptibility, and biofilm formation of Bacillus cereus isolated from ricotta processing. Int. Dairy J. 38, 16–23. doi: 10.1016/j.idairyj.2014.03.009
Freitas, D. B., Reis, M. P., Lima-Bittencourt, C. I., Costa, P. S., Assis, P. S., Chartone-Souza, E., et al. (2008). Genotypic and phenotypic diversity of Bacillus spp. isolated from steel plant waste. BMC Res. Notes 1:92. doi: 10.1186/1756-0500-1-92
Glasset, B., Herbin, S., Guillier, L., Cadel-Six, S., Vignaud, M. L., Grout, J., et al. (2016). Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterization. Euro Surveill. 21:30413. doi: 10.2807/1560-7917.es.2016.21.48.30413
Griffiths, M. W., and Schraft, H. (2002). “Bacillus cereus food poisoning,” in Foodborne Diseases, 2nd Edn, eds D. O. Cliver and H. P. Riemann (New York, NY: Academic Press), 261–270.
Gundogan, N., and Avci, E. (2014). Occurrence and antibiotic resistance of Escherichia coli, Staphylococcus aureus and Bacillus cereus in raw milk and dairy products in Turkey. Int. J. Dairy Technol. 67, 562–569. doi: 10.1111/1471-0307.12149
Hansen, B. M., and Hendriksen, N. B. (2001). Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67, 185–189. doi: 10.1128/aem.67.1.185-189.2001
Helgason, E., Tourasse, N. J., Meisal, R., Caugant, D. A., and Kolsto, A. B. (2004). Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70, 191–201. doi: 10.1128/aem.70.1.191-201.2004
Hill, K. K., Ticknor, L. O., Okinaka, R. T., Asay, M., Blair, H., Bliss, K. A., et al. (2004). Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70, 1068–1080. doi: 10.1128/aem.70.2.1068-1080.2004
Hsueh, P. R., Teng, L. J., Yang, P. C., Pan, H. L., Ho, S. W., and Luh, K. T. (1999). Nosocomial pseudoepidemic caused by Bacillus cereus traced to contaminated ethyl alcohol from a liquor factory. J. Clin. Microbiol. 37, 2280–2284.
Hunter, P. R., and Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 26, 2465–2466.
Jensen, G. B., Hansen, B. M., Eilenberg, J., and Mahillon, J. (2003). The hidden lifestyles of B. cereus and relatives. Environ. Microbiol. 5, 631–640. doi: 10.1046/j.1462-2920.2003.00461.x
Jessberger, N., Krey, V. M., Rademacher, C., Bohm, M. E., Mohr, A. K., Ehling-Schulz, M., et al. (2015). From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front. Microbiol. 6:560. doi: 10.3389/fmicb.2015.00560
Kim, C. W., Cho, S. H., Kang, S. H., Park, Y. B., Yoon, M. H., Lee, J. B., et al. (2015). Prevalence, genetic diversity, and antibiotic resistance of Bacillus cereus isolated from Korean fermented soybean products. J. Food Sci. 80, M123–M128. doi: 10.1111/1750-3841.12720
Kim, H. S., Choi, S. J., and Yoon, K. S. (2017). Efficacy evaluation of control measures on the reduction of Staphylococcus aureus in salad and Bacillus cereus in fried rice served at restaurants. Foodborne Pathog. Dis. doi: 10.1089/fpd.2017.2334 [Epub ahead of print].
Kumari, S., and Sarkar, P. K. (2016). Bacillus cereus hazard and control in industrial dairy processing environment. Food Control 69, 20–29. doi: 10.1016/j.foodcont.2016.04.012
Kuwana, R., Imamura, D., Takamatsu, H., and Watabe, K. (2012). Discrimination of the Bacillus cereus group members by pattern analysis of random amplified polymorphic DNA-PCR. Biocontrol Sci. 17, 83–86. doi: 10.4265/bio.17.83
Lan, X., Wang, J., Zheng, N., Zhao, S., Li, S., and Li, F. (2018). Prevalence and risk factors for Bacillus cereus in raw milk in Inner Mongolia, Northern China. Int. J. Dairy Technol. 71, 269–273. doi: 10.1111/1471-0307.12416
Lin, S., Schraft, H., Odumeru, J. A., and Griffiths, M. W. (1998). Identification of contamination sources of Bacillus cereus in pasteurized milk. Int. J. Food Microbiol. 43, 159–171. doi: 10.1016/S0168-1605(98)00105-6
Lopez, A. C., and Alippi, A. M. (2007). Phenotypic and genotypic diversity of Bacillus cereus isolates recovered from honey. Int. J. Food Microbiol. 117, 175–184. doi: 10.1016/j.ijfoodmicro.2007.03.007
Luna, V. A., King, D. S., Gulledge, J., Cannons, A. C., Amuso, P. T., and Cattani, J. (2007). Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 antimicrobials using Sensititre (R) automated microbroth dilution and Etest (R) agar gradient diffusion methods. J. Antimicrob. Chemother. 60, 555–567. doi: 10.1093/jac/dkm213
Lund, T., De Buyser, M. L., and Granum, P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38, 254–261. doi: 10.1046/j.1365-2958.2000.02147.x
Lund, T., and Granum, P. E. (1996). Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol. Lett. 141, 151–156. doi: 10.1016/0378-1097(96)00208-x
Lund, T., and Granum, P. E. (1999). The 105-kDa protein component of Bacillus cereus non-haemolytic enterotoxin (Nhe) is a metalloprotease with gelatinolytic and collagenolytic activity. FEMS Microbiol. Lett. 178, 355–361. doi: 10.1111/j.1574-6968.1999.tb08699.x
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug- resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mao, X., Hu, J., and Liu, X. (2010). Epidemiological analysis of 1060 bacterial foodborne diseases in China in 2003-2007. Chin. J. Food Hyg. 22, 224–228.
Merzougui, S., Lkhider, M., Grosset, N., Gautier, M., and Cohen, N. (2014). Prevalence, PFGE typing, and antibiotic resistance of Bacillus cereus group isolated from food in Morocco. Foodborne Pathog. Dis. 11, 145–149. doi: 10.1089/fpd.2013.1615
Miao, J., Peters, B. M., Li, L., Li, B., Zhao, X. H., Xu, Z. B., et al. (2016). Evaluation of ERIC-PCR for fingerprinting methicillin-resistant Staphylococcus aureus strains. Basic Clin. Pharmacol. Toxicol. 118(Suppl. 1), 3–117. doi: 10.1111/bcpt.12592
Miller, R. A., Beno, S. M., Kent, D. J., Carroll, L. M., Martin, N. H., Boor, K. J., et al. (2016). Bacillus wiedmannii sp nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int. J. Syst. Evol. Microbiol. 66, 4744–4753. doi: 10.1099/ijsem.0.001421
Nilsson, J., Svensson, B., Ekelund, K., and Christiansson, A. (1998). A RAPD-PCR method for large-scale typing of Bacillus cereus. Lett. Appl. Microbiol. 27, 168–172. doi: 10.1046/j.1472-765X.1998.00402.x
Notermans, S., Dufrenne, J., Teunis, P., Beumer, R., Giffel, M. T., and Weem, P. P. (1997). A risk assessment study of Bacillus cereus present in pasteurized milk. Food Microbiol. 14, 143–151. doi: 10.1006/fmic.1996.0076
Oltuszak-Walczak, E., and Walczak, P. (2013). PCR detection of cytK gene in Bacillus cereus group strains isolated from food samples. J. Microbiol. Methods 95, 295–301. doi: 10.1016/j.mimet.2013.09.012
Owusu-Kwarteng, J., Wuni, A., Akabanda, F., Tano-Debrah, K., and Jespersen, L. (2017). Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 17:65. doi: 10.1186/s12866-017-0975-9
Posfay-Barbe, K. M., Schrenzel, J., Frey, J., Studer, R., Korff, C., Belli, D. C., et al. (2008). Food poisoning as a cause of acute liver failure. Pediatr. Infect. Dis. J. 27, 846–847. doi: 10.1097/INF.0b013e318170f2ae
Rajkovic, A., Uyttendaele, M., Vermeulen, A., Andjelkovic, M., Fitz-James, I., In’t Veld, P., et al. (2008). Heat resistance of Bacillus cereus emetic toxin, cereulide. Lett. Appl. Microbiol. 46, 536–541. doi: 10.1111/j.1472-765X.2008.02350.x
Rather, M. A., Aulakh, R. S., Gill, J. P. S., Verma, R., and Rao, T. S. (2011). Enterotoxigenic profile of Bacillus cereus strains isolated from raw and pasteurized milk. Indian J. Anim. Res. 81, 448–452.
Reis, A. L., Montanhini, M. T., Bittencourt, J. V., Destro, M. T., and Bersot, L. S. (2013). Gene detection and toxin production evaluation of hemolysin BL of Bacillus cereus isolated from milk and dairy products marketed in Brazil. Braz. J. Microbiol. 44, 1195–1198. doi: 10.1590/S1517-83822013000400024
Rishi, E., Rishi, P., Sengupta, S., Jambulingam, M., Madhavan, H. N., Gopal, L., et al. (2013). Acute postoperative Bacillus cereus endophthalmitis mimicking toxic anterior segment syndrome. Ophthalmology 120, 181–185. doi: 10.1016/j.ophtha.2012.07.009
Saleh-Lakha, S., Leon-Velarde, C. G., Chen, S., Lee, S., Shannon, K., Fabri, M., et al. (2017). A study to assess the numbers and prevalence of Bacillus cereus and its toxins in pasteurized fluid milk. J. Food Prot. 80, 1085–1089. doi: 10.4315/0362-028x.jfp-16-521
Santé Publique France (2015). Data from: Data on Collective Food-Borne Outbreaks Reported in France in 2013. Available at: http://invs.Santepubliquefrance.fr/Dossiers-thematiques/Maladiesinfectieuses/Risquesinfectieux-d-originealimentaire/Toxi-infections-alimentaires-collectives/Donnees-epidemiologiques
Savic, D., Miljkovic-Selimovic, B., Lepsanovic, Z., Tambur, Z., Konstantinovic, S., Stankovic, N., et al. (2016). Antimicrobial susceptibility and beta-lactamase production in Bacillus cereus isolates from stool of patients, food and environment samples. Vojnosanit Pregl. 73, 904–909. doi: 10.2298/vsp150415134s
Schoeni, J. L., and Wong, A. C. (2005). Bacillus cereus food poisoning and its toxins. J. Food Prot. 68, 636–648. doi: 10.4315/0362-028X-68.3.636
Shangkuan, Y. H., Yang, J. F., Lin, H. C., and Shaio, M. F. (2000). Comparison of PCR-RFLP, ribotyping and ERIC-PCR for typing Bacillus anthracis and Bacillus cereus strains. J. Appl. Microbiol. 89, 452–462. doi: 10.1046/j.1365-2672.2000.01134.x
Shinagawa, K., Konuma, H., Sekita, H., and Sugii, S. (1995). Emesis of rhesus monkeys induced by intragastric administration with the HEp-2 vacuolation factor (cereulide) produced by Bacillus cereus. FEMS Microbiol. Lett. 130, 87–90. doi: 10.1016/0378-1097(95)00188-b
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Svensson, B., Monthan, A., Shaheen, R., Andersson, M. A., Salkinoja-Salonen, M., and Christiansson, A. (2006). Occurrence of emetic toxin producing Bacillus cereus in the dairy production chain. Int. Dairy J. 16, 740–749. doi: 10.1016/j.idairyj.2005.07.002
Tatara, R., Nagai, T., Suzuki, M., Oh, I., Fujiwara, S., Norizuki, M., et al. (2013). Sepsis and meningoencephalitis caused by Bacillus cereus in a patient with myelodysplastic syndrome. Intern. Med. 52, 1987–1990. doi: 10.2169/internalmedicine.52.0529
The Clinical, and Laboratory Standards Institute [CLSI] (2010/2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Approved Standard-M02-A12. Wayne, PA: The clinical and laboratory standards institute.
The Hygiene Ministry of China (2010a). National Food Safety Standard. Food Microbiological Examination: Bacillus cereus Test. Beijing: The Hygiene Ministry of China.
The Hygiene Ministry of China (2010b). National Food Safety Standard. Food Microbiological Examination: Milk and Milk Products. Beijing: The Hygiene Ministry of China.
Thorsen, L., Hansen, B. M., Nielsen, K. F., Hendriksen, N. B., Phipps, R. K., and Budde, B. B. (2006). Characterization of emetic Bacillus weihenstephanensis, a new cereulide-producing bacterium. Appl. Environ. Microbiol. 72, 5118–5121. doi: 10.1128/aem.00170-06
Torkar, K. G., Bedenic, B., and Plecko, V. (2016). Antimicrobial susceptibility and the in vitro postantibiotic effects of vancomycin and ciprofloxacin against Bacillus cereus isolates. J. Chemother. 28, 151–158. doi: 10.1179/1973947815y.0000000069
Versalovic, J., Koeuth, T., and Lupski, J. R. (1991). Distribution of repetitive DNA-sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Vicente-Martins, S., Oleastro, M., Domingues, F. C., and Ferreira, S. (2018). Arcobacter spp. at retail food from Portugal: prevalence, genotyping and antibiotics resistance. Food Control 85, 107–112. doi: 10.1016/j.foodcont.2017.09.024
Wasfi, R., Elkhatib, W. F., and Ashour, H. M. (2016). Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci. Rep. 6:38929. doi: 10.1038/srep38929
Yibar, A., Cetinkaya, F., Soyutemiz, E., and Yaman, G. (2017). Prevalence, enterotoxin production and antibiotic resistance of Bacillus cereus isolated from milk and cheese. Kafkas Univ. Vet. Fak. Derg. 23, 635–642. doi: 10.9775/kvfd.2017.17480
Yobouet, B. A., Kouame-Sina, S. M., Dadie, A., Makita, K., Grace, D., Dje, K. M., et al. (2014). Contamination of raw milk with Bacillus cereus from farm to retail in Abidjan, Cte d’Ivoire and possible health implications. Dairy Sci. Technol. 94, 51–60. doi: 10.1007/s13594-013-0140-7
Keywords: Bacillus cereus, pasteurized milk, risk assessment, virulence genes, antibiotic resistance, ERIC-PCR, genetic polymorphism, food-borne pathogen
Citation: Gao T, Ding Y, Wu Q, Wang J, Zhang J, Yu S, Yu P, Liu C, Kong L, Feng Z, Chen M, Wu S, Zeng H and Wu H (2018) Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus cereus Isolated From Pasteurized Milk in China. Front. Microbiol. 9:533. doi: 10.3389/fmicb.2018.00533
Received: 24 January 2018; Accepted: 08 March 2018;
Published: 26 March 2018.
Edited by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaReviewed by:
Jason Sahl, Northern Arizona University, United StatesKeping Ye, Nanjing Agricultural University, China
Yong Zhao, Shanghai Ocean University, China
Copyright © 2018 Gao, Ding, Wu, Wang, Zhang, Yu, Yu, Liu, Kong, Feng, Chen, Wu, Zeng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, wuqp203@163.com
†These authors have contributed equally to this work.
 Tiantian Gao
Tiantian Gao Yu Ding
Yu Ding Qingping Wu
Qingping Wu Juan Wang
Juan Wang Jumei Zhang
Jumei Zhang Shubo Yu
Shubo Yu Pengfei Yu
Pengfei Yu Chengcheng Liu
Chengcheng Liu Li Kong
Li Kong Zhao Feng
Zhao Feng Moutong Chen
Moutong Chen Shi Wu
Shi Wu Haiyan Zeng
Haiyan Zeng Haoming Wu
Haoming Wu