Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing
- 1Division of Gastroenterology and Hepatology, Department of Internal Medicine, Keio University School of Medicine, Tokyo, Japan
- 2Division of Research and Development for Minimally Invasive Treatment, Cancer Center, Keio University School of Medicine, Tokyo, Japan
Antibiotic resistance in Helicobacter pylori (H.pylori) is the main factor affecting the efficacy of current treatment methods against infection caused by this organism. The traditional culture methods for testing bacterial susceptibility to antibiotics are expensive and require 10–14 days. Since resistance to clarithromycin, fluoroquinolone, and tetracycline seems to be exclusively caused by specific mutations in a small region of the responsible gene, molecular methods offer an attractive alternative to the above-mentioned techniques. The technique of polymerase chain reaction (PCR) is an accurate and rapid method for the detection of mutations that confer antibiotic resistance. This review highlights the mechanisms of antibiotic resistance in H. pylori and the molecular methods for antibiotic susceptibility testing.
Introduction
H. pylori is identified as a Group 1 carcinogen by the World Health Organization International Agency for Research on Cancer (WHO/IARC), and is associated with the development of gastric cancer. Eradication of H. pylori infection has been reported as an effective strategy in the treatment of peptic ulcers and gastric mucosa-associated lymphoid tissue lymphoma as well as in the prevention of gastric cancer (Fukase et al., 2008).
Triple treatment including proton pump inhibitor-amoxicillin and clarithromycin or metronidazole proposed at the first Maastricht conference is globally accepted as the technique used to treat H. pylori infection. However, recent data show that efficacy of this combination has decreased, with successful cure in only 70% patients (Nishizawa et al., 2014). The 4th edition of the Maastricht consensus recommended a threshold of 15–20% to separate regions of high and low clarithromycin resistance (Malfertheiner et al., 2012). In areas of low clarithromycin resistance, treatments containing clarithromycin are recommended as a first-line empirical treatment. After failure of clarithromycin-containing therapy, either bismuth-containing quadruple therapy or levofloxacin-containing triple therapy is recommended. In areas of high clarithromycin resistance, bismuth-containing quadruple treatments (bismuth subsalicylate, PPI, tetracycline, and metronidazole) are recommended as a first-line empirical treatment. After failure of the quadruple therapy, levofloxacin-containing triple therapy is recommended. After failure of the second line of treatment, subsequent treatment methods should be guided by antimicrobial susceptibility testing whenever possible. The alternative candidates for third-line therapy are rifabutin, fluoroquinolones, tetracycline, furazolidone, and high-dose PPI/amoxicillin therapy (Nishizawa et al., 2008, 2012; Zhang et al., 2014). The traditional culture test for bacterial susceptibility to antibiotics is requires 10–14 days, and is not routinely performed in clinical practice. Minimal inhibitor concentration (MIC)-based individualized treatment for H. pylori infection is not prevalent among general practitioners. Molecular techniques for antibiotic susceptibility testing can determine bacterial susceptibility to some antibiotics within a few days. We reviewed the mechanisms of antibiotic resistance in H. pylori and the molecular techniques for antibiotic susceptibility testing.
Clarithromycin
Clarithromycin resistance possibly results from the use of the antibiotic in the pediatric, respiratory, and otorhinolaryngology fields (Kaneko et al., 2004). Global clarithromycin-resistance rates have increased from 9% in 1998 to 17.6% in 2008 in Europe and from 7% in 2000 to 27.7% in 2006 in Japan (Asaka et al., 2010). In patients with clarithromycin-resistant H. pylori, it has been reported that the eradication rate achieved with clarithromycin-based regimens shows a marked decrease (Nishizawa et al., 2014). Therefore, the choice of clarithromycin after drug susceptibility testing is promising approach.
The bacteriostatic activity of macrolides such as erythromycin and clarithromycin depends on the capacity to inhibit protein synthesis by binding to the 23S ribosomal subunit (23S rRNA). Extensive studies have demonstrated that point mutations in the peptidyltransferase region encoded in domain V of 23S rRNA are responsible for macrolide resistance. These mutations result in the inhibition of binding between clarithromycin and the ribosomal subunit dedicated to specific antibiotic-related protein synthesis (Versalovic et al., 1996). In particular, the main 23S rRNA mutations include an adenine-to-guanine transition at positions 2142 (11.7%) and 2143 (69.8%), and an adenine-to-cytosine transversion at position 2142 (2.6%).
Several other point mutations have been identified, such as A2115G, T2117C, G2141A, T2182C, G2224A, C2245T, T2289C, C2611A, and T2717C (Francesco et al., 2011). Besides their low frequency, the clinical relevance of A2115G, T2117C, G2141A, G2224A, T2289C, C2245T mutations is still not proven. The T2182C, C2611A, and T2717C have been associated with low resistance levels (Francesco et al., 2011).
Another relevant mechanism for macrolide resistance is attributed to the efflux pump system. At least 4 conserved families of efflux systems associated with bacterial resistance to antibiotics have been identified (Bina et al., 2000). One of these, widespread among gram-negative bacteria, is the resistance-nodulation-cell division (RND) family of efflux systems (Ma et al., 1995). Bina et al. identified the RND efflux system in H. pylori. HP0605 from H. pylori is a homolog of the E. coli gene (TolC) encoding the outer membrane protein, TolC, while HP0607 and HP0606 and are homologs of the E. coli genes acrA and acrB encoding the membrane fusion and RND cytoplasmic pump protein, respectively (Bina et al., 2000). Kutschke et al. reported that HP0607 knockout mutants exhibited increased susceptibility to penicillin G, cefotaxime, erythromycin, clarithromycin, tetracycline, clindamycin, novobiocin, and ethidium bromide (Kutschke and de Jonge, 2005). We previously investigated the efficacy of efflux pump inhibitor (Phe-Arg-beta-naphthylamide) in 15 clarithromycin resistant H. pylori. In all 15 strains, efflux pump mRNA was expressed, and the MIC of clarithromycin were decreased by using efflux pump inhibitor, despite possessing 23s rRNA point mutations. In addition, the MIC of clarithromycin was decreased by the efflux pump inhibitor in a concentration-dependent fashion (Hirata et al., 2010).
Most clarithromycin-resistant strains of H. pylori have a A2142G or A2143G mutation. Furuta et al. designed the forward primer FP2143G and the reverse primer RP2142G, which specifically anneal to A2143G and A2142G mutated sequences of the H. pylori 23S rRNA gene (Furuta et al., 2007). H. pylori strains with A2142G, A2143G, and wild-type genotype can be differentiated by amplicon size using the allele-specific, primer-polymerase chain reaction (PCR) method, which is useful and requires only a single PCR run.
Wang et al. reported multiplex sequence analysis (Wang et al., 1999), wherein positions 2142 and 2143 have an AA sequence in wild-type cells, while mutant cells show a change to GA (A2142G) and AG (A2143G), respectively. In the presence of dCTP, dATP, and ddGTP terminating DNA strand elongation results in products of unique length, depending on type of mutation.
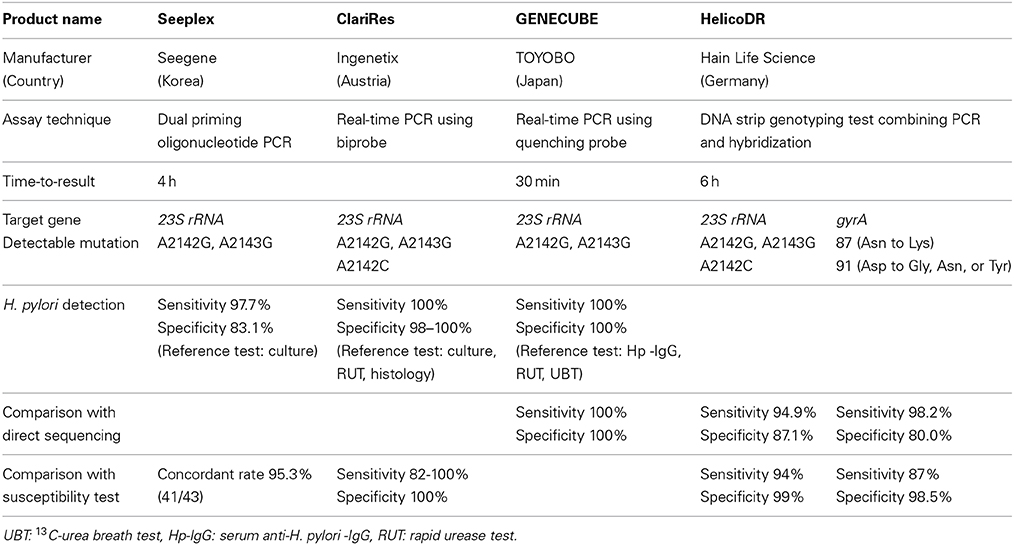
Dual priming oligonucleotide (DPO)-PCR is a multiplex PCR assay that increases specificity and sensitivity of detection compared to conventional PCR, blocking non-specific binding sites therefore eliminating imperfect primer annealing. Seeplex® ClaR-H. pylori ACE detection (Seegene, Inc., Seoul, Korea) is commercially available DPO-PCR kit to detect H. pylori and A2142G, A2143G mutations. Although Seeplex® ClaR-H. pylori ACE detection kit does not allow of detection of A2142C mutation, the mutation is less common (<5% of resistant isolates). Lehours et al. reported E-test and DPO-PCR were concordant with regard to clarithromycin susceptibility in 95.3% of the cases (41/43) (Lehours et al., 2011) (Table 1).
Versalovic et al. developed a method based on rapid restriction analysis of the amplicon obtained from H. pylori (Versalovic et al., 1996). Single point mutations at positions 2143, 2142, and 2717 generate specific restriction sites, namely BsaI, MboII, and HhaI, respectively, which can be used in rapid screening for clarithromycin resistance (Masaoka et al., 2004). Fontana et al. developed a new method involving semi-nested PCR and digestion by MboII, BsaI, and HhaI using stool samples. This method is non-invasive and easy to perform (Fontana et al., 2003).
Klesiewicz et al. evaluated the occurrence of A2143G and A2142G mutations in 21 H. pylori strains resistant to clarithromycin. The point mutations were detected by PCR followed by restriction fragment length polymorphism (RFLP) analysis. Nine H. pylori strains exhibited A2143G mutation and nine H. pylori strains exhibited A2142G mutation. The results of RFLP analysis of 3 clarithromycin-resistant strains were negative for both mutations (Klesiewicz et al., 2014).
Wu et al. evaluated the utility of the string test to detect genotypic clarithromycin-resistant H. pylori by PCR-RFLP. In the string test, a 90-cm nylon string coiled inside a gelatin capsule was used. A free-end looped string protrudes through a hole in the other end of the capsule. Before the capsule was swallowed, 10–20 cm of the free-end string was pulled out and its position was ensured by adhesion of a small piece of tape to the patient's cheek. It was swallowed with 300 mL of water after 8 h of fasting. One hour after swallowing, the string was retrieved. Approximately 0.5 mL of gastric juice with H. pylori attached by every 10 cm of the string was reasonable for molecular biological analysis. Forty three isolates were successfully cultured in 79 patients in whom 23S rRNA was successfully amplified. Of 5 patients with clarithromycin-resistant H. pylori, 23S rRNA of H. pylori isolates from 4 patients could be digested by BsaI. In 38 susceptible isolates, 23S rRNA of H. pylori isolates from 36 patients could not be digested by either BsaI or BbsI. The sensitivity and specificity of the string test to detect genotypic clarithromycin resistance were 66.7 and 97.3%, respectively (Wu et al., 2014).
The restriction enzyme is capable of identifying an A-to-G mutation by the creation of a restriction site, but if an A-to-C mutation occurs, the restriction enzyme may not restrict the DNA at that site. Stone et al. developed a rapid assay based on PCR followed by oligonucleotide ligation for rapid detection of these point mutations, which could differentiate between H. pylori strains with A2142G, A2143G, A2142C, and wild–type genotype (Stone et al., 1997).
Several quantitative PCR assays for the determination of clarithromycin susceptibility in H. pylori have subsequently been reported. These include a two-step process involving LightCycler PCR for the detection of H. pylori followed by melting curve analysis using probe hybridization to detect resistance (Oleastro et al., 2003).
GENECUBE® (TOYOBO Co., LTD. Japan) is a novel, fully automated rapid genetic analyzer capable of extracting nucleic acids from biological material, preparing reaction mixtures, and amplifying the target gene all within 30 min. The amplified target DNA is hybridized with a fluoresce-labeled oligonucleotide (a Qprobe). Upon binding to the target DNA, the Qprobe fluorescence is quenched by the guanine bases in the target. However, the fluorescence reappears as the Qprobe disassociated from the melting target. By detecting this change in fluorescence intensity, A2143G and A2142G mutations are detected. Furuta et al. reported the GENECUBE® genotyping results of the 23S rRNA gene from gastric tissue samples (n = 50) were in complete agreement with those for direct sequencing. Furthermore, gastric juice samples were collected during gastroduodenoscopy in 132 patients. Twenty six of the 132 samples were H. pylori-negative based on analysis of serum anti-H. pylori-IgG, urease and the 13C-urea breath test, and the remaining 106 were H. pylori-positive. The GENECUBE® could detect H. pylori infection in all patients infected with H. pylori based on analysis of serum anti-H. pylori-IgG, urease and the 13C-urea breath test. Thus, the sensitivity, specificity and validity of the GENECUBE® assay were all 100% (Furuta et al., 2013). GENECUBE® is commercially available, it is not approved for clinical diagnostic use (research use only).
ClariRes® assay (Ingenetix, Vienna, Austria) is a novel commercially available quantitative PCR assay allowing H. pylori detection and clarithromycin susceptibility testing in either gastric biopsy or stool specimens. In the biprobe quantitative PCR protocol, followed by hybridization melting point analysis, A2143G, A2142G, and A2142C mutations are detected.
Schabereiter et al. evaluated the clinical usefulness of ClariRes® test in 92 patients who underwent endoscopy. 45 were found to be H. pylori infected and invariably were also culture positive. With respect to the detection of H. pylori infection, ClariRes® test showed sensitivities of 100% and a specificity of 98%. Of the 45 isolates, 11 were shown to be resistant to clarithromycin by E-test. Compared to E-test, the sensitivity and specificity of ClariRes® test for clarithromycin resistance were 82% and 100% (Schabereiter-Gurtner et al., 2004). Scaletsky et al. evaluated the clinical usefulness of ClariRes® test in Brazilian children. Forty five of the 217 samples were H. pylori-positive based on analysis of culture, rapid urease test, or histological examination, and the remaining 172 were H. pylori-negative. The sensitivity, specificity and validity of the ClariRes® assay were all 100% for the detection of H. pylori infection. In the 45 culture positive patients, the ClariRes® genotyping results of the 23S rRNA gene from gastric tissue samples were in complete agreement with those for the E-test (Scaletsky et al., 2011).
Can et al. developed a fluorescent in situ hybridization (FISH) method to detect H. pylori and determine clarithromycin resistance in formalin-fixed, paraffin-embedded, gastric biopsy specimens (Can et al., 2005). Cerqueira et al. evaluated a peptide nucleic acid-FISH method for H. pylori clarithromycin resistance detection in paraffin-embedded gastric biopsy specimens. In the retrospective study (n = 30 patients), full agreement between peptide nucleic acid-FISH and PCR-sequencing was observed. Compared to the culture followed by E-test, the specificity and sensitivity of peptide nucleic acid-FISH were 90.9 and 84.2%, respectively. In the prospective cohort (n = 93 patients), 21 cases were positive by culture. For the patients harboring clarithromycin-resistant H. pylori, the method showed sensitivity of 80.0% and specificity of 93.8% (Cerqueira et al., 2013).
Xuan et al. developed an enzymatic colorimetric DNA chip (Xuan et al., 2009) including A2142G, A2142C, A2143G, A2143C, and G2224A mutations, where results were 96.8% (61/63) consistent with those of DNA sequencing. Due to its simplicity and rapidness, the colorimetric DNA chip might be technically feasible for use in general clinical practice.
Fluoroquinolones
The primary resistance of H. pylori to fluoroquinolones has been reported to range between 2–22% in different countries or regions (Suzuki et al., 2010). Resistance to fluoroquinolones is easily acquired, and the resistance rate is relatively high in countries with a high consumption of these drugs (Nishizawa et al., 2006).
Fluoroquinolones exert their antimicrobial activity by inhibiting the function of the enzyme DNA gyrase (Moore et al., 1995). The bacterial gyrase is essential for maintaining the DNA helicoidal structure in addition to being involved in DNA replication, recombination and transcription. The gyrase is a tetramer consisting of two A and two B subunits encoded by the gyrA and gyrB genes, respectively. The A subunit of DNA gyrase is responsible for DNA cleavage and rejoining, is also the site of action of fluoroquinolones (Matsuzaki et al., 2010). Point mutations in the Quinolones Resistance-Determining Region (QRDR) of gyrA prevent binding between the antibiotic and the enzyme, conferring antibiotic bacterial resistance (Nishizawa et al., 2009). H. pylori does not possess a gene encoding topoisomerase IV, an important fluoroquinolone target in other bacteria. Point mutations in the QRDR of the gyrA gene are mainly at amino acid 87 (Asn to Lys or Tyr) or 91 (Asp to Gly, Asn, or Tyr). We previously reported that the MICs of sitafloxacin in gyrA mutation-positive strains differed, depending on the position of the gyrA mutation (Suzuki et al., 2009a; Matsuzaki et al., 2012). The MICs were higher in N87-mutated strains (0.21 ± 0.16 μg/ml) than in D91-mutated strains (0.12 ± 0.11 μg/ml, P = 0.03) (Matsuzaki et al., 2012). Rimbara et al. proposed that mutation at position 463 in gyrB would be a novel mechanism of fluoroquinolone resistance in H. pylori (Rimbara et al., 2012).
We previously designed the allele-specific primer, which specifically annealed to the C261A, C261G (87 Asn to Lys), G271A (91 Asp to Asn), G271T (91 Asp to Tyr), and A272G (91 Asp to Gly) mutated sequences of the gyrA gene in H. pylori (Nishizawa et al., 2007). In the allele-specific PCR method, PCR amplification was performed using allele-specific primers in which the second nucleotide from the 3′ end was designed to match the site of the point mutation.
Glocker et al. developed a reliable fluorescence resonance energy transfer-based quantitative PCR method to detect mutations in the gyrA gene (Glocker and Kist, 2004). This method was developed on DNA extracts from H. pylori isolates from German patients. Because of the known genetic heterogeneity of H. pylori (Suerbaum, 2000), the assay may fail with strains isolated outside Germany, but the test could be altered to adapt to the genetic gyrA variants found in different geographical regions.
Cambau et al. developed HelicoDR®, a DNA strip genotyping test combining PCR and hybridization (Cambau et al., 2009) that allows the molecular detection of mutations in the gyrA gene and 23S rRNA within 6 h. The sensitivity and specificity of detecting resistance using this method were 94 and 99% for clarithromycin and 87% and 98.5% for levofloxacin, respectively. HelicoDR® (Hain Life Science, Germany) is commercially available.
Lee et al. evaluated the clinical usefulness of HelicoDR® test in Korea. Both DNA sequencing after MIC test and HelicoDR® test were performed in H. pylori isolates from the gastric mucosa of 101 patients. Among 42 isolates with A2143G mutation by HelicoDR®, 83.3% (35/42) of concordance rate was estimated with direct sequencing method and 85.7% (36/42) for MIC test. Among 43 isolates with amino acid 87 (Asn to Lys) mutation by HelicoDR®, 71.1% (31/43) of concordance rate was estimated with direct sequencing and 88.4% (38/43) for MIC test. Compared to direct sequencing, the sensitivity and specificity of HelicoDR® test for 23S rRNA mutation were 94.9 and 87.1%, and those for gyrA 98.2 and 80.0%. Compared to MIC test, the sensitivity and specificity of HelicoDR® test for clarithromycin resistance were 55.0 and 80.0% and those for fluoroquinolone were 74.4 and 70.0% (Lee et al., 2004).
Metronidazole
The prevalence of resistance to metronidazole in H. pylori has been reported to range between 8 and 80% in different countries (Suzuki et al., 2010). Metronidazole resistance is much higher in developing countries (more than 60%) than in developed countries (Banatvala et al., 1994). Metronidazole is a prodrug that needs to be activated by reduction of the nitro group attached to the imidazole ring. This reduction step leads to the production of DNA-damaging nitroso- and hydroxylamine-containing compounds. The reduction of metronidazole is mainly mediated by oxygen-insensitive NADPH nitroreductase (RdxA), NADPH-flavin-oxidoreductase (FrxA), and ferredoxin-like enzymes (FrxB) in H. pylori (Francesco et al., 2011). Different mutations involving the rdxA gene have been identified in metronidazole-resistant strains (Masaoka et al., 2006). In the rdxA gene, complex genetic events (insertions and deletions of transposons, and missense and frameshift mutations) could be simultaneously present. These mutations are recognized as the main mechanism conferring metronidazole resistance in H. pylori. Point mutations in frxA and frxB can increase bacterial resistance exclusively in the presence of mutations in the rdxA gene. We previously demonstrated a novel mechanism of metronidazole resistance in H. pylori, namely, aberrant increase of superoxide dismutase expression resulting from the mutation of the ferric uptake regulator (Fur) (Tsugawa et al., 2011). Superoxide dismutase is essential for protection against superoxide attack. Superoxide dismutase is derepressed by mutant-type Fur, which is associated with the development of metronidazole resistance.
Due to various mutations of rdxA, molecular antibiotic susceptibility testing is not applicable for metronidazole.
Amoxicillin
The prevalence of resistance to amoxicillin has fortunately remained low, where most studies have reported it as less than 2% in all countries, except in Bangladesh (6.6%) (Nahar et al., 2004). Amoxicillin acts by interfering with peptidoglycan synthesis, especially by blocking transporters, namely penicillin binding proteins (PBP). Multiple mutations in pbp1 gene are the major reason for amoxicillin resistance. Although amoxicillin resistance in H. pylori is rare, we previously reported that the MIC90 of amoxicillin showed a 2-fold increase with the failure of each eradication treatment (Nishizawa et al., 2011a). Low-level resistant strains (MIC: 0.06–0.25 μg/ml) had 0–2 substitutions, while high-level resistant strains had 1–3 substitutions. Low-level resistance to amoxicillin is linked to a point mutation on pbp1, and the accumulation of PBP1 mutations could result in a gradual increase in amoxicillin resistance. Although production of β-lactamase is rare and almost inactive in H. pylori, Tseng et al. reported that high-level amoxicillin resistance is associated with β-lactamase production in H. pylori (Tseng et al., 2009). Due to different mutations of PBP1, molecular antibiotic susceptibility testing is not applicable for amoxicillin.
Tetracycline
The prevalence of resistance to tetracycline also has fortunately remained low; it was reported to be less than 2% in most studies (Suzuki et al., 2010). Tetracyclines are bacteriostatic drugs that exert their antimicrobial effect on the 30S subunit of the ribosome and block the binding of aminoacyl-tRNA, resulting in impaired protein biosynthesis. The resistance of H. pylori to tetracyclines is reported to be caused by mutations in the 16S rRNA (Gerrits et al., 2002). Simultaneous triple point-mutations at positions 965–967 are recognized to be major responsible for tetracycline resistance. Levels of resistance are proportional to the number of changes in the AGA 965–967. Single and double point-mutations are associated with low and intermediate MIC values, respectively. High resistance levels are observed in the substitution of an AGA with a TTC triplet.
Ribeiro et al. developed a PCR-RFLP assay allowing rapid and reproducible identification of mutations mediating high-level tetracycline resistance in H. pylori (Ribeiro et al., 2004). The substitution of an AGA with a TTC triplet creates an additional HinfI restriction site. This PCR-RFLP assay distinguishes high-level tetracycline resistant isolates from low-level tetracycline resistant and tetracycline susceptible H. pylori strains.
Glocker et al. developed real-time PCR to detect 16S rRNA gene mutations that was capable of differentiating between wild-type strains and resistant strains exhibiting single-, double-, or triple-base-pair mutations (Glocker et al., 2005). Future studies need to address the question of whether additional mutations play a role in the resistance of H. pylori to tetracycline.
Rifabutin
The mean H. pylori rifabutin-resistance rate (calculated from 11 studies including 2982 patients) was 1.3% (Gisbert and Calvet, 2012). When only studies including patients naïve to H. pylori eradication treatment were considered, this figure was even lower (0.6%). We previously investigated the resistance to rifabutin of H. pylori isolated from both general hospital and a hospital specialized for chronic respiratory disease, including pulmonary tuberculosis. Among 94 strains tested, 7 (7.4%) were isolated from patients with a past rifampicin treatment. All these 7 strains showed high rifabutin resistance (Suzuki et al., 2009b). Rifabutin is an antituberculous agent derived from rifamycin-S, which is structurally similar to rifampicin. Rifabutin inhibits the expression of beta-subunit of DNA-dependent RNA polymerase of H. pylori, which is encoded by the rpoB gene (Nishizawa et al., 2011b). Rifabutin-resistant isolates of H. pylori showed mutations in codon 149, codons 525 to 545, or codon 586 (Heep et al., 2002). Due to the different mutations of rpoB, the molecular antibiotic susceptibility testing is not applicable for rifabutin.
Conclusion
Conventional methods used to assess the level of antibiotic-resistance of H. pylori are culture-based methods used in combination with agar dilution or the E-test. However, because of the slow growth and particular requirements of H. pylori culture, this approach is not reliable for use in most routine clinical laboratory. Since resistance to clarithromycin, fluoroquinolone, and tetracycline seems to be a result of specific mutations in a small region of the responsible gene, molecular methods offer an attractive alternative. Some reliable molecular methods are commercially available. However, large-scale prospective studies should be performed to assess the full clinical potential of these molecular methods and its economic feasibility. Users should keep in mind that whenever possible H. pylori culture should be performed and only in cases where standard microbiology fails, the use of molecular methods are really indicated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (B) (25293178 to Hidekazu Suzuki) from the Japan Society for the Promotion of Science (JSPS), a research fund from Daiwa Securities Health Foundation, Princess Takamatsu Cancer Research grants (to Hidekazu Suzuki), a grant from the Smoking Research Foundation (to Hidekazu Suzuki), and the Keio Gijuku Academic Development Fund (to Hidekazu Suzuki).
References
Asaka, M., Kato, M., Takahashi, S., Fukuda, Y., Sugiyama, T., Ota, H., et al. (2010). Guidelines for the management of Helicobacter pylori infection in Japan: 2009 revised edition. Helicobacter 15, 1–20. doi: 10.1111/j.1523-5378.2009.00738.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banatvala, N., Davies, G. R., Abdi, Y., Clements, L., Rampton, D. S., Hardie, J. M., et al. (1994). High prevalence of Helicobacter pylori metronidazole resistance in migrants to east London: relation with previous nitroimidazole exposure and gastroduodenal disease. Gut 35, 1562–1566. doi: 10.1136/gut.35.11.1562
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bina, J. E., Alm, R. A., Uria-Nickelsen, M., Thomas, S. R., Trust, T. J., and Hancock, R. E. (2000). Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob. Agents Chemother. 44, 248–254. doi: 10.1128/AAC.44.2.248-254.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cambau, E., Allerheiligen, V., Coulon, C., Corbel, C., Lascols, C., Deforges, L., et al. (2009). Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J. Clin. Microbiol. 47, 3600–3607. doi: 10.1128/JCM.00744-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Can, F., Yilmaz, Z., Demirbilek, M., Bilezikci, B., Kunefeci, G., Atac, F. B., et al. (2005). Diagnosis of Helicobacter pylori infection and determination of clarithromycin resistance by fluorescence in situ hybridization from formalin-fixed, paraffin-embedded gastric biopsy specimens. Can. J. Microbiol. 51, 569–573. doi: 10.1139/w05-035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cerqueira, L., Fernandes, R. M., Ferreira, R. M., Oleastro, M., Carneiro, F., Brandao, C., et al. (2013). Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J. Clin. Microbiol. 51, 1887–1893. doi: 10.1128/JCM.00302-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fontana, C., Favaro, M., Pietroiusti, A., Pistoia, E. S., Galante, A., and Favalli, C. (2003). Detection of clarithromycin-resistant Helicobacter pylori in stool samples. J. Clin. Microbiol. 41, 3636–3640. doi: 10.1128/JCM.41.8.3636-3640.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Francesco, V. D., Zullo, A., Hassan, C., Giorgio, F., Rosania, R., and Ierardi, E. (2011). Mechanisms of Helicobacter pylori antibiotic resistance: an updated appraisal. World J. Gastrointest. Pathophysiol. 2, 35–41. doi: 10.4291/wjgp.v2.i3.35
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukase, K., Kato, M., Kikuchi, S., Inoue, K., Uemura, N., Okamoto, S., et al. (2008). Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet 372, 392–397. doi: 10.1016/S0140-6736(08)61159-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Furuta, T., Soya, Y., Sugimoto, M., Nishino, M., Yamade, M., Uotani, T., et al. (2013). Rapid Automated Genotyping of CYP2C19 and Helicobacter pylori 23S rRNA Gene in Gastric Juice. J. Gastorenterol. Hepatol. Res. 2, 508–512. doi: 10.6051/j.issn.2224-3992.2013.02.221
Furuta, T., Soya, Y., Sugimoto, M., Shirai, N., Nakamura, A., Kodaira, C., et al. (2007). Modified allele-specific primer-polymerase chain reaction method for analysis of susceptibility of Helicobacter pylori strains to clarithromycin. J. Gastroenterol. Hepatol. 22, 1810–1815. doi: 10.1111/j.1440-1746.2007.04919.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gerrits, M. M., de Zoete, M. R., Arents, N. L., Kuipers, E. J., and Kusters, J. G. (2002). 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 46, 2996–3000. doi: 10.1128/AAC.46.9.2996-3000.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gisbert, J. P., and Calvet, X. (2012). Review article: rifabutin in the treatment of refractory Helicobacter pylori infection. Aliment. Pharmacol. Ther. 35, 209–221. doi: 10.1111/j.1365-2036.2011.04937.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glocker, E., Berning, M., Gerrits, M. M., Kusters, J. G., and Kist, M. (2005). Real-time PCR screening for 16S rRNA mutations associated with resistance to tetracycline in Helicobacter pylori. Antimicrob. Agents Chemother. 49, 3166–3170. doi: 10.1128/AAC.49.8.3166-3170.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glocker, E., and Kist, M. (2004). Rapid detection of point mutations in the gyrA gene of Helicobacter pylori conferring resistance to ciprofloxacin by a fluorescence resonance energy transfer-based real-time PCR approach. J. Clin. Microbiol. 42, 2241–2246. doi: 10.1128/JCM.42.5.2241-2246.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heep, M., Lehn, N., Brandstatter, B., Rieger, U., Senzenberger, S., and Wehrl, W. (2002). Detection of rifabutin resistance and association of rpoB mutations with resistance to four rifamycin derivatives in Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 21, 143–145. doi: 10.1007/s10096-001-0672-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hirata, K., Suzuki, H., Nishizawa, T., Tsugawa, H., Muraoka, H., Saito, Y., et al. (2010). Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J. Gastroenterol. Hepatol. 25 (Suppl. 1), S75–S79. doi: 10.1111/j.1440-1746.2009.06220.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaneko, F., Suzuki, H., Hasegawa, N., Kurabayshi, K., Saito, H., Otani, S., et al. (2004). High prevalence rate of Helicobacter pylori resistance to clarithromycin during long-term multiple antibiotic therapy for chronic respiratory disease caused by non-tuberculous mycobacteria. Aliment. Pharmacol. Ther. 20 (Suppl. 1), 62–67. doi: 10.1111/j.1365-2036.2004.01993.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klesiewicz, K., Nowak, P., Karczewska, E., Skiba, I., Wojtas-Bonior, I., Sito, E., et al. (2014). PCR-RFLP detection of point mutations A2143G and A2142G in 23S rRNA gene conferring resistance to clarithromycin in Helicobacter pylori strains. Acta Biochim. Pol. 61, 311–315.
Kutschke, A., and de Jonge, B. L. (2005). Compound efflux in Helicobacter pylori. Antimicrob. Agents Chemother. 49, 3009–3010. doi: 10.1128/AAC.49.7.3009-3010.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J. W., Kim, N., Nam, R. H., Park, J. H., Choi, Y. J., Kim, J. M., et al. (2004). GenoType HelicoDR test in the determination of antimicrobial resistance of Helicobacter pylori in Korea. Scand. J. Gastroenterol. 49, 1058–1067. doi: 10.3109/00365521.2014.894117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lehours, P., Siffre, E., and Megraud, F. (2011). DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterol. 11:112. doi: 10.1186/1471-230X-11-112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H., and Hearst, J. E. (1995). Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16, 45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malfertheiner, P., Megraud, F., O'Morain, C. A., Atherton, J., Axon, A. T., Bazzoli, F., et al. (2012). Management of Helicobacter pylori infection–the Maastricht IV/ Florence Consensus Report. Gut 61, 646–664. doi: 10.1136/gutjnl-2012-302084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Masaoka, T., Suzuki, H., Kurabayashi, K., Kamiya, A., and Ishii, H. (2004). Second-line treatment of Helicobacter pylori infection after dilution agar methods and PCR-RFLP analysis. Aliment. Pharmacol. Ther. 20 (Suppl. 1), 68–73. doi: 10.1111/j.1365-2036.2004.01994.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Masaoka, T., Suzuki, H., Kurabayashi, K., Nomoto, Y., Nishizawa, T., Mori, M., et al. (2006). Could frameshift mutations in the frxA and rdxA genes of Helicobacter pylori be a marker for metronidazole resistance? Aliment. Pharmacol. Ther. 24 (Suppl. 4), 81–87. doi: 10.1111/j.1746-6342.2006.00029.x
Matsuzaki, J., Suzuki, H., Nishizawa, T., Hirata, K., Tsugawa, H., Saito, Y., et al. (2012). Efficacy of sitafloxacin-based rescue therapy for Helicobacter pylori after failures of first- and second-line therapies. Antimicrob. Agents Chemother. 56, 1643–1645. doi: 10.1128/AAC.05941-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuzaki, J., Suzuki, H., Tsugawa, H., Nishizawa, T., and Hibi, T. (2010). Homology model of the DNA gyrase enzyme of Helicobacter pylori, a target of quinolone-based eradication therapy. J. Gastroenterol. Hepatol. 25 (Suppl. 1), S7–S10. doi: 10.1111/j.1440-1746.2010.06245.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, R. A., Beckthold, B., Wong, S., Kureishi, A., and Bryan, L. E. (1995). Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob. Agents Chemother. 39, 107–111. doi: 10.1128/AAC.39.1.107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nahar, S., Mukhopadhyay, A. K., Khan, R., Ahmad, M. M., Datta, S., Chattopadhyay, S., et al. (2004). Antimicrobial susceptibility of Helicobacter pylori strains isolated in Bangladesh. J. Clin. Microbiol. 42, 4856–4858. doi: 10.1128/JCM.42.10.4856-4858.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Maekawa, T., Watanabe, N., Harada, N., Hosoda, Y., Yoshinaga, M., et al. (2014). Clarithromycin Versus Metronidazole as First-line Helicobacter pylori Eradication: a multicenter, prospective, randomized controlled study in Japan. J. Clin. Gastroenterol. doi: 10.1097/MCG.0000000000000165. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., and Hibi, T. (2009). Quinolone-Based Third-Line Therapy for Helicobacter pylori Eradication. J. Clin. Biochem. Nutr. 44, 119–124. doi: 10.3164/jcbn.08-220R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Kurabayashi, K., Masaoka, T., Muraoka, H., Mori, M., et al. (2006). Gatifloxacin resistance and mutations in gyra after unsuccessful Helicobacter pylori eradication in Japan. Antimicrob. Agents Chemother. 50, 1538–1540. doi: 10.1128/AAC.50.4.1538-1540.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Maekawa, T., Harada, N., Toyokawa, T., Kuwai, T., et al. (2012). Dual therapy for third-line Helicobacter pylori eradication and urea breath test prediction. World J. Gastroenterol. 18, 2735–2738. doi: 10.3748/wjg.v18.i21.2735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Matsuzaki, J., Muraoka, H., Tsugawa, H., Hirata, K., et al. (2011b). Helicobacter pylori resistance to rifabutin in the last 7 years. Antimicrob. Agents Chemother. 55, 5374–5375. doi: 10.1128/AAC.05437-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Nakagawa, I., Iwasaki, E., Masaoka, T., and Hibi, T. (2008). Gatifloxacin-based triple therapy as a third-line regimen for Helicobacter pylori eradication. J. Gastroenterol. Hepatol. 23 (Suppl. 2), S167–S170. doi: 10.1111/j.1440-1746.2008.05407.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Tsugawa, H., Muraoka, H., Matsuzaki, J., Hirata, K., et al. (2011a). Enhancement of amoxicillin resistance after unsuccessful Helicobacter pylori eradication. Antimicrob. Agents Chemother. 55, 3012–3014. doi: 10.1128/AAC.00188-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, T., Suzuki, H., Umezawa, A., Muraoka, H., Iwasaki, E., Masaoka, T., et al. (2007). Rapid detection of point mutations conferring resistance to fluoroquinolone in gyrA of Helicobacter pylori by allele-specific PCR. J. Clin. Microbiol. 45, 303–305. doi: 10.1128/JCM.01997-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oleastro, M., Menard, A., Santos, A., Lamouliatte, H., Monteiro, L., Barthelemy, P., et al. (2003). Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J. Clin. Microbiol. 41, 397–402. doi: 10.1128/JCM.41.1.397-402.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ribeiro, M. L., Gerrits, M. M., Benvengo, Y. H., Berning, M., Godoy, A. P., Kuipers, E. J., et al. (2004). Detection of high-level tetracycline resistance in clinical isolates of Helicobacter pylori using PCR-RFLP. FEMS Immunol. Med. Microbiol. 40, 57–61. doi: 10.1016/S0928-8244(03)00277-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rimbara, E., Noguchi, N., Kawai, T., and Sasatsu, M. (2012). Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 17, 36–42. doi: 10.1111/j.1523-5378.2011.00912.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scaletsky, I. C., Aranda, K. R., Garcia, G. T., Goncalves, M. E., Cardoso, S. R., Iriya, K., et al. (2011). Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter 16, 311–315. doi: 10.1111/j.1523-5378.2011.00845.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schabereiter-Gurtner, C., Hirschl, A. M., Dragosics, B., Hufnagl, P., Puz, S., Kovach, Z., et al. (2004). Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J. Clin. Microbiol. 42, 4512–4518. doi: 10.1128/JCM.42.10.4512-4518.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stone, G. G., Shortridge, D., Versalovic, J., Beyer, J., Flamm, R. K., Graham, D. Y., et al. (1997). A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41, 712–714.
Suerbaum, S. (2000). Genetic variability within Helicobacter pylori. Int. J. Med. Microbiol. 290, 175–181. doi: 10.1016/S1438-4221(00)80087-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, H., Nishizawa, T., and Hibi, T. (2010). Helicobacter pylori eradication therapy. Future Microbiol. 5, 639–648. doi: 10.2217/fmb.10.25
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, H., Nishizawa, T., Muraoka, H., and Hibi, T. (2009a). Sitafloxacin and garenoxacin may overcome the antibiotic resistance of Helicobacter pylori with gyrA mutation. Antimicrob. Agents Chemother. 53, 1720–1721. doi: 10.1128/AAC.00049-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, S., Suzuki, H., Nishizawa, T., Kaneko, F., Ootani, S., Muraoka, H., et al. (2009b). Past rifampicin dosing determines rifabutin resistance of Helicobacter pylori. Digestion. 79, 1–4. doi: 10.1159/000191204
Tseng, Y. S., Wu, D. C., Chang, C. Y., Kuo, C. H., Yang, Y. C., Jan, C. M., et al. (2009). Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur. J. Clin. Invest. 39, 807–812. doi: 10.1111/j.1365-2362.2009.02166.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsugawa, H., Suzuki, H., Satoh, K., Hirata, K., Matsuzaki, J., Saito, Y., et al. (2011). Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid. Redox Signal. 14, 15–23. doi: 10.1089/ars.2010.3146
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Versalovic, J., Shortridge, D., Kibler, K., Griffy, M. V., Beyer, J., Flamm, R. K., et al. (1996). Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40, 477–480.
Wang, G., Rahman, M. S., Humayun, M. Z., and Taylor, D. E. (1999). Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 43, 683–685.
Wu, J. Y., Wang, S. S., Lee, Y. C., Yamaoka, Y., Graham, D. Y., Jan, C. M., et al. (2014). Detection of genotypic clarithromycin-resistant Helicobacter pylori by string tests. World J. Gastroenterol. 20, 3343–3349. doi: 10.3748/wjg.v20.i12.3343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xuan, S. H., Zhou, Y. G., Shao, B., Cui, Y. L., Li, J., Yin, H. B., et al. (2009). Enzymic colorimetry-based DNA chip: a rapid and accurate assay for detecting mutations for clarithromycin resistance in the 23S rRNA gene of Helicobacter pylori. J. Med. Microbiol. 58(Pt 11), 1443–1448. doi: 10.1099/jmm.0.010785-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Gao, W., Cheng, H., Zhang, X., and Hu, F. (2014). Tetracycline- and Furazolidone-containing Quadruple Regimen as Rescue Treatment for Helicobacter pylori Infection: a single center retrospective study. Helicobacter 19, 382–386. doi: 10.1111/hel.12143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Helicobacter pylori, 23S rRNA, gyrA, PBP1, 16S rRNA
Citation: Nishizawa T and Suzuki H (2014) Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front. Mol. Biosci. 1:19. doi: 10.3389/fmolb.2014.00019
Received: 23 August 2014; Accepted: 04 October 2014;
Published online: 24 October 2014.
Edited by:
Joao Inacio, University of Brighton, UKReviewed by:
Nuno F. Azevedo, Faculty of Engineering at the University of Porto, PortugalOsamu Handa, Kyoto Prefectural University of Medicine, Japan
Joao Inacio, University of Brighton, UK
Kengo Tokunaga, The Third Department of Internal Medicine Kyorin University School of Medicine, Japan
Copyright © 2014 Nishizawa and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hidekazu Suzuki, Division of Gastroenterology and Hepatology, Department of Internal Medicine, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan e-mail: hsuzuki@a6.keio.jp
 Toshihiro Nishizawa
Toshihiro Nishizawa Hidekazu Suzuki
Hidekazu Suzuki