Matricellular proteins of the Cyr61/CTGF/NOV (CCN) family and the nervous system
- Laboratory of Molecular and Cellular Neurobiology, International Institute of Molecular and Cell Biology, Warsaw, Poland
Matricellular proteins are secreted proteins that exist at the border of cells and the extracellular matrix (ECM). However, instead of playing a role in structural integrity of the ECM, these proteins, that act as modulators of various surface receptors, have a regulatory function and instruct a multitude of cellular responses. Among matricellular proteins are members of the Cyr61/CTGF/NOV (CCN) protein family. These proteins exert their activity by binding directly to integrins and heparan sulfate proteoglycans and activating multiple intracellular signaling pathways. CCN proteins also influence the activity of growth factors and cytokines and integrate their activity with integrin signaling. At the cellular level, CCN proteins regulate gene expression and cell survival, proliferation, differentiation, senescence, adhesion, and migration. To date, CCN proteins have been extensively studied in the context of osteo- and chondrogenesis, angiogenesis, and carcinogenesis, but the expression of these proteins is also observed in a variety of tissues. The role of CCN proteins in the nervous system has not been systematically studied or described. Thus, the major aim of this review is to introduce the CCN protein family to the neuroscience community. We first discuss the structure, interactions, and cellular functions of CCN proteins and then provide a detailed review of the available data on the neuronal expression and contribution of CCN proteins to nervous system development, function, and pathology.
Introduction
Matricellular proteins are secreted proteins that exist at the border of cells and the extracellular matrix (ECM). However, instead of playing a role in structural integrity of the ECM, these proteins, that act as modulators of various surface receptors, have a regulatory function and instruct a multitude of cellular responses. Thrombospondin-1 (TSP-1), SPARC, and Tenascin-C (TN-C) are considered canonical matricellular proteins, however numerous new ones have been described. Among them are members of the Cyr61/CTGF/NOV (CCN) protein family. Over the last two decades neurobiologist have started to appreciate role of ECM in the nervous system development, physiology and pathology. Accordingly, important contributions of canonical matricellular proteins were described. In contrast, the CCN proteins, although expressed in the nervous system, received relatively little attention in this context. Instead major research efforts focused on their importance for cardiovascular system and skeleton development as well as their links with cancer. Yet, over last few years evidence has accumulated that at least some of CCN family members play important role in the nervous system, especially during development. Thus, the major aim of this review is to introduce the CCN protein family to the neuroscience community. We first briefly introduce matricellular proteins in general. Next, we provide basic facts about structure, interactions, and cellular functions of CCN proteins that are needed as a background to understand the involvement of these proteins in the nervous system. The following parts of this article contain a detailed review of the available data on the neuronal expression and contribution of CCN proteins to nervous system development, function, and pathology.
Matricellular Proteins
The idea of matricellular proteins arose almost 20 years ago with the identification of pericellular matrix proteins, which were shown to not be directly involved in structuring the ECM but rather regulate cell function (Bornstein, 1995; Bornstein and Sage, 2002; Murphy-Ullrich and Sage, 2014). TSP-1, SPARC, and TN-C became prototypic matricellular proteins, and their properties served as selection criteria for the subsequent identification of other matricellular proteins (Bornstein and Sage, 2002; Murphy-Ullrich and Sage, 2014). Specifically, matricellular proteins should: (i) be highly expressed during development or upon injury; (ii) modulate cell-matrix interactions; (iii) bind to cell surface receptors, the ECM, growth factors, cytokines, or proteases; and (iv) induce de-adhesion or support states of “intermediate adhesion” (i.e., a condition characterized by the reorganization of F-actin stress fibers to dismantle focal adhesions). Consequently, the group of matricellular proteins expanded and now includes additional TSPs (2–5), tenascins (R, W, X, Y), osteopontin, and members of the CCN family (discussed in more detail below; Murphy-Ullrich and Sage, 2014). The role of matricellular proteins has been mostly studied in wound healing, cancer, and the production of connective tissue. Some of these proteins, however, have also been studied in the nervous system. The classic matricellular proteins TSP-1 and SPARC were shown to accelerate synaptogenesis when released by astrocytes (Christopherson et al., 2005; Jones et al., 2011). Osteopontin expression increases after ischemic or mechanical brain injury and likely contributes to regenerative processes (Yan et al., 2009; van Velthoven et al., 2011; Plantman, 2012). The use of TN-C knockout mice revealed that TN-C is important for neuromuscular junction formation and plasticity (Cifuentes-Diaz et al., 1998), hippocampus and cortex structure and electrical properties (Irintchev et al., 2005; Gurevicius et al., 2009), hippocampal and cerebellar plasticity (Evers et al., 2002; Andjus et al., 2005), olfactory detection (de Chevigny et al., 2006) and learning and memory (Strekalova et al., 2002). TN-C also positively regulates neurite outgrowth (Rigato et al., 2002; Michele and Faissner, 2009). Similarly, TN-R was shown to regulate multiple processes in the nervous system, including neurogenesis (Xu et al., 2014), neuronal migration (Saghatelyan et al., 2004), axon navigation and neurite growth (Becker et al., 2003, 2004; Zacharias and Rauch, 2006), the formation of perineuronal nets (Brückner et al., 2000; Morawski et al., 2014), and neuron excitability and plasticity (Saghatelyan et al., 2001; Nikonenko et al., 2003; Gurevicius et al., 2004). In contrast, the CCN protein family received relatively little attention from neurobiologists. Nonetheless, CCN matricellular proteins are expressed in the nervous system and some examples exist to confirm their importance for the proper development and function of the nervous system.
CCN Family
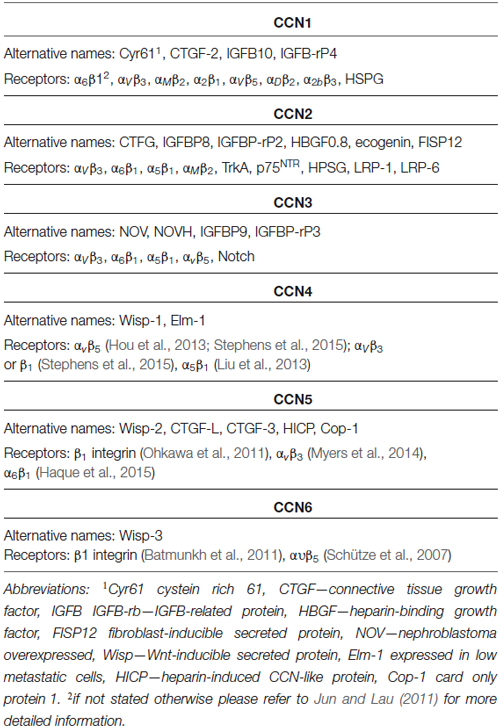
The CCN gene family (Cyr61/CTGF/NOV) consists of six members: CCN-1 (Cyr61), CCN-2 (CTGF), CCN-3 (NOV), CCN-4 (WISP1), CCN-5 (WISP2), and CCN-6 (WISP3). The family name was derived from early nomenclature for the first three identified proteins. These names often reflect the history of the discovery of particular CCN proteins or an initial view of their functions but may not necessarily reflect the current state of knowledge. For example, CCN1 (Cyr61) was identified as 61st gene among genes, the expression of which was induced by trophic factors, e.g., epidermal growth factor (EGF) in fibroblasts (Lau and Nathans, 1987). Because of its mitogenic activity in connective tissue, CCN2 was named connective tissue growth factor (CTGF; Bradham et al., 1991). CCN3 was cloned as a gene that is overexpressed in nephroblastoma because of proviral insertion sites, and it was consequently named after this observation, i.e., Nephroblastoma overexpressed (NOV; Joliot et al., 1992). CCN4 and CCN5, initially called expressed in low-metastatic type 1 cells (ELM-1; Hashimoto et al., 1998) and card-only protein 1 (rCOP-1; Zhang et al., 1998), respectively, were later identified as genes that were upregulated in response to Wnt-1 and consequently renamed WNT-inducible signaling pathway protein 1 (WISP1) and WISP2 (Pennica et al., 1998). Finally, CCN6 was identified as a WISP1 homolog and named WISP3. This diversity of names within the CCN gene family (Table 1) became a major source of confusion and difficulty in attempts to follow research progress in the CCN field. Therefore, after the first international workshop on the CCN Family of genes, CCN researchers proposed a unification of the nomenclature for CCN family members (Brigstock et al., 2003); see also http://ccnsociety.com/ccn_nomenclature/index.html), which will be used hereinafter in the present review (Table 1). CCN family genes encode relatively small proteins (~40 kDa) that share ~40–60% similarity in their primary structure. Their most prominent common features are their functional domain composition and conservation of 38 cysteins through the peptide (Figure 1). Consequently, CCN proteins have several similarities in their molecular mechanisms of action and types of regulated cellular processes (e.g., cell adhesion, migration, proliferation, survival, and differentiation). However, the activity of particular CCN members may vary, depending on the cellular/tissue context (see below). Several extensive reviews have been published on the molecular biology of CCN proteins and their function in physiology and pathology (Leask and Abraham, 2006; Holbourn et al., 2008; Chen and Lau, 2009; Katsube et al., 2009; Jun and Lau, 2011; Perbal, 2013).
Structure, Molecular Interactions, and Basic Modes of Action of CCN Proteins
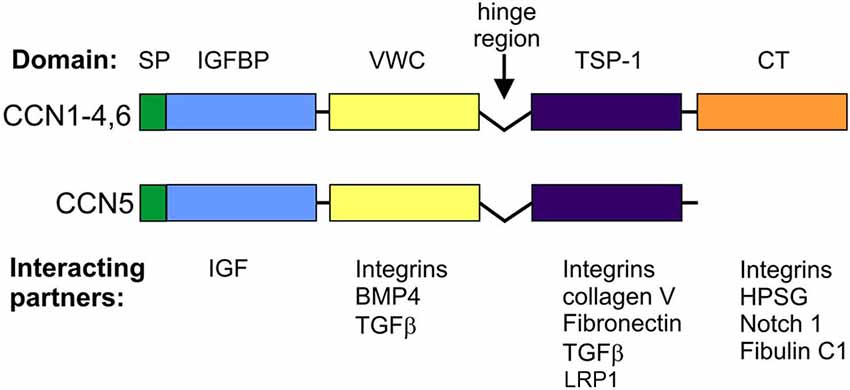
CCN proteins are structurally very similar. With the exception of CCN5, CCNs contain four functional domains that are preceded by an export signaling peptide at their N-terminus, namely insulin-like growth factor binding protein (IGFBP), a von Willebrand factor type C repeat (VWC), thrombospondin type-1 repeat (TSP-1 or TSR), and a cystein knot-containing domain (CT; Figure 1). The last of these is missing in CCN5. IGFBP and VWC constitute the N-terminal half of CCN proteins, which is separated from the C-terminal half that contains TSP-1 and CT by a “hinge” region. Additional forms of CCNs, which have different activities from full-length proteins, can be produced by proteolysis (Brigstock et al., 1997; Steffen et al., 1998) or alternative splicing (Perbal, 2009). Each of the CCN protein domains is responsible for interactions with different sets of molecules (Figure 1). Consequently, CCN proteins bridge a variety of cell surface receptors, signaling molecules, and the ECM. Thus, modular composition, which is unique for the CCN family, explains the functional distinctiveness of CCNs from other proteins that contain one of the domains but raises the issue of what makes them functionally different from each other. Three-dimensional structure modeling revealed subtle changes in electrostatic surfaces that might be responsible for the differential interactions of particular CCN family members (Holbourn et al., 2008). However, most experimental evidence points to other sources of diversity, namely differential, often non-overlapping CCN expression patterns and different sets of receptors and/or proteins that interact with CCNs and are expressed by target cells.
CCN proteins have two major modes of action: (i) interactions with cell surface receptors; and (ii) interactions with receptor ligands. To date, the best studied are the cellular effects of CCN protein interactions with cell adhesion receptors [i.e., integrins and heparan sulfate proteoglycans (HSPGs); Table 1]. At the molecular level, such binding results in the cellular context-specific activation of signaling cascades, including extracellular signal-regulated kinases (ERKs), phosphoinositide 3-kinase (PI3K), and small GTPases of the Rho family (Leask and Abraham, 2006; Chen and Lau, 2009; Jun and Lau, 2011). This in turn leads to myriad cellular responses, of which changes in gene expression and cytoskeleton dynamics have been the most extensively studied. Some non-canonical outcomes (e.g., sustained reactive oxygen species production and cell senescence in response to CCN1-induced Rac1 activation) have also been described (Chen et al., 2007). One of the characteristic futures of CCN members is an ability to interact with various integrins (Table 1). Specific CCN proteins can bind different integrins, triggering dissimilar cellular responses. For example, in fibroblasts, CCN1 interactions with α6β1 stimulate cell adhesion, whereas its binding to αvβ3 results in the induction of DNA synthesis. Moreover, the binding sites for different β-integrins do not overlap; therefore, simultaneous interactions with two integrins and a combined outcome are possible. However, cellular responses even to the same CCN protein-integrin couple may vary depending on the cell type (see below).
Integrins and HSPGs are currently the best described CCN receptors, but they are not the only ones. For example, the adhesion of rat hepatic stellate cells to a CCN2-coated surface requires binding to low-density lipoprotein receptor-related protein 1 (LRP-1) and HSPG (Gao and Brigstock, 2003). Binding to LRP-6 is likely responsible for the inhibitory effects of CCN2 on the canonical Wnt pathway in developing Xenopus laevis embryos. CCN2 binds tyrosine kinase receptor-A (TrkA) and p75 neurotrophin receptor (p75NTR; Wahab et al., 2005), whereas Notch is a receptor for CCN3 (Sakamoto et al., 2002; Minamizato et al., 2007). In addition to receptor binding, CCNs interact with receptor ligands and modulate their binding affinity or bioavailability. In the former case, the most thoroughly described interactions include those between CCNs and bone morphogenetic proteins (BMPs), which has an inhibitory effect, and between CCNs and transforming growth factor β (TGFβ), which has an activational effect (Abreu et al., 2002). CCN2 and CCN1 bind vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF), respectively, and modify their bioavailability (Chen and Lau, 2009).
General Functions of CCN Proteins
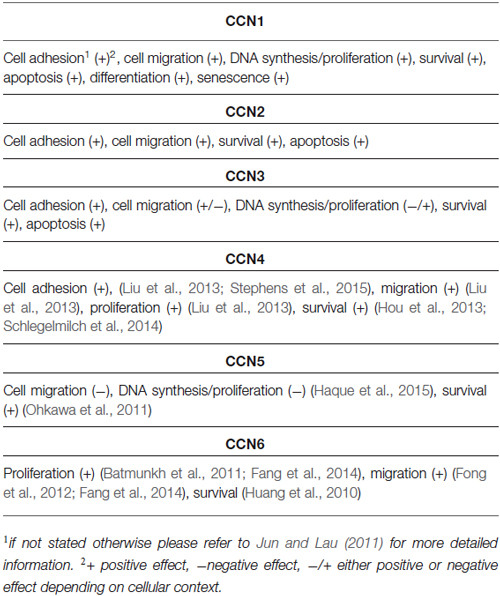
Detailed descriptions of the functions of CCN proteins have been provided in several reviews (Leask and Abraham, 2006; Holbourn et al., 2008; Chen and Lau, 2009; Katsube et al., 2009; Jun and Lau, 2011; Perbal, 2013). The diversity of CCNs’ modes of action results in a variety of cellular responses to CCN proteins. CCNs have been shown to regulate often converse cellular processes (e.g., cell proliferation/differentiation, cell death/survival, and cell adhesion/migration; for review see Jun and Lau, 2011; Table 2). Unsurprising is that different CCNs regulate opposite cellular processes. For example, CCN1, CCN2, and CCN3 are considered positive regulators of adhesion, whereas CCN5 and CCN6 are considered negative regulators (for review see Jun and Lau, 2011). However, opposing or very diverse cellular responses to the same CCN protein have also been reported. For example, CCN1 binds α6β1, αVβ3, α2Bβ3, and αMβ3 integrins and positively regulates the adhesion of fibroblasts and smooth muscle cells, activated endothelial cells, platelets, and monocytes and macrophages, respectively (for review see Jun and Lau, 2011). However, the binding of CCN1 to αVβ5 and α6β1 induces the migration of fibroblasts and smooth muscle cell chemotaxis, respectively (for review see Jun and Lau, 2011). As mentioned above, the binding of CCN1 to αvβ3 integrin in fibroblasts does not affect adhesion/migration but rather regulates DNA synthesis. This example shows that CCN1 can have pleiotropic activities, depending on the cell type, functional/developmental cell status, and type of accessible receptors (e.g., integrins). The same is true for other CCN proteins (for review see Jun and Lau, 2011; Table 2).
In addition to the intrinsic properties of a cell, the surrounding environment/tissue can also modulate responses to a particular CCN. Therefore, a deeper understanding of the physiological roles of CCNs and their links to human diseases became possible only with in vivo models. These in vivo studies point to a primary role for CCN family members in angiogenesis and cardiovascular and skeletal development. For example, CCN1, CCN2, and CCN3 stimulate in vivo blood vessel growth, whereas CCN1 knockout (CCN1−/−) embryos die from placental vascular deficiency (Mo et al., 2002). The surviving embryos exhibit atrioventricular septal defects. CCN1+/– mice are viable, but 20% of the adult mice have ostium primum atrial septal defects (Mo and Lau, 2006). Also CCN2-deficient mice have defects in vasculature remodeling (Hall-Glenn et al., 2012). CCN2, CCN3, CCN4, and CCN6 are mostly known for their importance in bone formation and/or remodeling. CCN2 knockout mice exhibit skeletal deformations, reduced ossification, and the expansion of mineralizing cartilage zones. In CCN3 knockout mice, Matsushita et al. (2013) observed accelerated bone regeneration. A study of transgenic mice that overexpress CCN4 in preosteoblasts reported that CCN4 stimulates osteogenesis (Ono et al., 2011). Finally, mutations in CCN6 were repeatedly found in progressive pseudorheumatoid dysplasia, an autosomal recessive disorder (Hurvitz et al., 1999). Surprisingly, however, CCN6 knockout mice have no obvious bone phenotype (Kutz et al., 2005; Hann et al., 2013), whereas some defects that may be relevant to human disease were observed in zebrafish morphants (Nakamura et al., 2007). In addition to the major research areas described above, CCNs have been extensively studied in the context of proper wound healing (Jun and Lau, 2011). Mounting evidence also indicates links between CCNs and tumorigenesis (Jun and Lau, 2011). In contrast, knowledge concerning the role of CCNs in the nervous system is relatively lagging. Part of the problem is that full, nonconditional knockout results in embryonic (CCN1, CCN5) or perinatal (CCN2) death. Often such early lethality precludes studies on several tissue/organ functions, including in the nervous system. In cases of CCN knockout animals that survive embryonic development, no apparent nervous system problems were reported, but usually deeper analyses in this regard have not been performed because different authors have different major interests (e.g., bone development). Another reason for the low recognition of CCNs’ roles in the nervous system stems from scattering of relevant information. To date, a comprehensive summary of the current state of knowledge concerning CCN expression patterns, the involvement of CCNs in neuronal development and physiology, and their links to nervous system pathology has been lacking.
Expression and Functions of CCN Family Members in the Nervous System
Major research on CCN proteins has not focused on the nervous system, but for some CCNs, the central nervous system (CNS) is one of the major sites of expression. This observation supports the hypothesis of a possible role for the CNN protein family in different aspects of CNS physiology and pathology. In fact, the role of some CNN family members in the CNS has already been demonstrated, and the number of reports concerning this issue is constantly growing.
CCN Expression in the Mammalian Central Nervous System
CCN proteins are expressed in non-neuronal tissues mainly during highly dynamic processes that involve tissue remodeling. CCN expression is thus observed during embryonic development and under pathological conditions (e.g., inflammation, injury, and cancer). Similarly, in the CNS, the expression of CCNs can be induced under such conditions. For example, both CCN1 and CCN2 were elevated in gliomas and glioblastomas (Pan et al., 2002; Xie et al., 2004a,b). However, CCN proteins are also expressed in the properly developing and adult CNS. Although knowledge of the pattern of CCN expression during embryogenesis and postnatal development and in the adult CNS is incomplete and restricted mainly to CCN1, CCN2, and CCN3, the number of studies that are investigating this issue is growing. Additional information concerning CCN expression patterns in the rodent and human brain can be extracted from large databases, including the collection of Allen Brain Atlases (ABA), e.g., Allen Mouse Brain Atlas (Lein et al., 2007)1 and Allen Developing Mouse Brain Atlas2 for in situ hybridization data, BrainStars* (BS*) for microarray data (Kasukawa et al., 2011)3, and the Human Protein Atlas (HPA) for immunohistochemistry data (Uhlén et al., 2015).4 The major advantages of these databases, which resulted from large-scale projects, are standardized procedures, reagents, and biological material. In this review, we use this knowledge to discuss previously published data and identify gaps in the literature.
CCN1 Expression in the Developing and Adult Central Nervous System
CCN1 mRNA has been detected in different areas of the developing and adult human and rat brain and in rat hippocampal and cortical neurons cultured in vitro (Albrecht et al., 2000; Malik et al., 2013). Interestingly, CCN1 mRNA levels in the rat hippocampus were higher during embryonic development and dropped postnatally, suggesting that this protein may play an important role during embryonic brain development (Malik et al., 2013). A similar developmental trend in CCN1 expression was observed in cultured hippocampal neurons (Malik et al., 2013). An analysis of ABA data showed fluctuations of CCN1 mRNA amount in embryonic brain but confirmed higher CCN1 mRNA levels in the mouse embryonic day 18 (E18) brain compared with early postnatal weeks. The ABA suggests partial expression recovery in the cortex from postnatal day 28 (P28) onward (Figure 2). In the adult brain (P56) the ABA provides evidence for CCN1 mRNA in deeper cortical layers (Figure 3). BrainStars* provides evidence of CCN1 mRNA in the adult mouse CNS, with the highest levels in the retina. In the adult human brain, CCN1 mRNA was detected throughout the CNS, with the strongest expression in the spinal cord, frontal, temporal, and occipital cortices, hippocampus, and caudate nucleus (Albrecht et al., 2000). This observation appears to be confirmed by the HPA, in which mild CCN1 protein levels were present in the cortex and hippocampus, but the highest levels were detected in the cerebellum (Figure 4). An analysis of the cell specificity of immunohistochemical signal showed that CCN1 protein can be detected mostly in neurons, whereas its expression in glia is lower or undetectable (Table 3).
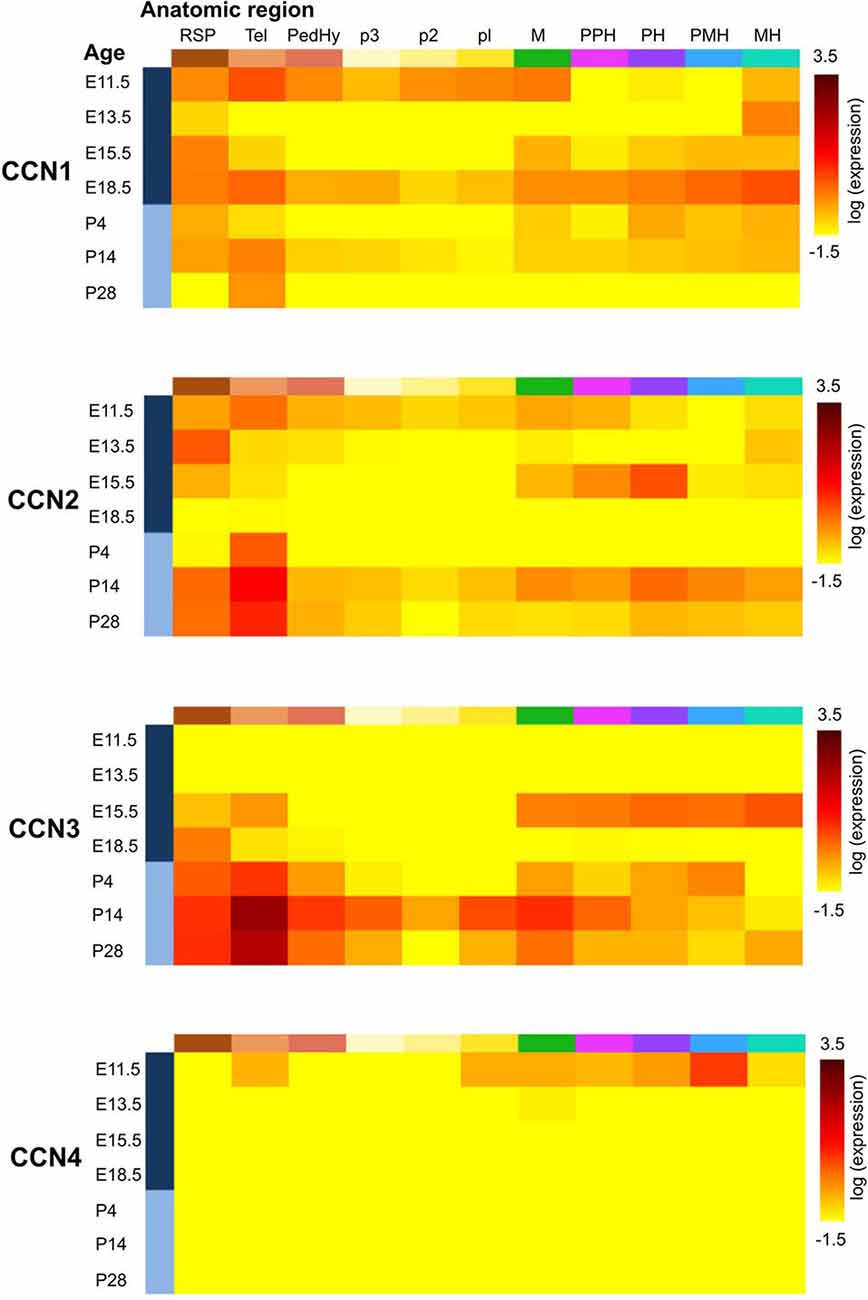
Figure 2. Expression of CCN1–4 genes in the developing mouse brain according to the Allen Developing Mouse Brain Atlas [© 2013 Allen Institute for Brain Science. Allen Developing Mouse Brain Atlas (Internet)]. Gene expression summary for selected brain structures were downloaded from http://developingmouse.brain-map.org/ and are expressed as a heatmap of expression energy (for more details please refer to http://help.brain-map.org/display/devmouse/In+Situ+Hybridization+(ISH)+Data#InSituHybridization(ISH)Data-CorrelationSearch). Heatmaps source: CCN1: http://developingmouse.brain-map.org/gene/show/15780; CCN2: http://developingmouse.brain-map.org/gene/show/13996; CCN3: http://developingmouse.brain-map.org/gene/show/17900; CCN4: http://developingmouse.brain-map.org/gene/show/22159. RSP—rostral secondary prosencephalon; Tel—telencephalic vesicle; PedHy—peduncular (caudal) hypothalamus; p3 prosomere 3; p2 prosomere 2; p1 prosomere 1; M—midbrain; PPH—prepontine hindbrain; PH—pontine hindbrain; PMH—pontomedullary hindbrain; MH—medullary hindbrain (medulla).
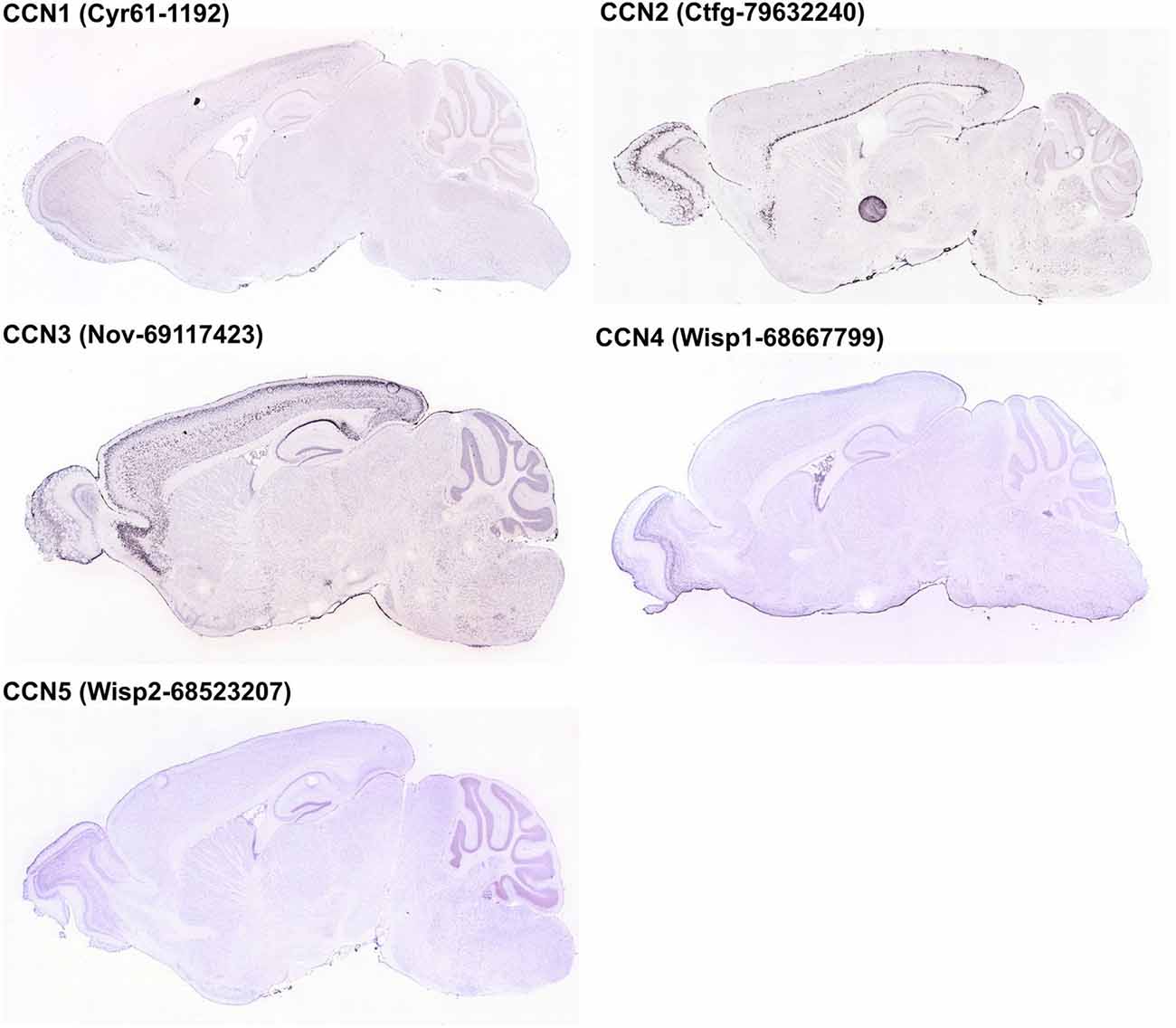
Figure 3. Expression of CCN1–5 genes in the adult mouse brain according to the Allen Brain Atlas [© 2014 Allen Institute for Brain Science. Allen Mouse Brain Atlas (Internet)]. Images of brain sections were downloaded from http://mouse.brain-map.org/. Experiment name is given in parentheses.
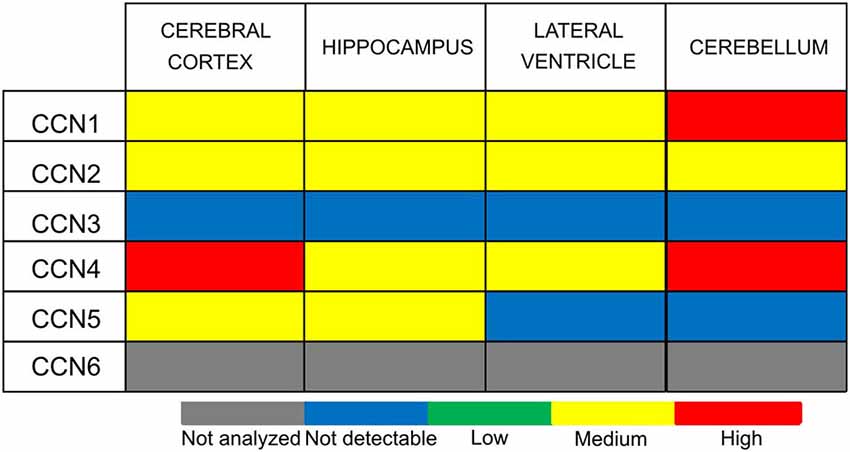
Figure 4. CCN protein abundance in the selected human brain regions according to the Human Protein Atlas (HPA).
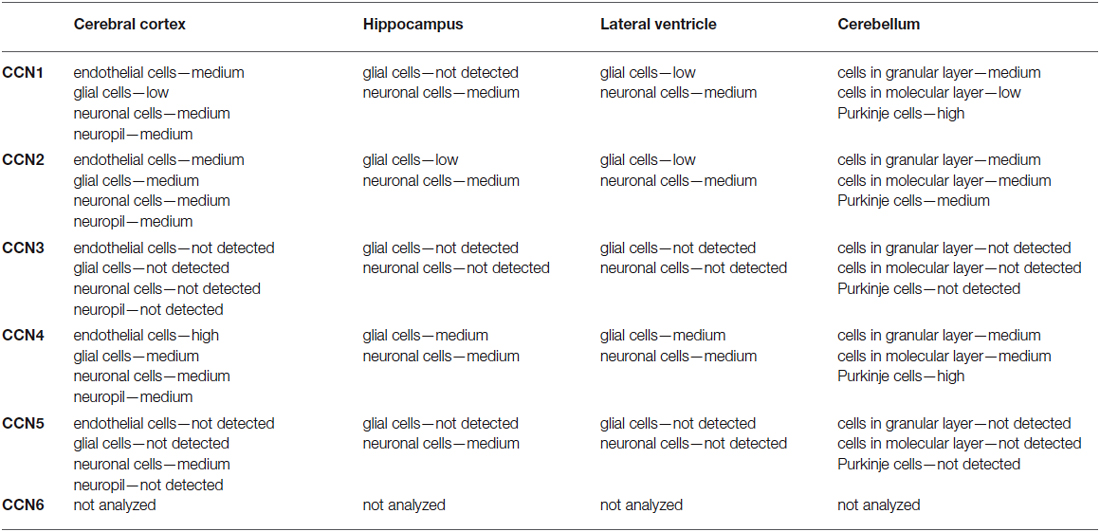
Table 3. Cellular specificity of CCN protein expression in selected regions of the CNS according to the Human Protein Atlas.
Our data from in vitro hippocampal neuron cultures showed that CCN1 expression can be modulated by various stimuli [e.g., brain-derived neurotrophic factor (BDNF), insulin, and bicuculline], both in developing and “mature” neurons. The effects of bicuculline suggest a role for enhanced excitatory transmission in the regulation of CCN1 mRNA levels. Similar results were obtained in organotypic hippocampal slices (Iacono et al., 2013). In vivo, CCN1 mRNA level increases with decreases in excitatory transmission that are caused by N-methyl-D-aspartate receptor inhibition (Ito et al., 2007). Nevertheless, the mechanism behind this phenomenon might be indirect and involve increases in the activation of other neurotransmitter systems. Elevations of CCN1 mRNA were also observed upon methamphetamine administration (Ito et al., 2007), administration of the dopamine receptor agonist clozapine (Sakuma et al., 2015), and the activation of muscarinic acetylcholine receptors (Chung and Ahn, 1998; Albrecht et al., 2000).
CCN2 Expression in the Developing and Adult Central Nervous System
Similar to CCN1, CCN2 is also expressed in different parts of the CNS and rat primary neuron cultures in vitro (Albrecht et al., 2000), and its levels are regulated developmentally. CCN2 mRNA is present in neural tissue in developing mouse embryos already at embryonic day 9.5 (Ivkovic et al., 2003). Williams et al. (2007) reported presence of CCN2 mRNA in olfactory bulb at embryonic days 14, 17.5 and at postnatal day 3. In the adult brain presence of both CCN2 mRNA and CCN2 protein was reported in the hippocampus (Hertel et al., 2000) and olfactory bulb (Williams et al., 2007). The ABA shows moderate levels of CCN2 mRNA between E11.5-E15.5 and a complete lack of CCN2 expression just before birth. However, CCN2 mRNA levels gradually increase from P4 until adulthood (Figure 2). Such dynamic regulation of CCN2 expression in early postnatal development was studied in detail in the rodent olfactory bulb. CCN2 mRNA in the glomerular and mitral cell layers could be detected around P5. In the glomerular layer, CCN2 mRNA level remained stable throughout postnatal development and in the adult brain, whereas it gradually decreased in the mitral cell layer and was barely detectable already on P12 (Khodosevich et al., 2013). In the adult rat brain, CCN2 mRNA showed a highly selective distribution pattern in the forebrain, with strong labeling that was restricted to specific regions of the olfactory bulb, endopiriform nucleus, and supracallosal layer in the cerebral cortex. Particularly strong staining was observed in large neurons of layer VI in the cerebral cortex (Heuer et al., 2003). In the adult mouse brain, CCN2 mRNA could be detected in layer VI of the cortex and mitral cell and glomerular layers of the olfactory bulb (Khodosevich et al., 2013). These observations are consistent with the ABA (Figure 3) and BS* data. Presence of CCN2 mRNA was also shown throughout the adult human CNS, including different brain regions and the spinal cord. In fact, the CCN2 expression pattern is similar to CCN1, with the exception that tissue levels of CCN2 transcripts were lower in the hippocampus, caudate nucleus, and corpus callosum (Albrecht et al., 2000). In the healthy human brain, CCN2 protein was predominantly detected in neurons and partially in subtypes of glial cells (Schwab et al., 2000, 2001; Table 3). The HPA also suggests the presence of CCN2 protein in glial cells, especially in cortical areas (Table 3). Supporting further glial expression of CCN2, its expression has been detected in rat CNS astrocytes and tanycytes (Kondo et al., 1999). In the normal human spinal cord, the low expression of CCN2 was observed in cells with astroglial morphology, particularly in white matter (Spliet et al., 2003). In the senile human brain, exclusively CCN2 neuronal expression was found throughout different brain areas, particularly in entorhinal layer pre-alpha neurons and cortical pyramidal cells, both in somatodendritic and axonal compartments (Ueberham et al., 2003). Expression of CCN2 also appears to be regulated by neuronal activity. Khodosevich et al. (2013) demonstrated that CCN2 protein level is dynamically regulated in response to changes in olfactory input.
CCN3 Expression in the Developing and Adult Central Nervous System
CCN3 is also expressed in the developing and adult CNS. In fact, the CNS appears to be the major site of CCN3 expression during development compared with other tissues. This was demonstrated in chick embryos (Joliot et al., 1992) and human embryos in the first trimester of embryogenesis, in which CCN3 mRNA and CCN3 were most abundantly present in motor neurons and the floor plate of the spinal cord (Kocialkowski et al., 2001). CCN3 expression undergoes dynamic changes during development, suggesting that CCN3 protein might play a role in maintaining or establishing specific brain functions. The abundance of CCN3 mRNA increases throughout the postnatal period during rat brain development. Increases in amount of CCN3 mRNA and CCN3 protein were detected in the developing rat brain after birth, with a pronounced peak between P15 and P150 (Su et al., 2001). A similar increase seems to occur in the developing murine brain according to the ABA (Figure 2) through a majority of brain areas and the highest levels of CCN3 mRNA are present in olfactory bulb, cortex and CA1 field of hippocampus (Figure 3). Intriguingly, the ABA also reports relatively low levels of CCN3 mRNA in the cerebellum. But CCN3 mRNA and CCN3 protein were detected in the rat cerebellum during the postnatal period, with the most prominent increase from P7 to P14 (Le Dréau et al., 2010b). In the human brain during early developmental stages, CCN3 mRNA was mainly observed in somatomotor neurons in the lower CNS. At later stages, however, CCN3 was expressed in the higher CNS (Su et al., 1998). High levels of CCN3 mRNA were reported in the cortex, hippocampus, amygdala, and spinal cord (Albrecht et al., 2000). Surprisingly, the HPA reveals no CCN3 protein expression in the analyzed areas (Figure 4).
CCN3 expression is particularly observed in neurons. For example, presence of CCN3 mRNA was shown in rat neuronal cultures in vitro (Albrecht et al., 2000) while CCN3 protein was detected in rat cerebellar Purkinje cells (Su et al., 2001; Le Dréau et al., 2010b), and neurons of dorsal root ganglia and the dorsal horn of the rat spinal cord (Kular et al., 2012). However, CCN3 immunoreactivity was also observed in astrocytes in the rat cerebral cortex, corpus callosum, and hippocampus (Le Dréau et al., 2010a). CCN3 mRNA and CCN3 protein are also expressed in specific structures in the developing chicken eye, including the lens, ciliary body, optic nerve, pecten, and retina (Laurent et al., 2012).
CCN4, CCN5, and CCN6 Expression in the Central Nervous System
In contrast to CCN1–3, very little is known about CCN4, CCN5, and CCN6 expression patterns. The data that are available are also somewhat contradictory. A study of CCN4, CCN5, and CCN6 mRNA levels in human embryonic and adult tissues showed no expression in the brain (Pennica et al., 1998), but other studies do not fully support this finding. Indeed, during rat embryonic development, CCN4 mRNA presence was reported exclusively in osteoblasts and osteoblastic progenitor cells of the perichondral mesenchyme (French et al., 2004). The ABA also does not reveal the presence of CCN4 mRNA during development after E11.5 (Figure 2). But, both the ABA and BS* suggest that CCN4 mRNA is present in the adult murine brain, although at relatively low levels, with the highest expression in the olfactory bulb (ABA, BS*) and retina (BS*; Figure 3). Also, the HPA shows presence of CCN4 protein in the adult human cerebral cortex and cerebellum (Figure 4), with the highest expression levels in cortical endothelial cells and Purkinje neurons (Table 3). Additionally, CCN4 protein was shown to be expressed in rat primary neurons during oxygen-glucose deprivation (Wang et al., 2012). CCN5 mRNA, according to the ABA, is not present in P56 murine brain (Figure 3). But the BS* reports low to moderate CCN5 mRNA levels throughout the adult mouse CNS, with the highest expression in the retina. Moreover, Ohkawa et al. (2011) detected some CCN5 mRNA in the cerebellum and spinal cord of 15 week old mice. In contrast, CCN5 immunostaining was reported in fetal mouse brain (E12–16) but not human brain (4 months; Jones et al., 2007). Cytoplasmic and nuclear CCN5 immunostaining was also reported in cortical neurons in the adult rat (Gray et al., 2007). The presence of CCN5 protein in the adult cortex is supported by the HPA (Figure 4), which additionally provides evidence of its hippocampal expression. Virtually nothing is known for CCN6. No data are reported concerning its expression in either the ABA or HPA. Only a BS* microarray analysis detected almost equal amounts of CCN6 transcript in all 51 analyzed brain regions.
Role of CCN Proteins in the Central Nervous System
CCNs can be detected in the CNS. Importantly, CCN levels dynamically change during development. This observation supports their possible role in CNS development and physiological functions. Here, we will discuss the available data that indicate the involvement of CCN proteins, mainly CCN1, CCN2, and CCN3, in CNS development and physiology.
CCN1
In 1998, CCN1 was suggested to play an important role in neuronal differentiation, in which it was induced by bFGF during the differentiation of immortalized hippocampal progenitor (H19–7) cells (Chung and Ahn, 1998). Indeed, we recently confirmed that CCN1 is required for proper development of the dendritic tree in rat hippocampal neurons in vitro and acts downstream of Ras, ERK, and PI3K. Moreover, CCN1 overexpression promoted dendritic branching, and this effect depended on β1-integrin (Malik et al., 2013).
CCN1 is secreted by retinal Muller glial (RMG) cells that are cultured in vitro in response to glial cell line-derived neurotrophic factor (GDNF) treatment, which is known to have pro-survival effects in the retina. Thus, the neuroprotective and pro-survival activity of CCN1 was studied in a mouse model of retinitis pigmentosa (Kucharska et al., 2014). In organotypic retinal cultures that were derived from these mice, CCN1 treatment increased the survival rates of photoreceptors. The authors showed that CCN1 activates the mitogen-activated protein kinase (MAPK)/Erk and Janus-associated kinase (JAK)/Stat pathways but not PI3K/Akt pathway in retinal explants from retinitis pigmentosa mice. Interestingly, in primary porcine cultures, CCN1 did not stimulate photoreceptors themselves but rather only RMG cells and retinal pigment epithelium (RPE) cells, suggesting that the protective effect on photoreceptors occurs indirectly. In disagreement with the results that were obtained in the retinitis pigmentosa model, CCN1 treatment in pure porcine RMG cultures stimulated not only the MAPK/Erk and JAK/Stat pathways but also the PI3K/Akt pathway, whereas it led to activation of the PI3K/Akt and MAPK/Erk pathways in RPE cultures.
The stimulation of muscarinic acetylcholine receptors (mAChRs) induced CCN1 mRNA expression in primary neurons and the rat brain, in which CCN1 mRNA was detected in cortical layers V and VI and thalamic nuclei. mAChRs modulate neuronal functions, including long-term potentiation and synaptic plasticity in neuronal circuits that are involved in learning and memory formation. The authors suggested a role for CCN1 in the cholinergic regulation of synaptic plasticity (Albrecht et al., 2000).
CCN2
Consistent with its expression pattern, CCN2 was shown to play an important role in the rodent olfactory bulb, where it acts as a proapoptotic factor that eliminates newborn neurons in an activity-dependent and locally restricted manner. CCN2 acts via glial-derived transforming growth factor β2 (TGF-β2) to promote the apoptosis of newly generated periglomerular interneurons in the glomerular layer (Khodosevich et al., 2013). CCN2 levels, which are dynamically regulated in response to changes in olfactory input, adjust the survival of postnatally born neurons in an odorant-specific fashion. Importantly, CCN2 levels decrease in the mouse olfactory bulb if the sensory input is suppressed (Khodosevich et al., 2013). In contrast, olfactory activity enhances CCN2 expression specifically in odor-activated glomeruli in the olfactory bulb. This phenomenon has important behavioral consequences, and CCN2 knockdown mice perform better in odorant detection and olfactory discrimination than controls.
As mentioned above, CCN2 binds TrkA and p75NTR, receptors that transduce neurotrophin signals (Wahab et al., 2005). The authors showed that CCN2 stimulates TrkA and induces its autophosphorylation. The nerve growth factor (NGF)-induced activation of TrkA is widely known to control neuronal cell survival and axonal growth (Kuruvilla et al., 2000; Atwal et al., 2003). Nonetheless, the physiological importance of CCN2’s influence on neurotrophin signaling in brain development, physiology, and disease remains to be evaluated.
CCN3
A role for CCN3 in brain development has also been reported. Specific patterns and dynamic changes in CCN3 expression in the rat cerebellum (Le Dréau et al., 2010b) likely reflect its function in postnatal cerebellum development. Supporting this possibility, an in vitro study showed that CCN3 reduces cerebellar granule neuron precursor (GNP) proliferation that is induced by Sonic Hedgehog (SHH). SHH maintains GNPs in a proliferative state and delays their differentiation, resulting in a decrease in the proportion of postmitotic neurons. By counteracting SHH-driven effects, CCN3 reduces the proliferation of GNPs and consequently promotes their differentiation. This anti-proliferative action of CCN3 requires glycogen synthase kinase 3β (GSK3-β) activity. Moreover, CCN3 stimulates the migration and chemotaxis of cerebellar GNPs in vitro (Le Dréau et al., 2010b). The role of CCN3 in the retina has also been studied. CCN3 is expressed in the chick retina during development in distinct cell types and is regulated by the Notch and BMP signaling pathways (Laurent et al., 2012). However, the ectopic expression of CCN3 had no effect on retinal development. These authors did not describe the effects of CCN3 depletion, and the possible role of CCN3 in the developing retina remains unclear (Laurent et al., 2012).
CCN5
A role for CCN5 in neuronal development has also been suggested. CCN5 was shown to enhance neurite outgrowth in Neuro2a cells. The underlying mechanism remains elusive, although the authors suggested the involvement of the CCN5-triggered, β1-integrin-dependent activation of Akt (Ohkawa et al., 2011). Thus, far no data are available concerning neuronal functions of CCN4 and CCN6. Most likely almost complete lack of data concerning role of CCN4, 5 and 6 in the nervous system reflects their low abundance in the nervous system and relatively poor repertoire of tools to reliably study their expression. Alternatively, those proteins may attract less attention when identified in large “omics” screens because of limited knowledge concerning their cellular functions.
CNN Proteins and Nervous System Dysfunction
As in non-neuronal tissues, CCN proteins play a role in responses to injury in the CNS. This role has been particularly attributed to CCN2. CCN2 appears to be involved in gliosis and glia scar formation in response to different types of brain injury, such as trauma, cerebral infarction, and excitotoxic brain damage. After kainic acid-induced lesions of the CA3 area of the hippocampus (i.e., a model of excitotoxic brain damage), CCN2 protein was detected in the CA1, CA3, and dentate gyrus, mainly in neurons that mostly died on subsequent days. At later stages when repair processes are active, CCN2 was found extracellularly and in GFAP-positive astrocytes (Hertel et al., 2000).
CCN2 upregulation has also been observed in reactive gliosis adjacent to the site of mechanical injury and in brain tissue in stroke patients and following traumatic brain injury (Schwab et al., 2000, 2001). A role in neuroinflammation has also been suggested for CCN3. CCN3 protein was shown to be expressed in astrocytes and regulate astrocyte chemokine synthesis in vitro and in vivo (Le Dréau et al., 2010a). CCN3 was suggested to attenuate inflammatory pain (Kular et al., 2012). CCN3 protein expression decreased in dorsal root ganglia and the dorsal horn of the spinal cord in a rat model of inflammatory pain. Interestingly, intrathecal administration of CCN3 siRNA during early stages of an inflammatory pain model resulted in a significant increase in mechanical allodynia. In contrast, CCN3 treatment significantly attenuated mechanical allodynia in rats but had no effect on basal pain perception in control animals. As demonstrated both in vitro and in vivo, CCN3 influences matrix metalloproteinase (MMP) expression, which might contribute to the possible mechanism of CCN3’s involvement in inflammatory pain (Kular et al., 2012).
The role of several CCN proteins has been suggested in neurodegenerative disease, based largely on the observation that they are expressed in patients’ brains in both neurons and astrocytes. Notably, however, a causal role for CCN proteins in neurodegeneration has not yet been proven, and increased levels of CCNs might result from inflammation that accompanies neurodegeneration. CCN2 appears to be involved in the progression and persistence of astrogliosis in neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD). In AD patients’ brains, CCN2 protein was detected in the entorhinal cortex, hippocampus, and temporal cortex, and the signal was associated with neurofibrillary tangles and neurons that are associated with amyloid plaques (Ueberham et al., 2003). Moreover, CCN2 was detected in perivascular astrocytes and astrocytes that were associated with plaques (Ueberham et al., 2003). Another study showed that CCN2 expression in the AD brain is correlated with the progression of clinical dementia in AD and amyloid plaques but not neurofibrillary tangle pathology. The authors suggested that CCN2 may play a role in the pathogenesis of AD by promoting amyloid β peptide levels. This hypothesis was supported by findings from a Tg2576 AD model in mice that were exposed to a diabetogenic diet. These mice developed insulin resistance, accompanied by elevations in CCN2 levels in the brain and the promotion of AD-type amyloid plaque burden (Zhao et al., 2005). The authors also found that CCN2 activated the MAPK and PI3K/Akt pathways in human H4-APP751 neuronal cells and suggested that this may lead to an increase in amyloid β peptide levels and contribute to AD pathology (Zhao et al., 2005).
CCN2 expression was also linked to ALS. In normal human spinal cord, the low expression of CCN2 protein is observed in cells with astroglial morphology, particularly in white matter. CCN2 protein levels are increased in ALS cases. It can be detected in vimentin-positive reactive astrocytes and remaining motor neurons in the ventral horn in long-term-surviving patients (Spliet et al., 2003).
CCN4 has been linked to neurodegeneration protection. Treatment with CCN4 blocked primary neuronal injury and apoptosis during oxygen-glucose deprivation, and this effect depended on PI3K-Akt signaling (Wang et al., 2012).
Conclusions and Perspectives
CCN proteins have not been studied vigorously or systematically in the nervous system, but their known functions in other tissues and organs support their potential involvement in the proper development and physiology of the nervous system. Such a role is also supported by a few discoveries of CCNs’ involvement in neuroprecursors proliferation, neuronal survival, and differentiation. Examples of nervous system pathologies that are linked to changes in CCN gene expression support the importance of CCNs in the nervous system. Yet, based on these few examples it would be premature to build a comprehensive model of CCN protein-dependent regulation of the nervous system development and physiology. As in case of other systems, it is very likely that the final outcome of CCN activities may depend on cell type and repertoire of receptors expressed by a given cell at a particular developmental/differentiation stage. What is more, similar to studies on skeletal development, studies in the context of the whole organism are key to understanding CCNs’ modulatory functions. Therefore, further progress in our understanding of the functions of CCNs in the nervous system will require the development of animal models using spatially and developmentally regulated knockouts of CCN family genes. Studies on the role of CCNs in the nervous system can accelerate in the future only with such models.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Polish National Science Center grant no. 2011/03/B/NZ3/01970. The work of AM is partly financed within the Parent-Bridge program of the Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund. The work of EL is financed by National Science Center “Sonata” grant no. 2013/11/D/NZ3/01079. JJ and AM are recipients of “Master” Professorial subsidy and “Master” fellowship of the Foundation for Polish Science, respectively.
Footnotes
- ^ http://www.brain-map.org
- ^ http://developingmouse.brain-map.org
- ^ http://www.brainstars.org
- ^ http://www.proteinatlas.org
References
Abreu, J. G., Ketpura, N. I., Reversade, B., and De Robertis, E. M. (2002). Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat. Cell Biol. 4, 599–604. doi: 10.1038/ncb826
Albrecht, C., von Der Kammer, H., Mayhaus, M., Klaudiny, J., Schweizer, M., and Nitsch, R. M. (2000). Muscarinic acetylcholine receptors induce the expression of the immediate early growth regulatory gene CYR61. J. Biol. Chem. 275, 28929–28936. doi: 10.1074/jbc.m003053200
Andjus, P. R., Bajić, A., Zhu, L., Schachner, M., and Strata, P. (2005). Short-term facilitation and depression in the cerebellum: some observations on wild-type and mutant rodents deficient in the extracellular matrix molecule tenascin C. Ann. N Y Acad. Sci. 1048, 185–197. doi: 10.1196/annals.1342.017
Atwal, J. K., Singh, K. K., Tessier-Lavigne, M., Miller, F. D., and Kaplan, D. R. (2003). Semaphorin 3F antagonizes neurotrophin-induced phosphatidylinositol 3-kinase and mitogen-activated protein kinase kinase signaling: a mechanism for growth cone collapse. J. Neurosci. 23, 7602–7609.
Batmunkh, R., Nishioka, Y., Aono, Y., Azuma, M., Kinoshita, K., Kishi, J., et al. (2011). CCN6 as a profibrotic mediator that stimulates the proliferation of lung fibroblasts via the integrin β1/focal adhesion kinase pathway. J. Med. Invest. 58, 188–196. doi: 10.2152/jmi.58.188
Becker, C. G., Schweitzer, J., Feldner, J., Becker, T., and Schachner, M. (2003). Tenascin-R as a repellent guidance molecule for developing optic axons in zebrafish. J. Neurosci. 23, 6232–6237.
Becker, C. G., Schweitzer, J., Feldner, J., Schachner, M., and Becker, T. (2004). Tenascin-R as a repellent guidance molecule for newly growing and regenerating optic axons in adult zebrafish. Mol. Cell. Neurosci. 26, 376–389. doi: 10.1016/j.mcn.2004.03.003
Bornstein, P. (1995). Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J. Cell Biol. 130, 503–506. doi: 10.1083/jcb.130.3.503
Bornstein, P., and Sage, E. H. (2002). Matricellular proteins: extracellular modulators of cell function. Curr. Opin. Cell Biol. 14, 608–616. doi: 10.1016/s0955-0674(02)00361-7
Bradham, D. M., Igarashi, A., Potter, R. L., and Grotendorst, G. R. (1991). Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J. Cell Biol. 114, 1285–1294. doi: 10.1083/jcb.114.6.1285
Brigstock, D. R., Goldschmeding, R., Katsube, K., Lam, S. C.-T., Lau, L. F., Lyons, K., et al. (2003). Proposal for a unified CCN nomenclature. Mol. Pathol. 56, 127–128. doi: 10.1136/mp.56.2.127
Brigstock, D. R., Steffen, C. L., Kim, G. Y., Vegunta, R. K., Diehl, J. R., and Harding, P. A. (1997). Purification and characterization of novel heparin-binding growth factors in uterine secretory fluids. Identification as heparin-regulated Mr 10,000 forms of connective tissue growth factor. J. Biol. Chem. 272, 20275–20282. doi: 10.1074/jbc.272.32.20275
Brückner, G., Grosche, J., Schmidt, S., Härtig, W., Margolis, R. U., Delpech, B., et al. (2000). Postnatal development of perineuronal nets in wild-type mice and in a mutant deficient in tenascin-R. J. Comp. Neurol. 428, 616–629. doi: 10.1002/1096-9861(20001225)428:4<616::aid-cne3>3.0.co;2-k
Chen, C.-C., and Lau, L. F. (2009). Functions and mechanisms of action of CCN matricellular proteins. Int. J. Biochem. Cell Biol. 41, 771–783. doi: 10.1016/j.biocel.2008.07.025
Chen, C. C., Young, J. L., Monzon, R. I., Chen, N., Todorović, V., and Lau, L. F. (2007). Cytotoxicity of TNFalpha is regulated by integrin-mediated matrix signaling. EMBO J. 26, 1257–1267. doi: 10.1038/sj.emboj.7601596
Christopherson, K. S., Ullian, E. M., Stokes, C. C. A., Mullowney, C. E., Hell, J. W., Agah, A., et al. (2005). Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433. doi: 10.1016/j.cell.2004.12.020
Chung, K. C., and Ahn, Y. S. (1998). Expression of immediate early gene cyr61 during the differentiation of immortalized embryonic hippocampal neuronal cells. Neurosci. Lett. 255, 155–158. doi: 10.1016/s0304-3940(98)00733-2
Cifuentes-Diaz, C., Velasco, E., Meunier, F. A., Goudou, D., Belkadi, L., Faille, L., et al. (1998). The peripheral nerve and the neuromuscular junction are affected in the tenascin-C-deficient mouse. Cell. Mol. Biol. (Noisy-le-grand) 44, 357–379.
de Chevigny, A., Lemasson, M., Saghatelyan, A., Sibbe, M., Schachner, M., and Lledo, P.-M. (2006). Delayed onset of odor detection in neonatal mice lacking tenascin-C. Mol. Cell. Neurosci. 32, 174–186. doi: 10.1016/j.mcn.2006.04.002
Evers, M. R., Salmen, B., Bukalo, O., Rollenhagen, A., Bösl, M. R., Morellini, F., et al. (2002). Impairment of L-type Ca2+ channel-dependent forms of hippocampal synaptic plasticity in mice deficient in the extracellular matrix glycoprotein tenascin-C. J. Neurosci. 22, 7177–7194.
Fang, F., Zhao, W.-Y., Li, R.-K., Yang, X.-M., Li, J., Ao, J.-P., et al. (2014). Silencing of WISP3 suppresses gastric cancer cell proliferation and metastasis and inhibits Wnt/β-catenin signaling. Int. J. Clin. Exp. Pathol. 7, 6447–6461.
Fong, Y.-C., Lin, C.-Y., Su, Y.-C., Chen, W.-C., Tsai, F.-J., Tsai, C.-H., et al. (2012). CCN6 enhances ICAM-1 expression and cell motility in human chondrosarcoma cells. J. Cell. Physiol. 227, 223–232. doi: 10.1002/jcp.22720
French, D. M., Kaul, R. J., D’Souza, A. L., Crowley, C. W., Bao, M., Frantz, G. D., et al. (2004). WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am. J. Pathol. 165, 855–867. doi: 10.1016/s0002-9440(10)63348-2
Gao, R., and Brigstock, D. R. (2003). Low density lipoprotein receptor-related protein (LRP) is a heparin-dependent adhesion receptor for connective tissue growth factor (CTGF) in rat activated hepatic stellate cells. Hepatol. Res. 27, 214–220. doi: 10.1016/s1386-6346(03)00241-9
Gray, M. R., Malmquist, J. A., Sullivan, M., Blea, M., and Castellot, J. J. (2007). CCN5 Expression in mammals. II. Adult rodent tissues. J. Cell Commun. Signal. 1, 145–158. doi: 10.1007/s12079-007-0013-z
Gurevicius, K., Gureviciene, I., Valjakka, A., Schachner, M., and Tanila, H. (2004). Enhanced cortical and hippocampal neuronal excitability in mice deficient in the extracellular matrix glycoprotein tenascin-R. Mol. Cell. Neurosci. 25, 515–523. doi: 10.1016/j.mcn.2003.12.001
Gurevicius, K., Kuang, F., Stoenica, L., Irintchev, A., Gureviciene, I., Dityatev, A., et al. (2009). Genetic ablation of tenascin-C expression leads to abnormal hippocampal CA1 structure and electrical activity in vivo. Hippocampus 19, 1232–1246. doi: 10.1002/hipo.20585
Hall-Glenn, F., De Young, R. A., Huang, B.-L., van Handel, B., Hofmann, J. J., Chen, T. T., et al. (2012). CCN2/connective tissue growth factor is essential for pericyte adhesion and endothelial basement membrane formation during angiogenesis. PLoS One 7:e30562. doi: 10.1371/journal.pone.0030562
Hann, S., Kvenvold, L., Newby, B. N., Hong, M., and Warman, M. L. (2013). A Wisp3 Cre-knockin allele produces efficient recombination in spermatocytes during early prophase of meiosis I. PLoS One 8:e75116. doi: 10.1371/journal.pone.0075116
Haque, I., Banerjee, S., De, A., Maity, G., Sarkar, S., Majumdar, M., et al. (2015). CCN5/WISP-2 promotes growth arrest of triple-negative breast cancer cells through accumulation and trafficking of p27(Kip1) via Skp2 and FOXO3a regulation. Oncogene 34, 3152–3163. doi: 10.1038/onc.2014.250
Hashimoto, Y., Shindo-Okada, N., Tani, M., Nagamachi, Y., Takeuchi, K., Shiroishi, T., et al. (1998). Expression of the Elm1 gene, a novel gene of the CCN (connective tissue growth factor, Cyr61/Cef10 and neuroblastoma overexpressed gene) family, suppresses In vivo tumor growth and metastasis of K-1735 murine melanoma cells. J. Exp. Med. 187, 289–296. doi: 10.1084/jem.187.3.289
Hertel, M., Tretter, Y., Alzheimer, C., and Werner, S. (2000). Connective tissue growth factor: a novel player in tissue reorganization after brain injury? Eur. J. Neurosci. 12, 376–380. doi: 10.1046/j.1460-9568.2000.00930.x
Heuer, H., Christ, S., Friedrichsen, S., Brauer, D., Winckler, M., Bauer, K., et al. (2003). Connective tissue growth factor: a novel marker of layer vii neurons in the rat cerebral cortex. Neuroscience 119, 43–52. doi: 10.1016/s0306-4522(03)00100-3
Holbourn, K. P., Acharya, K. R., and Perbal, B. (2008). The CCN family of proteins: structure-function relationships. Trends Biochem. Sci. 33, 461–473. doi: 10.1016/j.tibs.2008.07.006
Hou, C.-H., Tang, C.-H., Hsu, C.-J., Hou, S.-M., and Liu, J.-F. (2013). CCN4 induces IL-6 production through αvβ5 receptor, PI3K, Akt and NF-κB singling pathway in human synovial fibroblasts. Arthritis Res. Ther. 15:R19. doi: 10.1186/ar4151
Huang, W., Gonzalez, M. E., Toy, K. A., Banerjee, M., and Kleer, C. G. (2010). Blockade of CCN6 (WISP3) activates growth factor-independent survival and resistance to anoikis in human mammary epithelial cells. Cancer Res. 70, 3340–3350. doi: 10.1158/0008-5472.CAN-09-4225
Hurvitz, J. R., Suwairi, W. M., Van Hul, W., El-Shanti, H., Superti-Furga, A., Roudier, J., et al. (1999). Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat. Genet. 23, 94–98. doi: 10.1038/12699
Iacono, G., Altafini, C., and Torre, V. (2013). Early Phase of Plasticity-Related Gene Regulation and SRF Dependent Transcription in the Hippocampus. PLoS One 8:e68078. doi: 10.1371/journal.pone.0068078
Irintchev, A., Rollenhagen, A., Troncoso, E., Kiss, J. Z., and Schachner, M. (2005). Structural and functional aberrations in the cerebral cortex of tenascin-C deficient mice. Cereb. Cortex 15, 950–962. doi: 10.1093/cercor/bhh195
Ito, T., Hiraoka, S., Kuroda, Y., Ishii, S., Umino, A., Kashiwa, A., et al. (2007). Effects of schizophrenomimetics on the expression of the CCN1 (CYR 61) gene encoding a matricellular protein in the infant and adult neocortex of the mouse and rat. Int. J. Neuropsychopharmacol. 10, 717–725. doi: 10.1017/S1461145707007882
Ivkovic, S., Yoon, B. S., Popoff, S. N., Safadi, F. F., Libuda, D. E., Stephenson, R. C., et al. (2003). Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal. Development 130, 2779–2791. doi: 10.1242/dev.00505
Joliot, V., Martinerie, C., Dambrine, G., Plassiart, G., Brisac, M., Crochet, J., et al. (1992). Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol. Cell. Biol. 12, 10–21.
Jones, E. V., Bernardinelli, Y., Tse, Y. C., Chierzi, S., Wong, T. P., and Murai, K. K. (2011). Astrocytes control glutamate receptor levels at developing synapses through SPARC-beta-integrin interactions. J. Neurosci. 31, 4154–4165. doi: 10.1523/JNEUROSCI.4757-10.2011
Jones, J. A., Gray, M. R., Oliveira, B. E., Koch, M., and Castellot, J. J. Jr. (2007). CCN5 expression in mammals: I. Embryonic and fetal tissues of mouse and human. J. Cell Commun. Signal. 1, 127–143. doi: 10.1007/s12079-007-0012-0
Jun, J.-I., and Lau, L. F. (2011). Taking Aim at the Extracellular Matrix: CCN Proteins as Emerging Therapeutic Targets. Nat. Rev. Drug Discov. 10, 945–963. doi: 10.1038/nrd3599
Kasukawa, T., Masumoto, K., Nikaido, I., Nagano, M., Uno, K. D., Tsujino, K., et al. (2011). Quantitative Expression Profile of Distinct Functional Regions in the Adult Mouse Brain. PLoS One 6:e23228. doi: 10.1371/journal.pone.0023228
Katsube, K., Sakamoto, K., Tamamura, Y., and Yamaguchi, A. (2009). Role of CCN, a vertebrate specific gene family, in development. Dev. Growth Differ. 51, 55–67. doi: 10.1111/j.1440-169x.2009.01077.x
Khodosevich, K., Lazarini, F., von Engelhardt, J., Kaneko, H., Lledo, P.-M., and Monyer, H. (2013). Connective Tissue Growth Factor Regulates Interneuron Survival and Information Processing in the Olfactory Bulb. Neuron 79, 1136–1151. doi: 10.1016/j.neuron.2013.07.011
Kocialkowski, S., Yeger, H., Kingdom, J., Perbal, B., and Schofield, P. N. (2001). Expression of the human NOV gene in first trimester fetal tissues. Anat. Embryol. (Berl) 203, 417–427. doi: 10.1007/s004290100177
Kondo, Y., Nakanishi, T., Takigawa, M., and Ogawa, N. (1999). Immunohistochemical localization of connective tissue growth factor in the rat central nervous system. Brain Res. 834, 146–151. doi: 10.1016/s0006-8993(99)01517-6
Kucharska, J., Del Río, P., Arango-Gonzalez, B., Gorza, M., Feuchtinger, A., Hauck, S. M., et al. (2014). Cyr61 activates retinal cells and prolongs photoreceptor survival in rd1 mouse model of retinitis pigmentosa. J. Neurochem. 130, 227–240. doi: 10.1111/jnc.12704
Kular, L., Rivat, C., Lelongt, B., Calmel, C., Laurent, M., Pohl, M., et al. (2012). NOV/CCN3 attenuates inflammatory pain through regulation of matrix metalloproteinases-2 and -9. J. Neuroinflammation 9:36. doi: 10.1186/1742-2094-9-36
Kuruvilla, R., Ye, H., and Ginty, D. D. (2000). Spatially and functionally distinct roles of the PI3-K effector pathway during NGF signaling in sympathetic neurons. Neuron 27, 499–512. doi: 10.1016/s0896-6273(00)00061-1
Kutz, W. E., Gong, Y., and Warman, M. L. (2005). WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol. Cell. Biol. 25, 414–421. doi: 10.1128/mcb.25.1.414-421.2005
Lau, L. F., and Nathans, D. (1987). Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc. Natl. Acad. Sci. U S A 84, 1182–1186. doi: 10.1073/pnas.84.5.1182
Laurent, M., Le Dréau, G., Guillonneau, X., Lelièvre, E., Slembrouck, A., Goureau, O., et al. (2012). Temporal and spatial expression of CCN3 during retina development. Dev. Neurobiol. 72, 1363–1375. doi: 10.1002/dneu.20994
Leask, A., and Abraham, D. J. (2006). All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 119, 4803–4810. doi: 10.1242/jcs.03270
Le Dréau, G., Kular, L., Nicot, A. B., Calmel, C., Melik-Parsadaniantz, S., Kitabgi, P., et al. (2010a). NOV/CCN3 upregulates CCL2 and CXCL1 expression in astrocytes through beta1 and beta5 integrins. Glia 58, 1510–1521. doi: 10.1002/glia.21025
Le Dréau, G., Nicot, A., Bénard, M., Thibout, H., Vaudry, D., Martinerie, C., et al. (2010b). NOV/CCN3 promotes maturation of cerebellar granule neuron precursors. Mol. Cell. Neurosci. 43, 60–71. doi: 10.1016/j.mcn.2009.02.011
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. doi: 10.1038/nature05453
Liu, H., Dong, W., Lin, Z., Lu, J., Wan, H., Zhou, Z., et al. (2013). CCN4 regulates vascular smooth muscle cell migration and proliferation. Mol. Cells 36, 112–118. doi: 10.1007/s10059-013-0012-2
Malik, A. R., Urbanska, M., Gozdz, A., Swiech, L. J., Nagalski, A., Perycz, M., et al. (2013). Cyr61, a matricellular protein, is needed for dendritic arborization of hippocampal neurons. J. Biol. Chem. 288, 8544–8559. doi: 10.1074/jbc.M112.411629
Matsushita, Y., Sakamoto, K., Tamamura, Y., Shibata, Y., Minamizato, T., Kihara, T., et al. (2013). CCN3 protein participates in bone regeneration as an inhibitory factor. J. Biol. Chem. 288, 19973–19985. doi: 10.1074/jbc.M113.454652
Michele, M., and Faissner, A. (2009). Tenascin-C stimulates contactin-dependent neurite outgrowth via activation of phospholipase C. Mol. Cell. Neurosci. 41, 397–408. doi: 10.1016/j.mcn.2009.04.004
Minamizato, T., Sakamoto, K., Liu, T., Kokubo, H., Katsube, K., Perbal, B., et al. (2007). CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem. Biophys. Res. Commun. 354, 567–573. doi: 10.1016/j.bbrc.2007.01.029
Mo, F. E., and Lau, L. F. (2006). The matricellular protein CCN1 is essential for cardiac development. Circ. Res. 99, 961–969. doi: 10.1161/01.res.0000248426.35019.89
Mo, F.-E., Muntean, A. G., Chen, C.-C., Stolz, D. B., Watkins, S. C., and Lau, L. F. (2002). CYR61 (CCN1) Is Essential for Placental Development and Vascular Integrity. Mol. Cell. Biol. 22, 8709–8720. doi: 10.1128/mcb.22.24.8709-8720.2002
Morawski, M., Dityatev, A., Hartlage-Rübsamen, M., Blosa, M., Holzer, M., Flach, K., et al. (2014). Tenascin-R promotes assembly of the extracellular matrix of perineuronal nets via clustering of aggrecan. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20140046. doi: 10.1098/rstb.2014.0046
Murphy-Ullrich, J. E., and Sage, E. H. (2014). Revisiting the matricellular concept. Matrix Biol. 37, 1–14. doi: 10.1016/j.matbio.2014.07.005
Myers, R. B., Wei, L., and Castellot, J. J. Jr. (2014). The matricellular protein CCN5 regulates podosome function via interaction with integrin αvβ3. J. Cell Commun. Signal. 8, 135–146. doi: 10.1007/s12079-013-0218-2
Nakamura, Y., Weidinger, G., Liang, J. O., Aquilina-Beck, A., Tamai, K., Moon, R. T., et al. (2007). The CCN family member Wisp3, mutant in progressive pseudorheumatoid dysplasia, modulates BMP and Wnt signaling. J. Clin. Invest. 117, 3075–3086. doi: 10.1172/jci32001
Nikonenko, A., Schmidt, S., Skibo, G., Brückner, G., and Schachner, M. (2003). Tenascin-R-deficient mice show structural alterations of symmetric perisomatic synapses in the CA1 region of the hippocampus. J. Comp. Neurol. 456, 338–349. doi: 10.1002/cne.10537
Ohkawa, Y., Ohmi, Y., Tajima, O., Yamauchi, Y., Furukawa, K., and Furukawa, K. (2011). Wisp2/CCN5 up-regulated in the central nervous system of GM3-only mice facilitates neurite formation in Neuro2a cells via integrin-Akt signaling. Biochem. Biophys. Res. Commun. 411, 483–489. doi: 10.1016/j.bbrc.2011.06.118
Ono, M., Inkson, C. A., Kilts, T. M., and Young, M. F. (2011). WISP-1/CCN4 Regulates Osteogenesis by Enhancing BMP-2 Activity. J. Bone Miner. Res. 26, 193–208. doi: 10.1002/jbmr.205
Pan, L.-H., Beppu, T., Kurose, A., Yamauchi, K., Sugawara, A., Suzuki, M., et al. (2002). Neoplastic cells and proliferating endothelial cells express connective tissue growth factor (CTGF) in glioblastoma. Neurol. Res. 24, 677–683. doi: 10.1179/016164102101200573
Pennica, D., Swanson, T. A., Welsh, J. W., Roy, M. A., Lawrence, D. A., Lee, J., et al. (1998). WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc. Natl. Acad. Sci. U S A 95, 14717–14722. doi: 10.1073/pnas.95.25.14717
Perbal, B. (2009). Alternative splicing of CCN mRNAs …. it has been upon us. J. Cell Commun. Signal. 3, 153–157. doi: 10.1007/s12079-009-0051-9
Perbal, B. (2013). CCN proteins: A centralized communication network. J. Cell Commun. Signal. 7, 169–177. doi: 10.1007/s12079-013-0193-7
Plantman, S. (2012). Osteopontin is upregulated after mechanical brain injury and stimulates neurite growth from hippocampal neurons through β1 integrin and CD44. Neuroreport 23, 647–652. doi: 10.1097/WNR.0b013e328355380e
Rigato, F., Garwood, J., Calco, V., Heck, N., Faivre-Sarrailh, C., and Faissner, A. (2002). Tenascin-C promotes neurite outgrowth of embryonic hippocampal neurons through the alternatively spliced fibronectin type III BD domains via activation of the cell adhesion molecule F3/contactin. J. Neurosci. 22, 6596–6609.
Saghatelyan, A., de Chevigny, A., Schachner, M., and Lledo, P.-M. (2004). Tenascin-R mediates activity-dependent recruitment of neuroblasts in the adult mouse forebrain. Nat. Neurosci. 7, 347–356. doi: 10.1038/nn1211
Saghatelyan, A. K., Dityatev, A., Schmidt, S., Schuster, T., Bartsch, U., and Schachner, M. (2001). Reduced perisomatic inhibition, increased excitatory transmission and impaired long-term potentiation in mice deficient for the extracellular matrix glycoprotein tenascin-R. Mol. Cell. Neurosci. 17, 226–240. doi: 10.1006/mcne.2000.0922
Sakamoto, K., Yamaguchi, S., Ando, R., Miyawaki, A., Kabasawa, Y., Takagi, M., et al. (2002). The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J. Biol. Chem. 277, 29399–29405. doi: 10.1074/jbc.m203727200
Sakuma, K., Komatsu, H., Maruyama, M., Imaichi, S., Habata, Y., and Mori, M. (2015). Temporal and Spatial Transcriptional Fingerprints by Antipsychotic or Propsychotic Drugs in Mouse Brain. PLoS One 10:e118510. doi: 10.1371/journal.pone.0118510
Schlegelmilch, K., Keller, A., Zehe, V., Hondke, S., Schilling, T., Jakob, F., et al. (2014). WISP 1 is an important survival factor in human mesenchymal stromal cells. Gene 551, 243–254. doi: 10.1016/j.gene.2014.09.002
Schütze, N., Schenk, R., Fiedler, J., Mattes, T., Jakob, F., and Brenner, R. E. (2007). CYR61/CCN1 and WISP3/CCN6 are chemoattractive ligands for human multipotent mesenchymal stroma cells. BMC Cell Biol. 8:45. doi: 10.1186/1471-2121-8-45
Schwab, J. M., Beschorner, R., Nguyen, T. D., Meyermann, R., and Schluesener, H. J. (2001). Differential cellular accumulation of connective tissue growth factor defines a subset of reactive astrocytes, invading fibroblasts and endothelial cells following central nervous system injury in rats and humans. J. Neurotrauma 18, 377–388. doi: 10.1089/089771501750170930
Schwab, J. M., Postler, E., Nguyen, T. D., Mittelbronn, M., Meyermann, R., and Schluesener, H. J. (2000). Connective tissue growth factor is expressed by a subset of reactive astrocytes in human cerebral infarction. Neuropathol. Appl. Neurobiol. 26, 434–440. doi: 10.1046/j.1365-2990.2000.00271.x
Spliet, W. G. M., Aronica, E., Ramkema, M., Aten, J., and Troost, D. (2003). Increased expression of connective tissue growth factor in amyotrophic lateral sclerosis human spinal cord. Acta Neuropathol. (Berl) 106, 449–457. doi: 10.1007/s00401-003-0741-y
Steffen, C. L., Ball-Mirth, D. K., Harding, P. A., Bhattacharyya, N., Pillai, S., and Brigstock, D. R. (1998). Characterization of cell-associated and soluble forms of connective tissue growth factor (CTGF) produced by fibroblast cells in vitro. Growth Factors 15, 199–213. doi: 10.3109/08977199809002117
Stephens, S., Palmer, J., Konstantinova, I., Pearce, A., Jarai, G., and Day, E. (2015). A functional analysis of Wnt inducible signalling pathway protein -1 (WISP-1/CCN4). J. Cell Commun. Signal. 9, 63–72. doi: 10.1007/s12079-015-0267-9
Strekalova, T., Sun, M., Sibbe, M., Evers, M., Dityatev, A., Gass, P., et al. (2002). Fibronectin domains of extracellular matrix molecule tenascin-C modulate hippocampal learning and synaptic plasticity. Mol. Cell. Neurosci. 21, 173–187. doi: 10.1006/mcne.2002.1172
Su, B. Y., Cai, W. Q., Zhang, C. G., Martinez, V., Lombet, A., and Perbal, B. (2001). The expression of ccn3 (nov) RNA and protein in the rat central nervous system is developmentally regulated. Mol. Pathol. 54, 184–191. doi: 10.1136/mp.54.3.184
Su, B. Y., Cai, W. Q., Zhang, C. G., Su, H. C., and Perbal, B. (1998). A developmental study of novH gene expression in human central nervous system. C. R. Acad. Sci. III 321, 883–892. doi: 10.1016/s0764-4469(99)80002-x
Ueberham, U., Ueberham, E., Gruschka, H., and Arendt, T. (2003). Connective tissue growth factor in Alzheimer’s disease. Neuroscience 116, 1–6. doi: 10.1016/s0306-4522(02)00670-x
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419
van Velthoven, C. T. J., Heijnen, C. J., van Bel, F., and Kavelaars, A. (2011). Osteopontin enhances endogenous repair after neonatal hypoxic-ischemic brain injury. Stroke 42, 2294–2301. doi: 10.1161/STROKEAHA.110.608315
Wahab, N. A., Weston, B. S., and Mason, R. M. (2005). Connective tissue growth factor CCN2 interacts with and activates the tyrosine kinase receptor TrkA. J. Am. Soc. Nephrol. 16, 340–351. doi: 10.1681/asn.2003100905
Wang, S., Chong, Z. Z., Shang, Y. C., and Maiese, K. (2012). Wnt1 inducible signaling pathway protein 1 (WISP1) blocks neurodegeneration through phosphoinositide 3 kinase/Akt1 and apoptotic mitochondrial signaling involving Bad, Bax, Bim and Bcl-xL. Curr. Neurovasc. Res. 9, 20–31. doi: 10.2174/156720212799297137
Williams, E. O., Xiao, Y., Sickles, H. M., Shafer, P., Yona, G., Yang, J. Y., et al. (2007). Novel subdomains of the mouse olfactory bulb defined by molecular heterogeneity in the nascent external plexiform and glomerular layers. BMC Dev. Biol. 7:48. doi: 10.1186/1471-213x-7-48
Xie, D., Yin, D., Tong, X., O’Kelly, J., Mori, A., Miller, C., et al. (2004a). Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and beta-catenin-TCF/Lef signaling pathways. Cancer Res. 64, 1987–1996. doi: 10.1158/0008-5472.can-03-0666
Xie, D., Yin, D., Wang, H.-J., Liu, G.-T., Elashoff, R., Black, K., et al. (2004b). Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin. Cancer Res. 10, 2072–2081. doi: 10.1158/1078-0432.ccr-0659-03
Xu, J.-C., Xiao, M.-F., Jakovcevski, I., Sivukhina, E., Hargus, G., Cui, Y.-F., et al. (2014). The extracellular matrix glycoprotein tenascin-R regulates neurogenesis during development and in the adult dentate gyrus of mice. J. Cell Sci. 127, 641–652. doi: 10.1242/jcs.137612
Yan, Y.-P., Lang, B. T., Vemuganti, R., and Dempsey, R. J. (2009). Osteopontin is a mediator of the lateral migration of neuroblasts from the subventricular zone after focal cerebral ischemia. Neurochem. Int. 55, 826–832. doi: 10.1016/j.neuint.2009.08.007
Zacharias, U., and Rauch, U. (2006). Competition and cooperation between tenascin-R, lecticans and contactin 1 regulate neurite growth and morphology. J. Cell Sci. 119, 3456–3466. doi: 10.1242/jcs.03094
Zhang, R., Averboukh, L., Zhu, W., Zhang, H., Jo, H., Dempsey, P. J., et al. (1998). Identification of rCop-1, a New Member of the CCN Protein Family, as a Negative Regulator for Cell Transformation. Mol. Cell. Biol. 18, 6131–6141.
Zhao, Z., Ho, L., Wang, J., Qin, W., Festa, E. D., Mobbs, C., et al. (2005). Connective tissue growth factor (CTGF) expression in the brain is a downstream effector of insulin resistance- associated promotion of Alzheimer’s disease beta-amyloid neuropathology. FASEB J. 19, 2081–2082. doi: 10.1096/fj.05-4359fje
Keywords: nervous system, matricellular proteins, extracellular matrix, signal transduction, CCN
Citation: Malik AR, Liszewska E and Jaworski J (2015) Matricellular proteins of the Cyr61/CTGF/NOV (CCN) family and the nervous system. Front. Cell. Neurosci. 9:237. doi: 10.3389/fncel.2015.00237
Received: 12 April 2015; Accepted: 12 June 2015;
Published: 24 June 2015.
Edited by:
Jerzy W. Mozrzymas, Wroclaw Medical University, PolandReviewed by:
Ulkan Kilic, Istanbul Medipol University, TurkeyCarlo Di Cristo, University of Sannio, Italy
Copyright © 2015 Malik, Liszewska and Jaworski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacek Jaworski, Laboratory of Molecular and Cellular Neurobiology, International Institute of Molecular and Cell Biology, 4 Ks. Trojdena Street, 02-109 Warsaw, Poland, jaworski@iimcb.gov.pl
 Anna R. Malik
Anna R. Malik Ewa Liszewska
Ewa Liszewska  Jacek Jaworski
Jacek Jaworski