The effects of aging on the BTBR mouse model of autism spectrum disorder
- 1Metabolism Unit, Laboratory of Clinical Investigation, National Institutes of Health, National Institute on Aging, Baltimore, MD, USA
- 2Department of Neurology, Johns Hopkins University School of Medicine, Kennedy Krieger Institute, Baltimore, MD, USA
- 3Receptor Pharmacology Unit, Laboratory of Neurosciences, National Institute on Aging, Baltimore, MD, USA
- 4VIB-Department of Molecular Genetics, University of Antwerp, Antwerp, Belgium
Autism spectrum disorder (ASD) is a complex heterogeneous neurodevelopmental disorder characterized by alterations in social functioning, communicative abilities, and engagement in repetitive or restrictive behaviors. The process of aging in individuals with autism and related neurodevelopmental disorders is not well understood, despite the fact that the number of individuals with ASD aged 65 and older is projected to increase by over half a million individuals in the next 20 years. To elucidate the effects of aging in the context of a modified central nervous system, we investigated the effects of age on the BTBR T + tf/j mouse, a well characterized and widely used mouse model that displays an ASD-like phenotype. We found that a reduction in social behavior persists into old age in male BTBR T + tf/j mice. We employed quantitative proteomics to discover potential alterations in signaling systems that could regulate aging in the BTBR mice. Unbiased proteomic analysis of hippocampal and cortical tissue of BTBR mice compared to age-matched wild-type controls revealed a significant decrease in brain derived neurotrophic factor and significant increases in multiple synaptic markers (spinophilin, Synapsin I, PSD 95, NeuN), as well as distinct changes in functional pathways related to these proteins, including “Neural synaptic plasticity regulation” and “Neurotransmitter secretion regulation.” Taken together, these results contribute to our understanding of the effects of aging on an ASD-like mouse model in regards to both behavior and protein alterations, though additional studies are needed to fully understand the complex interplay underlying aging in mouse models displaying an ASD-like phenotype.
Introduction
Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disorder characterized by varying degrees of altered social functioning and engagement in repetitive, stereotyped behaviors accompanied by display of narrowed interests (American Psychiatric Association, DSM-V 2012). The most recent surveillance data collected in 2008 by the Center for Disease Control's Autism and Developmental Disabilities Monitoring Network indicates that approximately 1 in 88 children has an autism spectrum disorder (CDC, 2010). The United States Census Bureau in 2006 projected a doubling of the US population aged 65 and older by 2030. Assuming the life expectancy of individuals with ASD is similar to that of the general population, based on current prevalence rates in school aged children, this population increase would result in a prevalence of approximately 700,000 individuals with ASD aged 65 in the next 20 years (Piven and Rabins, 2011).
Despite the population statistics indicating a significant number of aged individuals with ASD, studies that investigate the effects of the neurodevelopmental disabilities including ASD, in an aged physiological context are lacking. Due to the paucity and limitations of studies and the expected increase in the aged individuals presenting ASD a multidisciplinary conference was held in March 2010 to identify gaps in knowledge regarding older adults with ASD. The panel concluded that descriptive studies of aging in humans and rodent models were needed (Piven and Rabins, 2011). A limited number of human studies have investigated the persistence of the ASD core deficits into adulthood (Ballaban-Gil et al., 1996; Seltzer et al., 2003; Shattuck et al., 2007), childhood factors that are associated with prognosis (Rumsey et al., 1985), and outcome studies (Rutter et al., 1967; Howlin et al., 2004; Renty and Roeyers, 2006). Major findings from these studies suggest that language development (Kanner et al., 1972) and Intelligence Quotient (IQ) predicts outcome (Lotter, 1974; Rumsey et al., 1985; Szatmari et al., 1989). The ASD diagnosis persists with inconsistent findings about which, if any, core deficits change (Ballaban-Gil et al., 1996; Piven et al., 1996; Seltzer et al., 2003), and that the adults with ASD remain dependent on others (Rutter et al., 1967) even if they have a normal IQ (Howlin et al., 2004). Despite persistent dependence some studies found that individuals with ASD had improved symptoms over time (Szatmari et al., 1995; Piven et al., 1996; Seltzer et al., 2003). However, a few limitations hinder these early studies of aging in ASD, including study individuals being only 20–30 years old, small sample sizes, and an inability to completely generalize the study findings given the heterogeneity of the population examined (Rumsey et al., 1985; Gillberg and Wing, 1999; Fombonne et al., 2001; Howlin and Moss, 2012).
Given the heterogeneous nature of ASD, mouse models have proven to be particularly useful and reliable in elucidating the mechanism of these disorders (Crawley, 2007; Moy and Nadler, 2008). There are several approaches to ASD mouse models: targeted gene mutations, defects in neurotransmitters or brain regions, inbred strains, and models based on the comorbidity of other human diseases with autism (Crawley, 2007). One strain in particular, BTBR T + tf/j, has been shown to be an especially relevant animal model of ASD (Moy et al., 2007). Extensive behavioral characterization of the BTBR (black and tan, brachyuric) mouse model has revealed low sociability compared to wildtype strains (Bolivar et al., 2007; Moy et al., 2007), resistance to change (Moy et al., 2007; Moy and Nadler, 2008), increased display of self-grooming behavior (Pobbe et al., 2010), display of repetitive behaviors (Pearson et al., 2011), and reduced display of territorial scent marking (Wohr et al., 2011). Furthermore, unusual vocalizations have also been extensively characterized in BTBR mice (Scattoni et al., 2008, 2011), in addition to instances of social avoidance and gaze aversion (Defensor et al., 2011). While the autistic-like behavioral phenotype of the young male BTBR mouse has been studied extensively, the effects of age on this ASD-like mouse model have not been elucidated. As aged human autistic tissue samples are limited and the BTBR mouse model is well characterized and widely accepted, we selected this mouse model to study the nature of the ASD phenotype in an aged context. To enhance our understanding of the aged BTBR mouse and to gain further insight into the effects of aging of ASD, we followed our extensive behavioral characterization of the aged BTBR mouse with quantitative proteomic analyses on cortical and hippocampal tissues as these two brain regions have been strongly associated with ASD (Mundy, 2003; Nadler et al., 2006). We hypothesized that both the behavioral phenotype and proteomic profile would be different between the aged ASD-like mice [or mice with an abnormal central nervous system (CNS)] and the aged wildtype mice (or mice with a presumed normal CNS). Our results contribute to our understanding of the effects of aging on an ASD-like mouse model, and it is our hope that further research will enhance our understanding of aging in an abnormal CNS.
Materials and Methods
Experimental Animals
All experimental animal procedures were approved by the Animal Care and Use Committee of the National Institute on Aging, National Institutes of Health. Male BTBR T + tf/J mice (n = 8, 15 months of age) and male control wildtype (WT) C57BL6J (n = 5, 15 months of age) were housed in the NIA animal facility on a 12 h light and dark cycle from 6 am to 6 pm and received ad libitum access to food and water throughout the duration of the study. All behavioral testing described in detail below was performed between the hours of 9 AM and 5 PM. Prior to behavioral testing, animals were moved from the vivarium to an experimental room and given a minimum of 30 min to habituate to the new environment. Between each experimental animal, all apparatuses were wiped down with 70% ethanol followed by D. I. water to prevent any scent retention that could possibly bias the performances of later mice.
Social Preference Assessment
Social behavior was assessed using a three-chambered sociability apparatus with a protocol modeled after previously established methods (Crawley, 2007). In brief, animals were first habituated for 10 min to the three-chambered sociability apparatus (60 × 40 × 22 cm, Stoelting, Wood Dale, IL) free of any stimuli and returned to their home cage for a minimum of 30 min. Prior to the return of the experimental mouse to the chamber for testing, another mouse of the same strain, age, and gender and an inanimate object both housed in identical wire mesh cylinders (15 cm tall, 7 cm in diameter) were placed in opposing compartments of the apparatus. Experimental animals were then returned to the center chamber of the apparatus and were free to choose to interact with either the mouse or object enclosed in the respective wire mesh containers. Interactions were recorded for 10 min and analyzed with ANY-Maze Video Tracking Software (Stoelting). A discrimination index (DI) was calculated that takes into account the relative exploration time spent with either the contained mouse or object: (animal time/total time—object time/total time) × 100. As socially-normal mice show a robust preference for another mouse over an inanimate object, a high DI is reflective of socially-normal behavior while a low DI is indicative of deficits in social functioning.
Novel Object Preference Test
The novel object preference test was performed as described previously (Dere et al., 2007; Park et al., 2010). In brief, mice were given 10 min to explore a 25 cm2 opaque walled box with two identical objects placed equidistant from each other in the center of the box. After 10 min of habituation to the two identical objects was completed, animals were returned to their home cages for a minimum of 30 min. After the habituation task, trained experimenters replaced one of the identical objects with a novel object that had never been seen by the experimental mouse. Experimental animals were then returned to the opaque box and given 10 min in which they were free to interact with either the familiar or novel object. Both the habituation session and testing session were videotaped and the animal's behavior automatically tracked by ANY-Maze Video Tracking Software (Stoelting). Discrimination indices were again calculated: (novel time/total time-same time/total time) × 100. A high discrimination index (DI) is indicative of an enhanced ability to preferentially recognize the novel object, suggesting intact memory functioning.
Open Field Test
A generalized view of the behavioral characteristics of both experimental and control mice was obtained via an open field test performed as described previously (Holmes et al., 2002). In brief, each mouse was placed in a square chamber approximately 0.14 m2 in size (Med Associates Inc, St. Albans, VT) and given 10 min to freely explore. Activity Monitor software (Med Associates Inc) automatically recorded multiple parameters over the duration of the 10 min test. Individual data points were then compiled and subsequently analyzed to produce the following measurements: distance traveled, time spent in the peripheral areas of the chamber, time spent in the center area of the chamber, number of ambulatory episodes, and number of jumping episodes. Relative time spent in the periphery vs. the center was calculated for each mouse by dividing the time spent in either the periphery or center by the total exploration time (600 s for all animals).
Light/Dark Exploration Test
The light-dark exploration test was performed as previously described (Chadwick et al., 2011) as a means to assess general anxiety behavior. The same square chamber used in the Open field test was again used to complete the Light/Dark exploration test, with the additional insertion of a dark chamber taking up 50% of the area of the entire apparatus (Dark box insert for mouse open field activity, Med Associates Inc). The dark chamber contains a small arched entrance to allow mice to access the dark environment; this entryway was placed toward the center of the apparatus so mice could easily access it. Time spent in either the light or dark compartment was automatically recorded by Activity Monitor software (Med Associates Inc). Relative time spent in the light chamber vs. the dark chamber was calculated for each mouse by dividing the time spent in either the light or the dark by the total exploration time (600 s for all animals).
Elevated Plus Maze
General anxiety behavior was also evaluated using an elevated plus maze test as described previously (Chadwick et al., 2011). In brief, animals were given 5 min to explore a plus shaped maze raised approximately 36 cm off of the ground. Two of the arms of the maze were enclosed on all sides; two arms remained open for the duration of the test. ANY-Maze Video Tracking Software (Stoelting) was used to record time spent in either the closed or open arms of the apparatus and the relative time spent in the closed or open arms was calculated by dividing the time spent in either arm by the total exploration time (300 s for all animals).
Rota-Rod Test
Motor coordination was evaluated using an accelerating Rota-Rod treadmill (Med. Associates, Inc) as described previously (Bohlen et al., 2009). Briefly, mice were given two training sessions on the spinning Rota-Rod apparatus on the day prior to the testing day (the first trial was administered in the morning and the second trial was administered in the afternoon). Each habituation training trial lasted 2 min [4 revolutions per minute (rpm)]. On the test day, the mice were placed on the Rota-Rod, which gradually accelerated from 4 to 40 rpm over a 5 min time interval. The test was performed twice per day and the latency to fall was measured and averaged.
Animal Euthanization and Tissue Collection
Following completion of behavioral analyses, animals were euthanized via isoflurane overdose (Butler Animal Health Supply, Dublin, OH) as described previously and in accord with approved animal procedures (Chadwick et al., 2011). Bodyweight data was collected immediately prior to euthanization for each animal. Hippocampal and cortical brain tissues were snap frozen on dry ice and stored at −80°C until used for further analyses.
Protein Digestion, iTRAQ Labeling and Western Blotting
Protein digestion and iTRAQ labeling were processed according to the iTRAQ protocol (iTRAQ Reagents—8plex: amine-modifying Labeling Reagents for Multiplexed Relative and Absolute Protein Quantitation, ABSciex). All tissue samples (Hippocampus and Cortex; WT and BTBR) were prepared in parallel throughout the labeling procedures. Briefly, 100 μg of protein in each condition was acetone precipitated and resuspended in 20 μL of iTRAQ dissolution buffer (0.5 M triethylammonium bicarbonate (TEAB), ABSciex) containing 0.1% ProteaseMAX detergent (Promega) to denature the proteins. The sample was then reduced by adding iTRAQ Reducing Reagent [Tris(2-carboxyethyl) phosphine (TCEP), ABSciex] to a final concentration of 5 mM and incubated at 60°C for 1 h. Subsequently, the sample was alkylated with iTRAQ Cysteine-Blocking Reagent (10 mM methyl methanethiosulfonate (MMTS), ABSciex) for 10 min at room temperature. The protein sample was then digested with 5 μg sequencing-grade trypsin (Promega) per 100 μg protein at 37°C overnight. Labeling of the samples with iTRAQ 8-plex labels was performed at room temperature for 2 h. After labeling, the samples to be compared were mixed and underwent an off-line strong cation exchange (SCX) fractionation (ICAT Cation Exchange Buffer Pack, ABSciex) to 16 fractions to reduce sample complexity. After reversed-phase desalting (C18 tips, Pierce), the samples were re-constituted in water with 0.1% formic acid, then stored at −20°C until LC/MS/MS analysis. Western blotting procedures as described previously (Maudsley et al., 2007; Martin et al., 2009a), were performed using the same protein lysates employed for iTRAQ labeling. Antibodies used were as follows: rabbit anti-BDNF [(brain-derived neurotrophic factor) Santa Cruz Biotechnology, Santa Cruz CA], rabbit antiphosphorylated TrkB and TrkB, rabbit anti-phosphorylated Akt and Akt, rabbit antiphosphorylated synapsin 1 and synapsin 1, rabbit anti-spinophilin, rabbit anti-PSD95 and rabbit anti-synaptophysin (Cell signaling technology, Inc., Danvers, MA), mouse anti-NeuN (EMD Millipore Corporation, Billerica, MA) and mouse anti-beta actin (Sigma-Aldrich, St. Louis, MO). Results were quantified and statistical analysis was performed using an unpaired student t-test. p < 0.05 was considered statistically significant.
LC/MS/MS Analysis
Samples were analyzed using an Eksigent NanoLC Ultra 2D (Dublin, CA) and Thermo Fisher Scientific LTQ Orbitrap XL (San Jose, CA). In brief, peptides were first loaded onto a trap cartridge (Agilent), then eluted onto a reversed phase PicoFrit column (New Objective, Woburn, MA) using a linear 120 min gradient of acetonitrile (2–62%) containing 0.1% formic acid at 250 nL/min flowrate. The eluted peptides were sprayed into the LTQ Orbitrap XL. The data-dependent acquisition mode was enabled, and each FTMS MS1 scan (60,000 resolution) was followed by 6 MS2 scans (alternating CID at unit resolution and HCD at 7500 resolution on 3 precursor ions). The spray voltage and ion transfer tube temperature were set at 1.8 kV and 180°C, respectively.
Database Search and iTRAQ Quantification
Proteome Discoverer 1.2 (Thermo Fisher Scientific) was used for protein identification and iTRAQ quantification using Sequest algorithms. The following criteria were followed: SwissProt mouse database; enzyme: trypsin; miscleavages: 2; static modifications: methylthio (+45.988 Da on C), iTRAQ8plex (+304.205 Da on N-terminus and K); dynamic modifications: oxidation (+15.995 Da on M), deamidation (+0.984 Da on N and Q); peptide tolerance as 25 ppm; MS2 tolerance as 0.8 Da. Peptides reported via all search engines were accepted only if they met the false discovery rate of 5%. For iTRAQ quantification, the reporter ion intensities in MS2 spectra (m/z 113–121, integration width tolerance 50 mmu) were used to calculate the expression ratios among the different conditions (Hippocampus and Cortex; WT and BTBR).
Bioinformatics Analysis
Functional annotational clustering of our proteomic data, i.e., gene ontology (GO), KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was performed using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/, 3/2014) as previously described (Chadwick et al., 2010a, 2011). In addition Ingenuity Pathway Analysis (http://Ingenuity.com, 3/2014) was employed for Canonical Signaling Pathway enrichment investigations. In brief, GO analysis allows for broad clustering of genes into functional groups, while KEGG pathways analysis allows for more specific clustering into signaling pathways (Maudsley et al., 2011). Inclusion criteria were set as follows: pathway groups needed to meet a minimum population of two genes from the input experimental set, and also needed to possess a probability significance of enrichment compared to a control background dataset of less than p < 0.05 (hypergeometric test of significance). In addition, Venn diagrams were also constructed to identify common and uniquely altered genes between hippocampus and cortex, using VennPlex (Cai et al., 2013), as described previously (Martin et al., 2009b).
Textrous! Latent Semantic Analysis-Based Investigation
Textrous! latent semantic analysis was performed as described previously (Chen et al., 2013). In brief, corresponding gene symbols were uploaded into Textrous! (http://textrous.irp.nia.nih.gov/, 3/2014) and then the relevant output indices, i.e., Cosine Similarity, Z-score and p-value were extracted. In addition to the direct list-based semantic output both the Collective processing mode (generating hierarchical word-clouds), as well the Individual processing mode (generating symbol-word heatmaps) were applied to provide more nuanced informatic appreciation of the specific input datasets.
Statistical Analysis
All statistical analyses were conducted using a Student's t-test (two-tailed with equal variances: GraphPad Prism, version 5.02). p < 0.05 was considered statistically significant throughout the study. Error bars represent 95% confidence interval. All data represent means ± s.e.m. (standard error of mean). *p < 0.05; **p < 0.01; ***p < 0.001.
Results
ASD-Like Behavioral Phenotype Persists in Aged BTBR Mice
Commensurate with an extant ASD phenotype aged BTBR mice demonstrated no sociability preference between an object or another mouse, while WT controls spent significantly more time with another mouse compared to an object (Figure 1A). Relative percentage of time spent with either the mouse or the object was used to calculate a social discrimination index [DI: (novel time/total time-same time/total time) × 100] (Figure 2B). WT and BTBR mice performed similarly in the novel object preference test of learning and memory, with both groups spending significantly more time with a novel object compared to a familiar object (Figures 1C,D). Both WT and BTBR mice significantly preferred to explore the peripheral areas of the chamber over the center of the chamber, though the BTBR mice did show a significantly higher preference for the periphery over the center (Figure 1E). No significant differences in total distance traveled (Figure 1F) or number of ambulatory episodes (Figure 1G) were found. BTBR mice however demonstrated significantly less jumping activity compared to WT (Figure 1H). In light-dark exploration tests BTBR mice spent significantly more time in the dark chamber compared to the lighted chamber, while WT control mice show no statistical difference in preference for either chamber (Figure 1I). In the elevated plus maze test of anxiety, both WT and BTBR mice significantly preferred closed arms to open arms (Figure 1J). Motor coordination, assessed using a Rotarod test was similar in both groups (Figure 1K).
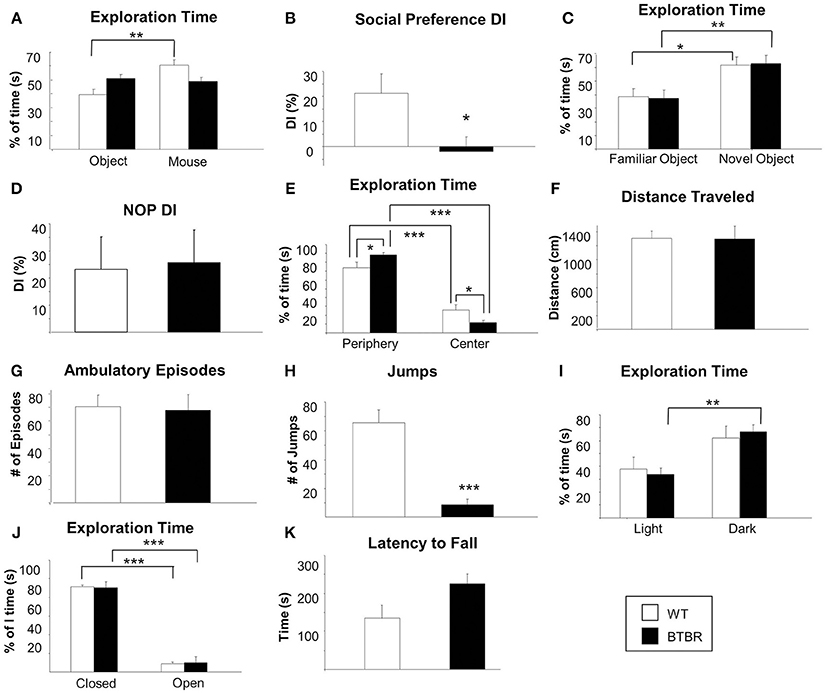
Figure 1. Social function, cognitive ability, general behavior, anxiety and motor coordination in an aged ASD mouse model. Social preference was assessed using a three-chambered sociability apparatus in aged male BTBR T + tf/j mice and age-sex-matched control mice (A, B). Percentage of time spent exploring both the contained inanimate object and the contained mouse was measured for male BTBR (n = 8) and control mice (n = 5) (A). A discrimination index (DI) was calculated (B) which represents the degree to which experimental or control animals displayed a preference for exploring the contained mouse over the contained inanimate object. Cognitive functioning was assessed via the novel object preference task (NOP) (C, D). Percentage of time spent exploring either a familiar or a novel object was again measured for male BTBR (n = 8) and control mice (n = 5) (C) and a DI was calculated (D) with a higher DI indicating an increased ability to discern a novel object from a familiar object. Exploration of an open field apparatus was assessed in male BTBR and control mice was measured with regard to the following parameters: time spent exploring the peripheral or central areas of the apparatus relative to the total amount of time spent exploring (E), total distance traveled (F), number of ambulatory episodes (G), and number of jumps (H). Exploration of the open field apparatus was repeated with a Light/Dark insert add-on and relative time spent in either the light half or the dark half of the chamber was recorded (I). General anxiety was assessed using the elevated plus maze task. For both BTBR T + tf/j mice and age-and sex-matched controls, relative spent in the closed vs. the open arm was recorded as a percentage of total time (J). Finally, motor coordination was assessed using a rotarod apparatus latency to fall was recorded in both BTBR T + tf/j and control mice (K, n = 5). Data are expressed as means ± s.e.m. Asterisks represent p-values as shown: *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance was measured using a Student's t-test.
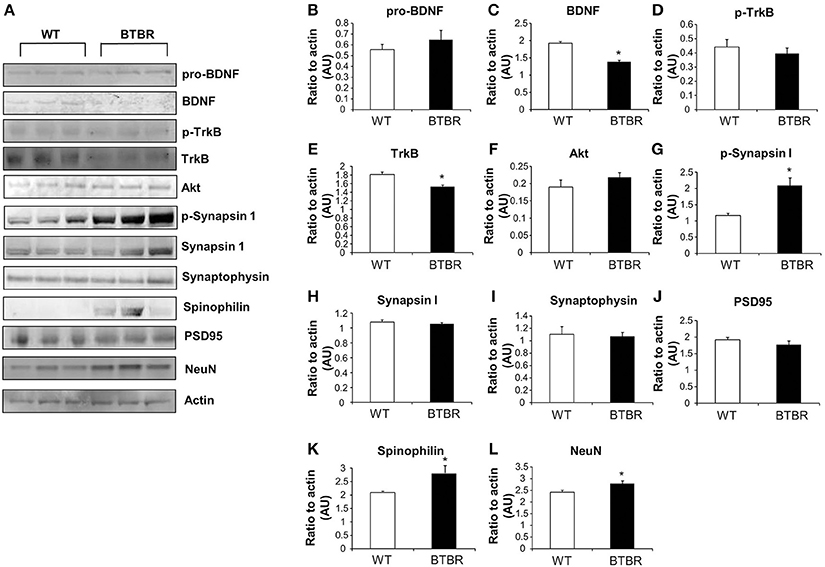
Figure 2. Protein expression in wild type and BTBR mouse cortex. Western blotting was performed using mouse cortical protein lysates (A). Protein levels were measured in BTBR and control mice with respect to the following proteins in the cortex: pro-BDNF (B), mature BDNF (C), phospho-TrkB (D), total TrkB (E), Akt (F) phospho-synapsin 1 (G), synapsin 1 (H) synaptophysin (I), PSD95 (J), spinophilin (K), Neuronal cell marker NeuN (L). Data are expressed as means ± s.e.m. Asterisks represent p-values as shown: *p < 0.05. Statistical significance was measured using a Student's t-test, n = 3 for each group.
Altered Protein Expression in CNS of Aged BTBR Mice
Cortical and hippocampal western blotting of proteins, selected for their ability to indicate alterations of neurosynaptic activity, was employed to detect potential expression differences between aged WT and BTBR mice. Our panel of neurosynaptic factors analyzed in cortical tissue is indicated in Figure 2A. No significant difference between WT and BTBR pro-BDNF (brain-derived neurotrophic factor) levels was detected (Figure 2B), whereas a significant decreased in BTBR mature BDNF was observed compared to WT (Figure 2C). Levels of phosphorylated TrkB receptor were similar in WT and BTBR cortex (Figure 2D) while the total TrkB expression was significantly lower in BTBR mice compared to WT (Figure 2E). No significant differences in Akt1 expression was noted in the BTBR model compared to WT (Figure 2F). Levels of phosphorylated synapsin 1 (Figure 2G), but not total synapsin 1 (Figure 2H) were significantly elevated in BTBR mice compared to WT. Expression of both synaptophysin (Figure 2I) and PSD95 (Figure 2J) was not affected in BTBR mice, while both spinophilin (Figure 2K) and NeuN (Figure 2L) levels were significantly elevated in BTBR mice compared to WT.
In the hippocampus we again found no significant differences in the levels of pro-BDNF between BTBR and WT (Figure 3B) that was coincident with a significant reduction in extant BDNF levels (Figure 3C). Phosphorylated TrkB expression was unchanged in BTBR mice compared to WT (Figure 3D), while there was a trend, similar to that in the cortex, to reductions of total TrkB receptor expression in the BTBR mice (Figure 3E). As with the cortex no significant alterations in Akt1 were observed (Figure 3F). Significant potentiation of both phosphorylated (Figure 3G) and non-phosphorylated synapsin 1 (Figure 3H) expression levels was seen in the BTBR hippocampus. In accordance with the cortical data we found no significant BTBR-induced changes in the hippocampal expression of synaptophysin (Figure 3I) or PSD95 (Figure 3J). Levels of both spinophilin (Figure 3K) and NeuN (Figure 3L) were again significantly potentiated in the BTBR mice, mirroring our previous cortical data.
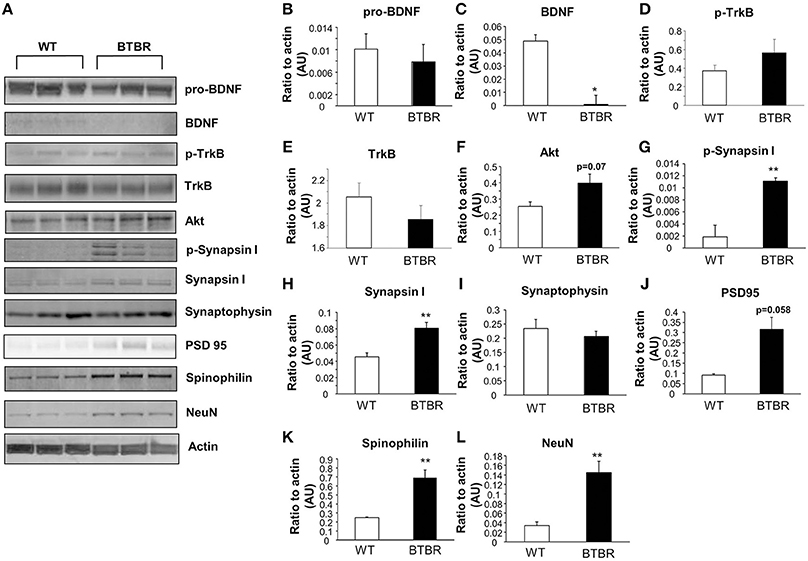
Figure 3. Protein expression in wild type and BTBR mouse hippocampus. Western blotting was performed using mouse hippocampal protein lysates (A). Protein levels were measured in BTBR and control mice with respect to the following proteins in the hippocampus: pro-BDNF (B), mature BDNF (C), phospho-TrkB (D), total TrkB (E), Akt (F) phospho-synapsin 1 (G), synapsin 1 (H) synaptophysin (I), PSD95 (J), spinophilin (K), Neuronal cell marker NeuN (L). Data are expressed as means ± s.e.m. Asterisks represent p-values as shown: *p < 0.05, **p < 0.01. Statistical significance was measured using a Student's t-test, n = 3 for each group.
Systemic Analysis of BTBR Central Nervous Protein Expression
To gain an unbiased appreciation of the multiple tissue protein expression pattern changes generated in the BTBR mice compare to WT we applied quantitative isobaric mass-tag labeling (iTRAQ) to cortical and hippocampal tissue extracts. In the cortex we identified and generated relative expression profiles for 674 proteins (Figure 4A: Table S1), while in the hippocampus 656 proteins were identified and quantified in BTBR mice (Figure 4B: Table S2). Log2-transformed iTRAQ expression ratios were employed to generate snake plot graphs of individual proteins. To initially validate some of the iTRAQ data we chose three random proteins, Rab3A, CaMKIID and Cplx1 and found that at the western level of detection our results were comparable to our iTRAQ expression ratio data (Figures 4C,D).
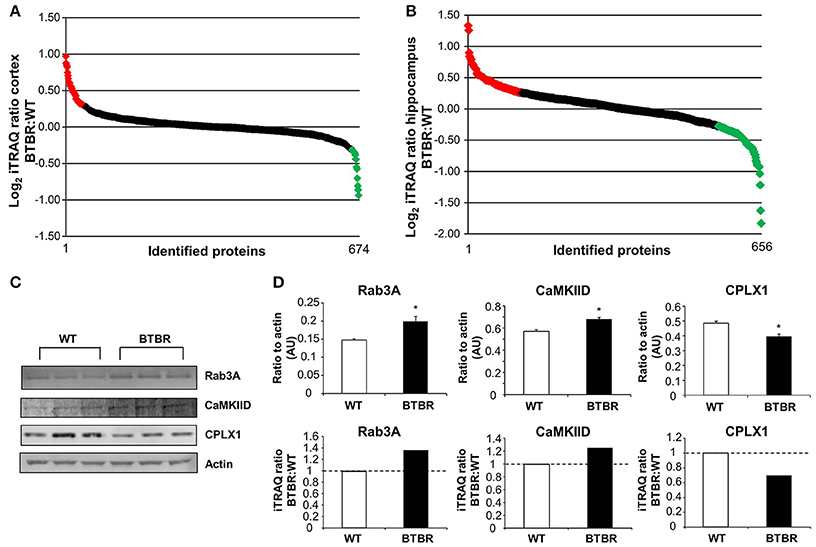
Figure 4. Quantitative proteomic and bioinformatics analysis of BTBR CNS tissues. Log2transformed iTRAQ ratio data (BTBR:WT) snake plot of the proteins identified in the cortex (A) and hippocampus (B) are depicted. Upregulated proteins are highlighted in red, while downregulated proteins are highlighted in green. Three proteins identified by iTRAQ were validated via western blot and results correlated to the iTRAQ results (C, D). Data are expressed as means ± s.e.m. Asterisks represent p-values as shown: *p < 0.05. Statistical significance was measured using a Student's t-test.
Gene Ontology Bioinformatic Analysis of BTBR-Specific Altered Proteins
Using a standard arbitrary cut-off of >1.2 (elevated in BTBR vs. WT) or <0.8 (reduced in BTBR vs. WT) for the iTRAQ ratios for differentially expressed proteins between BTBR and WT mice we investigated the functional clustering of the BTBR-specific protein changes into Gene Ontology term groups. Analysis of the differentially–regulated [elevated (<1.2) and reduced (<0.8)] proteins in the BTBR cortex we found a strong GO-biological process clustering of endoplasmic reticular, vesicular transport and neurotransmission related functions (Table 1). No GO-molecular function annotation was achieved with the cortical data however. With respect to GO-cellular compartment analysis we found the differential BTBR proteins were strongly associated with endocytic activity, neuronal outgrowth and architecture as well as mitochondrial metabolic activity (Table 1). GO-biological process analysis of the differential BTBR hippocampal protein set indicated a strong bias toward neuronal synaptic secretion, neuronal plasticity, metabolic activity and inter-cellular contact (Table 2). GO-cellular compartment interrogation of the hippocampal dataset revealed a strong bias for protein interactions in the synaptic cleft, clathrin-coated vesicles and mitochondrial-associated metabolic areas (Table 2). GO-molecular function annotation was evident with the hippocampal data and revealed the presence of GO groups associated with malate metabolism, redox activity, synaptic vesicle docking and structural modeling (Table 2).
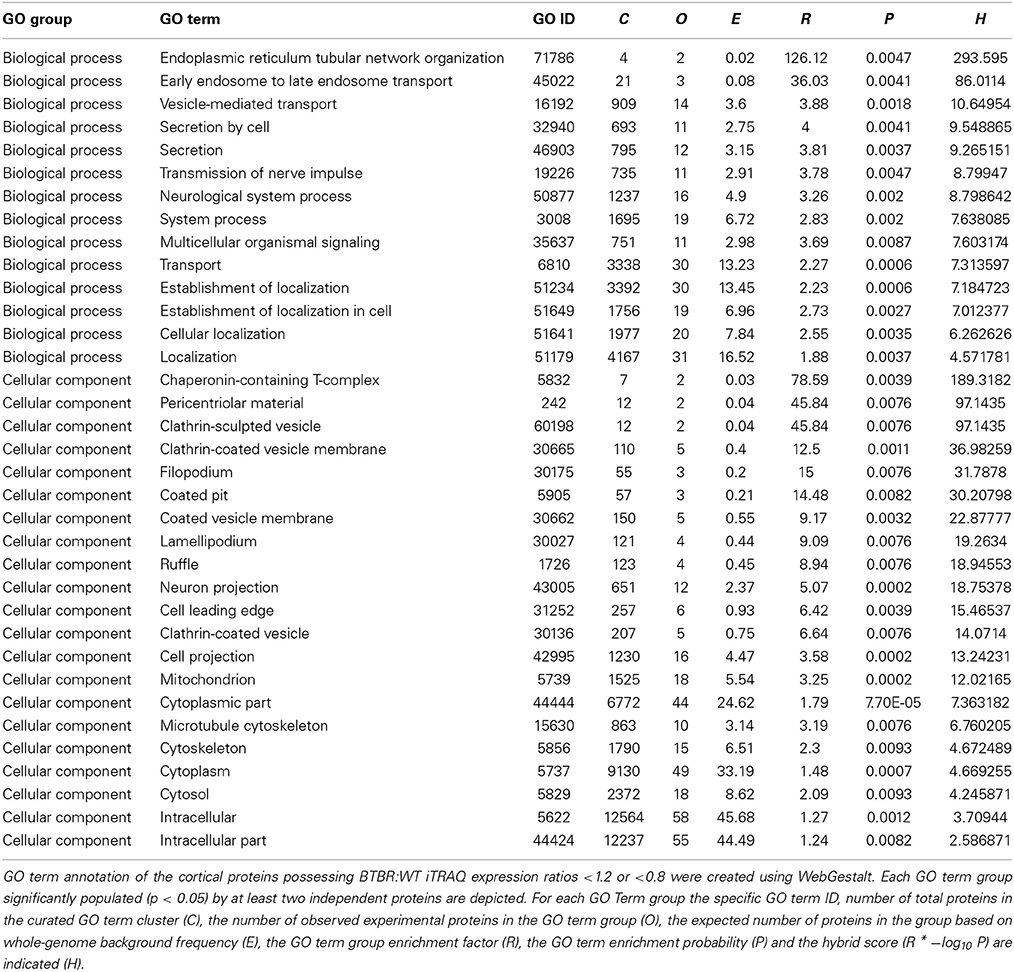
Table 1. GO term analysis for cortical protein alterations observed in aged BTBR compared to WT control.
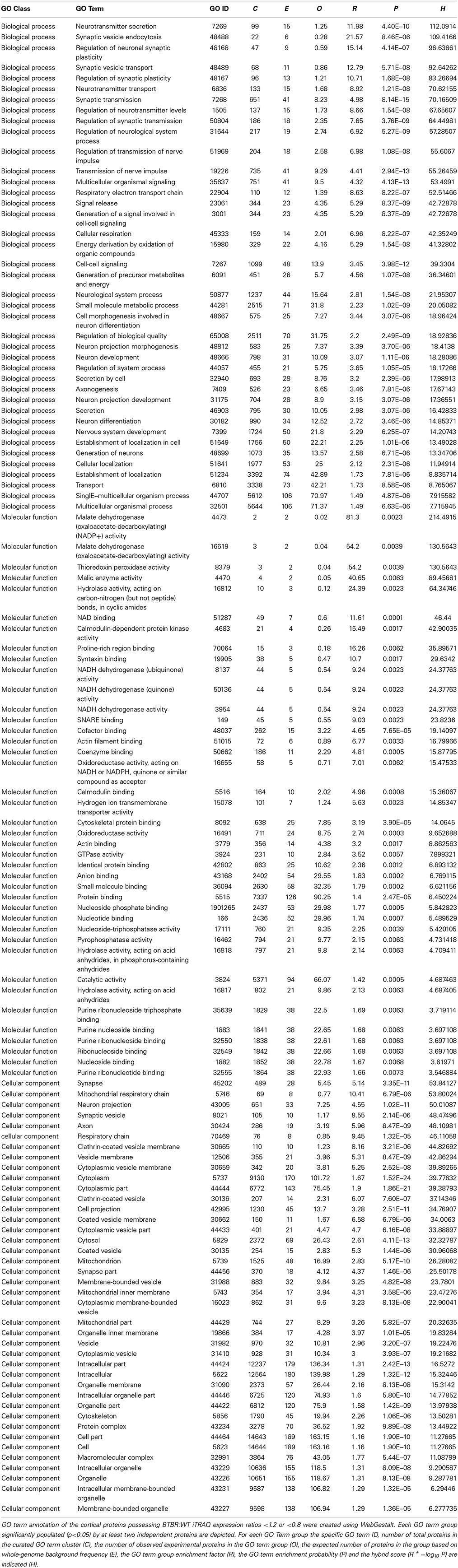
Table 2. GO term analysis for hippocampal protein alterations observed in aged BTBR compared to WT controls.
Signaling Pathway Analysis of Differentially Regulated BTBR-Specific Proteins
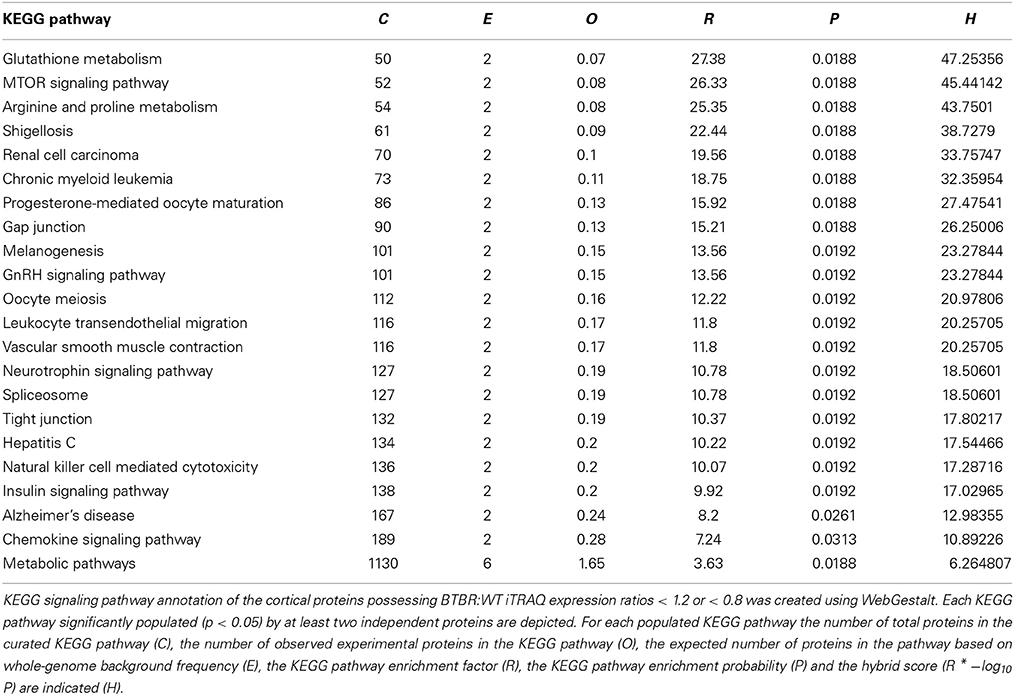
While GO annotation indicates some degree of functional commonality between groups of proteins, associating multiple proteins with classically-identified molecular signaling paradigms provides additional functional information regarding the potential physiological ramifications of specific group of differentially-regulated factors. We therefore performed both KEGG and Ingenuity-base pathway analysis upon our cortical and hippocampal BTBR protein sets. Inspecting the top ten highest scoring KEGG pathways from cortical proteins (Figure 5A: Table 3) a strong population of metabolic (Glutathione metabolism, Insulin signaling pathway, Metabolic pathways), neurotransmission/degenerative (Neurotrophic signaling pathway, Alzheimer's disease) and ultrastructural (Gap junction, Tight junction, Spliceosome) signaling pathways was evident. Applying the same analysis to the hippocampus an intense population of signaling systems linked to oxidative metabolism [Oxidative phosphorylation, Pyruvate metabolism, Citrate cycle (TCA cycle)] and central neurodegeneration (Alzheimer's disease, Huntington's disease, Parkinson's disease, Long-term potentiation) was clear (Figure 5B: Table 4). Extraction of potential physiological meaning from complex data sets is best performed using multiple informatics tools. Therefore, we also sought interpretation of the potential signaling pathways enriched in the BTBR tissues using canonical signaling pathway analysis (Ingenuity). Canonical signaling analysis of the cortex revealed a strong bias toward cytokine signaling (Leptin signaling in obesity, JAK family kinases in IL-6-type signaling), neurodegenerative disease related activity (Amyloid processing, CDK5 signaling, CNTF signaling) and energy metabolism (IGF-1 signaling) (Figure 5C: Table 5). Similar pathway processing of hippocampal data again revealed a strong prediction of metabolism-related activity (Oxidative phosphorylation, Mitochondrial dysfunction, G protein signaling mediated by Tubby), neurodegenerative activity (Huntington's disease signaling) and interestingly, given the strong presentation of, Melanocyte development and pigmentation signaling' in the cortex (Figure 5C), melatonin-related activity (Melatonin signaling) (Figure 5D: Table 6).
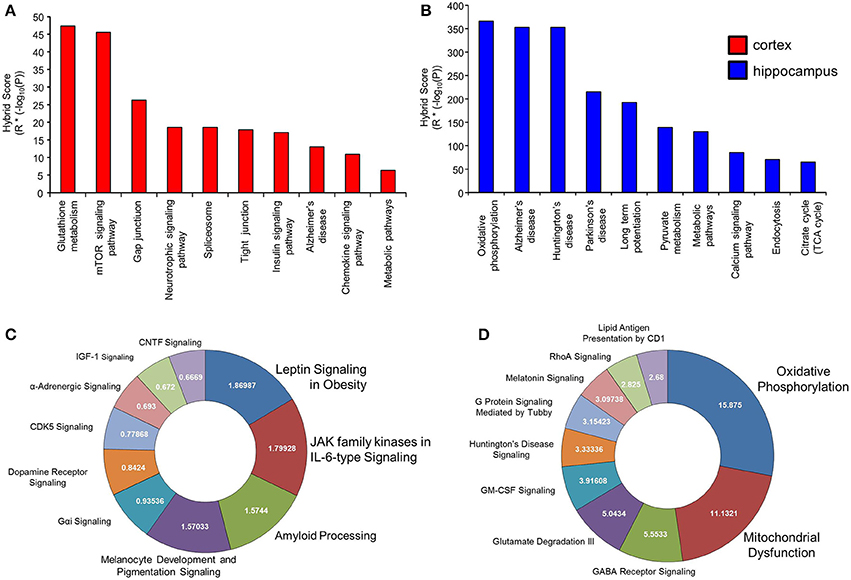
Figure 5. Signaling pathway analysis of cortical and hippocampal proteins differentially regulated BTBR and WT. Cortical and hippocampal proteins exhibiting differential regulation between BTBR and control mice were analyzed for potential functional pathway interactions using KEGG (A-cortex, B-hippocampus) and Ingenuity Canonical Signaling pathway analysis (C-cortex, D-hippocampus). For (A,B), the top 10 most significantly-populated (calculated with Hybrid scores: enrichment factor (R) * (−log10 enrichment probability, P) neuronally-specific KEGG or Canonical signaling pathways are shown.
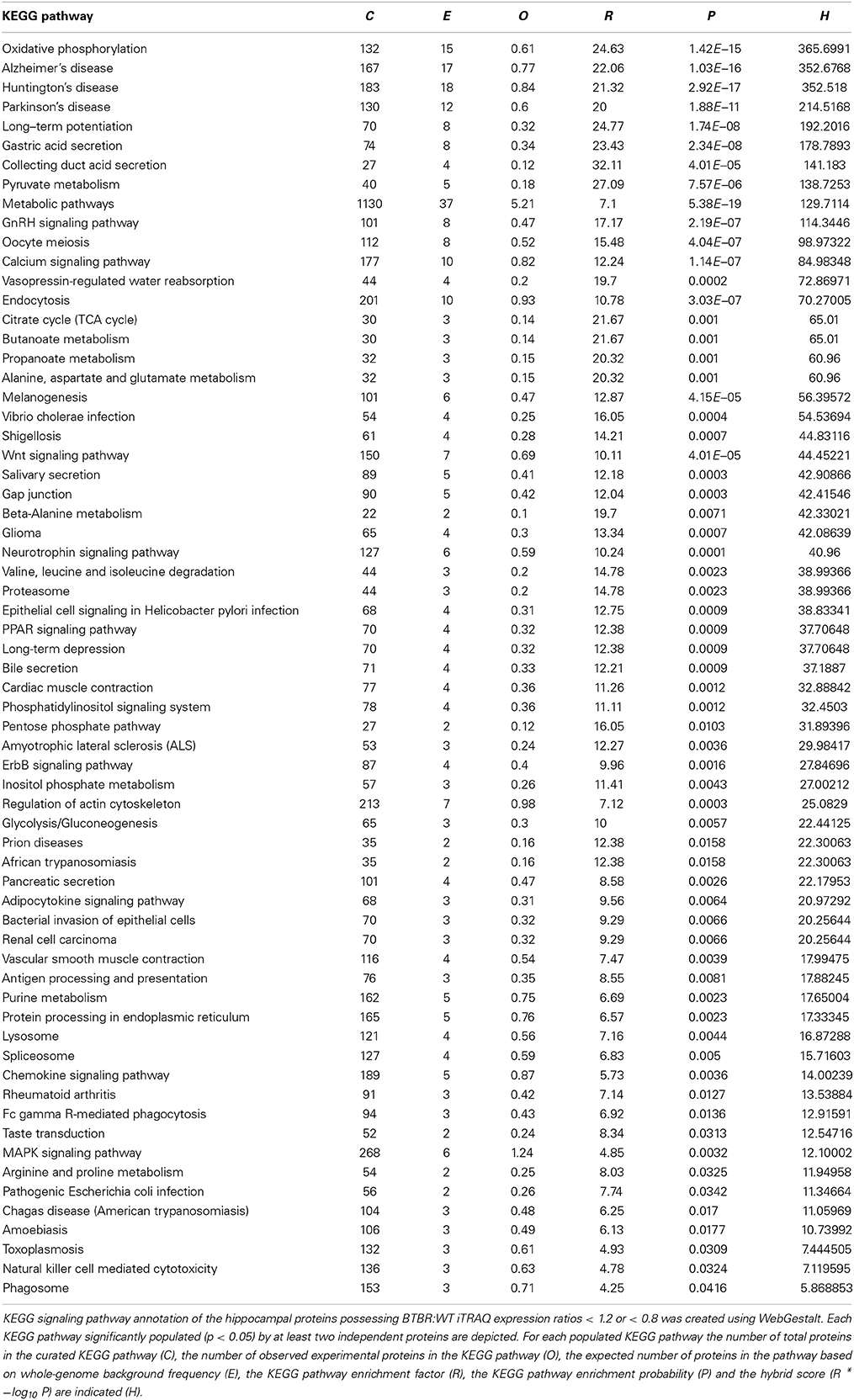
Table 4. KEGG Pathway analysis for hippocampal proteins altered in BTBR mice compared to WT controls.
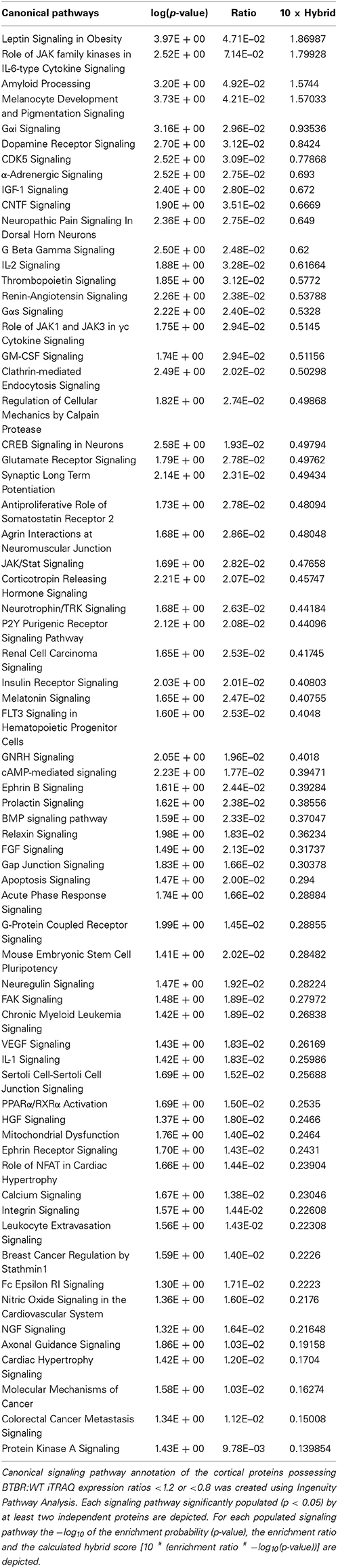
Table 5. Ingenuity Canonical Signaling Pathway analysis of proteins differentially regulated in the cortex of aged BTBR compared to WT controls.
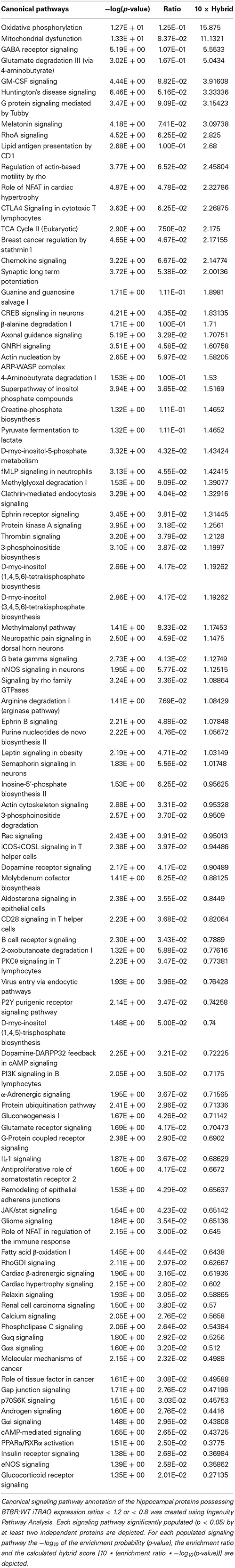
Table 6. Ingenuity Canonical Signaling Pathway analysis of proteins differentially regulated in the hippocampus of aged BTBR compared to WT controls.
BTBR-Regulated Coherent Protein Regulation Patterns
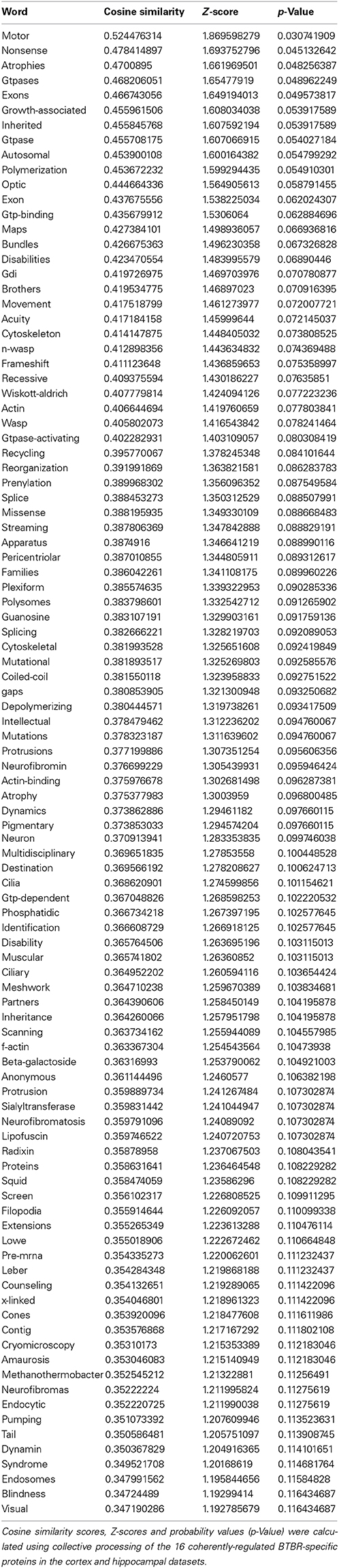
Along with our global analysis of protein set data from the two tissues we also investigated the predicted functional nature of the divergent expression polarity subsets of BTBR-specific proteins uniquely regulated in either the cortex or hippocampus. Using VennPlex (Figure 6A: Table 7) (Cai et al., 2013) we were able to identify multiple groups of coherently-controlled proteins across multiple CNS tissues. Using our previously developed informatics application, Textrous!, we generated physiological predictions extracted from multiple databases using latent semantic analysis (Chen et al., 2013). Using the collective processing capacity of Textrous!. which generates interactive hierarchical word-clouds, we found that the tissue-unique up or down regulated sets of proteins generated quite distinct functional signatures in the cortex or hippocampus. For example, the specifically upregulated proteins in the cortex were strongly focused into activity controlling excitatory amino acid synaptic activity (Figure S1A: Table S3), while the downregulated cortical only proteins demonstrated a strong link to a reduction of protein kinase activity (Figure S1B: Table S4). With respect to the hippocampus, the upregulated protein set appeared to be linked to increases in heat-shock factors and synaptic pathophysiology (Figure S2A: Table S5), while the downregulated hippocampal protein set suggested a reduction in oligodendrocyte generation and CNS myelination (Figure S2B: Table S6).
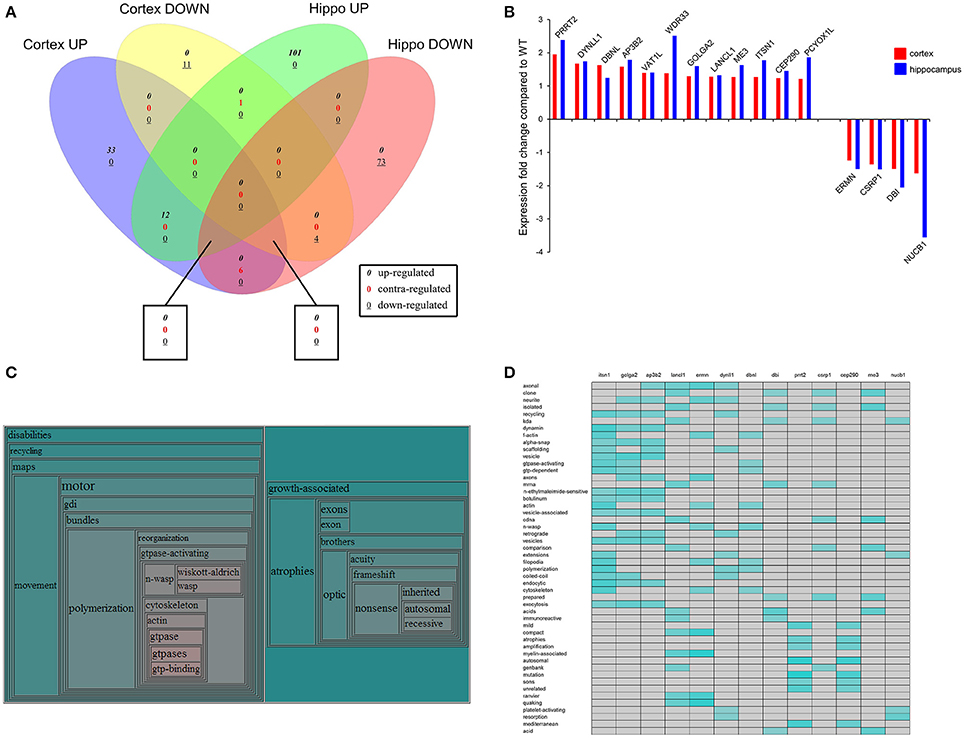
Figure 6. Identification of coherent regulatory protein signatures in BTBR CNS tissues. Vennplex diagram analysis of the upregulated (BTBR:WT iTRAQ ratio > 1.2) or downregulated (iTRAQ ratio < 0.8) proteins in both the hippocampus and cortex are depicted (A). A small coherently-regulated protein subset across both tissues was evident and comprised a focused protein set strongly linked to human ASD-like conditions (B). Textrous!-based collective processing interrogation of the coherently-regulated BTBR protein set revealed, in the resultant hierarchical word-cloud, a strong potential role of skeletal modeling activity in this protein set (C). This skeletal signaling dependence of ASD was confirmed using the individual processing module of Textrous! (D). In this heatmap output correlation strength between protein and functional term is indicated by the intensity of the teal-colored blocks.

Table 7. VennPlex Venn diagram analysis of up and downregulated proteins across the cortex and hippocampus of BTBR mice compared to WT controls.
In addition to our global data set informatics interpretation we also sought to investigate the function of coherently-regulated groups of BTBR-associated proteins observed across multiple tissues. We therefore focused on the subset of proteins (12 upregulated; 4 downregulated: Table 7) coherently altered in a BTBR-specific manner in both the cortex and hippocampus (Figure 6B). To explore the potential functional connection between these select proteins we applied our novel informatic platform Textrous! (Chen et al., 2013). Textrous! allows the extraction of functional data, using latent semantic analysis (LSA), from even the smallest of datasets. Textrous! is a unique LSA tool as it possesses two forms of functional data output, i.e., collective and individual processing. Collective processing of the coherent BTBR protein set results in the creation of a hierarchical word-cloud revealing a strong and novel link between the BTBR ASD phenotype and N-WASP (neuronal Wiskott–Aldrich Syndrome protein) mediated activity (Figure 6C: Table 8). In addition there was a second, less strong demonstration of the importance of inherited genomic activity in ASD phenotypes. The individual processing mode in Textrous! also investigates the strongest individual protein-term relationships within a specific protein subset (Figure 6D). Again with this distinct analytical output we observed a strong relationship between cytoskeletal remodeling activity (f-actin, scaffolding, n-wasp, filopodia) and proteins such as Itsn1 (intersectin 1), Ap3b2 (adaptor-related protein complex 3, beta 2 subunit), Dbnl (drebrinlike) and Ermn (Ermin, ERM-like protein) with the BTBR phenotype. Interestingly other tight relationships between BTBR coherently-regulated factors were found, e.g., myelin-associated activity [Lancl1 (LanC bacterial lantibiotic synthetase component C)-like 1 and Ermn] and inherited neuronal atrophy and impairment [Prrt2 (proline-rich transmembrane protein 2) and Cep290 (centrosomal protein 290)] (Figure 6D).
Discussion
In the present study we evaluated the behavior, synaptic protein alterations and targeted proteomic signatures in the hippocampus and cortex of 15 month old BTBR and WT mice to determine whether the ASD-like phenotype of BTBR mice was resistant to neuronal aging. Thus, we assessed both the behavioral phenotype and proteomic profiles between aged ASD-like mice (abnormal CNS) and aged wild type mice (with a relatively normal CNS). We found that several neurosynaptic proteins were significantly increased in both the hippocampus and cortex of the aged ASD-like mice in comparison to the aged wild type. In particular, the phosphorylated form of the pre-synaptic marker synapsin 1, and the post-synaptic marker spinophilin were increased in both the aged BTBR hippocampus and cortex; PSD95 was significantly elevated in aged BTBR hippocampus. Aged BTBR mice still presented a behavioral phenotype typically observed in young autistic mice, suggesting that the physiological actions underlying the ASD-like condition of the BTBR mice is relatively resistant to aging.
The maintenance of the ASD-phenotype into advanced age in BTBR mice is intriguing because in the normal developing CNS synapses are overproduced but then become pruned in response to neuronal activity during post-natal development (Petit, 1988; Cohen and Greenberg, 2008; Ebert and Greenberg, 2013). It is proposed that autism is caused by dysregulation of the machinery controlling synaptic messenger RNA translation leading to increased synthesis of synaptic proteins and lack of synaptic pruning, in turn inducing a hyperconnectivity and over-reinforcement of neuronal circuits. In fact it is one of the hallmark characteristics of ASD observed in the brains of both mouse models and humans with ASD (Kelleher and Bear, 2008; Gkogkas et al., 2013). Fragile X syndrome, for example which is the leading cause of inherited intellectual disability and ASD (Bhakar et al., 2012), is caused by mutations in FMR1 gene resulting in decreased expression of fragile X mental retardation protein (FMRP). This protein modulates the rate of translational elongation of mRNAs that encode synaptic proteins (Darnell et al., 2011). A decrease in FMRP impairs regulation of synaptic protein expression (Bhakar et al., 2012) and post-natal synapse elimination (Harlow et al., 2010).
As increased synaptic marker expression is associated with the autistic-like mouse phenotype it is not surprising that we found that the BTBR T + tf/j mice maintained their reduced social behavior into old age (Figure 1). This is consistent with the limited human literature that found that social impairments in individuals with ASD persist into adulthood (Ballaban-Gil et al., 1996; Szatmari, 1998; Howlin and Moss, 2012). Although increased synaptic markers have been found to be deleterious for the developing CNS, it is possible that the increased synapses and hyperconnected neuronal circuits be an effective advantage in the aging process where typically nervous system connectivity is degraded through accumulated damage. BDNF and TrkB mRNAs are localized in dendritic spines, thus age-related spine density decreases in older individuals suggest a link between BDNF and TrKB mRNA expression and impaired synaptic connections with aging (Tapia-Arancibia et al., 2008). Indeed decreases in both BDNF and TrkB expression have been documented in the hippocampus and in the neocortex during physiological aging (Phillips et al., 1991; Murray et al., 1994; Connor et al., 1997; Ferrer et al., 1999; Quartu et al., 1999; Hock et al., 2000; Holsinger et al., 2000; Fahnestock et al., 2002; Romanczyk et al., 2002; Tapia-Arancibia et al., 2008; Zhao et al., 2010).
We found that levels of mature BDNF were significantly decreased in the aged BTBR hippocampus and cortex compared with the aged WT (Figures 2A, 3A), suggesting an accelerated aging phenomena, but did not find a significant change in the overall level of active TrkB receptor (Figures 2D, 3D), in the BTBR mice. In addition, we also found that there was a non-significant trend for the increase in the expression in both cortical and hippocampal levels of Akt1, one of the most important downstream targets of TrkB signaling. Thus, via a potentially complex compensatory mechanism, the BTBR mice may have increased their BDNF signaling efficiency and downstream signal transduction capacity compared to WT mice.
Our findings of reduced BDNF in both the hippocampus and cortex in old BTBR mice compared to old WT mice despite seemingly intact memory functioning may be related to this efficiency potentiation of the BDNF-TrkB system in the BTBR mice. These effects may be related to changes in endocytic proteins such as Itsn1, Golga2 and Ap3b2 (Table 7) that may affect the signaling dynamics of the TrkB receptor (Mattson et al., 2004). In addition it is possible that the elevated levels of various synaptic markers served to protect the aging BTBR brain against age-related declines in cognition often associated with decreased levels of BDNF.
Given the complexity of the ASD, it is unlikely that BDNF signaling and various neuronal cell markers are the only protein products altered in the presence of ASD; thus we performed global quantitative iTRAQ proteomics, coupled to advanced bioinformatics in order to gain a comprehensive, systems-level understanding of potential changes in aged BTBR mice compared to WT counterparts. With respect to current relevant literature we discovered that many of these proteins differentially regulated in the brain of the old BTBR mice demonstrated strong correlations with those altered in humans with neurodevelopmental disabilities (Table S7). Nearly twenty percent of the coherently-regulated proteins across the hippocampus and cortex of aged BTBR mice also are strongly linked to human ASD phenotypes, suggesting that future investigation of the remaining factors may prove extremely fruitful for autism-related research (Figure 6B). With respect to the highly-focused, coherently-regulated protein set (Figure 6B: Table 7) Prrt2 was coherently altered in both the hippocampus and cortex of aged BTBR mice compared to aged WT (Table 7). Alterations in activity of Prrt2 have been found in the frontal cortex of individuals with autism (Ji et al., 2012) and have also been genetically-associated with ASD (Weber et al., 2013). Vat1l was up-regulated in both the hippocampus and cortex of the old BTBR mice.
We performed functional informatics data clustering to gain a wider appreciation of the signaling phenotype present in aged BTBR mice. Using GO term annotation it was evident in the cortex that considerable alterations in ER (endoplasmic reticulum), vesicular transport, cytoskeletal remodeling, and energy generation were apparent (Figures 4E,F). The functional GO term clustering of differentially expressed proteins in the BTBR hippocampus also indicated a role of energy generation, neurotransmitter release and synaptic activity but did not display a robust skeletal remodeling component (Table 2). We also employed KEGG and Ingenuity Canonical Signaling pathway analysis for the differentially-regulated cortex and hippocampal protein sets. At this higher level of functional interrogation (compared to GO term clustering) we also observed a subtle divergence in tissue behavior, i.e., cortical KEGG pathway analysis demonstrated a strong bias toward energy metabolic and neurotrophic/neuroprotective mechanisms (Figure 5A) while the hippocampus demonstrated a more degenerative phenotype associated with metabolic instabilities and calcium management factors (Figure 5B). Interestingly and to highlight the need for multidimensional informatics analyses for physiological interpretation, the canonical signaling pathway analysis of both tissue datasets revealed additional processes that could affect the ASD phenotype, i.e., the prominence of cytokine signaling activity in the cortex and the identification of the potential role of melatonin function in both CNS tissues (Figures 5C,D).
To further reinforce the importance of applying unbiased informatics for the analysis of complex data associated with a highly nuanced phenotype such as ASD we found that analysis of different subgroups of the data yielded interesting, tissue-specific insights into the effects of aging on ASD. Using our novel Textrous! platform we were able to identify differential tissue activity, i.e., enhanced glutamatergic cortical activity coincident with attenuated kinase signaling (Figure S1). These cortical changes occurred contemporaneously with an elevation of hippocampal protein misfolding/aggregation coincident with an attenuated CNS myelination and oligodendrocyte functionality (Figure S2). Both of these hippocampal processes have been recently associated with dysfunctional brain development (Mitew et al., 2013).
When approaching complex issues such as ASD with a systems-biology approach it is important to maintain this concept of multiple levels of protein interaction, function and hierarchical architecture, i.e., functions can be tissue specific, expression polarity specific while also existing contemporaneously at a level above tissue-or polarity-dependence. For example we found that inspection of the coherently-regulated multi-tissue protein set revealed an exciting and potentially novel functional association of neuronal Wasp (Wiskott-Aldrich syndrome protein) activity and ASD (Figures 6C,D). To our current knowledge this is the first example of such a functional correlation, however it is relatively simple to predict that the molecular activities of this skeletal remodeling factor could impact a disorder such as ASD in which a less synaptically plastic neurological system is present.
In conclusion, we found that both the social behavioral phenotype and the CNS proteomic profile of this ASD model are relatively age-resistant. Aged BTBR mice possessed elevations in hippocampal and cortical synaptic markers that likely play a role in the etiology of the autistic-like phenotype. Low levels of BDNF have been found inconsistently in the ASD literature but more consistently in the aging and Alzheimer Disease literature (Chadwick et al., 2010b). Diminished mature BDNF in the aged BTBR hippocampus/cortex were seen along with complementary alterations in the downstream signaling pathways compared to WT aged mice. These findings in-part reinforce the potential long-term neuroprotective capacity of the ASD phenotype with respect to cognitive aging. It is therefore interesting to posit that despite lower levels of neurosynaptic plasticity that the more robust and reinforced neuronal network in ASD phenotypes may indeed act prophylactically over long periods of time compared to normal CNS networks that degrade due to unrepaired damage and synaptic loss. The complex proteotype of the BTBR mice revealed molecular and functional signatures highly reminiscent of those found in humans with neurodevelopmental disabilities including ASD. Many of these identified processes play a role in neurodevelopment based on studies in murine models, and give more evidence for proteins that are playing a role in neuroprotection allowing for maintenance of cognition in the aged BTBR mice. Therefore, it is possible that the elevated levels of various synaptic markers and proteins identified proteomically such as Picalm, Itsn1, Mobp, Ina, as well as signaling actions involving Wasp functionality served to protect the aging BTBR brain against age-related declines in cognition often associated with decreased BDNF. In the current study we observed a robust maintenance of cognitive ability despite advanced age that may be due to unique neurosynaptic architecture and activity in the ASD phenotype. The role of the associated proteins and synaptic markers provide a foundation for further investigation into this intriguing mechanism.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was carried out with the support of the Intramural Research Program of the National Institute on Aging at the National Institutes of Health. Authors are grateful to Tatiahna Rivera, Vivian Cong, Dianna Augustyniak, Emily Maxwell, Shilpa Kadam for technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnagi.2014.00225/abstract
Figure S1. Collective Textrous! analysis of differentially-regulated cortical protein subsets. Functional hierarchical word-clouds were generated from the upregulated (BTBR:WT iTRAQ ratio > 1.2) (A) and downregulated (BTBR:WT iTRAQ ratio < 0.8) cortical protein data sets.
Figure S2. Collective Textrous! analysis of differentially-regulated hippocampal protein subsets. Functional hierarchical word-clouds were generated from the upregulated (BTBR:WT iTRAQ ratio >1.2) (A) and downregulated (BTBR:WT iTRAQ ratio < 0.8) hippocampal protein data sets.
Abbreviations
BDNF, brain derived neurotrophic factor; BTBR, black and tan, brachyuric; ASD, autism spectrum disorder.
References
American Psychiatric Association. (2012). Diagnostic and Statistical Manual of Mental Disorders: DSM-V. Washington, DC: American Psychiatric Association.
Anitha, A., Nakamura, K., Thanseem, I., Yamada, K., Iwayama, Y., Toyota, T., et al. (2012). Brain region-specific altered expression and association of mitochondria-related genes in autism. Mol. Autism 3:12. doi: 10.1186/2040-2392-3-12
Ballaban-Gil, K., Rapin, L., Tuchman, T., and Shinnar, S. (1996). Longitudinal examination of the behavioral, language, and social changes in a population of adolescent and young adults with autistic disorder. Pediatr. Neurol. 15, 217–223. doi: 10.1016/S0887-8994(96)00219-6
Bhakar, A. L., Dölen, G., and Bear, M. F. (2012). The pathophysiology of fragile X (and what it teaches us about synapses). Ann. Rev. Neurosci. 35, 417–443. doi: 10.1146/annurev-neuro-060909-153138
Blednov, Y. A., Walker, D. L., Iyer, S. V., Homanics, G., and Harris, A. R. (2010). Mice lacking Gad2 show altered behavioral effects of ethanol, flurazepam and gabaxadol. Addict. Biol. 15, 45–61. doi: 10.1111/j.1369-1600.2009.00186.x
Bohlen, M., Cameron, A., Metten, P., Crabbe, J. C., and Wahlsten, D. (2009). Calibration of rotational acceleration for the rotarod test of rodent motor coordination. J. Neurosci. Methods 178, 10–14. doi: 10.1016/j.jneumeth.2008.11.001
Bolivar, V. J., Walter, S. R., and Phoenix, J. L. (2007). Assessing autism-like behavior in mice: variations in social interactions among inbred strains. Behav. Brain Res. 176:2126. doi: 10.1016/j.bbr.2006.09.007
Cai, H., Chen, H., Yi, T., Daimon, C., Boyle, J., Peers, C., et al. (2013). VennPlex-a novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PLoS ONE 8:e53388. doi: 10.1371/journal.pone.0053388
Centers for disease control and prevention. (2010).Available online at: http://www.cdc.gov/ncbddd/dd/default.html [Accessed, 2013]
Chadwick, W., Brenneman, R., Martin, B., and Maudsley, S. (2010a). Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease. Int. J. Alzheimer's Disease 2010:604792. doi: 10.4061/2010/604792
Chadwick, W., Mitchell, N., Caroll, J., Zhou, Y., Park, S. S., Wang, L., et al. (2011). Amitriptylinemediated cognitive enhancement in aged 3 × Tg Alzheimer's disease mice is associated with neurogenesis and neurotrophic activity. PLoS ONE 6:e21660. doi: 10.1371/journal.pone.0021660
Chadwick, W., Zhou, Y., Park, S. S., Wang, L., Mitchell, N., Stone, M., et al. (2010b). Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS ONE 5:e14352. doi: 10.1371/journal.pone.0014352
Chao, H. T., Chen, H., Samaco, R. C., Xue, M., Chahrour, M., Yoo, J., et al. (2010). Dysfunction in GABA signaling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 468, 263–269. doi: 10.1038/nature09582
Chen, H., Martin, B., Daimon, C., Siddiqui, S., Luttrell, L., and Maudsley, S. (2013). Textrous!: extracting semantic textual meaning from gene sets. PLoS ONE 8:e62665. doi: 10.1371/journal.pone.0062665
Cohen, S., and Greenberg, M. E. (2008). Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209. doi: 10.1146/annurev.cellbio.24.110707.175235
Connor, B., Young, D., Yan, Q., Faull, R. L., Synek, B., and Dragunow, M. (1997). Brain-derived neurotrophic factor is reduced in Alzheimer's disease. Mol. Brain Res. 49:7181. doi: 10.1016/S0169-328X(97)00125-3
Coppieters, F., Casteels, I., Meire, F., De Jaegere, S., Hooghe, S., van Regemorter, N., et al. (2010). Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum. Mutat. 31, E1709–E1766. doi: 10.1002/humu.21336
Crawley, J. N. (2007). Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 17, 448–459. doi: 10.1111/j.1750-3639.2007.00096.x
Cukier, H. N., Dueker, N., Slifer, S., Lee, J. M., Whitehead, P. L., Lalanne, E., et al. (2014) J. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol. Autism 5:1. doi: 10.1186/2040-2392-5-1
Cukier, H. N., Lee, J. M., Ma, D., Young, J. I., Mayo, V., Butler, B. L., et al. (2012). The expanding role of MBD genes in autism: identification of a MECP2 duplication and novel alterations in MBD5, MBD6, and SETDB1. Autism Res. 5, 385–397. doi: 10.1002/aur.1251
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi: 10.1016/j.cell.2011.06.013
Defensor, E. B., Pearson, B. L., Pobbe, R. L. H., Bolivar, V. J., Blanchard, D. C., and Blanchard, R. J. (2011). A novel social proximity test suggests patterns of social avoidance and gaze aversion-like behavior in BTBR T + tf/J mice. Behav. Brain Res. 217, 302–308. doi: 10.1016/j.bbr.2010.10.033
Dere, E., Huston, J. P., and De Souza Silva, M. A. (2007). The pharmacology, neuroanatomy and neurogenetics of one-trial object recognition in rodents. Neurosci. Biobehav. Rev. 31, 673–704. doi: 10.1016/j.neubiorev.2007.01.005
Dickson, T. C., Chuckowree, J. A., Chuah, M. I., West, A. K., and Vickers, J. C. (2005). Alphainternexin immunoreactivity reflects variable neuronal vulnerability in Alzheimer's disease and supports the role of the beta-amyloid plaques in inducing neuronal injury. Neurobiol. Dis. 18, 286–295. doi: 10.1016/j.nbd.2004.10.001
Ebert, D., and Greenberg, M. (2013). Activity-dependent neuronal signaling and autism spectrum disorder. Nature 493, 327–337. doi: 10.1038/nature11860
Fahnestock, M., Garzon, D., Holsinger, R. M., and Michalski, B. (2002). Neurotrophic factors and Alzheimer's disease: are we focusing on the wrong molecule? J. Neural. Transm. Suppl. 62, 241–252. doi: 10.1007/978-3-7091-6139-5_22
Ferrer, I., Marin, C., Rey, M. J., Ribalta, T., Goutan, E., Blanco, R., et al. (1999). BDNF and full-length and truncated TrkB expression in Alzheimer disease: implications in therapeutic strategies. J. Neuropathol. Exp. Neurol. 58, 729–739. doi: 10.1097/00005072-199907000-00007
Feyder, M., Karlsson, R., Mathur, P., Lyman, M., Bock, R., Momenan, R., et al. (2010). Association of mouse D1g4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams' syndrome. Am. J. Psychiatry 167, 1508–1517. doi: 10.1176/appi.ajp.2010.10040484
Fombonne, E., Simmons, H., Ford, T., Meltzer, H., and Goodman, R. (2001). Prevalence of pervasive developmental disorders in the British nationwide survey of child mental health. J. Am. Acad. Child Adolesc. Psychiatry 40, 820–827. doi: 10.1097/00004583-200107000-00017
Fruhmesser, A., Blake, J., Haberlandt, E., Baying, B., Raeder, B., Runz, H., et al. (2013). Disruption of EXOC6B in a patient with developmental delay, epilepsy, and a de novo balanced t(2;8) translocation. Eur. J. Hum. Genet. 21, 1177–1180. doi: 10.1038/ejhg.2013.18
Gillberg, C., and Wing, L. (1999). Autism: not an extremely rare disorder. Acta Psychiatr. Scand. 99, 399–406. doi: 10.1111/j.1600-0447.1999.tb00984.x
Gkogkas, C. G., Khoutorsky, A., and Ran, I. (2013). Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377. doi: 10.1038/nature11628
Griswold, A. J., Ma, D., Cukier, H. N., Nations, L. D., Schmidt, M. A., Chung, R. H., et al. (2012). Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum. Mol. Genet. 21:3523. doi: 10.1093/hmg/dds164
Harlow, E. G., Till, S. M., Russell, T. A., Wijetunge, L. S., Kind, P., and Contractor, A. (2010). Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron 65, 385–398. doi: 10.1016/j.neuron.2010.01.024
Harold, D., Abraham, R., Hollingworth, P., Sims, R., Gerrish, A., Hamshere, M. L., et al. (2013). Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 45, 712. doi: 10.1038/ng0613-712a
Hedges, D. J., Hamilton-Nelson, K. L., Sacharow, S. J., Nations, L., Beecham, G. W., Kozhekbaeva, Z. M., et al. (2012). Evidence of novel fine-scale structural variation at autism spectrum disorder candidate loci. Mol. Autism 3:2. doi: 10.1186/20402392-3-2
Hock, C., Heese, K., Hulette, C., Rosenberg, C., and Otten, U. (2000). Region-specific neurotrophin imbalances in Alzheimer disease: decreased levels of brain derived neurotrophic factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch. Neurol. 57, 846–851. doi: 10.1001/archneur.57.6.846
Holmes, A., Wrenn, C. C., Harris, A. P., Thayer, K. E., and Crawley, J. N. (2002). Behavioral profiles of inbred strains on novel olfactory, spatial and emotional tests for reference memory in mice. Genes Brain Behav. 1, 55–59. doi: 10.1046/j.1601-1848.2001.00005.x
Holsinger, R. M., Schnarr, J., Henry, P., Castelo, V. T., and Fahnestock, M. (2000). Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: decreased levels in Alzheimer's disease. Brain Research. Mol. Brain Res. 76, 347–354. doi: 10.1016/S0169-328X(00)00023-1
Howlin, P., Goode, S., Hutton, J., and Rutter, M. (2004). Adult outcome for children with autism. J. Child Psychol. Psychiatry 45, 212–229. doi: 10.1111/j.1469-7610.2004.00215.x
Howlin, P., and Moss, P. (2012). Adults with autism spectrum disorders. Can. J. Psychiatry 57, 275–283.
Hunter, M. P., Nelson, M., Kurzer, M., Wang, X., Kryscio, R. J., Head, E., et al. (2011). Intersectin 1 contributes to phenotypes in vivo: implications for down syndrome. Neuroreport 22, 767–772. doi: 10.1097/WNR.0b013e32834ae348
Hussain, R. J., Stumpo, D. J., Blackshear, P. J., Lenox, R. H., Abel, T., and McNamara, R. K. (2006). Myristoylated alanine rich C kinase substrate (MARCKS) heterozygous mutant mice exhibit deficits in hippocampal mossy fiber-CA3 long-term potentiation. Hippocampus 16, 495–503. doi: 10.1002/hipo.20177
Ji, L., Chauhan, A., and Chauhan, V. (2012). Reduced activity of protein kinase C in the frontal cortex of subjects with regressive autism: relationship with developmental abnormalities. Int. J. Biol. Sci. 8, 1075–1084. doi: 10.7150/ijbs.4742
Jones, E. L., Mok, K., Hanney, M., Harold, D., Sims, R., Williams, J., et al. (2013). Evidence that PICALM affects age at onset of Alzheimer's dementia in down syndrome. Neurobiol. Aging 34, 2441.e1-5. doi: 10.1016/j.neurobiolaging.2013.03.018
Kanner, L., Rodriguez, A., and Ashenden, B. (1972). How far can autistic children go in matters of social adaptation? J. Autism Child. Schizophr. 19, 213–225.
Kelleher, R., and Bear, M. (2008). The autistic neuron: troubled translation? Cell 135:401406. doi: 10.1016/j.cell.2008.10.017
Khalifa, A. M., Watson-Siriboe, A., Shukry, S. G., Chiu, W. L., Nelson, M. E., Geng, Y., et al. (2012). Thr136Ile polymorphism of human vesicular monoamine transporter-1 (SLC18A1 gene) influences its transport activity in vitro. Neuro. Endocrinol. Lett. 33, 546–551.
Liu, C. C., Kanekiyo, T., Xu, H., and Bu, G. (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature Rev. Neurol. 9, 106–118. doi: 10.1038/nrneurol.2012.263
Lohoff, F. W., Weller, A. E., Bloch, P. J., Buono, R. J., Doyle, G. A., Ferraro, T. N., et al. (2008). Association between polymorphisms in the vesicular monoamine transporter 1 gene (VMAT1/SLC18A1) on chromosome 8p and schizophrenia. Neuropsychobiology 57, 56–60. doi: 10.1159/000129668
Lotter, V. (1974). Social adjustment and placement of autistic children in Middlesex: a follow-up study. J. Autism Child. Schizophr. 4, 11–32. doi: 10.1007/BF02104997
Martin, B., Brenneman, R., Golden, E., Walent, T., Becker, K., Prabhu, V., et al. (2009a). Growth factor signals in neural cells: coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J. Biol. Chem. 284, 2493–2511. doi: 10.1074/jbc.M804545200
Martin, B., Pearson, M., Brenneman, R., Golden, E., Wood, W. I., Prabhu, V., et al. (2009b). Gonadal transcriptome alterations in response to dietary energy intake: sensing the reproductive environment. PLoS ONE 4:e4146. doi: 10.1371/journal.pone.0004146
Mattson, M. P., Maudsley, S., and Martin, B. (2004). BDNF and 5-HT: a dynamic duo in age-related neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 27, 589–594. doi: 10.1016/j.tins.2004.08.001
Maudsley, S., Chadwick, W., Wang, L., Zhou, Y., Martin, B., and Park, S. S. (2011). Bioinformatic approaches to metabolic pathways analysis. Methods Mol. Biol. 756, 99–130. doi: 10.1007/978-1-61779-160-4_5
Maudsley, S., Naor, Z., Bonfil, D., Davidson, L., Karali, D., Pawson, A., et al. (2007). Proline-rich tyrosine kinase 2 mediates gonadotropin-releasing hormone signaling to a specific extracellularly regulated kinase-sensitive transcriptional locus in the luteinizing hormone beta-subunit gene. Mol. Endocrinol. 21, 1216–1233. doi: 10.1210/me.2006-0053
Mitew, S., Hay, C., Peckham, H., Xiao, J., Koenning, M., and Emery, B. (2013). Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. doi: 10.1016/j.neuroscience.2013.11.029. [Epub ahead of print].
Moy, S. S., and Nadler, J. J. (2008). Advances in behavioral genetics: mouse models of autism. Mol. Psychiatry 13, 4–26. doi: 10.1038/sj.mp.4002082
Moy, S. S., Nadler, J. J., Young, N. B., Perez, A., Holloway, L. P., Barbaro, R. P., et al. (2007). Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav. Brain Res. 176, 4–20. doi: 10.1016/j.bbr.2006.07.030
Mundy, P. (2003). The neural basis of social impairments in autism: the role of the dorsal medial-frontal cortex and anterior cingulate system. J. Child Psychol. Psychiatry 44, 793–809. doi: 10.1111/1469-7610.00165
Murray, K. D., Gall, C. M., Jones, E. G., and Isackson, P. J. (1994). Differential regulation of brain-derived neurotrophic factor and type II calcium/calmodulin dependent protein kinase messenger RNA expression in Alzheimer's disease. Neuroscience 60, 37–48. doi: 10.1016/0306-4522(94)90202-X
Nadler, J. J., Zou, F., Huang, H., Moy, S. S., Lauder, J., Crawley, J. N., et al. (2006). Large scale gene expression differences across brain regions and inbred strain correlate with behavioral phenotype. Genetics 174, 1229–1236. doi: 10.1534/genetics.106.061481
Nishimura, K., Lee, S. B., Park, J. H., and Park, M. H. (2012). Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 42, 703–710. doi: 10.1007/s00726-011-0986-z
Noor, A., Whibley, A., Marshall, C., Gianakopoulos, P., Piton, A., Carson, A., et al. (2010). Disruption at the PTCHDI locus on Xp22.11 in autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2:49ra68. doi: 10.1126/scitranslmed.3001267
Park, J., Shimojo, E., and Shimojo, S. (2010). Roles of familiarity and novelty in visual preference judgments are segregated across object categories. Proc. Natl. Acad. Sci. U.S.A. 107, 14552–14555. doi: 10.1073/pnas.1004374107
Pathania, M., Davenport, E., Muir, J., Sheehan, D., Lopez-Domenech, G., and Kittler, J. (2014). The autism and schizophrenia associated gene CYFIPI is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry 4:e374. doi: 10.1038/tp.2014.36
Pearson, B. L., Pobbe, R. L. H., Defensor, E. B., Osay, L., Bolivar, V. J., Blanchard, D. C., et al. (2011). Motor and cognitive stereotypies in the BTBR T+tf/J mouse model of autism. Genes Brain Behav. 10, 228–235. doi: 10.1111/j.1601-183X.2010.00659.x
Petit, T. L. (1988). Synaptic plasticity and the structural basis of learning and memory. Neurol. Neurobiol. Neural. Plast. 36, 201–234.
Phillips, H. S., Hains, J. M., Armanini, M., Laramee, G. R., and Johnson, S. A. (1991). BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer's disease. Neuron 7:695702.
Piven, J., Bailey, A. S., and Andreasen, N. (1996). Regional brain enlargement in autism: a magnetic resonance imaging study. J. Am. Acad. Child Adolesc. Psychiatry 35, 530–536. doi: 10.1097/00004583-199604000-00020
Piven, J., and Rabins, P. (2011). Autism spectrum disorders in older adults: toward defining a research agenda. J. Am. Geriatr. Soc. 59, 2151–2155. doi: 10.1111/j.1532-5415.2011.03632.x
Pobbe, R. L., Defensor, E. B., Pearson, B. L., Bolivar, V. J., and Blanchard, D. C. (2010). Expression of social behaviors of C57BL/6J versus BTBR inbred mouse strains in the visible burrow system. Behav. Brain Res. 214, 443–449. doi: 10.1016/j.bbr.2010.06.025
Quartu, M., Lai, M. L., and Del Fiacco, M. (1999). Neurotrophin-like immunoreactivity in the human hippocampal formation. Brain Res. Bull. 48, 375–382. doi: 10.1016/S0361-9230(99)00009-X
Renty, J. O., and Roeyers, H. (2006). Quality of life in high-functioning adults with autism spectrum disorder: the predictive value of disability and support characteristics. Autism 10, 511–524. doi: 10.1177/1362361306066604
Romanczyk, T. B., Weickert, C. S., Webster, M. J., Herman, M. M., Akil, M., and Kleinman, J. E. (2002). Alterations in TrkB mRNA in the human prefrontal cortex throughout the lifespan. Eur. J. Neurosci. 15, 269–280. doi: 10.1046/j.0953-816x.2001.01858.x
Rumsey, J. M., Rapoport, J. L., and Sceery, W. R. (1985). Autistic children as adults: psychiatric, social, and behavioral outcomes. J. Am. Acad. Child Psychiatry 24, 465473. doi: 10.1016/S0002-7138(09)60566-5
Rutter, M., Greenfeld, D., and Lockyer, L. (1967). A five to fifteen year follow-up study of infantile psychosis (II. Social and behavioural outcome). Br. J. Psychiatry 113, 1183–1199. doi: 10.1192/bjp.113.504.1183
Scattoni, M. L., Gandhy, S. U., Ricceri, L., and Crawley, J. N. (2008). Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS ONE 3:e3067. doi: 10.1371/journal.pone.0003067
Scattoni, M. L., Ricceri, L., and Crawley, J. N. (2011). Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 10:4446. doi: 10.1111/j.1601-183X.2010.00623.x
Seltzer, M. M., Krauss, M. W., Shattuck, P. T., Orsmond, G., Swe, A., and Lord, C. (2003). The symptoms of autism spectrum disorders in adolescence and adulthood. J. Autism Dev. Disord. 33, 565–581. doi: 10.1023/B:JADD.0000005995.02453.0b
Shattuck, P. T., Seltzer, M. M., Greenberg, J. S., Orsmond, G. I., Bolt, D., Kring, S., et al. (2007). Change in autism symptoms and maladaptive behaviors in adolescents and adults with an autism spectrum disorder. J. Autism Dev. Disord. 37, 1735–1747. doi: 10.1007/s10803-006-0307-7
Sokol, D. K., Maloney, B., Long, J. M., Ray, B., and Lahiri, D. K. (2011). Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links. Neurology 76:13441352. doi: 10.1212/WNL.0b013e3182166dc7
Stamova, B., Tian, Y., Nordahl, C., Shen, M., Rogers, S., Amaral, D., et al. (2013). Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders. Mol. Autism 4:30. doi: 10.1186/2040-2392-4-30
Stumpo, D. J., Bock, C. B., Tuttle, J. S., and Blackshear, P. J. (1995). MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc. Natl. Acad. Sci. U.S.A. 92, 944–948.
Suckow, A. T., Craige, B., Faundez, V., Cain, W. J., and Chessler, S. D. (2010). An AP-3dependent mechanism drives synaptic-like microvesicle biogenesis in pancreatic islet beta-cells. Am. J. Physiol. Endocrinol. Metab. 299, E23–E32. doi: 10.1152/ajpendo.00664.2009
Szatmari, P., Bartolucci, G., Bremner, R., Bond, S., and Rich, S. (1989). A follow up study of high-functioning autistic children. J. Autism Dev. Disord. 19, 213–225. doi: 10.1007/BF02211842
Szatmari, P., Archer, L., Fisman, S., Streiner, D. L., and Wilson, F. (1995). Asperger's syndrome and autism: differences in behavior, cognition, and adaptive functioning. J. Am. Acad. Child Adolesc. Psychiatry, 34, 1662–1671. doi: 10.1097/00004583-199512000-00017
Szatmari, P. (1998). Differential diagnosis of Asperger's disorder. Asperger Syndr. High Funct. Autism? 7, 61–76. doi: 10.1007/978-1-4615-5369-4_4
Tapia-Arancibia, L., Aliaga, E., Silhol, M., and Arancibia, S. (2008). New insights into brain BDNF function in normal aging and Alzheimer disease. Brain Res. Rev. 59, 201–220. doi: 10.1016/j.brainresrev.2008.07.007
Weber, A., Kohler, A., Hahn, A., Neubauer, B., and Muller, U. (2013). Benign infantile convulsions (IC) and subsequent paroxysmal kinesigenic dyskinesia (PKD) in a patient with 16pl 1.2 microdeletion syndrome. Neurogenetics 14, 251–253. doi: 10.1007/s10048-013-0376-7
Weimer, J. M., Yokota, Y., Stanco, A., Stumpo, D. J., Blackshear, P. J., and Anton, E. S. (2009). MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development 136, 2965–2975. doi: 10.1242/dev.036616
Wohr, M., Roullet, F. I., and Crawley, J. N. (2011). Reduced scent marking and ultrasonic vocalizations in the BTBR T+tf/J mouse model of autism. Genes Brain Behav. 10, 35–43. doi: 10.1111/j.1601-183X.2010.00582.x
Zhao, L., Yao, J., Mao, Z., Chen, S., Wang, Y., and Brinton, R. D. (2010). 17β-estradiol regulates insulin-degrading enzyme expression via an ERβ /PI3-K pathway in hippocampus: relevance to Alzheimer's prevention. Neurobiol. Aging 32, 1949–1963. doi: 10.1016/j.neurobiolaging.2009.12.010
Keywords: ASD, autism, BDNF, aging, synaptic marker, neuroprotection, neurodevelopmental disorder, BTBR
Citation: Jasien JM, Daimon CM, Wang R, Shapiro BK, Martin B and Maudsley S (2014) The effects of aging on the BTBR mouse model of autism spectrum disorder. Front. Aging Neurosci. 6:225. doi: 10.3389/fnagi.2014.00225
Received: 27 June 2014; Accepted: 08 August 2014;
Published online: 01 September 2014.
Edited by:
Fernanda Laezza, University of Texas Medical Branch, USACopyright © 2014 Jasien, Daimon, Wang, Shapiro, Martin and Maudsley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joan M. Jasien, Department of Neurology, Johns Hopkins University School of Medicine, Kennedy Krieger Institute, 251 Bayview Blvd., Suite 100, 716 North Broadway, # 441, Baltimore, MD 21205, USA e-mail: jasien@kennedykrieger.org;
Stuart Maudsley, VIB-Department of Molecular Genetics, University of Antwerp, Antwerp 2610, Belgium e-mail: stuart.maudsley@molgen.vib-ua.be
 Joan M. Jasien
Joan M. Jasien Caitlin M. Daimon
Caitlin M. Daimon Rui Wang
Rui Wang Bruce K. Shapiro
Bruce K. Shapiro Bronwen Martin
Bronwen Martin Stuart Maudsley
Stuart Maudsley