The hippocampal neuroproteome with aging and cognitive decline: past progress and future directions
- Department of Pharmacology, Penn State College of Medicine, Milton S. Hershey Medical Center, Hershey, PA, USA
Although steady progress on understanding brain aging has been made over recent decades through standard anatomical, immunohistochemical, and biochemical techniques, the biological basis of non-neurodegenerative cognitive decline with aging remains to be determined. This is due in part to technical limitations of traditional approaches, in which only a small fraction of neurobiologically relevant proteins, mRNAs or metabolites can be assessed at a time. With the development and refinement of proteomic technologies that enable simultaneous quantitative assessment of hundreds to thousands of proteins, neuroproteomic studies of brain aging and cognitive decline are becoming more widespread. This review focuses on the contributions of neuroproteomic investigations to advances in our understanding of age-related deficits of hippocampus-dependent spatial learning and memory. Accumulating neuroproteomic data demonstrate that hippocampal aging involves common themes of dysregulated metabolism, increased oxidative stress, altered protein processing, and decreased synaptic function. Additionally, growing evidence suggests that cognitive decline does not represent a “more aged” phenotype, but rather is associated with specific neuroproteomic changes that occur in addition to age-related alterations. Understanding if and how age-related changes in the hippocampal neuroproteome contribute to cognitive decline and elucidating the pathways and processes that lead to cognitive decline are critical objectives that remain to be achieved. Progress in the field and challenges that remain to be addressed with regard to animal models, behavioral testing, and proteomic reporting are also discussed.
Background
Age-related cognitive decline is a common complication of aging that impacts a variety of brain functions including perceptual speed, inductive reasoning, and perhaps most disabling, hippocampus-dependent spatial learning and memory (Hedden and Gabrieli, 2004). Impairment of these functions reduces quality of life for aging individuals by diminishing the health-span and decreasing independence. Currently, an estimated 40% of the otherwise healthy population over age 60 is affected by varying degrees of cognitive decline, which increases in prevalence and severity with co-morbid conditions such as Type 2 diabetes, hypertension, and heart disease (Qiu et al., 2005; Panza et al., 2008; Dahle et al., 2009; Crowe et al., 2010; Ding et al., 2010; Okonkwo et al., 2010). As the quality and availability of health care in developed countries continue to improve, the percentage of aged individuals in the population is expected to continue to rise. For example, as of the 2000 U.S. census, approximately 12% of the U.S. population was over age 60 (CensusScope, 2010), and population projections suggest that by 2050, this age group will account for more than one in five persons in the total U.S. population (Shrestha, 2006). The prevalence of age-related health conditions is expected to rise concomitantly with our increasing lifespan, and as such, a greater emphasis must be placed on understanding, preventing, and treating complications like cognitive decline.
Our knowledge of the molecular basis of brain function in general, and neurotransmission specifically, has grown exponentially over the last two decades. Although much has been learned from targeted studies focused on single proteins [e.g., determination of SNARE binding domains assessed cleavage/deletion-mediated protein inactivation (Schiavo et al., 1993; Deak et al., 2006)] or families of proteins [e.g., demonstration of the impact of multiple synapsin isoform-knockouts on the synaptic vesicle cycle and cognitive function (Corradi et al., 2008)], these traditional approaches provide circumscribed insight into the complexities of the neuroproteome and its role in mediating various brain functions (e.g., learning and memory). This is due in part to low throughput and the requirement that the identities of specific proteins of interest be determined a priori for functional and targeted studies. Recent advances in proteomic technologies and their increasing implementation in neuroscience research present an opportunity to expand our existing understanding and provide a deeper insight into the molecular components of cognition. Using neuroproteomic approaches, we can also better characterize brain region-specific alterations that contribute to neural dysfunction and cognitive impairments with conditions ranging from normal aging to neurodegenerative diseases. Of primary necessity is a detailed assessment of the effects of age on the neuroproteome and how they affect generalized cellular functions such as energy production and cytoskeletal maintenance, as well as neuron-specific functions including maintenance of vesicle trafficking, neurotransmitter release and reuptake, and synaptic remodeling. This is particularly true for the hippocampus, which is the primary brain structure implicated in spatial learning and memory, and which is profoundly affected by aging. This review will highlight progress made in proteomic studies of the hippocampus with aging and cognitive decline and not detail the technical aspects of various proteomic methods which have been discussed in several previous reviews (Freeman and Hemby, 2004; Drabik et al., 2007; Friedman et al., 2009; Lull et al., 2010). Aspects of the neuroproteome that remain to be examined and future directions for the field will also be discussed in the context of known physiological alterations in hippocampal function with aging and cognitive decline.
Hippocampal Function is Altered with Aging and Cognitive Decline
The hippocampus is the primary mediator of spatial learning and memory, which is impaired in aging subjects experiencing cognitive decline (Hedden and Gabrieli, 2004). Loss of hippocampus-dependent spatial cognition reduces independence, mobility, and activity, and diminishes health status and quality of life. Numerous anatomical and physiological alterations have been characterized in the hippocampus with aging that may contribute to cognitive decline, including decreased hippocampal volume without gross neuronal loss, vascular rarefaction, decreased trophic support, increased oxidative stress and inflammation, decreased metabolism and glucose utilization, dysregulation of protein synthesis, folding, and accumulation, and impaired neurotransmitter synthesis and release (Sonntag et al., 1999; Calabrese et al., 2004; Paz Gavilan et al., 2006; Poon et al., 2006; Rowe et al., 2007; Freeman et al., 2009; Pawlowski et al., 2009; Erickson et al., 2010). Broad dysregulation of these processes with increasing age impacts hippocampal function in a variety of ways ranging from cytoprotection to energy biogenesis to intracellular trafficking of proteins and organelles.
At the most basic level, cognitive function is a reflection of neuronal activity and synaptic signaling. Synaptic dysfunction, indicated by altered hippocampal neurochemistry, morphology, and electrophysiology (e.g., LTP, LTD, PPF), is also associated with aging and cognitive decline (Boric et al., 2008; Sierra-Mercado et al., 2008; Gureviciene et al., 2009). Electrophysiological correlates of hippocampal function are disrupted with aging and learning impairment, and are consistent with unstable encoding of spatial representations (Barnes et al., 1997; Rosenzweig and Barnes, 2003; Wilson et al., 2003; Kumar et al., 2007). This instability manifests in resistance to LTP induction, facilitation of LTD, and aberrant spatiotemporal activation of ensemble networks. These characteristics may be related to atypical synapse morphology, decreased neurotransmitter synthesis and receptor signaling, and changes in neuronal protein and gene expression occurring with increasing age (Burke and Barnes, 2006; Shi et al., 2007; Liu et al., 2008). Additionally, age-related cognitive decline is associated with a preferential loss of functional high-efficiency synapses (i.e., multiple spine bouton and perforated synapses) implicated in spatial learning and memory formation (Geinisman et al., 1986; Shi et al., 2005; Burke and Barnes, 2006). The molecular processes underlying these changes have been partially identified, but remain to be comprehensively investigated. Further, identification of both the specific proteins and the broader pathways and processes dysregulated specifically with cognitive decline is needed to delineate the general effects of aging from those implicated in decreasing cognitive ability. A full understanding of the hippocampal neuroproteome with aging and age-related cognitive deficits will be an important first step in development of therapies that target cognition-associated alterations to prevent or reverse cognitive decline.
Defining the Hippocampal Neuroproteome
Given the complexities of obtaining and analyzing human brain tissue, the well-described similarities between human and rodent neurophysiology, and the availability and practicality of rodent model organisms, rat and mouse models are the most common mammalian model systems in neuroscience research. As such, initial neuroproteomic studies focused on defining components of the whole-brain rodent proteome (Fountoulakis et al., 1999). Although profiling of the whole-brain proteome has yielded useful information, specific examination of the hippocampus is important in elucidating the molecular mechanisms underlying the formation and retrieval of spatial memory. A key early assessment of the human hippocampal proteome was reported in 2000 by Edgar et al. (1999), who applied 2DE protein separation, liquid chromatography–coupled mass spectrometry (LC–MS) and N-terminal sequencing to profile protein species present in the hippocampus of one middle aged female. This work identified nearly seventy proteins, including numerous cytoskeletal and mitochondrial elements. Subsequent work extended these findings with higher-throughput profiling of the hippocampal proteome in postmortem tissue isolated from adult males and females (mean age 61 years; Yang et al., 2004). This approach combined 2DE protein separation with tandem mass spectrometry (MS/MS) that yields more conclusive protein identifications. More than 160 unique proteins were identified, the majority of which are components of metabolic pathways comprising glycolysis and the citric acid cycle. Assessment of the rat hippocampus (Fountoulakis et al., 2005) further added to known components of the hippocampal neuroproteome and, in agreement with proteomic profiles of the human hippocampus (Edgar et al., 1999; Yang et al., 2004), cytosolic proteins and mitochondrial enzymes comprised approximately 50% of the identified proteins. These proteins are highly abundant and tend to represent the majority of proteins identified in unfractionated neural tissue. Together these, and other, early profiling studies provided a groundwork for future neuroproteomic studies by demonstrating the utility of proteomic technologies in characterizing the diversity of proteins expressed in the hippocampus. A challenge common to these reports is the limited detection of low-abundance and subcellular compartment-specific neuronal proteins (e.g., regulators of the synaptic vesicle cycle), which are frequently masked by highly expressed proteins (e.g., metabolic enzymes and cytoskeletal components). This challenge has been partially addressed through the development of approaches that increase the coverage (i.e., the diversity of proteins simultaneously examined) and the sensitivity (i.e., detection of low-abundance proteins) of neuroproteomic assessments.
An effective approach to increasing the insight provided by neuroproteomic studies is examination of hippocampal synaptosome, synaptic plasma membrane, and synaptic lipid raft preparations (Bai and Witzmann, 2007; Jiang et al., 2008; Fernandez et al., 2009; Wu et al., 2010). These subcellular fractionation techniques enrich for synaptic proteins typically masked by highly abundant metabolic and cytoskeletal proteins. The value of sample fractionation is demonstrated by a 2005 report describing mass spectrometric profiling of hippocampal plasma membrane fractions (Nielsen et al., 2005). This study used both standard LC–MS/MS and higher accuracy Fourier transform ion cyclotron resonance MS (FT-MS), which resolves ion mass-to-charge ratios by their rotational frequency in a strong magnetic field as they pass by the detection plates. This combinatorial approach described led to identification of numerous species, the majority of which, unlike whole-tissue profiling, were annotated as membrane proteins. Of these, only a small percentage originated from mitochondrial or endoplasmic reticulum membranes, and compared to whole hippocampus, mitochondrial content was nearly depleted which allowed identification of proteins previously not detected in proteomic analyses. Many integral and plasma membrane proteins identified were hippocampally expressed subunits of GABA and glutamate receptors, as well as voltage-gated potassium, sodium, and calcium channels. Proteomic characterization of hippocampal postsynaptic densities by LC–MS/MS profiling of affinity-purified, PSD95-associated proteins (Dosemeci et al., 2006) has also identified a number of synaptic proteins, including maguin, PSD93, Shank 1, and Shank 3, and signaling molecules such as CaMKII, SynGAP, and protein kinase C. Presynaptic “contaminants” (e.g., piccolo, bassoon, synapsin 1, and synapsin 2) and proteins associated with neuronal extracellular matrix (e.g., tenascin-R, proteoglycan-link protein, fibronectin) were also identified, perhaps complicating analysis of PSD fractions per se but also increasing our awareness of the complexity of the hippocampal neuroproteome. These reports contribute valuable understanding of the composition of hippocampal subproteomes, and demonstrates that pairing fractionation with proteomic approaches increases sensitivity of proteomic technologies to evaluate significant populations of low-abundance proteins in small amounts of neurologically derived starting material. Similar approaches have been broadly implemented in neuroscience research, and are seeing increasing usage in the aging research field.
The neuroproteomic studies discussed thus far have been broad-based profiling studies seeking to determine the identity and diversity of proteins present in hippocampal tissue. Although characterization of the hippocampal neuroproteome is important to our understanding of potential mechanisms of hippocampal function, integration of methods capable of quantitative comparisons of proteomic composition in health and disease are also necessary. Multiple methods of proteome quantitation exist, each with its strengths and limitations (Chen et al., 2007b; Wiese et al., 2007). One example of the capabilities of quantitative neuroproteomic assessment is the demonstration of enriched synaptic plasticity-associated protein (e.g., PSD-95, CaMKIIα, homer 1, NMDA, and AMPA receptors) content in hippocampal postsynaptic density preparations compared to those from other brain regions (e.g., cerebellum; Trinidad et al., 2008). With the advances made by the studies described above, it is clear that proteomic approaches are highly useful and are becoming more routinely implemented in neuroscience research. The ability to quantitatively characterize the hippocampal neuroproteome and subproteomes (either through anatomical fractionation or proteome enrichment) provides the aging research field with a powerful tool for investigating differential protein expression with aging and age-related impairment of spatial learning and memory. Examination of changes in protein abundance, modification, and localization across the lifespan, as described below and summarized in Table 1, has dramatically increased our understanding of brain aging. Further, these studies have also identified molecular correlates, and potential molecular mechanisms, of deficits of hippocampal function.
Characterization of the Hippocampal Proteome with Aging
The Human Proteome Organization initiated the Brain Pilot Project in 2003 with the goal of characterizing the neuroproteomes of humans and rodent models throughout the lifespan. Quantitative proteomic analyses of the aging hippocampus have identified numerous proteins altered in expression with increasing age and provide insight into processes and signaling pathways implicated in brain aging. While initial reports focused on quantitative comparisons of the whole-brain proteome between neonatal and adult rodents, they are informative of the temporal regulation of protein expression in neural tissue. For example, in agreement with known increases in glucose utilization in the adult brain, this study also identified upregulation metabolic enzymes including creatine kinase, alpha enolase, and triosephosphate dehydrogenase in adult (8 weeks) compared to neonatal (7 days) mice (Carrette et al., 2006). Similar work compared whole mouse brain proteomes of these age groups and an additional embryonic group (embryonic day 16), using two-dimensional difference gel electrophoresis (2-DIGE; Focking et al., 2006). The 2-DIGE technology fluorescently labels cysteine residues, allowing multiplexing of up to three protein samples on each gel. Each 2-DIGE gel typically combines two experimental samples and a normalization pool (containing equal protein from all study samples) used to standardize protein spot quantitation between gels. This screen enabled a broader assessment of temporal regulation of protein expression, and identified a variety of proteins peaking at different ages. For example, fascin, alpha-fetoprotein, tubulin isoforms, and actin were most highly expressed in embryonic brain, while neonatal brain most highly expressed fatty acid binding protein, syntaxin, peroxiredoxin 2, and dihydropyrimidinase-related protein-3. Metabolic proteins involved in ATP generation and glucose bioenergetics were most highly expressed in the adult brain, again concurring with the higher rate of glucose metabolism in adulthood. These studies, although focused on developmental stages rather than the aging process, provided valuable data regarding neuroproteomic regulation with increasing age.
In 2008, Yang et al. (2008) published a laudable effort to profile the rodent brain proteome over a broad time-course of natural aging by comparing protein isolated from whole brains of neonatal (4 days), young adult (3 months), mature adult (6 months), middle aged (12 months) and aged (15 months) male Kunming mice using 2DE and semi-quantitative densitometry. This analysis identified decreased expression of numerous protein regulators of protein quality (proteasome subunits, ubiquitin carboxyl-terminal esterase L3, calreticulin) and synaptic function (calbindin, amphiphysin, valosin-containing protein), among others, with increasing age. Proteins that increased with aging were mainly enzymes that mediate energy production and oxidative stress. A key feature of this report is the demonstration of multiple protein expression patterns across the lifespan. These changes included consistent increases (transketolase, mitochondrial creatine kinase) and consistent decreases (proteasome subunits alpha type 3, alpha type 6, 26S non-ATPase 14), as well as biphasic patterns in which proteins increased in expression through adulthood but decreased with aging (amphiphysin, dynamin, voltage-dependent anion channel 1). As is typical in proteomic analyses of unfractionated neural tissue, a high percentage of identified proteins represented abundant nuclear (30%) and mitochondrial (20%) species, underscoring the value of enrichment approaches to increase detection of less-abundant proteins.
These works assessed the proteomic composition of the entire brain rather than its functionally distinct structures, and focused on relatively young ages. To assess hippocampus-specific alterations with advanced age, the Youdim laboratory performed neuroproteomic profiling of female Wistar rats aged 8 and 27 months by 2DE with MS protein identification (Weinreb et al., 2007). Significantly higher hippocampal expression of neurofilament light chain, ferritin heavy polypeptide 1, T-complex protein 1, profilin 1, and isocitrate dehydrogenase 2α were evident in aged rats compared to adults. Expression of protein translation elongation factor Tu, phosphatidylethanolamine binding protein (PEBP), astrocytic phosphoprotein PEA15, peroxiredoxin 2, and the Va and Vb subunits of cytochrome c oxidase was decreased with aging. Differential expression of members of proposed classes of proteins represented by proteomic alterations [i.e., neurodegenerative disease (ferritin), binding proteins/chaperones (profilin 1), and metabolism/oxidation (peroxiredoxin 2)] was confirmed by targeted immunoblotting, which is becoming a routine component of neuroproteomic investigations. Several age-regulated proteins identified by this work have been associated with age-related decrements of hippocampus-dependent learning and memory and increased oxidative stress, suggesting a potential mechanistic role for dysregulation of these proteins in functional deficits with hippocampal aging (Chen et al., 2003; Feldmann et al., 2008; Ramos et al., 2009; Quintana and Gutierrez, 2010).
In addition to studying the expression of proteins, it is also important to identify changes in protein post-translational modifications as they play integral roles not only in protein localization and function, but also in the pathogenesis of a variety of conditions including age-related disease. In a study of hippocampal protein oxidation in aged male Wistar rats, neuroproteomic approaches were implemented to quantify carbonyl, 4-hydroxynonenal (HNE), and 3-nitrotyrosine (3-NT) protein adducts and to assess the effects of caloric restriction on protein modification by these reactive species (Poon et al., 2006). Rats either had free access to food or were calorically restricted to approximately 60% of control intake from 12 months of age, with food available every two days. After rats reached 28 months of age the hippocampal neuroproteome was quantitatively characterized by 2DE with MS identification of differentially expressed/modified proteins. Protein carbonylation and nitrosylation was determined by two-dimensional immunoblotting (often referred to as “far-western” blotting) while carbonyl, 3-NT, and HNE levels were determined by slot blot. These analyses determined that caloric restriction of aged rats reduces bulk hippocampal HNE and 3-NT levels, and also decreases nitrosylation of malate dehydrogenase, 14-3-3 zeta, and phosphoglycerate kinase 1. Further, compared to age-matched controls, calorically restricted rats exhibited increased expression of aconitase 2, dihydrolipoamide dehydrogenase, neuroprotective peptide H3, dynamin-like protein 1, and eukaryotic initiation factor 5a. These findings suggest a diverse mode of action of caloric restriction via potential processes ranging from metabolism and protein synthesis to neuronal integrity. Unfortunately, younger age groups were not included in this analysis, so it is not possible to conclude that caloric restriction produces a “less aged” phenotype, or whether its putative beneficial effects are diminished in aged rats compared to their younger counterparts.
The effects of aging on the proteomic composition of hippocampal synapses (i.e., the hippocampal synaptoproteome) have also been described. Using an approach combining synaptosome isolation from young (9 weeks) and aged (30 months) female Fischer rats with 2-DIGE proteomic quantitation and MS protein identification, Sato et al. (2005) identified a number of proteins regulated with aging. Proteins decreased in aged rats included T-complex 1, collapsin response mediator protein 4, hsp60, coronin 1A, and several metabolic enzymes, while proteins increased with age included GFAP, SNAP25, septin 2, and actin. Evaluation of female rats is less common than males for logistical purposes, and so the proteomic profiles generated here are beneficial for intersex comparisons of neuroproteomic composition and assessment of potential differential susceptibility to age-related processes in males versus females. Although the focus on lower abundance, synapse-specific proteins is often more valuable than approaches using unfractionated tissue, the small number of proteins identified in this study limits broader bioinformatic analysis and interpretation.
Our laboratory recently reported the results of an investigation of the hippocampal synaptoproteome that included male Fischer 344 × Brown Norway (F1) hybrid rats aged 3 (young-adult), 12 (adult), and 26 months (aged; VanGuilder et al., 2010). These rats are genetically identical, and exhibit the long lifespan of the Fischer 344 rat combined with the health-span of the Brown Norway, which minimizes potential age-related pathologies that can confound behavioral analyses. 2-DIGE quantitation of hippocampal synaptosomes determined differential expression of more than 250 proteins between the three age groups. Age-regulated proteins, as well as a large selection of stably expressed proteins, were identified by MS/MS. A subset of proteins observed in this work confirms previous findings of the Sato et al. (2005) investigation of hippocampal synaptosomes isolated from young-adult and aged female Fischer rats. Commonalities include decreased expression of isocitrate dehydrogenase, creatine kinase B, and protein disulfide isomerase, and increased expression of GFAP in aged rats, underscoring disruptions of glucose and protein metabolism with aging. Bioinformatic analysis of proteins regulated with increasing age identified a network of “nervous system function” proteins that was significantly over-represented among differentially expressed proteins. The majority of neurotransmission-related changes in synaptosomal protein expression occurred between adulthood and advanced age, and included downregulation of commonly identified structural proteins (F-, gamma- and beta-actin, numerous tubulin isoforms, internexin) as well as lower-abundance synaptic proteins (synapsin 1, syndapin 1, clathrin, tropomodulin 2, amphiphysin, syntaxin binding protein). Immunoblot confirmation of a number of proteins with well-characterized roles in neurotransmission confirmed expression decreases (14-3-3 zeta and gamma, synapsin 1, dynamin 1) and increases (synapsin 2, hippocalcin) with increasing age. Based on these neuroproteomic findings, immunoblot analyses were extended to include additional neurotransmission-regulating proteins, and determined a significant effect of age on synaptic expression of synaptophysin, PSD95, and the SNARE proteins VAMP2, SNAP25, and syntaxin 1, all of which decreased with increasing age. In total, 404 hippocampal synaptosomal proteins were identified with high confidence, contributing to our understanding of the complex proteomic composition of hippocampal synapses. The findings of this study suggest that expression of many proteins regulating hippocampal neurotransmission undergoes significant age-related alterations between adulthood and advanced age. The proteins affected are implicated in synaptic vesicle exocytosis and endocytosis, postsynaptic receptor aggregation, and activity-dependent synaptic maintenance, which may decrease stimulus-induced neurotransmission and could potentially impact learning and memory functions in the aged hippocampus.
The studies described above have examined effects of aging on the whole-brain, hippocampal, and hippocampal synaptosomal neuroproteomes, and demonstrate not only the complexity of the neuroproteome but also the high degree of regulation that occurs with natural aging. A limitation of the aging neuroproteomics research field is a lack of standard rodent models, ages, and technological approaches. The existing reports of age-related regulation of the neuroproteome have utilized multiple strains of mice and rats, assessed both male and female, and included time points ranging from embryonic and neonatal stages to advanced age, which limits direct comparison of these datasets. However, commonalities in processes regulated with aging have emerged from the findings of these studies.
Common Themes in Neuroproteomic Investigations of the Aging Hippocampus
Neuroproteomic investigations of the aging hippocampus have identified specific protein species as well as protein pathways and signaling networks that are dysregulated with aging and age-related cognitive decline. Additionally, this research has provided mechanistic insight into broader processes known to be altered with aging (Figure 1). These processes include glucose metabolism, protein processing, oxidative stress and inflammation, and synaptic function. For example, metabolic disruption and decreased glucose utilization are reproducibly demonstrated in aging humans and animal models of human aging (Tack et al., 1989; Sonntag et al., 2006; Kantarci et al., 2010). Quantitative assessments of the hippocampal proteome have revealed altered expression of numerous proteins that play both integral and regulatory roles in glycolysis and gluconeogenesis (e.g., enolase, aldolase, creatine kinase, lactate dehydrogenase, pyruvate kinase, phosphoglycerate mutase, isocitrate dehydrogenase, phosphofructokinase). A subset of these proteins is necessary for the ATP-generating stages of glucose metabolism, which converges with functional studies revealing decreased energy biogenesis in the aging brain. Interestingly, several of these proteins (e.g., alpha enolase) are differentially expressed with both aging and cognitive decline, indicating the complexity of intersecting processes regulated with increasing age and learning and memory. The identification of these differentially expressed proteins provides the opportunity for interventional studies seeking to normalize glucose metabolism and restore energy production in the aging hippocampus. Similarly, impaired protein processing is associated with hippocampal aging. This is commonly evident in the accumulation of protein aggregates and increased cellular stress caused by oxidized and misfolded proteins (Squier, 2001; Paz Gavilan et al., 2006). Age-related dysregulation of proteins driving protein folding, trafficking and degradation has been characterized by neuroproteomic studies of the unfractionated hippocampus as well as of hippocampal synapses. Decreased expression of proteins with chaperone and folding functions (e.g., heat shock proteins, T-complex protein 1, protein disulfide isomerase A3, protein phosphatase 1) as well as components of the protein degradation process (proteasome alpha subunits 1, 3, and 6, 26S proteasome non-ATPase regulatory subunit 14, ubiquitin C-terminal esterase L3, pyromycin-sensitive aminopeptidase) indicates a loss of protein quality regulation with aging that may contribute to the observed buildup of cytoskeletal proteins (e.g., increased neurofilament light chain and tubulin) with increasing age. Likewise, impairment of protein folding and degradation pathways may contribute to accumulation of misfolded and oxidized proteins, which are implicated in inflammation- and stress-response with aging (Keller et al., 2002).
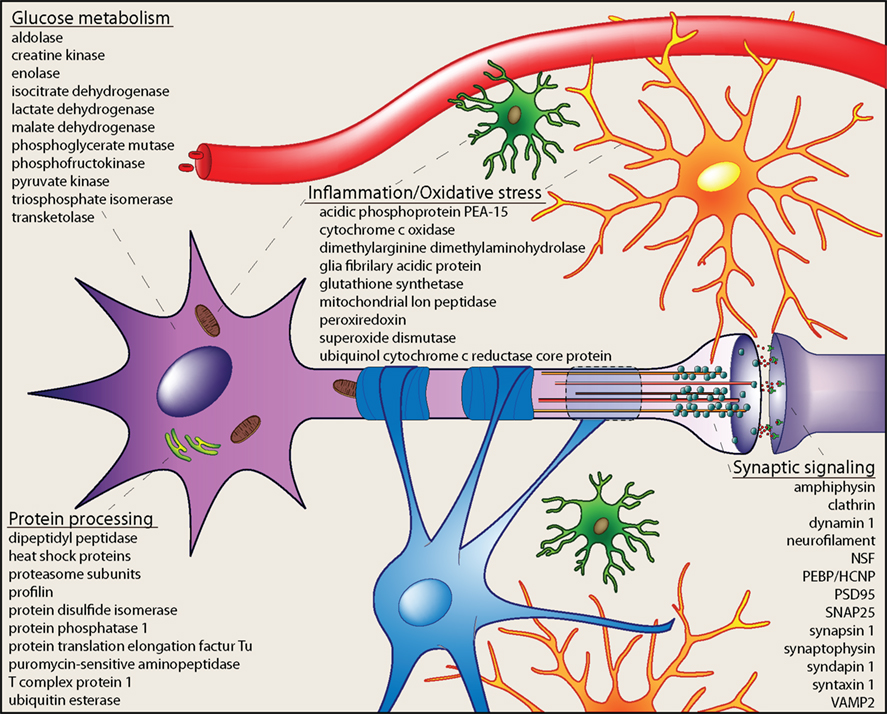
Figure 1. Cellular processes altered in the aging hippocampus. Integration of age-related neuroproteomic alterations in the hippocampus reveals dysregulation of cellular processes including glucose metabolism, protein processing, inflammation and oxidative stress, and synaptic signaling. Proteins differentially expressed with increasing age in neuroproteomic investigations are included as examples. These cellular processes suggest that numerous cell types [microglia (green), astrocytes (orange), oligodendrocytes (blue), and neurons (violet)] and subcellular components [mitochondria (brown), endoplasmic reticulum (green), cytoskeleton (orange/red), and synaptic machinery] are affected by brain aging, and may contribute to functional deficits related to the aging hippocampus.
Oxidative stress is typically associated with mitochondrial dysfunction and/or modification of proteins by reactive adducts. Neuroproteomic assessment of the aged hippocampus has revealed decreased expression of anti-oxidant proteins (e.g., peroxiredoxin 2 and 6) as well as oxido-reductase enzymes (cytochrome c oxidase subunits and ubiquinol-cytochrome core protein 1), suggesting an impairment of regulated mitochondrial energy production. Aberrant coupling of electron transfer via decreased expression of these proteins may not only impair energy production, but also increase mitochondrial stress load. Likely downstream effects of this include increased hippocampal oxidative stress and subsequent inflammation, both of which have been reported with normative aging as well as with age-related neurodegenerative diseases (Weinstock et al., 2009; Ownby, 2010). This is evident in upregulated expression of inflammation-response proteins GFAP and annexin A4 and decreased astrocytic phosphoprotein 15, an inhibitor of tumor necrosis factor receptor signaling in neuroproteomic studies of aging hippocampus. Together, dysregulation of these proteins suggests a mechanism of impaired anti-oxidant/anti-inflammatory activity and increased oxidative stress and inflammation response in the aged hippocampus.
Impaired hippocampal neuronal activity is a well-characterized effect of aging in humans, and often coincides with deficits of learning and memory (Daselaar et al., 2006; Dennis et al., 2008; Beeri et al., 2009). Age-related decrements in hippocampal activity and encoding of spatial field maps, demonstrated by ensemble recordings, also occur in aged rodents and correlate with impaired performance on behavioral tests of hippocampus-dependent spatial learning and memory (Norris et al., 1996; Barnes et al., 1997; Wilson et al., 2003). Impaired synaptic plasticity (i.e., long-term potentiation, long-term depression, paired-pulse facilitation) and deficits of synaptic transmission underlie these phenomena and have been reproducibly demonstrated in aged rodents compared to their younger counterparts (Norris et al., 1996; Boric et al., 2008; Sierra-Mercado et al., 2008; Gureviciene et al., 2009). Numerous neuroproteomic reports have identified decreased expression of proteins contributing to these processes with increasing age. For example, SNARE complex proteins (VAMP2, SNAP25, and syntaxin 1), vesicle-mobilizing proteins (synapsin 1, synaptophysin) and postsynaptic proteins (PSD95) are significantly decreased in hippocampal synapses of aged rats compared to mature adults, indicating a loss of effective neurotransmission-mediating machinery. Likewise, components of the synaptic vesicle cycle (amphiphysin, dynamin, septin, N-ethylmaleimide sensitive fusion protein) are decreased with age, which likely decreases vesicle recycling and synaptic efficacy. Mechanisms of cytoskeletal dynamics and neuronal signal transduction are also implicated in age-related hippocampal dysfunction by neuroproteomic findings demonstrating altered expression of regulatory proteins including coronin, spectrin, profilin, collapsin response mediator proteins, PEBP, tropomodulin, and syndapin, among others. Together, these results indicate a broad impairment of synaptic function with aging that likely leads to aberrant hippocampal activity and ultimately, to cognitive symptoms.
The Hippocampal Proteome with Learning and Memory
Given that age-related decrements in cognitive function occur heterogeneously in both human and animal model populations, and that cognitive decline occurs on a background of aging-related proteomic alterations, studies pairing behavioral assessments of cognitive function with quantitative proteomic analyses at multiple ages are needed. As the relationship between neurochemistry and behavior has moved to the forefront of neuroscience research, investigations of the effects of spatial learning and memory on the hippocampal proteome have been initiated (Table 1). The introduction of additional complexities of study design must be taken into consideration in these investigations to maximize the utility of resultant data. In addition to the intricacies of the proteomic techniques, clearly described animal model and behavioral testing methods are necessary; if these important details are lacking, interpretation of neuroproteomic datasets in a biological context is hindered, ultimately minimizing the impact of the findings. This is illustrated by a potentially influential report describing changes in protein–protein interactions induced by the formation of spatial memory (Nelson et al., 2004). Rats of undescribed strain, sex, and age were divided into three training groups (Morris Water Maze trained, yoked swim control, and naïve). Following training and demonstration of a spatially focused search pattern in the maze-trained animals, a complex combination of proteomic techniques was used to study differential protein interactions. Hippocampal protein samples were subjected to affinity-based enrichment of protein–protein complexes and depletion of non-interacting proteins. Identification of interacting proteins significantly regulated with spatial learning was performed by separation using 2DE, followed by LC–MS analysis. Of the nearly 250 proteins determined to participate in binding interactions, 32 were statistically increased in protein–protein interaction in the trained versus swim control group (e.g., stathmin, tropomyosin, tubulin, GFAP, spectrin, F-actin capping protein B) while eight decreased their interactions (e.g., actin, complexin 1). Seven proteins with differential binding between swim control and naïve groups were interpreted as indicators of water maze-induced stress. The majority of proteins with spatial memory-induced interaction alterations were classified as synaptic, structural, and signaling proteins. Five of the protein interactions determined to be regulated with spatial memory formation and for which a protein component was identified were selected for pull-down of binding partners. For this work, bait proteins were expressed in bacteria for capture of putative interacting proteins from hippocampal extracts The captured proteins were identified by LC–MS/MS and quantitated by immunoblotting, which demonstrated the following protein–protein interactions: complexin/p25α and complexin/drac1-like protein (decreased with training); CapZ/tubulin, adenylate kinase/actin, and ATP synthase/14-3-3 gamma (increased with training). Interestingly, these changes appeared to be specific to the protein–protein interactions themselves, as total expression of these individual species was not altered.
This study demonstrates the value of combining traditional biochemical and modern proteomic approaches with behavioral testing to provide information regarding neuronal protein–protein interactions that participate in spatial memory formation that could not have been obtained without prior knowledge of at least one binding partner in these interactions. The report, however, is rather technology-driven and lacks information regarding animal strain, age, sex, and behavioral training/testing, as well as a rationale for determining protein–protein interactions in bacteria instead of the hippocampal protein samples from these animals. Although significant confirmatory and control experiments are needed to validate the differential interactions reported with maze training in this study, future studies implementing this approach may provide further insight into the acute effects of spatial learning and memory on the hippocampal proteome and on specific protein–protein interactions that may affect cognitive function, particularly with increasing age.
Using a more traditional proteomic approach, Henninger et al. (2007) present a well-designed and highly informative study pairing a spatial learning task with 2DE-based quantitation of cytosolic proteins in the rat hippocampus. Forty young-adult male Wistar rats (mean age 3 months) were either trained to locate a hidden escape platform in the Morris Water Maze based on surrounding contextual cues, or were placed in the maze with no platform present to control for exercise and stress in the absence of spatial learning. Maze training was performed twice daily for 7 days, and all animals were sacrificed immediately following the last trial for 2DE quantitation of hydrophilic hippocampal proteins. Protein spots with significantly different expression between maze-trained and control groups were determined to be associated with spatial learning. Nearly 80 unique proteins, representing both training-induced increases and decreases, were identified by MS, and primarily represented metabolic, cytoskeletal, signal transduction, and degradation processes. Enolase, actin-interacting, and proteasome/lysosome subunits were among the subset of proteins increased after spatial learning, while numerous proteins related to synaptic function including NSF, synapsin 2, dynamin, and the 14-3-3 isoforms gamma and epsilon, were decreased. An added strength of this study is the inclusion of immunoblot confirmations to validate neuroproteomic data (i.e., protein identification and quantitation). As the authors point out, many proteins involved in the neuronal processes underlying learning and memory were absent from this analysis, likely because membrane-associated proteins were depleted by the isolation method used, and also because these low-abundance proteins fall below the detection threshold in traditional neuroproteomic techniques. One unfortunate limitation of this work is that proteins determined to be unchanged by spatial training were excluded from identification, which would have added to the growing coverage of the rodent hippocampal proteome and provided insight regarding proteins not necessarily critical for learning and memory.
In a well-designed study of plasticity-induced proteomic alterations, McNair et al. (2006) demonstrated both temporal and class-specific protein expression pattern changes in ex vivo hippocampal slices prepared from mice aged 4–8 weeks. Plasticity was simulated both electrophysiologically, using high-frequency stimulation of CA1–CA3 pyramidal cells to induce LTP, and pharmacologically, using glutamate-induced receptor activation. Both acute and chronic alterations in protein expression induced with these models of synaptic plasticity were examined by 2-DIGE and identified by MS and MS/MS. Four hours following glutamate treatment, long-lasting altered expression of 79 proteins was detected, 58 of which were sensitive to NMDA receptor blockade. Many of these proteins were related to glutamate receptor cycling, cytoskeleton regulation, and vesicle trafficking. For example, glutamate stimulation downregulated NSF and upregulated actin, which was confirmed by two-dimensional immunoblotting. More than 80% of hippocampal proteins identified were related to metabolism, cytoplasmic organization and biogenesis, and protein transport and modification. Electrophysiological LTP induction led to altered expression of energy metabolism-related proteins after 10 min, and of cytoplasmic organization- and biogenesis-related proteins after 240 min. LTP-mediated protein changes included calcineurin, VDAC1, MEK1, growth-associated protein 43, synapsin 2, syntaxin 19, and copine 6. Little overlap in proteins with altered expression was detected between the 10 and 240 min time points, although there were similarities between electrophysiological and pharmacological treatment groups. These findings indicate that, in ex vivo models of hippocampus-dependent learning and memory, distinct temporally regulated proteomic alterations are induced. These changes may represent neuromolecular processes underlying acquisition and retention of spatial memory.
Building on these results, a follow-up study was conducted to examine the hippocampal neuroproteome following environmental enrichment, which has been demonstrated to increase neuronal activity and induce spatial learning and memory formation, on the hippocampal neuroproteome (McNair et al., 2007). Eight-week old male hooded Lister rats were exposed to an open field (non-enriched) or enriched environment for six weeks, for a final age at sacrifice of 14 weeks (i.e., young-adult). The enriched regimen included toys, running wheels, and novel objects known to stimulate hippocampal plasticity and learning. Neural somata were segregated from their distal processes by dissecting CA1 stratum pyramidale and stratum radiate to allow examination of somatic and dendritic proteomic composition, respectively. Proteomic analyses were performed by 2-DIGE and highly sensitive quadrupole MS. Rats subjected to environmental enrichment exhibited altered expression of numerous proteins in both somatic and dendritic samples, with both inductions and reductions observed compared to non-enriched controls. In total, 50 proteins were identified regardless of expression, approximately 70% of which are involved in energy metabolism, signal transduction, and cytoplasmic organization and biogenesis. In stratum pyramidale, environmental enrichment increased expression of actin, dynamin, rab7, and proteasome subunits, and decreased expression of protein phosphatase 3, synapsin 1, PSD95, dynactin, collapsin response mediator protein 2, destrin, and ubiquitin. Interestingly, many proteins altered with enrichment were identified as housekeeping gene products, indicating that these species may play more complex roles in neuronal function than traditionally thought. In stratum radiate, exposure to the enriched environment was associated with increased phosphoglycerate mutase 1, ATP synthase subunits, VAMP-associated protein B, GFAP, and actin, while GFP58, GNBP alpha inhibiting protein 2, septin 5, cyclin-dependent kinase, and diacylglycerol kinase zeta were decreased. This study design did not control for cognitive or proteomic effects of increased exercise in rats exposed to the enriched environment versus the open field, but it demonstrates two important points. First, exposure to environmental enrichment alters components of the hippocampal proteome, some of which have been previously implicated in learning and memory while others are novel. Second, distinct proteomic changes are induced in CA1 neuronal processes versus cell bodies, indicating that fractionation approaches are necessary for detecting and differentiating these protein expression alterations. Although the findings of this study are quite valuable to the cognitive neuroscience field, additional work examining the effects of environmental manipulation and exercise mature adult and aged rats is needed.
Exercise is known to improve neuronal activity and cognitive function, and exerts neuroprotective effects in aging hippocampus (O’Callaghan et al., 2009; Gomes da Silva et al., 2010; Jedrziewski et al., 2010; Voss et al., 2010). The effects of exercise on the hippocampal neuroproteome has been evaluated in a noteworthy study of aged male Sprague Dawley rats reported by Chen et al. (2007a). Sedentary rats were compared voluntarily (running wheel) and involuntarily (treadmill) exercising rats after an 18-month exercise regimen started during early adulthood. At 23 months of age, the animals were sacrificed for 2DE-MS/MS analysis of the hippocampal neuroproteome. Surprisingly, of 74 signaling proteins identified, 15 species were regulated in expression with exercise, and only two of these (protein phosphatase 1 and guanine nucleotide-binding protein 1) were common to both voluntary and involuntary exercise compared to sedentary animals. The 15 differentially expressed proteins were represented in either involuntary exercise versus voluntary exercise or involuntary exercise versus sedentary rats, indicating a major effect of consistent treadmill running throughout adulthood. Interestingly, expression of signaling proteins such as PEBP, 14-3-3 zeta, septin 8, and the early endosome fusion protein EEA1 was significantly different between voluntarily and involuntarily exercising rats, suggesting that voluntary exercise impacts processes underlying learning and memory to a greater extent than involuntary exercise. In agreement with recent findings that exercise improves hippocampus-dependent spatial learning and memory, many of the exercise-regulated proteins identified here have been associated with cognitive function. Additional work pursuing exercise-mediated regulation of memory associated proteins with aging will no doubt increase our understanding of the role of exercise in improved cognition throughout the lifespan.
As illustrated by the comparison of voluntary and involuntary exercise above, implementation of neuroproteomic approaches in behavioral neuroscience research has led to the identification of task-dependent regulation of specific proteins. Differential effects of spatial learning paradigms have been demonstrated in young-adult (10–14 weeks) wild-type mice trained in common behavioral tests of hippocampus-dependent cognitive function (Zheng et al., 2009). In the Barnes Maze, mice were trained to locate a dark escape chamber hidden below one of 20 holes in a brightly lit circular platform based on fixed visual cues. Mice trained in the Multiple T-Maze learned to locate a goal box containing a food pellet in a maze designed with seven choice points. For both paradigms, yoked controls were included to minimize confounding effects of stress or food deprivation. Following a probe trial to ensure that trained mice had successfully acquired and retained the assigned spatial task, hippocampal proteomic composition was evaluated by 2DE with nano-LC–MS identification of proteins of interest. Several differentially expressed proteins were identified between mice exposed to the different training paradigms. Although most of these proteins are associated with metabolism and have not been previously linked to synaptic mechanisms of learning and memory, several proteins, including dual-specificity protein phosphatase 3, calmodulin, and NSF, are implicated in synaptic signaling, demonstrating the importance of synaptic function and maintenance in cognitive function. The maze-dependent specificity of proteomic profiles described here emphasizes the difficulty of comparing behavioral studies and even between identical rodent strains, and the need for careful reporting of testing paradigms in neuroproteomic evaluations in cognitive neuroscience research. As with age-based neuroproteomic investigations, comparing studies of the hippocampal neuroproteome with models of learning and memory formation is made difficult by study-to-study variation in study design. This is particularly true with regard to the behavioral testing methods used, potential confounding variables such as stress, food deprivation and exercise, and the age and species of study subjects.
It is interesting to note that several proteins implicated in hippocampus-dependent learning and memory by neuroproteomic investigations are differentially expressed with aging. For example, 14-3-3 signaling proteins, which decrease in expression with increasing age (VanGuilder et al., 2010), are regulated at the levels of protein content and protein–protein interaction following spatial training and exercise (Nelson et al., 2004; Chen et al., 2007a; Henninger et al., 2007). Similarly, members of the family of dihydropyrimidinase-related proteins (also known as collapsin response mediator proteins) are regulated both with aging (Carrette et al., 2006; Focking et al., 2006; VanGuilder et al., 2010) and in models of learning and memory formation (McNair et al., 2006, 2007; Henninger et al., 2007; Zheng et al., 2009). Dihydropyrimidinase-related proteins contribute to signal transduction and regulation of microtubule dynamics and cytoskeletal remodeling (Charrier et al., 2003; Brittain et al., 2009). Lastly, regulation of protein phosphatases and PEBP (also called hippocampal cholinergic neurostimulating peptide) has been identified in the aged hippocampal neuroproteome (Weinreb et al., 2007; Freeman et al., 2009; VanGuilder et al., 2010). These proteins have been previously implicated in learning and memory, and are among the proteins changed in expression following exercise, environmental enrichment, and training in spatial learning tasks (Chen et al., 2007a; McNair et al., 2007; Zheng et al., 2009), suggesting that age-related dysregulation of these proteins may contribute to deficits of spatial learning and memory.
The Hippocampal Neuroproteome with Age-Related Cognitive Decline
An important aspect of age-related cognitive impairment that is often overlooked in molecular analyses is the heterogeneity of its incidence and severity in both humans and rodent models of aging. A number of studies have established that declining spatial cognitive function affects a subset of aged individuals, while counterparts matched for age and health status retain cognitive abilities similar to younger adults (Schaie, 1996; Gallagher and Rapp, 1997; Schonknecht et al., 2005; Freeman et al., 2009; Mungas et al., 2010). Investigation of proteins (i.e., the functional products of gene transcripts) has been limited to targeted studies of specific pre-selected proteins or sets of proteins. Although valuable to our understanding of the potential functional roles and regulation of these proteins in age-related cognitive decline, larger-scale approaches are needed to provide broad quantitative assessment of the hippocampal neuroproteome since cognitive decline is a complex phenomenon comprising both age-related and cognition-specific processes that are not entirely understood. Whole-genome profiling approaches (e.g., high-density microarrays) investigating the effects of aging on cognitive function and hippocampal gene expression have identified novel subsets of genes associated with declining cognitive performance in aged animals (Blalock et al., 2003) as well as cognitive decline-specific genes altered between aged, cognitively impaired and aged unimpaired rats (Rowe et al., 2007). Bioinformatic analysis of these large datasets places findings within a biological context of known processes, pathways, and functional networks, enabling the identification of dysregulated cellular functions not previously associated with cognitive decline. To assess this phenomenon at the neuroproteomic level, we performed 2-DIGE quantitation and MS/MS identification of unfractionated hippocampal protein isolated from behaviorally characterized male Fischer 344 × Brown Norway rats (Freeman et al., 2009). Adult (10 months) and aged (27 months) were assessed for spatial cognitive performance in the Morris Water Maze, and aged animals were categorized as cognitively intact (performing similarly to adults) or impaired (performing outside the range of adults) prior to sacrifice and proteomic assessment. A principal components analysis (PCA) of the resulting 2-DIGE expression data revealed that two distinct shifts occur in the hippocampal proteome: one with age, which accounted for approximately 70% of proteomic variation between groups, and one specific to cognitive status. The latter component comprised 30% of study variance, and segregated aged cognitively impaired animals from both the adult and aged cognitively intact groups. In total, nearly 200 hippocampal proteins were confidently identified and included a broad range including enzymes and proteasome subunits as well as species implicated in learning and memory such as protein phosphatase 1E and PEBP. Interestingly, of the 19 identified proteins regulated with aging (aged versus adult) and nine identified proteins regulated specifically with cognitive decline (aged impaired versus aged intact) only enolase was common to both conditions. Numerous proteins involved in carbohydrate, protein, coenzyme, and nucleic acid metabolism were regulated with age, while regulators of protein quality control such as puromycin-sensitive aminopeptidase and heat shock proteins were altered with cognitive decline. This work identified an age-related dysregulation of multiple enzymes that mediate the ATP-generating steps of glycolysis, and more importantly, demonstrated that two distinct hippocampal proteomic shifts occur with age-related declines in spatial learning and memory that cannot be examined without performance-based stratification of aged animals into cognitively intact and impaired groups. Additional studies, including a more comprehensive range of time points and proteomic fractionation techniques, need to be performed to identify whether segregation of the hippocampal proteome occurs prior to the development of spatial learning and memory impairments, and when cognitive deficits and proteomic alterations first manifest.
Future Work
The work described here has contributed valuable understanding of the neuroproteome to the neuroscience and aging fields, and marks an important transition from targeted, single molecule studies to more systems-level evaluations of neuroproteomic profiles. A number of excellent reports describing alterations in hippocampal protein expression, as assessed by focused study of targets of interest, suggest that aging and cognitive decline are associated with altered synaptic, metabolic, and signaling functions (Shimohama et al., 1998; Baxter et al., 1999; Smith et al., 2000; Calabrese et al., 2004; Hwang et al., 2006; Nyffeler et al., 2007; Shi et al., 2007; Adams et al., 2008; Liu et al., 2008; Canas et al., 2009; Head et al., 2009). Neuroproteomic analyses of age-related cognitive decline demonstrate that brain aging involves dysregulation of metabolism, oxidative stress, protein trafficking, and synaptic function, and that many of these processes are also regulated with cognitive decline. Additionally, although increasing age is a primary risk factor for cognitive decline, the occurrence of cognitive impairment with aging is heterogeneous and likely reflects distinct neuroproteomic alterations. Growing evidence suggests that cognitive decline does not represent a “more aged” phenotype, but rather is associated with specific changes that occur in addition to age-related alterations. The current data are far from complete, however, as the processes specifically necessary and sufficient for age-related cognitive decline are not yet evident from existing neuroproteomic studies of the aging hippocampus (Table 1). Future work addressing cognition-specific neuroproteomic alterations in aging studies is needed to identify novel processes contributing to the cognitive decline phenotype. While a multitude of neuroproteomic reports demonstrate dysregulation of protein expression in human and rodent brain with age-related neurodegenerative conditions such as mild cognitive impairment and Alzheimer’s disease (Schonberger et al., 2001; Vercauteren et al., 2004; Butterfield and Poon, 2005; Sultana et al., 2007; Hartl et al., 2008; Martin et al., 2008; Wang et al., 2008; Owen et al., 2009), as well in transgenic models of accelerated aging (Butterfield and Poon, 2005; Diez-Vives et al., 2009; Perluigi et al., 2010), profiling efforts with normal aging and age-related alterations in cognitive status in mammalian model organisms remain relatively lacking while studies of human tissue are virtually nonexistent.
Conclusion
A major next step to addressing this incomplete data would be an aging research community-wide effort to comprehensively profile the neuroproteome in relevant brain regions of the most common rodent models across adulthood and advanced age. While laudable, the current distributed efforts suffer from disparate animal models and variations in technical methods that make reaching the necessary consensus difficult. In such an aging brain atlas effort, behavior phenotyping of the animals would also allow determination of those changes with age that correlate to cognitive differences and those that do not. Understanding the cumulative effects of aging- and cognition-related changes in the brain remains a major unanswered question in the field. While age-related changes in the neuroproteome likely play necessary roles in cognitive decline they are also clearly not sufficient to impair cognitive function as only a subset of aging individuals experience cognitive deficits. Determining which age-related changes in the neuroproteome play a causative role in cognitive decline and which are epiphenomenon is critical to understanding the heterogeneous presentation of cognitive decline in humans and rodent models. Nonetheless, a clear finding from the proteomic studies reviewed here and the transcriptomic efforts detailed elsewhere (Blalock et al., 2003; Pawlowski et al., 2009; Blalock et al., 2010) is that behavioral characterization of animals is critical to identifying cognition-specific changes which are otherwise disguised without behavioral phenotyping.
With the continued advances in proteomic technology, the study of the neuroproteome with aging and cognitive decline is entering a new phase of discovery. The full potential of this discovery work will be realized through the combining new technical approaches that allow broader analysis of the neuroproteome with systems biology-level data analysis, confirmation experiments, and localization of regulated targets to specific cell types. Additionally, carefully designed and well-executed animal experiments addressing both aging and cognitive decline are needed. While study design and reporting standards such as the Minimum Information About a Proteomic Experiment (MIAPE) guidelines (Taylor et al., 2007) have been developed and are increasingly adopted in the proteomic field, behavioral neuroscience research currently lacks clear standards. A limitation of some of the reports discussed here is that details regarding animal ages, care, and behavior assessments/measures are limited, which impedes cross-study comparison and meta-analyses.
The neurobiology of aging field is a pivotal stage where translation of basic research findings to humans is urgently needed. Even modest gains in maintaining cognitive function with aging would have major societal benefits. With the pressing demographic and economic challenges caused by an aging population, continued and concerted efforts in basic and applied research must be made to understand the biology of brain aging and cognitive decline.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge funding support by the NIH National Institute on Aging (grant # R01AG026607). Special thanks to Lois Sanborn, Colleen Van Kirk, and William Sonntag for valuable editorial guidance. The authors have no conflicts or competing interests to declare.
References
Adams, M. M., Shi, L., Linville, M. C., Forbes, M. E., Long, A. B., Bennett, C., Newton, I. G., Carter, C. S., Sonntag, W. E., Riddle, D. R., and Brunso-Bechtold, J. K. (2008). Caloric restriction and age affect synaptic proteins in hippocampal CA3 and spatial learning ability. Exp. Neurol. 211, 141–149.
Barnes, C. A., Suster, M. S., Shen, J., and McNaughton, B. L. (1997). Multistability of cognitive maps in the hippocampus of old rats. Nature 388, 272–275.
Baxter, M. G., Frick, K. M., Price, D. L., Breckler, S. J., Markowska, A. L., and Gorman, L. K. (1999). Presynaptic markers of cholinergic function in the rat brain: relationship with age and cognitive status. Neuroscience 89, 771–779.
Beeri, M. S., Lee, H., Cheng, H., Wollman, D., Silverman, J. M., and Prohovnik, I. (2009). Memory activation in healthy nonagenarians. Neurobiol. Aging 32, 515–523.
Blalock, E. M., Chen, K. C., Sharrow, K., Herman, J. P., Porter, N. M., Foster, T. C., and Landfield, P. W. (2003). Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 23, 3807–3819.
Blalock, E. M., Grondin, R., Chen, K. C., Thibault, O., Thibault, V., Pandya, J. D., Dowling, A., Zhang, Z., Sullivan, P., Porter, N. M., and Landfield, P. W. (2010). Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J. Neurosci. 30, 6058–6071.
Boric, K., Munoz, P., Gallagher, M., and Kirkwood, A. (2008). Potential adaptive function for altered long-term potentiation mechanisms in aging hippocampus. J. Neurosci. 28, 8034–8039.
Brittain, J. M., Piekarz, A. D., Wang, Y., Kondo, T., Cummins, T. R., and Khanna, R. (2009). An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. J. Biol. Chem. 284, 31375–31390.
Burke, S. N., and Barnes, C. A. (2006). Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 7, 30–40.
Butterfield, D. A., and Poon, H. F. (2005). The senescence-accelerated prone mouse (SAMP8): a model of age-related cognitive decline with relevance to alterations of the gene expression and protein abnormalities in Alzheimer’s disease. Exp. Gerontol. 40, 774–783.
Calabrese, V., Scapagnini, G., Ravagna, A., Colombrita, C., Spadaro, F., Butterfield, D. A., and Giuffrida Stella, A. M. (2004). Increased expression of heat shock proteins in rat brain during aging: relationship with mitochondrial function and glutathione redox state. Mech. Ageing Dev. 125, 325–335.
Canas, P. M., Duarte, J. M., Rodrigues, R. J., Kofalvi, A., and Cunha, R. A. (2009). Modification upon aging of the density of presynaptic modulation systems in the hippocampus. Neurobiol. Aging 30, 1877–1884.
Carrette, O., Burkhard, P. R., Hochstrasser, D. F., and Sanchez, J. C. (2006). Age-related proteome analysis of the mouse brain: a 2-DE study. Proteomics 6, 4940–4949.
CensusScope (2010). United States Age Distribution. Available at: http://www.censusscope org/us/chart_age.html
Charrier, E., Reibel, S., Rogemond, V., Aguera, M., Thomasset, N., and Honnorat, J. (2003). Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol. Neurobiol. 28, 51–64.
Chen, W., Ji, J., Xu, X., He, S., and Ru, B. (2003). Proteomic comparison between human young and old brains by two-dimensional gel electrophoresis and identification of proteins. Int. J. Dev. Neurosci. 21, 209–216.
Chen, W. Q., Viidik, A., Skalicky, M., Hoger, H., and Lubec, G. (2007a). Hippocampal signaling cascades are modulated in voluntary and treadmill exercise rats. Electrophoresis 28, 4392–4400.
Chen, X., Sun, L., Yu, Y., Xue, Y., and Yang, P. (2007b). Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics 4, 25–37.
Corradi, A., Zanardi, A., Giacomini, C., Onofri, F., Valtorta, F., Zoli, M., and Benfenati, F. (2008). Synapsin-I- and synapsin-II-null mice display an increased age-dependent cognitive impairment. J. Cell Sci. 121, 3042–3051.
Crowe, M., Sartori, A., Clay, O. J., Wadley, V. G., Andel, R., Wang, H. X., Sawyer, P., and Allman, R. M. (2010). Diabetes and cognitive decline: investigating the potential influence of factors related to health disparities. J. Aging Health 22, 292–306.
Dahle, C. L., Jacobs, B. S., and Raz, N. (2009). Aging, vascular risk, and cognition: blood glucose, pulse pressure, and cognitive performance in healthy adults. Psychol. Aging 24, 154–162.
Daselaar, S. M., Fleck, M. S., Dobbins, I. G., Madden, D. J., and Cabeza, R. (2006). Effects of healthy aging on hippocampal and rhinal memory functions: an event-related fMRI study. Cereb. Cortex 16, 1771–1782.
Deak, F., Shin, O. H., Kavalali, E. T., and Sudhof, T. C. (2006). Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J. Neurosci. 26, 6668–6676.
Dennis, N. A., Hayes, S. M., Prince, S. E., Madden, D. J., Huettel, S. A., and Cabeza, R. (2008). Effects of aging on the neural correlates of successful item and source memory encoding. J. Exp. Psychol. Learn. Mem. Cogn. 34, 791–808.
Diez-Vives, C., Gay, M., Garcia-Matas, S., Comellas, F., Carrascal, M., Abian, J., Ortega-Aznar, A., Cristofol, R., and Sanfeliu, C. (2009). Proteomic study of neuron and astrocyte cultures from senescence-accelerated mouse SAMP8 reveals degenerative changes. J. Neurochem. 111, 945–955.
Ding, J., Strachan, M. W., Reynolds, R. M., Frier, B. M., Deary, I. J., Fowkes, F. G., Lee, A. J., McKnight, J., Halpin, P., Swa, K., and Price, J. F. (2010). Diabetic retinopathy and cognitive decline in older people with type 2 diabetes: the Edinburgh type 2 diabetes study. Diabetes 59, 2883–2889.
Dosemeci, A., Tao-Cheng, J. H., Vinade, L., and Jaffe, H. (2006). Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem. Biophys. Res. Commun. 339, 687–694.
Drabik, A., Bierczynska-Krzysik, A., Bodzon-Kulakowska, A., Suder, P., Kotlinska, J., and Silberring, J. (2007). Proteomics in neurosciences. Mass Spectrom. Rev. 26, 432–450.
Edgar, P. F., Douglas, J. E., Knight, C., Cooper, G. J., Faull, R. L., and Kydd, R. (1999). Proteome map of the human hippocampus. Hippocampus 9, 644–650.
Erickson, K. I., Prakash, R. S., Voss, M. W., Chaddock, L., Heo, S., McLaren, M., Pence, B. D., Martin, S. A., Vieira, V. J., Woods, J. A., McAuley, E., and Kramer, A. F. (2010). Brain-derived neurotrophic factor is associated with age-related decline in hippocampal volume. J. Neurosci. 30, 5368–5375.
Feldmann, R. E. Jr., Maurer, M. H., Hunzinger, C., Lewicka, S., Buergers, H. F., Kalenka, A., Hinkelbein, J., Broemme, J. O., Seidler, G. H., Martin, E., and Plaschke, K. (2008). Reduction in rat phosphatidylethanolamine binding protein-1 (PEBP1) after chronic corticosterone treatment may be paralleled by cognitive impairment: a first study. Stress 11, 134–147.
Fernandez, E., Collins, M. O., Uren, R. T., Kopanitsa, M. V., Komiyama, N. H., Croning, M. D., Zografos, L., Armstrong, J. D., Choudhary, J. S., and Grant, S. G. (2009). Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5, 269.
Focking, M., Boersema, P. J., O’Donoghue, N., Lubec, G., Pennington, S. R., Cotter, D. R., and Dunn, M. J. (2006). 2-D DIGE as a quantitative tool for investigating the HUPO Brain Proteome Project mouse series. Proteomics 6, 4914–4931.
Fountoulakis, M., Schuller, E., Hardmeier, R., Berndt, P., and Lubec, G. (1999). Rat brain proteins: two-dimensional protein database and variations in the expression level. Electrophoresis 20, 3572–3579.
Fountoulakis, M., Tsangaris, G. T., Maris, A., and Lubec, G. (2005). The rat brain hippocampus proteome. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 819, 115–129.
Freeman, W. M., and Hemby, S. E. (2004). Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 29, 1065–1081.
Freeman, W. M., Vanguilder, H. D., Bennett, C., and Sonntag, W. E. (2009). Cognitive performance and age-related changes in the hippocampal proteome. Neuroscience 159, 183–195.
Friedman, D. B., Hoving, S., and Westermeier, R. (2009). Isoelectric focusing and two-dimensional gel electrophoresis. Meth. Enzymol. 463, 515–540.
Gallagher, M., and Rapp, P. R. (1997). The use of animal models to study the effects of aging on cognition. Annu. Rev. Psychol. 48, 339–370.
Geinisman, Y., de Toledo-Morrell, L., and Morrell, F. (1986). Loss of perforated synapses in the dentate gyrus: morphological substrate of memory deficit in aged rats. Proc. Natl. Acad. Sci. U.S.A 83, 3027–3031.
Gomes da Silva, S., Unsain, N., Masco, D. H., Toscano-Silva, M., de Amorim, H. A., Silva Araujo, B. H., Simoes, P. S., da Graça Naffah-Mazzacoratti, M., Mortara, R. A., Scorza, F. A., Cavalheiro, E. A., and Arida, R. M. (2010). Early exercise promotes positive hippocampal plasticity and improves spatial memory in the adult life of rats. Hippocampus, 21, doi: 10.1002/hipo.20903. [Epub ahead of print].
Gureviciene, I., Gurevicius, K., and Tanila, H. (2009). Aging and alpha-synuclein affect synaptic plasticity in the dentate gyrus. J. Neural Transm. 116, 13–22.
Hartl, D., Rohe, M., Mao, L., Staufenbiel, M., Zabel, C., and Klose, J. (2008). Impairment of adolescent hippocampal plasticity in a mouse model for Alzheimer’s disease precedes disease phenotype. PLoS One 3, e2759. doi: 10.1371/journal.pone.0002759
Head, E., Corrada, M. M., Kahle-Wrobleski, K., Kim, R. C., Sarsoza, F., Goodus, M., and Kawas, C. H. (2009). Synaptic proteins, neuropathology and cognitive status in the oldest-old. Neurobiol. Aging 30, 1125–1134.
Hedden, T., and Gabrieli, J. D. (2004). Insights into the ageing mind: a view from cognitive neuroscience. Nat. Rev. Neurosci. 5, 87–96.
Henninger, N., Feldmann, R. E. Jr., Futterer, C. D., Schrempp, C., Maurer, M. H., Waschke, K. F., Kuschinsky, W., and Schwab, S. (2007). Spatial learning induces predominant downregulation of cytosolic proteins in the rat hippocampus. Genes Brain Behav. 6, 128–140.
Hwang, I. K., Yoo, K. Y., Jung, B. K., Cho, J. H., Kim, D. H., Kang, T. C., Kwon, Y. G., Kim, Y. S., and Won, M. H. (2006). Correlations between neuronal loss, decrease of memory, and decrease expression of brain-derived neurotrophic factor in the gerbil hippocampus during normal aging. Exp. Neurol. 201, 75–83.
Jedrziewski, M. K., Ewbank, D. C., Wang, H., and Trojanowski, J. Q. (2010). Exercise and cognition: results from the National Long Term Care Survey. Alzheimers Dement. 6, 448–455.
Jiang, L., Fang, J., Moore, D. S., Gogichaeva, N. V., Galeva, N. A., Michaelis, M. L., and Zaidi, A. (2008). Age-associated changes in synaptic lipid raft proteins revealed by two-dimensional fluorescence difference gel electrophoresis. Neurobiol. Aging 3, 2146–2159.
Kantarci, K., Senjem, M. L., Lowe, V. J., Wiste, H. J., Weigand, S. D., Kemp, B. J., Frank, A. R., Shiung, M. M., Boeve, B. F., Knopman, D. S., Petersen, R. C., and Jack, C. R. Jr. (2010). Effects of age on the glucose metabolic changes in mild cognitive impairment. AJNR Am. J. Neuroradiol. 31, 1247–1253.
Keller, J. N., Gee, J., and Ding, Q. (2002). The proteasome in brain aging. Ageing Res. Rev. 1, 279–293.
Kumar, A., Thinschmidt, J. S., Foster, T. C., and King, M. A. (2007). Aging effects on the limits and stability of long-term synaptic potentiation and depression in rat hippocampal area CA1. J. Neurophysiol. 98, 594–601.
Liu, P., Smith, P. F., and Darlington, C. L. (2008). Glutamate receptor subunits expression in memory-associated brain structures: regional variations and effects of aging. Synapse 62, 834–841.
Lull, M. E., Freeman, W. M., Vanguilder, H. D., and Vrana, K. E. (2010). The use of neuroproteomics in drug abuse research. Drug Alcohol Depend. 107, 11–22.
Martin, B., Brenneman, R., Becker, K. G., Gucek, M., Cole, R. N., and Maudsley, S. (2008). iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer’s mice: understanding the interface between physiology and disease. PLoS One 3, e2750. doi: 10.1371/journal.pone.0002750
McNair, K., Broad, J., Riedel, G., Davies, C. H., and Cobb, S. R. (2007). Global changes in the hippocampal proteome following exposure to an enriched environment. Neuroscience 145, 413–422.
McNair, K., Davies, C. H., and Cobb, S. R. (2006). Plasticity-related regulation of the hippocampal proteome. Eur. J. Neurosci. 23, 575–580.
Mungas, D., Beckett, L., Harvey, D., Tomaszewski, F. S., Reed, B., Carmichael, O., Olichney, J., Miller, J., and Decarli, C. (2010). Heterogeneity of cognitive trajectories in diverse older persons. Psychol. Aging 25, 606–619.
Nelson, T. J., Backlund, P. S. Jr., and Alkon, D. L. (2004). Hippocampal protein-protein interactions in spatial memory. Hippocampus 14, 46–57.
Nielsen, P. A., Olsen, J. V., Podtelejnikov, A. V., Andersen, J. R., Mann, M., and Wisniewski, J. R. (2005). Proteomic mapping of brain plasma membrane proteins. Mol. Cell Proteomics 4, 402–408.
Norris, C. M., Korol, D. L., and Foster, T. C. (1996). Increased susceptibility to induction of long-term depression and long-term potentiation reversal during aging. J. Neurosci. 16, 5382–5392.
Nyffeler, M., Zhang, W. N., Feldon, J., and Knuesel, I. (2007). Differential expression of PSD proteins in age-related spatial learning impairments. Neurobiol. Aging 28, 143–155.
O’Callaghan, R. M., Griffin, E. W., and Kelly, A. M. (2009). Long-term treadmill exposure protects against age-related neurodegenerative change in the rat hippocampus. Hippocampus 19, 1019–1029.
Okonkwo, O. C., Cohen, R. A., Gunstad, J., Tremont, G., Alosco, M. L., and Poppas, A. (2010). Longitudinal trajectories of cognitive decline among older adults with cardiovascular disease. Cerebrovasc. Dis. 30, 362–373.
Owen, J. B., Di, D. F., Sultana, R., Perluigi, M., Cini, C., Pierce, W. M., and Butterfield, D. A. (2009). Proteomics-determined differences in the concanavalin-A-fractionated proteome of hippocampus and inferior parietal lobule in subjects with Alzheimer’s disease and mild cognitive impairment: implications for progression of AD. J. Proteome Res. 8, 471–482.
Panza, F., Capurso, C., D’Introno, A., Colacicco, A. M., Frisardi, V., Santamato, A., Ranieri, M., Fiore, P., Vendemiale, G., Seripa, D., Pilotto, A., Capurso, A., and Solfrizzi, V. (2008). Vascular risk factors, alcohol intake, and cognitive decline. J. Nutr. Health Aging 12, 376–381.
Pawlowski, T. L., Bellush, L. L., Wright, A. W., Walker, J. P., Colvin, R. A., and Huentelman, M. J. (2009). Hippocampal gene expression changes during age-related cognitive decline. Brain Res. 1256, 101–110.
Paz Gavilan, M., Vela, J., Castano, A., Ramos, B., del Rio, J. C., Vitorica, J., and Ruano, D. (2006). Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol. Aging 27, 973–982.
Perluigi, M., Di, D. F., Giorgi, A., Schinina, M. E., Coccia, R., Cini, C., Bellia, F., Cambria, M. T., Cornelius, C., Butterfield, D. A., and Calabrese, V. (2010). Redox proteomics in aging rat brain: involvement of mitochondrial reduced glutathione status and mitochondrial protein oxidation in the aging process. J. Neurosci. Res. 88, 3498–3507.
Poon, H. F., Shepherd, H. M., Reed, T. T., Calabrese, V., Stella, A. M., Pennisi, G., Cai, J., Pierce, W. M., Klein, J. B., and Butterfield, D. A. (2006). Proteomics analysis provides insight into caloric restriction mediated oxidation and expression of brain proteins associated with age-related impaired cellular processes: Mitochondrial dysfunction, glutamate dysregulation and impaired protein synthesis. Neurobiol. Aging 27, 1020–1034.
Qiu, C., Winblad, B., and Fratiglioni, L. (2005). The age-dependent relation of blood pressure to cognitive function and dementia. Lancet Neurol. 4, 487–499.
Quintana, C., and Gutierrez, L. (2010). Could a dysfunction of ferritin be a determinant factor in the aetiology of some neurodegenerative diseases? Biochim. Biophys. Acta 1800, 770–782.
Ramos, J. W., Townsend, D. A., Piarulli, D., Kolata, S., Light, K., Hale, G., and Matzel, L. D. (2009). Deletion of PEA-15 in mice is associated with specific impairments of spatial learning abilities. BMC Neurosci. 10, 134.
Rosenzweig, E. S., and Barnes, C. A. (2003). Impact of aging on hippocampal function: plasticity, network dynamics, and cognition. Prog. Neurobiol. 69, 143–179.
Rowe, W. B., Blalock, E. M., Chen, K. C., Kadish, I., Wang, D., Barrett, J. E., Thibault, O., Porter, N. M., Rose, G. M., and Landfield, P. W. (2007). Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J. Neurosci. 27, 3098–3110.
Sato, Y., Yamanaka, H., Toda, T., Shinohara, Y., and Endo, T. (2005). Comparison of hippocampal synaptosome proteins in young-adult and aged rats. Neurosci. Lett. 382, 22–26.
Schaie, K. W. (1996). Intellectual Development in Adulthood: The Seattle Longitudinal Study. Cambridge: Cambridge University Press.
Schiavo, G., Santucci, A., Dasgupta, B. R., Mehta, P. P., Jontes, J., Benfenati, F., Wilson, M. C., and Montecucco, C. (1993). Botulinum neurotoxins serotypes A and E cleave SNAP-25 at distinct COOH-terminal peptide bonds. FEBS Lett. 335, 99–103.
Schonberger, S. J., Edgar, P. F., Kydd, R., Faull, R. L., and Cooper, G. J. (2001). Proteomic analysis of the brain in Alzheimer’s disease: molecular phenotype of a complex disease process. Proteomics 1, 1519–1528.
Schonknecht, P., Pantel, J., Kruse, A., and Schroder, J. (2005). Prevalence and natural course of aging-associated cognitive decline in a population-based sample of young-old subjects. Am. J. Psychiatry 162, 2071–2077.
Shi, L., Adams, M. M., Linville, M. C., Newton, I. G., Forbes, M. E., Long, A. B., Riddle, D. R., and Brunso-Bechtold, J. K. (2007). Caloric restriction eliminates the aging-related decline in NMDA and AMPA receptor subunits in the rat hippocampus and induces homeostasis. Exp. Neurol. 206, 70–79.
Shi, L., Linville, M. C., Tucker, E. W., Sonntag, W. E., and Brunso-Bechtold, J. K. (2005). Differential effects of aging and insulin-like growth factor-1 on synapses in CA1 of rat hippocampus. Cereb. Cortex 15, 571–577.
Shimohama, S., Fujimoto, S., Sumida, Y., Akagawa, K., Shirao, T., Matsuoka, Y., and Taniguchi, T. (1998). Differential expression of rat brain synaptic proteins in development and aging. Biochem. Biophys. Res. Commun. 251, 394–398.
Shrestha, L. (2006). The Changing Demographic Profile of the United States; CRS Report for Congress. Available at: http://www.fas.org/sgp/crs/misc/RL32701.pdf
Sierra-Mercado, D., Dieguez, D. Jr., and Barea-Rodriguez, E. J. (2008). Brief novelty exposure facilitates dentate gyrus LTP in aged rats. Hippocampus 18, 835–843.
Smith, A. C., Gerrard, J. L., Barnes, C. A., and McNaughton, B. L. (2000). Effect of age on burst firing characteristics of rat hippocampal pyramidal cells. Neuroreport 11, 3865–3871.
Sonntag, W. E., Bennett, C., Ingram, R., Donahue, A., Ingraham, J., Chen, H., Moore, T., Brunso-Bechtold, J. K., and Riddle, D. (2006). Growth hormone and IGF-I modulate local cerebral glucose utilization and ATP levels in a model of adult-onset growth hormone deficiency. Am. J. Physiol. Endocrinol. Metab. 291, E604–E610.
Sonntag, W. E., Lynch, C. D., Bennett, S. A., Khan, A. S., Thornton, P. L., Cooney, P. T., Ingram, R. L., McShane, T., and Brunso-Bechtold, J. K. (1999). Alterations in insulin-like growth factor-1 gene and protein expression and type 1 insulin-like growth factor receptors in the brains of ageing rats. Neuroscience 88, 269–279.
Squier, T. C. (2001). Oxidative stress and protein aggregation during biological aging. Exp. Gerontol. 36, 1539–1550.
Sultana, R., Boyd-Kimball, D., Cai, J., Pierce, W. M., Klein, J. B., Merchant, M., and Butterfield, D. A. (2007). Proteomics analysis of the Alzheimer’s disease hippocampal proteome. J. Alzheimers. Dis. 11, 153–164.
Tack, W., Wree, A., and Schleicher, A. (1989). Local cerebral glucose utilization in the hippocampus of old rats. Histochemistry 92, 413–419.
Taylor, C. F., Paton, N. W., Lilley, K. S., Binz, P. A., Julian, R. K. Jr., Jones, A. R., Zhu, W., Apweiler, R., Aebersold, R., Deutsch, E. W., Dunn, M. J., Heck, A. J., Leitner, A., Macht, M., Mann, M., Martens, L., Neubert, T. A., Patterson, S. D., Ping, P., Seymour, S. L., Souda, P., Tsugita, A., Vandekerckhove, J., Vondriska, T. M., Whitelegge, J. P., Wilkins, M. R., Xenarios, I., Yates, J. R. III, and Hermjakob, H. (2007). The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893.
Trinidad, J. C., Thalhammer, A., Specht, C. G., Lynn, A. J., Baker, P. R., Schoepfer, R., and Burlingame, A. L. (2008). Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell Proteomics 7, 684–696.
VanGuilder, H. D., Yan, H., Farley, J. A., Sonntag, W. E., and Freeman, W. M. (2010). Aging alters the expression of neurotransmission-regulating proteins in the hippocampal synaptoproteome. J. Neurochem. 113, 1577–1588.
Vercauteren, F. G., Clerens, S., Roy, L., Hamel, N., Arckens, L., Vandesande, F., Alhonen, L., Janne, J., Szyf, M., and Cuello, A. C. (2004). Early dysregulation of hippocampal proteins in transgenic rats with Alzheimer’s disease-linked mutations in amyloid precursor protein and presenilin 1. Brain Res. Mol. Brain Res. 132, 241–259.
Voss, M. W., Prakash, R. S., Erickson, K. I., Basak, C., Chaddock, L., Kim, J. S., Alves, H., Heo, S., Szabo, A. N., White, S. M., Wojcicki, T. R., Mailey, E. L., Gothe, N., Olson, E. A., McAuley, E., and Kramer, A. F. (2010). Plasticity of brain networks in a randomized intervention trial of exercise training in older adults. Front. Aging Neurosci. 2, 32. doi: 10.3389/fnagi.2010.00032
Wang, Q., Liu, Y., Zou, X., Wang, Q., An, M., Guan, X., He, J., Tong, Y., and Ji, J. (2008). The hippocampal proteomic analysis of senescence-accelerated mouse: implications of Uchl3 and mitofilin in cognitive disorder and mitochondria dysfunction in SAMP8. Neurochem. Res. 33, 1776–1782.
Weinreb, O., Drigues, N., Sagi, Y., Reznick, A. Z., Amit, T., and Youdim, M. B. (2007). The application of proteomics and genomics to the study of age-related neurodegeneration and neuroprotection. Antioxid. Redox Signal. 9, 169–179.
Weinstock, M., Luques, L., Poltyrev, T., Bejar, C., and Shoham, S. (2009). Ladostigil prevents age-related glial activation and spatial memory deficits in rats. Neurobiol. Aging 32, 1069–1078.
Wiese, S., Reidegeld, K. A., Meyer, H. E., and Warscheid, B. (2007). Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350.
Wilson, I. A., Ikonen, S., McMahan, R. W., Gallagher, M., Eichenbaum, H., and Tanila, H. (2003). Place cell rigidity correlates with impaired spatial learning in aged rats. Neurobiol. Aging 24, 297–305.
Wu, H. I., Cheng, G. H., Wong, Y. Y., Lin, C. M., Fang, W., Chow, W. Y., and Chang, Y. C. (2010). A lab-on-a-chip platform for studying the subcellular functional proteome of neuronal axons. Lab. Chip 10, 647–653.
Yang, J. W., Czech, T., and Lubec, G. (2004). Proteomic profiling of human hippocampus. Electrophoresis 25, 1169–1174.
Yang, S., Liu, T., Li, S., Zhang, X., Ding, Q., Que, H., Yan, X., Wei, K., and Liu, S. (2008). Comparative proteomic analysis of brains of naturally aging mice. Neuroscience 154, 1107–1120.
Keywords: proteome, aging, cognitive decline, learning and memory, brain, hippocampus
Citation: VanGuilder H. D. and Freeman W. M (2011) The hippocampal neuroproteome with aging and cognitive decline: past progress and future directions. Front. Ag. Neurosci. 3:8. doi: 10.3389/fnagi.2011.00008
Received: 06 January 2011; Accepted: 12 May 2011;
Published online: 23 May 2011.
Edited by:
Emil C. Toescu, Birmingham University, UKReviewed by:
Junming Wang, University of Mississippi Medical Center, USALori Lynn Mcmahon, University of Alabama, USA
Aurel Popa Wagner, University of Greifswald, Germany
Copyright: © 2011 VanGuilder and Freeman. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Willard M. Freeman, Department of Pharmacology, Penn State College of Medicine, 500 University Drive, Hershey, PA 17033, USA. e-mail: wfreeman@psu.edu