- 1State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 2The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 3State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
Ascospores act as the primary inoculum of Fusarium graminearum, which causes the destructive disease Fusarium head blight (FHB), or scab. MicroRNAs (miRNAs) have been reported in the F. graminearum vegetative stage, and Fgdcl2 is involved in microRNA-like RNA (milRNA) biogenesis but has no major impact on vegetative growth, abiotic stress or pathogenesis. In the present study, we found that ascospore discharge was decreased in the Fgdcl1 deletion mutant, and completely blocked in the double-deletion mutant of Fgdcl1 and Fgdcl2. Besides, more immature asci were observed in the double-deletion mutant. Interestingly, the up-regulated differentially expressed genes (DEGs) common to ΔFgdcl1 and ΔFgdcl1/2 were related to ion transmembrane transporter and membrane components. The combination of small RNA and transcriptome sequencing with bioinformatics analysis predicted 143 novel milRNAs in wild-type perithecia, and 138 of these milRNAs partly or absolutely depended on Fgdcl1, while only 5 novel milRNAs were still obtained in the Fgdcl1 and Fgdcl2 double-deletion mutant. Furthermore, 117 potential target genes were predicted. Overall, Fgdcl1 and Fgdcl2 genes were partly functionally redundant in ascospore discharge and perithecium-specific milRNA generation in F. graminearum, and these perithecium-specific milRNAs play potential roles in sexual development.
Introduction
Fusarium graminearum (teleomorph Gibberella zeae) is one of the major pathogens that cause Fusarium head blight (FHB) in barley, maize and other grains (Bai and Shaner, 2004; Goswami and Kistler, 2004). Ascospores act as the primary inoculum of plant infections by releasing from perithecia and then falling on flowering spikelets (Trail et al., 2002). Therefore, sexual reproduction and ascospore discharge play a critical role in head blight epidemiology. In sexual reproduction, in addition to MAT genes, ion transport proteins (such as potassium, chloride, and calcium icon channels) and certain transcription factors are associated with perithecium formation and discharge mechanism (Hallen et al., 2007; Kim et al., 2015). Interestingly, Fgago2, which is related to the RNA interference pathway, is associated with meiosis and subsequent developmental pathways in F. graminearum (Kim et al., 2015). In Neurospora crassa, dcl-1, an RNA silencing regulator, also effected sexual development at early stage (Alexander et al., 2008). The RNAi component mutants of these ascomycetes affect sexual development, suggesting that certain endogenous short RNAs (esRNAs) play regulatory roles during sexual development.
Recently, several types of esRNAs have been described in fungi, such as exonic-small interfering RNAs (ex-siRNAs) in Mucor circinelloides and F. graminearum, tRNA-derived fragments (tRFs) in Magnaporthe oryzae, heterochromatin-derived short-interfering RNAs (siRNAs) in Schizosaccharomyces pombe, QDE-2-interacting sRNAs (qiRNAs), and microRNA-like RNAs (milRNAs) in N. crassa (Volpe et al., 2002; Lee et al., 2009, 2010; Nicolas et al., 2010; Nunes et al., 2011; Son et al., 2017). MicroRNAs (miRNAs), the most common example of esRNAs in plants and animals, also have been detected in several fungal organisms, such as Sclerotinia sclerotiorum, Cryptococcus neoformans, Trichoderma reesei, Fusarium oxysporum, F. graminearum, Penicillium marneffei, and Penicillium chrysogenum (Jiang et al., 2012; Zhou et al., 2012; Kang et al., 2013; Lau et al., 2013; Chen et al., 2014, 2015; Dahlmann and Kuck, 2015). miRNAs are produced by a canonical Dicer-dependent biogenesis pathway and control gene expression at the post-transcriptional level in plants and animals (Ambros et al., 2003). In N. crassa, Dicer, Argonaute, RNA-dependent RNA polymerase (RdRp), the RNase III domain-containing protein MRPL3, and RNA exosomes play important roles in milRNA biogenesis pathways (Lee et al., 2010; Xue et al., 2012). Animal miRNAs recognize their targets by the miRNA seed region; nevertheless, plant miRNAs nearly perfectly matched to their targets (Rhoades et al., 2002; Lewis et al., 2003). Although miRNAs have been found in various fungi, there is little information on their function and target recognition.
Fusarium graminearum, similar to major ascomycete fungi, contains two Dicer-like and two Argonaute-like protein coding genes (Nakayashiki, 2005). In the F. graminearum asexual stage, the RNAi components are not involved in vegetative development or pathogenesis (Chen et al., 2015). In stark contrast, Fgdcl1 and Fgago2 affect ascospore formation during the sexual phase (Son et al., 2017). Indeed, we speculated that certain sex-induced milRNAs exist in perithecium and play regulatory functions in sexual reproduction. To gain insights into these milRNAs, we predicted the wild-type strain PH-1 milRNAs before (0 dps) and after (7 dps) self-crossing and compared the perithecium-specific milRNAs of the Fgdcl-1 (FGRRES_09025) and Fgdcl1/2 (the gene id of Fgdcl2 is FGRRES_04408) deletion mutants with those in PH-1 via deep sequencing and bioinformatics analysis. To explore the function of milRNA, we also predicted and analyzed milRNA target genes.
Materials and Methods
Fungal Strains and Culture Conditions
The F. graminearum strain PH-1 (NRRL 31084) (Trail and Common, 2000) was used as the wild-type strain in the present study, and mutant strains were derived from the wild-type strain. All strains in this study were cultured on potato dextrose agar (PDA) plates at 25°C and stored in PDA slants at 4°C.
To characterize the biological properties of asexual stage, colony morphology, hyphal extensions, conidial production, and conidial germination were examined. The hyphal extension rate (mm day-1) was measured on PDA, complete medium (CM) and minimal medium (MM) (Correll et al., 1987) at 25°C between 1 and 3 days using three replicate plates per strain. Colony morphology was observed on PDA media at 25°C for 3 days. To assay the sensibility of various stresses, one mycelial plug was centrally inoculated in PDA medium containing 1 M glucose, 1 M sorbitol, 1 M NaCl, 1 M KCl, 0.5 M CaCl2, 0.02% SDS, 0.05% H2O2, or 0.2 mg/ml Congo red. For conidiation assays, one mycelial plug was inoculated into 20 ml of carboxymethyl cellulose (CMC) (1 g NH4NO3, 1 g KH2PO3, 0.5 g MgSO4.7H2O, 1 g yeast extract, 15 g CMC, and 1 L of water) liquid medium at 25°C for 5 days in a shaker (200 rpm), and the number of conidia was assessed using a hemocytometer (Cappellini and Peterson, 1965). For conidial germination, conidia were harvested and then inoculated into YEPD (0.3% yeast extract, 1% peptone, and 2% dextrose) liquid medium at 25°C in a shaker (200 rpm), and then conidial germination was observed and counted after 4 and 6 h of incubation.
Sexual Development and Ascospore Discharge
With certain modifications, the sexual development and ascospore discharge assays in F. graminearum were performed as previously described (Leslie et al., 2006; Cavinder et al., 2012). Briefly, one mycelial plug was inoculated onto carrot plats at 25°C for 7 days and then removed with the back of a surgical blade and added to 600 μl of 2.5% Tween 60 solution to induce sexual development. The sample was continuously incubated under near-UV light (wavelength, 365 nm; beauty bright Lighting Electrical Appliance Co., Ltd., Zhongshan, China) at 23°C. After induction for 7 days induction, perithecia and asci were observed under dissecting and light microscopes, respectively. The glycogen staining of asci was performed as previously reported (da R Campos and Costa, 2010). Ascospore discharge was detected in a coverslip and place in a closed box. A semicircular agar block (10 mm in diameter) covered with mature perithecia was placed on a coverslip for 24 h, and the images were captured on camera (Trail et al., 2005; Min et al., 2010; Cavinder et al., 2012).
Plant Infection Assays
Infection assays on flowering wheat heads were performed as previously described (Hou et al., 2002). To this end, 10 μl of 1 × 105 conidia/ml conidia suspension, collect from 5-day-old CMC cultures, was inoculated onto the fifth flowering spikelet of Zhengmai 9023. The symptomatic spikelets were examined and images were captured at 20 days after inoculation. Corn silk infection assay was performed according to Seong et al. (2005). Corn silks were inoculated with mycelia plugs (diameter = 6 mm) and counted at 6 dpi.
RNA Extraction and Quantitative Real Time (qRT)-PCR
In the asexual stage, the RNA samples were isolated from 24-h YEPD cultures (mycelium) and 24-h CMC cultures (sporulation). During sexual development, the RNA samples were collected from 7-day carrot plat hyphae (0 day post self-crossing, 0 dps) and 3-, 5-, 7-day perithecia (3, 5, and 7 days post self-crossing). Total RNA was isolated with the RNAiso Plus regent (Takara, Shiga, Japan) according to the manufacturer’s protocols. Potential DNA contamination was removed, and first-strand cDNA was synthesized by the EasyScript One-step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech, Beijing, China) according to the manufacturer’s instructions.
The gene transcript levels were assessed by quantitative real-time reverse transcriptase PCR (qRT-PCR). qRT-PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States) with the iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, United States USA). The F. graminearum actin gene was used as the internal control. The gene expression was calculated with the 2-ΔΔCt method, and the mean and standard deviation were calculated from three biological replicates (Livak and Schmittgen, 2001). The gene-specific primer pairs for qRT-PCR are listed in Supplementary Table S1.
DNA Manipulation and Fungal Transformations
The strategy for gene deletion is illustrated in Supplementary Figure S1 according to a split-marker system (see Supplementary Materials) (Catlett et al., 2003). PCR products for targeted gene deletion were constructed by a slightly modified double-joint (DJ) PCR. First, the upstream and the downstream fragments of Fgdcl2 were amplified from genomic DNA using the primer pair dcl2-1F1/1R and dcl2-2F/2R1, respectively (Supplementary Table S1). Next, the hygromycin resistance gene (HPH) was amplified from PUCH18 using the primer pair HYG-F/HYG-R (Supplementary Table S1; Yang et al., 2018). The three amplicons (the upstream fragment, the downstream fragment, and HPH) were fused after a second round of DJ-PCR. Finally, two DNA fragments were amplified from the second-round product with the primer pairs dc12-1F1/HY-R and YG-F/dcl2-2R1 (Supplementary Table S1). These two DNA fragments were subsequently transformed into protoplasts of wild-type strain PH-1 according to a previously published protocol (Kim et al., 2006; Han et al., 2007). For Fgdcl1 and Fgdcl2 double deletion, the geneticin resistance cassette (gen) was used as a selectable marker. Transformations were selected by using the PDA plates amended with 225 μg ml-1 hygromycin B (Sigma-Aldrich) or 300 μg ml-1 geneticin (Sigma-Aldrich). Transformants were purified by single conidium isolation and stored in 20% glycerol at -80°C.
For Southern blotting, the genomic DNAs of PH-1 and mutants were extracted according to the Fusarium laboratory manual and then digested with XbaI (Leslie et al., 2006). Southern blotting was performed using the Amersham AlkPhos Direct Labeling and Detection System (GE Healthcare, Little Chalfont, United Kingdom). The probe primers are listed in Supplementary Table S1 and the probes were labeled with alkaline phosphatase.
RNA Isolation and Library Construction
Mature perithecia (7 days post self-crossing) of the wild-type strain PH-1 and mutants (ΔFgdcl1 and ΔFgdcl1/2) were isolated to construct sexual stage small RNA and cDNA libraries. The mycelia of PH-1 were inoculated onto carrot plate for 7 days and collected as 0 day post self-crossing (0 dps) sample. Total RNA was extracted using the RNAiso Plus regent (Takara, Shiga, Japan) according to the manufacturer’s instructions and then treated with RNase-free DNase I. RNA quality was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States) and Agilent 2100 Bioanalyzer (Agilent, United States). Small RNAs (15∼30 nt) were extracted from total RNA on a 15% denaturing polyacrylamide gel and ligated to specific 5′ adaptor and 3′ adaptor samples. After reverse transcription, the cDNA libraries were sequenced on an Illumina HiSeq 2000 platform (BGI, Shenzhen, China). For each strain, one small RNA library and one transcriptome library were constructed. For each strain, two biological replicates were used. The RNA-seq and sRNA-seq data were deposited in the NCBI Sequence Read Archive database with accession codes SRP132245 and SRP131559, respectively.
Data Analysis of Small RNA and Transcriptome
The small RNA analysis and microRNA prediction were performed as previously reported (Zhou et al., 2012). After the removal of poor-quality reads and adaptor sequences, clean reads were obtained with the FASTX toolkit. Small sequence reads were completely mapped to the genome of PH-1 from the Ensembl Fungi database using bowtie (King et al., 2015). After removal of known non-coding RNAs (rRNA, tRNA, snRNA, and snoRNA) by BLAST or bowtie, the unannotated small RNAs were used for novel miRNA prediction by using MIREAP (Li et al., 2012). The raw abundance of milRNAs was normalized according to transcripts per million (TPM) normalization, where the n_base is 1,000,000 (Jeong et al., 2010; Belli Kullan et al., 2015). PsRobot (Wu et al., 2012) and TargetFinder (Lavorgna et al., 1999) were used for the prediction of miRNA according to plant-like target interactions, and based on the methods described by Allen with slightly modified: (1) less than four mismatches in total and no more than two adjacent mismatches; (2) In positions 1–12, no more than 2.5 mismatches; (3) after 17 position, one gap or bulge was permitted.
The transcriptome analysis in F. graminearum was performed as previously reported (Cao et al., 2016). The RNA-seq reads were mapped to the genome of F. graminearum strain PH-1 with Tophat2, and then the transcript levels were calculated by FeatureCounts (Kim et al., 2013; Liao et al., 2014). Differential expression analysis of genes was performed with the edgeRun package (Dimont et al., 2015). Genes with a false discovery rate (FDR) below 0.05 were considered differentially expressed. Gene ontology (GO) functions were enriched according to Young et al. (2010) reported, the p-value was calculated and subjected to Benjamini-Hochberg correction, with adjusted p-values ≤ 0.05 as a threshold. In addition, we summarized and visualized the GO terms employing REVIGO (Bauer et al., 2008; Supek et al., 2011).
Results
Fgdcl1 Partly Cooperates With Fgdcl2 in Ascospore Discharge
A previous study has shown that the RNAi components not involved in vegetative development and pathogenesis in F. graminearum (Chen et al., 2015). Therefore, we analyzed the expression patterns of Fgdcl genes in asexual and sexual stage. The results showed that the expression level of Fgdcl1 in sporulation was higher than those in mycelium (Figure 1A), similar to previous research (Chen et al., 2015). Meanwhile, compared to 0 dps (0 day post self-crossing), the expression level of Fgdcl1 was markedly increased after sexual induction for 3, 5, and 7 days, especially at 7 dps (Figure 1B). To explore whether Fgdcl plays a physiological role in sexual development, we constructed the gene deletion mutants ΔFgdcl1, ΔFgdcl2, and ΔFgdcl1/2. These mutants were confirmed by PCR amplification and Southern blotting (Supplementary Figure S1). In the asexual stage, no significant changes in hyphal growth, conidiation, conidia morphology, or virulence and under various environmental stress conditions were observed (Supplementary Figures S2, S3). During sexual development, normal-shaped perithecia were obtained from these mutants at 7 days after fertilization. In contrast to the perithecia in PH-1 and ΔFgdcl2, few cirrhi were produced in ΔFgdcl1, and ΔFgdcl1/2 (Figure 2A). The discharged ascospores were decreased in ΔFgdcl1 and almost abolished in ΔFgdcl1/2, compared with those in PH-1 (Figure 2B). The dissection of perithecia showed that less than 20% perithecia had normal asci (type I) containing eight spindle-shaped ascospores in ΔFgdcl1 and ΔFgdcl1/2, while 63% and 45% type I asci were observed in PH-1 and ΔFgdcl2, respectively (Figure 2C). In ΔFgdcl1 and ΔFgdcl1/2, most of the asci were defective, such as abnormal asci with fewer ascospores (type II) and smaller asci (type III) (Figure 2C). Indeed, the ascospore discharges was partly decreased in ΔFgdcl1 and arrested in ΔFgdcl1/2 due to the defective asci. Moreover, similar results were obtained from other deletion mutants (Supplementary Figure S4). Taken together, these findings reveal that Fgdcl1 plays a more important role in sexual reproduction than in the asexual stage and that Fgdcl1 partly cooperates with Fgdcl2 in ascospore discharge.
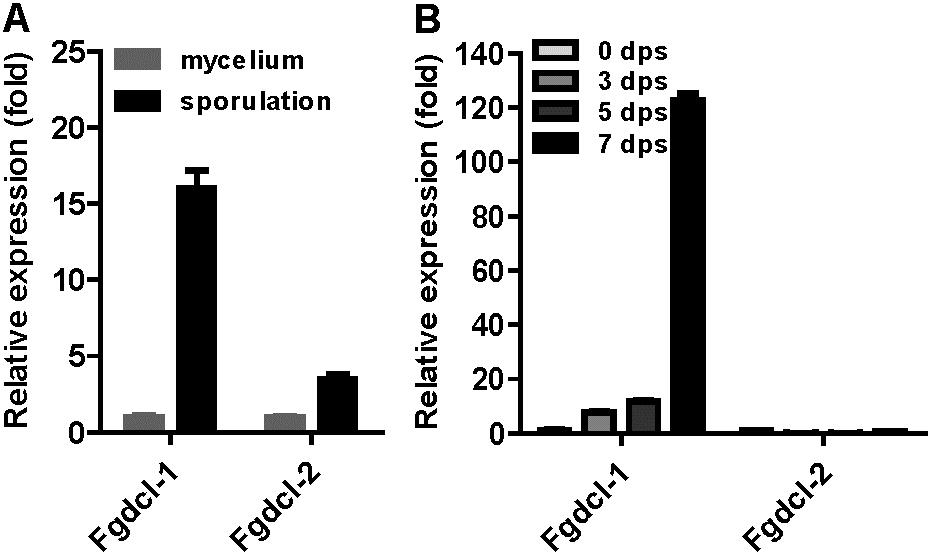
FIGURE 1. The expression patterns of Fgdcl gene in asexual and sexual stages. (A) The expression patterns of Fgdcl gene in asexual stages. In asexual stage, cultures were collected from YEPD and carboxymethyl cellulose (CMC) liquid medium after 24 h inoculation as mycelium and sporulation samples, respectively. The expression of Fgdcl genes in mycelium was set as the control. (B) The expression patterns of Fgdcl gene in sexual stage. The perithecium was collected from carrot plate after 3, 5, and 7 days of self-crossing. The mycelium collected from carrot plate for 7-day inoculation was considered as the 0 day post self-crossing (0 dps) sample. The expression of Fgdcl genes at 0 dps was set as the control. The relative mRNA expression of the Fgdcl genes was determined by quantitative RT-PCR. Bars indicate standard the deviation from three repeated experiments.
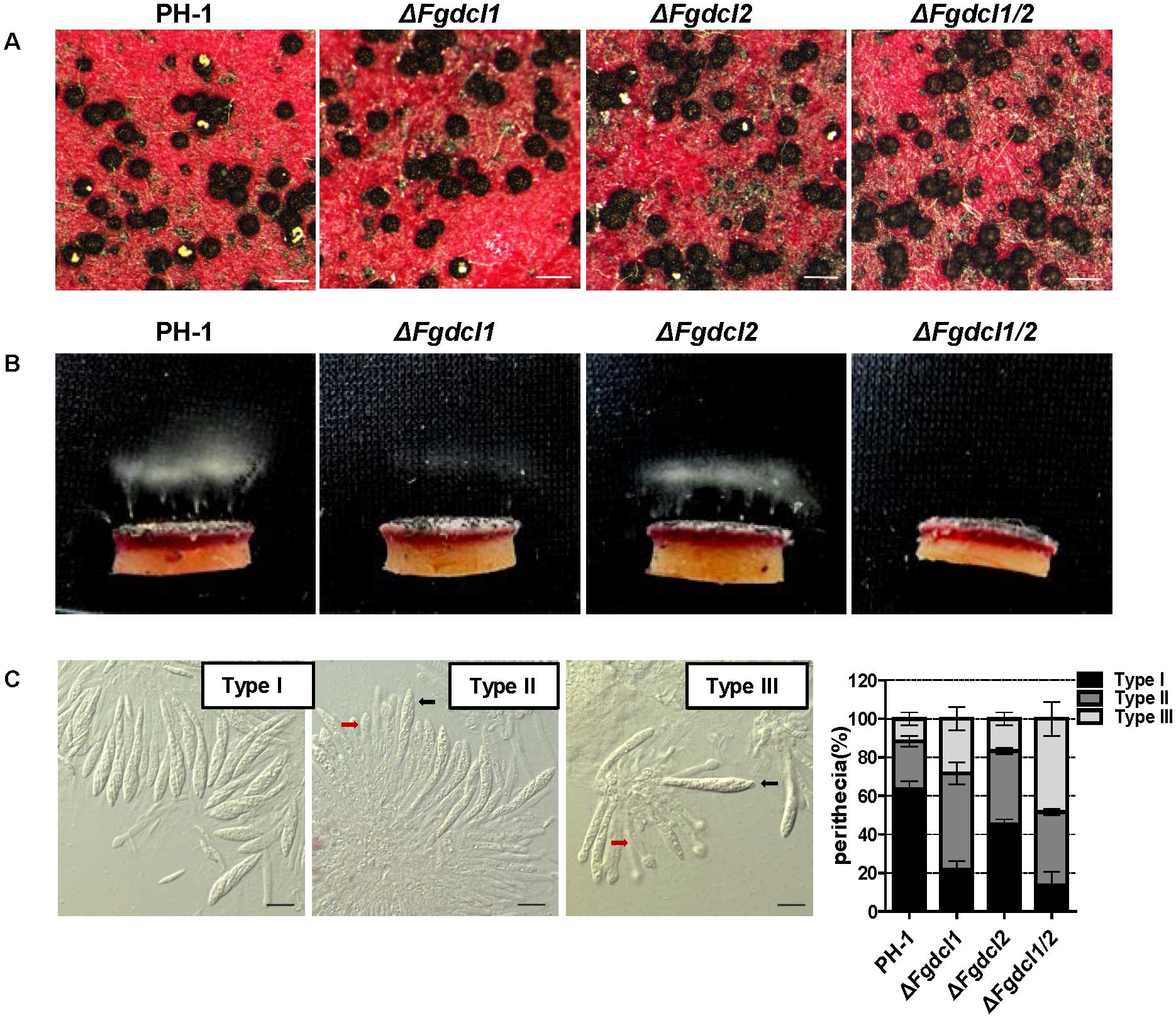
FIGURE 2. Comparison of the sexual development among the wild-type strain PH-1, ΔFgdcl1, ΔFgdcl2 and ΔFgdcl1/2. (A) Comparison of the cirrhi of wild-type strain PH-1, ΔFgdcl1, ΔFgdcl2 and ΔFgdcl1/2. Each strain was inoculated on carrot medium, and the images were captured at 12 days post self-crossing (dps). The yellow cirrhi was the overflowed ascospores from the ostiole of perithecia. Scale bar = 200 μm. (B) Ascospore discharge was assayed with 7-day-old perithecia. The white cloud is the accumulation of discharged ascospores. The images were captured after incubation for 20 h. (C) Morphology and statistical analysis of asci rosettes of 7-day-old perithecia. The morphology of type I asci rosettes was selected from the wild-type strain PH-1; type II and type III were selected from ΔFgdcl1/2. The black and red arrows indicate the normal and defective asci, respectively. Bar = 20 μm. The statistical analysis of ascus morphology was conducted after 7 days post self-crossing. A total of 50 asci rosettes were observed and classified in each strain with light microscopy. Line bars indicate standard errors from three repeated experiments.
The Common Up-Regulated DEGs in ΔFgdcl1 and ΔFgdcl1/2 Involved in Ascospore Discharge
To identify the genes regulated by Fgdcl1 and investigate the molecular mechanisms underlying ascospore discharge, the transcriptomes at 7 dps perithecia of the wild-type strain PH-1, as well as ΔFgdcl1 and ΔFgdcl1/2 were obtained using the Illumina HiSeqTM 2000 platform (BGI, Shenzhen, China), and the reads were mapped to F. graminearum genome (Supplementary Table S2). Genes with a FDR of less than 0.05 and a fold-change greater than 2 compared with the wild-type strain PH-1 were considered differentially expressed genes (DEGs). In total, 846 DEGs were identified in ΔFgdcl1, including 376 down-regulated and 470 up-regulated genes (Figure 3A and Supplementary Table S3). More DEGs (2372 genes) were obtained in ΔFgdcl1/2, including 1170 down-regulated and 1202 up-regulated genes (Figure 3A and Supplementary Table S3). Additionally, the common up-regulated DEGs in ΔFgdcl1 and ΔFgdcl1/2 showed a mild positive correlation (R2 = 0.6119), but this was not the case in common down-regulated DEGs, suggesting that the common up-regulated DEGs between ΔFgdcl1 and ΔFgdcl1/2 may share certain pathways related to ascospore discharge (Figure 3B). The GO annotation of 351 common up-regulated DEGs showed that 11 GO terms were enriched (adjusted p-value less than 0.05), such as “nucleoside metabolic” and “glycosyl compound metabolic process” in biological process; “membrane” in cellular component; and “sodium ion transmembrane transporter activity,” “symporter activity,” “galactose oxidase activity,” and “oxidoreductase activity” in molecular function (Figure 3C). Moreover, the network of GO terms indicated that the up-regulated DEGs were involved in transporter activity, nucleoside metabolism, transmembrane transport and membrane components (Supplementary Figure S5). Furthermore, certain enriched GO terms provided useful information for sexual development and ascospore discharge, including galactose oxidase activity, sodium ion trans membrane transporter activity, and solute: cation symporter activity, which may relate to the buildup of turgor pressure, an important process for ascospore discharge (Trail et al., 2002). The expression patterns of discharge-related terms gene were verified by qRT-PCR, and the results were consistent with the sequencing results (Supplementary Figure S6A). In addition, no significant terms were enriched in the down-regulated DEGs. Taken together, these results suggested that the common up-regulated DEGs of ΔFgdcl1 and ΔFgdcl1/2 may have a relationship with defective asci and ascospore discharge.
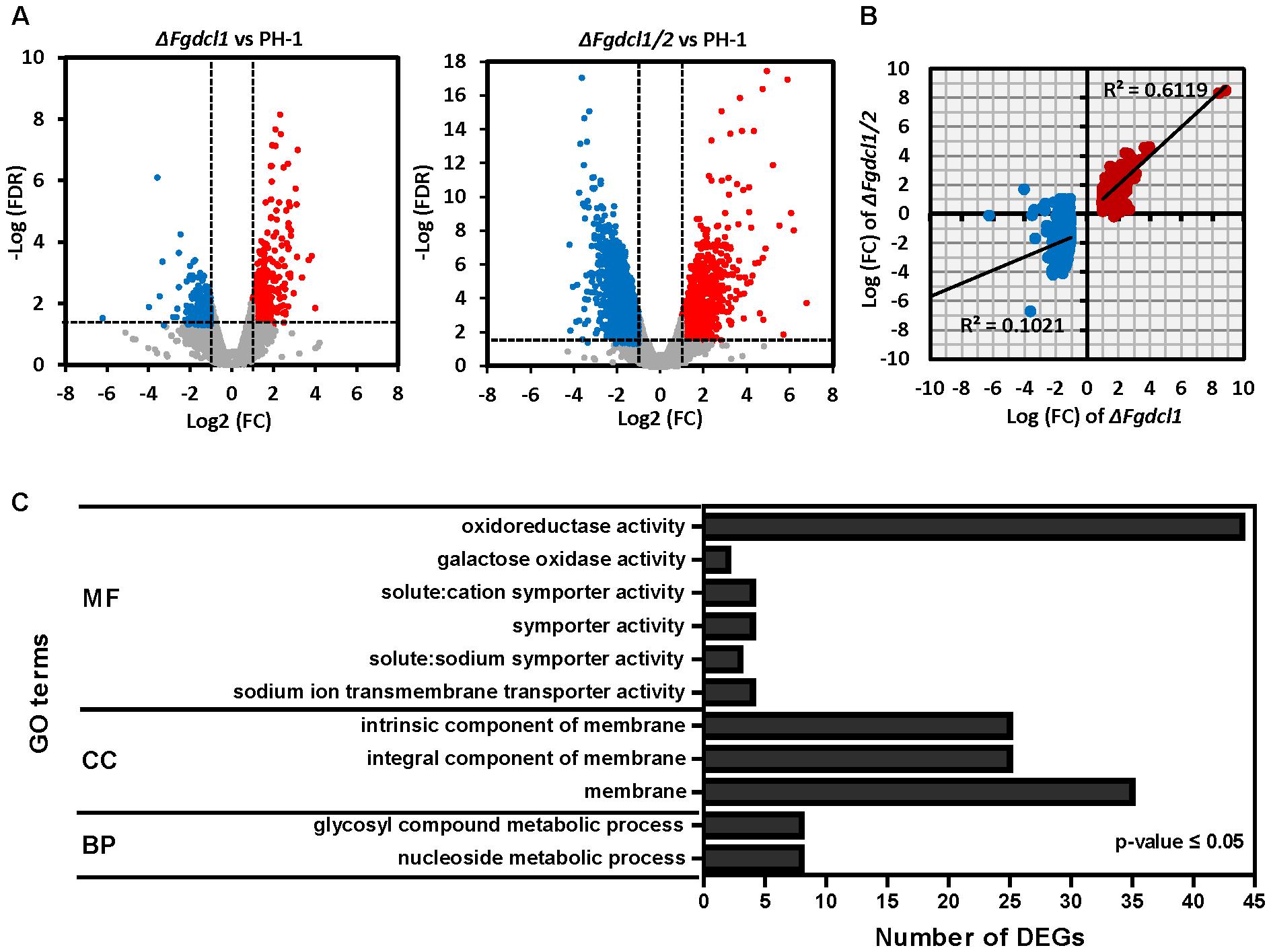
FIGURE 3. Transcriptomics analysis of the wild-type strain PH-1, ΔFgdcl1, and ΔFgdcl1/2 in sexual stage. (A) Volcano plots of up/down-regulated differentially expressed genes (DEGs) and non-differentially expressed genes between ΔFgdcl1 and PH-1, and between ΔFgdcl1/2 and PH-1. The red and blue dots indicate the up-regulated and down-regulated DEGs, respectively. The gray dots indicated non-differentially expression genes. Genes with an false discovery rate (FDR) below 0.05 and fold changes greater than two were considered as DEG. (B) Correlation analyses of DEGs of ΔFgdcl1 and ΔFgdcl1/2. The red and blue dots indicate the up-regulated and down-regulated DEGs in ΔFgdcl1, respectively. (C) The Gene ontology (GO) enrichment of common up-regulated DEGs between ΔFgdcl1 and ΔFgdcl1/2. The shown GO terms act as the significant enrichment terms, with p-values less than 0.05, adjusted by Benjamini-Hochberg correction. BP, biological process; CC, cellular component; MF, molecular function.
Prediction of F. graminearum Perithecium-Specific milRNAs
In the present study and previous studies, Fgdcl1 was induced and found to play a crucial role during sexual development (Figure 1; Son et al., 2017). Fgdcl2 and Fgago1 were involved in the RNAi pathway, and the accumulation of siRNA was reduced in the Fgdcl2 deletion mutant in the asexual stage (Chen et al., 2015). To explore whether Fgdcl1 is involved in perithecium-specific sRNAs biogenesis, the sRNA libraries of PH-1, ΔFgdcl1 and ΔFgdcl1/2 were constructed and sequenced (Supplementary Table S4). By comparing sRNA sequences between 0 and 7 dps, 2.71% unique sRNAs were common, and 88.75% unique sRNAs were detected only in 7 dps, it means most of the sRNAs classes were obtain only from perithecia (Figure 4A). These results indicated that the sRNAs at 7 dps markedly differed from those at 0 dps, and most of these molecules were perithecium-specific. Analyses of the length distribution of these sRNAs revealed that these sRNAs ranged from 18–30 nt, as previously reported in fungi. Additionally, the number of 22–24 nt sRNAs at 7 dps was higher than that at 0 dps and peaked at 24 nt (Figure 4B).
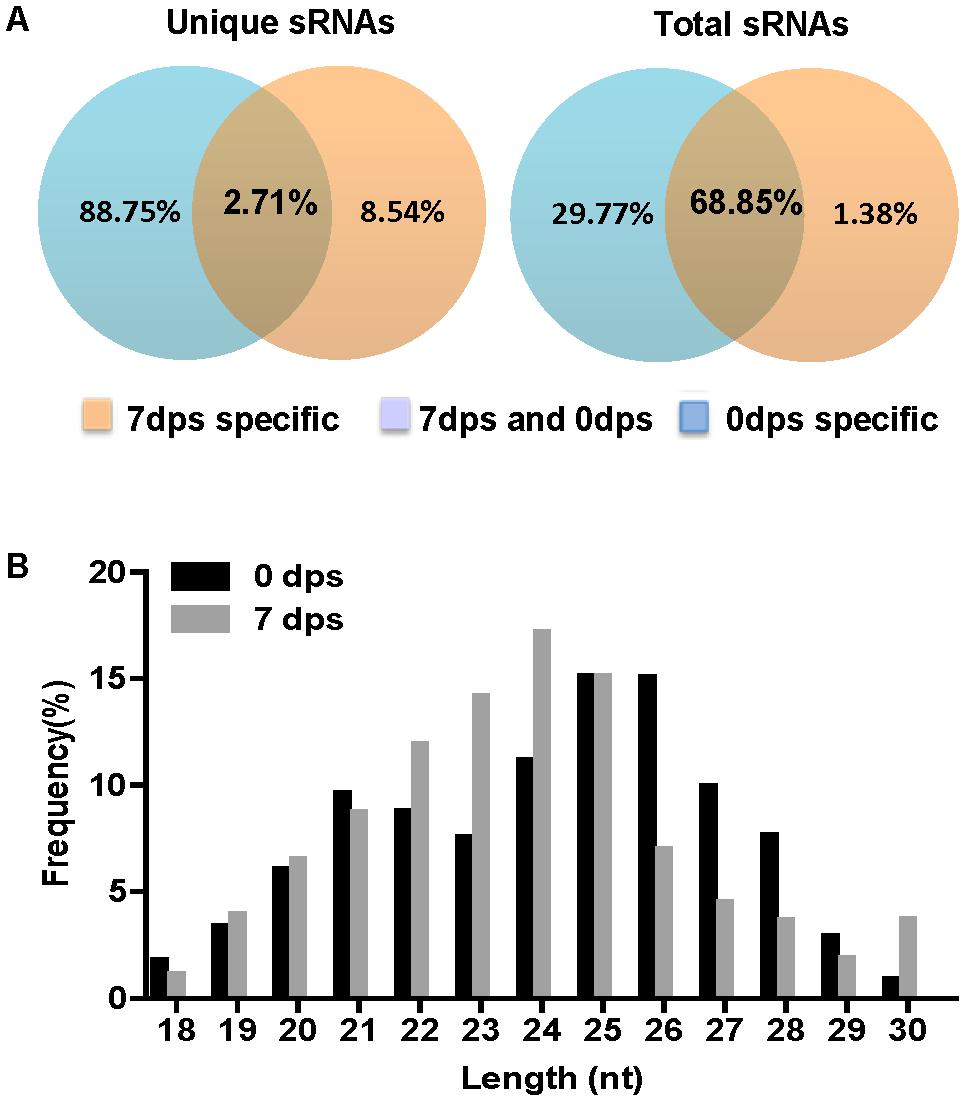
FIGURE 4. Comparison of small RNA reads among 0 day post self-crossing (dps) and 7 dps. (A) Venn diagram showing the public and special sRNAs between 0 dps and 7 dps. (B) Length distribution of the small RNAs from 0 dps and 7 dps of wild-type strain PH-1.
To predict novel miRNAs in F. graminearum, the small RNA sequences were mapped and annotated to the F. graminearum genome (Supplementary Tables S4, S5). Then, we predicted the micro-like RNAs from exon antisense, intron and unannotated RNAs using MIREAP (Li et al., 2012). The data showed that only 20 novel milRNAs were identified at 0 dps, while 143 novel milRNAs were generated from the mature perithecia of PH-1 (7 dps), and 139 milRNAs were detected only at 7 dps (Figure 5A and Supplementary Table S6), indicating that more milRNAs were induced during sexual reproduction. Among these perithecium-specific milRNAs, 75 (54%) were located in intergenic regions, 61 (44%) were located in antisense exon regions, and 3 (2%) were located in introns (Figure 5B and Supplementary Table S6). Compared to asexual stage milRNAs, the percentage of exon antisense loci was higher (∼two fold) in the sexual stage milRNAs (Chen et al., 2015). In addition, the unique perithecium-specific milRNAs ranged from 19 to 24 nt and were enriched in 22 and 23 nt molecules (Figure 5C). Moreover, a majority of 22–23 nt FgmilRNAs were located on gene antisense and intergenic regions (Figure 5C). Similar to most milRNAs in fungi, 73.61% of perithecium-specific milRNAs start with uracil (Figure 5D).
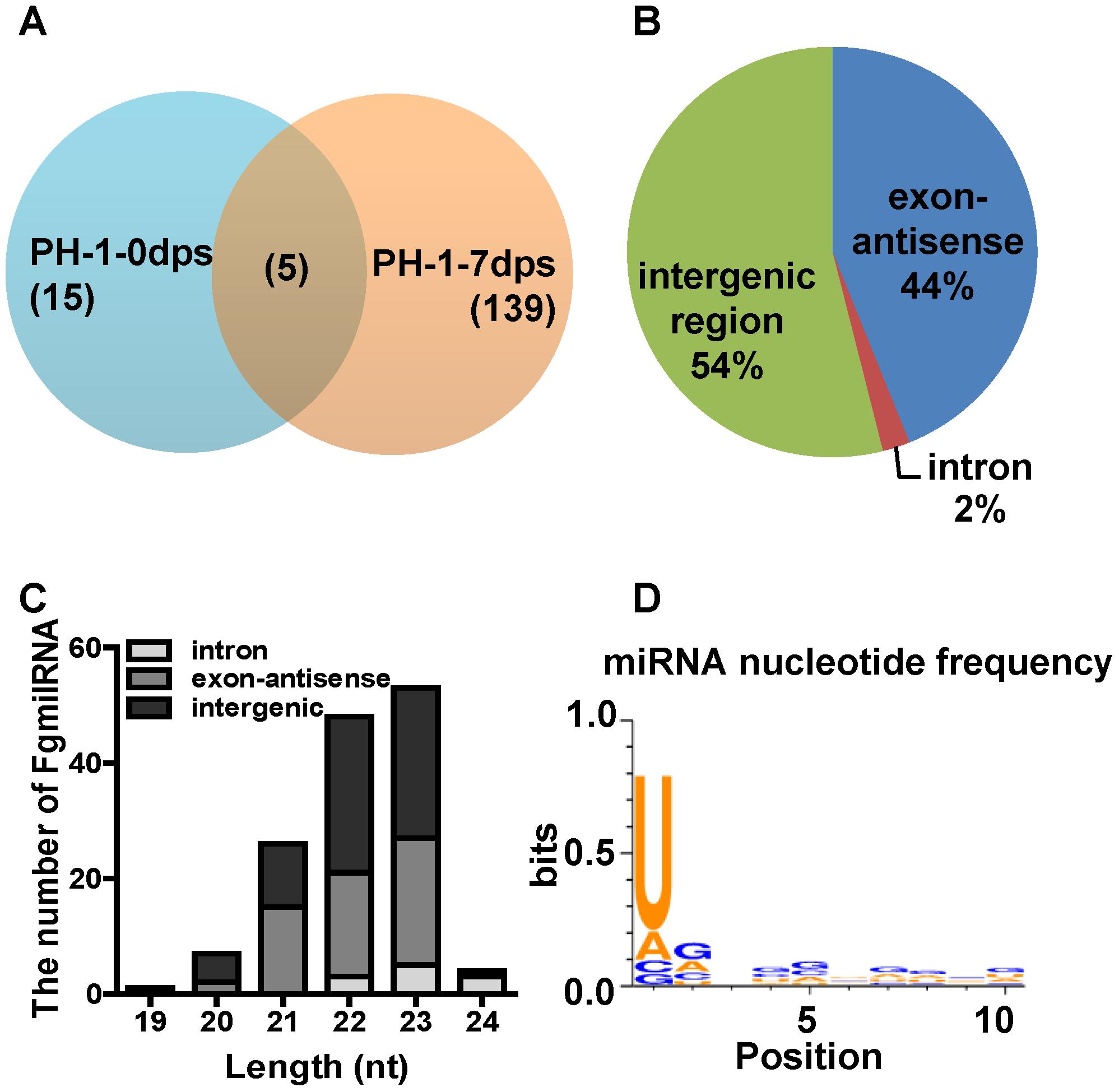
FIGURE 5. Characterization of microRNA-like RNAs (milRNAs) from Fusarium graminearum wild-type strain PH-1 sexual phase. (A) Venn diagram showing the summary of milRNAs between 0 dps and 7 dps of wild-type strain PH-1. (B) Distribution of pri-milRNA loci of perithecium-specific milRNAs in wild-type strain. (C) The length distribution of unique milRNAs and their locations. (D) The nucleotide frequency at 1-10 nt of unique perithecium-specific milRNAs in wild-type strain PH-1.
To examine whether Dicer processes these perithecium-specific milRNAs, we constructed and sequenced the sRNA libraries of ΔFgdcl1 and ΔFgdcl1/2. A comparison of the sRNA libraries of PH-1, ΔFgdcl1 and ΔFgdcl1/2 revealed that the proportions of 23–25 nt reads were decreased in ΔFgdcl1 and ΔFgdcl1/2, especially in 24 nt reads (Figure 6A). This finding implied that Fgdcl1 and Fgdcl2 may process this class of esRNAs. The prediction of milRNA showed that 113 and 44 milRNAs were obtained in ΔFgdcl1 and ΔFgdcl1/2, respectively (Figure 6B). Nevertheless, only 18 and 4 perithecium-specific milRNAs of PH-1 could also be induced in ΔFgdcl1 and ΔFgdcl1/2, respectively, and the expressions of these 18 milRNAs was lower than those in the wild-type strain (Figure 6C and Supplementary Table S6). These results indicate that most of these milRNAs may be processed by two Dicer proteins. However, Fgdcl1-dependent or two Fgdcl-dependent milRNAs showed low expression (Figure 6C and Supplementary Table S6). Furthermore, certain novel milRNAs were identified from ΔFgdcl1 or ΔFgdcl1/2 but not from the wild type strain PH-1.
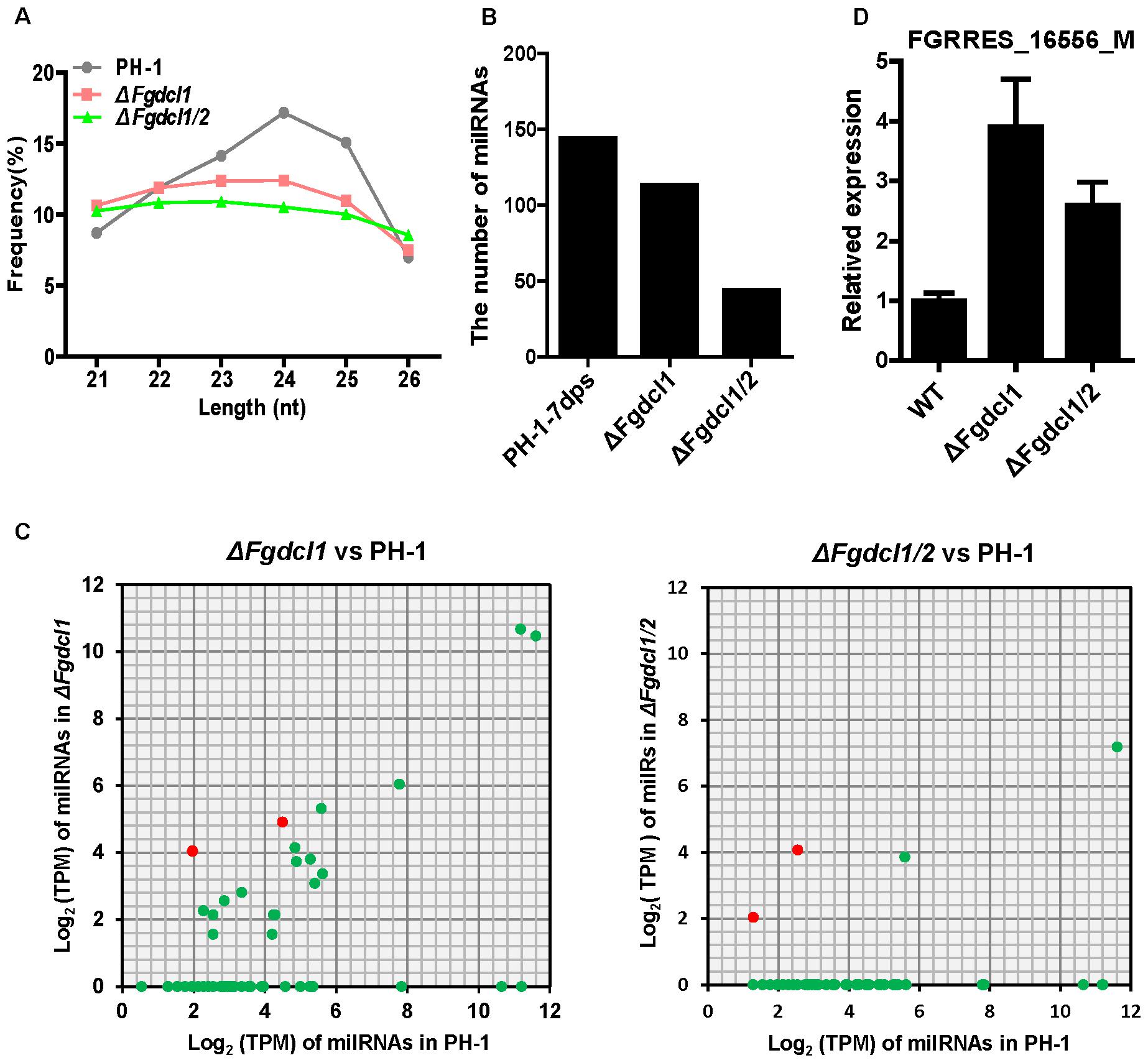
FIGURE 6. The expression patterns of milRNAs and their targets in the wild-type strain PH-1, ΔFgdcl1 and ΔFgdcl1/2. (A) Length distribution of 21–26 nt small RNAs from 7-day-old perithecia of the wild-type strain PH-1, ΔFgdcl1 and ΔFgdcl1/2. (B) The number of unique milRNA in the wild-type strain PH-1, ΔFgdcl1 and ΔFgdcl1/2. (C) Comparison of the expression patterns of perithecium-specific milRNAs candidates between ΔFgdcl1 and the wild-type PH-1, and between ΔFgdcl1/2 and the wild-type PH-1. The expression level of milRNAs was normalized by TPM normalized formula. The milRNAs with transcripts per million (TPM) values higher than one are represented. (D) Relative expression of FgmilR-7dps-61 putative target gene in the wild-type strain PH-1, ΔFgdcl1 and ΔFgdcl1/2. The total RNAs were isolated from the 7-day-old perithecia of PH-1, ΔFgdcl1 and ΔFgdcl1/2. The expression levels of these target genes in PH-1 served as control. Line bars indicate standard errors from three repeated experiments.
Role of Perithecium-Specific milRNA in the Regulation of Sexual Development of F. graminearum
The discharge of ascospores was reduced in ΔFgdcl1 and arrested in ΔFgdcl1/2 (Figure 2), and the common up-regulated DEGs of ΔFgdcl1 and ΔFgdcl1/2 paly more important roles in ascospore discharge than those common down-regulated DEGs (Figure 3). To explore whether FgmilRNAs negatively regulate their targets and have a relationship with the physiological changes of ΔFgdcl1 and ΔFgdcl1/2, the novel perithecium-specific milRNAs, which TPM values higher than 10, were selected to predict their targets by psRobot and TargetFinder. After duplicates were removed, 117 putative target genes were identified with the target penalty scores no more than 4 (Supplementary Table S7). Statistical results of GO classification and KEGG pathway revealed that no significant GO categories or KEGG pathways were enriched (Supplementary Figures S7, S8). Nevertheless, certain KEGG pathways attracted our attention because of their vital roles in sexual reproduction. For example, the peroxisome category (pathway ID: ko04146) is involved in the formation of fruiting bodies and the maturation and germination of sexual spores (Peraza-Reyes and Berteaux-Lecellier, 2013). Fructose and mannose metabolism (pathway ID: ko00051) affects ascospore discharge (Trail et al., 2002; Supplementary Figure S8 and Supplementary Table S7).
MicroRNAs directly degrade their target mRNA in plants, and mediate suppressed mRNA translation in plants and animals. Indeed, based on the transcriptomic data, we calculated the expression level of putative target genes in ΔFgdcl1 and ΔFgdcl1/2 in compared to those in the wild-type strain and the correlation between putative targets and FgmiRNAs. In total, 54 putative targets was negatively (r < 0) correlated with FgmiRNAs, of which 34 was strong negatively correlation (r ≤-0.7) (Supplementary Table S7). Besides, the expression patterns of several putative target genes were verified by qRT-PCR (Supplementary Figure S6B). Interestingly, mtlD (FGRRES_16556_M, the putative target gene of FgmilR-7dps-61, r = -0.93), which belongs to the fructose and mannose metabolism pathway, was induced in ΔFgdcl1 and ΔFgdcl1/2. The mRNA transcript level of FGRRES_16556_M was notably increased in ΔFgdcl1 and ΔFgdcl1/2, compared with the wild-type strain (Figure 6D). These results suggested that perithecium-specific milRNA potentially regulate the sexual development of F. graminearum.
Discussion
RNAi has been found in animals, plants and filamentous fungi, and the components of the RNAi pathway are conserved in a wide range of eukaryotic genomes, while the biological function of these components varies with fungal species. In Trichoderma atroviride, Botrytis cinerea, M. circinelloides, M. oryzae, C. neoformans, and Colletotrichum higginsianum, the components of the RNAi pathway are involved in the regulation of growth or conidiation (Zamore et al., 2000; Kadotani et al., 2004; Nicolas et al., 2007; de Haro et al., 2009; Janbon et al., 2010; Carreras-Villasenor et al., 2013; Weiberg et al., 2013; Campo et al., 2016). Nevertheless, no biological function has been reported in Aspergillus nidulans, Saccharomyces castellii, and Cryphonectria parasitica (Segers et al., 2007; Hammond et al., 2008; Drinnenberg et al., 2009). F. graminearum also has a functional RNA silencing pathway and contains five RdRP, two Argonaute, and two Dicer genes in its genome (Chen et al., 2015). Previous studies have shown that Fgdcl1 and Fgago2 were induced during the sporulation stage, while the deletion of these genes had no effect on the phenotype of F. graminearum during the asexual phase (Chen et al., 2015). In the present study, Fgdcl1 was expressed at a high level in perithecia, and the ascospore discharge of ΔFgdcl1 was less than that of the wild type strain (Figures 1, 2), similar to N. crassa dcl-1, which also plays an important role during sexual development (Alexander et al., 2008). Additionally, conidia germination of ΔFgdcl1 was mildly delayed at the early stage (Supplementary Figure S2C). In addition, Fgago2 is also essential for ascospore discharge (Kim et al., 2015). A previous study showed that perithecia induction inevitably induced asexual sporulation, which may contribute to the high expression of Fgdcl1 and Fgago2 during sporulation (Chen et al., 2015; Son et al., 2016). Taken together, these findings show that the ascospore discharge of ΔFgdcl1 was decreased, and it was blocked in ΔFgdcl1/2, suggesting that Fgdcl1 partly cooperates with Fgdcl2.
To explore the mechanism of the inhibitory ascospore discharge of Fgdcl, the RNA-sequencing of ΔFgdcl1 and ΔFgdcl1/2 revealed common up-regulated DEGs enriched in membrane and transmembrane transport (Figure 3C and Supplementary Figure S5). The discharge-deficient mutant Fgamd1, similar to ΔFgdcl1/2, which showed no significant changes in growth and conidiation, also has more than two-thirds up-regulated DEGs related to membrane and transporter activity (Cao et al., 2017). In F. graminearum, the turgor pressure of asci is necessary for ascospore discharge, and ion fluxes and sugar alcohol accumulation are related to the generation of turgor pressure, especially in K+, Na+, Cl- and Ca++ ion channels (Trail et al., 2002, 2005; Min et al., 2010). Indeed, this process may contribute to the up-regulated DEGs in sodium ion transmembrane transporter activity, solute: cation symporter activity, and the decrease in glycogen accumulation by glycogen staining (Figure 3C and Supplementary Figures S3A, S5; da R Campos and Costa, 2010).
Small non-coding RNAs (sRNAs), which range from 17 to 29 nt, act as the core factors mediating the RNAi pathway and are widely identified in eukaryotic organisms (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Zamore et al., 2000). According to extrinsic origins and biological pathways, sRNAs can be grouped into three major categories: miRNA, siRNA and piwi-interacting RNA (piRNA). In addition to these three major small RNAs, new sRNA, such as tasiRNA, natsiRNA, ex-siRNA, endo-siRNA, have also been discovered, and these sRNAs differ in their biogenesis and function. In F. graminearum, milRNA and ex-siRNA have been detected in the asexual and sexual stages, respectively (Chen et al., 2015; Son et al., 2017). In asexual stage, 49 milRNAs were detected and 64% loci distributed on intergenic regions (Chen et al., 2015). In the present study, 143 FgmilRNAs were enriched in sexual phases with a strong preference for uracil in the first position and having most of their loci distributed on intergenic regions (54%) (Figure 5). Unlike perithecium-specific milRNAs, the asexual stage milRNAs started with 5′ G, and the percentage of exon loci (26%) was decreased compared to that in the sexual stage (44%) (Chen et al., 2015). This finding suggested that the difference between asexual and sexual sRNAs. Moreover, the expression of 4 asexual stage milRNAs have no change in ΔFgdcl2, implying the potential role of Fgdcl1 in asexual stage. In sexual development, ex-siRNAs, another class of sRNA, also have been reported in F. graminearum (Son et al., 2017). Comparing to milRNAs, ex-siRNAs were exonic-specific. Interestingly, certain perithecium-specific milRNAs and ex-siRNAs shared common gene loci and started with 5′ U, implying that the perithecium-specific milRNAs and ex-siRNAs may be similar in biogenesis (Supplementary Table S6). MSUD, a silencing mechanism of meiosis, has also been demonstrated in F. graminearum, and Fgsad-1 was required (Son et al., 2011). However, due to the homothallic mating type of F. graminearum, less invasive DNA were produced and MSUD-associated siRNAs (masiRNAs) have been reported less often. Indeed, masiRNAs were dispensable for the regulation of sexual development. To the best of our knowledge, Fgdcl1 and Fgago2 processed the ex-siRNAs, while Fgdcl2 and Fgago1 processed the milRNAs at the asexual stage. In the present study, 19 (13.7%) perithecium-specific milRNAs were detected in ΔFgdcl1 but with lower expression than in the wild-type, and most of perithecium-specific milRNAs were missing in ΔFgdcl1/2 (Figure 6C). Hence, we suspected that Fgdcl1 may cooperate with Fgdcl2 in the biogenesis of perithecium-specific milRNAs. In N. crassa, diverse pathways for small RNA biogenesis were discovered, and other factors also play important roles in these pathways, such as RNase III domain-containing protein MRPL3, the exonuclease QIP, and RNA exosome (Lee et al., 2010; Xue et al., 2012). Taken together, these finding make a reasonable case that in addition to pathways dependent to Fgdcl2 and Fgago1, and Fgdcl1 and Fgago2, more pathways for small RNA biogenesis exist, and additional components are involved in these process. A similar relationship of the two dicer genes in perithecium-specific milRNAs biogenesis and sexual development revealed that the milRNAs may play a role in sexual development. MilRNAs negatively regulated gene targeting by seed-region matching and near-perfect matching in animals and plants, respectively (Rhoades et al., 2002; Lewis et al., 2003). In the present study, the bioinformatic prediction of milRNA targets showed that certain target genes related to sexual development and the ascospore discharge pathway. In fungi, the research on target recognization of milRNAs has been less reported. Indeed, the transcription levels of putative targets were identified using qRT-PCR, The results indicated that the putative target genes were increased in ΔFgdcl1 and ΔFgdcl1/2, whilst the expression of milRNAs were decreased or absent (Figure 6D, Supplementary Figure S6B, and Supplementary Table S7). Hence, this condition may be the reason why more up-regulated DEGs were obtained and may function in ascospore discharge in ΔFgdcl1 and ΔFgdcl1/2, especially in ΔFgdcl1/2. The regulation of the ex-siRNA by degrading mRNA transcripts may also be a factor (Son et al., 2017). In addition, no milRNA target genes were found in the ex-siRNA gene loci, indicating that ex-siRNAs have no effect on milRNA targets. Taken together, our results suggested that perithecium-specific milRNAs play important roles in the regulation of sexual development in F. graminearum, and the mechanism needs to be further studied.
Author Contributions
WZ and JC designed the research and wrote the paper. WZ, JW, YW, and JL executed the experiments. WZ, YF, JX, DJ, TC, and JC performed the data and bioinformatic analyses. All authors read and approved the final manuscript.
Funding
This research was supported by the Fundamental Research Funds for the Central Universities (2662017PY010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZM and handling Editor declared their shared affiliation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00818/full#supplementary-material
FIGURE S1 | Verification of deletion mutants using Southern blotting and PCR. (A) Strategic map of gene disruption construction and restriction map. WT, Fusarium graminearum wild-type strain PH-1; genr, geneticin resistance gene cassette; hygr, hygromycin B resistance gene cassette; X, XbaI; P1: the fragment P1, used to identify deletion mutant; P2: the fragment P2, used to identify deletion mutant; P3: the fragment P3, used to identify deletion mutant. The blue line indicates the region of PCR amplification with primers. (B) PCR amplification. P1, P2, P3, indicate the fragment P1, P2, P3, respectively. M, marker; the sizes from top to bottom: 5 kb, 3 kb, 2 kb, 1.5 kb, 1 kb, 750 bp, 500 bp, 250 bp, and 100 bp. (C) Southern blotting analysis. Genomic DNA from all strains was digested by XbaI. The probe 1 (from hygromycin B resistance gene) and probe 2 (from geneticin resistance gene) labeled with alkaline phosphatase according to the manual. The sizes of the marker are indicated on the left of the blot.
FIGURE S2 | The assays of growth, conidiation, conidial germination, and virulence of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. (A) Growth rate of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. The mycelial growth rate based on the colony diameter of cultures incubated at 25°C for 3 days. PDA, potato dextrose agar; MM, minimal medium; CM, complete medium. Mean and standard deviation were calculated from three independent replicates, and analyzed with Fisher’s least significant difference (LSD) test. (B) Statistics of conidiation of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. The conidial production in carboxymethyl cellulose (CMC) liquid culture at 25°C on a rotary shaker (200 rpm) for 5 days determined with a hemocytometer. Mean and standard deviation were calculated from three independent replicates, and analyzed with Fisher’s LSD test. (C) Comparison of conidial germination among F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants in YEPD at 25°C for 4 and 6 h. hpi, hour post-inoculation. The lines indicate the mean value. The mean values were calculated from five independent replicates. (D) Flowering wheat heads were inoculated with the conidial suspension of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. The infected wheat heads were imaged after 21 days of inoculation. (E) Corn silks were inoculated with the mycelial plugs of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. Images were captured after 6 days of inoculation.
FIGURE S3 | Comparison of colony morphology, conidia morphology, perithecia, asci, and the stress procession of F. graminearum wild-type strain PH-1 and Fgdcl deletion mutants. (A) Comparison of the colony morphology, conidia, perithecia, asci. The colony morphology was imaged after 3 days of incubation on PDA at 25°C. One milliliter of a conidia suspension of each strain was observed after incubation in CMC liquid culture at 25°C on a rotary shaker (200 rpm) for 5 days; scale bar = 10 μm. The images of the morphology of perithecia and asci were captured at 7 dps, and the asci were stained by KI-I2; scale bar: 200 μm and 20 μm, respectively. (B) The tolerance to osmotic, kalium, calcium, peroxide, and cell wall damaging agents stress among the wild-type strain PH-1 and mutants. PH-1 and mutants were incubated on the PDA plates containing 1 M glucose, 1 M sorbitol, 1 M NaCl, 1 M KCl, 0.5 M CaCl2, 0.02% SDS, 0.05% H2O2, and 0.2 mg/ml Congo red. The colony morphology of PDA plates and PDA containing 1 M glucose, 1 M sorbitol, 1 M NaCl, and 1 M KCl were imaged after 2 days, the others were imaged after 3 days.
FIGURE S4 | Comparison of sexual development among the wild-type strain PH-1,ΔFgdcl1-1, ΔFgdcl1-2, ΔFgdcl1/2-1, and ΔFgdcl1/2-2. (A) Comparsion of the cirrhi and ascospore discharge of wild-type strain PH-1, ΔFgdcl1-1, ΔFgdcl1-2, ΔFgdcl1/2-1, and ΔFgdcl1/2-2. Each strain was inoculated on carrot medium, and the cirrhi were captured at 12 days post self-crossing, Scale bar = 500 μm. Ascospore discharge was assayed with 7-day-old perithecia. The images were captured 14 h after ascospores had been released. (B) Statistical analysis of asci rosettes of 7-day-old perithecia. Ascus morphology was conducted after 7 days post self-crossing. A total of 20 asci rosettes were observed and classified in each strain with light microscopy. Line bars indicate standard errors from three repeated experiments.
FIGURE S5 | The network of enriched Gene ontology (GO) terms of common up-regulated DEGs between ΔFgdcl1 and ΔFgdcl1/2 according to REVIGO. Bubble color and size indicate the adjusted p-value and the frequency of the GO term in the underlying GO database, respectively. Highly similar GO terms are linked by edges in the graph, where the line width indicates the degree of similarity.
FIGURE S6 | Verification of the transcript levels of enriched GO terms genes (A) and the putative target genes of microRNA-like RNAs (milRNAs) (B) using qRT-PCR. The black bars reflect the results of qRT-PCR, and the gray lines indicate the data of transcriptome sequencing. FGRRES_17029_M, FGRRES_00220, FGRRES_04240, and FGRRES_02426 belong to transmembrane transporter; FGRRES_11032 and FGRRES_03569 involve in galactose oxidase activity; FGRRES_06554 and FGRRES_06942 are peroxisome (k04146); FGRRES_09366, FGRRES_16556_M class into starch and sucrose metabolism (k00500) and fructose and mannose metabolism (k00051), respectively.
FIGURE S7 | Gene ontology classification of target genes. No GO term was significantly enriched due to the adjusted p-values greater than 0.05. BP, biological process; CC, cellular component; MF, molecular function.
FIGURE S8 | KEGG classification of target genes. The bracketed number indicates the target number in these KEGG pathways. No KEGG pathways were significantly enriched due to adjusted p-values greater than 0.05.
TABLE S1 | PCR primers used in this study.
TABLE S2 | Statistics of the clean reads and the mapped reads in transcriptome of the wild-type strain PH-1, ΔFgdcl1, and ΔFgdcl1/2.
TABLE S3 | The differentially expressed genes (DEGs) in ΔFgdcl1 and ΔFgdcl1/2.
TABLE S4 | Summary statistics of the clean reads and the mapped reads in small RNA libraries of the wild-type strain PH-1, ΔFgdcl1, and ΔFgdcl1/2.
TABLE S5 | The annotations of mapped small reads of the wild-type strain, ΔFgdcl1 and ΔFgdcl1/2.
TABLE S6 | The prediction and expression of novel perithecium-specific microRNA-like RNA (milRNAs).
TABLE S7 | The prediction, expression, and annotation of target genes.
References
Alexander, W. G., Raju, N. B., Xiao, H., Hammond, T. M., Perdue, T. D., Metzenberg, R. L., et al. (2008). DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45, 719–727. doi: 10.1016/j.fgb.2007.10.006
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., et al. (2003). A uniform system for microRNA annotation. RNA 9, 277–279. doi: 10.1261/rna.2183803
Bai, G., and Shaner, G. (2004). Management and resistance in wheat and barley to fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi: 10.1146/annurev.phyto.42.040803.140340
Bauer, S., Grossmann, S., Vingron, M., and Robinson, P. N. (2008). Ontologizer 2.0–a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24, 1650–1651. doi: 10.1093/bioinformatics/btn250
Belli Kullan, J., Lopes Paim Pinto, D., Bertolini, E., Fasoli, M., Zenoni, S., Tornielli, G. B., et al. (2015). miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16:393. doi: 10.1186/s12864-015-1610-5
Campo, S., Gilbert, K. B., and Carrington, J. C. (2016). Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 12:e1005640. doi: 10.1371/journal.ppat.1005640
Cao, S., He, Y., Hao, C., Xu, Y., Zhang, H., Wang, C., et al. (2017). RNA editing of the AMD1 gene is important for ascus maturation and ascospore discharge in Fusarium graminearum. Sci. Rep. 7:4617. doi: 10.1038/s41598-017-04960-7
Cao, S., Zhang, S., Hao, C., Liu, H., Xu, J. R., and Jin, Q. (2016). FgSsn3 kinase, a component of the mediator complex, is important for sexual reproduction and pathogenesis in Fusarium graminearum. Sci. Rep. 6:22333. doi: 10.1038/srep22333
Cappellini, R. A., and Peterson, J. L. (1965). Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57, 962–966. doi: 10.2307/3756895
Carreras-Villasenor, N., Esquivel-Naranjo, E. U., Villalobos-Escobedo, J. M., Abreu-Goodger, C., and Herrera-Estrella, A. (2013). The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol. Microbiol. 89, 96–112. doi: 10.1111/mmi.12261
Catlett, N. L., Lee, B.-N., Yoder, O. C., and Turgeon, B. G. (2003). Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Rep. 50, 9–11. doi: 10.4148/1941-4765.1150
Cavinder, B., Sikhakolli, U., Fellows, K. M., and Trail, F. (2012). Sexual development and ascospore discharge in Fusarium graminearum. Jove J. Vis. Exp. 61:3895. doi: 10.3791/3895
Chen, R., Jiang, N., Jiang, Q., Sun, X., Wang, Y., Zhang, H., et al. (2014). Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum. PLoS One 9:e104956. doi: 10.1371/journal.pone.0104956
Chen, Y., Gao, Q., Huang, M., Liu, Y., Liu, Z., Liu, X., et al. (2015). Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 5:12500. doi: 10.1038/srep12500
Correll, J. C., Klittich, C. J. R., and Leslie, J. F. (1987). Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77, 1640–1646. doi: 10.1094/Phyto-77-1640
da R Campos, A. N., and Costa, M. D. (2010). Histochemistry and storage of organic compounds during basidiosporogenesis in the ectomycorrhizal fungus Pisolithus microcarpus. World J. Microbiol. Biotechnol. 26, 1745–1753. doi: 10.1007/s11274-010-0353-3
Dahlmann, T. A., and Kuck, U. (2015). Dicer-dependent biogenesis of small RNAs and evidence for MicroRNA-like RNAs in the penicillin producing fungus Penicillium chrysogenum. PLoS One 10:e0125989. doi: 10.1371/journal.pone.0125989
de Haro, J. P., Calo, S., Cervantes, M., Nicolas, F. E., Torres-Martinez, S., and Ruiz-Vazquez, R. M. (2009). A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell 8, 1486–1497. doi: 10.1128/EC.00191-09
Dimont, E., Shi, J. T., Kirchner, R., and Hide, W. (2015). edgeRun: an R package for sensitive, functionally relevant differential expression discovery using an unconditional exact test. Bioinformatics 31, 2589–2590. doi: 10.1093/bioinformatics/btv209
Drinnenberg, I. A., Weinberg, D. E., Xie, K. T., Mower, J. P., Wolfe, K. H., Fink, G. R., et al. (2009). RNAi in budding yeast. Science 326, 544–550. doi: 10.1126/science.1176945
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/j.1364-3703.2004.00252.x
Hallen, H. E., Huebner, M., Shiu, S. H., Guldener, U., and Trail, F. (2007). Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet. Biol. 44, 1146–1156. doi: 10.1016/j.fgb.2007.04.007
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Hammond, S. M., Bernstein, E., Beach, D., and Hannon, G. J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. doi: 10.1038/35005107
Hammond, T. M., Bok, J. W., Andrewski, M. D., Reyes-Dominguez, Y., Scazzocchio, C., and Keller, N. P. (2008). RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 7, 339–349. doi: 10.1128/EC.00355-07
Han, Y. K., Kim, M. D., Lee, S. H., Yun, S. H., and Lee, Y. W. (2007). A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol. Microbiol. 63, 768–779. doi: 10.1111/j.1365-2958.2006.05557.x
Hou, Z. M., Xue, C. Y., Peng, Y. L., Katan, T., Kistler, H. C., and Xu, J. R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15, 1119–1127. doi: 10.1094/Mpmi.2002.15.11.1119
Janbon, G., Maeng, S., Yang, D. H., Ko, Y. J., Jung, K. W., Moyrand, F., et al. (2010). Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet. Biol. 47, 1070–1080. doi: 10.1016/j.fgb.2010.10.005
Jeong, D. H., German, M. A., Rymarquis, L. A., Thatcher, S. R., and Green, P. J. (2010). Abiotic stress-associated miRNAs: detection and functional analysis. Methods Mol. Biol. 592, 203–230. doi: 10.1007/978-1-60327-005-2_14
Jiang, N., Yang, Y., Janbon, G., Pan, J., and Zhu, X. (2012). Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans. PLoS One 7:e52734. doi: 10.1371/journal.pone.0052734
Kadotani, N., Nakayashiki, H., Tosa, Y., and Mayama, S. (2004). One of the two dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J. Biol. Chem. 279, 44467–44474. doi: 10.1074/jbc.M408259200
Kang, K., Zhong, J., Jiang, L., Liu, G., Gou, C. Y., Wu, Q., et al. (2013). Identification of microRNA-Like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PLoS One 8:e76288. doi: 10.1371/journal.pone.0076288
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/Gb-2013-14-4-R36
Kim, H. K., Jo, S. M., Kim, G. Y., Kim, D. W., Kim, Y. K., and Yun, S. H. (2015). A large-scale functional analysis of putative target genes of mating-type loci provides insight into the regulation of sexual development of the cereal pathogen Fusarium graminearum. PLoS Genet. 11:e1005486. doi: 10.1371/journal.pgen.1005486
Kim, J. E., Jin, J. M., Kim, H., Kim, J. C., Yun, S. H., and Lee, Y. W. (2006). GIP2, a putative transcription factor that regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae. Appl. Environ. Microbiol. 72, 1645–1652. doi: 10.1128/Aem.72.2.1645-1652.2006
King, R., Urban, M., Hammond-Kosack, M. C., Hassani-Pak, K., and Hammond-Kosack, K. E. (2015). The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genomics 16:544. doi: 10.1186/s12864-015-1756-1
Lau, S. K., Chow, W. N., Wong, A. Y., Yeung, J. M., Bao, J., Zhang, N., et al. (2013). Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl. Trop. Dis. 7:e2398. doi: 10.1371/journal.pntd.0002398
Lavorgna, G., Guffanti, A., Borsani, G., Ballabio, A., and Boncinelli, E. (1999). TargetFinder: searching annotated sequence databases for target genes of transcription factors. Bioinformatics 15, 172–173. doi: 10.1093/bioinformatics/15.2.172
Lee, H. C., Chang, S. S., Choudhary, S., Aalto, A. P., Maiti, M., Bamford, D. H., et al. (2009). qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459, 274–277. doi: 10.1038/nature08041
Lee, H. C., Li, L., Gu, W., Xue, Z., Crosthwaite, S. K., Pertsemlidis, A., et al. (2010). Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38, 803–814. doi: 10.1016/j.molcel.2010.04.005
Leslie, J. F., Summerell, B. A., and Bullock, S., (2006). The Fusarium Laboratory Manual. Ames, IA: Blackwell Publishing. doi: 10.1002/9780470278376
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. doi: 10.1016/S0092-8674(03)01018-3
Li, Y., Zhang, Z., Liu, F., Vongsangnak, W., Jing, Q., and Shen, B. (2012). Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 40, 4298–4305. doi: 10.1093/nar/gks043
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Min, K., Lee, J., Kim, J. C., Kim, S. G., Kim, Y. H., Vogel, S., et al. (2010). A novel gene, ROA, is required for normal morphogenesis and discharge of ascospores in Gibberella zeae. Eukaryot. Cell 9, 1495–1503. doi: 10.1128/EC.00083-10
Nakayashiki, H. (2005). RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579, 5950–5957. doi: 10.1016/j.febslet.2005.08.016
Nicolas, F. E., de Haro, J. P., Torres-Martinez, S., and Ruiz-Vazquez, R. M. (2007). Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44, 504–516. doi: 10.1016/j.fgb.2006.09.003
Nicolas, F. E., Moxon, S., de Haro, J. P., Calo, S., Grigoriev, I. V., Torres-Martinez, S., et al. (2010). Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 38, 5535–5541. doi: 10.1093/nar/gkq301
Nunes, C. C., Gowda, M., Sailsbery, J., Xue, M., Chen, F., Brown, D. E., et al. (2011). Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics 12:288. doi: 10.1186/1471-2164-12-288
Peraza-Reyes, L., and Berteaux-Lecellier, V. (2013). Peroxisomes and sexual development in fungi. Front. Physiol. 4:244. doi: 10.3389/Fphys.2013.00244
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Segers, G. C., Zhang, X., Deng, F., Sun, Q., and Nuss, D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906. doi: 10.1073/pnas.0702500104
Seong, K., Hou, Z. M., Tracy, M., Kistler, H. C., and Xu, J. R. (2005). Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 95, 744–750. doi: 10.1094/Phyto-95-0744
Son, H., Lim, J. Y., Lee, Y., and Lee, Y. W. (2016). Utilization of a conidia-deficient mutant to study sexual development in Fusarium graminearum. PLoS One 11:e0155671. doi: 10.1371/journal.pone.0155671
Son, H., Min, K., Lee, J., Raju, N. B., and Lee, Y. W. (2011). Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 115, 1290–1302. doi: 10.1016/j.funbio.2011.09.006
Son, H., Park, A. R., Lim, J. Y., Shin, C., and Lee, Y. W. (2017). Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet. 13:e1006595. doi: 10.1371/journal.pgen.1006595
Supek, F., Bosnjak, M., Skunca, N., and Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800
Trail, F., and Common, R. (2000). Perithecial development by Gibberella zeae: a light microscopy study. Mycologia 92, 130–138. doi: 10.2307/3761457
Trail, F., Gaffoor, I., and Vogel, S. (2005). Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 42, 528–533. doi: 10.1016/j.fgb.2005.03.008
Trail, F., Xu, H., Loranger, R., and Gadoury, D. (2002). Physiological and environmental aspects of ascospore discharge in Gibberella zeae (anamorph Fusarium graminearum). Mycologia 94, 181–189. doi: 10.1080/15572536.2003.11833223
Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I., and Martienssen, R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. doi: 10.1126/science.1074973
Weiberg, A., Wang, M., Lin, F. M., Zhao, H. W., Zhang, Z. H., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wu, H. J., Ma, Y. K., Chen, T., Wang, M., and Wang, X. J. (2012). PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40, W22–W28. doi: 10.1093/nar/gks554
Xue, Z., Yuan, H., Guo, J., and Liu, Y. (2012). Reconstitution of an Argonaute-dependent small RNA biogenesis pathway reveals a handover mechanism involving the RNA exosome and the exonuclease QIP. Mol. Cell 46, 299–310. doi: 10.1016/j.molcel.2012.03.019
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/Gb-2010-11-2-R14
Zamore, P. D., Tuschl, T., Sharp, P. A., and Bartel, D. P. (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. doi: 10.1016/S0092-8674(00)80620-0
Keywords: microRNAs, sexual development, perithecium-specific milRNAs, dicer-like protein, Fusarium graminearum
Citation: Zeng W, Wang J, Wang Y, Lin J, Fu Y, Xie J, Jiang D, Chen T, Liu H and Cheng J (2018) Dicer-Like Proteins Regulate Sexual Development via the Biogenesis of Perithecium-Specific MicroRNAs in a Plant Pathogenic Fungus Fusarium graminearum. Front. Microbiol. 9:818. doi: 10.3389/fmicb.2018.00818
Received: 10 February 2018; Accepted: 10 April 2018;
Published: 26 April 2018.
Edited by:
Weiguo Fang, Zhejiang University, ChinaReviewed by:
Alfredo Herrera-Estrella, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoBo Huang, Anhui Agricultural University, China
Zhonghua Ma, Zhejiang University, China
Copyright © 2018 Zeng, Wang, Wang, Lin, Fu, Xie, Jiang, Chen, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiasen Cheng, jiasencheng@mail.hzau.edu.cn
 Wenping Zeng
Wenping Zeng Jie Wang1,2
Jie Wang1,2 Jing Lin
Jing Lin Yanping Fu
Yanping Fu Jiatao Xie
Jiatao Xie Daohong Jiang
Daohong Jiang Tao Chen
Tao Chen Huiquan Liu
Huiquan Liu Jiasen Cheng
Jiasen Cheng