- 1Laboratory of Ruminant Nutrition, Institute of Dairy Science, College of Animal Sciences, Zhejiang University, Hangzhou, China
- 2Department of Animal Sciences, The Ohio State University, Columbus, OH, United States
The rumen and the hindgut represent two different fermentation organs in herbivorous mammals, with the former producing much more methane than the latter. The objective of this study was to elucidate the microbial underpinning of such differential methane outputs between these two digestive organs. Methane production was measured from 5 adult sheep and 15 adult rabbits, both of which were placed in open-circuit respiratory chambers and fed the same diet (alfalfa hay). The sheep produced more methane than the rabbits per unit of metabolic body weight, digestible neutral detergent fiber, and acid detergent fiber. pH in the sheep rumen was more than 1 unit higher than that in the rabbit cecum. The acetate to propionate ratio in the rabbit cecum was more than threefold greater than that in the sheep rumen. Comparative analysis of 16S rRNA gene amplicon libraries revealed distinct microbiota between the rumen of sheep and the cecum of rabbits. Hydrogen-producing fibrolytic bacteria, especially Butyrivibrio, Succiniclastium, Mogibacterium, Prevotella, and Christensenellaceae, were more predominant in the sheep rumen, whereas non-hydrogen producing fibrolytic bacteria, such as Bacteroides, were more predominant in the rabbit cecum. The rabbit cecum had a greater predominance of acetogens, such as those in the genus Blautia, order Clostridiales, and family Ruminococcaceae. The differences in the occurrence of hydrogen-metabolizing bacteria probably explain much of the differential methane outputs from the rumen and the cecum. Future research using metatranscriptomics and metabolomics shall help confirm this premise and understand the factors that shape the differential microbiota between the two digestive organs. Furthermore, our present study strongly suggests the presence of new fibrolytic bacteria in the rabbit cecum, which may explain the stronger fibrolytic activities therein.
Introduction
Mammalian herbivores do not synthesize the enzymes needed to digest cellulose or hemicellulose. They depend on a symbiotic relationship with a community of microbes (primarily bacteria) with fibrolytic ability in either their foregut (i.e., the rumen of ruminants and the pseudo-ruminants) or their hindgut (i.e., the cecum and colon of non-ruminant herbivores) for fiber digestion (Furness et al., 2015). Both foregut and hindgut fermenters produce methane (CH4) as an inevitable by-product during feed fermentation. As a greenhouse gas, CH4 is 23 times more potent than carbon dioxide (CO2) (IPCC, 2014). A significant portion of the ingested feed energy is also lost as CH4, ranging from 1.5 to 12% of the gross energy intake in cattle (Johnson and Johnson, 1995; Franz et al., 2010). Ruminants are the main producing animals of meat and milk, but they also produce more CH4 than monogastric animals per unit of BW0.75 or product (Franz et al., 2010). Indeed, up to 20% of the global anthropogenic CH4 is emitted by ruminants (Bhatta et al., 2007). Intensive research has aimed to mitigate CH4 emission to ensure sustainable production of beef, lamb, and dairy products.
Methanogenesis in the rumen and hindgut is predominately driven via the hydrogenotrophic pathway using hydrogen (H2) and CO2 (also formate) as the substrates (Liu and Whitman, 2008) though some CH4 is also produced through the methylotrophic methanogenesis pathway using methanol and methylamines as the substrates (Poulsen et al., 2013). The genus Methanobrevibacter is the most ubiquitous and predominant hydrogenotrophic methanogens found in the foregut and hindgut of herbivores (Kušar and Avguštin, 2010; St-Pierre and Wright, 2013) although several species of Methanomassiliicoccales can use methyl substrates (Poulsen et al., 2013). Although affected by several factors, such as pH, the rate and CH4 output from herbivores are primarily determined by the availability of methanogenic substrates (i.e., H2 and CO2), which are in turn determined by the rates of production and consumption. Fermentative acetate production accompanied with H2 production is thermodynamically favored, especially when forage-based diets are fed because more ATP is synthesized (Russell and Rychlik, 2001). CH4 output can vary among cows or sheep fed the same diet (Kittelmann et al., 2014; Wallace et al., 2015), and CH4 output was found to be positively associated with bacterial populations that ferment ingested feed to relatively more hydrogen in sheep (Kittelmann et al., 2014). Furthermore, hydrogenotrophic methanogens are thermodynamic favored than acetogens when competing for hydrogen in rumen (Cord-Ruwisch et al., 1988; Joblin, 1999), but in the foregut (tubular) of kangaroos, acetogens outcompete methanogens for CO2 and H2 and can synthesize acetate via the acetyl-CoA pathway, providing a significant energetic benefit to the host animal (Attwood and McSweeney, 2008). Similar hydrogen disposal pathway was thought to be present in the cecum of rabbits, but no acetogens were reported (Piattoni et al., 1996). In a recent study, species of Blautia including B. coccoides, B. hydrogenotrophica, and B. schinikii, which are known acetogens, were found at high predominance in rabbit cecum (Yang et al., 2016). We hypothesized that the hindgut of hindgut fermenters probably also has a distinct microbiota than the rumen of ruminants, and such difference may be the main reason for the differential CH4 outputs between these two types of herbivores. In the present study, we tested this hypothesis using sheep as ruminants and New Zealand White (NZW) rabbits as a non-ruminant herbivore, with alfalfa hay as the only diet. Feed consumption, fermentation characteristics, CH4 emission, and the microbiota in the sheep rumen and the rabbit cecum were comparatively analyzed. The differences determined in the above measurements will help understand the physiological and microbial underpinnings of differential CH4 production between ruminat and non-ruminant herbivores, and the knowledge on correlations between the microbiota and CH4 production might be useful for targeted intervention of rumen microbiota to mitigate CH4 production from ruminants.
Materials and Methods
Animals, Diets, and Experimental Design
All experiments involving animals and the animal use protocols were approved by the Animal Care Committee of Zhejiang University (Hangzhou, China). Five 1.5-years old healthy male sheep (63.91 ± 6.18 kg BW) each with a permanent ruminal cannula were each allocated to an open circuit respiration chamber, which was constructed using aluminum frames and resin sheets, allowing animals in neighboring chambers see each other. Temperature and humidity inside the chambers were respectively maintained at 25°C and 60%. Before gas determination, both the door and the food hopper of each chamber were kept open. Fifteen 1-year old healthy male NZW rabbits (3.14 ± 0.14 kg BW) were each housed in an indoor cage (60 × 50 × 35 cm in dimensions). Both the chambers and the cages were placed in a temperature-regulated room (24–26°C) with a natural light-dark cycle (approximately 13 h of light and 11 h of dark). Both the sheep and the rabbits were fed the same diet consisting of only alfalfa hay (18.5% CP, 46.0% NDF and 33.0% ADF) and had ad libitum access to fresh drinking water during the whole feeding experiment of 23 days for the sheep and 24 days for the rabbits. The feeding experiment consisted of 15 days for acclimation, 7 days for sample collection, and 1 day for gas measurement.
Measurement of Feed Intake and Digestibility
At the beginning of the experiment, the rabbits were blocked by BW (5 blocks, 3 rabbits per block), and each block was transferred to an indoor metabolism cage. The amount of feed offered and refused was recorded daily, and all feces were collected using fecal collection plates at 8:30 AM daily from individual sheep and blocks of rabbits during the 7 days of sample collection. Daily feed and orts samples were pooled by experimental unit (individual sheep and rabbit blocks). Fecal output was weighed, and 100 g wet feces were added to 10 mL 10% hydrochloric acid to preserve the samples for nitrogen analysis. All the feed and the fecal samples were dried in a forced-air oven at 65°C for 72 h, then ground through a 1 mm screen, and stored in sealed plastic containers at 4°C until analysis. Standard analysis methods (Official Methods of Analysis [AOAC], 2015) were used for analysis for dry matter (DM, method 930.15), CP (method 990.03), and ADF (method 973.18). NDF contents were analyzed following the procedure of Van Soest et al. (1991) without sodium sulfite and amylase added. The BW was weighted before measuring gas production at day 22 of the feeding experiment.
Determination of Gas Production
Gas production was determined using open circuit chambers (1.16 m3 interior volume each). Each chamber was completely airtight but received a continuous air flow at 8.0 m3 h-1. Total air flow was recorded using a flow meter (model number: SY-LWD-B-20; Shi Yi Automation Equipment Co., Ltd., Hangzhou, China), and concentration of CH4 and CO2 was determined using a gas detector (model number: Photoacoustic Gas Monitor INNOVA 1412; Innova AirTech Instruments A/S, Ballerup, Denmark). The alfalfa hay diet was provided to the animals twice daily at 09:00 and 16:00 using a food hopper that was reloaded outside of the chambers via a lid without opening the whole chamber. After the sample collection period, gas production from each sheep was directly determined for 24 h at day 23 of the feeding experiment. Each block of the rabbits was transferred to a chamber at day 23 of the feeding experiment for 24 h acclimation before continuous gas measurement for 24 h at day 24. CH4 production was expressed as CH4 yield per kg of BW0.75, DMI, digestible NDF intake, and digestible ADF intake, while CO2 production was expressed as CO2 yield per kg of BW0.75.
Collection of Ruminal and Cecal Samples
Ruminal content samples (about 50 mL each) were taken from individual sheep through their rumen cannula immediately after completing the gas determination. All the rabbits were sacrificed following euthanasia that was administered by a licensed animal technician following the procedures described by Yang et al. (2016). A quiet environment was provided to individual rabbit on a table with a slight angle to avoid stress and minimize pressure on the diaphragm. Rabbits were intravenous injected phenobarbital sodium (Sigma, Saint Louis, MO, United States) with a dose of 100 mg/kg BW. Once losing toe pinch and leg withdrawal reflex, each rabbit received ear intravenous injection of 20 mL of air. Then, cecal content samples were immediately collected (Yang et al., 2016). Briefly, each cecum was delineated into its proximal, middle, and distal segments, which were tied at the boundaries with a nylon string to prevent the cecal digesta from moving longitudinally. Each of the three cecal segments was cut separated, and its digesta content was squeezed into one 50-ml sterile Falcon tubes within 30 min of decease. One composite sample was prepared for each rabbit by combining about the same amount of digesta from each cecal segment. After immediate pH measurement using a pH meter (PB-10; Sartorius, Göettingen, Germany), the cecum samples were stored in liquid nitrogen and transported to the laboratory. Approximately 20 g of each rumen content and cecal content sample were freeze-dried for 30 h using a freeze-dryer (model number: BETA 1-8 LD; Martin Christ Gefriertrocknungsanlagen GmbH, Osterode, Germany). Each of the freeze-dried samples was crushed into fine particles manually and stored at -80°C until further analysis.
Measurement of Volatile Fatty Acid (VFA) Concentrations
An aliquot of each ruminal and cecal sample was subjected to analysis for VFAs as described by Yang et al. (2015). Briefly, approximately 2 g of each ruminal content sample and 1 g of each cecal content sample were added to 5 mL and 3 mL sterile phosphate buffered saline (PBS, pH 7.0), respectively, and mixed, and the mixture was centrifuged at 13,000 ×g at 4°C for 15 min. To 1 mL of each supernatant were added 20 μL of 85% orthophosphate acid and centrifuged again as described above to obtain the final supernatant. The VFAs concentration in the supernatant was determined using a gas chromatograph (model number: GC-2010; Shimadzu Corp., Kyoto, Japan) against external standards purchased from Aladdin (China, Shanghai).
Measurement of Microbial Enzyme Activity
The activities of CMCase, MCCase, xylanase, and pectinase of each sample was determined essentially as described previously (Wang et al., 2015), using carboxymethyl cellulose sodium (Sigma-Aldrich, Saint Louis, MO, United States), microcrystalline cellulose (Sigma-Aldrich), beechwood xylan (Sigma-Aldrich), and pectin from citrus peel (Fluka, St. Louis, MO, United States) as respective substrates, according to the dinitrosalicylic acid (DNS) method expounded by Bailey et al. (1992). Briefly, approximately 0.5 g of each freeze-dried ruminal content sample or cecal content sample was vortexed in 6 mL sterile PBS (pH 7.0), and the sample suspension was then sonicated (20 kHz, 195 W, 10 min) using a JY92-IIN Ultrasonic Cell Mixer (Ningbo Scientz, Ningbo, China) and centrifuged at 12,000 ×g at 4°C for 10 min. Then, 0.2 mL of the supernatant of each sample was mixed with 0.2 mL corresponding substrates (0.01 g mL-1 in phosphate buffer, pH 6.6) and then incubated at 39°C for 30 min. The enzyme activity was expressed as μmoL of glucose (for CMCase and MCCase), xylose (for xylanase), or D-galacturonic acid (for pectinase) released min-1 g-1 of the freeze-dried samples or their microbial crude protein (MCP).
Measurement of Microbial Crude Protein
Approximately 0.5 g of each freeze-dried ruminal content sample or cecal content sample was vortexed in 6 mL sterile PBS buffer solution (pH 7.0) to get the microbes in the gut fluid and most of the microbes adhering to the feed particles, and the suspension was then centrifuged at 408 ×g for 5 min to remove protozoa and remain feed particles. Then, 1 mL of each supernatant was centrifuged at 25,000 × g at 4°C for 20 min. The supernatants were discarded, and the pelleted microbial cells were suspended in 3 mL of 0.25 N sodium hydroxide and heated in boiling water for 10 min to lyse the microbial cells. The cell lysate samples were centrifuged at 25,000 ×g for 30 min, and the supernatants were subjected to protein assay with bovine serum albumin as the standard using the Coomassie brilliant blue (CBB) method (Makkar et al., 1982). The content of MCP was expressed as mg g-1 freeze-dried ruminal content or cecal content samples.
DNA Extraction and Real-Time PCR Quantification
Metagenomic DNA was extracted from 0.1 g each of freeze-dried ruminal content sample or cecal content sample using the CTAB (cetyltrimethylammonium bromide) method but with bead-beating (Gagen et al., 2010). The quality of the DNA extracts was evaluated using agarose (1%) electrophoresis, while the DNA concentration was determined using the Qubit dsDNA BR Assay Kit (Invitrogen Corporation, United States) on a Qubit 2.0 fluorometer (Invitrogen Corporation, United States). Standards for qPCR assay were prepared for individual groups of targeted microbes or targeted genes (the primers were listed in Supplementary Table S1) using cloning of PCR amplicons with a pGEM®T Easy kit (Promega, Shanghai, China) following the method of Koike et al. (2007). The abundance of each species or group of microbes was quantified using real-time PCR as described previously (Liu et al., 2014) and expressed as log10 copies of 16S rRNA gene (or 18S rRNA gene in the case of protozoa, and ITS1 in the case of fungi) per g of freeze-dried ruminal content or cecal content samples.
Analysis of Microbiota
One amplicon library each was separately prepared for archaea and bacteria from each of the metagenomic DNA samples using the primers M86F/M448R and 515F/806R, respectively. All amplicons were pooled in equal molar ratio and sequenced using the 2 × 250 bp paired-end protocol on an Illumina MiSeq system. The raw sequences were de-multiplexed, quality-filtered, and analyzed using QIIME (v 1.9.0) (Caporaso et al., 2010). Briefly, bases from each sequencing read with a Q score less than 25 were trimmed off, then the paired reads (R1 and R2) were merged to form single sequences using the fastq-join script (Aronesty, 2011). Sequences shorter than 352 bp for archaea and 281 bp for bacteria were discarded, and the primers were further trimmed off. Chimera checking was performed using the ChimeraSlayer algorithm (Haas et al., 2011). The quality-checked sequences were clustered into species-equivalent operational taxonomic units (OTUs) by comparison to the Greengenes database 13.5 (DeSantis et al., 2006) using the open-reference OTU picking option (pick_open_reference_otus.py). The OTUs were taxonomically classified by comparison to the Greengenes database 13.5. Minor OTUs were filtered out if they were each represented by less than 0.005% of the total sequences (Bokulich et al., 2013) or appeared in less than 60% of each experimental animal species. The sequences of each sample were rarefied to the same number (46,609 sequences/sample for archaea and 19,170 sequences/sample for bacteria) before diversity analysis. Alpha diversity measurements including Chao1 richness estimate, Shannon diversity index, and observed number of OTUs were calculated for each sample. The microbiota were compare as beta diversity using the distance matrices generated from weighted UniFrac analysis (Lozupone and Knight, 2005) and principal coordinates analysis (PCoA). The raw sequence data were deposited in the Sequence Read Archive of NCBI under accession no. SRP108266.
Statistical Analysis
Statistical analysis of the data was performed using one-way ANOVA, with means separation using t-test at the level of significance of 0.05 using the SAS software package (SAS Institute Inc., 2000). Pearson correlation coefficients were calculated to examine the correlation between animal performances and relative abundance of microbial groups. The data were expressed as Mean ± SD in the Tables.
Results
Feed Digestibility and Gas Yields
The two species of animals used in this study consumed a similar amount of feed (DM) per unit of BW0.75 daily (Table 1). However, the sheep had a higher apparent digestibility of DM, NDF, and ADF than the rabbits (P < 0.05). Each of the sheep emitted substantially more CH4 than each rabbit per day per unit of BW0.75, DMI, digestible NDF, or digestible ADF (Figures 1A–D). Per unit of BW0.75, however, the sheep emitted less CO2, resulting in a 6.4 times higher CH4 to CO2 ratio than the rabbits (Figures 1E,F).
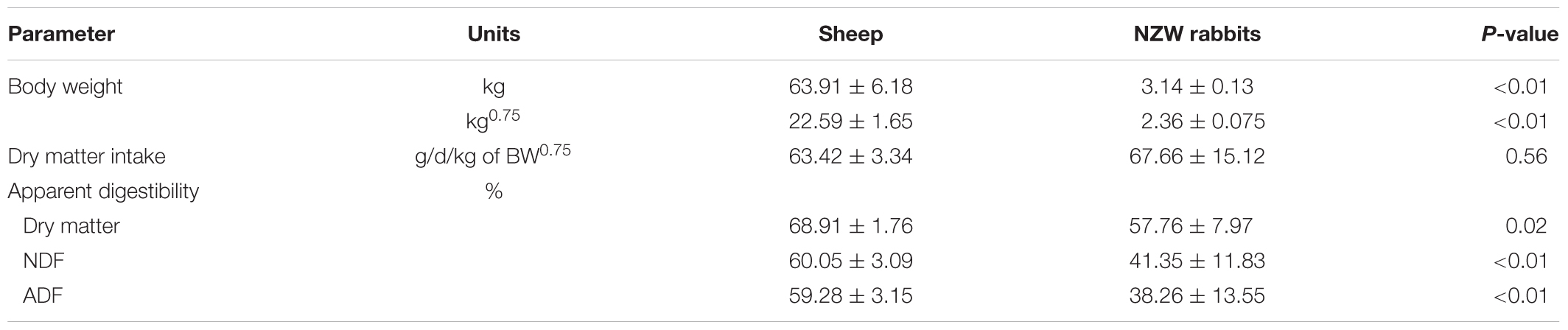
TABLE 1. Body weight, feed dry matter intake, and digestibility of the sheep (n = 5) and the rabbits (n = 15).
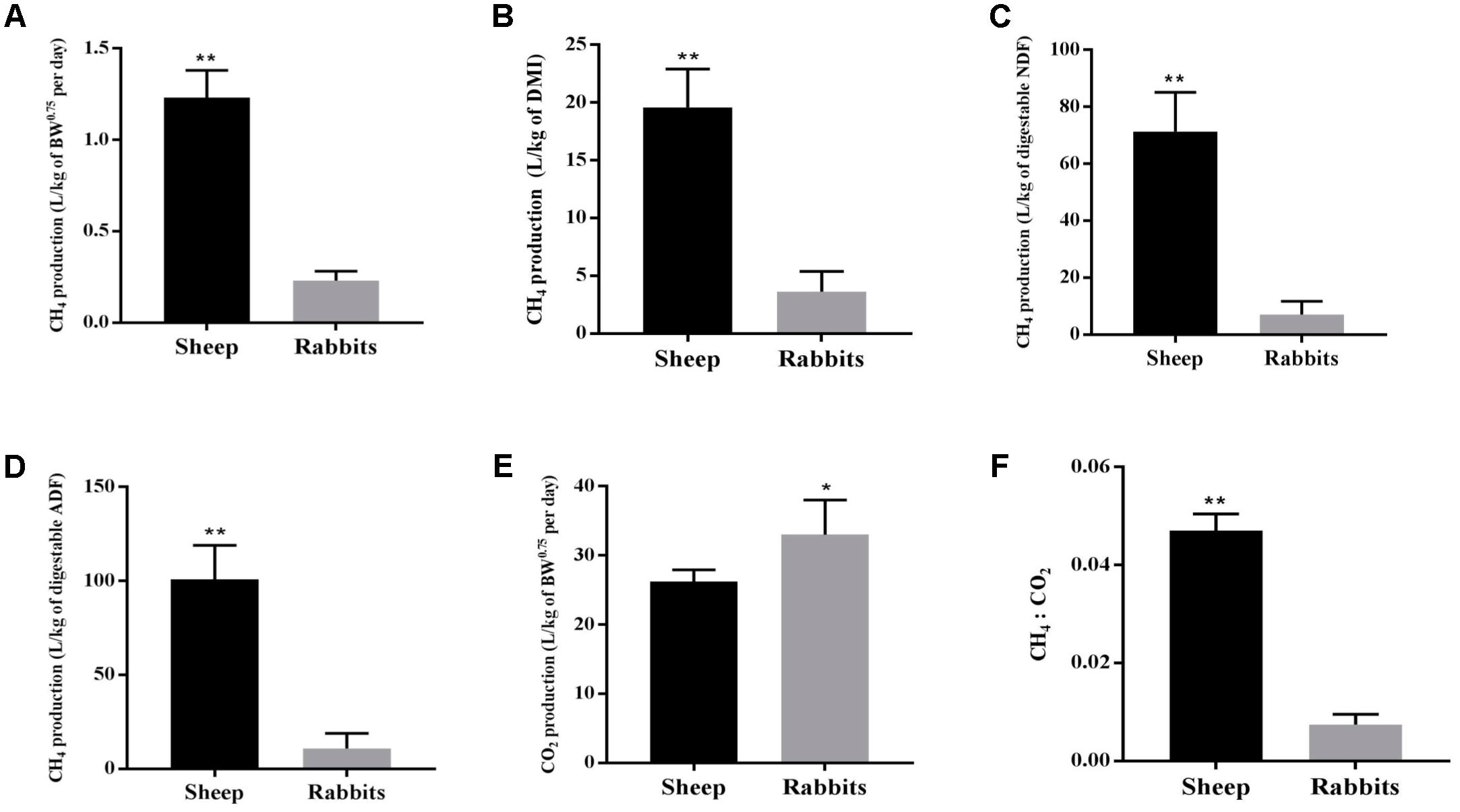
FIGURE 1. Yields of CH4 and CO2 and their ratio of the sheep (n = 5) and the rabbits (n = 15). CH4 yields per unit of metabolic body weight (A), dry matter intake (B), digestible NDF (C), and digestible ADF (D); CO2 yields per unit of metabolic body weight (E) and CH4 : CO2 ratio (F) of the sheep and the rabbits. ∗∗P < 0.01; ∗P < 0.05.
Fermentation Characteristics
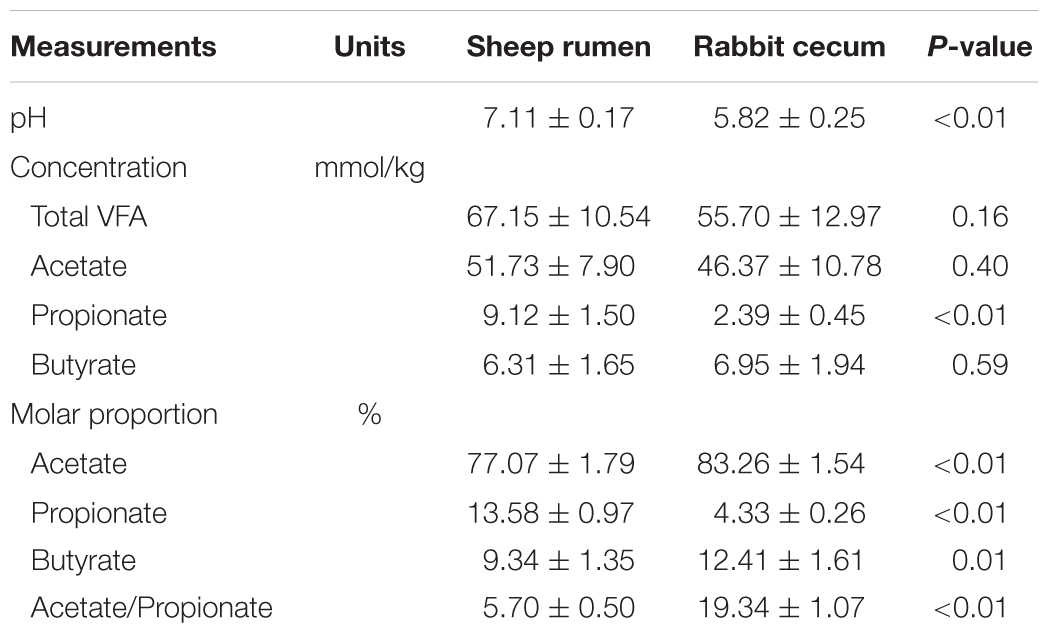
The main fermentation characteristics, including pH, concentrations of VFA, and their molar proportions, in the sheep rumen and the rabbit cecum are presented in Table 2. The pH in the sheep rumen was more than 1 unit higher (P < 0.01) than that in the rabbit cecum. No significant difference (P = 0.16) in total VFA concentration was observed between the two digestive organs. However, a much lower propionate concentration was seen in the rabbit cecum than in the sheep rumen. The two digestive organs also differed (P ≤ 0.01) in molar proportions of VFA, with the rabbit cecum having a higher value for acetate and butyrate but a lower value for propionate. The acetate to propionate (A: P) ratio in the rabbit cecum was more than threefold greater than that in the sheep rumen.
Microbial Crude Protein Yields and Enzymes Activity
The rabbit cecal content had a higher (P < 0.01) concentration of MCP than the sheep rumen content (Table 3). A higher (P < 0.01) activity of CMCase, MCCase, and pectinase was observed in the rabbit cecum than in the sheep rumen either per g content or mg MCP. Xylanase activity was similar between the two digestive organs (Table 3).

TABLE 3. Microbial crude protein (MCP) and enzyme activities in the sheep rumen (n = 5) and the rabbit cecum (n = 15).
Abundance of Select Microbes and Genes Involved in Hydrogen Metabolism
The total bacterial population (log10 16S rRNA gene copies/g sample) was larger in the rabbit cecum than in the sheep rumen (Figure 2A). The abundance of Ruminococcus albus, R. flavefaciens, Fibrobacter succinogenes, and Butyrivibrio fibrisolvens was greater in the sheep rumen than in the rabbit cecum. The same holds true for the abundance of fungi and protozoa. The abundance of Clostridium Cluster XIVa was similar between the two digestive organs, while that of Clostridium Cluster IV was greater in the rabbit cecum than in the sheep rumen (Figure 2B). The copy number of mcrA gene per g sample was greater in the sheep rumen than in the rabbit cecum, while that of fhs gene and frdA gene was smaller (Figure 2C). The sheep rumen had a greater abundance of RCC methanogens and non-RCC methanogens (including Methanobrevibacter, Methanomicrobium, Methanobacterium, Methanomicrococcus, and Methanosphaera) than the rabbit cecum. The archaea : bacteria ratio differed between the two different digestive organs, 0.089 in the sheep rumen and 2.30E-05 in the rabbit cecum.
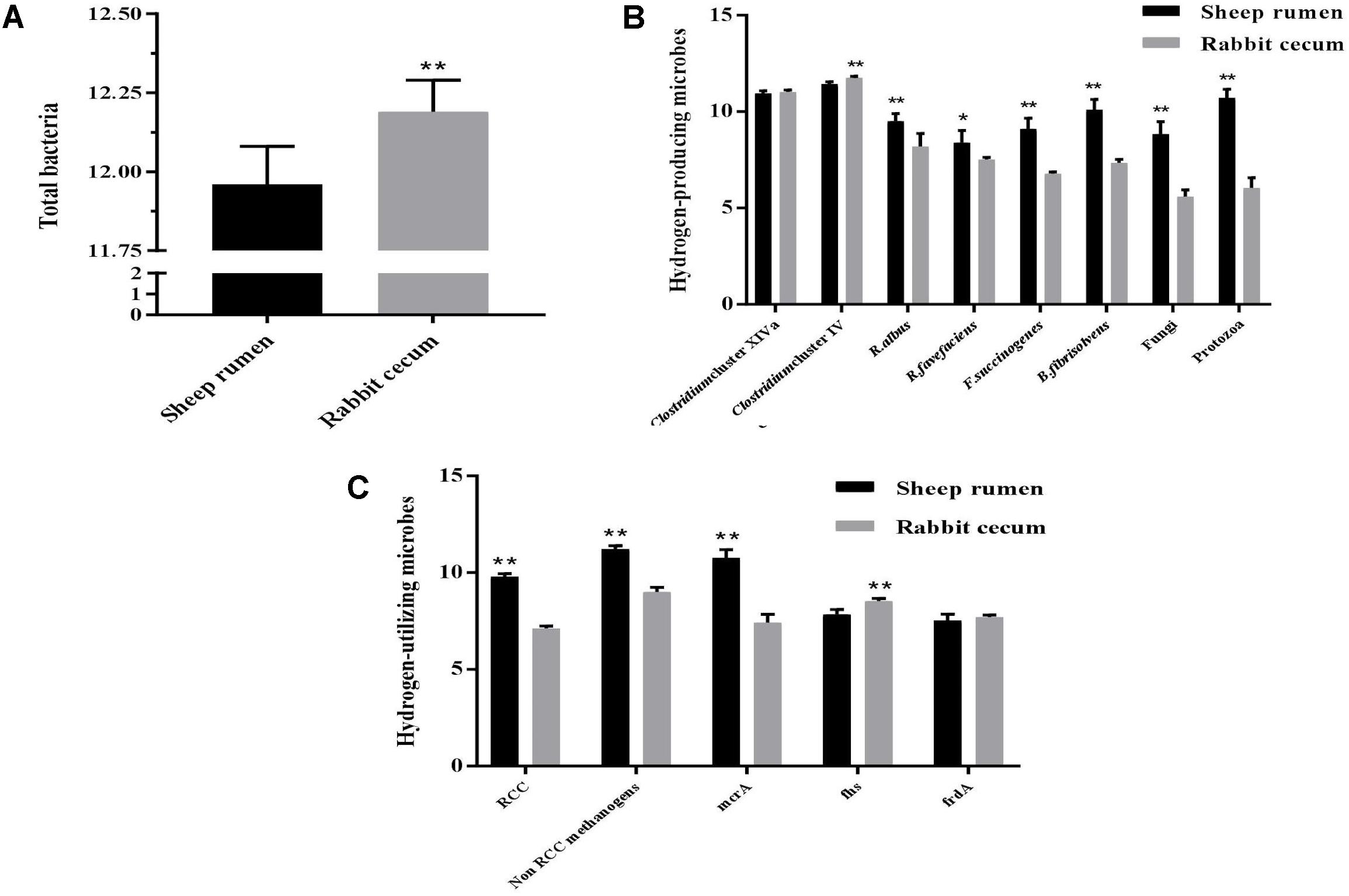
FIGURE 2. Abundance of select microbes in the sheep rumen (n = 5) and the rabbit cecum (n = 15). (A) Total bacteria; (B) Select hydrogen-producing microbes; (C) Select hydrogen-utilizing microbes. Abundance was expressed as log10 copies of target gene/g freeze-dried digesta sample. Target gene: 16S rRNA gene for bacteria and methanogens (except for mcrA); ITS1 for fungi; 18S rRNA for protozoa; mcrA, methyl-CoA reductase α subunit gene; fhs, Formyltetrahydrofolate synthetase gene; frdA, Fumarate reductase gene α subunit gene. ∗∗P < 0.01; ∗P < 0.05.
Diversity, Species Richness, and Composition of Archaeal Microbiota
The Chao1 richness estimate was similar between the two digestive organs, but the rabbit cecum had a lower Shannon diversity index and Simpson index of diversity than the sheep rumen (Table 4A). The archaeal microbiota of the two digestive organs clustered separately along the PC1 that explained greater than 84% of total variation. The rabbit cecal archaeal microbiotas clustered relatively tightly, while those of the sheep rumen quite scattered along PC2 that explained less than 10% total variation (Figure 3A).
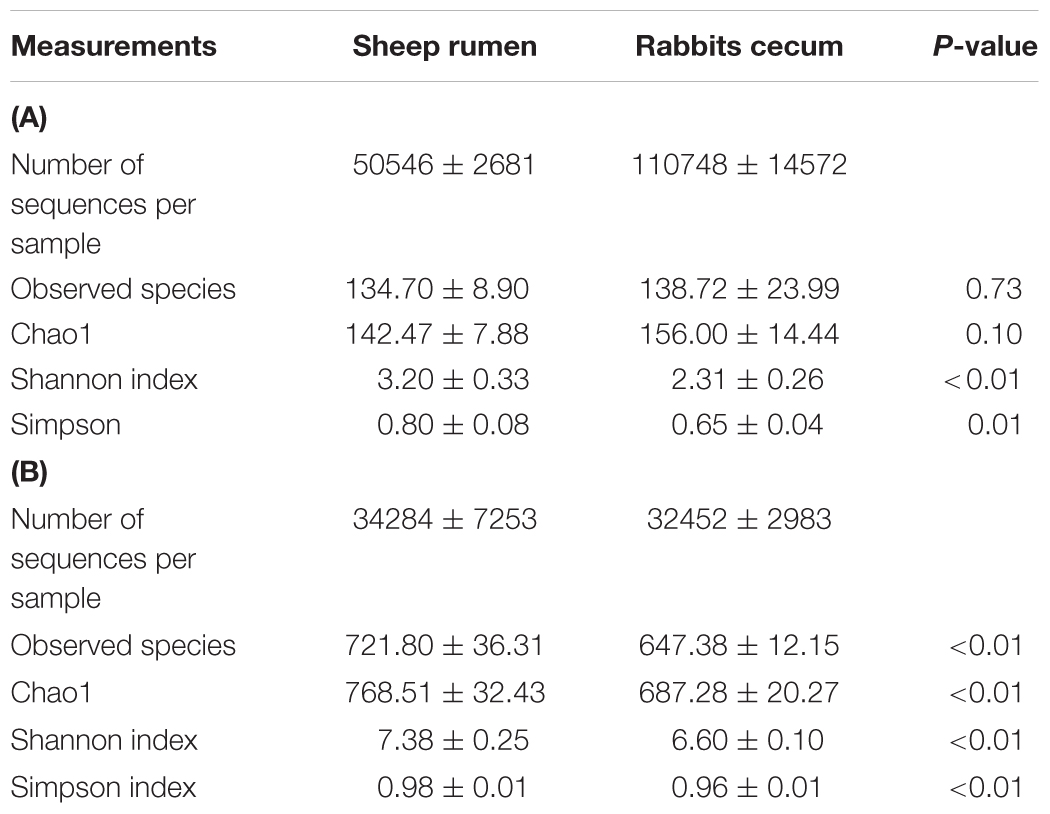
TABLE 4. Alpha diversity measurements of archaeal (A) and bacterial (B) microbiota of the sheep rumen (n = 5) and the rabbit cecum (n = 5 blocks).
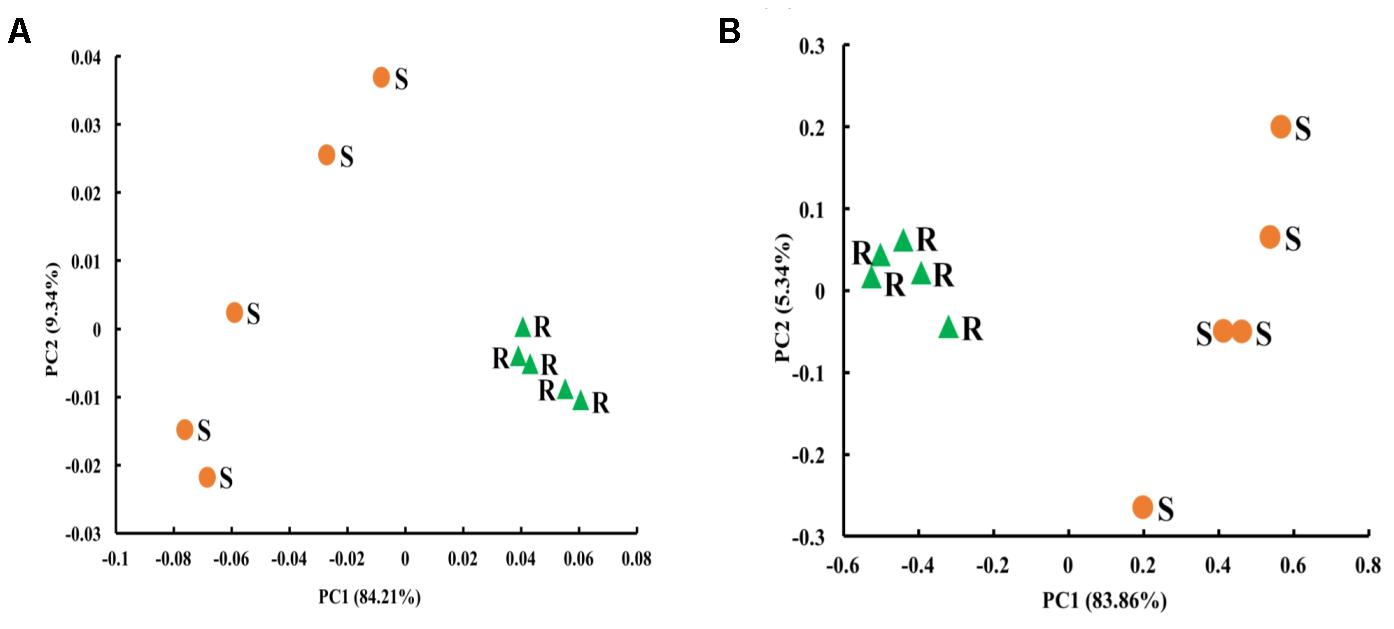
FIGURE 3. Principal coordinates analysis (PCoA) plots in the sheep rumen (n = 5) and the rabbit cecum (n = 5 blocks). (A) Archaeal microbiota; (B) Bacterial microbiota. The PCoA were based on distance matrices generated using weighted UniFrac. S, sheep rumen samples; R, rabbits cecal samples.
Approximately 99.7% of the sequences obtained from both the digestive organs were assigned to known archaeal genera (Figure 4). More than 95% of the archaeal sequences from the rabbit cecum were assigned to the genus Methanobrevibacter, while the archaeal sequences from the sheep rumen were assigned to Methanobrevibacter (68.3%), Methanosphaera (17.3%), and unidentified achaeon vadinCA11 (14.2%). The two digestive organs shared 120 archaeal OTUs besides their unique archaeal OTUs. The 30 OTUs unique to the sheep rumen were assigned to Methanobrevibacter (8 OTUs), vadinCA11 (10 OTUs), and Methanosphaera (8 OTUs), together accounting for 3.6% of total archaeal sequences identified therein. Of the 59 OTUs only found in the rabbit cecum, 53 were classified to Methanobrevibacter, and these 53 Methanobrevibacter OTUs accounted for only 1.1% of total archaeal sequences identified in the rabbit cecum. A significant portion of the archaeal sequences was assigned to known species, with 36.3% assigned to ∗∗∗M. thaueri, 14.6% to M. woesei, and 10.8% to M. millerae for the sheep rumen sequences, while for the rabbit cecal sequences, 74.6% to M. woesei, and 13.9% to M. thaueri.
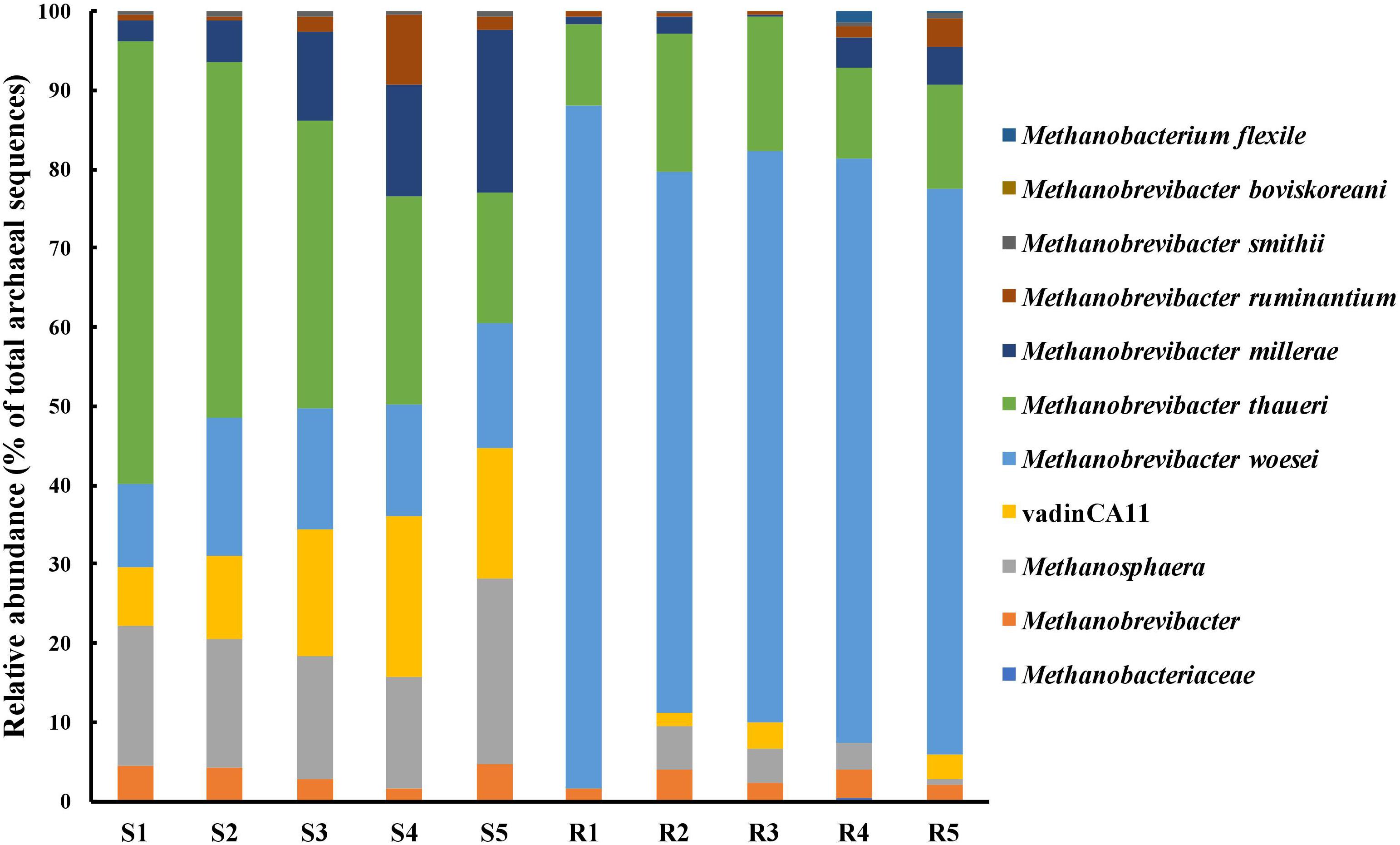
FIGURE 4. Comparison of the relative abundance of species and genera of methanogens identified in the two digestive organs. S1–S5, the five sheep; R1–R5, the five blocks of rabbits.
Diversity, Species Richness, and Composition of Bacterial Microbiota
The number of OTUs, Chao1 richness estimate, Shannon diversity index, and Simpson index of diversity in the sheep rumen were all significantly greater than those in the rabbit cecum (Table 4B). When compared using weighted UniFrac distance, the rabbit cecal bacterial microbiotas were separated, as a tight cluster, from those of the sheep rumen along the PC1 that explained greater than 83% of total variation (Figure 3B). The sheep rumen bacterial microbiotas of the five sheep exhibited considerable scattering along PC2, but it only explained less than 6% of total variation.
Almost all the sequences obtained from both the sheep rumen and the rabbit cecum were assigned to known bacterial phyla, with Firmicutes (48.7 vs. 56.1%) and Bacteroidetes (47.4 vs. 36.1%) being represented by more sequences than other phyla (Figure 5A). Seven bacterial phyla were identified in both the sheep rumen and the rabbit cecum, which included, in addition to the above two predominant phyla, Actinobacteria, Proteobacteria, Tenericutes, Verrucomicrobia, and Synergistetes. However, another five bacterial phyla, i.e., Spirochaetes, Chloroflexi, Fibrobacteres, Planctomycetes, and candidate phylum SR1 were only found in the sheep rumen. Of the common bacterial phyla, the relative abundance of Verrucomicrobia was significantly greater in the rabbit cecum than in the sheep rumen (5.7 vs. 0.6%, P < 0.01).
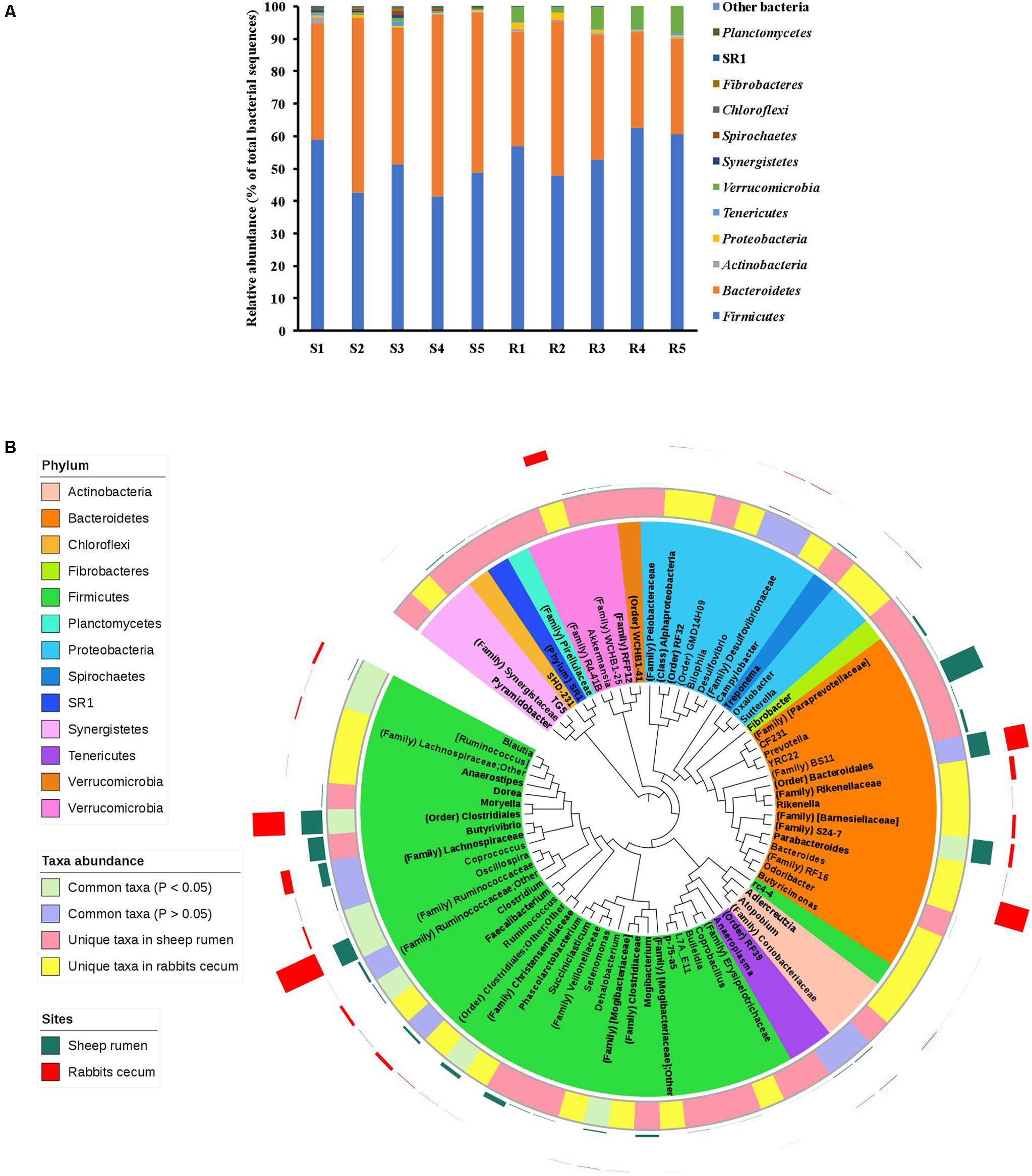
FIGURE 5. Comparison of the bacterial microbiota in the sheep rumen and the rabbit cecum. (A) The relative abundance of bacterial phyla; (B) A neighbor-joining phylogenetic tree of the representative sequences of the OTUs that were graphically displayed in the Interactive Tree of Life. S1–S5, the five sheep; R1–R5, the five blocks of rabbits.
In total, 840 and 728 OTUs were respectively observed in the sheep rumen and the rabbit cecum, and only 51 OTUs were found in both digestive organs. These common OTUs were assigned, at the lowest taxonomic rank, to the candidate order RF39 (3 OTUs), Clostridiales (23 OTUs), Bacteroidales (2 OTUs), candidate family S24-7 (1 OTUs), Ruminococcaceae (7 OTUs), Lachnospiraceae (4 OTUs), Ruminococcus (3 OTUs), Oscillospira (3 OTUs), Blautia (3 OTUs), and Bacteroides (1 OTUs). These common OTUs represented 9.6% and 16.9% of the total bacterial sequences in the sheep rumen and the rabbit cecum, respectively.
The OTUs found in the sheep rumen and the rabbit cecum was assigned to 49 and 45 lowest possible taxa, respectively. However, only 25 (for the sheep rumen) and 24 (for the rabbit cecum) of them are recognized genera. The two digestive organs shared 18 common taxa, leaving 31 taxa being unique in the sheep rumen and 27 taxa found only in the rabbit cecum (Figure 5B). The predominant taxa (each represented by >1.0% of total bacterial sequences in at least 3 of the 5 experiment units) common to the sheep rumen and the rabbit cecum included Bacteroidales (11.1 vs. 12%, P = 0.83), Lachnospiraceae (4.9 vs. 5.3%, P = 0.82), and Ruminococcus (1.4 vs. 1.9%, P = 0.24). A small number of taxa had different relative abundance between the sheep rumen and the rabbit cecum, with Ruminococcaceae (10.1 vs. 24.1%, P < 0.01), Clostridiales (12 vs. 18%, P = 0.03), Oscillospira (0.1 vs.1.2%, P < 0.01), Blautia (0.4 vs. 1.7%, P = 0.01), and Clostridium (0.5 vs. 1.7%, P = 0.01) being less predominant, while Christensenellaceae (2.2 vs. 0.1%, P < 0.01) and candidate family S24-7 (11.3 vs. 2.1%, P = 0.02) being more predominant in the sheep rumen than in the rabbit cecum. The major unique taxa (with a relative abundance of >1.0% in at least 3 of the 5 experiment units) in the sheep rumen included Prevotella (21.3%), Butyrivibrio (9%), Succiniclasticum (3%), candidate family BS11 (1.7%), candidate family [Paraprevotellaceae] (1.4%), and Mogibacterium (1.3%). Bacteroides (16.9%), Akkermansia (5.7%), Rikenellaceae (2.9%), and candidate family [Barnesiellaceae] (2%) were the major taxa unique to the rabbit cecum.
Pearson Correlation Between Chemical Parameters and Dominant Bacterial Taxa
Pearson correlation coefficients were calculated to reveal correlations between the animal phenotypic data and the predominant bacterial taxa (Figure 6A). The relative abundance of Butyrivibrio, Prevotella, Succiniclasticum, Mogibacterium, Christensenellaceae, candidate family [Mogibacteriaceae], and candidate family S24-7 appeared to be positively correlated (P < 0.05) with both CH4 yield and feed digestibility, whereas that of Oscillospira, Ruminococcaceae, Clostridiales, candidate genus [Ruminococcus], Blautia, Clostridium, Bacteroides, Rikenellaceae and Akkermansia was negatively (P < 0.05) correlated with these two measurements. A positive correlation was also seen between A:P ratio and the relative abundance of some taxa, including Oscillospira, Ruminococcaceae, Clostridiales, candidate genus [Ruminococcus], Blautia, Clostridium, Bacteroides, Akkermansia, and Rikenellaceae.
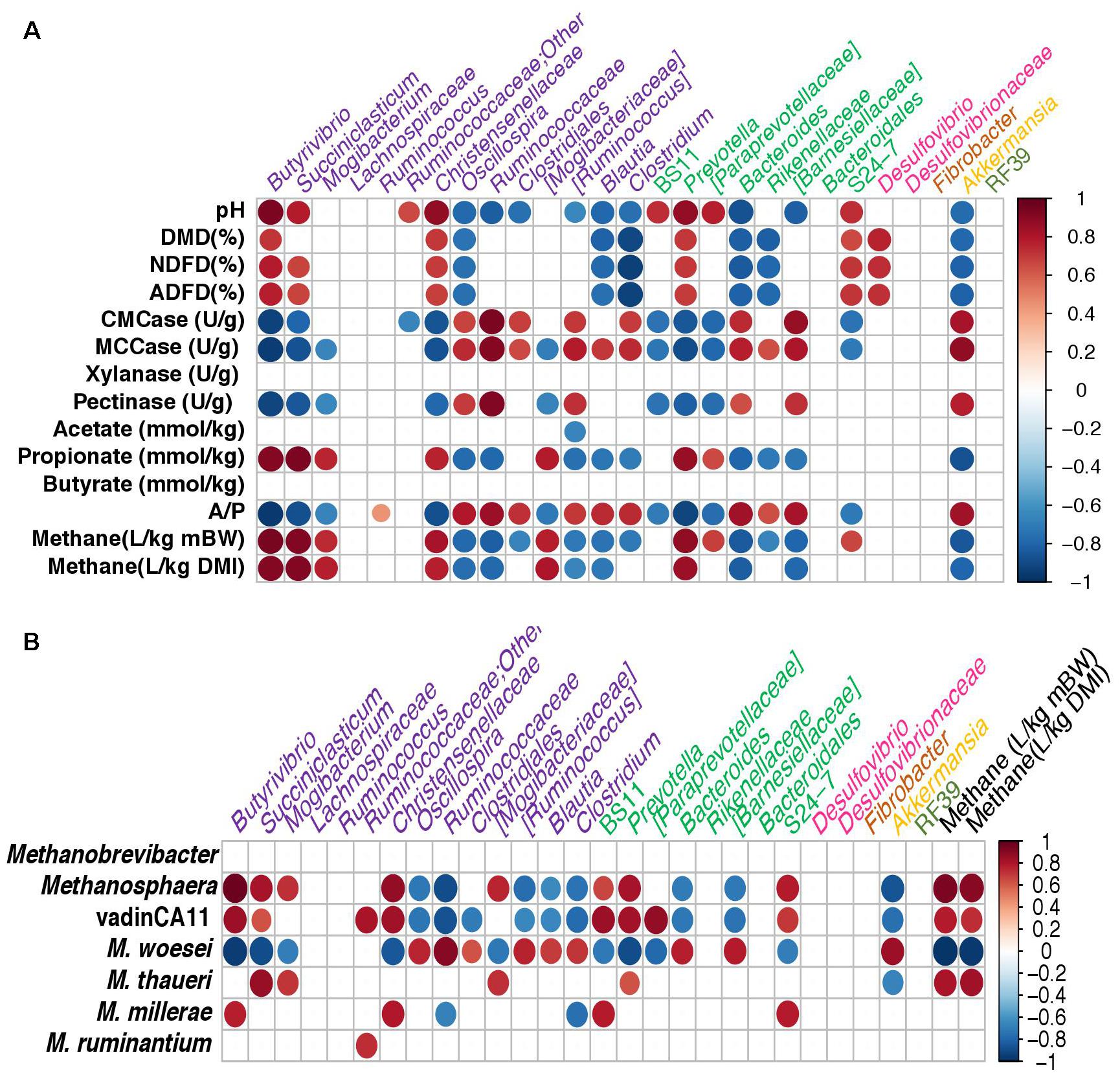
FIGURE 6. Pearson correlation between (A) chemical parameters and the relative abundance of bacterial taxa; (B) Archaeal taxa and bacterial taxa in relative abundance for the sheep rumen (n = 5) and the rabbit cecum (n = 5 blocks) in combination. Taxa were included in the matrix only if they were found in at least 3 of the five sheep or five rabbit blocks and they were each represented by at least 1.0% of the total bacterial sequences. Size of the filled circle reflects the strength of the correlations. Only significant correlation (P < 0.05) were shown with colors. The scale colors denote the direction of correlation (1 to –1). The taxa assigned to different phyla were color coded: purple, Firmicutes; green, Bacteroidetes; pink, Proteobacteria; brown, Fibrobacteres; yellow, Verrucomicrobia; and olive, Tenericutes.
Pearson Correlation Between the Archaeal Taxa and Bacterial Taxa in Relative Abundance
Pearson correlation coefficients were calculated to reveal correlations between the relative abundance of archaea and bacteria (Figure 6B). The relative abundance of M. thaueri appeared to be positively correlated (P < 0.05) with Succiniclasticum, Mogibacterium, candidate family [Mogibacteriaceae], Prevotella, and CH4 yields, and was negatively (P < 0.05) correlated with Akkermansia. The relative abundance of Methanosphaera and unidentified achaeon vadinCA11 appeared to be positively correlated (P < 0.05) with Butyrivibrio, Succiniclasticum, Christensenellaceae, candidate family [Mogibacteriaceae], BS11, Prevotella, S24-7, as well as CH4 yields, whereas were negatively (P < 0.05) correlated with the relative abundance of Oscillospira, Ruminococcaceae, candidate genus [Ruminococcus], Blautia, Clostridium, Bacteroides, and Akkermansia. The opposite correlations of M. woesei with both CH4 yields and those bacteria were showed.
Discussion
Both ruminants and non-ruminant herbivores emit CH4, but the former emits much more CH4 than the latter (Franz et al., 2010, 2011; Cabezas Garcia, 2017). It has been speculated that such difference in CH4 emission is probably attributable primarily to the differences in the microbiota of the rumen and the hindgut of non-ruminant herbivores (Yang et al., 2016). However, the microbiological peculiarity for the different CH4 emissions by these two groups of herbivores is largely unknown. Identification of these responsible microbes and the relationship to CH4 emission and the fermentation characteristics of the rumen and the cecum will help understand the factors that affect CH4 production in the rumen and develop dietary strategies to effectively mitigate CH4 emission from ruminants. In the present study, we comparatively characterized the microbiota and the fermentation characteristics in the rumen of sheep and the cecum of rabbits when fed the same diet. This approach allowed us to quantitatively determine and compare CH4 production by a representative species of ruminants and non-ruminants on the basis of feed intake, feed digestibility, and metabolic BW0.75. This approach overcomes the limitation of using the same ruminant animal in which CH4 emission from the rumen and large intestines cannot be independently determined, and the two digestive organs received different fermentation substrates.
The rabbits produced no more than ¼ of the amount of CH4 produced by the sheep when compared on per unit of BW0.75, DMI, digestible NDF or ADF. Franz et al. (2010, 2011) proposed a linear relationship between BW and CH4 production by both ruminants and non-ruminants. However, the magnitude of different CH4 outputs between the sheep and the rabbits probably suggests physiological and microbiological peculiarities of these two digestive organs. First, the pH inside the rabbit cecum was nearly 1.3 units lower than that in the sheep rumen. It is well documented that methanogenesis is inhibited at low pH, as exemplified by no CH4 production at pH 5.5 or below in in-vitro cultures inoculated with rumen fluid from roughage-fed cows (Russell, 1998). Thus, the lower pH in the rabbit cecum (pH 5.8) than in the sheep rumen (pH 7.1) is probably a major chemical factor attributable to the low CH4 output from the rabbits. The rumen receives a large volume of saliva (about 1.31 L of saliva is secreted from one parotid gland per day for an adult sheep), which buffers the acidity from VFA (McDougall, 1948), while cecum receives no saliva. The lack of saliva secretion to the cecum is probably one of the reasons for the lower pH in the rabbit cecum than in the sheep rumen. Second, the Eh in the rabbit cecum ranges from -160 to -210 mV (Kimsé et al., 2010; Michelland et al., 2010), while the rumen Eh ranges from -268 to -318 mV (Mathieu et al., 1996). We did not analyze the Eh in the present study, but it should be within the above range. The tubiform and small in diameter of the rabbit cecum may explain its relatively higher Eh. Apparently, the Eh of the rabbit cecum is not optimal for hydrogenotrophic methanogenesis, which requires -238 mV (Cord-Ruwisch et al., 1988). Indeed, CH4 production in a Methanothermobacter thermautotrophicus culture was suppressed at Eh higher than -200 mV (Hirano et al., 2013). Future research using in vitro cultures of both digestive organs is warranted to verify if Eh is a primary factor determining the different CH4 production in the rumen and the cecum. The Eh of in vitro cultures can be regulated using bioelectrochemical systems that can control Eh without using oxidative and reducing agents (Hirano et al., 2013). Furthermore, digesta passage rate through the rumen has been found reversely correlated with CH4 production therein (Janssen, 2010; Goopy et al., 2014; Shi et al., 2014). The cecum is a tubiform tract, while the rumen is a large sac. Such structural difference can causes a faster digesta passage rate through the rabbit cecum than through the sheep rumen, contributing to the less CH4 output from the rabbits than from the sheep. This premise is consistent with the high passage rate and low CH4 production by kangaroos, a group of tubiform foregut fermenters (Von Engelhardt et al., 1978).
In the present study, we analyzed the diversity and structure of the archaeal microbiota and quantified the abundance of methanogens to understand the archaeal underpinning of the different CH4 yields between the two digestive organs. The rabbit cecum had a lower abundance of RCC methanogens, non-RCC methanogens, and total methanogens (as quantified as mcrA gene copies/g sample) than the sheep rumen. This is consistent with the finding in sheep (Popova et al., 2013), reindeer (Wedlock et al., 2013), and Chinese roe deer (Li et al., 2014), in which a greater abundance of methanogens was found in the rumen than in the cecum. The low abundance of methanogens in the rabbit cecum may be explained partially by the low pH, probably a higher Eh, and a greater passage. These three factors might have directly decreased methanogenesis in the rabbit cecum. Although the abundance of methanogens in the rumen does not necessarily linearly correlate to CH4 output (Danielsson et al., 2012; Patra and Yu, 2014), the greater abundance of methanogens in the sheep rumen than in the rabbit cecum corroborates the more CH4 produced by the former than by the latter.
The two digestive organs each harbored a distinct archaeal microbiota, with M. woesei being the dominant species in the rabbit cecum, whereas the sheep rumen containing M. thaueri as the most predominant known species followed by M. millerae and M. woesei. Based on a literature search of the Pubmed, only one study has analyzed the archaeal microbiota in the cecum of rabbits, and M. woesei was represented by more cloned 16S rRNA gene sequences than other species (Kušar and Avguštin, 2010). M. ruminantium, one of the two species (M. ruminantium and M. Olleyae) in the Methanobrevibacter RO clade (Janssen and Kirs, 2008; Kittelmann et al., 2013), was found in both the digestive organs at low relative abundance and low correlation with CH4 yield. The RO clade was found associated with low CH4 yield in the rumen (Danielsson et al., 2012). The dominance of M. woesei, which has only been reported in the chicken cecum (Saengkerdsub et al., 2007) other than rabbit cecum, is of further research interest. It is also interesting to note that M. thaueri and M. millerae, two of the four species (M. smithii, M. gottschalkii, M. Millerae, and M. thaueri) in the Methanobrevibacter SGMT clade (Janssen and Kirs, 2008; Kittelmann et al., 2013), were more predominant in the sheep rumen than in the rabbit cecum. The Methanobrevibacter SGMT clade, which possesses methyl coenzyme M reductase isozymes Mcr I and Mcr II and are competitive at high hydrogen concentrations (Leahy et al., 2010), has been reported to have a positive association with CH4 emissions from ruminants (Tapio et al., 2017). The positive correlation between M. thaueri and CH4 yield was found in the present study. It is not known if the differential predominance of these methanogen species is one explanation of the different CH4 production seen between the two animal species. Several studies revealed a strong correlation between CH4 yields and archaea: bacteria ratio (Wallace et al., 2014), and a similar finding was found in the present study. However, the previous study indicated that it was gene expression rather than gene abundance of methanogens that was strongly correlated with CH4 yields from sheep (Shi et al., 2014). Metatranscriptomic studies will help determine the contribution of each methanogen species to the overall CH4 yield in these two digestive organs.
Methane is produced by methanogens, but other members of the microbiota can determine or profoundly affects the rate and yield of methanogenesis (Kittelmann et al., 2014; Danielsson, 2016). In the present study, we characterized the bacterial microbiota for diversity and structure and quantified the population of microbes that produce hydrogen, including anaerobic fungi, protozoa, select hydrogen-producing bacteria, and acetogens to help understand the role and significance of these microbes in determining the different CH4 yields between the two digestive organs. Expectedly, the sheep rumen and the rabbit cecum differed in the communities of the above microbes. Different microbiota were also reported between the rumen and the cecum of growing bulls (Popova et al., 2017), reindeer (Wedlock et al., 2013), and Chinese roe deer (Li et al., 2014). Such difference may be attributable to the combined effect of a host of factors, including pH, saliva (present in the rumen but not in cecum), passage rate, nutrients (lack of non-structural carbohydrates and protein due to digestion and absorption in the foregut), mixing, Eh (higher in the cecum), mucosa and antimicrobial peptides (both present in the rabbit cecum but not in the sheep rumen).
Hydrogen-producing microbes provide the reducing power for hydrogenotrophic methanogenesis, and indeed, sheep producing more hydrogen also produced more CH4 than those that produced less hydrogen (Kittelmann et al., 2014). As determined by qPCR, the sheep rumen did have a greater abundance of R. albus, R. flavefaciens, B. fibrisolvens, fungi, and protozoa, all of which can produce hydrogen during feed fermentation, than the rabbit cecum. Combined analysis of the bacterial microbiota also revealed correlations between several animal phenotypic measurements and individual bacterial groups. Among the bacterial genera whose relative abundance was strongly and positively correlated with CH4 yield, Prevotella and related bacteria, Butyrivibrio, and Succiniclasticum are unique and/or predominant hydrogen-producing bacteria in the rumen (Leahy et al., 2013). The bacterial genera whose relative abundance appeared to be negatively correlated with CH4 yield are either acetogens (e.g., Blautia) (Müller and Frerichs, 2013), butyrate producers (e.g., Oscillospira) (Gophna et al., 2017), or succinate producers (e.g., Bacteroides) (Song et al., 2015). In the rumens of high methane-emitting sheep, members of Ruminococcaceae and Lachnospiraceae were found at higher relative abundance, while the rumens of low methane-emitting sheep were enriched with Erysipelotrichaceae, especially Sharpea spp. (Kamke et al., 2016). However, the low correlation between CH4 yield and the relative abundance of Ruminococcus and Lachnospiraceae were found when comparing the two different the digestive organs in the present study. The relationship between the abundance of F. succinogenes and CH4 production cannot be explained also, nor the contribution of Clostridium cluster IV. It should be noted that the correlation of a few bacterial genera with CH4 yield might be due to their occurrence in only one of the two digestive organs, such as Mogibacterium and Succiniclasticum that were only detected in the rumen, and Akkermansia that was only detected in the large intestines. Nevertheless, the positive correlation between CH4 yield and the relative abundance of Mogibacterium, which does not ferment carbohydrate (Nakazawa et al., 2015), is consistent with its ability to produce phenylacetate, a metabolite that is needed for the degradation of cellulose by some R. albus strains (Morrison et al., 1990).
Constant hydrogen disposal is essential for sustained fermentation in the rumen and the large intestines (Moss et al., 2010). Thus, alternative hydrogen utilization pathways must exist in the rabbit cecum. We analyzed two genes, fhs and frd, involved in two different [H]-utilizing pathways to understand the alternative hydrogen utilization potential in the two digestive organs. The rabbit cecum had a higher abundance of fhs, which encodes formyltetrahydrofolate synthetase, a key enzyme in the homoacetogenesis, consistent with the lower CH4 production and higher molar proportion of acetate therein. A strong positive correlation was also found between A: P ratio and some bacterial taxa, including some taxa of Clostridiales, Lachnospiraceae, Ruminococcaceae, and Blautia, all of which contain known acetogens (Yang et al., 2016). All these results suggest acetate is not just a hydrogen donor, but a hydrogen sink. Indeed, homoacetogenesis might be predominant in the rabbit cecum than in the sheep rumen. The dominance of homoacetogenesis has been reported in some tubiform gut ecosystems, such as rabbits cecum (Yang et al., 2016), kangaroos foregut (Gagen et al., 2010), and termite hindgut (Ottesen and Leadbetter, 2011). It has been suggested that pH might determine the predominance of hydrogen disposal pathways, with relatively neutral pH favoring methanogens and acidic pH favoring acetogens (Gibson et al., 1990). The relatively lower pH in the cecum of rabbits probably suppress hydrogenotrophic methanogenesis, allowing homocaetogenesis to increase. It is interesting to note that more CO2 was produced from the rabbits than from the sheep per kg of BW0.75. It is likely that less CO2 is consumed during homoacetogenesis in the cecum than during methanogenesis in the sheep rumen. This premise is consistent with the less thermodynamic feature of homoacetogenesis than hydrogenotrophic methanogenesis and the higher Ks of the former (Kohn and Boston, 2000). The lower pH in the rabbit cecum could also decrease the CO2 solubility and thus more CO2 emission.
Surprisingly, significantly greater activities of CMCase, MCCasse, and pectinase were detected in the rabbit cecum than in the sheep rumen, suggesting an enrichment of fibrolytic and pectinolytic microbes in the rabbit cecum. Along with the significantly different bacterial microbiota structures between the sheep rumen and the rabbit cecum, our findings indicate that rabbit cecum probably harbors novel and uncharacterized cellulolytic bacteria and glycoside hydrolases. These novel microbes and enzymes can be identified in future studies using functional metagenomics and transcriptomics.
Conclusion
The present study demonstrates that different methane production between the sheep and the rabbits can be explained by the different physiological environments of their respective digestive organs and the microbiota residing therein. Lower abundance of hydrogen-producing microbes (bacteria, fungi, and protozoa) and methanogens, and increased homoacetogenesis as an alternative hydrogen utilization pathway in the rabbit cecum might result in lower CH4 yield from the rabbits. The cecum of rabbits is potentially a rich resource to fibrolytic bacteria and hence novel cellulolytic enzymes. Future studies using functional approaches, such as functional metagenomics and transcriptomics, will help reveal the potential and functionality of metabolic pathways involved in fiber digestion, methanogenesis, and acetogenesis and help develop new strategies to achieve effective CH4 mitigation for ruminal livestock.
Author Contributions
JW, LM, and JL conceived and designed the study. LM performed both animal feeding and laboratory experiments, analyzed the sequencing data, interpreted the data, prepared the figures and tables, and wrote the manuscript. ZY and BY helped analyzing the sequencing data. XH and YL participated in the animal feeding experiments. ZY and JW helped interpret the data and write and revise the paper. All authors read and approved the final manuscript.
Funding
This research was partially supported by the National Natural Science Foundation of China (award number: 31372337). LM’s tenure at The Ohio State University was supported by a scholarship from Zhejiang University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Bo He, Chunlei Yang, Dan Feng, and Shanshan Wang for their assistance with feeding and caring of the animals and sample collection. The authors are also grateful to Dr. Lingling Wang for assistance in sequence data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00575/full#supplementary-material
Abbreviations
A, acetate; ADF, acid detergent fiber; BW, body weight; BW0.75, metabolic body weight; CMCase, carboxymethyl cellulase; CP, crude protein DM, dry matter; DMI, dry matter intake; Eh, redox potential; fhs, formyltetrahydrofolate synthetase gene; frdA, fumarate reductase gene α subunit; MCCase, microcrystalline cellulose cellulase; MCP, crude microbial protein; mcrA, methyl CoM reductase gene α subunit; NDF, neutral detergent fiber; NZW rabbit, New Zealand white rabbit; P, propionate; RCC, rumen cluster C; VFA, volatile fatty acid.
References
Aronesty, E. (2011). ea-utils: Command-line Tools for Processing Biological Sequencing Data. Available at: https://github.com/ExpressionAnalysis/ea-utils
Attwood, G., and McSweeney, C. (2008). Methanogen genomics to discover targets for methane mitigation technologies and options for alternative H2 utilisation in the rumen. Aust. J. Exp. Agric. 48, 28–37. doi: 10.1071/EA07203
Bailey, M. J., Biely, P., and Poutanen, K. (1992). Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23, 257–270. doi: 10.1016/0168-1656(92)90074-J
Bhatta, R., Enishi, O., and Kurihara, M. (2007). Measurement of methane production from ruminants. Asian Aust. J. Anim. Sci. 20, 1305–1318. doi: 10.5713/ajas.2007.1305
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Cabezas Garcia, E. H. (2017). Methane Production in Dairy Cows. Doctoral thesis, Swedish University of Agricultural Sciences, Umeå.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cord-Ruwisch, R., Seitz, H. J., and Conrad, R. (1988). The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149, 350–357. doi: 10.1007/BF00411655
Danielsson, R. (2016). Methane Production in Dairy Cows. Doctoral thesis, Swedish University of Agricultural Sciences, Uppsala.
Danielsson, R., Schnürer, A., Arthurson, V., and Bertilsson, J. (2012). Methanogenic population and CH4 production in Swedish dairy cows fed different levels of forage. Appl. Environ. Microbiol. 78, 6172–6179. doi: 10.1128/AEM.00675-12
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Franz, R., Soliva, C. R., Kreuzer, M., Hummel, J., and Clauss, M. (2011). Methane output of rabbits (Oryctolagus cuniculus) and guinea pigs (Cavia porcellus) fed a hay-only diet: implications for the scaling of methane production with body mass in non-ruminant mammalian herbivores. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 158, 177–181. doi: 10.1016/j.cbpa.2010.10.019
Franz, R., Soliva, C. R., Kreuzer, M., Steuer, P., Hummel, J., and Clauss, M. (2010). Methane production in relation to body mass of ruminants and equids. Evol. Ecol. Res. 12, 727–738.
Furness, J. B., Cottrell, J. J., and Bravo, D. M. (2015). Comparative gut physiology symposium: comparative physiology of digestion. J. Anim. Sci. 93, 485–491. doi: 10.2527/jas.2014-8481
Gagen, E. J., Denman, S. E., Padmanabha, J., Zadbuke, S., Al Jassim, R., Morrison, M., et al. (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microbiol. 76, 7785–7795. doi: 10.1128/AEM.01679-10
Gibson, G. R., Cummings, J. H., Macfarlane, G. T., Allison, C., Segal, I., Vorster, H. H., et al. (1990). Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 31, 679–683. doi: 10.1136/gut.31.6.679
Goopy, J. P., Donaldson, A., Hegarty, R., Vercoe, P. E., Haynes, F., Barnett, M., et al. (2014). Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br. J. Nutr. 111, 578–585. doi: 10.1017/S0007114513002936
Gophna, U., Konikoff, T., and Nielsen, H. B. (2017). Oscillospira and related bacteria–From metagenomic species to metabolic features. Environ. Microbiol. 19, 835–841. doi: 10.1111/1462-2920.13658
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hirano, S., Matsumoto, N., Morita, M., Sasaki, K., and Ohmura, N. (2013). Electrochemical control of redox potential affects methanogenesis of the hydrogenotrophic methanogen Methanothermobacter thermautotrophicus. Lett. Appl. Microbiol. 56, 315–321. doi: 10.1111/lam.12059
IPCC (2014). IPCC, 2014: Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of Intergovernmental Panel on Climate Change [Core Writing Team, eds R. K. Pachauri and L. A. Meyer]. Geneva: Geneva Press, 151.
Janssen, P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160, 1–22. doi: 10.1016/j.anifeedsci.2010.07.002
Janssen, P. H., and Kirs, M. (2008). Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625. doi: 10.1128/AEM.02812-07
Joblin, K. N. (1999). Ruminal acetogens and their potential to lower ruminant methane emissions. Aust. J. Agr. Res. 50, 1307–1314. doi: 10.1071/AR99004
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kamke, J., Sandra, K., Priya, S., Yang, L., Michael, T., Siva, G., et al. (2016). Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 4:56. doi: 10.1186/s40168-016-0201-2
Kimsé, M., Monteils, V., Bayourthe, C., and Gidenne, T. (2010). A new method to measure the redox potential (Eh) in rabbit caecum: relationship with pH and fermentation pattern. World Rabbit Sci. 17, 63–70. doi: 10.4995/wrs.2009.659
Kittelmann, S., Pinares-Patiño, C. S., Seedorf, H., Kirk, M. R., Ganesh, S., McEwan, J. C., et al. (2014). Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171
Kittelmann, S., Seedorf, H., Walters, W. A., Clemente, J. C., Knight, R., Gordon, J. I., et al. (2013). Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879
Kohn, R. A., and Boston, R. C. (2000). “The role of thermodynamics in controlling rumen metabolism,” in Modelling Nutrient Utilization in Farm Animals, ed. J. P. McNamara (Wallingford, OX: CABI Press), 11–24.
Koike, S., Yabuki, H., and Kobayashi, Y. (2007). Validation and application of real-time polymerase chain reaction assays for representative rumen bacteria. J. Anim. Sci. 78, 135–141. doi: 10.1111/j.1740-0929.2007.00417.x
Kušar, D., and Avguštin, G. (2010). Molecular profiling and identification of methanogenic archaeal species from rabbit caecum. FEMS Microbiol. Ecol. 74, 623–630. doi: 10.1111/j.1574-6941.2010.00980.x
Leahy, S. C., Kelly, W. J., Altermann, E., Ronimus, R. S., Yeoman, C. J., Pacheco, D. M., et al. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5:e8926. doi: 10.1371/journal.pone.0008926
Leahy, S. C., Kelly, W. J., Ronimus, R. S., Wedlock, N., Altermann, E., and Attwood, G. T. (2013). Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal 7, 235–243. doi: 10.1017/S1751731113000700
Li, Z., Zhang, Z., Xu, C., Zhao, J., Liu, H., Fan, Z., et al. (2014). Bacteria and methanogens differ along the gastrointestinal tract of Chinese roe deer (Capreolus pygargus). PLoS One 9:e114513. doi: 10.1371/journal.pone.0114513
Liu, J., Wang, J. K., Zhu, W., Pu, Y. Y., Guan, L. L., and Liu, J. X. (2014). Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol. Ecol. 87, 576–585. doi: 10.1111/1574-6941.12246
Liu, Y., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Makkar, H. P. S., Sharma, O. P., Dawra, R. K., and Negi, S. S. (1982). Simple determination of microbial protein in rumen liquor. J. Dairy Sci. 65, 2170–2173. doi: 10.3168/jds.S0022-0302(82)82477-6
Mathieu, F., Jouany, J. P., Senaud, J., Bohatier, J., Bertin, G., and Mercier, M. (1996). The effect of Saccharomyces cerevisiae and Aspergillus oryzae on fermentations in the rumen of faunated and defaunated sheep; protozoal and probiotic interactions. Reprod. Nutr. Dev. 36, 271–287. doi: 10.1051/rnd:19960305
McDougall, E. I. (1948). Studies on ruminant saliva. 1. the composition and output of sheep’s saliva. Biochem. J. 43, 99–109. doi: 10.1042/bj0430099
Michelland, R. J., Combes, S., Monteils, V., Cauquil, L., Gidenne, T., and Fortun-Lamothe, L. (2010). Molecular analysis of the bacterial community in digestive tract of rabbit. Anaerobe 16, 61–65. doi: 10.1016/j.anaerobe.2009.05.002
Morrison, M., Mackie, R. I., and Kistner, A. (1990). 3-Phenylpropanoic acid improves the affinity of Ruminococcus albus for cellulose in continuous culture. Appl. Environ. Microbiol. 56, 3220–3222.
Moss, R. H., Edmonds, J. A., Hibbard, K. A., Manning, M. R., Rose, S. K., Van Vuuren, D. P., et al. (2010). The next generation of scenarios for climate change research and assessment. Nature 463, 747–756. doi: 10.1038/nature08823
Müller, V., and Frerichs, J. (2013). “Acetogenic bacteria,” in eLS, ed. J. Battista (Chichecster, NJ: John Wiley and Sons Ltd.). doi: 10.1002/9780470015902.a0020086.pub2
Nakazawa, F., Sato, M., Poco, S. E., Hashimura, T., Ideka, T., Kalfas, S., et al. (2015). “Mogibacterium,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. B. W. William (New York, NY: The Editorial Board Press), 1–5.
Official Methods of Analysis [AOAC] (2015). Official Methods of Analysis. Gaithersburg, MD: Association of Official Analytical Chemists Press.
Ottesen, E. A., and Leadbetter, J. R. (2011). Formyltetrahydrofolate synthetase gene diversity in the guts of higher termites with different diets and lifestyles. Appl. Environ. Microbiol. 77, 3461–3467. doi: 10.1128/AEM.02657-10
Patra, A. K., and Yu, Z. T. (2014). Combinations of nitrate, saponin, and sulfate additively reduce methane production by rumen cultures in vitro while not adversely affecting feed digestion, fermentation or microbial communities. Bioresour. Technol. 155, 129–135. doi: 10.1016/j.biortech.2013.12.099
Piattoni, F., Demeyer, D., and Maertens, L. (1996). In vitro study of the age-dependent caecal fermentation pattern and methanogenesis in young rabbits. Reprod. Nutr. Dev. 36, 253–261. doi: 10.1051/rnd:19960303
Popova, M., McGovern, E., McCabe, M. S., Martin, C., Doreau, M., Arbre, M., et al. (2017). The structural and functional capacity of ruminal and cecal microbiota in growing cattle was unaffected by dietary supplementation of linseed oil and nitrate. Front. Microbiol. 8:937. doi: 10.3389/fmicb.2017.00937
Popova, M., Morgavi, D. P., and Martin, C. (2013). Methanogens and methanogenesis in the rumens and ceca of lambs fed two different high-grain-content diets. Appl. Environ. Microbiol. 79, 1777–1786. doi: 10.1128/AEM.03115-12
Poulsen, M., Schwab, C., Jensen, B. B., Engberg, R. M., Spang, A., Canibe, N., et al. (2013). Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 4:1428. doi: 10.1038/ncomms2432
Russell, J. B. (1998). The importance of pH in the regulation of ruminal acetate to propionate ratio and methane production in vitro. J. Dairy Sci. 81, 3222–3230. doi: 10.3168/jds.S0022-0302(98)75886-2
Russell, J. B., and Rychlik, J. L. (2001). Factors that alter rumen microbial ecology. Science 292, 1119–1122. doi: 10.1126/science.1058830
Saengkerdsub, S., Anderson, R. C., Wilkinson, H. H., Kim, W. K., Nisbet, D. J., and Ricke, S. C. (2007). Identification and quantification of methanogenic archaea in adult chicken ceca. Appl. Environ. Microbiol. 73, 353–356. doi: 10.1128/AEM.01931-06
SAS Institute Inc. (2000). SAS User’s Guide: Statistics. Version 9.2. (Cary, NC: SAS Institute Inc.)
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Song, Y., Liu, C., and Finegold, S. M. (2015). “Bacteroides,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. B. W. William (New York, NY: The Editorial Board Press), 1–24.
St-Pierre, B., and Wright, A. D. G. (2013). Diversity of gut methanogens in herbivorous animals. Animal 7, 49–56. doi: 10.1017/S1751731112000912
Tapio, I., Snelling, T. J., Strozzi, F., and Wallace, R. J. (2017). The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 8:7. doi: 10.1186/s40104-017-0141-0
Van Soest, P. V., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Von Engelhardt, W., Wolter, S., Lawrenz, H., and Hemsley, J. A. (1978). Production of methane in two non-ruminant herbivores. Comp. Biochem. Physiol. Part A Physiol. 60, 309–311. doi: 10.1016/0300-9629(78)90254-2
Wallace, R. J., Rooke, J. A., Duthie, C. A., Hyslop, J. J., Ross, D. W., McKain, N., et al. (2014). Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4:5892. doi: 10.1038/srep05892
Wallace, R. J., Rooke, J. A., McKain, N., Duthie, C. A., Hyslop, J. J., Ross, D. W., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0
Wang, J. K., He, B., Du, W., Luo, Y., Yu, Z. T., and Liu, J. X. (2015). Yeast with surface displayed xylanase as a new dual purpose delivery vehicle of xylanase and yeast. Anim. Feed Sci. Technol. 208, 44–52. doi: 10.1016/j.anifeedsci.2015.07.002
Wedlock, D. N., Janssen, P. H., Leahy, S. C., Shu, D., and Buddle, B. M. (2013). Progress in the development of vaccines against rumen methanogens. Animal 7, 244–252. doi: 10.1017/S1751731113000682
Yang, B., He, B., Wang, S. S., Liu, J. X., and Wang, J. K. (2015). Early supplementation of starter pellets with alfalfa improves the performance of pre-and postweaning Hu lambs. J. Anim. Sci. 93, 4984–4994. doi: 10.2527/jas.2015-9266
Keywords: acetogen, cecum, fibrolytic bacteria, hydrogen, methane, microbiota, pH, rumen
Citation: Mi L, Yang B, Hu X, Luo Y, Liu J, Yu Z and Wang J (2018) Comparative Analysis of the Microbiota Between Sheep Rumen and Rabbit Cecum Provides New Insight Into Their Differential Methane Production. Front. Microbiol. 9:575. doi: 10.3389/fmicb.2018.00575
Received: 18 December 2017; Accepted: 13 March 2018;
Published: 27 March 2018.
Edited by:
Diana Elizabeth Marco, National Scientific and Technical Research Council (CONICET), ArgentinaReviewed by:
Wang Min, Institute of Subtropical Agriculture (CAS), ChinaSanjay Kumar, University of Pennsylvania, United States
Stephan Schmitz-Esser, Iowa State University, United States
Copyright © 2018 Mi, Yang, Hu, Luo, Liu, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongtang Yu, yu.226@osu.edu Jiakun Wang, jiakunwang@zju.edu.cn
 Lan Mi
Lan Mi Bin Yang
Bin Yang Xialu Hu1
Xialu Hu1 Yang Luo
Yang Luo Jianxin Liu
Jianxin Liu Zhongtang Yu
Zhongtang Yu Jiakun Wang
Jiakun Wang