The role of microRNAs in regulating neuronal connectivity
- Division of Developmental Biology, Cincinnati Children’s Hospital Research Foundation, Cincinnati, OH, USA
The assembly of functional neural circuits is critical for complex thoughts, behavior and general brain function. Precise construction of neural circuits requires orderly transition of sequential events from axon outgrowth, pathfinding, branching, to synaptogenesis. Each of these steps is required to be tightly regulated in order to achieve meticulous formation of neuronal connections. MicroRNAs (miRNAs), which silence gene expression post-transcriptionally via either inhibition of translation or destabilization of messenger RNAs, have emerged as key regulators of neuronal connectivity. The expression of miRNAs in neurons is often temporally and spatially regulated, providing critical timing and local mechanisms that prime neuronal growth cones for dynamic responses to extrinsic cues. Here we summarize recent findings of miRNA regulation of neuronal connectivity in a variety of experimental platforms.
The diverse behaviors of organisms rely on fast information processing performed by the brain. Trillions of neurons comprising the brain form complex networks that enable animals to exhibit consciousness, accumulate memories, engage in learning, and adopt behaviors. The complexity of brain networks is greatly increased by the facts that one neuron can influence its target through multiple pathways (Gutierrez et al., 2013), and that common neurons shared by divergent circuits can modulate reciprocal inhibition between two mutually exclusive behaviors in response to environmental stimuli (Mann et al., 2013). The former case is achieved by the direct and indirect connections between two neurons via two or more synaptic routes and the latter depends on the appropriate information flow from sensory neurons to interneurons as well as from interneurons to motor neurons (Gutierrez et al., 2013; Mann et al., 2013). Both cases indicate the importance of precise connections between neurons to brain function. Precise neuronal connectivity is established through transition of sequential events from axon growth to synapse formation. Axons are attracted to targets, but upon arrival they must switch their responsiveness to guidance cues at the targets such that they are no longer sensitive to these cues, in order to stop outgrowth and form synaptic contacts (Tessier-Lavigne and Goodman, 1996; Stein and Tessier-Lavigne, 2001; Dickson, 2002; Chen et al., 2008). Miswiring of the nervous system can result in serious neurological deficits, such as autism, Parkinson’s, and Alzheimer’s diseases (Hoogland et al., 2003; Lesnick et al., 2007; Zikopoulos and Barbas, 2010). Thus, studying how neurons connect with each other to establish functional circuitry can help us better understand how animal behaviors go awry and may provide insights into potential therapeutic targets for neurological disorders.
Neural Circuit Assembly
Neurons connect with targets through a series of events: axon initiation, axon pathfinding, axon branching, and synapse formation (Carmeliet and Tessier-Lavigne, 2005; Kolodkin and Tessier-Lavigne, 2011). A single axon grows out from the cell body, and forms a highly motile structure called the growth cone at its tip. The growth cone navigates along the stereotypical pathway via interactions with a variety of guidance cues present in the environment and travels a long distance to reach the target with remarkable precision. Upon reaching the target, the growth cone turns into a presynaptic terminal to form a connection with the target, and the axon starts branching extensively to establish an intricate pattern of connectivity. Each step of neuronal circuit assembly involves local protein synthesis (Campbell and Holt, 2001; Jung et al., 2012). Specific mRNAs are anterogradely transported from the cell body to the axon and to the growth cone to construct local transcriptomes (Zivraj et al., 2010; Gumy et al., 2011; Willis et al., 2011; Cajigas et al., 2012). The expression of mRNAs is tightly controlled by many post-transcriptional regulatory mechanisms, of which microRNA (miRNA)-mediated gene repression is one of them (Bushati and Cohen, 2007; Deglincerti and Jaffrey, 2012; Jung et al., 2012). The fast and dynamic changes in the local proteome enable growth cones to respond rapidly to diverse environmental cues, resulting in elongation, turning, or collapse of growth cones (Campbell and Holt, 2001; Hengst et al., 2009; Andreassi et al., 2010; Zivraj et al., 2010). The post-transcriptional regulators, therefore, play a pivotal role in the establishment of neuronal connections.
miRNAs as Versatile and Reversible Regulators of Gene Expression in Neurons
MiRNA-mediated gene regulation is involved in many aspects of neuronal development and function (Kosik, 2006; Schratt, 2009; McNeill and Van Vactor, 2012). Recent studies have worked out a few molecular mechanics of miRNA-mediated gene silencing (Fabian and Sonenberg, 2012). These small non-coding RNAs bind to the 3′UTR of target mRNAs and repress gene expression by interfering with stability or inhibiting translation of mRNAs (Bartel, 2009). The pleiotropy, speed, and reversibility are features that contribute to the unique regulatory niche of miRNA-based gene regulation in the nervous system (Hobert, 2008). First, individual miRNAs can target multiple genes at the same time to cause broad and significant changes in neuronal transcriptomes (Brennecke et al., 2005; Giraldez et al., 2006). On the other hand, each gene can be targeted by different miRNAs, and/or may contain more than one binding site of the same miRNA, allowing miRNAs to “fine-tune” the level of gene expression (Bartel and Chen, 2004; Hon and Zhang, 2007). Second, the effect of miRNA-mediated gene repression is instant because miRNAs can shut down protein synthesis of the target genes at ribosomes (Pillai et al., 2005). The small size and non-coding nature also allows fast production of miRNAs as compared to transcription factors (Hobert, 2008). Lastly, the miRNA-mediated gene repression can be easily relieved by translocating the targeted mRNA from a miRNA-hijacked ribosome to an active one (Bhattacharyya et al., 2006). In summary, miRNAs provide a local, versatile, fast and reversible mechanism that is sensitive to extracellular stimuli and provides exquisite control of local protein dynamics and axon development.
Spatial and Temporal Roles of miRNAs in Neuronal Connectivity
MiRNA expression is either spatially restricted or temporally regulated in neuronal development. The spatially restricted expression of miRNAs within neurons suggests roles for miRNAs in diverse differentiation events from axon pathfinding to synapse formation. Many miRNAs are expressed specifically in the nervous system, and even in distinct neuronal subsets, implying potentially unique roles in specific cell types. (Lagos-Quintana et al., 2002; He et al., 2012). Furthermore, miRNAs can be localized to subcellular compartments, such as axons, growth cones or synapses, to rapidly alter local gene expression profiles, but not much is known about the transport mechanisms that provide that specificity (Corbin et al., 2009; Natera-Naranjo et al., 2010; Kaplan et al., 2013; Sasaki et al., 2013). The temporally regulated expression of miRNAs within neurons suggests a role in the orderly transition of sequential differentiation events (Johnston and Hobert, 2003; Chang et al., 2004b; Hsieh et al., 2012; Zou et al., 2012, 2013; Chiu and Chang, 2013). Recent evidence indeed showed that miRNAs can provide timing mechanisms for orderly developmental events in neurons (Zou et al., 2012). Significant changes in the expression level of some brain-specific miRNAs during cortical neurogenesis have been reported previously (Krichevsky et al., 2003), suggesting that gene expression profiles may be controlled by the up- or down-regulation of certain miRNAs at each stage of brain development. Thus, the compartmentalized expression of miRNAs provides subcellular control of local gene expression for specific neuronal differentiation events while the temporal constraint of miRNA expression allows for the correct transition timing of sequential differentiation events.
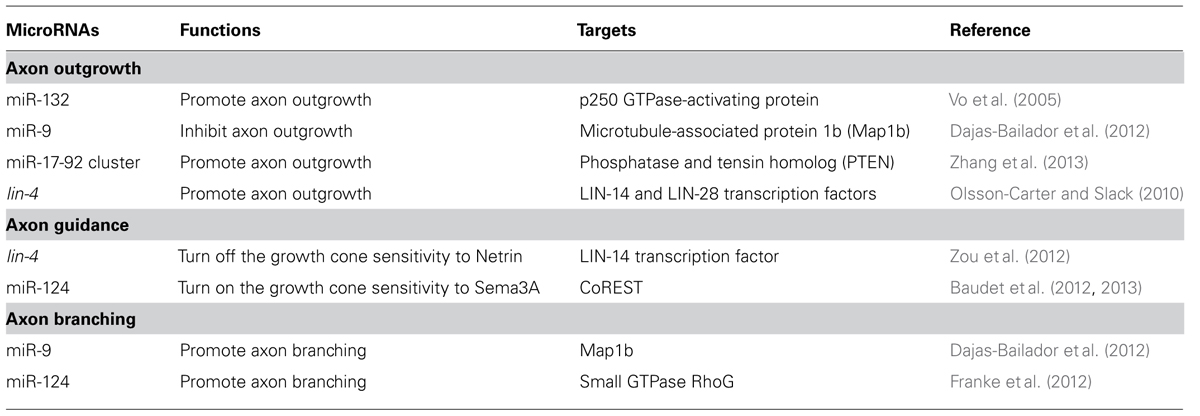
Control of Axon Outgrowth by miRNAs
The neural circuit assembly begins with axon outgrowth. In the early phase of neuronal development, a neuron first forms multiple naïve neurites around the soma. One of the neurites will be specified as an axon and extend further while the remaining become dendrites (Craig and Banker, 1994). Actin and microtubule dynamics are required for the neurite formation and elongation (Bradke and Dotti, 1999; Inagaki et al., 2001). Therefore, it is not surprising that miRNAs affect axon initiation or elongation by targeting regulators of the cytoskeleton (Table 1). miR-132, for example, induces neurite sprouting of cortical neurons. It does so by inhibiting the p250 GTPase-activating protein that acts upstream of small GTPases Cdc42 and RhoA, to regulate neuronal morphogenesis (Nakazawa et al., 2003; Vo et al., 2005). miRNAs can also regulate axon outgrowth by modulating local protein synthesis. miR-9 locally represses the translation of the microtubule-associated protein 1b (Map1b) in axons to control axon elongation of mouse cortical neurons (Dajas-Bailador et al., 2012). Overexpression of miR-9 decreases the Map1b regulatory effects on axonal microtubules, resulting in the reduction of axon length (Dajas-Bailador et al., 2012; Tymanskyj et al., 2012). miR-19a, a major member of the miR-17-92 cluster, acts in axons to down-regulate the protein level of phosphatase and tension homolog (PTEN) and activate phosphorylated mammalian target of rapamycin (mTOR) pathway (Zhang et al., 2013). mTOR activity is known to be required for local protein synthesis in axonal development and regeneration (Campbell and Holt, 2001; Park et al., 2008). Thus, miR-17-92 cluster promotes axon outgrowth by activating local protein synthesis (Zhang et al., 2013). Although several miRNAs have been shown to affect axon development, few have been shown to play a role specifically in the axon compartment (Aschrafi et al., 2008; Dajas-Bailador et al., 2012; Zhang et al., 2013). In addition, miRNAs also contribute to the temporal regulation of axon outgrowth. In Caenorhabditis elegans, the hermaphrodite specific neuron (HSN) projects a single axon to the ventral nerve cord in the fourth larva (L4) stage (Adler et al., 2006). The heterochronic miRNA lin-4 acts cell-autonomously in HSN neurons to control the timing of axon formation (Olsson-Carter and Slack, 2010). No axon is extended at the L4 stage in lin-4 mutants while precocious axon outgrowth at the third larval stage occurs in animals over-expressing lin-4 in HSN neurons (Olsson-Carter and Slack, 2010). The targets of the lin-4 miRNA, lin-14 and lin-28, inhibit differentiation of HSN neurons. Thus, the lin-4 miRNA signals axon outgrowth only after HSN neuronal fate is committed (Olsson-Carter and Slack, 2010).
miRNA Regulation of Axon Pathfinding
Axons are directed to their synaptic targets via the guidance of attractive or repellent cues presented in the environment (Tessier-Lavigne and Goodman, 1996). The sensitivity of axons to guidance cues is determined by the expression of the corresponding receptors in growth cones (Dickson, 2002). To prevent the axon from stalling at intermediate targets or overshooting, the receptor expression needs to be tightly controlled in a timely manner (Zou et al., 2000; Stein and Tessier-Lavigne, 2001). Here we discuss the role of miRNAs in regulating the growth cone responsiveness to two prominent guidance molecules, netrins and semaphorins, during axon pathfinding.
Netrins are highly conserved guidance molecules that can function as both attractants and repellents in many stage-dependent biological events, such as axon guidance and motile cell migration (Hedgecock et al., 1990; Ishii et al., 1992; Chan et al., 1996; Kolodkin and Tessier-Lavigne, 2011), but the timing mechanism that controls the responsiveness of growth cones or migrating cells to Netrins at precise times is not fully understood. The axon of the C. elegans anterior ventral microtubule (AVM) sensory neurons is guided to the ventral midline through combined actions of Slit repulsion from the dorsal body wall muscles and netrin attraction to the ventral nerve cord (Chang et al., 2004a). Once reaching the ventral midline, the AVM axon projects anteriorly to the nerve ring where it stops outgrowth and forms synapses (Figure 1A). An unexpected role was recently reported for the conserved heterochronic miRNA lin-4 and its target the LIN-14 transcription factor in AVM neuronal connectivity (Zou et al., 2012). Through genetic analysis of a well-characterized AVM axon ventral guidance event and a less characterized AVM synapse formation event, it was shown that lin-4 functions as a potent and specific negative regulator of netrin signaling in AVM neuronal connectivity by targeting the LIN-14 transcription factor to control the availability of the netrin receptor UNC-40/DCC (Deleted in Colorectal Cancer; Figure 1B; Zou et al., 2012). It was well known that heterochronic genes are used in timing mitotic cell development required for molting in worms and embryonic stem cells self-renewal in mice. These results show that these heterochronic genes are re-used in postmitotic neurons to time their differentiation events.
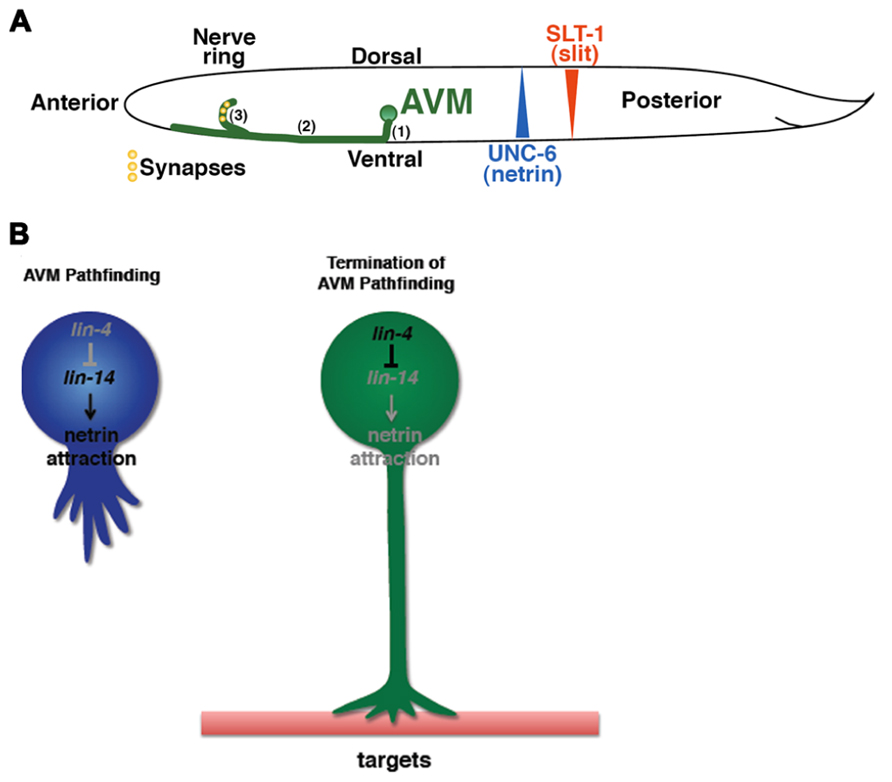
FIGURE 1. Identification of the timing mechanism for orderly AVM axon connectivity. (A) Sequential events of AVM axon outgrowth and synapse formation in the first larval (L1) stage of C. elegans. (1) Axon grows ventrally. (2) Axon grows anteriorly. (3) End of axon pathfinding and initiation of synapse formation in the nerve ring. (B) The lin-4–lin-14 regulatory circuit signals the end of axon pathfinding.
The Semaphorin family contains both secreted and transmembrane proteins that can function in long- or short-range guidance (Yazdani and Terman, 2006; Kolodkin and Tessier-Lavigne, 2011). Many Semaphorins bind the major receptor, Plexins, solely to mediate axonal repulsion. However, some of the secreted Semaphorins, such as Sema3A, bind to the co-receptor Neuropilins, instead (Kolodkin and Tessier-Lavigne, 2011). It has been shown that Sema3A is able to induce growth cone collapse at a specific stage of retinal ganglion cell (RGC) development in Xenopus (Campbell et al., 2001). The Sema3A responsiveness depends on the up-regulation of neuropilin-1 (NPR-1) receptor, which is indirectly controlled by the miRNA, miR-124, in a timely manner (Campbell et al., 2001; Baudet et al., 2012, 2013). The Repressor element-1 silencing transcription factor (REST) works with the REST corepressor 1 (CoREST) to inhibit the NPR-1 expression in young RGCs (Baudet et al., 2012, 2013). As the development progresses, the up-regulation of miR-124 accelerates the decay of CoREST mRNA, thereby attenuating the repression of NPR-1 expression (Baudet et al., 2012, 2013). The onset of Sema3A sensitivity of RGC growth cones is delayed in miR-124 knockdown, which is attributed to the retarded expression of NPR-1 (Baudet et al., 2012, 2013). Thus, miR-124 acts as an intrinsic timer to turn on the responsiveness of RGC growth cone to Sema3A by up-regulating NPR-1.
Slit is another key guidance molecule that is able to elicit axonal repulsion from a distance (Kolodkin and Tessier-Lavigne, 2011). The miRNA regulation of Slit signaling has been reported in the context of vascular patterning, but whether miRNAs control Slit signaling at specific times is unknown (Small et al., 2010). Even though no evidence has been shown that the Slit signaling is subject to the miRNA regulation in the context of axon guidance, it is likely that miRNAs may modulate Slit-mediated axon guidance since many molecular mechanisms are shared between vascular and axonal patterning (Carmeliet and Tessier-Lavigne, 2005).
Unique Roles of miRNAs in Axon Branching
Once axons are in the vicinity of synaptic targets, they slow down outgrowth and branch out to form elaborate neural networks (Acebes and Ferrús, 2000). Similar to axon outgrowth, axon branching relies on cytoskeleton rearrangement, so cytoskeletal regulators likely coordinate both events in response to environmental cues. As mentioned above, miR-9 regulates microtubule dynamics by targeting the Map1b to control axon outgrowth (Dajas-Bailador et al., 2012). The miR-9 level in mouse cortical neurons is regulated by the brain-derived neurotrophic factor (BDNF) that functions as a chemoattractant and also a branching factor (Hoshino et al., 2010; Panagiotaki et al., 2010). In the initial phase of cortical neuron development in vitro, shorter exposure to BDNF results in reduced miR-9 expression, which leads to high level of Map1b expression and steady axon outgrowth. Prolonged exposure to BDNF, which could mimic the target recognition phase in vivo, results in elevated miR-9 expression, which leads to axon branching as a result of the repressed Map1b expression (Dajas-Bailador et al., 2012). In hippocampal neurons, it was shown that miR-124 regulates axon and dendrite branching by targeting the small GTPase RhoG (Franke et al., 2012). The RhoG activity inhibits axon branching via the ELMO/Dock180/Rac1 pathway, while reducing dendrite branching through the Cdc42 signaling. Therefore, expression of miR-124 in hippocampal neurons promotes axon and dendrite branching (Franke et al., 2012).
Concluding Remarks
The exquisite precision with which neural circuits are assembled is crucial for proper brain function, as inappropriate wiring in the nervous system results in various debilitating neurological diseases. Neurons are able to form appropriate connections using processes that involve spatial and temporal regulatory mechanisms, which ensure rapid responses to diverse environmental cues and faithful transition of sequential events in neuronal connectivity. miRNAs have proven to be essential and efficient regulators at several steps of this integral process due to their spatiotemporal specificity, versatile targeting, speed of gene repression, and ease of reversibility. Further study on how miRNAs contribute to the formation of neural circuits will ultimately provide insights into how mis-wiring by miRNA mis-regulation can lead to diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by grants from the Whitehall Foundation, the March of Dimes Foundation, and by NSF grant IOS-1257023 to Chieh Chang.
References
Acebes, A., and Ferrús, A. (2000). Cellular and molecular features of axon collaterals and dendrites. Trends Neurosci. 23, 557–565. doi: 10.1016/S0166-2236(00)01646-5
Adler, C. E., Fetter, R. D., and Bargmann, C. I. (2006). UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9, 511–518. doi: 10.1038/nn1666
Andreassi, C., Zimmermann, C., Mitter, R., Fusco, S., De Vita, S., Saiardi, A., et al. (2010). An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 13, 291–301. doi: 10.1038/nn.2486
Aschrafi, A., Schwechter, A. D., Mameza, M. G., Natera-Naranjo, O., Gioio, A. E., and Kaplan, B. B. (2008). MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 28, 12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bartel, D. P., and Chen, C. Z. (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5, 396–400. doi: 10.1038/nrg1328
Baudet, M. L., Bellon, A., and Holt, C. E. (2013). Role of microRNAs in Semaphorin function and neural circuit formation. Semin. Cell Dev. Biol. 24, 146–155. doi: 10.1016/j.semcdb.2012.11.004
Baudet, M. L., Zivraj, K. H., Abreu-Goodger, C., Muldal, A., Armisen, J., Blenkiron, C., et al. (2012). miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 15, 29–38. doi: 10.1038/nn.2979
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124. doi: 10.1016/j.cell.2006.04.031
Bradke, F., and Dotti, C. G. (1999). The role of local actin instability in axon formation. Science 283, 1931–1934. doi: 10.1126/science.283.5409.1931
Brennecke, J., Stark, A., Russell, R. B., and Cohen, S. M. (2005). Principles of microRNA-target recognition. PLoS Biol. 3:e85. doi: 10.1371/journal.pbio.0030085
Bushati, N., and Cohen, S. M. (2007). MicroRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205. doi: 10.1146/Annurev.Cellbio.23.090506.123406
Cajigas, I. J., Tushev, G., Will, T. J., Tom Dieck, S., Fuerst, N., and Schuman, E. M. (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466. doi: 10.1016/j.neuron.2012.02.036
Campbell, D. S., and Holt, C. E. (2001). Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026. doi: 10.1016/S0896-6273(01)00551-7
Campbell, D. S., Regan, A. G., Lopez, J. S., Tannahill, D., Harris, W. A., and Holt, C. E. (2001). Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J. Neurosci. 21, 8538–8547.
Carmeliet, P., and Tessier-Lavigne, M. (2005). Common mechanisms of nerve and blood vessel wiring. Nature 436, 193–200. doi: 10.1038/nature03875
Chan, S. S., Zheng, H., Su, M. W., Wilk, R., Killeen, M. T., Hedgecock, E. M., et al. (1996). UNC-40, a C. elegans homolog of DCC (Deleted in Colorectal Cancer), is required in motile cells responding to UNC-6 netrin cues. Cell 87, 187–195. doi: 10.1016/S0092-8674(00)81337-9
Chang, C., Yu, T. W., Bargmann, C. I., and Tessier-Lavigne, M. (2004a). Inhibition of Netrin-mediated axon attraction by a receptor protein tyrosine phosphatase. Science 305, 103–106. doi: 10.1126/science.1096983
Chang, S., Johnston, R. J., Frokjaer-Jensen, C., Lockery, S., and Hobert, O. (2004b). MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430, 785–789. doi: 10.1038/Nature02752
Chen, Z., Gore, B. B., Long, H., Ma, L., and Tessier-Lavigne, M. (2008). Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58, 325–332. doi: 10.1016/j.neuron.2008.02.016
Corbin, R., Olsson-Carter, K., and Slack, F. (2009). The role of microRNAs in synaptic development and function. BMB Rep. 42, 131–135.
Craig, A. M., and Banker, G. (1994). Neuronal polarity. Annu. Rev. Neurosci. 17, 267–310. doi: 10.1146/annurev.ne.17.030194.001411
Dajas-Bailador, F., Bonev, B., Garcez, P., Stanley, P., Guillemot, F., and Papalopulu, N. (2012). microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15, 697–699. doi: 10.1038/nn.3082
Deglincerti, A., and Jaffrey, S. R. (2012). Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2, 120079. doi: 10.1098/rsob.120079
Dickson, B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964. doi: 10.1126/science.1072165
Fabian, M. R., and Sonenberg, N. (2012). The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593. doi: 10.1038/nsmb.2296
Franke, K., Otto, W., Johannes, S., Baumgart, J., Nitsch, R., and Schumacher, S. (2012). miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 31, 2908–2921. doi: 10.1038/emboj.2012.130
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. doi: 10.1126/science.1122689
Gumy, L. F., Yeo, G. S. H., Tung, Y.-C. L., Zivraj, K. H., Willis, D., Coppola, G., et al. (2011). Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA 17, 85–98. doi: 10.1261/rna.2386111
Gutierrez, G. J., O’Leary, T., and Marder, E. (2013). Multiple mechanisms switch an electrically coupled, synaptically inhibited neuron between competing rhythmic oscillators. Neuron 77, 845–858. doi: 10.1016/j.neuron.2013.01.016
He, M., Liu, Y., Wang, X., Zhang, M. Q., Hannon, G. J., and Huang, Z. J. (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73, 35–48. doi: 10.1016/j.neuron.2011.11.010
Hedgecock, E. M., Culotti, J. G., and Hall, D. H. (1990). The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4, 61–85. doi: 10.1016/0896-6273(90)90444-K
Hengst, U., Deglincerti, A., Kim, H. J., Jeon, N. L., and Jaffrey, S. R. (2009). Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat. Cell Biol. 11, 1024–1030. doi: 10.1038/ncb1916
Hobert, O. (2008). Gene regulation by transcription factors and microRNAs. Science 319, 1785–1786. doi: 10.1126/science.1151651
Hon, L. S., and Zhang, Z. (2007). The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 8, R166. doi: 10.1186/gb-2007-8-8-r166
Hoogland, P. V., Van Den Berg, R., and Huisman, E. (2003). Misrouted olfactory fibres and ectopic olfactory glomeruli in normal humans and in Parkinson and Alzheimer patients. Neuropathol. Appl. Neurobiol. 29, 303–311. doi: 10.1046/j.1365-2990.2003.00459.x
Hoshino, N., Vatterott, P., Egwiekhor, A., and Rochlin, M. W. (2010). Brain-derived neurotrophic factor attracts geniculate ganglion neurites during embryonic targeting. Dev. Neurosci. 32, 184–196. doi: 10.1159/000313902
Hsieh, Y. W., Chang, C., and Chuang, C. F. (2012). The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 8:e1002864. doi: 10.1371/journal.pgen.1002864
Inagaki, N., Chihara, K., Arimura, N., Menager, C., Kawano, Y., Matsuo, N., et al. (2001). CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci. 4, 781–782. doi: 10.1038/90476
Ishii, N., Wadsworth, W. G., Stern, B. D., Culotti, J. G., and Hedgecock, E. M. (1992). UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron 9, 873–881. doi: 10.1016/0896-6273(92)90240-E
Johnston, R. J., and Hobert, O. (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845–849. doi: 10.1038/nature02255
Jung, H., Yoon, B. C., and Holt, C. E. (2012). Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 13, 308–324. doi: 10.1038/nrn3210
Kaplan, B. B., Kar, A. N., Gioio, A. E., and Aschrafi, A. (2013). MicroRNAs in the axon and presynaptic nerve terminal. Front. Cell. Neurosci. 7:126. doi: 10.3389/fncel.2013.00126
Kolodkin, A. L., and Tessier-Lavigne, M. (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3, pii: a001727. doi: 10.1101/cshperspect.a001727
Kosik, K. S. (2006). The neuronal microRNA system. Nat. Rev. Neurosci. 7, 911–920. doi: 10.1038/nrn2037
Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K., and Kosik, K. S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281. doi: 10.1261/rna.5980303
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. doi: 10.1016/S0960-9822(02)00809-6
Lesnick, T. G., Papapetropoulos, S., Mash, D. C., Ffrench-Mullen, J., Shehadeh, L., De Andrade, M., et al. (2007). A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 3:e98. doi: 10.1371/journal.pgen.0030098
Mann, K., Gordon, M. D., and Scott, K. (2013). A pair of interneurons influences the choice between feeding and locomotion in Drosophila. Neuron 79, 754–765. doi: 10.1016/j.neuron.2013.06.018
McNeill, E., and Van Vactor, D. (2012). MicroRNAs shape the neuronal landscape. Neuron 75, 363–379. doi: 10.1016/j.neuron.2012.07.005
Nakazawa, T., Watabe, A. M., Tezuka, T., Yoshida, Y., Yokoyama, K., Umemori, H., et al. (2003). p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol. Biol. Cell 14, 2921–2934. doi: 10.1091/mbc.E02-09-0623
Natera-Naranjo, O., Aschrafi, A., Gioio, A. E., and Kaplan, B. B. (2010). Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16, 1516–1529. doi: 10.1261/rna.1833310
Olsson-Carter, K., and Slack, F. J. (2010). A developmental timing switch promotes axon outgrowth independent of known guidance receptors. PLoS Genet. 6:pii: e1001054. doi: 10.1371/journal.pgen.1001054
Panagiotaki, N., Dajas-Bailador, F., Amaya, E., Papalopulu, N., and Dorey, K. (2010). Characterisation of a new regulator of BDNF signalling, Sprouty3, involved in axonal morphogenesis in vivo. Development 137, 4005–4015. doi: 10.1242/dev.053173
Park, K. K., Liu, K., Hu, Y., Smith, P. D., Wang, C., Cai, B., et al. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966. doi: 10.1126/science.1161566
Pillai, R. S., Bhattacharyya, S. N., Artus, C. G., Zoller, T., Cougot, N., Basyuk, E., et al. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576. doi: 10.1126/science.1115079
Sasaki, Y., Gross, C., Xing, L., Goshima, Y., and Bassell, G. J. (2013). Identification of axon-enriched MicroRNAs localized to growth cones of cortical neurons. Dev. Neurobiol. doi: 10.1002/dneu.22113 [Epub ahead of print].
Small, E. M., Sutherland, L. B., Rajagopalan, K. N., Wang, S., and Olson, E. N. (2010). MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ. Res. 107, 1336–1344. doi: 10.1161/CIRCRESAHA.110.227926
Stein, E., and Tessier-Lavigne, M. (2001). Hierarchical organization of guidance receptors: silencing of netrin attraction by slit through a Robo/DCC receptor complex. Science 291, 1928–1938. doi: 10.1126/science.1058445
Tessier-Lavigne, M., and Goodman, C. S. (1996). The molecular biology of axon guidance. Science 274, 1123–1133. doi: 10.1126/science.274.5290.1123
Tymanskyj, S. R., Scales, T. M., and Gordon-Weeks, P. R. (2012). MAP1B enhances microtubule assembly rates and axon extension rates in developing neurons. Mol. Cell. Neurosci. 49, 110–119. doi: 10.1016/j.mcn.2011.10.003
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431. doi: 10.1073/pnas.0508448102
Willis, D. E., Xu, M., Donnelly, C. J., Tep, C., Kendall, M., Erenstheyn, M., et al. (2011). Axonal Localization of transgene mRNA in mature PNS and CNS neurons. J. Neurosci. 31, 14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011
Yazdani, U., and Terman, J. R. (2006). The semaphorins. Genome Biol. 7, 211. doi: 10.1186/gb-2006-7-3-211
Zhang, Y., Ueno, Y., Liu, X. S., Buller, B., Wang, X., Chopp, M., et al. (2013). The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J. Neurosci. 33, 6885–6894. doi: 10.1523/JNEUROSCI.5180-12.2013
Zikopoulos, B., and Barbas, H. (2010). Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. 30, 14595–14609. doi: 10.1523/JNEUROSCI.2257-10.2010
Zivraj, K. H., Tung, Y. C. L., Piper, M., Gumy, L., Fawcett, J. W., Yeo, G. S. H., et al. (2010). Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 30, 15464–15478. doi: 10.1523/jneurosci.1800-10.2010
Zou, Y., Chiu, H., Domenger, D., Chuang, C. F., and Chang, C. (2012). The lin-4 microRNA targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction. Sci. Signal. 5, ra43. doi: 10.1126/scisignal.2002437
Zou, Y., Chiu, H., Zinovyeva, A., Ambros, V., Chuang, C. F., and Chang, C. (2013). Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340, 372–376. doi: 10.1126/Science.1231321
Keywords: miRNAs, neuronal connectivity, axon pathfinding, axon branching, timing mechanisms, temporal regulation, heterochronic miRNAs, axon outgrowth
Citation: Chiu H, Alqadah A and Chang C (2014) The role of microRNAs in regulating neuronal connectivity. Front. Cell. Neurosci. 7:283. doi: 10.3389/fncel.2013.00283
Received: 01 October 2013; Accepted: 16 December 2013;
Published online: 03 January 2014.
Edited by:
Tommaso Pizzorusso, Università degli Studi di Firenze, ItalyReviewed by:
Jay Gibson, The University of Texas Southwestern Medical Center, USAFederico Dajas-Bailador, University of Nottingham, UK
Copyright © 2014 Chiu, Alqadah and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chieh Chang, Division of Developmental Biology, Cincinnati Children’s Hospital Research Foundation, 240 Albert Sabin Way, S3.419, Cincinnati, OH 45229-3039, USA e-mail: chieh.chang1@gmail.com
†Present address: Hui Chiu, Division of Biology and Biological Engineering and Howard Hughes Medical Institute, California Institute of Technology, Pasadena, CA 91125, USA
 Hui Chiu
Hui Chiu Amel Alqadah
Amel Alqadah Chieh Chang
Chieh Chang