- 1Research Center of Avian Diseases, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
To investigate tetracycline resistance and resistant genotype in Riemerella anatipestifer, the tetracycline susceptibility of 212 R. anatipestifer isolates from China between 2011 and 2017 was tested. The results showed that 192 of 212 (90.6%) R. anatipestifer isolates exhibited resistance to tetracycline (the MICs ranged from 4 to 256 μg/ml). The results of PCR detection showed that, 170 of 212 (80.2%) R. anatipestifer isolates possessed the tet(X) gene. Other genes, including tet(A), tet(M), tet(Q), tet(O), tet(B), and tet(O/W/32/O), were found at frequencies of 20.8, 4.7, 1.4, 0.9, 0.9, and 0.5%, respectively. However, tet(C), tet(E), tet(G), tet(K), and tet(W) were not detected in any isolate. In these tet gene positive strains, 31 (14.6%), 2 (0.9%), 5 (2.4%), 1 (0.5%), 3 (1.4%) were detected containing tet(A)/tet(X), tet(M)/tet(O), tet(M)/tet(X), tet(O)/tet(X), and tet(Q)/tet(X) simultaneously, respectively. One isolates, R131, unexpectedly contained three tet genes, i.e., tet(M), tet(O), and tet(X). Sequence analysis of the tet gene ORFs cloned from R. anatipestifer isolates confirmed that tet(A), tet(B), tet(M), tet(O), tet(Q) and an unusual mosaic tet gene tet(O/W/32/O) were present in R. anatipestifer. The MIC results of R. anatipestifer ATCC 11845 transconjugants carrying tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) genes exhibited tetracycline resistance with MIC values ranging from 4 to 64 μg/ml. Additionally, the tet(X) gene could transfer into susceptible strain via natural transformation (transformation frequencies of ~10−6). In conclusion, the tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) genes were found and conferred tetracycline resistance in R. anatipestifer isolates. Moreover, the tet(X) is the main mechanism of tetracycline resistance in R. anatipestifer isolates. To our knowledge, this is the first report of tet(A), tet(B), tet(M), tet(O), tet(Q), and mosaic gene tet(O/W/32/O) in R. anatipestifer.
Introduction
Tetracyclines are one of the cheapest broad-spectrum antibacterial agents, with activity against a wide range of host, including aerobic, anaerobic, gram-positive and gram-negative bacteria (Chopra and Roberts, 2001; Roberts, 2003). The antibiotic activity of tetracycline exerts through targeting the 30S ribosome subunit, resulting in inhibition of protein synthesis (Walsh, 2003). Due to overuse of tetracycline antibiotics in clinics, tetracycline-resistant strains and tetracycline resistance genes (tet) have occurred in bacteria with increasing frequency (Chopra and Roberts, 2001; Zhang et al., 2012). The major mechanisms of tetracycline resistance include: active efflux pumps, ribosomal protection and enzymatic modification (Thaker et al., 2010). For most bacteria, the predominant mechanisms of tetracycline resistance are active efflux pumps and ribosomal protection proteins. Currently, there are 59 different genes found in various genera of bacteria conferring tetracycline resistance, including: efflux genes (n = 33), ribosomal protection genes (n = 12) and enzymatic inactivation genes (n = 13) (http://faculty.washington.edu/marilynr/). In addition, an unknown gene tet(U) was reported to be located on the pKq10 plasmid in Enterococcus faecium (Ridenhour et al., 1996).
R. anatipestifer is an avian pathogen found worldwide, primarily infecting domestic ducks, geese, and turkeys, and results in characterized serositis and septicaemia (Ruiz and Sandhu, 2013). Tetracycline is widely used for the prevention and treatment of pathogen infections and the resistance phenomenon was quite serious in the avian industry (Zhang et al., 2012; Zhong et al., 2013). However, little is known about the mechanisms of tetracycline resistance in R. anatipestifer, except that the tet(X) gene was reported to be located on the pRA0511 plasmid in R. anatipestifer (Chen et al., 2010), and the resistance pump gene tet(C) was reported to be the main mechanism of tetracycline resistance in R. anatipestifer isolates (Zhong et al., 2013).
To determine the tetracycline resistance mechanisms of R. anatipestifer, we investigated the tetracycline susceptibility and the prevalence of 12 tet genes of 212 R. anatipestifer field isolates. In this study, we first reported that tet(A), tet(B), tet(O), tet(O/W/32/O), tet(Q) in R. anatipestifer and their resistant function were confirmed by transferring into tetracycline-susceptible R. anatipestifer ATCC 11845.
Materials and Methods
Bacteria, Plasmids, and Growth Conditions
The bacteria and plasmids used in this study are listed in Table S1. The R. anatipestifer field isolates were isolated from 58 large-scale duck farms in different regions of China between 2011 and 2017, including Sichuan, Jiangsu, Guizhou, Anhui, Guangdong, Chongqing, Guangxi, Hainan, Jiling, Henan and Beijing provinces. Under sterile conditions, the brains, hearts or livers were collected from infected or died ducks, and samples were streak-inoculated on tryptic soybean agar plates (TSA, Oxoid Ltd, Basingstoke, Hampshire, England) supplementary with 10% sheep-blood. A single colony from one duck was purified and cultured repeatedly. A total of 212 isolates were identified as R. anatipestifer by PCR amplifying 16S rRNA and sequencing and biochemical analyses. R. anatipestifer strains were cultured at 37°C in GC broth (GCB) or on GC agar (GCA) plates (Liu et al., 2017) when prepared for natural transformation, and were grown at 37°C in tryptic soybean broth (TSB; Oxoid Ltd, Basingstoke, Hampshire, England) or on TSA plates when prepared for susceptibility testing. Escherichia coli (E. coli) strains DH5α and S17-1 were grown at 37°C in Luria-Bertani (LB; Oxoid Ltd, Basingstoke, Hampshire, England) broth or on LB agar. When required, antibiotics were added as follows: 5 μg/ml tetracycline (TET); 2 μg/ml cefoxitin (FOX); 40 μg/ml kanamycin (KAN) or 100 μg/ml ampicillin (AMP). All antibiotics were obtained from Dalian Meilun Biotech Co., Ltd. (Dalian, China).
Tetracycline Susceptibility Testing
The 212 isolates were tested for tetracycline susceptibility in TSB. The micro-dilution method was used to determine the minimal inhibitory concentration (MIC) in 96-well microtiter-plates (Corning, NY, USA) according to Clinical and Laboratory Standards Institute (CLSI) guideline specific for bacteria isolated from animals (CLSI, 2013). The final concentrations of tetracycline ranged from 0.25 to 512 μg/ml. The concentration of the bacterial inoculum was about 106 CFU/ml (100 μl/well). The inoculated micro-plates were incubated at 37°C for 24 h. E. coli ATCC 25922 was used as a quality control strain. The experiments were repeated at least three times. Due to the lack of CLSI-approved tetracycline breakpoints applicable to R. anatipestifer (Nhung et al., 2017), and moreover, the tetracycline MIC of reference strain R. anatipestifer ATCC 11845 was only 0.25 μg/ml, the R. anatipestifer isolates were tentatively classified as resistance to the tetracycline on the basis of the CLSI-approved criteria for Streptococcus spp. (CLSI, 2016), i.e., strains with an MIC of tetracycline of ≤ 1 μg/ml were considered susceptible, 2 μg/ml as intermediate, and ≥4 μg/ml as resistant.
Detection of the tet Genes in R. anatipestifer Field Isolates
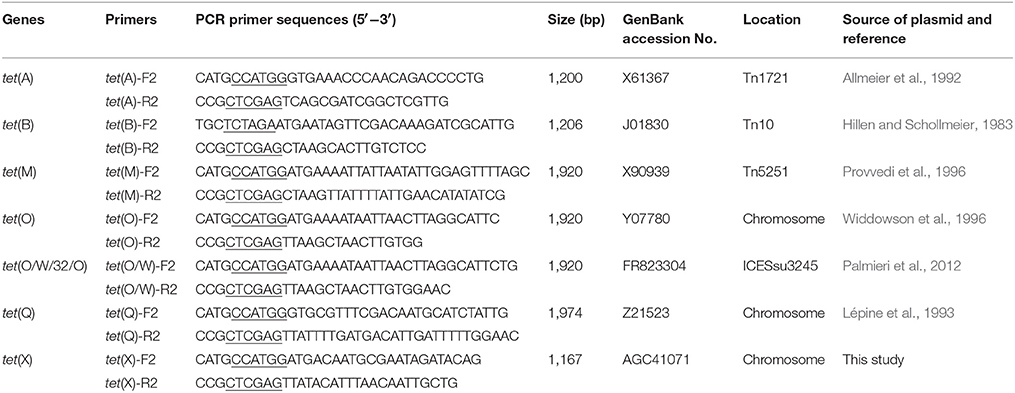
The PCR templates were prepared by treating the bacteria with lysis buffer (1% Triton, 20 mM Tris-HCl, pH 8.0; 2 mM EDTA, pH 8.0) and boiled for 10 min. The presence of the tetracycline resistance genes (tet) characterized previously in other bacteria were determined in all R. anatipestifer field isolates by PCR (Ng et al., 2001; Wu et al., 2010). These detected genes included 6 efflux pump genes [tet(A), tet(B), tet(C), tet(E), tet(G), tet(K)], 4 ribosomal protection genes [tet(M), tet(O), tet(Q), tet(W)], enzymatic inactivation gene tet(X), and a mosaic tetracycline resistance gene tet(O/W/32/O). The primers and protocols described previously (Ng et al., 2001; Ghosh et al., 2009; Wu et al., 2010; Koo and Woo, 2011; Palmieri et al., 2012) are listed in Table S2. The R. anatipestifer CH-2 containing tet(X) was used as positive control. Since the absence of available positive controls for other tet genes, all PCR products were size-confirmed by electrophoresis on a 1.5% agarose gel. All amplicons of tet(B), tet(M), tet(O), tet(O//W/32/O), and tet(Q) from R. anatipestifer isolates were verified by sequencing (BGI Tech Solutions Co., Ltd. Shenzhen, China), while only 12 amplicons of tet(A) were sequenced.
Cloning, Sequencing, and Sequence Analysis of tet Gene ORFs
The ORF of tet(X) gene was amplified by PCR from R. anatipestifer CH-2 using the primer sets tet(X)-F2/tet(X)-R2, listed in Table 1. At the same time, the ORFs of tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) gene were also amplified from tet-positive isolates confirmed above using the additional primers that were designed based on the previously described tet sequences in other bacteria (Table 1). The number of amplified ORFs were 6, 2, 5, 2, 1, and 3 for tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) genes, respectively. The purified amplicons were sequenced and digested with restriction enzymes, and then ligated to the same digested shuttle vector pLMF03 (Liu et al., 2016). Then the corrected recombinant plasmids were introduced into E. coli S17-1. These sequences were analyzed by BLAST in NCBI.
Transfer Experiment
The correct recombinant plasmids were transferred into R. anatipestifer ATCC 11845 by conjugative transfer as previously described (Neela et al., 2009; Luo et al., 2015). Briefly, the E. coli S17-1 containing recombinant plasmid was served as donor strain, while the reference strain R. anatipestifer ATCC 11845 was served as recipient strain, which does not carry any tet genes and is sensitive to tetracycline (Wang et al., 2012). Log-phase donor and recipient strains were mixed in 10 mM MgSO4 with 7:3 ratio and filtered through 0.22-μm membrane filter in the conjugation experiment. Finally, the transconjugants were selected on TSA plates supplemented with FOX (2 μg/ml) and KAN (40 μg/ml) and identified by PCR. At the same time, the negative control empty vector pLMF03 was transferred into ATCC 11845, resulting in the transconjugant ATCC 11845 (pLMF03). The MICs of tetracycline for the wild-type and transconjugants were measured as described above.
Transfer of the tet(X) Gene by Natural Transformation
The transferability of the tet(X) gene was verified by natural transformation as described previously (Liu et al., 2017). Briefly, the R. anatipestifer CH-2 genome (0.25, 0.5, 1, 2, and 5 μg) were respectively used as donor DNA and transferred into the recipient strain R. anatipestifer ATCC 11845. The transformants, designated ATCC 11845 [tet(X)], were screened using GCB plates supplemented with TET (5 μg/ml). The insertion of the transferred tet(X) genes was verified by PCR and sequencing. The tetracycline resistance phenotypes of transformant was determined as described above.
Nucleotide Sequence Accession Numbers
The sequences of tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) genes in R. anatipestifer isolates R100, R98, R131, R96, and R159 in this study have been deposited in GenBank under the accession numbers from MF969099 to MF969104, respectively.
Ethics Statements
All animals studies were conducted in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals, National Research Council. The animal-use procedures were approved by the Animal Ethics Committee of the Sichuan Agricultural University (approval No. 2015-017).
Results
Tetracycline Susceptibility of R. anatipestifer Field Isolates
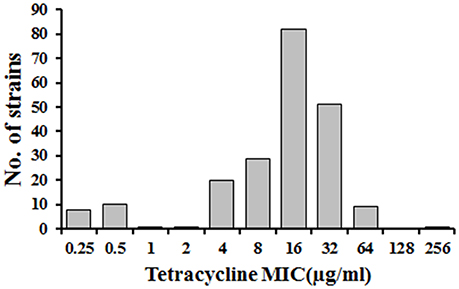
The R. anatipestifer field isolates were resistance to tetracycline by susceptibility test. The MICs of tetracycline in 212 R. anatipestifer field isolates ranged from 0.25 to 256 μg/ml (Figure 1). The results showed that 192 (90.6%) out of 212 tested strains exhibited tetracycline resistance (≥4 μg/ml), while 19 (9.0%) were susceptible (≤ 1 μg/ml). Only one isolate showed intermediate resistance (2 μg/ml). Most importantly, we noticed that 76.4% of isolates exhibited tetracycline resistance with MICs in range of 8–32 μg/ml.
Prevalence of the tet Genes in R. anatipestifer Field Isolates
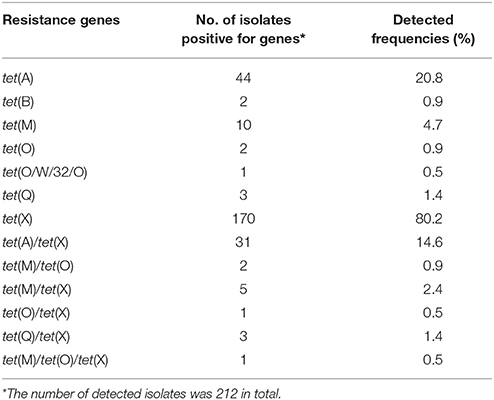
To assess the prevalence of the tet genes in R. anatipestifer, the existence of these genes in 212 R. anatipestifer field isolates was detected by PCR and verified by sequencing in this study. The results showed that among the 12 tet genes, tet(X) was detected in 170 of 212 isolates, exhibiting the highest rate of occurrence among tet genes (80.2%). The detected frequency of other tet genes was as followed: tet(A) (20.8%), tet(M) (4.7%), tet(Q) (1.4%), tet(O) (0.9%), tet(B) (0.9%), and tet(O/W/32/O) (0.5%). However, tet(C), tet(E), tet(G), tet(K), and tet(W) were not detected in any of 212 tested R. anatipestifer field isolates (Table 2). In these positive strains, many isolates contained multiple tet genes simultaneously (Table 2). Namely, tet(X) was detected simultaneously along with tet(A), tet(M), tet(O), and tet(Q) in 31(14.6%), 5 (2.4%), 1 (0.5%), and 3 (1.4%) isolates, respectively. Two isolates (0.9%) carried tet(M) and tet(O) genes simultaneously. Moreover, one isolate (0.5%) had three commensal tet genes, tet(M), tet(O), and tet(X), respectively. The R. anatipestifer field isolates containing one or more tet genes exhibited tetracycline resistance with MICs ranging from 4 to 256 μg/ml, while no tet genes were detected in 19 tetracycline-susceptible isolates and 1 intermediately resistant isolate. Although several isolates carried more than one tet genes in this study, they did not display higher MIC values. Thus, the positive rate of genotype was in line with that of tetracycline resistance phenotype.
ORF Sequences Analysis of the tet Genes in R. anatipestifer
The tetracycline resistance pheotype and genotype of R. anatipestifer isolates selected for ORFs sequencing were listed in Table 3. The ORFs of the tet(A) gene cloned from six different R. anatipestifer isolates shared 99–100% identity (Table S3). The one in R100 isolate (GenBank accession no. MF969099) showed 99% sequence identity with the tet(A) gene from transposon Tn1721 (GenBank accession no. X61367; Allmeier et al., 1992). Two ORFs of tet(B) gene cloned from R. anatipestifer isolates showed 99% identity each other. The one in R98 isolate (GenBank accession no. MF969100) showed 100% identity with tet(B) gene from transposon Tn10 (GenBank accession no. J01830; Hillen and Schollmeier, 1983). The ORFs of tet(M) genes from five different R. anatipestifer isolates shared 99–100% sequence homology. The one in R133 isolate (GenBank accession no. MF969101) exhibited 96 and 95% sequence identity with tet(M) genes from Streptococcus pneumoniae (GenBank accession no. X90939; Provvedi et al., 1996), and Enterococcus faecalis (GenBank accession no. M85225; Su et al., 1992), respectively. Two ORFs of tet(O) genes cloned from R. anatipestifer isolates were 100% identity. The tet(O) gene in R131 isolate (GenBank accession no. MF969102) exhibited 99% sequence identity with the tet(O) genes from Streptococcus mutans DL5 (GenBank accession no. M20925; Leblanc et al., 1988), Campylobacter jejuni (GenBank accession no. M18896; Manavathu et al., 1988), and Streptococcus pneumoniae SA40300 (GenBank accession no. Y07780; Widdowson et al., 1996). The tet(O/W/32/O) from R. anatipestifer isolate R96 (GenBank accession no. MF969103) was a mosaic tetracycline resistance gene derived from tet(O) and tet(W) genes. Sequence alignment result showed that it had 97 and 99% sequence identity with the tet(W/32/O) gene from Bifidobacterium thermophilum (GenBank accession no. AM710601) (van-Hoek et al., 2008) and tet(O/W/32/O) gene from Streptococcus suis integrative conjugative element ICESsu32457 (GenBank accession no. FR823304) (Palmieri et al., 2012; Figure S1), respectively. The ORFs of tet(Q) gene from three different R. anatipestifer isolates shared 100% identity. The tet(Q) gene in R159 isolate (GenBank accession no. MF969104) exhibited 97% sequence identity with the tet(Q) gene from Bacteroides thetaiotaomicron (GenBank accession no.X58717; Nikolich et al., 1992) or Bacteroides fragilis 1126 (GenBank accession no. Z21523; Lépine et al., 1993).
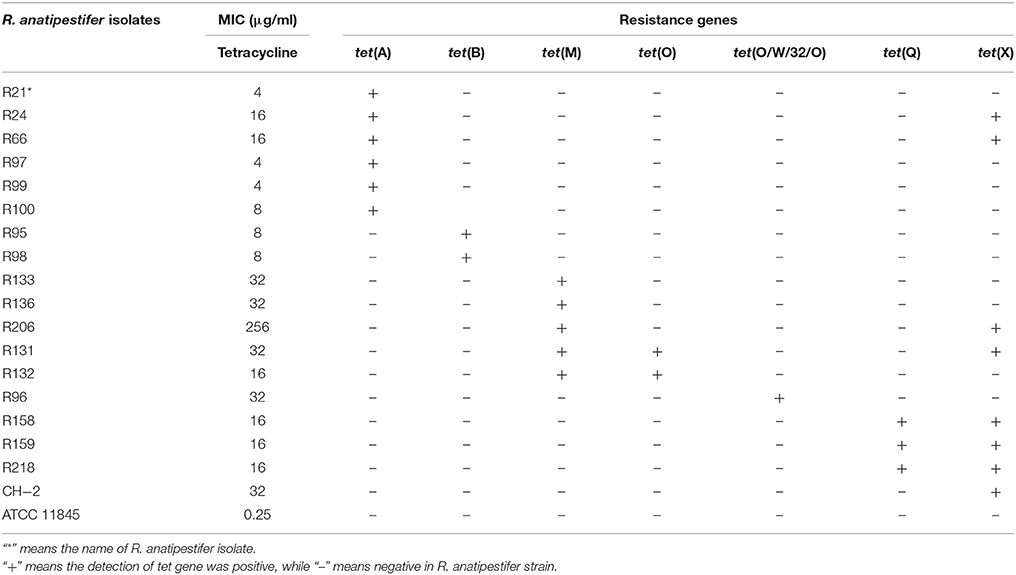
Table 3. The MICs of tetracycline and detected tet genes in R. anatipestifer isolates selected for sequencing ORFs.
Moreover, sequence analysis found that there were two copies of chromosomal tet(X) gene in R. anatipestifer CH-2 (Wang X. et al., 2014), which exhibited 92% sequence identity to the tet(X) gene previously reported on the pRA0511 plasmid in R. anatipestifer (Chen et al., 2010).
MIC of Tetracyclines for R. anatipestifer ATCC 11845 and Other Transconjugants
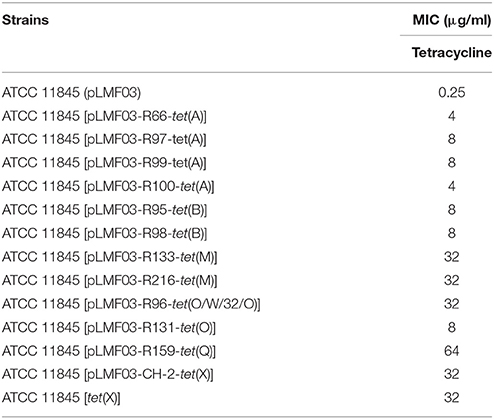
To further verify whether the identified tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) genes were responsible for tetracycline resistance in R. anatipestifer, recombinant plasmids carrying these genes amplified from different R. anatipestifer isolates were transferred into the tetracycline-susceptible R. anatipestifer ATCC 11845 by conjugative transfer. The tetracycline MICs for all transconjugants ranged from 4 to 64 μg/ml (Table 4). The transconjugants containing tet(A), tet(B), or tet(O) genes showed low-level tetracycline resistance, and their MIC values were 4–8 μg/ml. The transconjugants containing tet(M), tet(O/W/32/O), or tet(X) genes exhibited tetracycline resistance of 32 μg/ml. The tetracycline resistance of ATCC 11845 [pLMF03-R159-tet(Q)] was the highest among all transconjugants (the MIC value was 64 μg/ml).
Natural Transformation
To study the transferability of the tet(X) gene, we transferred the tet(X) gene from R. anatipestifer CH-2 to ATCC 11845 by natural transformation. The results showed that the tet(X) gene could be successfully transferred into tetracycline-susceptible ATCC 11845. The maximum transformants were obtained with transformation frequency about ~10−6 for 5 μg R. anatipestifer CH-2 genome. Compared with wild-type strain ATCC11845, the transformant ATCC 11845 [tet(X)] exhibited 128-fold increased tetracycline resistance (from 0.25 to 32 μg/ml; Table 4). The results indicated that the tet(X) gene could confer tetracycline resistance and be easily transferred by natural transformation in R. anatipestifer.
Discussion
Tetracyclines have been widely used for disease treatment and growth promotion in livestock (Roberts, 2003; Cheng et al., 2013). In this study, we investigated the tetracycline resistance and resistant genotypes in R. anatipestifer isolates in China. The results of antimicrobial susceptibility showed that tetracycline resistance in R. anatipestifer was widespread in China between 2011 and 2017, although the usage of this antibiotic treatment was decreased in the avian industry. By PCR detection, the genotypes of tetracycline resistance were abundant in our investigated R. anatipestifer isolates, including efflux genes [tet(A) and tet(B)], ribosomal protection genes [tet(M), tet(O), and tet(Q)], enzymatic gene tet(X), and mosaic tetracycline resistance gene tet(O/W/32/O). The total rate of positive resistance genes was as high as 90.6%. The tet(X) gene had the highest occurrence frequency and was the dominant mechanism conferring tetracycline resistance in R. anatipestifer isolates. Unexpectedly, this conclusion was in contrast to previous reports that ribosomal protection and efflux pumps were the classical tetracycline resistance mechanisms in other bacteria (Thaker et al., 2010). Meanwhile, no tet(C) detected in this study was also in contrast to the previous conclusion that tet(C) was the main mechanism of tetracycline resistance in R. anatipestifer isolates (Zhong et al., 2013). Although the positive rates were much lower than tet(X), tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) genes were detected in R. anatipestifer.
Currently, single plasmid may carry multiple different tet genes or an isolate may contain different tet genes on different plasmids or some tet genes on plasmid and other tet genes in the chromosome (Roberts, 2012). For example, the tet(B) or tet(S) gene was reported to accompany with tet(M) gene (Kim et al., 2004; Roberts, 2012). The E. coli isolates from cows and pigs in slaughterhouse setting carried two different tet genes simultaneously [tet(A)/tet(B), tet(A)/tet(C), and tet(A)/tet(D)] (Cho and Kim, 2008). The R. anatipestifer isolates were found containing two different tet genes [tet(A)/tet(C), and tet(C)/tet(M)]. In this study, multiple tet genes were also found in one R. anatipestifer isolate, such as tet(A)/tet(X), tet(M)/tet(O), tet(M)/tet(X), tet(O)/tet(X), tet(Q)/tet(X), and tet(M)/tet(O)/tet(X). This phenomenon might be due to strong selective pressure and horizontal gene transfer among the various bacteria (Bryan et al., 2004).
The tet genes were located on conjugative, nonconjugative, and mobilizable plasmid, transposons, conjugative transposons, and Salmonella genomic island 1 (Roberts, 2012). In general, the efflux genes are located in chromosome, while the ribosome protection genes are often found on conjugative transposons. These mobile elements can lead to the lateral transfer of tet genes within and between bacteria. As no plasmid had been extracted from R. anatipestifer CH-2 and R. anatipestifer isolates R100, R98, R133, R131, R96, and R159 by several attempts using plasmid Mini kit (OMEGA), the tet(X), tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) genes were most likely located in the chromosome of R. anatipestifer. We confirmed that the tet(X) gene could be transferred by natural transformation, and this transferability might contribute to its wide dissemination in R. anatipestifer.
Finally, through transferring the tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) genes to susceptible strain, we verified that all tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) genes were functional and conferred tetracycline resistance in R. anatipestifer. Interestingly, tet genes detected in this study have high identities with others reported previously, while they exhibited comparatively lower tetracycline resistance in R. anatipestifer (Takahashi et al., 2002; Wang W. et al., 2014). For example, tet(A), tet(B), and tet(O) conferred the tetracycline resistance with the MIC values from 4 to 8 μg/ml. We speculated that the low-level of resistance in R. anatipestifer might result from the low-level of tet gene expression, which needed to be further illustrated.
To the best of our knowledge, this is the first report of the presence of tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), and tet(Q) tetracycline resistance genes in R. anatipestifer. We confirmed that all the tet(A), tet(B), tet(M), tet(O), tet(O/W/32/O), tet(Q), and tet(X) confer the tetracycline resistance in R. anatipestifer. The predominant tetracycline resistance mechanism in R. anatipestifer is conferred by tet(X) gene.
Author Contributions
D-KZ, M-SW, and A-CC conceived and designed the project. H-YL, M-FL, and M-SW performed the tetracycline susceptibility test and prevalence of tet genes in R. anatipestifer isolates. D-KZ, X-XZ, and R-YJ cloned and sequenced the tet genes. SC, K-FS, and QY analyzed the sequences of tet genes. H-YL, YW, and X-YC transferred the shuttle vectors. D-KZ, H-YL, and M-SW performed the natural transformation of tet(X) gene. HL, D-KZ, and A-CC drafted and revised the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under Grant No.31372468; National Science and Technology Support Program under Grant No. 2015BAD12B05; China Agricultural Research System under Grant No. CARS-43-8; Program for International Cooperation Project of Science and Technology Department of Sichuan Province under Grant No. 2017HH0026.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank professor Francis Biville, Département Infection et Epidémiologie, Institut Pasteur, for his helpful suggestions which have improved the quality of this paper.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00585/full#supplementary-material
Figure S1. Sequence alignment of the tet(O/W/32/O) genes using DNAMAN 8.0 software (Lynnon-BioSoft, Ontario, Canada). GenBank accession No. annotation: MF969103, tet(O/W/32/O) from R. anatipestifer isolate R96; FR823304, tet(O/W/32/O) from S. suis integrative conjugative element ICESsu32457; AM710601, tet(W/32/O) from B. thermophilum.
Table S1. The strains and plasmids used in this study.
Table S2. Primers used in this study for PCR detection of the tet genes.
Table S3. The tet gene sequence analyses in this study.
References
Allmeier, H., Cresnar, B., Greck, M., and Schmitt, R. (1992). Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20. doi: 10.1016/0378-1119(92)90597-I
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004
Chen, Y.-P., Tsao, M.-Y., Lee, S.-H., Chou, C.-H., and Tsai, H.-J. (2010). Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol. 39, 333–338. doi: 10.1080/03079457.2010.507761
Cheng, W., Chen, H., Su, C., and Yan, S. (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ. Int. 61, 1–7. doi: 10.1016/j.envint.2013.08.023
Cho, J., and Kim, K. (2008). Antimicrobial resistance and distribution of tetracycline resistance genes of Escherichia coli isolated from human and livestock in slaughterhouse. Korean J. Vet. Public Health. 32, 213–223.
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
CLSI. (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard, 4th Edn. CLSI document VET01-A4. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI. (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement. CLSI document M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
Ghosh, S., Sadowsky, M. J., Roberts, M. C., Gralnick, J. A., and LaPara, T. M. (2009). Sphingobacterium sp. strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 106, 1336–1342. doi: 10.1111/j.1365-2672.2008.04101.x
Hillen, W., and Schollmeier, K. (1983). Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 11, 525–539. doi: 10.1093/nar/11.2.525
Kim, S. R., Nonaka, L., and Suzuki, S. (2004). Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol. Lett. 237, 147–156. doi: 10.1111/j.1574-6968.2004.tb09690.x
Koo, H. J., and Woo, G. J. (2011). Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 145, 407–413. doi: 10.1016/j.ijfoodmicro.2011.01.003
Leblanc, D. J., Lee, L. N., Titmas, B. M., Smith, C. J., and Tenover, F. C. (1988). Nucleotide sequence analysis of tetracycline resistance gene tetO from Streptococcus mutans DL5. J. Bacteriol. 170, 3618–3626. doi: 10.1128/jb.170.8.3618-3626.1988
Lépine, G., Lacroix, J. M., Walker, C. B., and Progulskefox, A. (1993). Sequencing of a tet(Q) gene isolated from Bacteroides fragilis 1126. Antimicrob. Agents Chemother. 37:2037. doi: 10.1128/AAC.37.9.2037
Liu, M. F., Wang, M. Y., Zhu, D. K., Wang, M. S., Jia, R. Y., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. doi: 10.1038/srep37159
Liu, M., Zhang, L., Huang, L., Biville, F., Zhu, D., Wang, M., et al. (2017). Natural transformation and use of it to establish an easy knockout method in Riemerella anatipestifer. Appl. Environ. Microbiol. 83:e00127-17. doi: 10.1128/AEM.00127-17
Luo, H., Liu, M., Wang, L., Zhou, W., Wang, M., Cheng, A., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. doi: 10.1080/03079457.2015.1019828
Manavathu, E. K., Hiratsuka, K., and Taylor, D. E. (1988). Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene 62, 17–26. doi: 10.1016/0378-1119(88)90576-8
Neela, F. A., Nonaka, L., Rahman, M. H., and Suzuki, S. (2009). Transfer of the chromosomally encoded tetracycline resistance gene tet(M) from marine bacteria to Escherichia coli and Enterococcus faecalis. World J. Microbiol. Biotechnol. 25, 1095–1101. doi: 10.1007/s11274-009-0004-8
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215. doi: 10.1006/mcpr.2001.0363
Nhung, N. T., Chansiripornchai, N., and Carrique-Mas, J. J. (2017). Antimicrobial resistance in bacterial poultry pathogens: a review. Front. Vet. Sci. 4:126. doi: 10.3389/fvets.2017.00126
Nikolich, M. P., Shoemaker, N. B., and Salyers, A. A. (1992). A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob. Agents Chemother. 36, 1005–1012. doi: 10.1128/AAC.36.5.1005
Palmieri, C., Magi, G., Mingoia, M., Bagnarelli, P., Ripa, S., Varaldo, P. E., et al. (2012). Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob. Agents Chemother. 56, 4697–4702. doi: 10.1128/AAC.00629-12
Provvedi, R., Manganelli, R., and Pozzi, G. (1996). Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 135, 231-236. doi: 10.1111/j.1574-6968.1996.tb07994.x
Ridenhour, M. B., Fletcher, H. M., Mortensen, J. E., and Daneo-Moore, L. (1996). A novel tetracycline-resistant determinant, tet(U), is encoded on the plasmid pKq10 in Enterococcus faecium. Plasmid 35, 71–80. doi: 10.1006/plas.1996.0009
Roberts, M. C. (2003). Tetracycline therapy: update. Clin. Infect. Dis. 36, 462–467. doi: 10.1086/367622
Roberts, M. C. (2012). Acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 543–568. doi: 10.1007/978-1-4614-1400-1_16
Ruiz, J., and Sandhu, T. S. (2013). “Riemerella anatipestifer infection,” in Diseases of Poultry, 13th Edn., eds Swayne D. E., Glisson J. R., McDougald L. R., Nolan L. K., Suarez D. L., Nair V. L (Hoboken, NJ: John Wiley and Sons, Inc.), 823–828.
Su, Y. A., He, P., and Clewell, D. B. (1992). Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36, 769–778. doi: 10.1128/AAC.36.4.769
Takahashi, H., Watanabe, H., Kuroki, T., Watanabe, Y., and Yamai, S. (2002). Identification of tet(B), encoding high-level tetracycline resistance, in Neisseria meningitidis. Antimicrob. Agents Chemother. 46, 4045–4046. doi: 10.1128/AAC.46.12.4045-4046.2002
Thaker, M., Spanogiannopoulos, P., and Wright, G. D. (2010). The tetracycline resistome. Cell. Mol. Life Sci. 67, 419–431. doi: 10.1007/s00018-009-0172-6
van-Hoek, A., Mayrhofer, S., Domig, K., Florez, A., Ammor, M., Mayo, B., et al. (2008). Mosaic tetracycline resistance genes and their flanking regions in Bifidobacterium thermophilum and Lactobacillus johnsonii. Antimicrob. Agents Chemother. 52, 248–252. doi: 10.1128/AAC.00714-07
Wang, W., Guo, Q., Xu, X., Sheng, Z. K., Ye, X., and Wang, M. (2014). High-level tetracycline resistance mediated by efflux pumps Tet(A) and Tet(A)-1 with two start codons. J. Med. Microbiol. 63 (Pt 11), 1454–1459. doi: 10.1099/jmm.0.078063-0
Wang, X., Liu, W., Zhu, D., Yang, L., Liu, M., Yin, S., et al. (2014). Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:479. doi: 10.1186/1471-2164-15-479
Wang, X., Zhu, D., Wang, M., Cheng, A., Jia, R., Zhou, Y., et al. (2012). Complete genome sequence of Riemerella anatipestifer reference strain. J. Bacteriol. 194, 3270–3271. doi: 10.1128/JB.00366-12
Widdowson, C. A., Klugman, K. P., and Hanslo, D. (1996). Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40, 2891–2893.
Wu, N., Qiao, M., Zhang, B., Cheng, W. D., and Zhu, Y. G. (2010). Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 44, 6933–6939. doi: 10.1021/es1007802
Zhang, T., Wang, C. G., Lv, J. C., Wang, R. S., and Zhong, X. H. (2012). Survey on tetracycline resistance and antibiotic-resistant genotype of avian Escherichia coli in North China. Poult. Sci. 91, 2774–2777. doi: 10.3382/ps.2012-02453
Keywords: Riemerella anatipestifer, tetracycline resistance, resistance gene, PCR, mosaic gene
Citation: Zhu D-K, Luo H-Y, Liu M-F, Zhao X-X, Jia R-Y, Chen S, Sun K-F, Yang Q, Wu Y, Chen X-Y, Cheng A-C and Wang M-S (2018) Various Profiles of tet Genes Addition to tet(X) in Riemerella anatipestifer Isolates From Ducks in China. Front. Microbiol. 9:585. doi: 10.3389/fmicb.2018.00585
Received: 03 July 2017; Accepted: 14 March 2018;
Published: 27 March 2018.
Edited by:
Satoru Suzuki, Ehime University, JapanReviewed by:
Yoshikazu Ishii, Toho University, JapanRahman Md. Habiur, University of Rajshahi, Bangladesh
Copyright © 2018 Zhu, Luo, Liu, Zhao, Jia, Chen, Sun, Yang, Wu, Chen, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: An-Chun Cheng, chenganchun@vip.163.com
Ming-Shu Wang, mshwang@163.com
†These authors have contributed equally to this work.
 De-Kang Zhu
De-Kang Zhu Hong-Yan Luo1,2†
Hong-Yan Luo1,2† Ma-Feng Liu
Ma-Feng Liu Xin-Xin Zhao
Xin-Xin Zhao An-Chun Cheng
An-Chun Cheng