- 1Laboratory of Synthetic Microbiology, School of Chemical Engineering & Technology, Tianjin University, Tianjin, China
- 2Key Laboratory of Systems Bioengineering, Ministry of Education of China, Tianjin, China
- 3Collaborative Innovation Center of Chemical Science and Engineering, Tianjin, China
- 4Center for Biosafety Research and Strategy, Tianjin University, Tianjin, China
The heterotrophic microalga Crypthecodinium cohnii has attracted considerable attention due to its capability of accumulating lipids with a high fraction of docosahexaenoic acid (DHA). In our previous study, ethanolamine (ETA) was identified as an effective chemical modulator for lipid accumulation in C. cohnii. In this study, to gain a better understanding of the lipid metabolism and mechanism for the positive effects of modulator ETA, metabolic flux analysis was performed using 13C-labeled glucose with and without 1 mM ETA modulator. The analysis of flux distribution showed that with the addition of ETA, flux in glycolysis pathway and citrate pyruvate cycle was strengthened while flux in pentose phosphate pathway was decreased. In addition, flux in TCA cycle was slightly decreased compared with the control without ETA. The enzyme activity of malic enzyme (ME) was significantly increased, suggesting that NADP+-dependent ME might be the major source of NADPH for lipid accumulation. The flux information obtained by this study could be valuable for the further efforts in improving lipid accumulation and DHA production in C. cohnii.
Introduction
Docosahexaenoic acid (DHA) is a polyunsaturated fatty acid (PUFA) belonging to the ω-3 group. In recent years, DHA has attracted much attention because of its broad beneficial effects on human health (Glaser et al., 2011). As an important component in cellular membranes of human nervous, visual, and reproductive tissues, DHA is considered essential for the neurological development of infants. In addition, DHA plays significant roles in alleviating cardiovascular diseases, hypertension, diabetes, and neuropsychiatric disorders (Karr et al., 2011; Chitranjali et al., 2015). Therefore, DHA has been widely used in food, pharmaceutical and feed industries. The traditional source of DHA is fish oil, as ocean fish can accumulate ω-3 PUFAs by consuming DHA-rich algae as food (Ward and Singh, 2005; Walsh et al., 2016). However, the use of fish oil as a food additive is limited due to problems associated with its typical fishy smell, unpleasant taste, poor oxidative stability and difficult purification (Hajjaji et al., 2011). It is thus necessary to develop alternative approaches for commercial DHA production directly using marine DHA-producing microorganisms (Ganuza et al., 2008; Mendes et al., 2008; Ji et al., 2015). Efforts to explore microalgae as an alternative source of DHA have been made in recent years, such as adaptive evolution, strain improvement by mutation, and culture condition optimization (Guo et al., 2016, 2017; Sun et al., 2016, 2018; Liu et al., 2017; Ren et al., 2017).
Crypthecodinium cohnii, a flagellated marine microalga, has been considered as a prolific producer of DHA, which contains 25–60% DHA while less than 1% of other types of PUFAs in its total fatty acids (TFAs) (Pei et al., 2017). However, up to now, no genome sequence is available for C. cohnii, and the DHA biosynthetic pathway in C. cohnii remains elusive (de Swaaf et al., 2003; Mendes et al., 2008). Previous studies suggested that C. cohnii might utilize the polyketide synthases (PKS) route for the biosynthesis and accumulation of DHA, which requires no oxygen or NADPH-dependent desaturases (Ratledge, 2004). Meanwhile, Pei et al. (2017) recently applied de novo transcriptome analysis to characterize central carbohydrate and fatty acid biosynthesis in C. cohnii, which suggested that C. cohnii might utilize a combination of PKS systems and desaturase steps for DHA biosynthesis. In addition, metabolomics analysis has also been employed to understand the possible mechanisms responsible for the increased lipid accumulation (Li et al., 2015). These studies have broadened our understanding of the molecular and biochemical mechanisms underlying lipid accumulation in C. cohnii. However, until now the knowledge of the C. cohnii metabolic network is still very limiting due to lack of quantitative analysis of metabolic fluxes. Thus, it is urgently required to develop methodologies of quantitative metabolic analysis of C. cohnii in order to better understand the intracellular distribution of carbon fluxes as well as fluxes in response to extracellular stimuli. On the other hand, many efforts have been made to improve the production of lipids and DHA by C. cohnii, which can be achieved by the application of mutation, culture condition optimization and chemical modulators (Sijtsma and de Swaaf, 2004; Li et al., 2015; Liu et al., 2017). For example, early studies showed that antioxidants, such as sesamol had great potential to enhance cell growth and the biosynthesis of lipids in C. cohnii (Liu et al., 2015, 2017). Our previous study showed that ethanolamine (ETA) as chemical modulator, could increase lipid accumulation in C. cohnii by 18.78% (Li et al., 2015). Meanwhile, another study also showed that ETA was able to enhance lipid accumulation in Scenedesmus obliquus by 22% and has been considered as a potential inducer for improving lipid accumulation in model photosynthetic organisms (Cheng et al., 2012). Therefore, it will be valuable to determine the mechanism of ETA to modulate the lipid synthesis and promote the future modification on lipid accumulation and DHA production in the industry-important C. cohnii.
13C-labeling based metabolic flux analysis as an integrated experimental and computational method is an important approach to determine the dynamics of biochemical networks and to provide quantitative insights into the in vivo distribution of molecular fluxes throughout central metabolism (Zamboni et al., 2009). Recent studies showed that 13C-labeled metabolic flux analysis (13C-MFA) could be a powerful analytical technology for understanding lipid accumulation mechanisms in various oleaginous microorganisms, such as oleaginous microalga Chlorella protothecoides (Xiong et al., 2010; Zhao et al., 2015), oleaginous yeast Yarrowia lipolytica and Trichosporon cutaneum (Liu et al., 2013, 2016; Wasylenko et al., 2015; Zhang et al., 2016), and oleaginous fungus Mucor circinelloides (Zhao et al., 2015). However, no report is available on 13C-MFA in C. cohnii up to now. In order to systematically understand the mechanisms underlying lipid accumulation as well as quantitative metabolic information in C. cohnii, in this study, we utilized gas chromatography-mass spectrometry to analyze the 13C labeling patterns of the amino acids in biomass hydrolysates of C. cohnii grown in an optimized chemically defined medium with and without 1 mM ETA addition. By integrating these labeling measurement data with metabolite balancing, the intracellular flux distributions in C. cohnii were further quantitated. The study provided valuable information to promote the future modification on lipid accumulation and DHA production in C. cohnii.
Materials and Methods
Strain and Growth Conditions
Crypthecodinium cohnii ATCC 30556 was obtained from American Type Culture Collection (ATCC), and grown on chemically defined medium composed of (g/L): glucose, 9; K2HPO4, 0.1; MgCl2.6H2O, 10.6; CaCl2, 1.1; KCl, 0.7; Na2SO4, 3.9; SrCl2.6H2O, 0.1; KBr, 0.1; NaCl, 23.5; NaHCO3, 0.2; disodium glycerophosphate, 0.15; 3 mL Tris buffer; 5 mL of metal mixture; 1 mL vitamin solution and nitrogen source. Metal mixture composed of (g/L): FeCl3.6H2O, 0.5; Na2EDTA, 10; H3BO3, 10; CoCl2.6H2O, 0.01; MnCl2∘4H2O, 1.6; ZnCl2, 0.1. The vitamin solution composed of (mg/L): biotin, 3; and thiamine, 100. The different inorganic nitrogen sources at a final concentration of 36 mM including NH4Cl, (NH4)2SO4, NaNO3, Ca(NO3)2, and KNO3. The organic nitrogen sources were 2 g/L yeast exact and various concentration of glutamate (0.5–5 g/L). The culture conditions of propagation were the same as those used in our previous study (Li et al., 2015). Briefly, the cells were grown in 250 mL Erlenmeyer flasks each containing 50 mL of medium. Cultures were maintained at 25°C and incubated in a reciprocal shaker shifting statically at 180 rpm. ETA was added at 36 h when the culture entering the early exponential phase and each concentration experiment was carried out in triplicate. Carbon isotope: [U-13C] glucose was purchased from Cambridge Isotope Laboratories, Inc. (>98%, Cambridge Isotope Laboratories, Inc., Andover, MA, United States). Labeling experiment: the carbon source was 20% [U-13C] glucose/L and 80% unlabeled glucose/L. ETA were purchased from Sigma (St. Louis, MO, United States) and other reagents used in the study were purchased from Sinopharm Chemical Reagent Co., Ltd., China.
Determination of Physiological Parameters
Cell density was measured on an ELx808 Absorbance Microplate Reader (BioTek, Winooski, VT, United States) at OD490. For the determination of dry cell weight (DW), triplicate samples of the culture were collected, washed with double-distilled water and freeze dried overnight. The cell density corresponded to OD490 by the regression equation y = 1.941x-0.178 (r2 = 0.9988, p < 0.05), where y is the cell density (g dry cell weight L-1) and x is the absorbance of the cell suspension at 490 nm. The specific growth rate (μ) in the log phase was calculated by using the equation μ = (ln X2-ln X1)/(t2-t1), where X1 and X2 are the cell density at OD490 at time t1 and t2, respectively. The glucose concentrations in the culture supernatants were determined according to the glucose oxidase method as previously described (de Swaaf et al., 1999). To determine the macromolecular composition of C. cohnii, the Lowry method (Holdsworth et al., 1988) was used to measure protein content, and amino acid composition was obtained with an Amino Acid Analyzer (L-8900, Hitachi, Tokyo, Japan). The phenol-sulfuric acid method was used to determine intracellular carbohydrate and starch contents (Masuko et al., 2005). The KOH/UV method (Benthin et al., 1991) and the modified Schneider method (Herbert et al., 1971) were used to determine RNA and DNA concentrations, respectively. The total lipids were extracted using a previous method as described below (Yang et al., 2009). A statistical t-test model was applied for the comparative analysis, and p-value less than 0.05 were considered statistically significant.
Total Lipid Extraction and Lipid Profile Analysis
Two methods were used to determine the lipid accumulation in C. cohnii cells. The first protocol involves fluorescence intensity measurements with excitation and emission wavelengths of 510 and 585 nm after Nile Red staining. A fluorescence spectrophotometer (F-2700FL, Hitachi, Tokyo, Japan) was used for the assay (Sui et al., 2014). The second protocol involves direct lipid extraction using a modified method described previously (Yang et al., 2009). Briefly, C. cohnii cells were collected at 60 h by centrifugation (3550 × g) for 5 min and freeze-dried to generate a lyophilized algal powder. 15–25 mg of lyophilized algal powder was used for extraction using a chloroform-methanol solution (2:1, v/v) with 0.01% butylated hydroxytoluene. The extraction process was repeated three to four times. The above extracts were washed with 1.0 mL of 1.0 M KCl followed by 1.0 mL of double-distilled water. The solvents were removed using a vacuum concentrator system (ZLS-1, Hunan, China). The lipid profile was analyzed using an Agilent 5975 MSD/7890 instrument (Agilent Corp., Santa Clara, CA, United States) according to previous publications (Xiong et al., 2008; Pei et al., 2017).
GC-MS Analyses of Protein Hydrolysates
To ensure the cells are being cultured under steady-state conditions, the cells were harvested at 60 h by centrifugation (15,800 × g, 4°C) for 2 min. Proteinogenic amino acid preparation for GC-MS analysis was performed following standard protocols (Zamboni et al., 2009). Approximately 25 mg of lyophilized biomass was hydrolyzed with 2 mL of 6 M HCl at 110°C for 24 h. The hydrolysate was dried in an oven at 80°C for 12 h. The hydrolysate was re-suspended in water-free dimethylformamide (DMF) and then centrifuged at 12000 × g for 10 min. The supernatant was added with 50 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (TBDMS, Sigma-Aldrich, St. Louis, MO, United States) and derivatived at 85°C for 1 h. The derivatived samples were analyzed by GC-MS using an Agilent 5975 MSD/7890 instrument (Agilent Corp, Santa Clara, CA, United States). The column was a HP-5MS (Restek, Bellefonte, PA, United States). The oven temperature was initially held at 60°C for 2 min and reached 180°C at 5°C per min, then raised to 260°C at 10°C per min, and finally held at 260°C for 5 min.
Flux Analysis
For metabolic flux ratio analysis, a mass isotopomer distribution vector, MDVα [Eq. (1)], is assigned on the basis of well-developed mathematical methodology (Nanchen et al., 2007).
Where m0 is the fractional abundance of molecules with monoisotopic mass and mi > 0 is the abundance of fragments with molecules with higher masses. iMS2Flux software was used to correct the factional labeling distribution of the amino acids for natural isotopic abundance (Hart et al., 2012). The resulting MDVαvalues from iMS2Flux software are used to assess the fractional labeling (FL) enrichment of each fragment using Eq. (2).
Where n represents the number of amino acid carbon atoms in the considered fragment and i is the different mass isotopomers. OpenFLUX software was utilized under MATLAB environment (Mathworks, Inc., Massachusetts) to solve for the fluxes (Quek et al., 2009). The application is based on the elementary metabolite unit (EMU) framework (Antoniewicz et al., 2007). Stoichiometric data on growth, substrate uptake rate, storage formation, and on the cellular composition of C. cohnii together with mass isotopomer distribution data of the labeled amino acids that were produced using the iMS2Flux software were used as model input.
Enzyme Activities Analysis
The cells were harvested at 60 h by centrifugation at 3000 × g for 5 min, and the pellet was resuspended in extraction buffer (containing 100 mM KH2PO4/KOH (pH 7.5), 20% (v/v) glycerol, 1 mM benzamidine⋅HCl, and 1 mM DTT). HNX-2 cell disruptor (Honour, Tianjin, China) was used to rupture cells. The supernatant was collected by centrifugation at 10,000 × g, 4°C for 15 min. The supernatant containing cytoplasmic and mitochondrial enzymes was subjected to the following enzyme activity analysis. The activities of NADP+-dependent ME and NADP+-dependent isocitrate dehydrogenase (ICDH) were determined using continuous spectrophotometric assays following the increase of NADPH (Hsu and Lardy, 1969; Liu et al., 2015; Safdar et al., 2017). The absorbance of the cuvettes at 340 nm was determined using a UV-1750 instrument (Shimadzu, Kyoto, Japan). The enzyme activity was defined as the reducing amount of NADP+ (nM) catalyzed by the enzyme solution with 1 mg of protein in 1 min (nM/min/mg protein). The negative controls were set as without the substrate (ME or isocitrate) and without the cell exact. The enzyme activity was normalized by negative controls. Standard Bradford method was used to determine protein concentration (Bradford, 1976).
Statistical Analysis
In this study, each experiment was performed in three biological replicates. All data were reported as means ± standard deviations and were analyzed with a t-test.
Results and Discussion
Optimization of Chemically Defined Medium for 13C Metabolic Flux Analysis
Medium composition is a key factor for flux analysis (Zamboni et al., 2009). The yeast extract composition in natural medium is complicated and provided part of carbon source, which could increase the number of carbon substrates and might compromise flux calculability. Therefore, we first developed a synthetic medium suitable for 13C metabolic flux analysis based on the ATCC 460 A2E6 medium (Pleissner and Eriksen, 2012). In addition, the effects of five inorganic nitrogen sources (i.e., ammonium chloride, ammonium sulfate, sodium nitrate, potassium nitrate and calcium nitrate) and two organic nitrogen sources (i.e., glutamate and yeast exact) on C. cohnii growth were investigated in flask cultures. As shown in Figure 1A, the growth of C. cohnii in the inorganic nitrogen sources was significantly inhibited when compared to that in organic nitrogen sources, suggesting that the inorganic nitrogen sources were less efficient to promote cell growth. This was consistent with the previous study that Chlorella protothecoides grew slower in inorganic nitrogen sources than organic nitrogen sources (Xiong et al., 2008). The results showed that growth rates of C. cohnii were 0.051 and 0.054 h-1 in the medium supplied with 2 g/L glutamate and yeast extract, respectively, suggesting that C. cohnii was able to grow normally in the medium containing glutamate as nitrogen source. To further investigate the most optimal concentration of glutamate, different concentrations of glutamate ranging from 0.5 to 5 g/L were added. As shown in Figure 1B, no significant differences were observed when the concentration ranging from 0.8 to 2 g/L. However, obvious inhibition was observed when the concentration reached 3–5 g/L. Notably, high concentration of glutamate would dilute the 13C labeling substrate. Thus, 1 g/L of glutamate was chosen and used as the optimal nitrogen concentration in the following analysis.
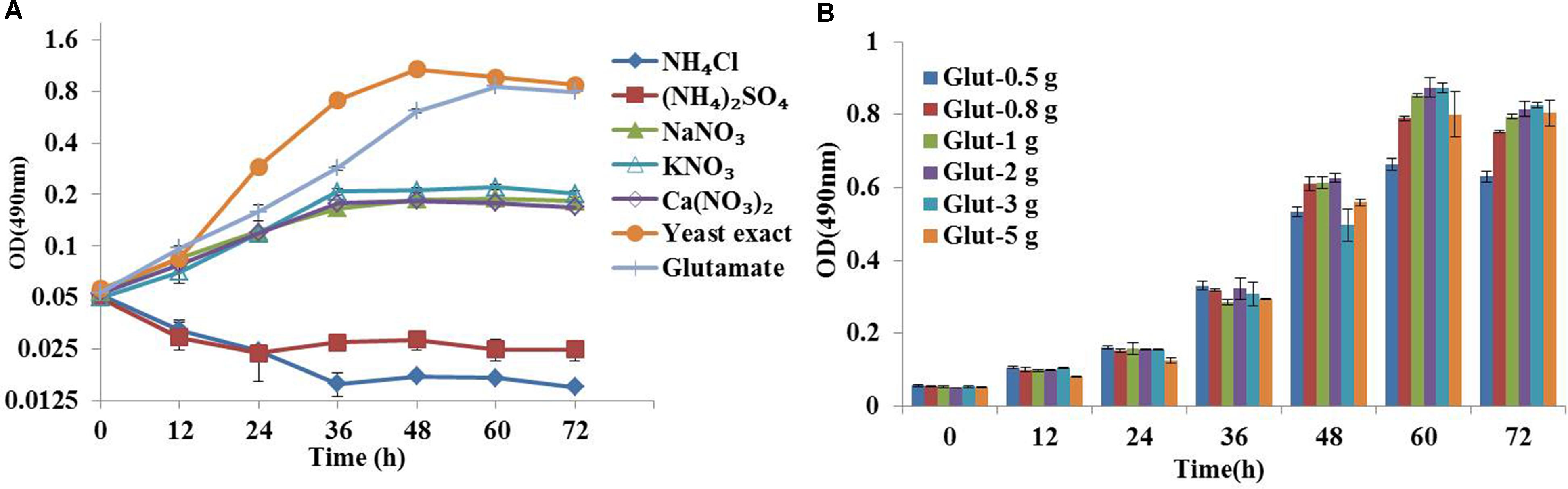
FIGURE 1. Optimization of chemically defined medium for 13C metabolic flux analysis. (A) Growth curves of Crypthecodinium cohnii under different inorganic nitrogen sources at a final concentration of 36 mM, 2 g/L yeast exact and glutamate. (B) Time courses of cell growth under different concentrations of glutamate (0.5, 0.8, 1, 2, 3, and 5 g/L).
Influence of ETA Concentration on the Growth and Lipid Accumulation of C. cohnii in the Optimized Chemically Defined Medium
Previous studies had suggested that chemical modulators were able to enhance lipid accumulation in a diverse of microorganisms, and the roles of fourteen chemicals selected from five chemical groups had been established in C. cohnii (Li et al., 2015). Specifically, ETA as an amine could increase lipid accumulation in C. cohnii by 18.78%, which had the most significant effort on lipid accumulation among all chemical modulators evaluated so far. Here, influence of ETA concentration on the growth and lipid accumulation of C. cohnii in the optimized chemically defined medium was further investigated here. As shown in Figure 2A, the growth rates were 0.051, 0.048, 0.047, 0.043, and 0.035 h-1 in the optimized chemically defined medium with the addition of 0, 0.5, 1, 2.5 and 5 mM ETA, respectively. The results suggested that in this synthetic medium the growth rate was not significantly affected upon 0.5–5 mM ETA addition. Meanwhile, the neutral lipid content of C. cohnii supplemented with different concentrations of ETA was determined using the lipophilic stain Nile Red approach (Figure 2B). The results showed that the lipid content was increased by 25.4% with addition of 1 mM ETA compared with control, which was the most significant effect on lipid accumulation among all concentrations evaluated. Early studies on plant amine indicated that amines as cellular signals have an important role in metabolic regulation. In plants, the production of hydrogen peroxide (H2O2) deriving from amine oxidation has been correlated with cell wall maturation and reinforcement during pathogen invasion (Alcázar et al., 2010; Pál et al., 2015). Furthermore, increasing evidence indicated that reactive oxygen species (ROS) signaling might act as a mediator of lipid accumulation, which was associated with dramatic changes in the transcriptome, proteome, and metabolome in oleaginous microorganisms (Yilancioglu et al., 2014; Yu et al., 2015; Shi et al., 2017). In C. cohnii, it could also be assumed that ETA as a kind of signal molecule might cause the change of ROS which is responsible for the increase of lipid accumulation.
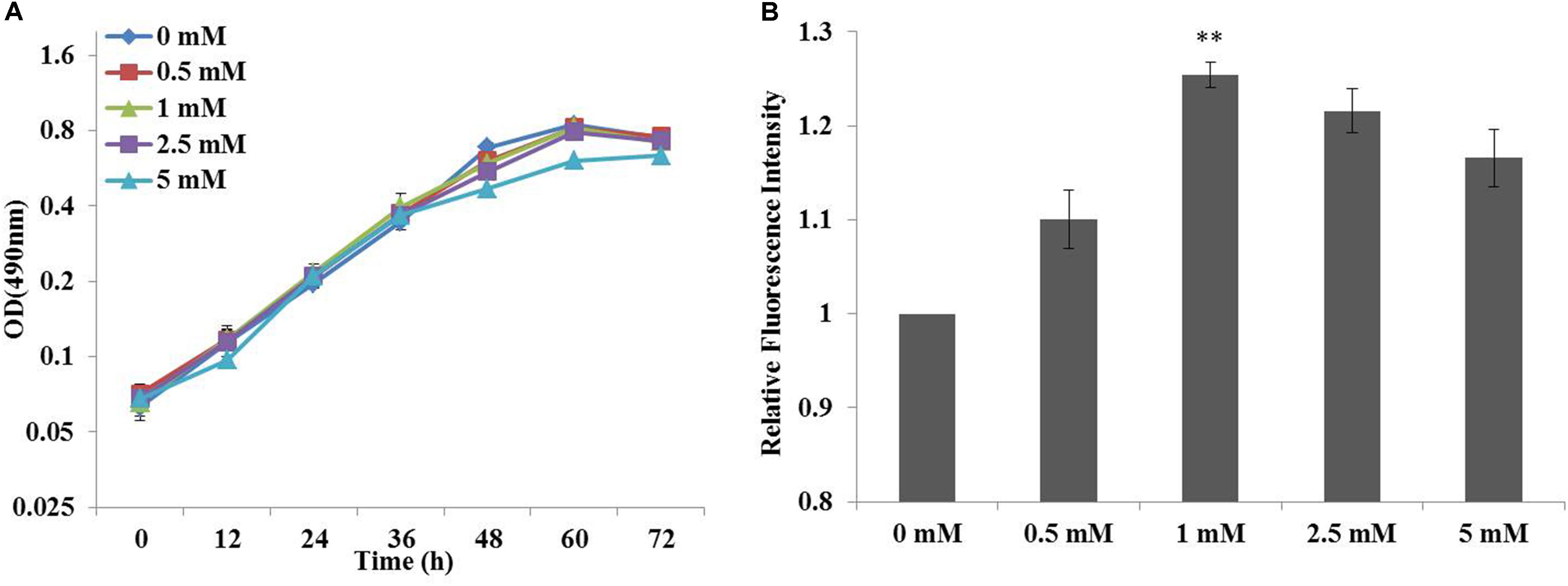
FIGURE 2. Effects of cell growth and lipid accumulation with ethanolamine (ETA) addition. (A) Growth curves of C. cohnii under different ETA concentrations. (B) The relative fluorescence intensity of C. cohnii stained with Nile Red after treated with different ETA concentrations. Statistical analysis was conducted as described in the text, as statistical significance indicated by ∗∗p < 0.01; ∗p < 0.05.
The growth characteristics of C. cohnii were then further determined in the optimized chemically defined medium. During the exponential growth phase, the specific growth rates of C. cohnii were 0.051 and 0.047 h-1 with and without 1 mM ETA (Table 1). The biomass yield didn’t show significant differences between both conditions while the glucose uptake rate with 1 mM ETA addition was slightly lower than the control.
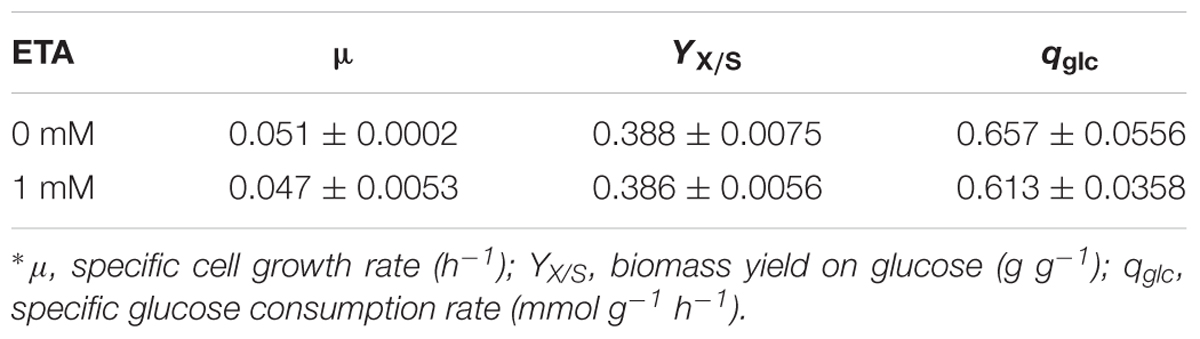
TABLE 1. Growth parameters of Crypthecodinium cohnii in medium with and without 1 mM ethanolamine (ETA) addition∗.
Metabolic Model Construction and Biomass Composition Analysis
Based on the de novo transcriptome data obtained recently (Pei et al., 2017), we constructed a primary metabolic network of C. cohnii. As shown in Supplementary Table S1, central carbohydrate metabolism of C. cohnii includes the following core metabolic pathways: glycolysis pathway (EMP), pentose phosphate (PP) pathway, tricarboxylic acid (TCA) cycle and citrate pyruvate cycle (Pei et al., 2017). All involved reactions were assumed localized into two main compartments: cytosol and mitochondria, where most of the common metabolic reactions take place. All the biochemical reactions involved in the network are listed in Supplementary Table S1.
To determine the coefficients in this biomass formation reaction, the biomass composition of C. cohnii was determined experimentally in this study. As shown in Figure 3, lipids were the most abundant component of C. cohnii (43.9% of dry weight), followed by carbohydrates (21.8%), proteins (12.8%), RNA (7.9%) and DNA (4.5%). The lipids content reached 50.45% in the presence of 1 mM ETA, which was 14.9% higher than the control culture, in accordance with the previous study that C. cohnii was treated with ETA on the medium consisted of glucose, yeast extract and sea salt (Li et al., 2015). The contents of carbohydrates and proteins were decreased by 20.7% and 18.1% in the presence of 1 mM ETA, respectively. Meanwhile, no significant difference of the DNA and RNA contents and the TFA content was observed between the cultures with and without 1 mM ETA. The relative distribution of amino acids in biomass was similar, with the notable exception of lysine, which was significantly lower with 1 mM ETA addition compared to the control culture (Figure 3C).
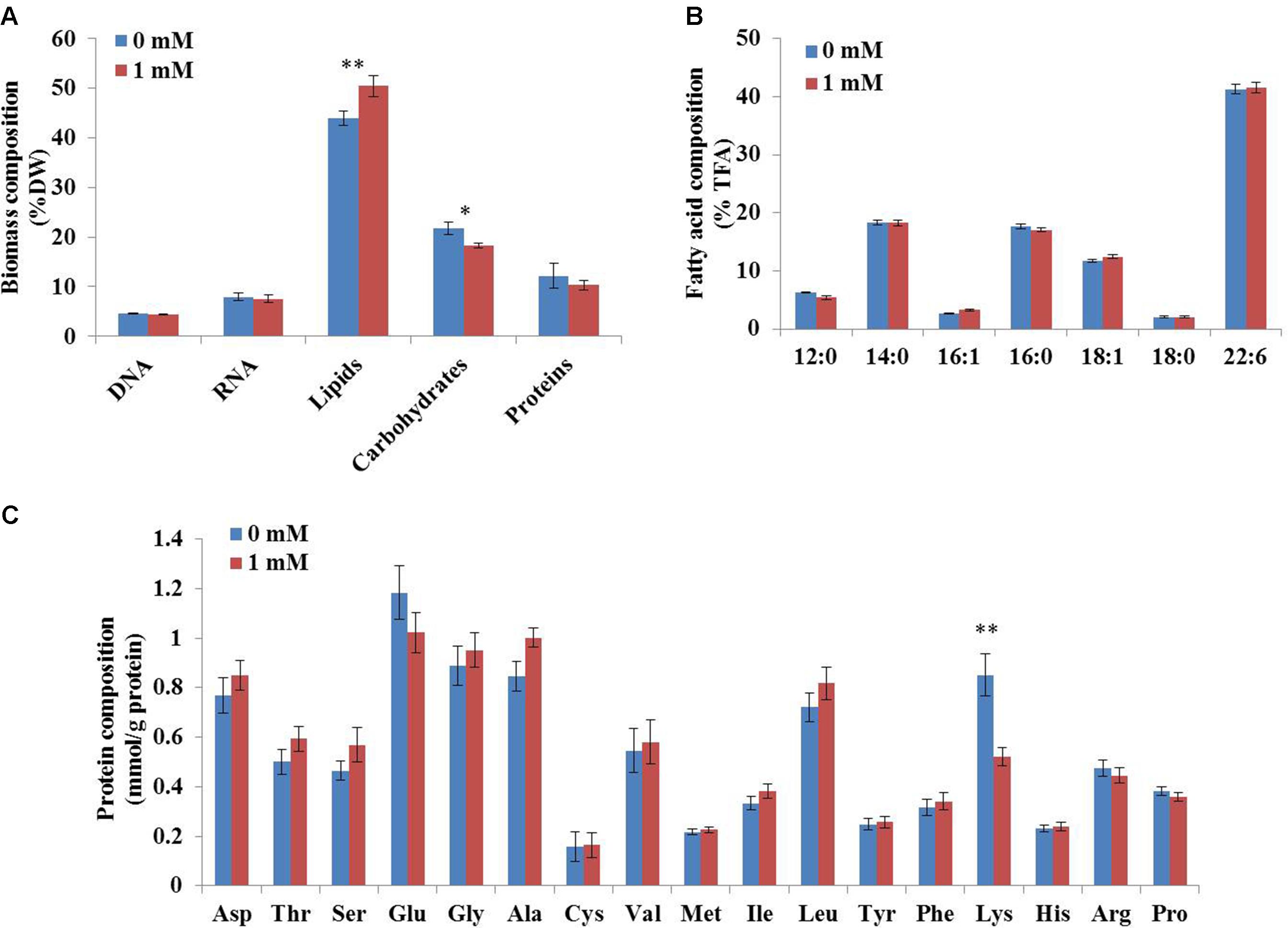
FIGURE 3. Biomass composition of C. cohnii with and without 1 mM ETA addition. (A) The fractional amounts of major biomass components. (B) The distribution of fatty acid composition. (C) The distribution of protein composition. Statistical significance was indicated by ∗∗p < 0.01; ∗p < 0.05.
13C Metabolic Flux Analysis
Following the standard GC-MS measurement and amino acid biosynthesis schemes, the labeling patterns of both amino acids and their carbon backbone precursors in central carbohydrate metabolism were subsequently acquired. We then adopted a methodology named metabolic flux ratio analysis (Nanchen et al., 2007) to reveal directly the flux distribution of C. cohnii grown under the tested conditions. The estimated metabolic fluxes of C. cohnii with and without ETA were shown schematically in Figure 4. Fluxes were normalized to glucose uptake rate, which was given a value of 100%. As shown in Figure 4, in control culture, the carbon flux through the EMP and PP pathway accounted for 88.4 and 2.18% of the glucose uptake, suggesting that less carbon flux went through the PP pathway. As G-6-PDH and 6-phosphogluconate dehydrogenase in the PP pathway served as the primary alternative for producing NADPH (Ratledge, 2014), our results suggested that the PP pathway might not be the main way to supply NADPH for lipid biosynthesis, which was consistent with the previous study that no G-6-PDH activity was detected in any samples of C. cohnii (Liu et al., 2015). Most flux of the glyceraldehyde 3-phosphate was generated from the EMP. Pyruvate was synthesized by two different routes: pyruvate kinase and ME (Liu et al., 2013). Approximately 60.8% flux was generated from the EMP and 39.2% was generated from citrate pyruvate cycle, respectively. The citrate pyruvate cycle was responsible for conversion of pyruvate to oxaloacetate, catalyzed by pyruvate carboxylase. Then, oxaloacetate was converted to citrate, which was transported outside the mitochondria and degraded into oxaloacetate and acetyl-CoA in the cytoplasm. Malate obtained from oxaloacetate could then be decarboxylated to generate pyruvate via the NADP+-dependent cytosolic ME, playing an important role in lipid accumulation in oleaginous microorganism (Ronnebaum et al., 2006). About 63.7% flux of pyruvate was directed toward acetyl-CoA, and 36.3% was routed to the formation of oxaloacetate, respectively. In addition, the total flux through citrate was 179%, among which 62% flux of the citrate entered the lipogenesis pathway and 38% flux was catabolized through TCA cycle, respectively, suggesting that there were more flux through the lipogenesis pathway during lipid accumulation stage. In the TCA cycle, the flux through α-ketoglutarate was from iso-citrate (85%) and glutamate (15%), respectively. In this case, the results showed that glutamate was not only the nitrogen source, but also the carbon source. The fragments from proline, glutamate and aspartate were shown with low FL values (less than 0.15) (Supplementary Table S2), indicating that the labeling enrichments of these three amino acids were severely diluted by unlabeled glutamate in the medium. In order to avoid impairing accurate estimation of fluxes, the glutamate was set as a substrate during the flux analysis (Supplementary Table S1). The flux distribution obtained here provided quantitative metabolic information on C. cohnii and can be used to better elucidate and understand mechanisms involved in lipid metabolism.
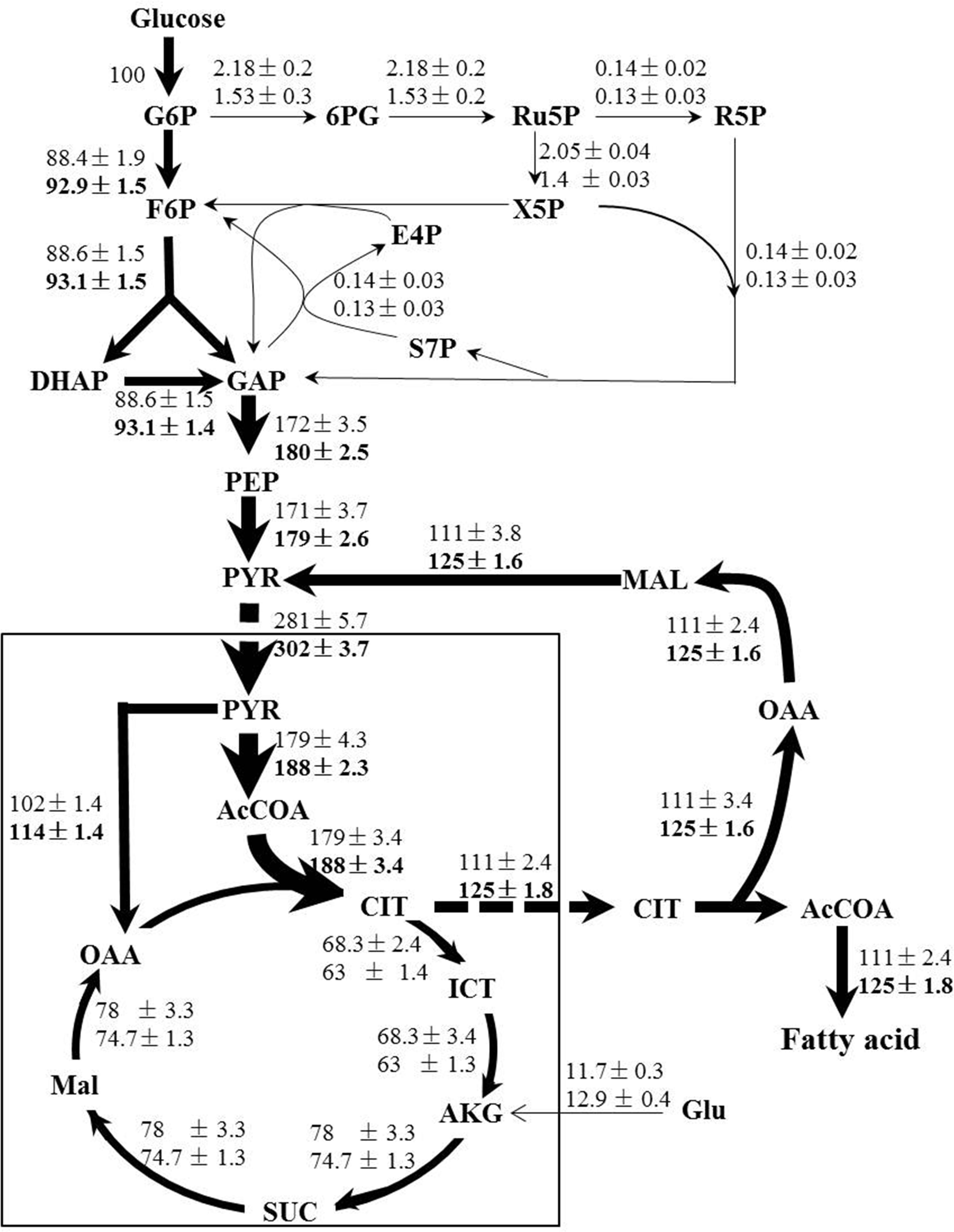
FIGURE 4. Flux maps of central carbon metabolism of C. cohnii in the presence of 0 and 1 mM ETA. The estimated fluxes were percentages of the relative rates normalized to the glucose uptake rates. The first and second line numbers represent relative flux when cultured without and with 1 mM ETA. The up-regulation fluxes with 1 mM ETA addition were in bold. G6P, glucose-6-phosphate; 6PG, 6-phospho-gluconate; Ru5P, ribulose 5-phosphate; F6P, fructose-6-phosphate; E4P, erythrose-4-phosphate; S7P, seduheptulose-7-phosphate; DHAP: dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl-CoA; OAA, oxaloacetate; CIT, citrate; ICT, isocitrate; AKG, a-ketoglutarate; SUC, succinate; MAL, malate; Glu, glutmate.
When compared with condition of 1 mM ETA addition, our results showed that the flux ratio of EMP/PP pathway was increased to 92.9/0.53 when cultured with 1 mM ETA from 88.4/2.18 in control. The flux of EMP was increased by 5% and the flux in PP pathway was decreased by 29%, respectively. The results were in accordance with the previous metabolomics analysis, which suggested that the EMP metabolites were up-regulated and PP pathway metabolites were down-regulated, respectively (Li et al., 2015). Interestingly, the whole citrate pyruvate cycle was enhanced during lipid accumulation with 1 mM ETA addition. We thus speculated that the citrate pyruvate cycle might be enhanced by specific metabolic regulation that was triggered by ETA addition, which further promoted the lipid accumulation in C. cohnii. NADP+-dependent ME is located in cytoplasm and known to play a key role in lipid biosynthesis in oleaginous yeasts (Liu et al., 2013). Our results suggested that the flux through NADP+-dependent ME was increased by 12.6% with ETA addition. The results obtained in this study therefore pointed to the possibility that reaction via the cytoplasmic ME was the primary source to supply reducing equivalent for lipid biosynthesis in C. cohnii, which was in good agreement with several previous studies (Liu et al., 2015; Pei et al., 2017). For example, Liu et al. (2015) showed that C. cohnii, to a large degree, utilizes ME rather than ICDH or G-6-PDH to produce NADPH for the de novo fatty acid biosynthesis (Liu et al., 2015). In addition, the transcript of ME was found up-regulated 1.7-fold during lipid and DHA accumulation in C. cohnii (Pei et al., 2017). Furthermore, with 1 mM ETA addition, the flux of ICDH was decreased by 7.7%, suggesting that ICDH was slightly weakened, consistent with previous report that the activities of NADP+-ICDH and NAD+-ICDH in nitrogen-starved culture were reduced by 8 and 31%, respectively, which suggested that more carbon flux flowed to lipogenesis rather than to TCA cycle (Safdar et al., 2017).
Activities of Key Enzymes
It is well-known that the supply of reducing power in the form of NADPH plays an important role in fatty acid biosynthesis in oil-rich microorganisms. High content of lipid accumulation in C. cohnii requires enough supplies of acetyl-CoA as the precursor and reducing equivalent (NADPH) as the cofactor for fatty acid synthesis. In order to further confirm whether the activity of ME was increased upon 1 mM ETA addition, the activity of ME and ICDH were then measured. As shown in Figure 5, the activity of ME was 360.9 nmol/min/mg protein at 60 h with 1 mM ETA addition, which was increased by 17.2% compared with no 1 mM ETA addition. The result was consistent with the flux analysis result, which suggested that ME played an important role in lipid accumulation. Furthermore, the activity of ICDH was determined to be 370 nmol/min/mg protein at 60 h with 1 mM ETA addition, which didn’t have a significant change compared with no ETA addition. It was assumed that the ETA as a kind of signal molecule might cause the change of ROS responsible for the increase of lipid accumulation, which was still yet to be investigated and the improved ME productivity might contribute to the improved lipid accumulation. ME has been reported to be a major provider of the reducing power NADPH required for the lipid biosynthesis in oleaginous fungi (Hao et al., 2014). Overexpression of ME resulted in significant increase in lipid accumulation in yeast, fungi, microalga (Zhang et al., 2007; Li et al., 2013; Hao et al., 2014; Jiao et al., 2015). For example, overexpression of ME in Mucor circinelloides led to a 2.5-fold increase in lipid accumulation (Zhang et al., 2007). Heterologous expression of NADP+-dependent ME from Mucor circinelloides in oleaginous yeast Rhodotorula glutinis resulted in a 2.0-fold increase in lipid production (Li et al., 2013). Jiao et al. (2015) reported that the overexpression of ME (PtME) from Phaeodactylum tricornutum markedly increased the total lipid content in transgenic cells by 2.5-fold and reached a record of 57.8% increase of dry cell weight with a similar growth rate to wild type (Jiao et al., 2015). However, whether the improved ME activity was directly resulted from the addition of ETA, or by unknown secondary responses, was still yet to be determined. Collectively, it can be hypothesized that the flux through cytoplasmic ME might be associated with the lipid accumulation in C. cohnii and therefore, it could be a potential target for genetic modification to further improve the lipid content in C. cohnii in the future.
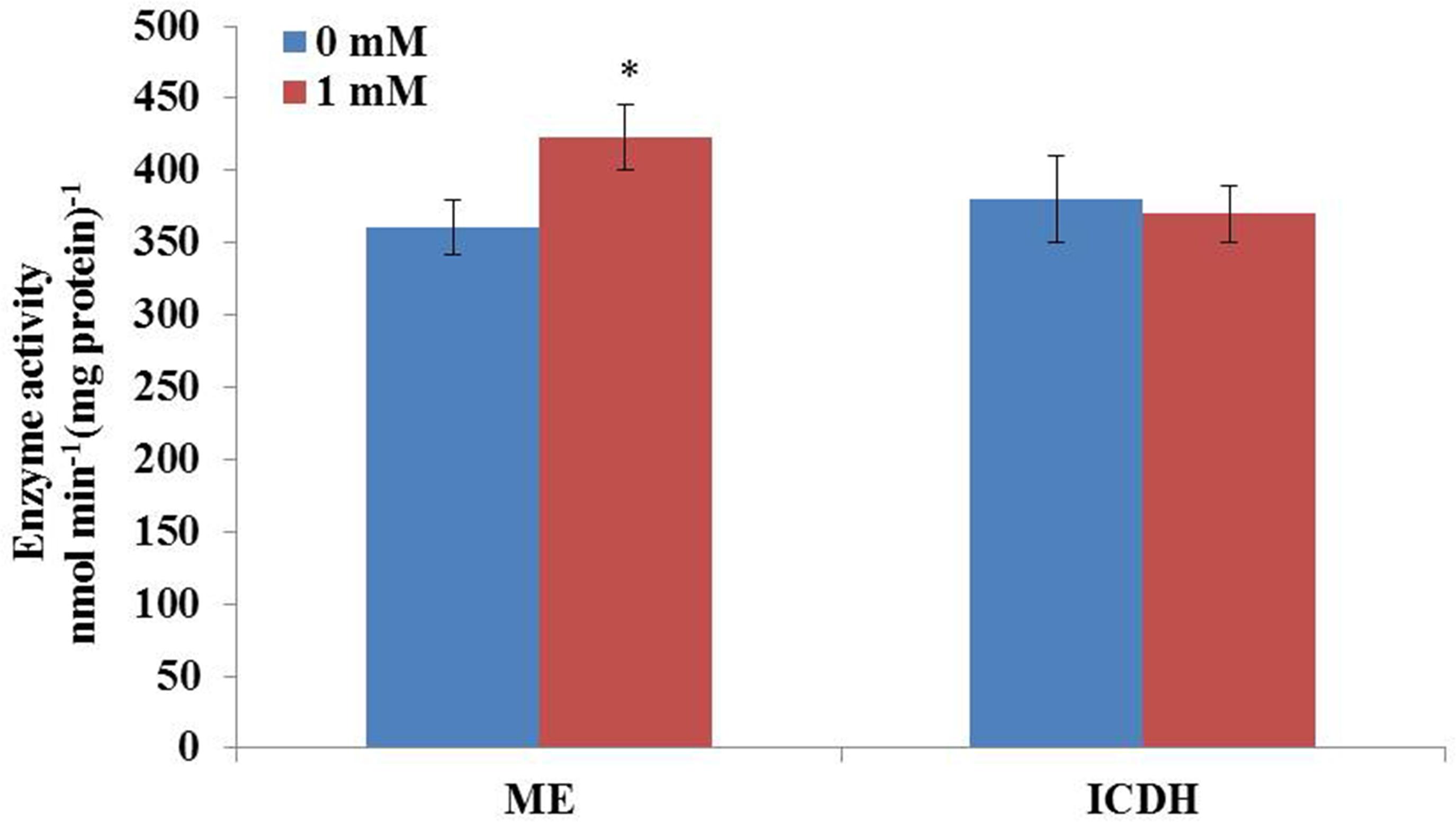
FIGURE 5. Effect of ETA on the activity of ME and IDCH of C. cohnii cultured with and without 1 mM ETA. Statistical significance was indicated by ∗∗p < 0.01; ∗p < 0.05.
Conclusion
In this study, the first 13C-metabolic flux analysis was performed in DHA-producing C. cohnii. Our results showed that with the addition of chemical modulator ETA, the flux through ME as well as the activity of ME were significantly increased, suggesting that NADP+-dependent ME might be the major source of NADPH for lipid accumulation. The analysis also suggested that in C. cohnii the whole citrate pyruvate cycle played an essential role in the lipid biosynthesis pathway. This study provided valuable information necessary for future genetic engineering of C. cohnii for improved lipid accumulation and DHA production.
Author Contributions
LC and WZ conceived and designed the study. JC performed the experiments. JC, JD, TS, MS, LL, LC, and WZ analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
The research was supported by grants from the Tianjin Municipal Science and Technology Commission (No. 15JCZDJC32500), the National Natural Science Foundation of China (Nos. 31770100, 21621004, 31370115 and 91751102), the National Basic Research Program of China (“973” Program, Project No. 2014CB745101), the Doctoral Program of Higher Education of China (Nos. 20120032110020 and 20130032120022), and Zaoneng Biotechnology Inc.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Guangsheng Pei of our laboratory for his help with metabolic model construction of C. cohnii.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00956/full#supplementary-material
References
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. doi: 10.1007/s00425-010-1130-0
Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007). Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86. doi: 10.1016/j.ymben.2006.09.001
Benthin, S., Nielsen, J., and Villadsen, J. (1991). A simple and reliable method for the determination of cellular RNA content. Biotechnol. Tech. 5, 39–42. doi: 10.1007/BF00152753
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cheng, J., Niu, Y., Lu, S., and Yuan, Y. (2012). Metabolome analysis reveals ethanolamine as potential marker for improving lipid accumulation of model photosynthetic organisms. J. Chem. Technol. Biotechnol. 87, 1409–1418. doi: 10.1002/jctb.3759
Chitranjali, T., Anoop Chandran, P., and Muraleedhara Kurup, G. (2015). Omega-3 fatty acid concentrate from Dunaliella salina possesses anti-inflammatory properties including blockade of NF-kB nuclear translocation. Immunopharmacol. Immunotoxicol. 37, 81–89. doi: 10.3109/08923973.2014.981639
de Swaaf, M. E., de Rijk, T. C., Eggink, G., and Sijtsma, L. (1999). Optimisation of docosahexaenoic acid production in batch cultivations by Crypthecodinium cohnii. J. Biotechnol. 70, 185–192. doi: 10.1016/S0168-1656(99)00071-1
de Swaaf, M. E., de Rijk, T. C., van der Meer, P., Eggink, G., and Sijtsma, L. (2003). Analysis of docosahexaenoic acid biosynthesis in Crypthecodinium cohnii by 13C labelling and desaturase inhibitor experiments. J. Biotechnol. 103, 21–29. doi: 10.1016/S0168-1656(03)00070-1
Ganuza, E., Benítez-Santana, T., Atalah, E., Vega-Orellana, O., Ganga, R., and Izquierdo, M. S. (2008). Crypthecodinium cohnii and Schizochytrium sp. as potential substitutes to fisheries-derived oils from seabream (Sparus aurata) microdiets. Aquaculture 277, 109–116. doi: 10.1016/j.aquaculture.2008.02.005
Glaser, C., Lattka, E., Rzehak, P., Steer, C., and Koletzko, B. (2011). Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern. Child Nutr. 7(Suppl. 2), 27–40. doi: 10.1111/j.1740-8709.2011.00319.x
Guo, D.-S., Ji, X.-J., Ren, L.-J., Li, G.-L., and Huang, H. (2017). Improving docosahexaenoic acid production by Schizochytrium sp. using a newly designed high-oxygen-supply bioreactor. AIChE J. 63, 4278–4286. doi: 10.1002/aic.15783
Guo, D. S., Ji, X. J., Ren, L. J., Li, G. L., Yin, F. W., and Huang, H. (2016). Development of a real-time bioprocess monitoring method for docosahexaenoic acid production by Schizochytrium sp. Bioresour. Technol. 216, 422–427. doi: 10.1016/j.biortech.2016.05.044
Hajjaji, N., Schubnel, V., and Bougnoux, P. (2011). Determinants of DHA incorporation into tumor tissue during dietary DHA supplementation. Lipids 46, 1063–1069. doi: 10.1007/s11745-011-3573-x
Hao, G., Chen, H., Wang, L., Gu, Z., Song, Y., Zhang, H., et al. (2014). Role of malic enzyme during fatty acid synthesis in the oleaginous fungus Mortierella alpina. Appl. Environ. Microbiol. 80, 2672–2678. doi: 10.1128/AEM.00140-14
Hart, P. C., Jan, H., Christian, K., Mathias, F., Yair, S. H., and Junker, B. H. (2012). iMS2Flux – a high–throughput processing tool for stable isotope labeled mass spectrometric data used for metabolic flux analysis. BMC Bioinformatics 13:295. doi: 10.1186/1471-2105-13-295
Herbert, D., Phipps, P. J., and Strange, R. E. (1971). “Chapter III Chemical analysis of microbial cells,” in Methods in Microbiology, eds J. R. Norris and D. W. Ribbons (New York, NY: Academic Press), 209–344.
Holdsworth, J. E., Veenhuis, M., and Ratledge, C. (1988). Enzyme activities in oleaginous yeasts accumulating and utilizing exogenous or endogenous lipids. J. Gen. Microbiol. 134, 2907–2915. doi: 10.1099/00221287-134-11-2907
Hsu, R. Y., and Lardy, H. A. (1969). Malic enzyme. Methods Enzymol. 13, 230–235. doi: 10.1016/0076-6879(69)13042-6
Ji, X. J., Ren, L. J., and Huang, H. (2015). Omega-3 biotechnology: a green and sustainable process for omega-3 fatty acids production. Front. Bioeng. Biotechnol. 3:158. doi: 10.3389/fbioe.2015.00158
Jiao, X., Ying-Fang, N., Tan, H., Wei-Dong, Y., Jie-Sheng, L., and Hong-Ye, L. (2015). Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. doi: 10.1016/j.ymben.2014.10.002
Karr, J. E., Alexander, J. E., and Winningham, R. G. (2011). Omega-3 polyunsaturated fatty acids and cognition throughout the lifespan: a review. Nutr. Neurosci. 14, 216–225. doi: 10.1179/1476830511Y.0000000012
Li, J., Niu, X., Pei, G., Sui, X., Zhang, X., Chen, L., et al. (2015). Identification and metabolomic analysis of chemical modulators for lipid accumulation in Crypthecodinium cohnii. Bioresour. Technol. 191, 362–368. doi: 10.1016/j.biortech.2015.03.068
Li, Z., Sun, H., Mo, X., Li, X., Xu, B., and Tian, P. (2013). Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 97, 4927–4936. doi: 10.1007/s00253-012-4571-5
Liu, B., Liu, J., Sun, P., Ma, X., Jiang, Y., and Chen, F. (2015). Sesamol enhances cell growth and the biosynthesis and accumulation of docosahexaenoic acid in the microalga Crypthecodinium cohnii. J. Agric. Food Chem. 63, 5640–5645. doi: 10.1021/acs.jafc.5b01441
Liu, J., Pei, G., Diao, J., Chen, Z., Liu, L., Chen, L., et al. (2017). Screening and transcriptomic analysis of Crypthecodinium cohnii mutants with high growth and lipid content using the acetyl-CoA carboxylase inhibitor sethoxydim. Appl. Microbiol. Biotechnol. 101, 6179–6191. doi: 10.1007/s00253-017-8397-z
Liu, N., Qiao, K., and Stephanopoulos, G. (2016). 13C Metabolic Flux Analysis of acetate conversion to lipids by Yarrowia lipolytica. Metab. Eng. 38, 86–97. doi: 10.1016/j.ymben.2016.06.006
Liu, Z., Gao, Y., Chen, J., Imanaka, T., Bao, J., and Hua, Q. (2013). Analysis of metabolic fluxes for better understanding of mechanisms related to lipid accumulation in oleaginous yeast Trichosporon cutaneum. Bioresour. Technol. 130, 144–151. doi: 10.1016/j.biortech.2012.12.072
Masuko, T., Minami, A., Iwasaki, N., Majima, T., Nishimura, S.-I., and Lee, Y. C. (2005). Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal. Biochem. 339, 69–72. doi: 10.1016/j.ab.2004.12.001
Mendes, A., Reis, A., Vasconcelos, R., Guerra, P., and Lopes da Silva, T. (2008). Crypthecodinium cohnii with emphasis on DHA production: a review. J. Appl. Phycol. 21, 199–214. doi: 10.1007/s10811-008-9351-3
Nanchen, A., Fuhrer, T., and Sauer, U. (2007). “Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data,” in Metabolomics: Methods and Protocols, ed. W. Weckwerth (Totowa, NJ: Humana Press), 177–197.
Pál, M., Szalai, G., and Janda, T. (2015). Speculation: polyamines are important in abiotic stress signaling. Plant Sci. 237, 16–23. doi: 10.1016/j.plantsci.2015.05.003
Pei, G., Li, X., Liu, L., Liu, J., Wang, F., Chen, L., et al. (2017). De novo transcriptomic and metabolomic analysis of docosahexaenoic acid (DHA)-producing Crypthecodinium cohnii during fed-batch fermentation. Algal Res. 26, 380–391. doi: 10.1016/j.algal.2017.07.031
Pleissner, D., and Eriksen, N. T. (2012). Effects of phosphorous, nitrogen, and carbon limitation on biomass composition in batch and continuous flow cultures of the heterotrophic dinoflagellate Crypthecodinium cohnii. Biotechnol. Bioeng. 109, 2005–2016. doi: 10.1002/bit.24470
Quek, L.-E., Wittmann, C., Nielsen, L. K., and Krömer, J. O. (2009). OpenFLUX: efficient modelling software for 13C-based metabolic flux analysis. Microb. Cell Fact. 8:25. doi: 10.1186/1475-2859-8-25
Ratledge, C. (2004). Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 86, 807–815. doi: 10.1016/j.biochi.2004.09.017
Ratledge, C. (2014). The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems. Biotechnol. Lett. 36, 1557–1568. doi: 10.1007/s10529-014-1532-3
Ren, L. J., Sun, X. M., Ji, X. J., Chen, S. L., Guo, D. S., and Huang, H. (2017). Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour. Technol. 223, 141–148. doi: 10.1016/j.biortech.2016.10.040
Ronnebaum, S. M., Ilkayeva, O., Burgess, S. C., Joseph, J. W., Lu, D., Stevens, R. D., et al. (2006). A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J. Biol. Chem. 281, 30593–30602. doi: 10.1074/jbc.M511908200
Safdar, W., Zan, X., Shamoon, M., Sharif, H. R., Mukama, O., Tang, X., et al. (2017). Effects of twenty standard amino acids on biochemical constituents, docosahexaenoic acid production and metabolic activity changes of Crypthecodinium cohnii. Bioresour. Technol. 238, 738–743. doi: 10.1016/j.biortech.2017.04.024
Shi, K., Gao, Z., Shi, T. Q., Song, P., Ren, L. J., Huang, H., et al. (2017). Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: the state of the art and future perspectives. Front. Microbiol. 8:793. doi: 10.3389/fmicb.2017.00793
Sijtsma, L., and de Swaaf, M. E. (2004). Biotechnological production and applications of the omega-3 polyunsaturated fatty acid docosahexaenoic acid. Appl. Microbiol. Biotechnol. 64, 146–153. doi: 10.1007/s00253-003-1525-y
Sui, X., Niu, X., Shi, M., Pei, G., Li, J., Chen, L., et al. (2014). Metabolomic analysis reveals mechanism of antioxidant butylated hydroxyanisole on lipid accumulation in Crypthecodinium cohnii. J. Agric. Food Chem. 62, 12477–12484. doi: 10.1021/jf503671m
Sun, X. M., Ren, L. J., Bi, Z. Q., Ji, X. J., Zhao, Q. Y., Jiang, L., et al. (2018). Development of a cooperative two-factor adaptive-evolution method to enhance lipid production and prevent lipid peroxidation in Schizochytrium sp. Biotechnol. Biofuels 11:65. doi: 10.1186/s13068-018-1065-4
Sun, X. M., Ren, L. J., Ji, X. J., Chen, S. L., Guo, D. S., and Huang, H. (2016). Adaptive evolution of Schizochytrium sp. by continuous high oxygen stimulations to enhance docosahexaenoic acid synthesis. Bioresour. Technol. 211, 374–381. doi: 10.1016/j.biortech.2016.03.093
Walsh, T. A., Bevan, S. A., Gachotte, D. J., Larsen, C. M., Moskal, W. A., Merlo, P. A., et al. (2016). Canola engineered with a microalgal polyketide synthase-like system produces oil enriched in docosahexaenoic acid. Nat. Biotechnol. 34, 881–887. doi: 10.1038/nbt.3585
Ward, O. P., and Singh, A. (2005). Omega-3/6 fatty acids: alternative sources of production. Process Biochem. 40, 3627–3652. doi: 10.1016/j.procbio.2005.02.020
Wasylenko, T. M., Ahn, W. S., and Stephanopoulos, G. (2015). The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 30, 27–39. doi: 10.1016/j.ymben.2015.02.007
Xiong, W., Li, X., Xiang, J., and Wu, Q. (2008). High-density fermentation of microalga Chlorella protothecoides in bioreactor for microbio-diesel production. Appl. Microbiol. Biotechnol. 78, 29–36. doi: 10.1007/s00253-007-1285-1
Xiong, W., Liu, L., Wu, C., Yang, C., and Wu, Q. (2010). 13C-tracer and gas chromatography-mass spectrometry analyses reveal metabolic flux distribution in the oleaginous microalga Chlorella protothecoides. Plant Physiol. 154, 1001–1011. doi: 10.1104/pp.110.158956
Yang, S., Lu, S.-H., and Yuan, Y.-J. (2009). Cerium elicitor-induced phosphatidic acid triggers apoptotic signaling development in Taxus cuspidata cell suspension cultures. Chem. Phys. Lipids 159, 13–20. doi: 10.1016/j.chemphyslip.2009.02.004
Yilancioglu, K., Cokol, M., Pastirmaci, I., Erman, B., and Cetiner, S. (2014). Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS One 9:e91957. doi: 10.1371/journal.pone.0091957
Yu, Q., Liu, Z., Xu, H., Zhang, B., Zhang, M., and Li, M. (2015). TiO2 nanoparticles promote the production of unsaturated fatty acids (UFAs) fighting against oxidative stress in Pichia pastoris. RSC Adv. 5, 41033–41040. doi: 10.1039/c5ra02366a
Zamboni, N., Fendt, S. M., Ruhl, M., and Sauer, U. (2009). 13C-based metabolic flux analysis. Nat. Protoc. 4, 878–892. doi: 10.1038/nprot.2009.58
Zhang, H., Wu, C., Wu, Q., Dai, J., and Song, Y. (2016). Metabolic flux analysis of lipid biosynthesis in the yeast Yarrowia lipolytica using 13C-labled glucose and gas chromatography-mass spectrometry. PLoS One 11:e0159187. doi: 10.1371/journal.pone.0159187
Zhang, Y., Adams, I. P., and Ratledge, C. (2007). Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153(Pt 7), 2013.
Keywords: metabolic flux analysis, chemical modulators, ethanolamine, lipid accumulation, NADP+-dependent malic enzyme, Crypthecodinium cohnii
Citation: Cui J, Diao J, Sun T, Shi M, Liu L, Wang F, Chen L and Zhang W (2018) 13C Metabolic Flux Analysis of Enhanced Lipid Accumulation Modulated by Ethanolamine in Crypthecodinium cohnii. Front. Microbiol. 9:956. doi: 10.3389/fmicb.2018.00956
Received: 01 March 2018; Accepted: 24 April 2018;
Published: 15 May 2018.
Edited by:
Qiang Wang, Institute of Hydrobiology (CAS), ChinaCopyright © 2018 Cui, Diao, Sun, Shi, Liu, Wang, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Chen, lchen@tju.edu.cn
 Jinyu Cui
Jinyu Cui Jinjin Diao
Jinjin Diao Tao Sun
Tao Sun Mengliang Shi
Mengliang Shi Liangsen Liu1,2,3
Liangsen Liu1,2,3 Lei Chen
Lei Chen Weiwen Zhang
Weiwen Zhang