- 1Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 2Department of Theoretical and Applied Biology, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
In the current study, we identified plasmids carrying antimicrobial resistance genes in draft whole genome sequences of 16 selected Salmonella enterica isolates representing six different serovars from humans in Ghana. The plasmids and the location of resistance genes in the genomes were predicted using a combination of PlasmidFinder, ResFinder, plasmidSPAdes and BLAST genomic analysis tools. Subsequently, S1-PFGE was employed for analysis of plasmid profiles. Whole genome sequencing confirmed the presence of antimicrobial resistance genes in Salmonella isolates showing multidrug resistance phenotypically. ESBL, either blaTEM52−B or blaCTX−M15 were present in two cephalosporin resistant isolates of S. Virchow and S. Poona, respectively. The systematic genome analysis revealed the presence of different plasmids in different serovars, with or without insertion of antimicrobial resistance genes. In S. Enteritidis, resistance genes were carried predominantly on plasmids of IncN type, in S. Typhimurium on plasmids of IncFII(S)/IncFIB(S)/IncQ1 type. In S. Virchow and in S. Poona, resistance genes were detected on plasmids of IncX1 and TrfA/IncHI2/IncHI2A type, respectively. The latter two plasmids were described for the first time in these serovars. The combination of genomic analytical tools allowed nearly full mapping of the resistance plasmids in all Salmonella strains analyzed. The results suggest that the improved analytical approach used in the current study may be used to identify plasmids that are specifically associated with resistance phenotypes in whole genome sequences. Such knowledge would allow the development of rapid multidrug resistance tracking tools in Salmonella populations using WGS.
Introduction
The increased and widespread use of antimicrobials in human and veterinary medicine has resulted in increased levels of antimicrobial resistance (World Health Organization, 2014). Global trade and travel mean that antimicrobial resistant bacteria that emerge in one country easily spread to other countries (Aarestrup et al., 2007; Hendriksen et al., 2011; Le Hello et al., 2011), and in recent years, multidrug and cephalosporin-resistant Salmonella have become a public health concern worldwide (Su et al., 2004; Yan et al., 2005; Anjum et al., 2011). This is problematic, since third generation or extended-spectrum cephalosporins are the drugs of choice in the treatment of Salmonella infections when fluoroquinolone treatment is unsuccessful (Parry and Threlfall, 2008).
The most common mean of antibiotic resistance development in Salmonella is through the acquisition of mobile genetic elements such as plasmids, transposons and integrons carrying resistance genes. The major concern is that these mobile elements, in particular plasmids, spread rapidly in the population, thus posing a risk of emergence of highly resistant clones of Salmonella within both human and animal hosts (Hoffmann et al., 2017).
Complete plasmid sequencing from Salmonella isolates from various sources is the ideal method to understand the nature and transfer of antibiotic resistance in bacteria. Though plasmid sequencing remains the main strategy for complete plasmid characterization, plasmid extraction from whole genome sequences is becoming increasingly common, and to date, a number of bioinformatic tools have been developed for in silico detection and characterization of plasmids in whole genome sequences (Arredondo-Alonso et al., 2017; Orlek et al., 2017). However, none of these tools (e.g., PlasmidFinder, PlasmidSPAdes, Recycler, PLACNET, etc.) are sufficiently powerful if used alone, and there is a need to develop combined approaches to allow confident identification and characterization of plasmids in silico.
In Ghana, Salmonella and other Enterobacterioceae which are resistant to cephalosporins have been reported in humans (Mills-Robertson et al., 2003; Obeng-Nkrumah et al., 2013; Andoh et al., 2017), but little is known about the plasmids that carry genes coding for such resistances. In this study, a strategic combination of bioinformatics tools were applied on whole genome sequence data to identify plasmids harboring genes associated with extended-spectrum β-lactamases (ESBLs) and other antimicrobial resistance phenotypes in multidrug-resistant and putatively ESBL-producing Salmonella enterica isolates from humans in Ghana.
Methods
Strain Collection
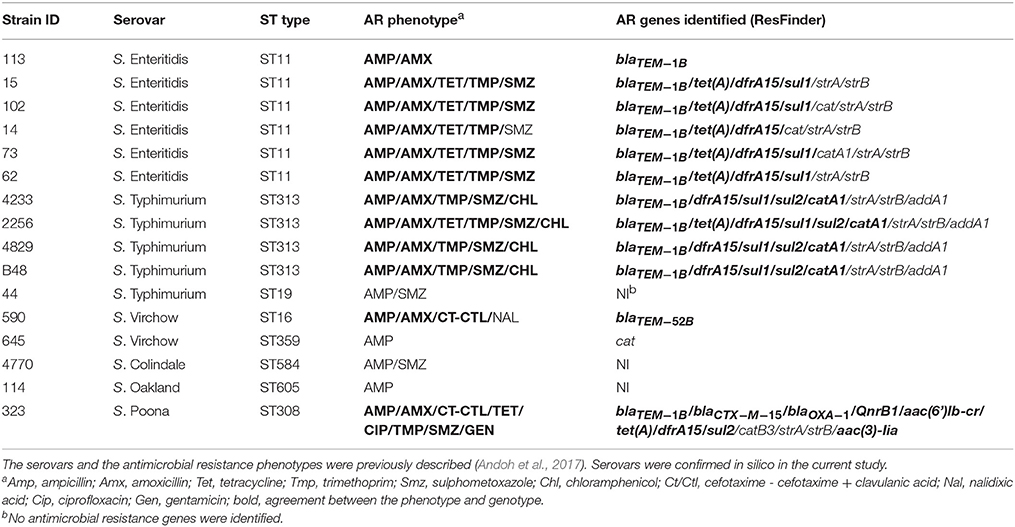
Sixteen antibiotic resistant Salmonella isolates were obtained from a previous study on human salmonellosis in Ghana. Strains were previously assigned to a specific serovar and antimicrobial resistance pattern (Andoh et al., 2017). Strains and their relevant characteristics are listed in Table 1.
Phenotypic Test for ESBL
ESBL phenotype was determined using ESBL CT/CTL 16/1 ETEST® for Antimicrobial Resistance Detection (BioMerieux, France). Manufacturer's instructions were followed to perform the test and to interpret the results.
Whole Genome Sequencing
The 16 Salmonella isolates were grown in Luria broth (LB) (240230; Difco, USA) for 16 h at 37°C while shaking, and genomic DNA was isolated using a blood and tissue kit (Qiagen, Stockach, Germany) according to the instructions of the supplier. Genomes were sequenced at a 300-bp paired-end-read format using the Nextera XT library preparation kit and the MiSeq instrument (Illumina, San Diego, California, USA) as previously reported (Madoshi et al., 2016). CLC Genomic Workbench v6.5 software package (https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/) was used for sequence-reads quality-assessment and adapter trimming. Draft genomes assembling was performed with SPAdes v3.10.1 (Bankevich et al., 2012), and the quality of the assembly was evaluated with QUAST v.2.3 (Gurevich et al., 2013). The draft genome sequences from the strains were submitted to GenBank (BioProject ID: PRJN335586).
In Silico Analysis of Serovar, Serogroup and Antigenic Profile
SISTR (Yoshida et al., 2016) and SeqSer (Zhang et al., 2015) bioinformatics tools were used to identify the sequence type (ST type), serovar, serogroup and antigenic profile in the 16 draft genomes of Salmonella.
In Silico Plasmid Identification
The presence of plasmids in the genomes was identified using a combination of the following bioinformatic tools: PlasmidFinder (Carattoli et al., 2014), plasmidSPAdes v3.10.1 (Antipov et al., 2016), BlastN at NCBI and locally within the CLC Genomics Workbench v6.5. Briefly, in the first step of the analysis, PlasmidFinder was used to identify plasmid replicon location in the whole genome assembly generated by SPAdes. To improve plasmid detection and to identify all contigs representing a specific plasmid, the plasmidSPAdes tool was then employed. The algorithm in plasmidSPAdes is able to predict which contigs belong to plasmid DNA, and to assign those contigs into components. Each component is considered as a putative plasmid composed of one or more contigs. In the third step, components (putative plasmids, SPAdes output) containing specific plasmid replicons and their combinations (PlasmidFinder output) from one selected strain were used to search for the most similar/reference plasmids at NCBI nr-database using BlastN. Finally, the sequences of the identified reference plasmids were downloaded from GenBank and searched back in all strains using local BlastN in CLC Genomics Workbench and by progressive MAUVE (Darling et al., 2010). This allowed the identification of plasmid DNA (contigs), which could not be assigned to a specific plasmid replicon type and/or its component by PlasmidFinder and plasmidSPAdes. A graphic overview of the approach giving an example of the analysis of one selected isolated is shown in Supplementary Figure 1.
In Silico Identification of Antibiotic Resistance Genes and Their Association With Plasmids
ResFinder (Zankari et al., 2012), available on the Centre for Biological Sequence analysis (CBS) server1 was employed for the in silico prediction of acquired antibiotic resistance genes in the genomes under study. For a hit to be reported by ResFinder, it had to cover at least 60% of the length of the gene sequence in the database with a sequence identity of 60%. The prediction of location of antibiotic resistance genes in plasmids was performed in a step-wise procedure by combining the results of resistance prediction and plasmid identification steps (Supplementary Figure 1). The output from ResFinder gives the contig IDs on which antimicrobial resistance genes are located in the whole genome assembly generated by SPAdes. Contigs identified by ResFinder were searched to determine whether they were the same as those carrying plasmid replicons identified using PlasmidFinder or those assigned to specific components created by plasmidSPAdes. Contigs, which carried antimicrobial resistance genes, but which were not assigned to plasmids by using the above tools, were blasted against reference plasmids locally.
S1-PFGE Plasmid Profiling
Plasmid profiling was performed according to the protocol described by Barton et al. (1995). Briefly, plasmids were digested with the S1 nuclease, and their linear forms were visualized using Pulsed-field gel electrophoresis (PFGE). The plasmid sizes were estimated by comparing their electrophoretic mobility with the electrophoretic mobility of fragments generated by XbaI digestion of Salmonella Braenderup strain H98122.
Results
In Silico Confirmation of Serovar and Predicition of Sequence Type
The 16 strains under study belonged to six serovars [S. Enteritidis (n = 6), S. Typhimurium (n = 5), S. Virchow (n = 2), S. Colindale (n = 1), S. Poona (n = 1), S. Oakland (n = 1)], of Salmonella enterica according to the phenotypic characterization previously performed (Andoh et al., 2017). All serovars were confirmed genotypically using the combination of SeqSero and draft genome assemblies analysis tools available on the SISTR platform (Table 1 and Supplementary Table 1).
Based on sequence type prediction by SISTR, all S. Enteritidis isolates were assigned to the same sequence type (ST11). The majority of S. Typhimurium isolates (n = 4/5) belonged to ST313, and only the S. Typhimurium strain 44 was assigned to ST19. The two S. Virchow, one S. Colindale, one S. Poona, and one S. Oakland isolates were predicted to belong to distinct ST types (Table 1).
In Silico Prediction of Antimicrobial Resistance Genes
The majority (81.3%) of the 16 strains selected for the whole genome sequence analysis were phenotypically resistant to more than one antimicrobial (Andoh et al., 2017). With regards to resistance to β-lactams, two out of the 16 strains showed resistance to cefotaxime/cefotaxime + clavulanic acid, and thus were ESBL-positive. The remaining 14 strains were only resistant to amino-penicillins (Table 1). The susceptibility of these strains to antimicrobials, as determined phenotypically (Andoh et al., 2017) was related to the presence of resistance genes in the whole genome sequences using the ResFinder tool.
Among the 16 Salmonella strains analyzed, ResFinder detected the presence of blaTEM−1B in 11 amino-penicillin resistant isolates. Two isolates associated with an ESBL phenotype carried recognized ESBL genes blaTEM−52B and blaCTX−M−15, respectively (Table 1). Five or more other antimicrobial resistance genes were identified per isolate from the genome sequences, with the most common genes encoding resistance to trimethoprim (dfrA1/dfrA15), aminoglycosides (strA/strB/aadA1), sulfamethoxazole (sul1/sul2), chloramphenicol (cat/catA1/catB3) and tetracycline (tetA). The majority of the phenotypic resistances were confirmed by the genome sequence analysis [Sul (9/12; 75.0%), Tmp (10/10; 100.0%), Tet (7/7; 100.0%), Amp-Amx (12/16; 75.0%), Chl (4/4; 100%), Cip (1/1; 100.0%), Nal (0/1; 0.0%), Gen (1/1; 100.0%)] (Table 1). The streptomycin-resistance genes strA/strB and the streptomycin/spectinomycin resistance gene aadA1 were detected in 10 and 4 out of the 16 strains, respectively. These antimicrobials were not screened phenotypically.
In Silico Prediction of Plasmids and Plasmid Replicons
PlasmidFinder was first used to identify plasmid replicons in the 16 Salmonella enterica strains under study. All genomes of S. Enteritidis (n = 6) and S. Typhimurium (n = 5) carried plasmid replicons of the IncFII(S) and IncFIB(S) types (Table 2). In addition, IncN and IncX1 type plasmid replicons were found in five and three isolates of S. Enteritidis, respectively, and all isolates of S. Typhimurium, except one, carried plasmid replicons of the ColRNAI type. Four isolates of S. Typhimurium harbored a plasmid replicon of the IncQ1 type. One isolate of S. Virchow showed a plasmid replicon of the IncX1 type. The S. Poona strain and another S. Virchow isolate carried three plasmid replicons of the IncHI2, IncHI2A and TrfA type (Table 2). PlasmidFinder did not detect any plasmid replicons in the isolates of S. Colindale and S. Oakland.
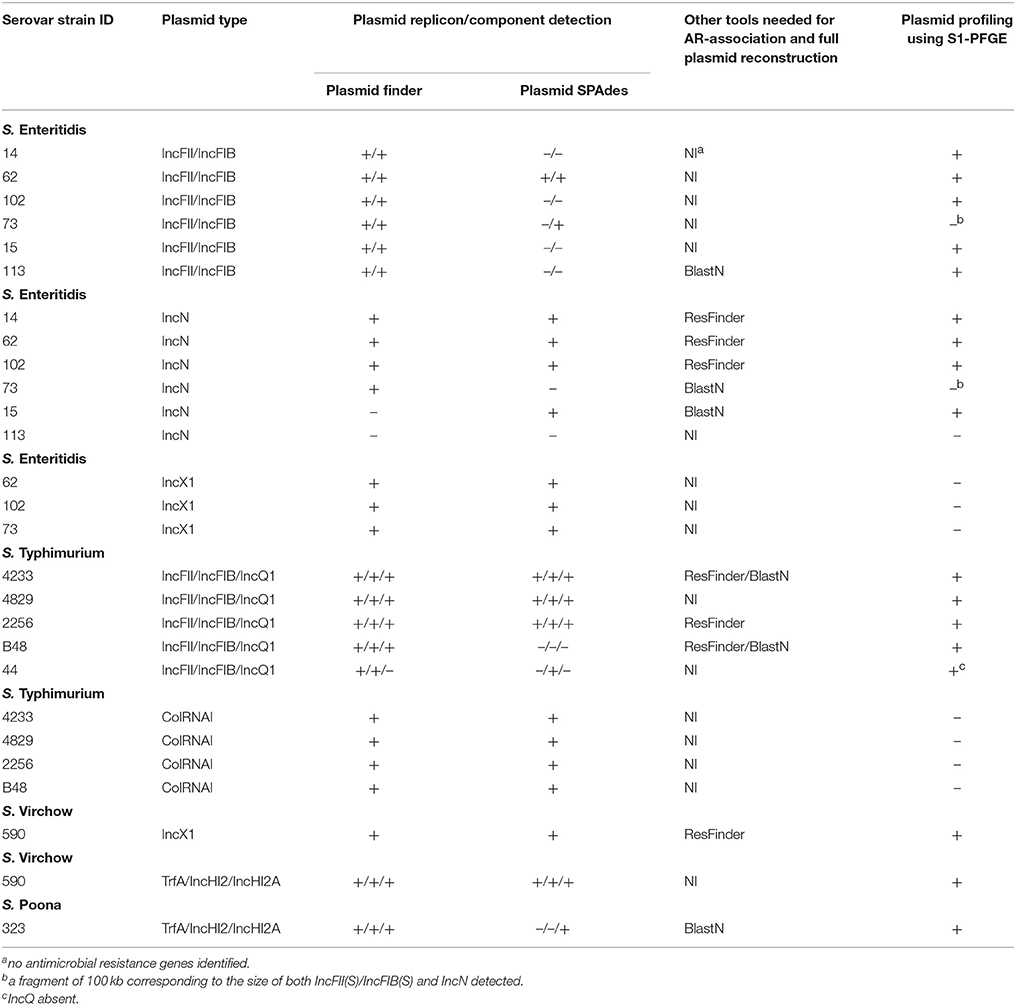
Table 2. Performance of bioinformatic tools in respect to plasmid reconstruction and detection of antimicriobial resistance gene location in the whole genome sequences of the Salmonella strains under study.
PlasmidFinder does not report plasmid replicons, unless they are present in the PlasmidFinder database. In an attempt to overcome this limitation, plasmids were also searched with the plasmidSPAdes tool, which uses the read coverage of contigs to assist in distinguishing between plasmids and the chromosome. In S. Enteritidis, the contigs predicted by PlasmidFinder to carry either IncFII(S) or IncFIB(S) plasmid replicons were confirmed in only two strains with SPAdes (Table 2). In one strain, both replicons were connected into one component, which indicated that they originated from the same plasmid (data not shown). Plasmid contigs with replicons of IncN type were confirmed in four strains, but failed in one strain. In two strains, IncN and IncX1 replicons were found on the same SPAdes component, whereas in other two strains IncN was found alone (data not shown). In S. Typhimurium, plasmid replicons IncFII(S) and IncFIB(S) were confirmed by SPAdes in four strains (Table 2). In one strain, sequences containing these replicons were connected into the same component, in two strains they were connected together with the contigs carrying IncQ1 plasmid replicon, and in one strain only the IncFIB(S) replicon was confirmed. PlasmidSPAdes confirmed the presence of ColRNAI containing contigs in all strains. In S. Virchow isolate 590, one of the putative plasmids predicted by SPAdes contained two plasmid replicons (TrfA and IncHI2), one putative plasmid contig harbored IncX1, and one putative plasmid harbored IncHI2A (Table 2). Three putative plasmids were predicted using SPAdes in the S. Oakland strain, yet no plasmid replicons were previously identified using PlasmidFinder tool. In S. Poona, only a contig carrying the IncHI2A replicon type was confirmed. No putative replicon plasmids were found in S. Virchow isolate 648, and in the S. Colindale strain.
PlasmidSPAdes components containing specific plasmid replicons were used to search for the most similar reference plasmids at NCBI nr-database using BlastN. The local BlastN and the alignment of the identified reference plasmids-sequences against the draft genome sequences allowed full reconstruction of plasmids in all isolates (Tables 2, 3). This analysis resulted in the detection of three plasmids in the strains of S. Enteritidis. The first one carrying plasmid replicons of IncFII(S) and IncFIB(S) was homologous (length coverage of 100%, sequence identity of 92–94%) to a plasmid of S. Enteritidis of 59,369 bp in length (GenBank: CP007529). This plasmid corresponds to the spv-encoding virulence plasmid of S. Enteritidis (Figure 1) (Allard et al., 2013). The second one carrying an IncN plasmid replicon was most similar (length coverage of 100%, sequence identity of 94–96%) to a plasmid of an uncultured bacterium from a wastewater treatment plant in Germany (GenBank: JN102343) of 47,905 bp in length. The third plasmid carried a replicon of the IncX1 type and was similar (length coverage of 100%, sequence identity of 100%) to an E. coli plasmid of 1,552 bp (GenBank: CP011143).
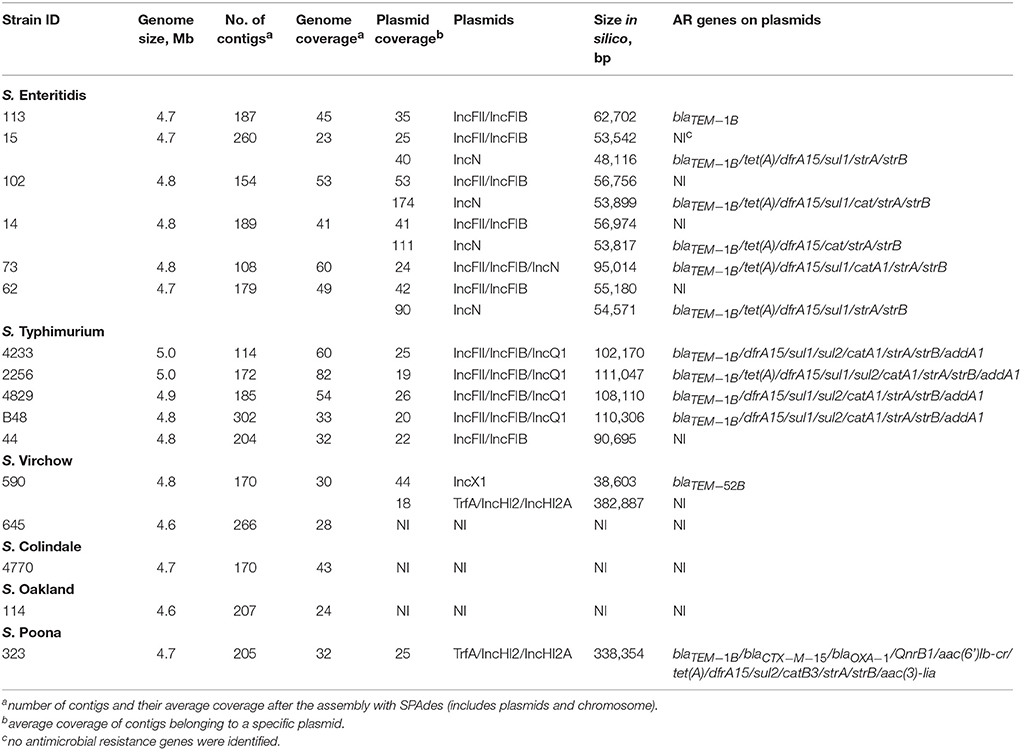
Table 3. Genome assembly characteristics of Salmonella enterica serovars, resistance plasmids and resistance genes identified.
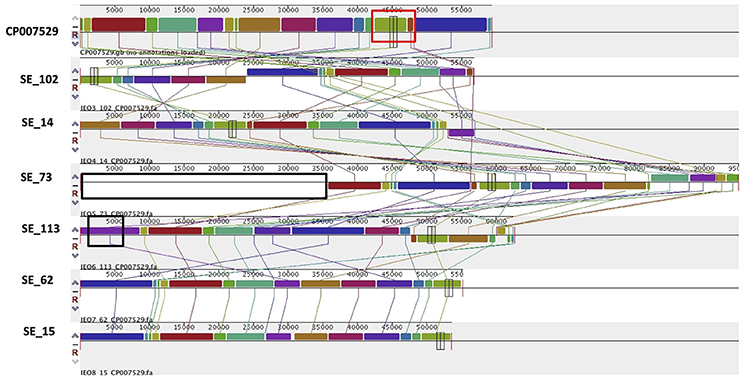
Figure 1. Alignment of IncFII(S)/IncFIB(S) plasmid homologs of Salmonella Enteritidis to a reference plasmid CP007529. The alignment was created with MAUVE. Top row shows the reference plasmid, and the rows below the reconstructed plasmids in Salmonella Enteritidis from the study. Each of the colored blocks outlines a region of the genome sequence that aligned to part of another genome, and is presumably homologous. Areas with no blocks were not aligned and contain sequence elements specific to a particular genome. The red box indicates the spv virulence- associated region in the reference plasmid. Black boxes surrounding a block in SE_113 and in SE_73 indicate antibiotic resistance gene blaTEM-1 and predicted IncN plasmid incorporation, respectively.
In S. Typhimurium, two plasmids were identified. One plasmid carried three plasmid replicon types (IncFII(S)/IncFIB(S)/IncQ1) and was found to be homologous (length coverage of 80–100%, sequence identity of 86–96%) to a previously characterized plasmid of S. Typhimurium of 117,047 bp in length (GenBank: FN432031). This plasmid carried spv-genes (Figure 2). The second one was of ColRNAI type. This plasmid was most similar (length coverage of 18–65%, sequence identity of 71–87%) to a plasmid of S. Typhi of 5,569 bp in length (GenBank: HQ730898). Two plasmids were detected in S. Virchow. The first plasmid carried three plasmid replicons (TrfA/IncHI2/IncHI2A) and was homologous (length coverage of 84%, sequence identity of 85%) to a plasmid from S. Typhimurium of 300,375 bp in length (GenBank: LN794248). The plasmid of S. Poona was also found to be most similar (length coverage of 100%, sequence identity of 87%) to the same plasmid. The second plasmid of S. Virchow carried a plasmid replicon of the IncX1 type and was homologous (length coverage of 100%, sequence identity of 94%) to a plasmid from E. coli of 38,611 bp in length (GenBank: JQ269336).
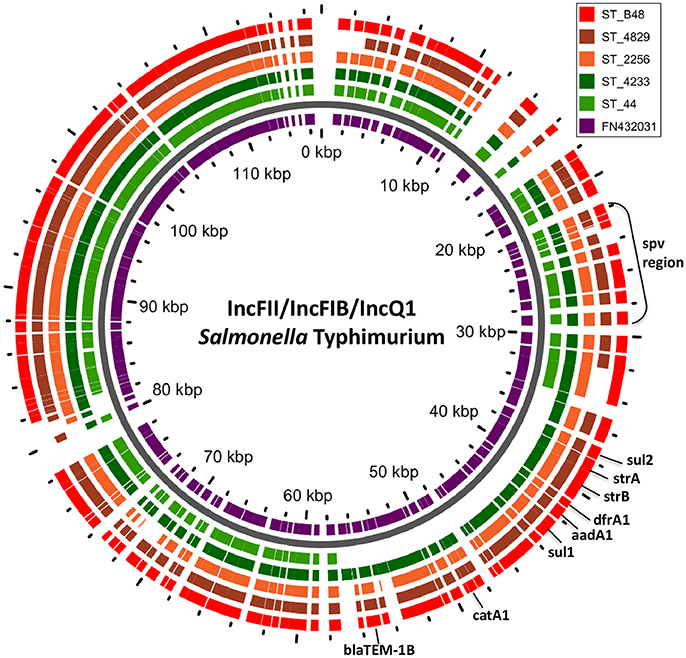
Figure 2. BLAST ring of IncFII(S)/IncFIB(S)/IncQ1 plasmid homologs of Salmonella Typhimurium and reference plasmid FN432031. Blast ring was created with GView. The middle ring is a reference plasmid, around which BLAST lanes are shown. Every lane corresponds to plasmid in each genome. The location of spv virulence-associated region, and antibiotic resistance genes are indicated outside the ring.
Prediction of the Antimicrobial Resistance Gene Location in the Plasmids
Combined analysis usingPlasmidFinder, ResFinder, plasmidSPAdes, and BlastN further allowed us to predict antimicrobial resistance gene location on the specific types of plasmids in all the 16 isolates under study (Tables 2, 3). In S. Enteritidis, the antimicrobial resistance genes were located on IncN plasmid (Figure 3). The transposable elements carrying resistance genes were inserted between the genes nuc and fipA. No transposon was found to be associated with the streptomycin resistance genes strA and strB, yet they are known to be the remnants of transposon Tn5393 (Eikmeyer et al., 2012). As previously described, transposon Tn1721 was linked to the carriage of tetracycline resistance genes tetR and tetA, and Tn3 to the ampicillin resistance gene blaTEM-1 (Eikmeyer et al., 2012). In addition, all isolates under study carried sul1 and dfr15, which were not present in reference plasmid JN102343. Based on BlastN at NCBI, these genes and their flanking regions were most similar (length coverage of 100%, sequence identity of 100%) to the region found in Enterobacter cloacae plasmid (GenBank: CP021897) and with a lower similarity to the regions found in plasmids of other Enterobacteriaceae. The insertion of the region was associated with the XerC superfamily integrase. In the IncN replicon type negative S. Enteritidis isolate 113, the gene blaTEM−1B gene was found to be located on an IncFII(S)/IncFIB(S) plasmid (Figure 1) associated to transposon Tn3.
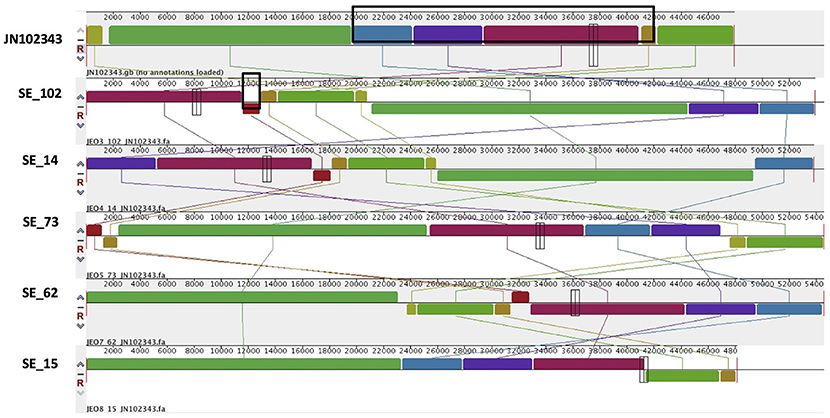
Figure 3. Alignment of IncN plasmid homologs of Salmonella Enteritidis to a reference plasmid JN102343. The alignment was created with MAUVE. The top row shows the reference plasmid, and the rows below the reconstructed plasmids in Salmonella Enteritidis from the study. Each of the colored blocks outlines a region of the genome sequence that aligned to part of another genome, and is presumably homologous. Areas with no blocks were not aligned and contain sequence elements specific to a particular genome. The black box in the reference plasmid surrounds a block found to contain antibiotic resistance genes strA, strB, tet(A), blaTEM-1B found in all isolates. The box in SE_102 indicate an insertion of sul1, dfrA15, which was found to be absent reference plasmid and in SE_15 isolate.
In S. Typhimurium, resistance genes were carried on a IncFII(S)/IncFIB(S)/IncQ1 plasmid (Figure 2). Interestingly, S. Typhimurium strain 44 did not show resistance to multiple classes of antimicrobials, except for amino-penicillin and sulphometaxazole, and carried only plasmid replicons of the IncFII(S)/IncFIB(S) type, but not IncQ1. The BlastN analysis of this plasmid against the reference plasmid showed the absence of a whole contig carrying the IncQ1 replicon type, where resistance genes to various antimicrobials were present in all other isolates (Figure 2). In accordance with previous observations, all resistance genes found in the plasmid of S. Typhimurium ST313 isolates under study, were located within a composite Tn21-like transposon (Kingsley et al., 2009).
As mentioned previously, a TrfA/IncHI2/IncHI2A plasmid was present in both S. Virchow and S. Poona strains (Figure 4), however only the plasmid found in S. Poona carried a multiple number of antimicrobial resistance genes including blaCTX−M−15. As previously mentioned this plasmid was homologous to a previously described IncHI2 plasmid of S. Typhimurium ST313 (GenBank: LN794248) isolated during an outbreak in Kenya (Kariuki et al., 2015). The majority of the resistance genes in this plasmid were located within a class 1 integron, while blaCTX−M−15 was associated with ISEcp1. In addition to the resistance genes that were present in the reference plasmid, the plasmid of S. Poona under study carried the quinolone resistance gene, qnrB1. The qnrB1-encoding region was found to be most similar (length coverage of 100%, sequence identity of 99%) to a region within the plasmid of Klebsiella pneumoniae (GenBank: CP023922), and was associated with the transposon Tn3.
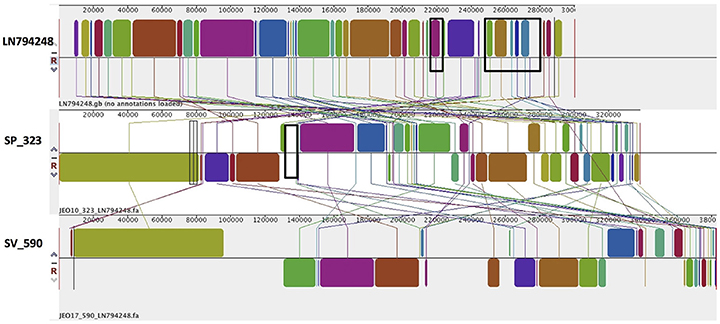
Figure 4. Alignment of TrfA/IncHI2/IncHI2A plasmid homologs of Salmonella Virchow and Salmonella Poona to a reference plasmid LN794248. The alignment was created with MAUVE. The top row shows the reference plasmid, and the rows below the reconstructed plasmids in Salmonella Virchow and Salmonella Poona from the study. Each of the colored blocks outlines a region of the genome sequence that aligned to part of another genome, and is presumably homologous. Areas with no blocks were not aligned and contain sequence elements specific to a particular genome. Black boxes in the reference plasmid surround blocks found to contain antibiotic resistance genes strA, strB, blaTEM-1B, sul2, tet(A), blaCTX-M-15, aac(3)-Iia, catB3, aac(6')Ib-cr, blaOXA-1, and dfrA14, which were present in S. Poona isolate from the study. The black box in the reconstructed plasmid in S. Poona indicates an insertion of QnrB1, which was absent in the reference plasmid.
The only antimicrobial resistance gene identified in S. Virchow, blaTEM−52B, was located on the IncX1 plasmid linked to the transposon Tn3 (Johnson et al., 2012) (Figure 5).
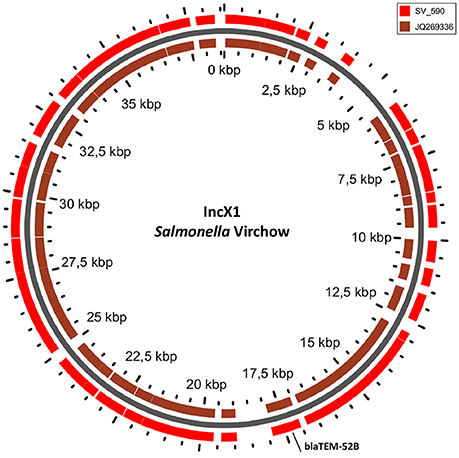
Figure 5. BLAST ring of IncX1 plasmid homolog of Salmonella Virchow and a reference plasmid JQ269336. Blast ring was created with GView. The middle ring is the reference plasmid, around which a BLAST lane of the plasmid in S. Virchow is shown. The location of antibiotic resistance gene is indicated outside the ring.
S1-PFGE Plasmid Analysis
S1-PFGE allowed visualization of plasmids with sizes ranging from 20 to 452 kb (Figure 6). The analysis revealed the presence of zero to up to three plasmids in the 16 strains characterized. In S. Enteritidis, the fragment of approximately 50 kb, corresponding to the IncN plasmid carrying resistance genes, was detected in 4 out of 5 isolates, which were shown to harbor this plasmid in silico (Table 2). According to the in silico prediction results, all six S. Enteritidis isolates carried the virulence plasmid IncFII(S)/IncFIB(S). In three of these strains, a plasmid of approximately 60 kb, corresponding to the size of the latter plasmid was present on the gel. A slightly bigger fragment of approximately 65–70 kb was found in the strain 113. According to our in silico results, this IncFII(S)/IncFIB(S) plasmid harbored the gene blaTEM−1B, which was absent in the IncFII(S)/IncFIB(S) plasmid from the three other strains. This resulted in an increase of the original virulence plasmid size, and it was classified as a virulence-resistance plasmid (Table 3). In the last two strains (73 and 14) of S. Enteritidis carrying the IncFII(S)/IncFIB(S) plasmid, the fragment of approximately 60 kb was not present, however a much larger fragment of approximately 100 kb was observed in both strains. Interestingly, two S. Enteritidis isolates carried additional plasmid fragments, which were not detected with the analysis in silico. Two such fragments of approximately 79 and 35 kb were found in strain 113, and one of approximately 50 kb in strain 62.
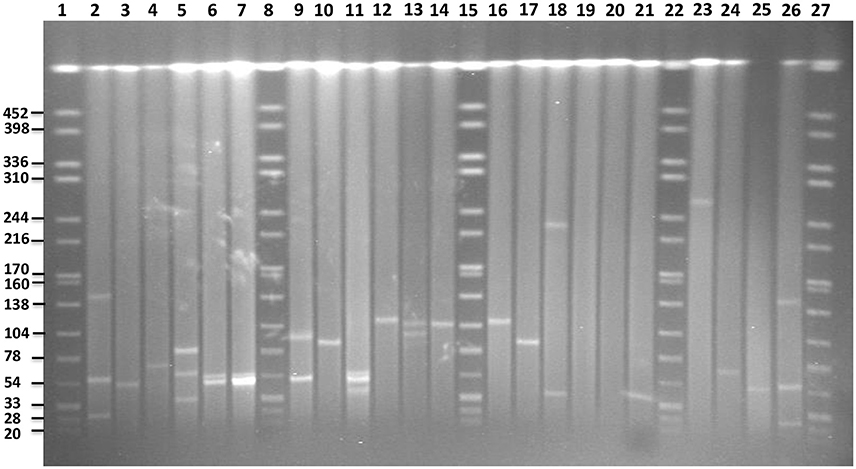
Figure 6. PFGE gel of plasmid profiles in the examined Salmonella serovars. Lanes 1, 8, 15, 22, and 27 represent fragments generated by XbaI digestion of Salmonella Braenderup strain H9812. Fragment sizes are indicated on the left side of the gel. Lanes 2, 3, 4, 24, 25, and 26 represent plasmids of known sizes in Salmonella isolates from our own collection. Lanes 5, 6, 7, 9, 10, and 11 represent plasmids profiles of S. Enteritidis isolates 113, 15, 102, 14, 73, and 62, respectively. Lanes 12, 13, 14, 16, and 17 represent plasmid profiles of S. Typhimurium isolates 4233, 2256, 4829, B48, and 44, respectively. Lanes 18, 19 represent plasmid profiles of S. Virchow 590 and 645, respectively. Lanes 20, 21 and 23 represent plasmid profiles of S. Colindale 4770, S. Oakland 114, and S. Poona 323, respectively.
In four out of five isolates of S. Typhimurium, a fragment of approximately 120 kb corresponding to the in silico detected IncFII(S)/IncFIB(S)/IncQ plasmid carrying the spv-region and multidrug resistance genes was detected on the gel. In the isolate 44, shown to be lacking the IncQ part in silico, a smaller fragment of approximately 85 kb was present. In addition, one fragment of approximately 100 kb was found to be present in the strain 2256 using S1-PFGE, yet it was not found in silico.
In comparison to in silico analyses of the genomes, fragments corresponding to TrfA/IncHI2/IncHI2were found to be smaller using S1-PFGE. The presence of a fragment of approximately 300 kb in S. Poona (n = 1) and 230 kb in S. Virchow (n = 1) strains were detected. Besides, a fragment of approximately 40 kb corresponding to the IncX plasmid carrying the gene blaTEM-52B was observed in the S. Virchow isolate 590. Overall the findings were in agreement with the in silico analysis of these strains, and no more additional fragments were found with S1-PFGE. Plasmid fragments were also absent in the S. Oakland and S. Colindale strains.
Discussion
Salmonella enterica Strains Analyzed From Ghana Belong to Common Multidrug—Associated MLST Types
The majority of the S. Typhimurium strains in this study were assigned to ST313 (n = 4) with only one strain typed as ST19. S. Typhimurium ST313 has been reported to be the major cause of invasive disease in sub-Saharan Africa (Kingsley et al., 2009). Recently, it has been also detected in India and Scotland (Crump et al., 2015). The reason behind an increased pathogenicity of ST313 strains compared to other isolates of S. Typhimurium is unclear, yet it may be associated with structural changes in their genome resulting in better host specialization or invasiveness (Crump et al., 2015). In addition, many S. Typhimurium non-typhoidal invasive isolates, including ST313, exhibit a multidrug resistance phenotype, which is encoded in the virulence plasmid of this serovar. It was suggested that antimicrobial usage may be selecting for the maintenance of these virulence-resistance plasmids in such isolates (Herrero et al., 2008; Kingsley et al., 2009). The S. Typhimurium strain assigned to ST19 did not exhibit a multidrug resistance pattern, and did not carry antimicrobial resistance genes. The ST19 genotype is also a common sequence type observed in Africa, however it is related to classical gastroenteritis, with no reported invasive infections (Kingsley et al., 2009; Boyle et al., 2011; Okoro et al., 2012). The fact that our study has an over-representation of ST313 strains may be related to the selection of strains in the original study (Andoh et al., 2017), which included a disproportional number of isolates from hospitalized patients, and thus were likely to originate from patients with systemic salmonellosis.
All isolates of S. Enteritidis were assigned to ST11, which is the most common genotype associated to this serovar (Achtman et al., 2012), and which is present in both humans and food production animals globally (Ghaderi et al., 2015; Papadopoulos et al., 2016). Invasive variants of S. Enteritidis are increasingly reported in developing countries and have been associated with virulence plasmids carrying multidrug resistant genes (Rodríguez et al., 2012). The association of IncI or IncN plasmids with the presence of cephalosporin and fluoroquinolones resistance determinants have been also shown recently in several clones worldwide (Antunes et al., 2011; Bado et al., 2012; Wong et al., 2015).
A strain of S. Virchow was assigned to the sequence type ST16. This type was not listed in the original publication on MLST by Achtman et al. (2012), but was later described as the most common ST linked to this serovar in a large collection of ESBL producing S. Virchow strains from Korea (Kim et al., 2016). A strain of S. Colindale was assigned to ST584. According to the original publication on Salmonella MLST (Achtman et al., 2012), this ST is one out of the two associated to this serovar. Finally, the strain of S. Poona was assigned to the genotype ST308 which is one out of six STs related to this serovar in the original MLST database (Achtman et al., 2012). So far, there have been no reports on the association of the latter serovar with plasmid mediated multidrug or cephalosporin resistance.
Multiple Resistance Genes Present in Salmonella Isolates
The majority of the strains analyzed were resistant to more than one antimicrobial. Multidrug resistant Salmonella has been increasing worldwide over the years, and has been mostly associated with the acquisition of β-lactamases encoded on mobile genetic elements. These plasmid-mediated resistances have been detected in Europe, USA and some countries in Asia and Africa (Valenzuela de Silva et al., 2006; Sjölund-Karlsson et al., 2011; Barua et al., 2013; Obeng-Nkrumah et al., 2013).
The majority (n = 12/16) of the amino-penicillin and suspected ESBL positive strains in our study, harbored blaTEM genes only. The blaTEM−1B and blaTEM−52B genes were found in 11 and one blaTEM positive strains, respectively. blaTEM−1B hydrolyses narrow spectrum cephalosporins, cefamandole, cefoperazone and penicillin (Fonzé et al., 1995), and is the most common beta lactamase encoding-gene among Enterobacteriaceae (Briñas et al., 2002; Olesen et al., 2004).
The gene blaCXT−M−15 was detected in only one cephalosporin resistant S. Poona strain. Salmonella isolates harboring blaCTX−M genes have not been previously reported in humans or food animals in Ghana, but they have been reported in other regions of the world (Barua et al., 2013; Akinyemi et al., 2014). Strains positive for blaCTX−M−15 have been associated with bloodstream infections in children and patients from intensive care units in Tanzania (Blomberg et al., 2005; Ndugulile et al., 2005) Senegal (Weill et al., 2004) and in other countries (Karim et al., 2001; Mushtaq et al., 2003; Pagani et al., 2003; Tawfik et al., 2011).
Plasmids carrying blaCTX−M genes often harbor genes encoding resistance to other antimicrobials (aminoglycosides, tetracycline, sulphonamides, trimethoprim and quinolones) (Diop et al., 2016) and therefore may confer co-resistance. This is also the case for the S. Poona strain under study, which was shown to carry blaCTX−M−15 together with genes encoding resistance to trimethoprim, sulphonamide, ampicillin and tetracycline on the same plasmid.
Interestingly, the plasmid-associated antimicrobial resistance gene profile varied between strains harboring the same plasmid type. In all cases, the plasmids regions carrying antimicrobial resistance genes were located within other mobile genetic elements such as transposons and integrons. This suggests that different recombination processes linked to mobile genetic elements might have occurred leading to acquisition and/or loss of antimicrobial resistance determinants in the plasmids as previously shown (Herrero-Fresno et al., 2014; García et al., 2016).
Distinct Plasmids Carry Antimicrobial Resistant Genes in Different Salmonella Serovars
Virulence-associated FII(S)/IncFIB(S)/IncQ plasmids carrying multidrug resistance were identified in four isolates of S. Typhimurium belonging to ST313. This kind of virulence-resistance plasmids were identified in previous studies (Guerra et al., 2002; Villa et al., 2010). The ST313-associated plasmid, pSLT-BT (EMBL: FN432031) (Kingsley et al., 2009), was also found in the ST313 isolates analyzed in this study where the MDR genes were located on a composite Tn21-like transposon, as was the case in previous investigation of ST313 strains (Kingsley et al., 2009). Interestingly, one S. Typhimurium isolate assigned to ST19 harbored a pSLT-BT-like plasmid as well, however the strain was sensitive to all antibiotics tested, and the region containing the IncQ replicon together with all the resistance genes was missing. This suggests that this region may be specific to ST313, and it would be of relevance to investigate whether this specific pSLT-BT region contributes to its invasive phenotype. Only one ST19 isolate was analyzed in our study, and the lack of such region should be confirmed in plasmids from a larger collection of non-invasive S. Typhimurium isolates.
In the S. Enteritidis strains under study, multiple resistance genes were located on a IncN plasmid. All the six S. Enteritidis isolates analyzed were assigned to ST11, but one of them did not carry the multidrug resistant IncN plasmid. Instead, only the blaTEM gene was located on the FII(S)/IncFIB(S) S. Enterititids virulence plasmid (Allard et al., 2013). S. Enteritidis ST11 is highly prevalent worldwide and is associated with genetically diverse isolates. Based on this study and on previous reports, it appears that multidrug resistance in ST11 strains can be located on both virulence-associated plasmids and on non-virulence plasmids of other replicon types, and this may be geographical location or ST11 sub-lineage dependent (Antunes et al., 2011; Bado et al., 2012; Rodríguez et al., 2012; Wong et al., 2015).
In our study, the S. Poona isolate carried multiple resistance genes (including blaCTX−M−15) on a TrfA/IncHI2/IncHI2A plasmid that has not been previously described in this serovar. IncHI2 plasmids carrying the blaCTX−M−15 gene have been reported in S. Virchow strains from South Korea (Kim et al., 2016). IncHI2 plasmids have also previously been associated with carriage of blaCTX−M genes in S. Typhimurium ST313 strains in Kenya (Kariuki et al., 2015), and in S. Virchow in the UK (Hopkins et al., 2006). A TrfA/IncHI2/IncHI2A plasmid similar to the one observed in S. Poona, however with no antimicrobial resistance genes, was also detected in the other phenotypically cephalosporin and ampicillin resistant S. Virchow isolate. In this S. Virchow isolate, the only antimicrobial resistance gene, blaTEM−52B, was found on an IncX1 plasmid. The plasmid of IncX type was previously reported to be highly associated with the carriage of blaTEM−52B gene sometimes together with resistance genes to other antimicrobials in a diverse Salmonella serovars from animals, meat products and humans in Europe, Canada and Korea (Bielak et al., 2011).
A combination of bioinformatics tools and S1-PFGE used in the present study enabled nearly full mapping of antibiotic resistance plasmids in different Salmonella enterica serovars. In most cases in silico prediction was concordant with the findings by S1-PFGE, however, there were some exceptions in particular related to possible plasmid fusion events. Thus, in two cases, in silico prediction and S1-PFGE analysis showed that either the IncFII(S)/IncFIB(S) or the IncN plasmid may have fused with other plasmids in two S. Enteritidis strains (73 and 14). In strain 73, the IncN and the IncFII/IncFIB were found to be fused in silico, and in agreement with this, only one fragment corresponding to the size of both plasmids was found in the S1-PFGE analysis. It is generally expected that after the fusion the loss of transfer genes occurs, however, blastN did not show differences in conjugal regions of these plasmids in comparison to these regions in the reference plasmids. In strain 14, a fragment of approximately 55 kb was present together with the fragment of 100 kb. According to in silico results, no indication of IncFII/IncFIB and IncN fusion was detected. Due to this, it remains unclear what kind of insertion/fusion occurred in the plasmids within this strain. However, the loss of two conjugative genes (TraM and X) were detected in the IncF plasmid of this strain in comparison to reference plasmid, which indicated the possible fusion event (García et al., 2007). Presently there is no method which could confirm these results from WGS (Arredondo-Alonso et al., 2017; Orlek et al., 2017), therefore only the sequencing of purified plasmids would enable to confirm plasmid fusion in these strains.
Finally, it is important to note that the approach that has been used in this study has a number limitations. Firstly, the library construction for sequencing of the genomes in the study was performed using Nextera XT kit, which includes a PCR-enrichment step, which biases the coverage of the reads belonging to different parts of the genome, and thus interferes with the ability of plasmidSPAdes to detect plasmids in some isolates. To overcome this limitation, blastN against reference and MAUVE were used for plasmid detection in such isolates. However, the method could be improved by using PCR-free sequencing protocols.
In summary, in the present study we have characterized plasmids from multidrug resistant Salmonella strains from Ghana. In doing so, we have demonstrated the successful implementation of a systematic approach to characterize plasmids directly from whole genome sequences and to predict whether antimicrobial resistance genes are located on such plasmids. In addition, previously unreported resistant or virulence-resistant plasmids in different Salmonella serovars were identified. This approach may be very valuable in future studies on how plasmids contribute to the development of novel R-plasmids and on plasmid evolution. The results of the detailed mapping of the few strains elucidate that homologous plasmids may be found in different bacteria originating from different regions of the world, and also that specific plasmid types may be associated with the antibiotic resistance in different Salmonella strains and serovars.
Author Contributions
JO, AD, and KO-D contributed conception and design of the study; EK performed bioinformatic analysis and wrote the first draft of the manuscript; LA performed ESBL testing and wrote the sections of the manuscript; SA prepared samples for whole genome sequencing; AH-F wrote the sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This research was supported by Danida (Danish International Development Assistance) through the Antimicrobial Drug Monitoring and Evaluation of Resistance (ADMER) project (http://admerproject.org).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Gitte Petersen and Katrine Aagaard, for excellent technical assistance in the laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01010/full#supplementary-material
Footnotes
References
Aarestrup, F. M., Hendriksen, R. S., Lockett, J., Gay, K., Teates, K., McDermott, P. F., et al. (2007). International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg. Infect. Dis. 13, 726–731. doi: 10.3201/eid1305.061489
Achtman, M., Wain, J., Weill, F. X., Nair, S., Zhou, Z., Sangal, V., et al. (2012). S. enterica MLST study group. multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. doi: 10.1371/journal.ppat.1002776
Akinyemi, K. O., Iwalokun, B. A., Agboyinu, J. A., Ogunyemi, O., and Fasure, A. K. (2014). Emergence of third generation cephalosporin resistance and typing by Randomly amplified polymorphic DNA (RAPD) among clinical Salmonella isolates from Lagos, Nigeria. Br. Microbiol. Res. J. 4, 668–677. doi: 10.9734/BMRJ/2014/8507
Allard, M. W., Luo, Y., Strain, E., Pettengill, J., Timme, R., Wang, C., et al. (2013). On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX01.0004. PLoS ONE 8:e55254. doi: 10.1371/journal.pone.0055254
Andoh, L. A., Ahmed, S., Olsen, J. E., Obiri-Danso, K., Newman, M. J., Opintan, J. A., et al. (2017). Prevalence and characterization of Salmonella among humans in Ghana. Trop. Med. Health. 45:3. doi: 10.1186/s41182-017-0043-z
Anjum, M. F., Choudhary, S., Morrison, V., Snow, L. C., Mafura, M., Slickers, P., et al. (2011). Identifying antimicrobial resistance genes of human clinical relevance within Salmonella isolated from food animals in Great Britain. J. Antimicrob. Chemother. 66, 550–559. doi: 10.1093/jac/dkq498
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Antunes, P., Mourão, J., Machado, J., and Peixe, L. (2011). First description of qnrS1-IncN plasmid in a ST11 Salmonella Enteritidis clinical isolate from Portugal. Diagn. Microbiol. Infect. Dis. 69, 463–465. doi: 10.1016/j.diagmicrobio.2010.11.004
Arredondo-Alonso, S., Willems, R. J., van Schaik, W., and Schürch, A. C. (2017). On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb. Genom. 3:e000128. doi: 10.1099/mgen.0.000128
Bado, I., García-Fulgueiras, V., Cordeiro, N. F., Betancor, L., Caiata, L., Seija, V., et al. (2012). First human isolate of Salmonella enterica serotype Enteritidis harboring blaCTX-M-14 in South America. Antimicrob. Agents Chemother. 56, 2132–2134. doi: 10.1128/AAC.05530-11
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Barua, H., Biswas, P. K., Olsen, K. E., Shil, S. K., and Christensen, J. P. (2013). Molecular characterization of motile serovars of Salmonella enterica from breeder and commercial broiler poultry farms in Bangladesh. PLoS ONE 8:e57811. doi: 10.1371/journal.pone.0057811
Bielak, E., Bergenholtz, R. D., Jørgensen, M. S., Sørensen, S. J., Hansen, L. H., and Hasman, H. (2011). Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 66, 2465–2474. doi: 10.1093/jac/dkr331
Blomberg, B., Jureen, R., Manji, K. P., Tamim, B. S., Mwakagile, D. S., Urassa, W. K., et al. (2005). High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J. Clin. Microbiol. 43, 745–749. doi: 10.1128/JCM.43.2.745-749.2005
Boyle, F., Healy, G., Hale, J., Kariuki, S., Cormican, M., and Morris, D. (2011). Characterization of a novel extended-spectrum β-lactamase phenotype from OXA-1 expression in Salmonella Typhimurium strains from Africa and Ireland. Diagn. Microbiol. Infect. Dis.. 70, 549–553. doi: 10.1016/j.diagmicrobio.2011.04.007
Briñas, L., Zarazaga, M., Sáenz, Y., Ruiz-Larrea, F., and Torres, C. (2002). Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46, 3156–3163. doi: 10.1128/AAC.46.10.3156-3163.2002
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. doi: 10.1371/journal.pone.0011147
Diop, A., Sambe-Ba, B., Seck, A., Dia, M. L., Timbiné, L. G., Niang, A. A., et al. (2016). First description of the extended spectrum-beta-lactamase gene blaCTX-M-109 in Salmonella grumpensis strains isolated from neonatal nosocomial infections in dakar, senegal. PLoS ONE 11:e0157683. doi: 10.1371/journal.pone.0157683
Eikmeyer, F., Hadiati, A., Szczepanowski, R., Wibberg, D., Schneiker-Bekel, S., Rogers, L. M., et al. (2012). The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 68, 13–24. doi: 10.1016/j.plasmid.2012.01.011
Fonzé, E., Charlier, P., To'th, Y., Vermeire, M., Raquet, X., Dubus, A., et al. (1995). TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr. D Biol. Crystallogr. 51(Pt 5), 682–694. doi: 10.1107/S0907444994014496
García, A., Navarro, F., Miró, E., Villa, L., Mirelis, B., Coll, P., et al. (2007). Acquisition and diffusion of bla CTX-M-9 gene by R478-IncHI2 derivative plasmids. FEMS Microbiol. Lett. 271, 71–77. doi: 10.1111/j.1574-6968.2007.00695.x
García, V., García, P., Rodríguez, I., Rodicio, R., and Rodicio, M. R. (2016). The role of IS26 in evolution of a derivative of the virulence plasmid of Salmonella enterica serovar Enteritidis which confers multiple drug resistance. Infect. Genet. Evol. 45, 246–249. doi: 10.1016/j.meegid.2016.09.008
Ghaderi, R., Tadayon, K., Khaki, P., and Mosavari, N. (2015). Iranian clonal population of Salmonella enterica serovar Enteritidis, characterized by multi-locus sequence typing (MLST) method. Iran. J. Microbiol. 7, 251–259.
Guerra, B., Soto, S., Helmuth, R., and Mendoza, M. C. (2002). Characterization of a self-transferable plasmid from Salmonella enterica serotype typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob. Agents Chemother. 46, 2977–2981. doi: 10.1128/AAC.46.9.2977-2981.2002
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hendriksen, R. S., Vieira, A. R., Karlsmose, S., Lo Fo Wong, D. M., Jensen, A. B., Wegener, H. C., et al. (2011). Global monitoring of Salmonella serovar distribution from the World health organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 8, 887–900. doi: 10.1089/fpd.2010.0787
Herrero, A., Mendoza, M. C., Rodicio, R., and Rodicio, M. R. (2008). Characterization of pUO-StVR2, a virulence-resistance plasmid evolved from the pSLT virulence plasmid of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52, 4514–4517. doi: 10.1128/AAC.00563-08
Herrero-Fresno, A., Wallrodt, I., Leekitcharoenphon, P., Olsen, J. E., Aarestrup, F. M., and Hendriksen, R. S. (2014). The role of the st313-td gene in virulence of Salmonella Typhimurium ST313. PLoS ONE 9:e84566. doi: 10.1371/journal.pone.0084566
Hoffmann, M., Pettengill, J. B., Gonzalez-Escalona, N., Miller, J., Ayers, S. L., Zhao, S., et al. (2017). Comparative sequence analysis of multidrug-resistant IncA/C plasmids from Salmonella enterica. Front. Microbiol. 7:1459. doi: 10.3389/fmicb.2017.01459
Hopkins, K. L., Liebana, E., Villa, L., Batchelor, M., Threlfall, E. J., and Carattoli, A. (2006). Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50, 3203–3206. doi: 10.1128/AAC.00149-06
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Karim, A., Poirel, L., Nagarajan, S., and Nordmann, P. (2001). Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201, 237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x
Kariuki, S., Okoro, C., Kiiru, J., Njoroge, S., Omuse, G., Langridge, G., et al. (2015). Ceftriaxone-resistant Salmonella enterica serotype typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob. Agents Chemother. 59, 3133–3139. doi: 10.1128/AAC.00078-15
Kim, J. S., Yun, Y. S., Kim, S. J., Jeon, S. E., Lee, D. Y., Chung, G. T., et al. (2016). Rapid emergence and clonal dissemination of CTX-M-15-producing Salmonella enterica serotype Virchow, South Korea. Emerging Infect. Dis. 22, 68–70. doi: 10.3201/eid2201.151220
Kingsley, R. A., Msefula, C. L., Thomson, N. R., Kariuki, S., Holt, K. E., Gordon, M. A., et al. (2009). Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279–2287. doi: 10.1101/gr.091017.109
Le Hello, S., Hendriksen, R. S., Doublet, B., Fisher, I., Nielsen, E. M., Whichard, J. M., et al. (2011). International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204, 675–684. doi: 10.1093/infdis/jir409
Madoshi, B. P., Kudirkiene, E., Mtambo, M. M., Muhairwa, A. P., Lupindu, A. M., and Olsen, J. E. (2016). Characterisation of commensal Escherichia coli isolated from apparently healthy cattle and their attendants in Tanzania. PLoS ONE 15:e0168160. doi: 10.1371/journal.pone.0168160
Mills-Robertson, F., Crupper, S. S., Addy, M. E., and Mensah, P. (2003). Antibiotic resistance and genotyping of clinical group B Salmonella isolated in Accra, Ghana. J. Appl. Microbiol. 94, 289–294. doi: 10.1046/j.1365-2672.2003.01833.x
Mushtaq, S., Woodford, N., Potz, N., and Livermore, D. M. (2003). Detection of CTX-M-15 extended-spectrum beta-lactamase in the United Kingdom. J. Antimicrob. Chemother. 52, 528–529. doi: 10.1093/jac/dkg353
Ndugulile, F., Jureen, R., Harthug, S., Urassa, W., and Langeland, N. (2005). Extended spectrum beta-lactamases among Gram-negative bacteria of nosocomial origin from an intensive care unit of a tertiary health facility in Tanzania. BMC Infect. Dis. 5:86. doi: 10.1186/1471-2334-5-86
Obeng-Nkrumah, N., Twum-Danso, K., Krogfelt, K. A., and Newman, M. J. (2013). High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am. J. Trop. Med. Hyg. 89, 960–964. doi: 10.4269/ajtmh.12-0642
Okoro, C. K., Kingsley, R. A., Quail, M. A., Kankwatira, A. M., Feasey, N. A., Parkhill, J., et al. (2012). High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin. Infect. Dis. 54, 955–963. doi: 10.1093/cid/cir1032
Olesen, I., Hasman, H., and Aarestrup, F. M. (2004). Prevalence of beta-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb. Drug Resist. 10, 334–340. doi: 10.1089/mdr.2004.10.334
Orlek, A., Stoesser, N., Anjum, M. F., Doumith, M., Ellington, M. J., Peto, T., et al. (2017). Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8:182. doi: 10.3389/fmicb.2017.00182
Pagani, L., Dell'Amico, E., Migliavacca, R., D'Andrea, M. M., Giacobone, E., Amicosante, G., et al. (2003). Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 41, 4264–4269. doi: 10.1128/JCM.41.9.4264-4269.2003
Papadopoulos, T., Petridou, E., Zdragas, A., Mandilara, G., Nair, S., Peters, T., et al. (2016). Comparative study of all Salmonella enterica serovar Enteritidis strains isolated from food and food animals in Greece from 2008 to 2010 with clinical isolates. Eur. J. Clin. Microbiol. Infect. Dis. 35, 741–746. doi: 10.1007/s10096-016-2591-2
Parry, C. M., and Threlfall, E. J. (2008). Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21, 531–538. doi: 10.1097/QCO.0b013e32830f453a
Rodríguez, I., Rodicio, M. R., Guerra, B., and Hopkins, K. L. (2012). Potential international spread of multidrug-resistant invasive Salmonella enterica serovar enteritidis. Emerg. Infect. Dis. 18, 1173–1176. doi: 10.3201/eid1807.120063
Sjölund-Karlsson, M., Howie, R., Krueger, A., Rickert, R., Pecic, G., Lupoli, K., et al. (2011). CTX-M-producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg. Infect. Dis. 17, 97–99. doi: 10.3201/eid1701.100511
Su, L. H., Chiu, C. H., Chu, C., and Ou, J. T. (2004). Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin. Infect. Dis. 39, 546–551. doi: 10.1086/422726
Tawfik, A. F., Alswailem, A. M., Shibl, A. M., and Al-Agamy, M. H. (2011). Prevalence and genetic characteristics of TEM, SHV, and CTX-M in clinical Klebsiella pneumoniae isolates from Saudi Arabia. Microb. Drug Resist. 17, 383–388. doi: 10.1089/mdr.2011.0011
Valenzuela de Silva, E. M., Mantilla Anaya, J. R., Reguero Reza, M. T., González Mejía, E. B., Pulido Manrique, I. Y., Darío Llerena, I., et al. (2006). Detection of CTX-M-1, CTX-M-15, and CTX-M-2 in clinical isolates of Enterobacteriaceae in Bogota, Colombia. J. Clin. Microbiol. 44, 1919–1920. doi: 10.1128/JCM.44.5.1919-1920.2006
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Weill, F. X., Perrier-Gros-Claude, J. D., Demartin, M., Coignard, S., and Grimont, P. A. (2004). Characterization of extended-spectrum-beta-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 38, 353–358. doi: 10.1016/j.femsle.2004.07.058
Wong, M. H., Kan, B., Chan, E. W., Yan, M., and Chen, S. (2015). IncI1 plasmids carrying various blaCTX-M genes contribute to ceftriaxone resistance in Salmonella enterica rerovar Enteritidis in China. Antimicrob. Agents Chemother. 7, 982–989. doi: 10.1128/AAC.02746-15
World Health Organization (2014). Antimicrobial Resistance: Global Report on Surveillance. WHO Library Cataloguing-in-Publication Data. Available online at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1
Yan, J. J., Chiou, C. S., Lauderdale, T. L., Tsai, S. H., and Wu, J. J. (2005). Cephalosporin and ciprofloxacin resistance in Salmonella, Taiwan. Emerging Infect. Dis. 11, 947–950. doi: 10.3201/eid1106.041153
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft salmonella genome assemblies. PLoS ONE 11:e0147101. doi: 10.1371/journal.pone.0147101
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: whole genome sequencing, Salmonella, multidrug resistance, plasmids, Ghana
Citation: Kudirkiene E, Andoh LA, Ahmed S, Herrero-Fresno A, Dalsgaard A, Obiri-Danso K and Olsen JE (2018) The Use of a Combined Bioinformatics Approach to Locate Antibiotic Resistance Genes on Plasmids From Whole Genome Sequences of Salmonella enterica Serovars From Humans in Ghana. Front. Microbiol. 9:1010. doi: 10.3389/fmicb.2018.01010
Received: 15 January 2018; Accepted: 30 April 2018;
Published: 17 May 2018.
Edited by:
Teresa M. Coque, Instituto Ramón y Cajal de Investigación Sanitaria, SpainReviewed by:
María de Toro, Centro de Investigación Biomédica de La Rioja, SpainJean-Yves Madec, Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES), France
Copyright © 2018 Kudirkiene, Andoh, Ahmed, Herrero-Fresno, Dalsgaard, Obiri-Danso and Olsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John E. Olsen, jeo@sund.ku.dk
 Egle Kudirkiene
Egle Kudirkiene Linda A. Andoh
Linda A. Andoh Shahana Ahmed
Shahana Ahmed Ana Herrero-Fresno
Ana Herrero-Fresno Anders Dalsgaard1
Anders Dalsgaard1 John E. Olsen
John E. Olsen