- 1Institute for Chemistry and Biology of the Marine Environment, Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
- 2Institute of Microbiology, Braunschweig University of Technology, Braunschweig, Germany
Dinoroseobacter shibae is an aerobic anoxygenic phototroph and able to utilize light energy to support its aerobic energy metabolism. Since the cells can also grow anaerobically with nitrate and nitrite as terminal electron acceptor, we were interested in how the cells profit from photosynthesis during denitrification and what the steps of chemiosmotic energy conservation are. Therefore, we conducted proton translocation experiments and compared O2-, NO3-, and NO2- respiration during different light regimes and in the dark. We used wild type cells and transposon mutants with knocked-out nitrate- and nitrite- reductase genes (napA and nirS), as well as a mutant (ppsR) impaired in bacteriochlorophyll a synthesis. Light had a positive impact on proton translocation, independent of the type of terminal electron acceptor present. In the absence of an electron acceptor, however, light did not stimulate proton translocation. The light-driven add-on to proton translocation was about 1.4 H+/e- for O2 respiration and about 1.1 H+/e- for NO3- and NO2-. We could see that the chemiosmotic energy conservation during aerobic respiration involved proton translocation, mediated by the NADH dehydrogenase, the cytochrome bc1 complex, and the cytochrome c oxidase. During denitrification the last proton translocation step of the electron transport was missing, resulting in a lower H+/e- ratio during anoxia. Furthermore, we studied the type of light-harvesting and found that the cells were able to channel light from the green–blue spectrum most efficiently, while red light has only minor impact. This fits well with the depth profiles for D. shibae abundance in the ocean and the penetration depth of light with different wavelengths into the water column.
Introduction
Dinoroseobacter shibae is an aerobic anoxygenic phototroph and has become a model organism for the Roseobacter group (Buchan et al., 2000; Martens et al., 2006; Wagner-Döbler and Biebl, 2006; Simon et al., 2017). This physiologically heterogeneous group of bacteria is highly abundant in photic zones of marine environments worldwide (Biebl and Wagner-Döbler, 2006). D. shibae maintains a fine-tuned regulatory network, which allows for coping with anoxic conditions. It is also capable of using nitrate and nitrite as an alternative electron acceptor (Ebert et al., 2017). All of the genes necessary for the stepwise reduction of nitrate to nitrite (via periplasmic nitrate reductase NapABC), nitric oxide (via nitrite reductase NirS), nitrous oxide (NorBC) and finally dinitrogen (NosZ) were found tightly clustered on the chromosome of D. shibae (Wagner-Döbler et al., 2010; Laass et al., 2014).
Dinoroseobacter shibae possesses a photosystem containing bacteriochlorophyll a and the carotenoid spheroidenone. Although photosynthesis in aerobic anoxygenic phototrophs involves cyclic electron transport only, the presence of a terminal electron acceptor is essential for the utilization of light (Shiba et al., 1979; Harashima et al., 1987; Holert et al., 2011). Under reducing conditions in the absence of terminal electron acceptors, photosynthesis is not performed, as the primary electron acceptor pheophytin is unable to take up electrons.
Dinoroseobacter shibae can utilize light energy to quickly regenerate, with the help of an in increased membrane potential, its energetic charge after suffering from short-term anoxia (Holert et al., 2011; Kirchhoff and Cypionka, 2017). Light exposure can also enhance survival of the cells during starvation (Soora and Cypionka, 2013). However, whether D. shibae can also benefit from light under anoxic conditions while performing denitrification was unclear.
In this study we wanted to assess the contribution of photosynthesis to chemiosmotic energy conservation during aerobic and anaerobic respiration. For this purpose, we measured the proton translocation under oxic conditions in the light and in the dark and compared this with the same process during denitrification. To examine whether the results are specific for the nitrate- and nitrite- reductase, two insertion mutants (napA and nirS) were analyzed for their potential in proton translocation (Ebert et al., 2013).
Furthermore, we were interested in the light-harvesting and channeling capabilities of the antenna complex of D. shibae. Therefore, we compared the effect of different light colors on proton translocation during aerobic respiration.
Materials and Methods
Bacterial Strains and Cultivation
Dinoroseobacter shibae DFL 12T (Biebl et al., 2005) was grown in artificial seawater medium (SWM) with 10 mM succinate and 25 mM nitrate in a volume of 150 ml (see Supplement 1). Cells were cultivated anaerobically in a diurnal light/dark rhythm (12 h/12 h, 12 μmol photons m-2 s-1) in a shaker at 25°C and 125 rpm (Soora and Cypionka, 2013). The expression of the photopigments is inhibited by light and therefore occurs predominantly in the dark (Endres et al., 2015), which makes phototrophy dependent on a day–night cycle, different to anaerobic phototrophs (Harashima et al., 1980; Yurkov and van Gemerden, 1993). The transposon insertion mutants napA (DSTn2943) and nirS (DSTn3258) (Ebert et al., 2013) lack the nitrate- and nitrite- reductase genes, respectively and are therefore not able to utilize NO3- or NO2- as terminal electron acceptor. Both were grown within the identical medium as the wild type, but under oxic conditions to provide comparability between both mutants. In control experiments, the napA and nirS mutants were also successfully grown under anoxic conditions with NO3- or NO2- as electron acceptor, which resulted in different ODmax and generation times. Additionally, the transposon mutant ppsR (DSTn4634, Ebert et al., 2013), which does not produce bacteriochlorophyll a (see Supplement 2), was used as a control strain.
Proton Translocation Measurements
The proton translocation experiments were conducted after a modified method after Fitz and Cypionka (1989). Cells were grown for 18 h to an OD436 of approximately 0.8 and subsequently harvested by centrifugation (150 ml culture, 10,000 ×g, 10 min, 4°C, Beckman J2-HS). The supernatant was discarded and the pellet was resuspended in 6 ml of non-buffered solution (300 mM NaCl, pH 7.4) and stored on ice. Proton translocation measurements were performed in a small reaction tube (3 ml) at 30°C. The tube was filled with 2 ml of cell suspension, 200 μl of KSCN (1 M in H2O) and 10 μl succinate (500 mM in H2O). Afterward the tube was closed with a rubber stopper and constantly mixed with a magnetic stirrer. The stopper had an in- and out-let for the N2-gassing to provide constant anoxic conditions, an opening for the pH electrode (type Inlab Micro, Mettler Toledo) and for the addition of various electron acceptors. For the establishment of anoxic conditions, the suspension was gassed for 30 min with N2. This gives the cell enough time to establish the denitrification apparatus. During subsequent steps the anoxic conditions were kept by continuous N2-flushing. Small amounts of O2 (10–20 μl O2-saturated H2O, 12.5–25 nmol), NO3- (10–20 μl 1 mM NO3- in H2O, 10–20 nmol) NO2- (10–20 μl 1 mM NO3- in H2O, 10–20 nmol) were added to the suspension and pH changes were recorded (Figure 1). For this purpose, a pH electrode was connected to an AD converter (ADC-16, pico Technology) which was handled by software (MPwin version 2008.08.25, Cypionka, 2005). KSCN was added to decrease the membrane potential of the cells by slowing down the backflow of the protons along the electrochemical gradient. The observed changes in pH were compared to calibration pulses (10 μl of 10 mM HCl) at the end of each experiment (Figure 1). This allowed for the calculation of the amount of protons translocated per added electron acceptor. Each value was extrapolated back to the time of electron acceptor addition, which usually resulted in an increase of about 10%. Exemplary proton translocation experiments with NO3- or NO2- can be found in Supplement 3.
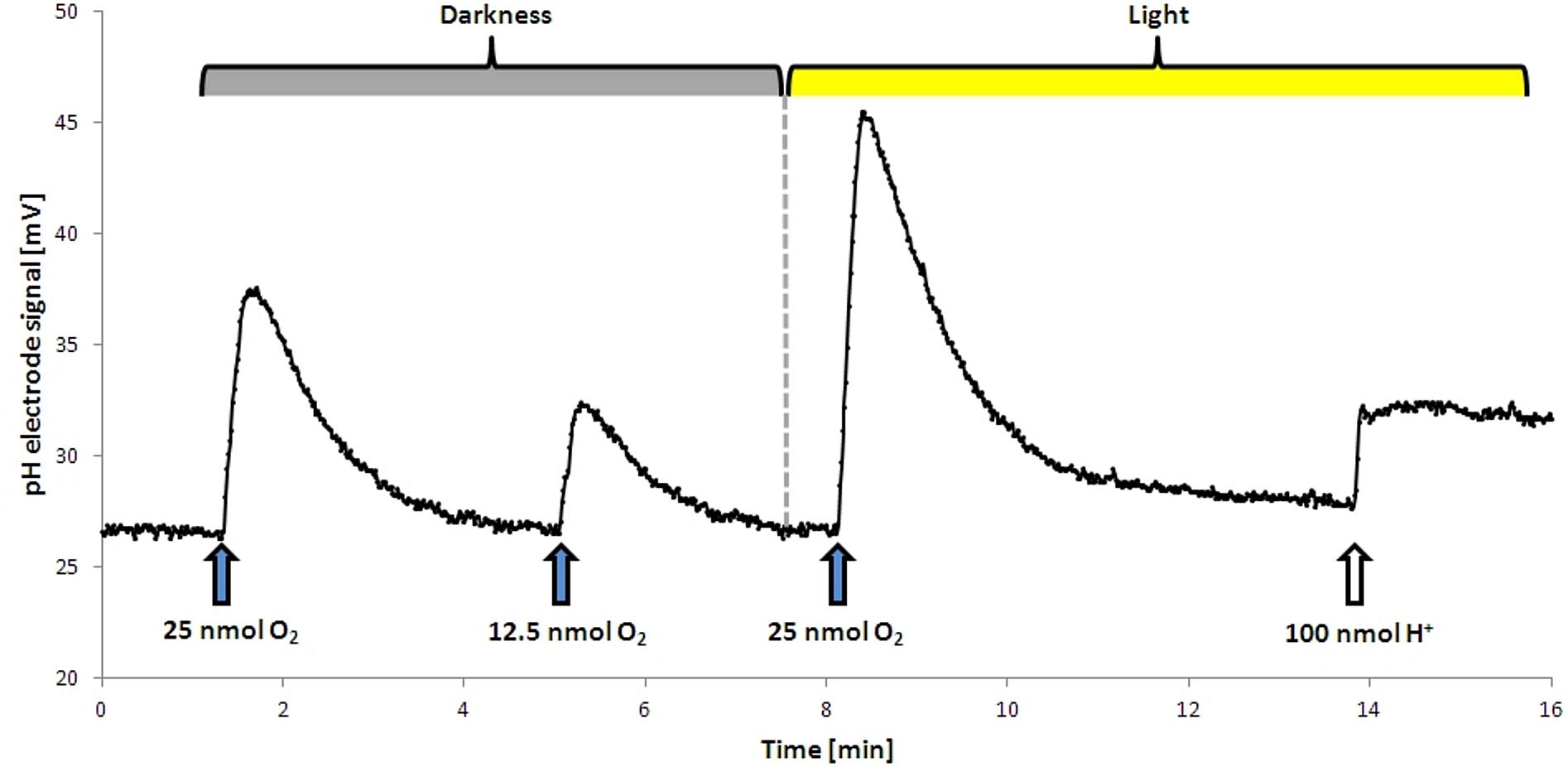
FIGURE 1. Proton translocation during aerobic respiration in the dark and in the light. Upon addition of O2 protons were released into the medium, resulting in a temporary increase in mV. This effect was stronger in the light. The dashed line shows the start point of illumination. No H+ translocation upon illumination alone was observed, the addition of a terminal electron acceptor was required. For calibration a defined pulse of HCl was added at the end of the analysis.
Set-Up for Light Quality Studies
To compare the proton translocation under different light conditions, we constructed a light cabinet with an interior coated by white paper. A lid allowed the operation of experiments from the outside. The cabinet held three types of LEDs (type 18418 1–3, Barthelme, Germany, 628, 515, and 476 nm), which allowed for the adjustment of defined conditions of red, green, and blue light. The intensity was set to 9.6 μmol photons m-2 s-1 with the help of a dimmer (Light meter model LI-189 by LI-COR, United States) For white light, all three LED types were activated simultaneously, with an intensity of 9.6 μmol photons m-2 s-1 each. In pre-experiments this intensity was shown to be non-saturating for a cell suspension of OD436 25.
Results
Light Supports Proton Translocation Independent of the Terminal Electron Acceptor of Respiration
Washed cells of D. shibae, previously grown aerobically with succinate in the presence of nitrate, were able to utilize O2, NO3-, and NO2- as terminal electron acceptors, when tested in an unbuffered cell suspension after harvesting. The quantitative measurement of proton translocation required the addition of KSCN to the suspension in order to lower the membrane potential of the cells and slow down the backflow of protons. The cells were able to translocate protons in the light and in the dark, although the amount of measured translocated protons per electron acceptor was different (Figure 2). In the dark, 8.1 H+ were translocated per O2 (mol/mol), followed by 3.9 H+ per NO3- and 2.3 H+ per NO2-. In the light, these values increased significantly for all tested electron acceptors, with O2 now allowing for the translocation of 13.7 H+, followed by 8.7 H+ per NO3- and 5.5 H+ per NO2-.
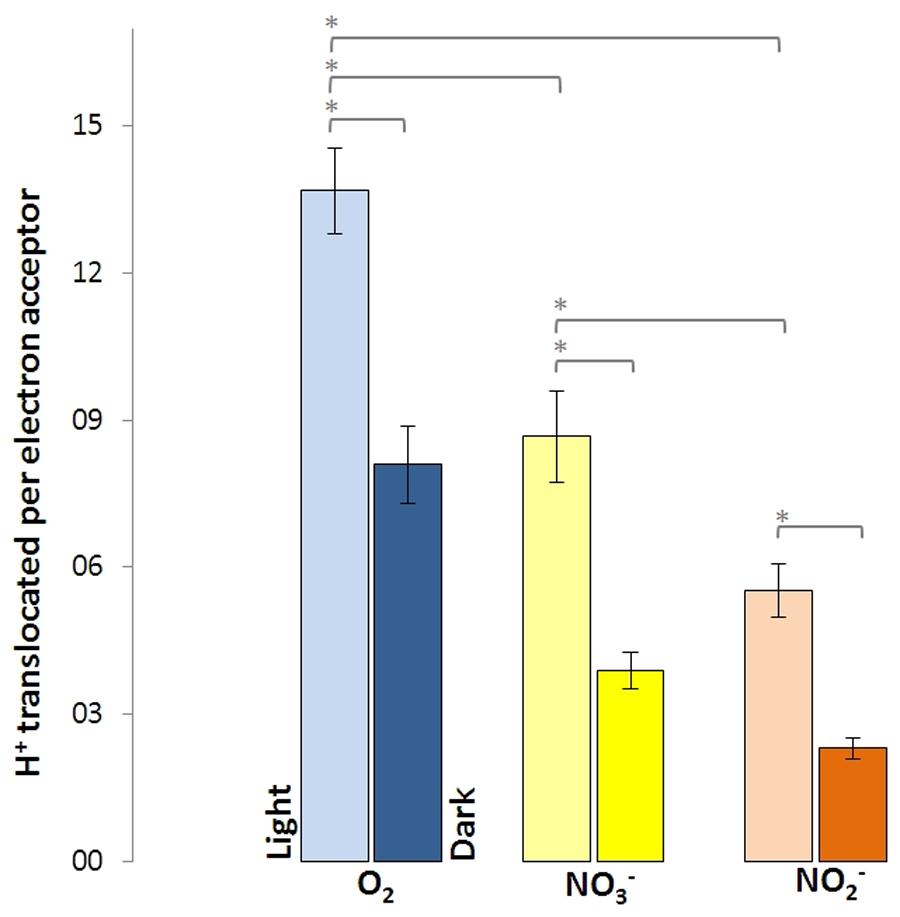
FIGURE 2. Protons translocated per electron acceptor molecule measured in white light and in the dark for Dinoroseobacter shibae wild type cells. Utilization of O2 as electron acceptor resulted in the most protons translocated, followed by NO3- and NO2-. Error bars indicate standard errors. P-values were determined using ANOVA and post hoc t-test. Asterisk indicates P-values ≤ 0.05. n = 6–8.
The nitrate reductase-deficient mutant napA and the nitrite-deficient mutant nirS translocated more protons per molecule O2 than with NO3- and NO2-. For the napA mutant strain almost no proton translocation was observed upon addition of NO3-. O2- and NO2- respiration were still operative in the napA mutant and significantly more H+ were translocated in the light than in the dark (Figure 3, left). Vice versa, the nirS mutant strain did not translocate H+ upon NO2- addition, while O2- and NO3- respiration was still operative (Figure 3, right). However, this time less H+ were translocated upon NO3- addition compared to the wild type and napA strain. This is likely due to the accumulation of NO2-.
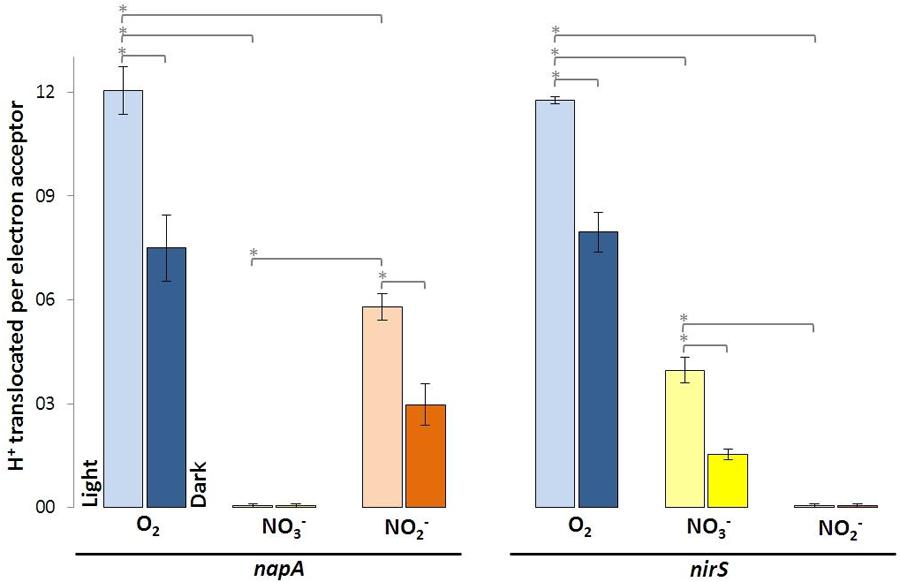
FIGURE 3. Protons translocated per electron acceptor molecule determined in white light and in the dark using indicated D. shibae transposon mutants. Error bars indicate standard errors. P-values were determined using ANOVA and post hoc t-test. Asterisk indicates P-values ≤ 0.05. n = 5–6.
Green Light Enhances Proton Translocation Most Efficiently, While Red Light Has Only Minimal Impact
Light-driven proton translocation depended on the applied wavelength. Red light (10.5 ± 0.3) had only little impact on the H+/O2 ratio, while green and blue light significantly enhanced the H+/O2 ratio (Figure 4). In the green light (14.5 ± 0.3) the cells translocated slightly more protons than in the blue light (12.7 ± 1.4). The bacteriochlorophyll a-deficient D. shibae ppsR mutant did not show any increase in proton translocation in the light at all tested wavelengths (see Supplement 4).
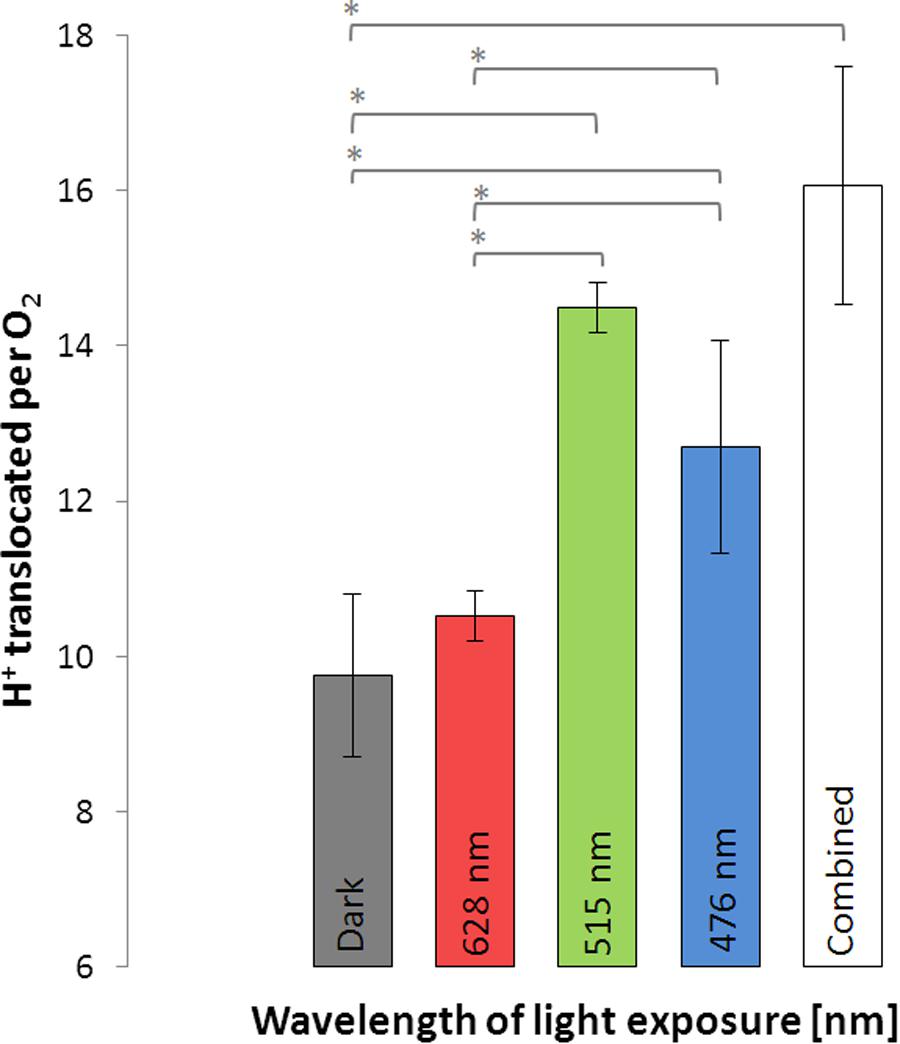
FIGURE 4. Protons translocated per molecule O2 during illumination with light of different wavelengths and in the dark using D. shibae wild type. Error bars indicate standard errors. P-values were determined using ANOVA and post hoc t-test. Asterisk indicates P-values ≤ 0.05. n = 6.
Discussion
In the present study, we have assessed proton translocation driven by aerobic respiration and denitrification with- and without illumination. We were able to differentiate the steps of chemiosmotic energy conservation in the electron transport chain of D. shibae and determined the influence of light of different wavelengths on them.
Light Supports Aerobic and Anaerobic Proton Translocation of D. shibae
As D. shibae is an aerobic anoxygenic phototrophic organism the photosystem can only support proton translocation when a terminal electron acceptor is present. This was confirmed in control experiments (Figure 1). Our observations suggested that light adds always the same surplus to the proton translocation during respiratory electron transduction, independent of the electron acceptor added. Apparently, the photosystem of D. shibae channels electrons into the electron transport chain for aerobic and anaerobic respiration in a comparative manner. This might be achieved by the reduction of the ubiquinone pool, as it was described for closely related purple bacteria (Klamt et al., 2008) and other Alphaproteobacteria (Wagner-Döbler and Biebl, 2006). The presence of cytochrome c2 genes in the genome of D. shibae (Wagner-Döbler et al., 2010) indicates a cyclic, light-driven electron transport between cytochrome c2 and the cytochrome bc1 complex (Figure 5), which might also work in the same way during denitrification. When no electron acceptor is available and the electron transport chain is fully reduced, the cyclic electron transport cannot proceed.
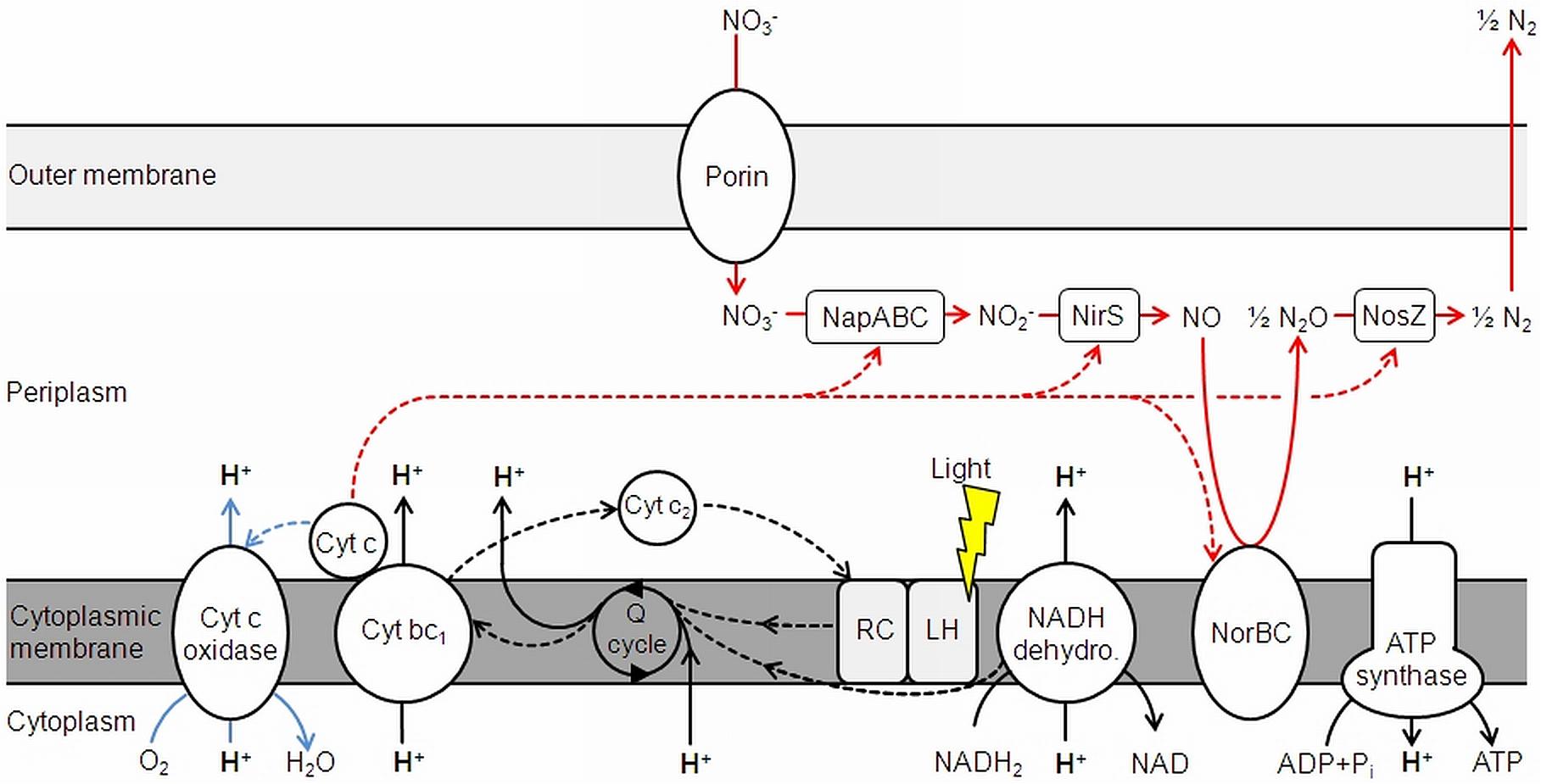
FIGURE 5. Electron transduction (dashed lines) and proton translocation in D. shibae. Denitrification is shown in red, aerobic respiration in blue. NapABC, nitrate reductase; NirS, nitrite reductase; NorBC, nitric oxide reductase; NosZ, nitrous oxide reductase; Cyt, cytochrome; Q cycle, quinone cycle. Modified and extended after Wagner-Döbler et al. (2010) and Laass et al. (2014).
It has to be taken into account that the measured proton translocation values are possibly underestimated, since the whole system is affected by the depression of the membrane potential by the addition KSCN to slow down the proton reflux (see section “Materials and Methods”). This might affect the H+/e- ratios. Anyhow, the differences in proton translocation between O2, NO3-, and NO2- can still be clearly distinguished.
When the results of the proton translocation were calculated as H+/e- ratio, O2 respiration fall from about 3.5 H+/e- in the light to 2.1 H+/e- in the dark, while both NO3- and NO2- were reduced from 1.9 to 0.8. It is interesting that the H+/e- ratio of both NO3- and NO2- were equal, although both are electron acceptors of different redox potential. This might be explained by the fact that both NapABC and NirS are not proton pumping. Still, the proportion between light and dark, as well as between O2, NO3-, and NO2- is reliable. This is on the basis that the reduction of O2 to H2O takes up 4 e- and the complete reduction of NO3- to N2 takes up 5 e-.
At a non-saturating intensity, light supported the proton translocation during aerobic respiration for 1.4 H+/e- and for 1.1 H+/e- during denitrification. Light generates a cyclic electron transport, while respiration relies on a linear electron transport. Consequently, a stoichiometric coupling between respiration-driven and light-driven proton translocation cannot be expected. Considering, that the respiratory rate decreases during illumination (Holert et al., 2011), the impact of light on proton translocation can be considered even higher than observed. The overall excess in translocated protons during aerobic respiration compared to denitrification, regardless of light exposure, leads to the conclusion that this is the result of the cytochrome c-oxidase, translocating additional protons across the membrane. This step only occurs in aerobic respiration, since O2 is reduced to H2O in the process (Figure 5). As outlined above, NapABC and NirS are not contributing to the proton gradient.
The steps of chemiosmotic energy conservation via translocated protons across the membrane during aerobic and anaerobic respiration are the NADH-dehydrogenase and the cytochrome bc1 complex. The succinate dehydrogenase also channels electrons into the Q cycle, but does not directly translocate H+ and is therefore excluded from Figure 5. The cytochrome c oxidase also adds to proton translocation, but only during aerobic respiration.
Nitrate Reduction Is Impaired When the Nitrite Cannot Be Further Metabolized
The transposon insertion mutants napA and nirS demonstrate the essential function of NapABC and NirS for the terminal reduction steps of the denitrification pathway. In the nitrite reductase deficient mutant nirS NO2- is accumulating. This obviously affects further nitrate reduction after short time, since the reaction balance is disturbed. Hence, the decrease in proton translocation compared to the wild type and the napA mutant strain can be explained (Figure 3, right).
Channeling of Blue–Green Light Is Reflected in the Environmental Distribution
Putting our results into context with the in vivo absorption spectrum of D. shibae (Biebl and Wagner-Döbler, 2006), the observed action spectrum of proton translocation efficiency (green > blue > > red) fits exactly the original data. It is build up by the combined absorption of bacteriochlorophyll a and the carotenoid spheroidenone within the light harvesting complex of D. shibae (Yurkov and Beatty, 1998; Biebl et al., 2005; Niedzwiedzki et al., 2017). Photon capturing by the present pigments and subsequent channeling into the reaction center was most efficient for light of the blue–green part of the spectrum and almost missing for red light. The adaptation of D. shibae to green–blue light is an adaptation to the highest penetration depth in the ocean. This is reflected by the depth profiles of aerobic anoxygenic phototroph abundance, which decreases from near sea surface to approximately 150 m depths (Ritchie and Johnson, 2012), following the penetration depths of preferred wavelengths of light.
Conclusion
The present study demonstrates the impact of light on the chemiosmotic energy conservation of D. shibae. It was documented that (1) proton translocation for both aerobic and anaerobic respiration is supported the almost identical degree by a light-driven cyclic electron transport. Furthermore, we have (2) identified the steps involved in proton translocation at the cytoplasmatic membrane for both types of respiration in darkness and light. Additionally, it turned out that (3) the light-harvesting antenna complex of D. shibae effectively captures photons from the blue–green spectrum to support proton translocation, while red light is almost not utilized. This fits with the environmental abundance of aerobic anoxygenic phototroph within the water column.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This study has been funded by the German Science Foundation within the Collaborative Research Centres SFB-TRR51 (Roseobacter).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the technical assistance by Frank Meyerjürgens and Jana Schmidt.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00903/full#supplementary-material
References
Biebl, H., Allgaier, M., Tindall, B. J., Koblizek, M., Lünsdorf, H., Pukall, R., et al. (2005). Dinoroseobacter shibae gen. nov., sp. nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int. J. Syst. Evol. Microbiol. 55, 1089–1096. doi: 10.1099/ijs.0.63511-0
Biebl, H., and Wagner-Döbler, I. (2006). Growth and bacteriochlorophyll a formation in taxonomically diverse aerobic anoxygenic phototrophic bacteria in chemostat culture: influence of light regimen and starvation. Process. Biochem. 41, 2153–2159. doi: 10.1016/j.procbio.2006.06.029
Buchan, A., Collier, L., Neidle, E., and Moran, M. (2000). Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl. Environ. Microbiol. 66, 4662–4672. doi: 10.1128/AEM.66.11.4662-4672.2000
Cypionka, H. (2005). Mpwin Software. Vers. 2008.08.25. Available at: www.pmbio.icbm.de/downlist.htm
Ebert, M., Laaß, S., Burghartz, M., Petersen, J., Koßmehl, S., Wöhlbrand, L., et al. (2013). Transposon mutagenesis identified chromosomal and plasmid genes essential for adaptation of the marine bacterium Dinoroseobacter shibae to anaerobic conditions. J. Bacteriol. 195, 4769–4777. doi: 10.1128/JB.00860-13
Ebert, M., Laaß, S., Thürmer, A., Roselius, L., Eckweiler, D., Daniel, R., et al. (2017). FnrL and three Dnr regulators are used for the metabolic adaptation to low oxygen tension in Dinoroseobacter shibae. Front. Microbiol. 8:642. doi: 10.3389/fmicb.2017.00642
Endres, S., Granzin, J., Circolone, F., Stadler, A., Krauss, U., Drepper, T., et al., (2015). Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae. BMC Microbiol. 15:30. doi: 10.1186/s12866-015-0365-0
Fitz, R., and Cypionka, H. (1989). A study on electron transport-driven proton translocation in Desulfovibrio desulfuricans. Arch. Microbiol. 152, 369–376. doi: 10.1007/BF00425175
Harashima, K., Hayasaki, J., Ikari, T., and Shiba, T. (1980). O2-stimulated synthesis of bacteriochlorophyll and carotenoids in marine bacteria. Plant Cell Physiol. 21, 1283–1294. doi: 10.1093/oxfordjournals.pcp.a076127
Harashima, K., Kawazoe, K., Yoshida, I., and Kamata, H. (1987). Light-stimultated aerobic growth of Erythrobacter species OCh 114. Plant Cell Physiol. 28, 365–374. doi: 10.1093/oxfordjournals.pcp.a077304
Holert, J., Hahnke, S., and Cypionka, H. (2011). Influence of light and anoxia on chemiosmotic energy conservation in Dinoroseobacter shibae. Environ. Microbiol. Rep. 3, 136–141. doi: 10.1111/j.1758-2229.2010.00199.x
Kirchhoff, C., and Cypionka, H. (2017). Boosted membrane potential as bioenergetic response to anoxia in Dinoroseobacter shibae. Front. Microbiol. 8:695. doi: 10.3389/fmicb.2017.00695
Klamt, S., Grammel, H., Straube, R., Ghosh, R., and Gilles, E. D. (2008). Modeling the electron transport chain of purple non-sulfur bacteria. Mol. Syst. Biol. 4:156. doi: 10.1038/msb4100191
Laass, S., Kleist, S., Bill, N., Druppel, K., Kossmehl, S., Wohlbrand, L., et al. (2014). Gene regulatory and metabolic adaptation processes of Dinoroseobacter shibae DFL12T during oxygen depletion. J. Biol. Chem. 289, 13219–13231. doi: 10.1074/jbc.M113.545004
Martens, T., Heidorn, T., Pukall, R., Simon, M., Tindall, T. J., and Brinkhoff, T. (2006). Reclassification of Roseobacter gallaeciensis Ruiz-Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uchino et al. 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera. Int. J. Syst. Evol. Microbiol. 56, 1293–1304. doi: 10.1099/ijs.0.63724-0
Niedzwiedzki, D., Dilbeck, P., Tang, Q., Martin, E., Bocian, D., Hunter, C., et al. (2017). New insights into the photochemistry of carotenoid spheroidenone in light-harvesting complex 2 from the purple bacterium Rhodobacter sphaeroides. Photosynth. Res. 131, 291–304. doi: 10.1007/s11120-016-0322-2
Ritchie, A., and Johnson, Z. (2012). Abundance and genetic diversity of aerobic anoxygenic phototrophic bacteria of coastal regions of the pacific ocean. Appl. Environ. Microbiol. 78, 2858–2866. doi: 10.1128/AEM.06268-11
Shiba, T., Simidu, U., and Taga, N. (1979). Distribution of aerobic bacteria which contain bacteriochlorophyll a. Appl. Environ. Microbiol. 38, 43–45.
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Soora, M., and Cypionka, H. (2013). Light enhances survival of Dinoroseobacter shibae during long-term starvation. PLoS One 8:e57487. doi: 10.1371/journal.pone.0057487
Wagner-Döbler, I., and Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Wagner-Döbler, I., Ballhausen, B., Berger, M., Brinkhoff, T., Buchholz, I., Bunk, B., et al. (2010). The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J. 4, 61–77. doi: 10.1038/ismej.2009.94
Yurkov, V., and van Gemerden, H. (1993). Impact of light/dark regimen on growth rate, biomass formation and bacteriochlorophyll synthesis in Erythromicrobium hydrolyticum. Arch. Microbiol. 159, 84–89. doi: 10.1007/BF00244268
Keywords: Roseobacter group, aerobic anoxygenic phototrophs, light-harvesting and channeling, napA, nirS
Citation: Kirchhoff C, Ebert M, Jahn D and Cypionka H (2018) Chemiosmotic Energy Conservation in Dinoroseobacter shibae: Proton Translocation Driven by Aerobic Respiration, Denitrification, and Photosynthetic Light Reaction. Front. Microbiol. 9:903. doi: 10.3389/fmicb.2018.00903
Received: 12 December 2017; Accepted: 18 April 2018;
Published: 09 May 2018.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, United StatesReviewed by:
Hiroyuki Arai, The University of Tokyo, JapanShinji Masuda, Tokyo Institute of Technology, Japan
Copyright © 2018 Kirchhoff, Ebert, Jahn and Cypionka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heribert Cypionka, heribert.cypionka@icbm.de; Cypionka@icbm.de
 Christian Kirchhoff
Christian Kirchhoff Matthias Ebert
Matthias Ebert Dieter Jahn
Dieter Jahn Heribert Cypionka
Heribert Cypionka