- 1College of Agro-grassland Science, Nanjing Agricultural University, Nanjing, China
- 2Department of Natural Resource Ecology and Management, Oklahoma State University, Stillwater, OK, United States
- 3Jiangsu Provincial Key lab for Organization, National Engineering Research Center for Organic-based Fertilizers, Jiangsu Collaborative Innovation, Nanjing Agricultural University, Nanjing, China
- 4Department of Grassland Science, China Agricultural University, Beijing, China
In grasslands, forage and livestock production results in soil nutrient deficits as grasslands typically receive no nutrient inputs, leading to a loss of grassland biomass. The application of mature compost has been shown to effectively increase grassland nutrient availability. However, research on fertilization regime influence and potential microbial ecological regulation mechanisms are rarely conducted in grassland soil. We conducted a two-year experiment in meadow steppe grasslands, focusing on above- and belowground consequences of organic or Trichoderma biofertilizer applications and potential soil microbial ecological mechanisms underlying soil chemistry and microbial community responses. Grassland biomass significantly (p = 0.019) increased following amendment with 9,000 kg ha−1 of Trichoderma biofertilizer (composted cattle manure + inoculum) compared with other assessed organic or biofertilizer rates, except for BOF3000 (fertilized with 3,000 kg ha−1 biofertilizer). This rate of Trichoderma biofertilizer treatment increased soil antifungal compounds that may suppress pathogenic fungi, potentially partially responsible for improved grassland biomass. Nonmetric multidimensional scaling (NMDS) revealed soil chemistry and fungal communities were all separated by different fertilization regime. Trichoderma biofertilizer (9,000 kg ha−1) increased relative abundances of Archaeorhizomyces and Trichoderma while decreasing Ophiosphaerella. Trichoderma can improve grassland biomass, while Ophiosphaerella has the opposite effect as it may secrete metabolites causing grass necrosis. Correlations between soil properties and microbial genera showed plant-available phosphorus may influence grassland biomass by increasing Archaeorhizomyces and Trichoderma while reducing Ophiosphaerella. According to our structural equation modeling (SEM), Trichoderma abundance was the primary contributor to aboveground grassland biomass. Our results suggest Trichoderma biofertilizer could be an important tool for management of soils and ultimately grassland plant biomass.
Introduction
Grasslands are valuable ecological resources due to their biodiversity and ecosystem functions (Soussana et al., 2007; Hermann et al., 2016). In recent years, grasslands have been over-exploited for grazing with consequences such as soil carbon loss and desertification (Kosmas et al., 2014; Conant et al., 2017). Aboveground plant biomass is the foundation of grassland animal husbandry, which is often co-limited by both N and P in semi-arid grasslands (Zhan et al., 2017). Globally, many grasslands receive little nutrient input, and grassland fertility declines are linked to reduced grassland provisioning services, such as primary productivity (Sattari et al., 2016). Herbivore manure is important in grassland nutrient cycling; however, intensified livestock production can lead to excess manure. Deposition of this waste can contaminate groundwater and increase nitrate and phosphate concentrations, causing soil hardening and/or salinization, negatively impacting soil physicochemical properties and soil microbial communities (Groenigen et al., 2005; Sun et al., 2015; Xu et al., 2017). To mitigate these issues, manure can be processed into organic fertilizer via composting (Moral et al., 2009; Tang et al., 2011). When applied in grasslands, compost effectively increases soil fertility, supporting soil microbial communities and improving grassland productivity (van der Heijden et al., 2008; Wagg et al., 2014).
Trichoderma spp. are well-known plant growth-promoting fungi (Masunaka et al., 2011) that enhance plant nutrient uptake, production of growth hormones, and protect plants from pathogen infection (De Souza et al., 2008; Contreras-Cornejo et at., 2009; Zhang et al., 2013a). Many plant growth-promoting fungi are effective in vitro, but do not significantly benefit plants in field studies because of their inability to colonize root systems in situ. The survival rate and population density of plant growth-promoting fungi are prerequisites for their effectiveness in the rhizosphere (Raaijmakers et al., 2009; Zhang et al., 2013b). Formulation of inoculum carriers is necessary to ensure survival and maintenance of a viable Trichoderma population in field soils. Combination of organic fertilizers (compost) and Trichoderma strains as biofertilizers (BOF) may be an effective way to facilitate greater plant biomass compared to amendments of organic fertilizers or Trichoderma separately (Huang et al., 2011; Zhang et al., 2013a). Organic matter in BOF improves soil fertility while being an excellent substrate for the growth of Trichoderma. Zhang et al. (2013b) showed application of Trichoderma biofertilizer significantly altered fungal diversity and community in the rhizosphere of cucumber, resulting in improved yields. Additional researches demonstrated Bacillus and Trichoderma biofertilizers improved banana growth and increased soil microbial diversity (Shen et al., 2015; Fu et al., 2017; Xiong et al., 2017). However, to the best of our knowledge, microbial ecological mechanisms influencing soil microbial genera and subsequent stimulation of root exudation, which supports plant growth responses to biofertilizers are rarely researched in grasslands.
Rhizosphere chemistry is the sum of root exudates, their breakdown products, and the microbial products of soil-derived chemicals (Pierre et al., 2017). Based on this, we defined extracted chemicals in bulk soil as “soil chemistry.” Grasslands harbor diverse plant species with diverse root exudates. Soil chemistry composition plays an important role mediating plant and soil microbial interactions (Oburger and Schmidt, 2016). Dessaux et al. (2016) demonstrated plant roots convert their associated soil into complex mesotrophic environments, supporting highly diverse microbial communities. Root exudates provide a major food source for soil microbes and directly affect their assemblage and reproduction (Raaijmakers et al., 2009). The studies of Ng et al. (2014) and Salles et al. (2009) also reported soil carbon composition strongly influences soil microbial community composition. Hence, plant roots may drive multitrophic interactions in soil via root exudation. Furthermore, some chemicals with antifungal activities may produce a chemical barrier to pathogenic fungi (Yuan et al., 2012; Raza et al., 2015), thereby indirectly improving grassland biomass. Beneficial plant-microbe interactions have received less attention in grassland systems, but advances in microbial genetics are improving our ability to link key microbial genera with soil health and ecosystem productivity (Finkel et al., 2017).
In our study, we applied organic fertilizer (composted cattle manure) or Trichoderma biofertilizer (composted cattle manure + inoculum) at different rates (0 kg ha−1, 3,000 kg ha−1, 6,000 kg ha−1, or 9,000 kg ha−1) to meadow steppe grassland plots for two growing seasons. We focused on the potential microbial ecological mechanisms underlying observed grassland biomass. This investigation addresses the following questions: (i) How do different fertilization regimes impact grassland biomass? (ii) Does addition of Trichoderma further increase grassland biomass above the effects of cattle manure alone? (iii) Do different fertilization regimes shift soil chemistry and soil microbial communities? (iv) Is soil chemistry significantly correlated with specific microbial genera? (v) Are soil microbial diversities or communities significantly correlated with grassland plant biomass? (vi) How do different variables (i.e., key microbial genera, soil properties, soil chemistry, and soil microbial communities) contribute to grassland plant biomass? Our findings will provide a better understanding of above- and belowground responses to alternative grassland fertilization and may ultimately improve management of grassland soils for key provisioning services.
Materials and Methods
Trichoderma Strain, Culture Conditions, and Conidia Suspension
We utilized a strain of Trichoderma rossicum, NAU-18 (CCTCC No. AF2017008, China Center for Type Culture Collection), isolated from soil at the Grassland Agro-ecosystems Station (49°26′12″N, 120°8″52″E, 695 m altitude). NAU-18 was identified based on morphological characteristics and internal transcribed spacer (ITS) sequence analysis (Figures S1A,B). The strain was stored at −80°C in 30% glycerol before use and routinely cultured on potato dextrose agar (PDA) at 28°C. NAU-18 conidia suspension was prepared according to the procedure of Zhang et al. (2013a). The final concentration was 3.2 × 108 colony-forming units (CFU) ml−1, based on hemocytometer counts. This conidia suspension was used as inoculum for solid-state fermentation.
Preparation of Biofertilizers (BOF)
The organic fertilizer (OF) used in our study was composted cattle manure, supplied by TeniheFarm (Hulunbuir, China). Cattle manure was composted at 30 to 70°C for ~20 days and contained 36.8% organic matter, 3.1% N, 2.5% P2O5, and 1.9% K2O, with moisture < 30%.
Wheat straw was utilized as a substrate for NAU-18 production. Sterile wheat straw and 1% urea were stirred thoroughly (moisture = 70%), after which NAU-18 conidia suspension was added to the substrate (9% of the total [v/w]), and solid-state fermentation was maintained at 30°C for 8 days in a shallow tray. The final dry weight density of NAU-18 was 1.1 × 1010 CFU g−1. Biofertilizer (BOF) was a mixture of 1:10 (w/w) Trichoderma NAU-18 fermentation products and composted cattle manure.
Study Site Description and Experimental Design
Our in situ fertilization experiment was initiated in June 2015 at the Grassland Agro-ecosystems Station, located in northeastern Inner Mongolia, Hulunbuir, China. This region is characterized by a continental temperate semi-arid climate typically with 350–400 mm annual precipitation (Inner Mongolia Meteorological service, http://www.imwb.gov.cn). The growing season is from late June to early September with an average temperature range of 16–21°C. The soil in this region is classified as a chernozem (IUSS WG WRB, 2015) with pH 6.59, 44.5 g kg−1 soil organic matter, 189.3 mg kg−1 plant-available N, 3.3 mg kg−1 plant-available P, and 134.2 mg kg−1plant-available K. The grassland plant community is dominated by Poapratensis L., Leymus chinensis (Trin.) Tzvelev, Bromusinermis Leyss, Stipabaicalensis Rochev., and Potentilla bifurca L.
Two fertilizer types and three amendment rates were utilized, plus a non-amended control, resulting in seven treatments in a randomized block design with three replicates per treatment. Each grassland plot was 5 m long and 3 m wide, and the study was conducted for two growing seasons. The seven treatments were: (1) CK: non-amended; (2) OF 3,000: 3000 kg ha−1 organic fertilizer; (3) OF 6000: 6000 kg ha−1organic fertilizer; (4) OF 9000: 9,000 kg ha−1 organic fertilizer; (5) BOF 3000: 3,000 kg ha−1 Trichoderma biofertilizer; (6) BOF 6000: 6,000 kg ha−1 Trichoderma biofertilizer; and (7) BOF 9000: 9,000 kg ha−1 Trichoderma biofertilizer. All treatments were applied on July 10, 2015, and June 2, 2016. All experimental plots were completely dependent on precipitation during our study.
Soil and Vegetation Sampling
Soil and vegetation samples were collected on August 25, 2016. Five cylindrical soil cores (0–10 cm depth, 6 cm diameter) were randomly collected and mixed to form a composite soil sample for each plot. All soil samples were sieved through 2.0-mm mesh and thoroughly homogenized. Each composite soil sample was then divided into three sub-samples: the first sub-sample was stored at 4°C for soil chemistry analysis, another was air dried at room temperature for 7 days for analysis of soil properties, and the final sub-sample was stored at −20°C for DNA extraction. Two random subplots (0.25 × 1.0 m) within each plot were destructively harvested for aboveground biomass. Shoots were cut at the soil surface and oven dried at 65°C for 72 h before weighing. Shoot dry weights were expressed as total grassland biomass per m2. Plant tissue crude protein was determined by Kjeldahl method (Da Silva et al., 2016).
Extraction and Identification of Soil Chemistry
For each replicate soil sub-sample, 5.0 g of soil were combined with 50 ml of ethyl acetate (1:10 ratio) in a 150 ml Erlenmeyer flask and shaken on a table agitator at 30°C for 2 h. The suspension was filtered through a 0.45-μm Millipore filter and concentrated to 0.5 ml at 35°C using a vacuum rotary evaporator (Yarong Model RE-52A, Shanghai, China). The concentrated solution analyzed by gas chromatography-mass spectrometry (GC-MS). The following protocol was used for GC-MS detection: 60°C for 3 min, followed by increasing the temperature at a rate of 5°C min−1 to 240°C, held at 240°C for 5 min, increased to 280°C at a rate of 20°C min−1, and held at 280°C for 2 additional minutes. The mass spectrometry detector (MSD) was operated in electron ionization mode at 70 eV, with a source temperature of 230°C. A continuous scan from 45 to 500 m/z was applied. Helium was the carrier gas at a linear velocity of 1.0 ml min−1. The chromatographic peaks of the components were compared with entries in the National Institute of Standards and Technology (NIST) database (Version 2.0) to characterize the variation in the chemical composition of soil sub-samples.
Determination of Soil Physicochemical Properties
Soil analyses were performed by the soil-testing lab in Qiyang at the red soil experimental station of the Chinese Academy of Agricultural Sciences. Soil pH was measured with soil-water (1:2.5, w/v) slurry using a compound electrode (PE-10; Sartorious, Germany). Soil organic C (SOC) and total N (TN) were determined with an Elementar Vario EL III (Germany). Plant-available N was hydrolyzed with 1.0 mol l−1 NaOH and measured according to the methods of Shi (1996). Total P was determined by perchloric acid digestion (Olsen and Sommers, 1982). Plant-available phosphorus (Olsen-P) was extracted with sodium bicarbonate and measured using the molybdenum-blue method (Watanabe and Olsen, 1965). Total and plant-available K were measured by flame photometry (Knudsen et al., 1982).
DNA Extraction, High-Throughput Sequencing, and Bioinformatics
Total soil genomic DNA was extracted using a Power Soil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) following the manufacturer's instructions. The concentration and quality (A260/A280 ratio) of the DNA samples were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Pyrosequencing analyses of the 16S rRNA gene and ITS region were performed to determine the diversity and composition of bacterial and fungal communities, respectively. The V4 region of the bacterial 16S rRNA gene was amplified using the gene-specific primers 520F (5′-AYTGGGYDTAAAGNG-3′) and 802R (5′-TACNVGGGTATCTAATCC-3′), while the ITS1 region of the fungal ITS was targeted by the primers ITS1F (5′- CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5-GCTGCGTTCTTCATCGATGC-3). PCR amplification of the bacterial 16S rRNA and fungal ITS sequences was conducted in a volume of 30 μl containing15 μl of 2 × Master Mix (Thermo Scientific® Phusion High-Fidelity PCR Master Mix, New England Biolabs, UK), 0.5 μM each primer, 10 ng of soil DNA template and nuclease-free water to bring the total to 30 μl. The obtained PCR products were purified using a PCR Purification Kit (Axygen Bio, USA) and quantified with PicoGreen® dsDNA reagent (Promega, USA). The purified amplicons were then pooled in equimolar concentrations as a single aliquot and employed for library construction using the NEB Next® Ultra ™ DNA Library Prep Kit for Illumina (New England Biolabs, UK). All library preparation was performed on the Illumina MiSeq platform at Total Genomics Solutions Biotechnology Co., Ltd. (Shenzhen, China).
Raw sequences were assembled for each sample based on unique barcodes using Quantitative Insights Into Microbial Ecology (QIIME) after removal of the adaptor and primer sequences (Caporaso et al., 2011). Split sequences for each sample were merged using FLASH V1.2.7 (Magoc and Salzberg, 2011), and low-quality sequences were then discarded using QIIME. The sequences retained for each sample were analyzed following the UPARSE pipeline, using USEARCH and Perl scripts to generate an operational taxonomic units (OTUs) table and pick representative sequences (Edgar, 2013). Sequences with a quality score < 0.5 or a length < 200 nt and singletons were discarded. After that, retained sequences were assigned to OTUs at 97% similarity. We chose a representative sequence from each OTU, and the Ribosomal Database Project (RDP) classifier (the RDP Bacterial 16S database for 16S rRNA data and the UNITE Fungal ITS database for ITS data) was used to assign taxonomic information (Kõljalg et al., 2013). The MOTHUR (version 1.25.1) standard operating procedure (SOP) was employed for further analyses of pyrosequencing data (Wang et al., 2007; Schloss et al., 2009). To correct for sampling effects, randomly selected subsets of 27,942 sequences per sample for 16S and 30,629 sequences per sample for ITS were chosen for further bacterial and fungal community analyses.
Statistical Analyses and Data Accessibility
Statistical analyses of all parameters were performed using the IBM SPSS statistical software package version 20 (IBM Corporation, New York, USA). Data from each treatment were analyzed using one-way analysis of variance (ANOVA), and Duncan's multiple range tests (p < 0.05) were performed for multiple comparisons. The multiple comparison correction is according to (Benjamini and Hochberg, 1995) Non-metric multidimensional scaling (NMDS) plots and a principal component analysis (PCA) were performed in R with the vegan package (Version 3.0.2, Oksanen et al., 2009). Differences in soil chemistry and bacterial and fungal communities between treatments were tested by analysis of similarities (ANOSIM) (Yan et al., 2017).
SEM is a powerful tool to examine relationships among inter-correlated variables, in which the net effects of an experimental treatment can be partitioned into direct and indirect effects. Observed variables rather than latent variables were used in the models directly. In our study, SEM was employed to test the direct and indirect pathways between soil organic matter, total N, soil chemistry, bacterial and fungal communities, Trichoderma abundance, and aboveground grassland biomass, providing a comprehensive analysis of all relationships between factors. We performed SEM using AMOS software (IBM SPSS AMOS 20.0.0). The fit of the model was tested using the maximum likelihood (χ2) goodness-of-fit test with p < 0.05 along with root mean square error of approximation (RMSEA). Figures were generated with Sigmaplot 12.0 (Systat Software Inc., CA, USA). Raw bacterial 16S and fungal ITS sequence data are available at the National Center for Biotechnology Information (NCBI) under accession number PRJNA393964.
Results
Grassland Biomass and Crude Protein
Plots fertilized with 9,000 kg ha−1 BOF had the greatest aboveground biomass of any treatment and were significantly more productive than non-amended plots and plots fertilized with any rate of OF or BOF, except for BOF3000 treatment (Figure 1). However, there was no significant difference in plant tissue crude protein between treatments or the CK, which was similar between all amended and CK plots (mean % ranged from 6.52 ± 0.27 to 7.31 ± 0.37).
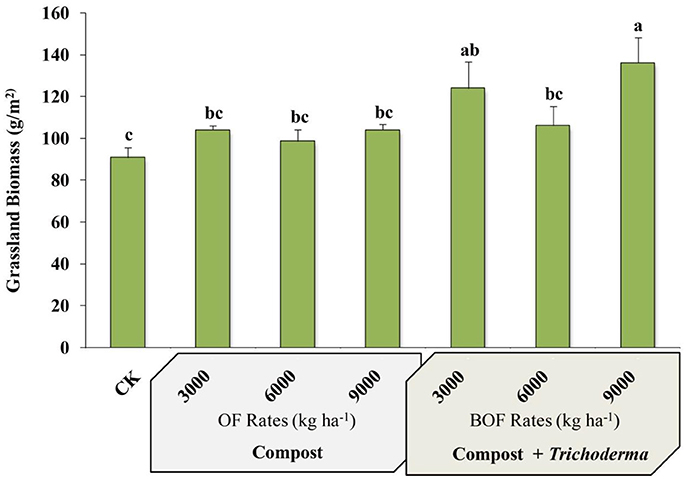
Figure 1. Grassland biomass by fertilization regime. CK: non-amended; OF 3000: 3,000 kg ha−1 organic fertilizer (composted cattle manure); OF 6000: 6,000 kg ha−1 organic fertilizer; OF 9000: 9,000 kg ha−1 organic fertilizer; BOF: 3,000 kg ha−1 biofertilizer (composted cattle manure + Trichoderma inoculum); BOF 6000: 6,000 kg ha−1 biofertilizer; BOF 9000: 9,000 kg ha−1 biofertilizer. Data from each treatment were analyzed using one-way analysis of variance (ANOVA). Bars represent mean values of three replicates ± SE. Bars that do not share a letter are significantly different (p < 0.05).
Soil Chemistry and Microbial Communities
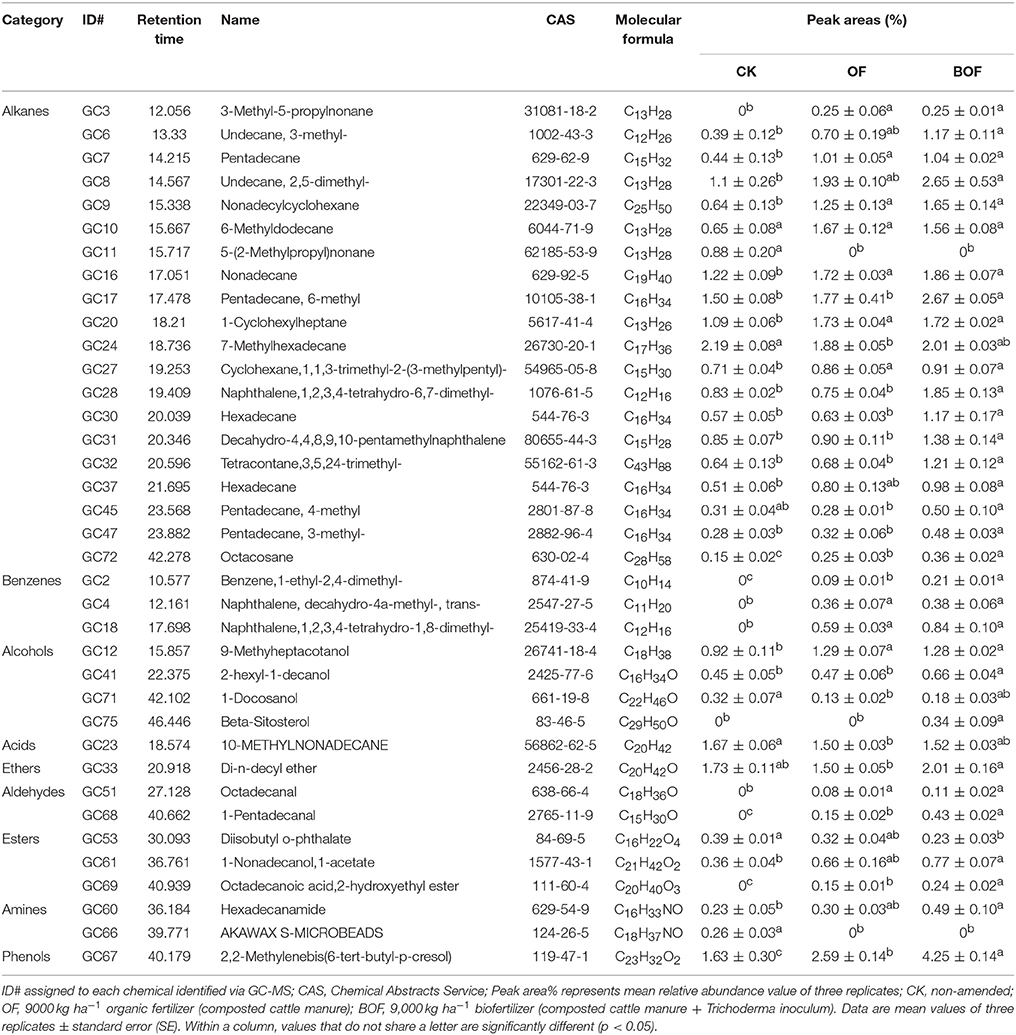
To identify potential soil microbial ecological mechanisms underlying observed differences in grassland biomass, we focused on three representative treatments (CK, OF 9000 and BOF 9,000, hereafter referred to as CK, OF, BOF). Ethyl acetate extracts from these soil samples were analyzed by GC-MS. The GC-MS analysis revealed 37 compounds, which showed significant (p < 0.05) differences in relative abundance between treatments (Table 1), including 20 alkanes, 3 benzenes, 4 alcohols, 1 acid, 1 ether, 2 aldehydes, 3 esters, 2 amines, and 1 phenol. The relative abundance of most compounds was significantly (p < 0.05) greater in plots amended with BOF, compared to non-amended control plots, and in several cases compared to OF plots.
Two-dimensional NMDS plots revealed how representative treatments related to differences in soil chemistry and microbial communities (Figure 2). For soil chemistry, pairwise contrasts indicate CK, OF, and BOF plots significantly (p < 0.05) separated by axis 1 (Figure 2A). Similarly, fungal communities showed significant (p < 0.05) variations by treatment (Figure 2B). However, bacterial communities were relatively similar across all plots (Figure 2C). As for soil microbial community compositions, we observed fewer bacterial genera (4) with significant (p < 0.05) differences in relative abundance by treatment compared to fungal genera (14) at the genus level (Table S1). Plots amended with BOF had greater abundance of fungal genera Archaeorhizomyces and Trichoderma but reduced abundance of Ophiosphaerella compared to OF or non-amended plots (Figure 3).
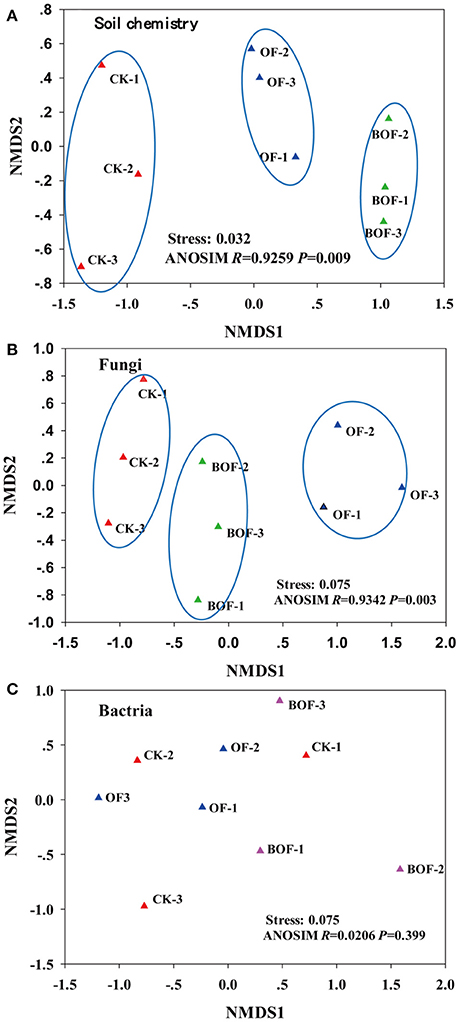
Figure 2. The first two non-metric multidimensional scaling (NMDS) axes of (A) soil chemistry community, (B) fungal community and (C) bacterial community. CK, non-amended; OF: 9,000 kg ha−1 organic fertilizer (composted cattle manure); BOF, 9,000 kg ha−1 biofertilizer (composted cattle manure + Trichoderma inoculum). Each treatment had three replications (CK-1, CK-2, CK-3; OF-1, OF-2, OF-3; BOF-1, BOF-2, BOF-3).
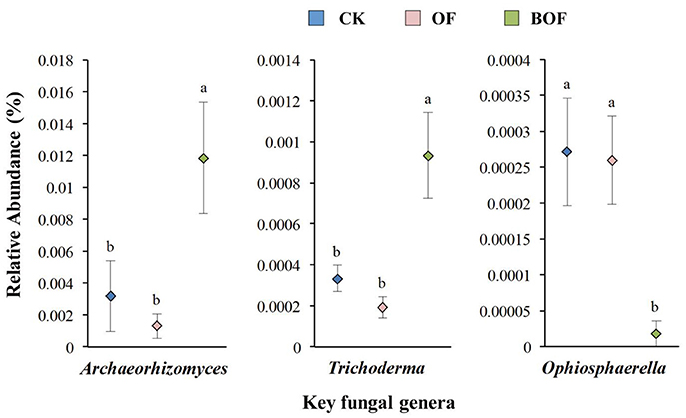
Figure 3. Key fungal genera by fertilization regime. Y-axis scale for figures is based on relative abundance (%) range for different genera. CK (blue): non-amended; OF (pink): 9,000 kg ha−1 organic fertilizer (composted cattle manure); BOF (green): 9,000 kg ha−1 biofertilizer (composted cattle manure + Trichoderma inoculum). Diamonds and lines represent mean values of three replicates ± SE. Values that do not share a letter are significantly different (p < 0.05).
Soil Properties, Microbes, and Grassland Biomass Interactions
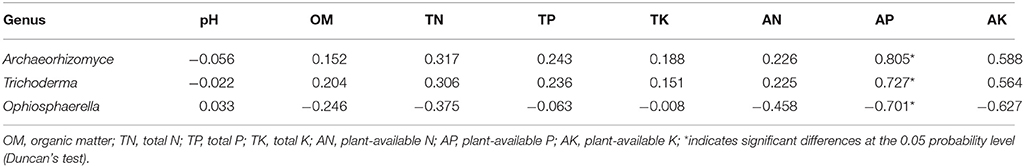
Soil properties differed by fertilization regime (Table S2). Application of any rate of OF or BOF significantly increased soil organic matter and total soil N, compared to non-amended control plots. However, no patterns were observed regarding differences in plant-available P, and there were no significant difference in other soil properties. According to Pearson's correlations (Table 2), the key fungal genera abundances of Archaeorhizomyces (p = 0.027) and Trichoderma (p = 0.035) were just positively correlated with higher plant-available P, while Ophiosphaerella (p = 0.035) was negatively correlated. As shown in Figure 4, positive correlations between grassland biomass and Archaeorhizomyces (p = 0.0068) and Trichoderma (p = 0.0048) were found, while Ophiosphaerella (p = 0.0432) was negatively correlated with aboveground grassland biomass.
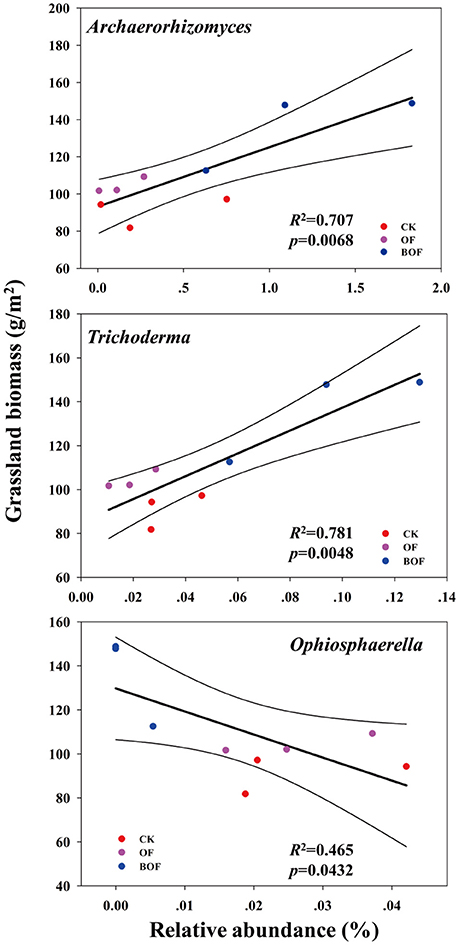
Figure 4. Correlations (p < 0.05) between key fungal genera (from Figure 3) and aboveground grassland biomass.
SEM Pathways and Grassland Biomass
Structural equation modeling for grassland biomass was a strong fit with the data (χ2 = 4.779, DF = 7, P = 0.687, NFI = 0.946, RFI = 0.838, IFI = 1.027, RMSEA = 0.000, Table S3). As shown in Figure 5, the explanation of the variance in grassland biomass was directly dependent on soil organic matter, Trichoderma abundance, bacterial and fungal communities, as affected by fertilization regime. Trichoderma abundance had the strongest overall effect (path coefficient = 0.879) on grassland biomass, and the effects of soil organic matter (path coefficient = 0.329), fungal community (path coefficient = 0.245), and bacterial community (path coefficient = 0.287) were also positive. However, the soil chemistry had no significant effect on grassland biomass. Fungal communities were directly mediated by total soil N (path coefficient = 1.250) and the soil chemistry (path coefficient = −1.226). Soil bacterial communities were mainly effected by the soil chemistry (path coefficient = 1.030).
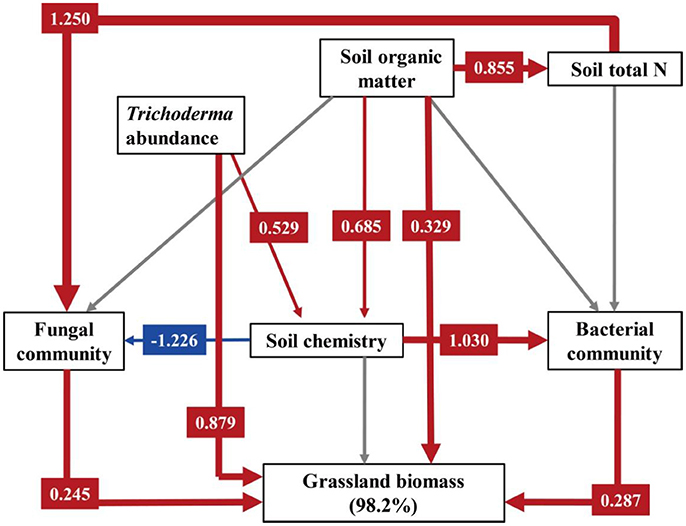
Figure 5. Structural equation modeling (SEM) for grassland biomass. Numbers following variables show the percentage of its variance explained by its predictors. We utilized PCA to demonstrate variations in fungal community across different treatments, and PC1 was selected as the parameter describing the fungal community (PC1 explained 87.25% of total variation in the fungal community). Similarly, PC1 explained 44.85 and 62.77% of total variation in the bacterial community and the soil chemistry, respectively. A path coefficient is analogous to a partial correlation coefficient and describes the strength and sign of the relationship between two variables. Negative pathways are shown as blue lines, positive pathways are shown as red lines, and line thickness represents the intensity of influence. Non-significant pathways are shown in gray. Models are a good fit of our data. Model fits are given in Table S3, and significance levels are provided in Table S4.
Discussion
Plant and microbial community structure and biodiversity are crucial for maintaining ecosystem sustainability and productivity (Bell et al., 2005; Cardinale et al., 2006). In our study, we found the application of organic fertilizer or Trichoderma biofertilizers resulted in improved aboveground grassland biomass, with optimal results from applying 9,000 kg ha−1 Trichoderma biofertilizer. Multiple mechanisms may be responsible for increasing grassland biomass. Our results indicate different fertilization regimes drove differences in soil chemistry and edaphic properties, with concomitant shifts in key soil fungal genera. This trend was most pronounced for high inputs of Trichoderma biofertilizer.
Differentiation of soil chemistry between selected fertilization regimes (CK, OF, and BOF), presumably formed distinct resource niches in our grassland plots. Many of the identified chemicals were most abundant in plots amended with BOF, compared with CK or OF. Previous studies indicate some of these substances, such as pentadecane and nonadecane, have antifungal properties (Yuan et al., 2012; Raza et al., 2015). In addition, antifungal activity has been reported for the majority of benzenes, alcohols, and phenols (Raza et al., 2015). Based on our findings, we propose antifungal substances were more abundant in the BOF9000 treatment, potentially protecting plants from fungal pathogens. Future studies should focus on how soil chemical profiles influence plant pathogens and could be managed to provide greater plant protection.
Various soil chemical structures are consumed by microbes and can facilitate microbial diversity (Mwafulirwa et al., 2016). We observed a corresponding differentiation of the fungal community but not the bacterial community, based on soil chemistry, driven by fertilization regime. Martin et al. (2015) reported application of compost is a key factor explaining differences in soil microflora. Sun et al. (2015) also demonstrated the role of organic fertilizer in facilitating soil microbial community stability and diversity, which provides a foundation for grassland biomass production. Based on our correlations between microbial taxa and grassland biomass (Figure 5), we propose that Archaeorhizomyces and Trichoderma may promote plant growth, while Ophiosphaerella had the opposite effect. Trichoderma spp. are well-known for their capacity to improve plant growth and promote health in agricultural systems (Harman et al., 2004; Gravel et al., 2007; Bae et al., 2009). Examples of these mechanisms include: improving secretion plant stimulatory compounds, such as growth hormones (indole acetic acids, cytokinins, gibberellins, and zeatins; Gravel et al., 2007; Contreras-Cornejo et at., 2009), enhancing solubilization of soil nutrients (Yedidia et al., 2001; Kapri and Tewari, 2010), increasing root length and number of root hairs to absorb nutrients by exploring larger spaces of soil (Bjorkman, 2004; Samolski et al., 2012). The life cycle, ecology, and evolution of Archaeorhizomycetes remain largely unknown. However, it is understood that Archaeorhizomycetes are non-pathogenic (Rosling et al., 2011). Evidence suggests Ophiosphaerella are phytopathogens (Kaminski and Hsiang, 2008). Hciii et al. (2007) reported three Ophiosphaerella species (O. herpotricha, O. korrae, O. narmari) can cause spring dead spot in Bermudagrass (Cynodondactylon [L.] Pers.). Venkatasubbaiah et al. (1994) demonstrated metabolites produced by O. Herpotricha can cause necrosis in Bermudagrass and other plant species. Archaeorhizomyces and Trichoderma were mainly positively correlated with greater plant-available P, while Ophiosphaerella presented a negative correlation (Table 2). Taken together, we conclude greater plant-available P was beneficial to aboveground plant biomass by increasing the relative abundances of Archaeorhizomyces and Trichoderma while decreasing Ophiosphaerella. Previous research has reported Trichoderma can increase plant P-uptake by increasing P-solubilization in soils (López-Bucio et al., 2015). This increased plant growth was greatest when plant-available P from composted cattle manure was provided along with Trichoderma inoculum (BOF). While our current research shows strong correlative linkages between plant, soil, and microbial factors, future research should identify if plant-available P increased plant growth directly, or via indirect effects on the abundance of Archaeorhizomyces and Trichoderma, and explore methods to better demonstrate a clear mechanistic pathway between microbial community composition, soil chemistry, and grassland biomass.
According to our SEM, soil organic matter, Total N, Trichoderma abundance, bacterial community, fungal community, and soil chemistry make a good explain for aboveground plant biomass in meadow steppe grasslands. Christian et al. (2008) demonstrated that fungal communities were most closely associated with changes in soil nutrient status, while soil pH was the best predictor of bacterial communities. This supports our results indicating total soil N had a significant influence on the fungal community and no significant effect on the bacterial community. However, Trichoderma had the greatest influence on grassland biomass. This supports the findings of Sivan et al. (1987), who demonstrated plant survival and yield can be increased, following Trichoderma inoculation in greenhouse or field settings.
Trichoderma biofertilizer (9,000 kg ha−1) effectively regulated soil chemistry and microbial communities, driving substantially improved aboveground plant biomass compared to organic fertilizer not containing Trichoderma. Certain soil compounds with antifungal activity may ensure individual plant fitness and increase grassland biomass. Plant-available P was beneficial to grassland biomass production, presumably by increasing the relative abundances of beneficial microbes and decreasing phytopathogenic microbes. Soil organic matter, Trichoderma abundance, and bacterial and fungal communities have direct influences on grassland biomass, while soil chemistry indirectly increases grassland biomass through alterations in bacterial and fungal communities. Among these factors, Trichoderma was the primary contributor to improve grassland biomass. Our study helps to provide a basis for grassland biomass production from the perspective of soil microbial ecology, and may ultimately improve management of grassland soils for key provisioning services. While the selected regimes in our study suggest potential underlying microbial ecological mechanisms, continuous monitoring and analyses of a gradient of fertilization rates will allow a more comprehensive mechanism understanding.
Author Contributions
FZ, GY, and YZ conceived and designed the experiments. FZ and YH performed the experiments. FZ and GL analyzed the data. FZ and YZ contributed reagents, materials, and analysis tools: FZ, AC, JZ, GW, and YZ wrote the paper. All authors reviewed and contributed to the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Elizabeth Bach for her helpful manuscript suggestions, and thank Dr. Xiao Sun and Dr. Jian Hu for their contribution to our revising work. This study was funded by the National Natural Science Foundation of China (31602006), the Natural Science Foundation of Jiangsu Province (BK20160735), the Nanjing Agricultural University Foundation (Y0201600442), the China Postdoctoral Science Foundation (2015M581815) and the Postdoctoral Science Foundation of Jiangsu Province (1601265C).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00848/full#supplementary-material
References
Bae, H., Sicher, R. C., Kim, M. S., Kim, S. H., Strem, M. D., Melnick, R. L., et al. (2009). The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. doi: 10.1093/jxb/erp165
Bell, T., Newman, J. A., Silverman, B. W., Turner, S. L., and Lilley, A. K. (2005). The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160. doi: 10.1038/nature03891
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57, 289–300.
Bjorkman, T. (2004). Effect of Trichoderma colonization on auxin-mediated regulation of root elongation. Plant Growth Regul. 43, 89–92. doi: 10.1023/B:GROW.0000038260.85276.82
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Cardinale, B. J., Srivastava, D. S., Duffy, J. E., Wright, J. P., Downing, A. L., Sankaran, M., et al. (2006). Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443, 989–992. doi: 10.1038/nature05202
Christian, L. L., Michael, S. S., Mark, A. B., and Noah, F. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415. doi: 10.1016/j.soilbio.2008.05.021
Conant, R. T., Cerri, C. E., Osborne, B. B., and Paustian, K. (2017). Grassland management impacts on soil carbon stocks: a new synthesis. Ecol. Appl. 27, 662–668. doi: 10.1002/eap.1473
Contreras-Cornejo, H., Macias-Rodriguez, L., Cortes-Penagos, C., and Lopez-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Da Silva, T. E., Detmann, E., Franco M de, O., Nobre, P. M. N., and Rocha, G. C. (2016). Evaluation of digestion procedures in Kjeldahl method to quantify total nitrogen in analyses applied to animal nutrition. Acta Sci. Anim. Sci. 38, 45–51. doi: 10.4025/actascianimsci.v38i1.29171
De Souza, J. T., Bailey, B. A., Pomella, A. W. V., Erbe, E. F., Murphy, C. A., Bae, H., et al. (2008). Colonization of cacao seedlings by Trichoderma stromaticumamycoparasite of the witches' broom pathogen, and its influence on plant growth and resistance. Biol. Control. 46, 36–45. doi: 10.1016/j.biocontrol.2008.01.010
Dessaux, Y., Grandclément, C., and Faure, D. (2016). Engineering the rhizosphere. Trends. Plant. Sci. 21, 566–578. doi: 10.1016/j.tplants.2016.01.002
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Finkel, O. M., Castrillo, G., Paredes, S. H., González, I. S., and Dang, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Fu, L., Penton, C. R., Ruan, Y., Shen, Z., Xue, C., Li, R., et al. (2017). Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil. Biol. Biochem. 104, 39–48. doi: 10.1016/j.soilbio.2016.10.008
Gravel, V., Antoun, H., and Tweddell, R. J. (2007). Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biol. Biochem. 39, 1968–1977. doi: 10.1016/j.soilbio.2007.02.015
Groenigen, J. W. V., Velthof, G. L., Bolt, F. J. E. V. D., An, V., and Kuikman, P. J. (2005). Seasonal variation in N2O emissions from urine patches: Effects of urine concentration, soil compaction and dung. Plant Soil 273, 15–27. doi: 10.1007/s11104-004-6261-2
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hciii, W., Skinner, D. Z., and Tisserat, N. A. (2007). Geographic distribution and genetic diversity of three Ophiosphaerella species that cause spring dead spot of bermudagrass. Plant Dis. 83, 1160–1166. doi: 10.1094/PDIS.1999.83.12.1160
Hermann, J. M., Lang, M., Gonçalves, J., and Hasenack, H. (2016). Forest-grassland biodiversity hotspot under siege: land conversion counteracts nature conservation. Ecosyst. Health Sustainabil. 2:e01224. doi: 10.1002/ehs2.1224
Huang, X., Chen, L., Ran, W., Shen, Q., and Yang, X. (2011). Trichoderma harzianum strain SQR-T37 and its bio-organic fertilizer could control Rhizoctoniasolani damping-off disease in cucumber seedlings mainly by the mycoparasitism. Appl. Microbiol. Biot. 91, 741–755. doi: 10.1007/s00253-011-3259-6
IUSS WG WRB, (2015). World Reference Base for Soil Resources 2014, update 2015. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. World Soil Resources Reports No. 106. FAO, Rome, 153–154.
Kaminski, J. E., and Hsiang, T. (2008). First report of Ophiosphaerella agrostis infecting creeping bentgrass in Canada. Plant Dis. 90:1114. doi: 10.1094/PD-90-1114B
Kapri, A., and Tewari, L. (2010). Phosphate solubilization potential and phosphatase activity of rhizospheric Trichoderma spp. Braz. J. Microbiol. 41, 787–795. doi: 10.1590/S1517-83822010005000001
Knudsen, D., Peterson, G., and Pratt, P. (1982). “Lithium, sodium, and potassium,” in Methods of Soil Analysis, Part 2, Chemical and Microbiological Properties. eds A. L. Page, R. H. Miller, and D. R. Kenney (Madison: American Society of Agronomy, Soil Science Society of American), 225–246.
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Komas, C., Kairis, O., Karavitis, C. H., Ritsema, C., Salvati, L., Acikalin, S., et al. (2014). Evaluation and selection of indicators for land degradation and desertification monitoring: methodological approach. Environ. Manage. 54, 951–970. doi: 10.1007/s00267-013-0109-6
López-Bucio, J., Pelagio-Flores, R., and Herrera-Estrella, A. (2015). Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic. 196, 109–123. doi: 10.1016/j.scienta.2015.08.043
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martin, H., Beat, F., Jochen, M., Paul, M., and Franco, W. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME. J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Masunaka, A., Hyakumachi, M., and Takenaka, S. (2011). Plant growth-promoting fungus, Trichoderma koningi suppresses isoflavonoid phytoalexin vestitol production for colonization on/in the roots of Lotus japonicus. Microbes Environ. 26, 128–134. doi: 10.1264/jsme2.ME10176
Moral, R., Paredes, C., Bustamante, M. A., Marhuenda-Egea, F., and Bernal, M. P. (2009). Utilisation of manure composts by high-value crops: safety and environmental challenges. Bioresour. Technol. 100, 5454–5460. doi: 10.1016/j.biortech.2008.12.007
Mwafulirwa, L., Baggs, E. M., Russell, J., George, T., Morley, N., Sim, A., et al. (2016). Barley genotype influences stabilization of rhizodeposition-derived C and soil organic matter mineralization. Soil Biol. Biochem. 95, 60–69. doi: 10.1016/j.soilbio.2015.12.011
Ng, E. L., Patti, A. F., Rose, M. T., Schefe, C. R., Wilkinson, K., Smernik, R. J., et al. (2014). Does the chemical nature of soil carbon drive the structure and functioning of soil microbial communities? Soil Biol. Biochem. 70, 54–61. doi: 10.1016/j.soilbio.2013.12.004
Oburger, E., and Schmidt, H. (2016). New methods to unravel rhizosphere processes. Trends Plant. Sci. 21, 243–255. doi: 10.1016/j.tplants.2015.12.005
Oksanen, J., Kindt, R., Legendre, P., O'Hara, B., et al. (2009). Vegan: Community Ecology Package. R. package. Version. 1.15–12.
Olsen, S. R., and Sommers, L. E. (1982). “Phosphorous,” in Methods of Soil Analysis. Part 2. Agronomy, 2nd Edn, ed A. L Page et al.(Madison: American Society of Agronomy), 403–429.
Pierre, P., Alex, W., Anne, C., Alexander, E. M., Stephen, A. R., and Jurriaan, T. (2017). Metabolite profiling of non-sterile rhizosphere soil. Plant. J. 92, 147–162. doi: 10.1111/tpj.13639
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moenne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soil borne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Raza, W., Yuan, J., Wu, Y. C., Rajer, F. U., Huang, Q., and Shen, Q. R. (2015). Biocontroltraits of two Paenibacilluspolymyxa strains SQR-21 and WR-2 inresponse to fusaric acid, a phytotoxin produced by Fusarium species. Plant Pathol. 64, 1041–1052. doi: 10.1111/ppa.12354
Rosling, A., Cox, F., Cruz-Martinez, K., Ihrmark, K., Grelet, G. A., Lindahl, B. D., James, T. Y., et al. (2011). Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333, 876–879. doi: 10.1126/science.1206958
Salles, J. F., Poly, F., Schmid, B., and Le Roux, X. (2009). Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 90, 3324–3332. doi: 10.1890/09-0188.1
Samolski, I., Rincón, A. M., Pinzón, L. M., Viterbo, A., and Monte, E. (2012). The qid74 gene from Trichoderma harzianum has a role in root architecture and plant biofertilization. Microbiol 158, 129–138. doi: 10.1099/mic.0.053140-0
Sattari, S. Z., Bouwman, A. F., Rodríguez, R. M., Beusen, A. H. W., and Van Ittersum, M. K. (2016). Negative global phosphorus budgets challenge sustainable intensification of grasslands. Nat. Commun. 7:10696. doi: 10.1038/ncomms10696
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shen, Z. Z., Ruan, Y. Z., Chao, X., Zhang, J., Li, R., and Shen, Q. R. (2015). Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fert. Soils. 51, 553–562. doi: 10.1007/s00374-015-1002-7
Shi, R. (1996). Agricultural Chemistry Analyses of Soils, 2nd Edn. Beijing: China Agricultural Press (in Chinese), 37–39.
Sivan, A., Ucko, O., and Chet, I. (1987). Biological control of Fusarium crown rot of tomato by Trichoderma harzianum under field conditions. Plant. Dis. 71, 587–592. doi: 10.1094/PD-71-0587
Soussana, J. F., Allard, V., Pilegaard, K., Ambus, P., Amman, C., Campbell, C., et al. (2007). Full accounting of the greenhouse gas (CO2, N2O, CH4) budget of nine European grassland sites. Agric. Ecosyst. Environ. 121, 121–134. doi: 10.1016/j.agee.2006.12.022
Sun, R. B., Zhang, X. X., Guo, X. S., Wang, D. Z., and Chu, H. Y. (2015). Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 88, 9–18. doi: 10.1016/j.soilbio.2015.05.007
Tang, Z., Yu, G., Liu, D., Xu, D., and Shen, Q. (2011). Different analysis techniques for fluorescence excitation-emission matrix spectroscopy to assess compost maturity. Chemosphere 82, 1202–1208. doi: 10.1016/j.chemosphere.2010.11.032
Venkatasubbaiah, P., Tisserat, N. A., and Chilton, W. S. (1994). Metabolites of Ophiosphaerella herpotricha, a cause of spring dead spot of bermudagrass. Mycopathologia 128, 155–159. doi: 10.1007/BF01138477
van der Heijden, M. G., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 111, 5266–5270. doi: 10.1073/pnas.1320054111
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Watanabe, F. S., and Olsen, S. R. (1965). Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soil. Soil. Sci. Soc. Am. J. 22, 677–678. doi: 10.2136/sssaj1965.03615995002900060025x
Xiong, W., Guo, S., Jousset, A., Zhao, Q., Wu, H., Li, R., et al. (2017). Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 114, 238–247. doi: 10.1016/j.soilbio.2017.07.016
Xu, W., Cai, Y. P., Yang, Z. F., et al. (2017). Microbial nitrification, denitrification and respiration in the leached cinnamon soil of the upper basin of Miyun Reservoir. Sci. Rep. 7:42032. doi: 10.1038/srep42032
Yan, Y., Kuramae, E. E., de Hollander, M., Klinkhamer, P. G., and van Veen, J. A. (2017). Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME. J. 11, 56–66. doi: 10.1038/ismej.2016.108
Yedidia, I., Srivastva, A. K., Kapulnik, Y., and Chet, I. (2001). Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant. Soil. 235, 235–242. doi: 10.1023/A:1011990013955
Yuan, J., Raza, W., Shen, Q. R., and Huang, Q. W. (2012). Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. Cubense. Appl. Environ. Micro. 78:5942. doi: 10.1128/AEM.01357-12
Zhan, S., Wang, Y., Zhu, Z., Li, W., and Bai, Y. (2017). Nitrogen enrichment alters plant N: P stoichiometry and intensifies phosphorus limitation in a steppe ecosystem. Environ. Exp. Bot. 134, 21–32. doi: 10.1016/j.envexpbot.2016.10.014
Zhang, F. G., Yuan, J., Yang, X. M., Cui, Y. Q., Chen, L. H., Ran, W., et al. (2013a). Putative Trichoderma harzianummutant promotes cucumber growth by enhanced production of indole acetic acid and plant colonization. Plant Soil. 368, 433–444. doi: 10.1007/s11104-012-1519-6
Keywords: in situ fertilization experiment, high-throughput sequencing, soil chemistry, key fungal genera, structure equation modeling
Citation: Zhang F, Huo Y, Cobb AB, Luo G, Zhou J, Yang G, Wilson GWT and Zhang Y (2018) Trichoderma Biofertilizer Links to Altered Soil Chemistry, Altered Microbial Communities, and Improved Grassland Biomass. Front. Microbiol. 9:848. doi: 10.3389/fmicb.2018.00848
Received: 20 October 2017; Accepted: 12 April 2018;
Published: 30 April 2018.
Edited by:
Etienne Yergeau, Institut National de la Recherche Scientifique (INRS), CanadaReviewed by:
Maaike Van Agtmaal, Imperial College London, United KingdomDavid Levy-Booth, University of British Columbia, Canada
Copyright © 2018 Zhang, Huo, Cobb, Luo, Zhou, Yang, Wilson and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingjun Zhang, zhangyj@cau.edu.cn
 Fengge Zhang
Fengge Zhang Yunqian Huo1
Yunqian Huo1 Adam B. Cobb
Adam B. Cobb Jiqiong Zhou
Jiqiong Zhou Gaowen Yang
Gaowen Yang Gail W. T. Wilson
Gail W. T. Wilson Yingjun Zhang
Yingjun Zhang