Ontology-based approach for in vivo human connectomics: the medial Brodmann area 6 case study
- Medicis, UMR 1099 LTSI, INSERM, University of Rennes 1, Rennes, France
Different non-invasive neuroimaging modalities and multi-level analysis of human connectomics datasets yield a great amount of heterogeneous data which are hard to integrate into an unified representation. Biomedical ontologies can provide a suitable integrative framework for domain knowledge as well as a tool to facilitate information retrieval, data sharing and data comparisons across scales, modalities and species. Especially, it is urgently needed to fill the gap between neurobiology and in vivo human connectomics in order to better take into account the reality highlighted in Magnetic Resonance Imaging (MRI) and relate it to existing brain knowledge. The aim of this study was to create a neuroanatomical ontology, called “Human Connectomics Ontology” (HCO), in order to represent macroscopic gray matter regions connected with fiber bundles assessed by diffusion tractography and to annotate MRI connectomics datasets acquired in the living human brain. First a neuroanatomical “view” called NEURO-DL-FMA was extracted from the reference ontology Foundational Model of Anatomy (FMA) in order to construct a gross anatomy ontology of the brain. HCO extends NEURO-DL-FMA by introducing entities (such as “MR_Node” and “MR_Route”) and object properties (such as “tracto_connects”) pertaining to MR connectivity. The Web Ontology Language Description Logics (OWL DL) formalism was used in order to enable reasoning with common reasoning engines. Moreover, an experimental work was achieved in order to demonstrate how the HCO could be effectively used to address complex queries concerning in vivo MRI connectomics datasets. Indeed, neuroimaging datasets of five healthy subjects were annotated with terms of the HCO and a multi-level analysis of the connectivity patterns assessed by diffusion tractography of the right medial Brodmann Area 6 was achieved using a set of queries. This approach can facilitate comparison of data across scales, modalities and species.
1. Introduction
The human brain is constituted of a vast amount of interconnected neurons forming structural circuits which transmit information. Multi-scale analysis of this anatomical connectivity (Caspers et al., 2013) from synaptic connections between individual neurons (microscopic scale), to brain regions interconnected via white matter fiber bundles (macroscopic scale) is fundamental to better apprehend the link between structure and function in diseased and healthy brains (Honey et al., 2010). A promising way for studying brain connectivity is to compile a coherent mapping of the network of elements and connections forming the human brain and defined as the human connectome (Sporns et al., 2005).
Recent advances in Magnetic Resonance Imaging (MRI) and brain networks have opened new possibilities to map and analyse anatomical and functional long-range connectivities in the living brain, giving birth to a new field of research: human connectomics (Behrens and Sporns, 2012). Currently, diffusion MRI (dMRI) and functional MRI (fMRI) are the most popular modalities to assess non invasively anatomical and functional connectivities, respectively (Craddock et al., 2013). dMRI estimates the local fiber bundles orientations at millimeter voxel resolution as the directions of least hindrance to water diffusion in brain. Then, tractography aims at reconstructing white matter fiber bundles using algorithmic approaches based on local fiber bundles orientations (Basser et al., 2000). fMRI uses temporal correlations in the fluctuations of the Blood-Oxygenation-Level-Dependent (BOLD) signal to infer functional connectivity (Smith et al., 2011). After reconstruction of anatomical or functional connectivities from MRI (Jbabdi and Johansen-Berg, 2011), in vivo neuroimaging data can be modeled and analyzed using connectomics in order to produce brain networks at macroscopic scale (~ 1 cm3 or greater) (Hagmann et al., 2007; Zalesky et al., 2011; Sporns, 2013). However, there is a great diversity in methodological approaches, especially no consensus currently exists on how to best define nodes for charting in vivo human connectome, i.e., subdividing the brain into macroscopic regions in an anatomo-functional coherent way (Craddock et al., 2013; Fortino et al., 2013). Indeed, depending on the scope of the study, nodes can represent small regions (~ 1 cm) or larger brain areas as a specific gyrus. Moreover comparing data across scales, modalities, and species remains challenging (Essen and Ugurbil, 2012; Leergaard et al., 2012). A real need exists of new neuroinformatics tools for in vivo human connectomics that allow different levels of granularity of multi-modal connectivity data to be described, shared, integrated and compared.
Semantic annotation of brain images consists in associating meaningful metadata using terms of an ontology in order to describe and share information related to that resource such as acquisition protocol, anatomical content, diagnosis etc. (Mechouche et al., 2009; Turner et al., 2010). Biomedical ontologies are structured vocabularies representing classes of entities which are of biomedical significance in reality. They focus on the definition of the entities of the domain being modeled and on the relations between them, especially the subtype relation used to organize the entities in a taxonomy. Ontologies also specify other relations, such as the “part of” relation, or any other relation that is relevant in the domain of interest. Specifying the set of relations (called axioms) that apply to all the instances of a class contributes to capture knowledge about this entity (Gruber, 1995). Axioms can be expressed in the OWL1 ontology language standard (Web Ontology Language, defined by the W3C), and especially the OWL DL2 sublanguage, based on Description Logics (DL) (Baader et al., 2003). OWL DL provides a good compromise between expressivity and computational complexity (and decidability). Moreover, it allows reasoning on formal knowledge and infering automatically new axioms using description logic reasoning engines such as FaCT++3. Ontology-based systems and reasoning engines are particularly relevant in the human connectomics realm as they provide the capability to apprehend consistently multi-scale knowledge, to describe heterogeneous data with semantic annotations, and finally to facilitate data querying, sharing and interoperability.
In recent years, different efforts were reported to specify computer models and ontologies to represent, collate, process and share human brain anatomical connectivity (OBO Relation Ontology, Smith et al., 2005; Swanson and Bota, 2010; Larson and Martone, 2013; Bota et al., 2014; Nichols et al., 2014). On one hand, the Foundational Model of Anatomy (FMA) was developed to provide a reference ontology for human anatomy. It includes many terms from Terminologia Anatomica (Federative Committee on Anatomical Terminology, 1998), which itself founds its origin in Nomina Anatomica (International Anatomical Nomenclature Committee, 1989). The foundamental difference between these terminologies and ontologies like FMA is that the former provide organizations of terms that enhance part of the intrinsic meaning of each term, in an implicit way, whereas ontologies such as FMA relate terms using relationships bearing explicit semantics such as subsumption links and “part of” links. FMA specifies anatomical connectivity relationships at different levels of granularity (Rosse and Mejino, 2003; Nichols et al., 2014). On the other hand, the Foundational Model of Connectivity (FMC) provides a high level conceptual framework suitable for modeling “structural architecture of nervous connectivity in all animals at all resolutions” (Swanson and Bota, 2010). In particular, this model influenced and is compatible with BAMS, the Brain Architecture Management System built by Mihail Bota and co., a neuroinformatics system to store, mine and model structural connectivity in multiple species such as mouse, rat, cat, macaque and human. Most connectivity data concern pathway-tracing experiments in animals, techniques based on injection of a tracer and tracing of neural connections either from their source to their point of termination (anterograde tracing) or the opposite (retrograde tracing). However, although these biological ontologies and conceptual models aim at representing anatomical connectivity, none of them can be used to represent connectivity assessed by diffusion tractography, yet. Indeed, diffusion tractography can only provide limited insight on the organization of in vivo white matter fiber bundles at the present time (cf. Section 4): for example it cannot determine polarity of connections, nor synaptic connections. Moreover, cytoarchitecture of the cerebral cortex cannot be rendered using MRI due to limited spatial resolution, so that concepts of gray matter region defined using criteria based on spatial distribution of a set of neuron types are not relevant for in vivo connectomics. So, a real need is emerging of new ontology in order to bridge the gap between experimental neurobiology and in vivo human connectomics observations provided by MRI.
An important and complementary field of research in neuroimaging concerns the development of digital atlases providing both a template brain and neuroanatomical labels in a conformed space. Individual brain datasets are aligned to the atlas using volumetric or surface-based registration approaches in order to propagate the neuroanatomical labels of the atlas to brain regions. A great number of in vivo neuroimaging datasets are currently annotated using brain atlases such as the Talairach Atlas (Talairach and Tournoux, 1988), the Montreal Neurological Institute (MNI) atlas (Tzourio-Mazoyer et al., 2002), or the atlases embedded in software tools such as Freesurfer4 (Fischl et al., 2004; Desikan et al., 2006) or the JHU white matter tractography atlas5 (Wakana et al., 2007; Hua et al., 2008). Recently, FMA provided a mapping between several terminologies used in brain atlases, such as Freesurfer or the JHU white matter tractography atlas, and the corresponding neuroanatomical concepts defined in FMA (Nichols et al., 2014). This effort facilitates the use of FMA as a reference and pivotal terminology for the annotation of brain segmentation results such as cortical or subcortical gray matter regions, and different white matter fiber bundles.
The main contribution of this paper was to create a generic neuroanatomical ontology called “Human Connectomics Ontology”6 (HCO) in order to represent macroscopic regions defined on MRI datasets connected via fiber bundles assessed by diffusion tractography in the living human brain. Grounded on the FMA reference ontology, HCO was expressed in OWL and used the OWL DL sublanguage in order to be processable by usual reasoning engines. The latter provide highly optimized implementations of reasoning algorithms to process and correctly answer arbitrarily complex queries, such as those involving, e.g., transitive part-whole and spatial relationships. Moreover, an experimental work was achieved in order to show how the HCO could be effectively used to address complex queries concerning in vivo MRI connectomics datasets: a multi-level analysis of the connectivity pattern of the right medial Brodmann Area 6 (BA6) reconstructed by diffusion tractography was achieved, using a set of queries on annotated neuroimaging datasets of five healthy subjects. The medial BA6 region is a cortical region defined using a set of cytoarchitectural criteria (Zilles and Amunts, 2010) and is part of the medial frontal cortex located on the midline surface of the hemisphere just in front of the primary motor cortex. This region of interest was chosen because different studies showed how it could be subdivided into different sub-regions in a reproducible way using criteria based on long-range connectivity assessed by diffusion tractography (Johansen-Berg et al., 2004; Anwander et al., 2007; Jbabdi et al., 2009). We believe that this approach can facilitate comparison of data across scales, modalities and species.
The following of the paper is organized as follows. Section 2 describes how the HCO was designed and achieved. Section 3 is related to the experimental work. Finally, Sections 4 and 5 are dedicated to the discussion and conclusion.
2. Materials and Methods
2.1. A Neuroanatomical Ontology for in vivo Human Connectomics
2.1.1. Requirements and design
Competency questions (Neuhaus and Vizedom, 2013) are more and more used to specify the domain that an ontology should cover. Therefore, we designed a set of competency questions pertaining to a use case inspired by the medial BA6 connectivity-based parcellation (Johansen-Berg et al., 2004), in order to assess how the Human Connectomics Ontology (HCO) can support hypothesis-driven analysis of connectomics datasets at different levels of granularity:
• Which gray matter parts of the right superior frontal gyrus have a connectivity pattern passing through the corticospinal tract or through some gray matter parts of the right precentral gyrus?
• Which gray matter parts of the right superior frontal gyrus have a connectivity pattern passing through some gray matter parts of the right medial parietal cortex or through some gray matter parts of the right inferior frontal cortex?
• Which gray matter parts of the right superior frontal gyrus have a connectivity pattern passing through the corticospinal tract or through some gray matter parts of the right precentral gyrus or through some gray matter parts contiguous with the right precentral gyrus?
• Which anatomical white matter fiber bundles connect some gray matter parts of the right superior frontal gyrus to some gray matter parts of the right temporal lobe?
In order to meet these requirements, a neuroanatomical ontology module, called “NEURO-DL-FMA,” was first constituted in order to annotate gross anatomy of the brain (i.e., gray matter regions, white matter fiber bundles). NEURO-DL-FMA was based on a subset of FMA which is an open source reference ontology representing the phenotypic structure of the human body at different scales. FMA contains more than 85,000 classes and 140 relationships between entities (Rosse and Mejino, 2003; Golbreich et al., 2013). Finally, the HCO was based on NEURO-DL-FMA and aimed at representing nodes connected with fiber bundles assessed by diffusion tractography. Moreover, nearest neighbor topology between gray matter regions was also represented.
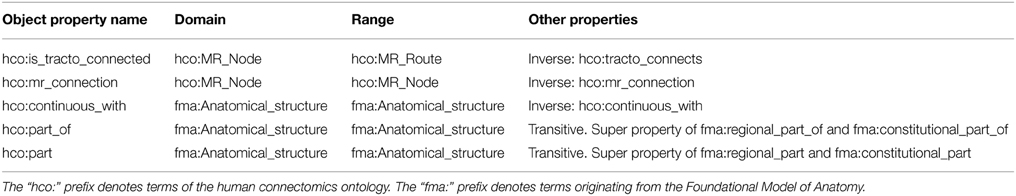
Figure 1 depicts a scenario of information retrieval concerning in vivo connectivity patterns assessed by diffusion tractography. Investigators can pose a wide range of queries using terms (i.e., classes and object properties) of the HCO. For example, an investigator could be interested in retrieving all cortical parcels of the right medial BA6 which have a connectivity pattern similar to the right Supplementary Motor Area (connectivity pattern passing through the right corticospinal tract or connected to gray matter parts of the right precentral gyrus). The query is submitted to a reasoning engine that infers automatically part-whole, connectivity and spatial relationships at different levels of granularity.
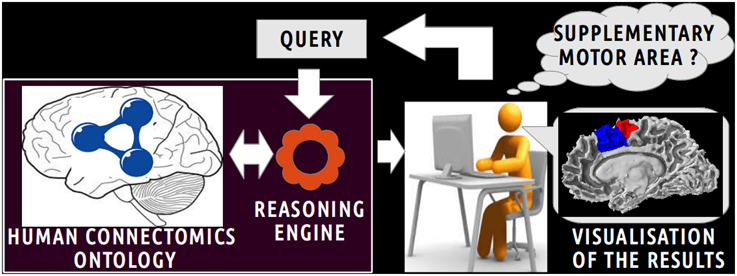
Figure 1. Scenario of information retrieval using the Human Connectomics Ontology (HCO). On the central part of the figure, an investigator can pose a wide range of queries using terms of the HCO: as an illustration, it could be to retrieve all cortical parcels belonging to the supplementary motor area. On the left part of the figure, the query is submitted to a reasoning engine that infers automatically part-whole, connectivity and spatial relationships at different level of granularity. On the right side of the figure, the results of the query can be easily visualized.
2.1.2. NEURO-DL-FMA: a Neuroanatomical Gross Anatomy Ontology
The NEURO-DL-FMA is a neuroanatomical gross anatomy ontology that was achieved in two steps. First all useful entities and relations were extracted as a “view” from the FMA reference ontology (OWL Full 3.2.1 version) (Noy and Rubin, 2008). This view was then translated into OWL DL, which was necessary since most commonly used reasoning engines do not support OWL Full.
As the FMA contains more than 85,000 anatomical concepts, the first step was to extract a “view” from the FMA in order to focus only on concepts and relationships of interest. This was achieved using vSparQL queries (Shaw et al., 2011) and the entities were extracted from the more specific to the more general ones. This view was constituted of (1) all concepts denoting a gray matter structure mapping a Freesurfer cortical region, (2) all concepts denoting a white matter bundle mapping an entity in the JHU white matter tractography atlas. A one-to-one mapping between FMA and the Freesurfer and JHU white matter tractography atlas terminologies was available in the 3.2.1 version of the FMA (Nichols et al., 2014). Then all the entities related using the fma:regional_part_of or fma:constitutional_part_of object properties were extracted recursively (the “fma” prefix denotes terms originating from the FMA). The Brodmann areas, the hippocampus parts, and the other object and data properties were excluded. All the entities that subsume the entities present in the current view were included recursively. All the meta-classes of FMA (Dameron et al., 2005) included in our view were then discarded as they did not contain useful information for our gross anatomy ontology. All concepts that did not concern the domain of neuroanatomical gross anatomy such as fma:Human_body were also discarded. In order to reuse nearest neighbor topology knowledge represented in the FMA, all the entities related using the fma:attributed_continuous_with object property of type fma:Continuous_with_relation were included in our view. Finally, only the following three object properties and their inverse (if exists) were kept: fma:constitutional_part, fma:regional_part, fma:attributed_continuous_with.
This view was achieved using a web service implementation developed by the University of Washington's Structural Informatics Group7. In this implementation, the FMA (OWL Full 3.2.1 version) is embedded in a MySQL8 relational database. This web service based on Apache Jena9 accepts VSparQL queries allowing portions of the FMA to be extracted by recursively following complex pathways within the ontology graph (Shaw et al., 2011). Figure 2 shows an example in which all entities related to the fma:Right_precentral_gyrus using the fma:regional_part_of or fma:constitutional_part_of object properties were extracted from the FMA using the following vSparQl request:
CONSTRUCT { ?x ?y ?z }
FROM <http://purl.org/sig/fma>
FROM NAMEDEV <rpo_precentral_gyrus> [
CONSTRUCT
{ temp:set temp:member ?x. }
FROM <http://purl.org/sig/fma>
WHERE {
fma:Right_precentral_gyrus gleen:OnPath(“([fma:regional_part_of]|[fma:constitutional_part_of])*” ?x). }]
WHERE GRAPH { <rpo_precentral_gyrus> { ?x ?y ?z }}.
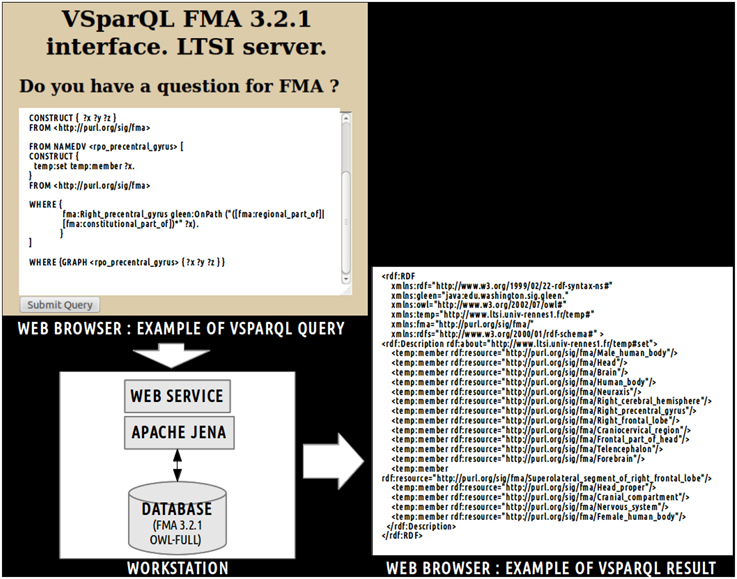
Figure 2. Example of vSparQL query submitted to a web service based on Apache Jena that permitted to extract portions of the Foundational Model of Anatomy (FMA) embedded in a MySQL database by recursively following complex pathways within the ontology graph. On the superior part of the figure, a vSparQL query aimed to extract all anatomical entities related to the fma:Right_precentral_gyrus (the “fma” prefix denotes that the entity was part of the FMA) following the fma:regional_part_of or fma:constitutional_part_of object properties. The right part of the figure shows the result of the query, expressed in RDF (Resource Description Framework).
This request was processed by a web service based on a local server at the university of Rennes 1. The result of this query (cf. Figure 2) lists all anatomical concepts from the right precentral gyrus to the human body entities illustrating part-whole relationships in FMA and was expressed using the Resource Description Framework10 (RDF).
Finally, a translation into OWL DL was necessary in order to enable the subsequent use of reasoning engines. This was achieved using a local java program based on the OWL API package11. All classes and object properties of the view were included in the ontology. As object properties were expressed at the individuals' level in the view, all object properties were translated at the classes' level using existential restrictions. Figure 3 depicts an example of translation of the fma:Precentral_gyrus entity from the view expressed in OWL Full (cf. part 1 of the figure) into NEURO-DL-FMA expressed in the OWL DL formalism (cf. part 2). In the view, entities such as fma:Precentral_gyrus appear both as a class and as an individual (cf. part 1). While part-whole relationships such as fma:constitutional_part or fma:regional_part_of were expressed at the individuals' level in the view (cf. part 1), the same object properties were expressed at the classes' level using existential restrictions in NEURO-DL-FMA (cf. part 2).
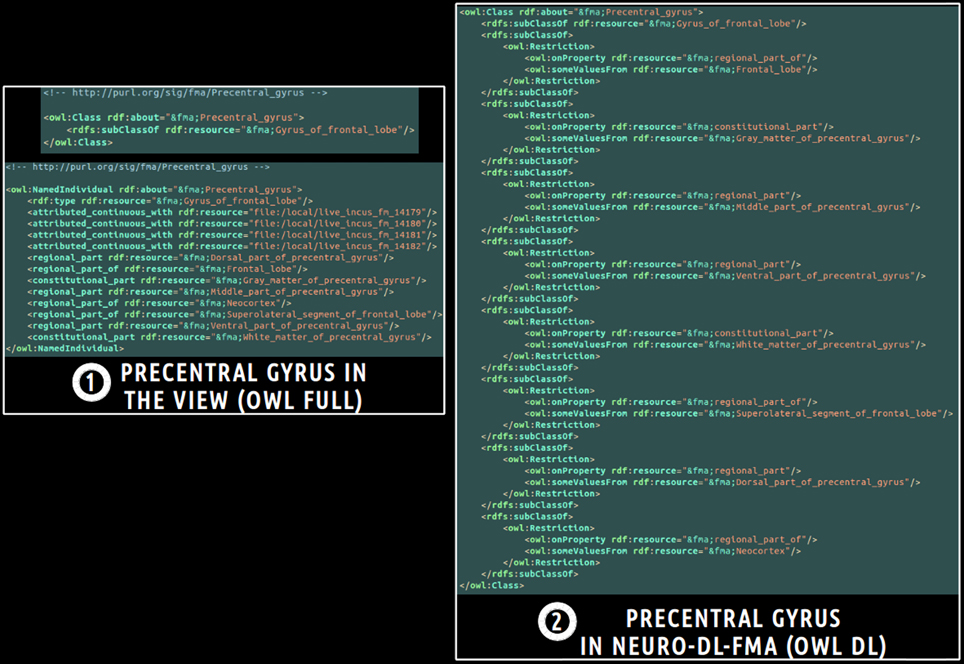
Figure 3. Example of translation of an entity from a subset (or a “view”) of the Foundational Model of Anatomy (FMA) expressed in OWL Full (cf. part 1) into the corresponding NEURO-DL-FMA entity expressed using the OWL DL sublanguage (cf. part 2). The “fma” prefix denotes that the entity was part of the FMA. In the left part of the figure (cf. part 1), concepts such as fma:Precentral_gyrus were both class and instance. Moreover, part-whole relationships such as fma:constitutional_part or fma:regional_part_of were expressed at the individuals' level. In the right part of the figure (cf. part 2), the same relationships were represented at the classes' level using existential restrictions.
2.1.3. The Human Connectomics Ontology
The “Human Connectomics Ontology” (HCO) was created in order to represent brain regions, nearest neighbor topology and connectivity relationships assessed by diffusion tractography.
Different classes and object properties were defined in the HCO (cf. Tables 1, 2):
• hco:Gray_matter_part: Any cell part cluster constituting a part of (i.e., fma:constitutional_part_of, fma:regional_part_of) a gray matter region. This concept is more general than the gray-matter-region12 term defined in the FMC thesaurus (Swanson and Bota, 2010), because it is not grounded on criteria based on cytoarchitecture as the spatial distribution of a specific set of neuron types.
• hco:MR_Node: This concept denotes any hco:Gray_matter_part in brain images where a connection assessed by diffusion tractography begins or ends. This concept is an adaptation of the node13 term, defined in the FMC thesaurus (Swanson and Bota, 2010), dedicated to MRI datasets.
• hco:White_matter_part: Any cell part cluster constituting a part (i.e., fma:constitutional_part_of, fma:regional_part_of) of a white matter region.
• hco:MR_Route: Any physical route of white matter fiber bundles reconstructed by diffusion tractography that links two hco:MR_Node in brain images. This concept is an adaptation of the route14 term, defined in the FMC thesaurus (Swanson and Bota, 2010), dedicated to the brain images domain as diffusion tractography does not probe the path of white matter bundles directly, but water diffusion in brain.
• hco:is_tracto_connected: Object property that links an hco:MR_Node to an hco:MR_Route. The inverse property is hco:tracto_connects.
• hco:mr_connection: Symmetric object property that denotes the existence of a pathway assessed by diffusion tractography linking two different hco:MR_Node. If a hco:is_tracto_connected b and b hco:tracto_connects c, then a hco:mr_connection c. This property is an adaptation of the connection15 term, defined in the FMC thesaurus (Swanson and Bota, 2010), dedicated to connections assessed by diffusion tractography.
• hco:continuous_with: Symmetric object property that links an fma:Anatomical_structure to some nearest neighbor fma:Anatomical_structure.
Figure 4 illustrates how a fiber bundle reconstructed by diffusion tractography that connected two cortical parcels via the corpus callosum white matter fiber bundle were represented using terms of the HCO. The hco:s1_gray_matter_of_right_superior_frontal_gyrus_17 instance of the hco:MR_Node class denotes a high resolution cortical parcel. Part-whole relationships were represented thanks to the fma:regional_part_of and the fma:constitutional_part_of object properties: this latter cortical parcel was a regional part of some gray matter of the right superior frontal gyrus which was a constitutional part of the right superior frontal gyrus which was a regional part of the right frontal lobe. The parcel in the right hemisphere was linked to the other hemisphere via the hco:s1_mr_route_118 instance of the hco:MR_Route class. This instance was related to the anterior part of the corpus callosum via a fma:regional_part_of object property. Finally the two different cortical parcels were linked via the hco:mr_connection object property.
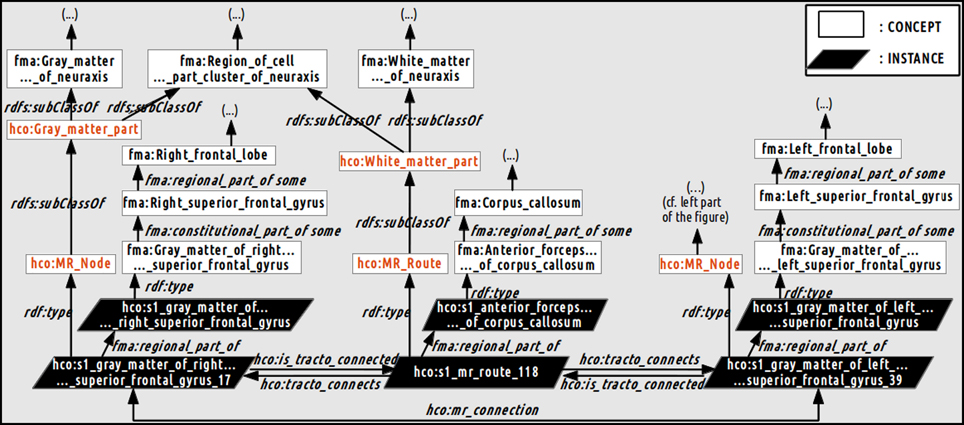
Figure 4. Schematic illustration of how the relationships between the gray and white matter entities were represented in the Human Connectomics Ontology (HCO). The “fma:” prefix denotes concepts (black) and object properties of the Foundational Model of Anatomy (FMA). The “hco:” prefix denotes concepts (red), instances and object properties of the HCO. The hco:s1_gray_matter_of_right_superior_frontal_gyrus_17 instance of the hco:MR_Node class denotes a high resolution cortical parcel linked to the left hemisphere via the hco:s1_mr_route_118 instance of the hco:MR_Route class. This instance was related to the anterior part of the corpus callosum via the fma:regional_part_of object property. The two different cortical parcels were linked via the hco:mr_connection object property. Part-whole relationships were represented thanks to the fma:regional_part_of and fma:constitutional_part_of object properties.
2.2. Experimental Work
The aim of this experimental work was to assess how the HCO can effectively be used to enhance multi-level hypothesis-driven analysis of connectomics datasets. Figure 5 depicts an overview of the main steps of this experimental work.
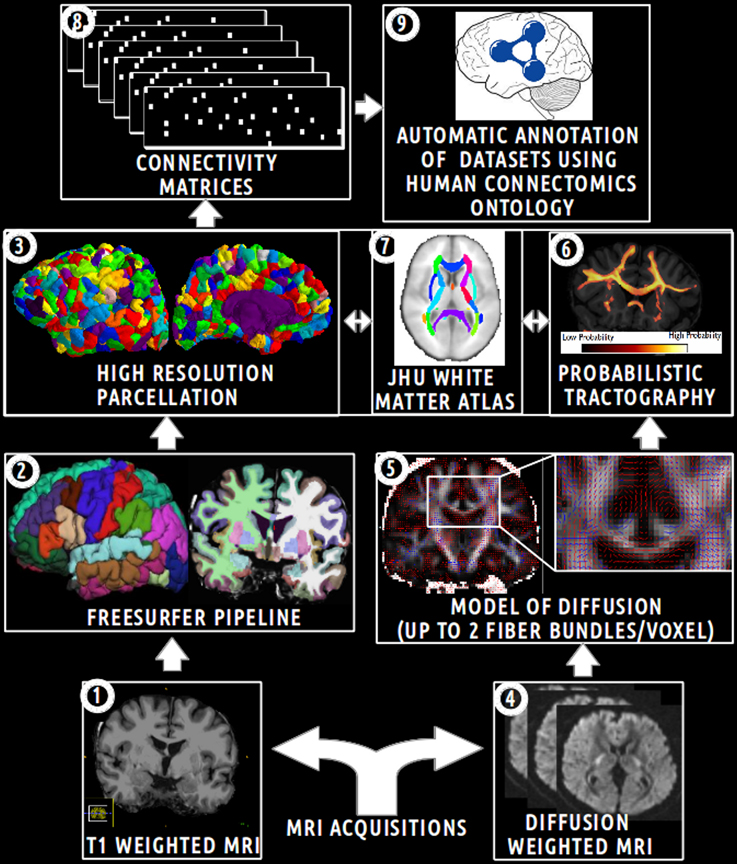
Figure 5. Schematic overview of the main steps of the experimental work. (1) Acquisition of T1 weighted MR images. (2) Automatic segmentation of brain regions using the Freesurfer pipeline. (3) Computation of an high resolution parcellation (CMTK toolkit). (4) Acquisition of diffusion weighted MR images. (5) Computation of diffusion model (FSL toolkit). (6) Computation of probabilistic tractography. (7) Automatic segmentation of anatomical fiber bundles based on the JHU white atlas (FSL toolkit). (8) Computation of connectivity matrices. (9) Automatic annotation of MRI connectomics datasets using terms of the Human Connectomics Ontology (HCO).
2.2.1. Neuroimaging Data and Preprocessing
The analysis was performed for five subjects of the NMR public database (Poupon et al., 2006). This database provided T1 (voxel size 0.9 × 0.9 × 1.2 mm) and diffusion-weighted datasets (voxel size of 1.9 × 1.9 × 2.0 mm) acquired with a GE Healthcare Signa 1.5 Tesla Excite II scanner. The diffusion datasets presented a high angular resolution (HARDI) based on 200 directions and a b-value of 3000 s/mm2. The use of a twice refocusing spin echo technique was used to compensate the echoplanar distortions due to eddy currents (Reese et al., 2003), at the first order. Susceptibility artifacts were corrected using a phase map acquisition. See “MRI acquisitions” on Figure 5.
The Freesurfer pipeline was applied on T1-weighted datasets producing cortical (Desikan et al., 2006) and sub-cortical segmentations (Dale et al., 1999). Then a high resolution cortical parcellation was performed for each subject. Each of the Freesurfer cortical regions was arbitrarily subdivided into a set of small and compact parcels of about 1.5 cm2 (Hagmann et al., 2008), resulting in 1000 parcels covering the entire cortex thanks to the connectome mapping toolkit16 (CMTK) (Daducci et al., 2012). The nearest neighbors of each parcel were assessed for each subject using a dilatation-based strategy. Thus, a total of 1000 cortical parcels and other regions (i.e., thalamus, caudate, putamen, pallidum, accumbens area, amygdala, hippocampus in both hemispheres, and brain-stem) were defined in the Freesurfer structural space. See “Freesurfer pipeline” and “high resolution parcellation” on Figure 5.
Two target masks were defined for the tractography in the Freesurfer structural space. The first target mask was defined as the set of high resolution cortical parcels included in the right medial Brodmann Area 6 (BA6). This right medial BA6 region of interest was defined in restricting the Freesurfer segmentation corresponding to the cortical region of the right superior frontal gyrus (i.e., “ctx-rh-superiorfrontal”) from y = −22 to y = 30 (MNI coordinates in the anteroposterior direction) and from the cingulate sulcus to the dorsal surface of the brain in order to include only voxels belonging to the gray matter on the medial wall (Johansen-Berg et al., 2004). The registration between the Freesurfer conformed space and the MNI space was computed using a linear registration (12 DOF) based on mutual information. This was achieved using the FLIRT tool of the FSL toolbox17. Finally, the second target mask was defined as the remaining cortical parcels or regions.
All the 20 white matter tracts of the JHU white matter tractography probabilistic atlas based on diffusion tensor imaging (Wakana et al., 2007; Hua et al., 2008) were segmented using a threshold at 25: anterior radiation of thalamus, corticospinal tract, anterior segment of cingulum bundle, anterior forceps of corpus callosum, posterior forceps of corpus callosum, inferior occipitofrontal fasciculus, inferior longitudinal fasciculus, uncinate fasciculus, superior longitudinal fasciculus in both hemispheres. A total of 22 white matter masks (one for each fiber bundle and two for the rest of the white matter in both hemispheres) were defined as seed masks for the tractography. An automatic linear (12 DOF) registration based on the correlation ratio between the JHU white matter tractography atlas and the Freesurfer structural space was computed using the FLIRT tool. See “JHU white matter atlas” on Figure 5.
A registration between the Freesurfer structural space and the diffusion dataset space was computed using a rigid registration (6 DOF) based on mutual information implemented in FLIRT. The registration was performed considering the average of five B0 volumes (i.e., volumes with b-value = 0) and the brain volume in the Freesurfer structural space.
2.2.2. Connectivity Assessed by Diffusion Tractography
The aim of the probabilistic tractography was to characterize the connectivity pattern of each structural element, denoted by seeds, in probing the Brownian movement of water molecules within white matter fiber bundles. Probabilistic tractography was performed using the bedpostX and probtrackX2 tools, part of the FSL toolbox (Behrens et al., 2007). BedpostX uses a Monte Carlo Markov chain sampling to estimate the diffusion parameters at each voxel. The probabilistic tractography could model up to two fiber bundles in each voxel. The burn-in of the Markov chains was set to 3000 in order to ensure convergence of the model. See “model of diffusion” on Figure 5.
A whole brain probabilistic tractography was achieved in order to assess gray-to-gray connectivity between the two target masks defined above. The probtrackX2 tractography tool drew 5000 probabilistic streamlines that were sent in both directions from the distribution connectivity of each white matter seed voxel. The 22 white matter masks defined above were used successively as seed for the tractography. If the streamline hitted the two target masks at two locations along either sides of the streamline, then the corresponding row and column of the connectivity matrix was filled. Streamlines that stopped before reaching a length of 30 mm or that passed through an exclusion mask (i.e., ventricles, cortico-spinal fluid (CSF), the choroid-plexus) were discarded. A distance correction was used in order to correct the fact that connectivity distribution drops with distance from the seed voxel. See “probabilistic tractography” on Figure 5.
Each of the 22 voxel-wise connectivity matrices was converted into a region-wise connectivity matrix (11 × 1004) between the 11 cortical regions in the first target mask and the 1004 cortical and other brain regions in the second target mask. The connectivity between two regions in the region-wise connectivity matrix was computed as the mean of the connectivities between the voxels belonging to the corresponding regions. After a logarithmic transformation of the region-wise connectivity matrix, a normalization of the values of each row was achieved. Finally, connectivity values in the region-wise connectivity matrices were thresholded at 0.7 in order to keep only connections reconstructed by diffusion tractography with a high probability. See “connectivity matrices” on Figure 5.
2.2.3. Automatic Annotation of MRI Connectomics Datasets
The HCO was populated with instances describing fiber bundles assessed by diffusion tractography between different gray matter regions of the five healthy subjects. This was achieved using a Java program based on OWL API. First, each gray matter region was represented as an instance of the hco:Gray_matter_part class. If the gray matter region was a high resolution cortical parcel, then the cortical parcel was related to the instance of the overlapping gyrus via the fma:regional_part_of object property. If two instances of fma:Anatomical_structure were found to be nearest neighbors then they were related together using the hco:continuous_with object property. Finally, each binary region-wise connectivity matrix (11 × 1004) was used to encode the connectivity reconstructed by diffusion tractography between the 11 cortical regions defined in the first target mask and the 1004 brain regions defined in the second target mask (cf. Figure 5, “automatic annotation of datasets using HCO”):
• The two corresponding structural elements of the row and column were represented as instances of the hco:MR_Node class and were related together using the hco:mr_connection object property.
• An instance of the hco:MR_Route class was created and related to the two corresponding instances of the hco:MR_Node class using the hco:tracto_connects object property.
• If the connectivity matrix was associated with an anatomical white matter fiber bundle, then the instance of the hco:MR_Route class was related to the instance of the overlapping fiber bundle using the fma:regional_part_of object property.
2.2.4. Answering Competency Questions through the Human Connectomics Ontology
Table 3 presents each competency question translated into terms of the HCO before submission to the FaCT++ reasoning engine via the “DL Query” tab of the ontology editor Protégé (Rubin et al., 2007). FaCT++ is an efficient tableaux-based reasoner implemented using C++ that supports OWL DL. It is used as one of the default reasoners in Protégé (version 4). In order to find all parts of (fma:regional_part_of, fma:constitutional_part_of) an anatomical structure, the hco:part_of transitive object property was used. The right medial parietal cortex was expressed using a conjunction of terms from the ontology: fma:Cortex_of_right_parietal_lobe and fma:Medial_segment_of_cerebral_hemisphere. The inferior frontal cortex was translated into the fma:Orbitobasal_segment_of_right_frontal_lobe term.
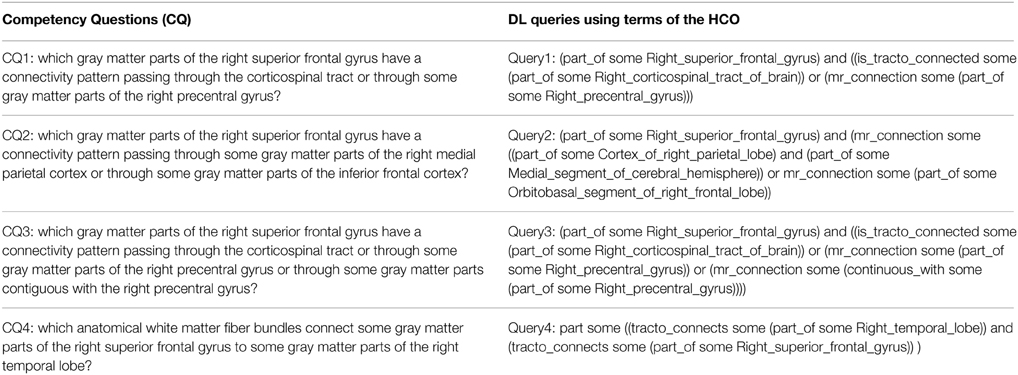
Table 3. Translation of the four competency questions into Description Logic (DL) queries using terms of the Human Connectomics Ontology (HCO).
3. Results
3.1. Right Medial Brodmann Area 6 Region of Interest
Our definition of the right medial BA6 was decomposed into eleven high resolution cortical parcels numbered: 20, 12, 32, 9, 17, 13, 42, 18, 24, 25, 38 in all five subjects. Columns 1 and 2 of the Figure 8 depict a map of these parcels on a medial view of the gray/white interface of the right hemisphere.
3.2. Structural Connectivity Assessed by Diffusion Tractography
Table 4 summarizes the set of regions that were found connected to the right medial BA6 via fiber bundles assessed by diffusion tractography for each subject. This set of regions was expressed using FMA terms denoting regions at a high level of granularity (i.e., gyrus level, or other anatomical structures): as an illustration the hco:s1_gray_matter_of_left_superior_frontal_gyrus_39 entity, which was a cortical region of the left superior frontal gyrus (cf. Figure 4), was denoted using the fma:Left_superior_frontal_gyrus concept. 23.7% of the total of anatomical terms that were connected to the right medial BA6 in our data were found common to the five subjects. 38.9% (69.5% resp.) of these anatomical terms were found common to at least 4 (3 resp.) subjects.
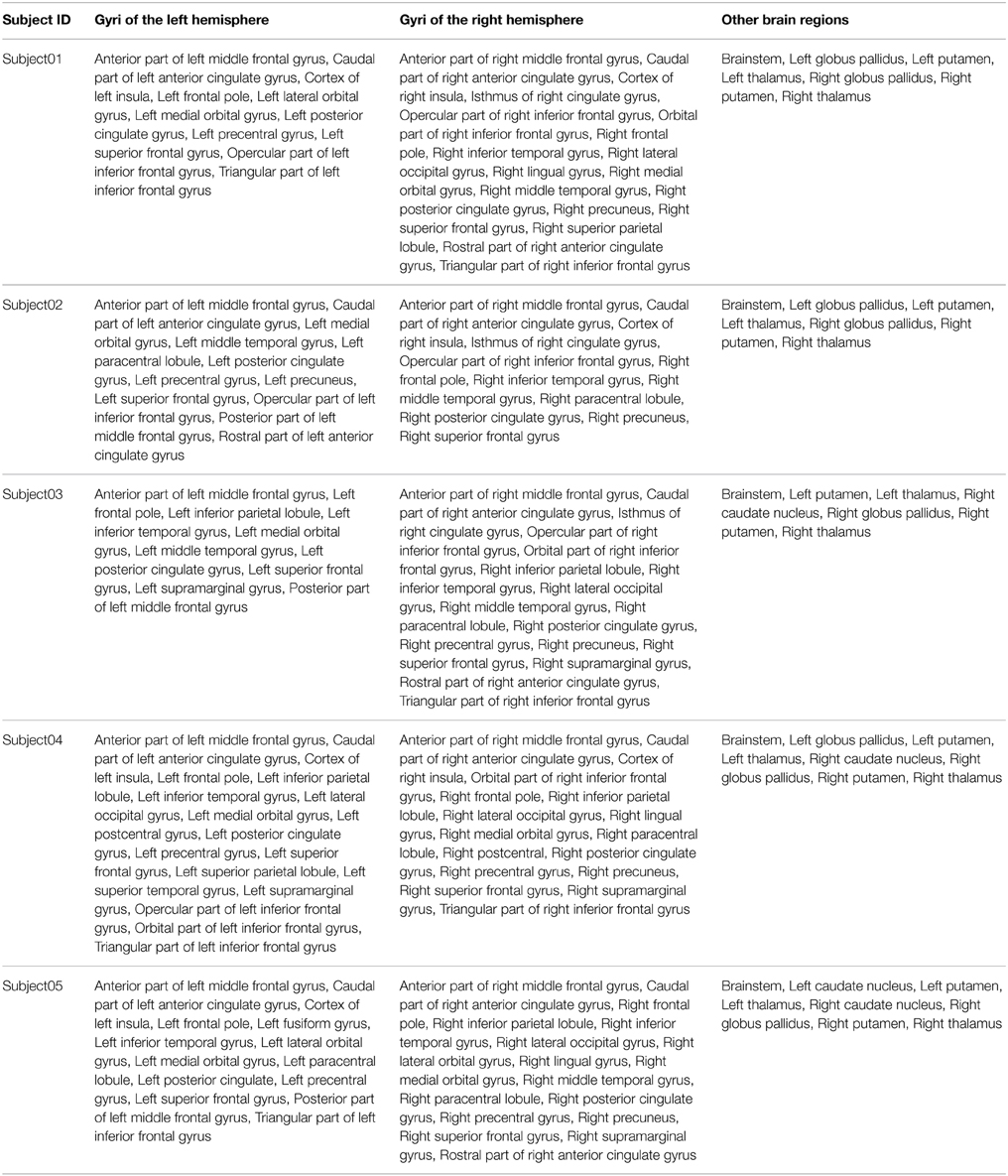
Table 4. Connectivity assessed by diffusion tractography between the right medial Brodmann area 6 (BA6) and other regions of the brain using the FMA terminology for the 5 subjects.
3.3. Semantic Annotation of MRI Connectomics Datasets
The HCO and its NEURO-DL-FMA module contained 811 classes, 11 object properties, no data property and a mean of 2321 instances per subject. The HCO was classified in less than 6 s per subject using the FaCT++ reasoning engine on a dual core processor, 3.06 GHz, 3.9 Go RAM workstation. The reasoning engine was used both to keep ontologies in a logically consistent state, and to infer new axioms between brain regions.
An example of the use of some part-whole, spatial and connectivity relationships using the HCO terms is provided on Figure 6. The part-whole relationship was expressed using the fma:regional_part_of object property denoting the fact that the high resolution cortical parcel 10 (i.e., hco:s1_gray_matter_of_left_superior_frontal_gyrus_10) was a regional part of the gray matter of the left superior frontal gyrus of the subject 01. The spatial relationship was expressed thanks to the hco:continuous_with object property denoting the fact that the cortical parcel 10 had several nearest neighbors in the gray matter of the left frontal gyrus, namely parcels number 16, 24, 32, 35, 39, and 8. Finally, the cortical parcel 10 was linked to the instance number 117 of the MR_Route class with the hco:is_tracto_connected object property denoting the existence of a fiber bundle reconstructed by diffusion tractography.
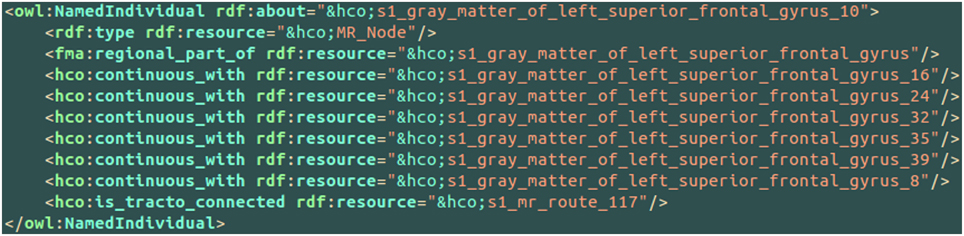
Figure 6. Example of use of some part-whole (i.e., fma:regional_part_of), spatial (i.e., hco:continuous_with) and connectivity (i.e., hco:is_tracto_connected) relationships between different cortical parcels (i.e., hco:MR_Node) and a fiber bundle reconstructed by diffusion tractography (i.e., s1_mr_route_117) with terms of the Human Connectomics Ontology (HCO). The “fma” and “hco” prefixes denote entities of the Foundational Model of Anatomy (FMA) and of the HCO, respectively.
Figure 7 aimed at illustrating the use of the hco:tracto_connects connectivity relationship using HCO terms. The s1_mr_route_117 instance of the MR_Route class denoted a fiber bundle assessed by diffusion tractography belonging to the subject 01. This route 117 was linked to two cortical parcels in the left (number 10) and right (number 25) superior frontal gyri using the hco:tracto_connects object property. The fma:regional_part_of object property expressed the fact that the route 117 was a part of the anterior forceps of the corpus callosum.
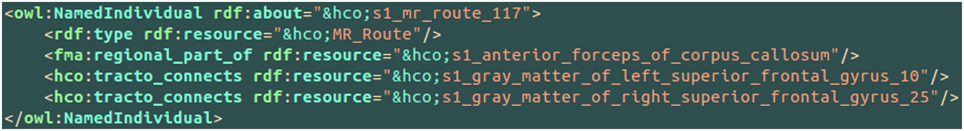
Figure 7. Illustration of the use of the hco:tracto_connects connectivity relationship between a fiber bundle assessed by diffusion tractography (i.e., hco:MR_Route) and different cortical parcels (i.e., hco:MR_Node) using terms of the Human Connectomics Ontology (HCO). The “fma” and “hco” prefixes denote entities of the Foundational Model of Anatomy (FMA) and of the HCO, respectively. The object property fma:regional_part_of denotes a part-whole relationship.
3.4. Ontology Application Testing: the Medial BA6 Case Study
Table 5 summarizes the results of the queries expressing our competency questions (cf. 2.1.1) and using terms of the HCO before submission to the FaCT++ reasoning engine. These results are expressed using the high resolution cortical parcel identifiers for columns 2, 3, and 4 (i.e., 17 was the identifier of the cortical parcel denoted by the following instance: hco:s1_gray_matter_of_right_superior_frontal_17). Column 5 of the table lists a set of instances denoting the white matter fiber bundles that were found to match the criteria of the last competency question.
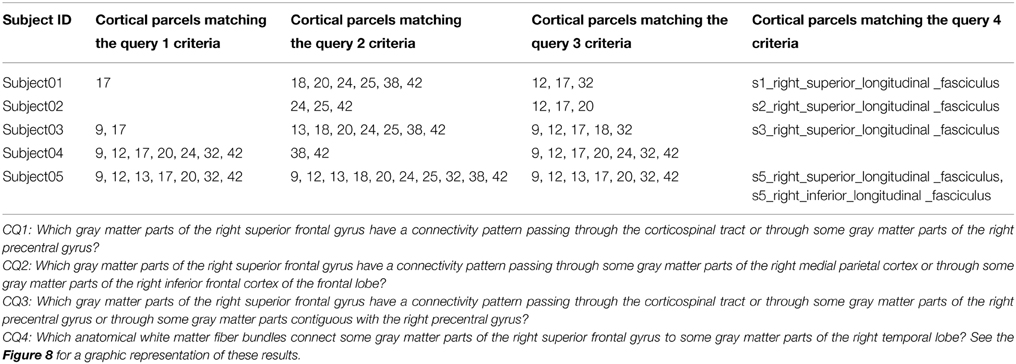
Table 5. Results of the different queries corresponding to the Competency Questions (CQ) that were translated into terms of the Human Connectomics Ontology (HCO) and submitted to the FaCT++ reasoning engine (cf. Table 3).
A map of the different high resolution cortical parcels presented in Table 5 in columns 2, 3, and 4 were plotted on the gray/white interface of the right hemisphere for each subject (cf. Figure 8). The first column of this figure represents (in red) the right medial BA6 region of interest for each subject. The second column of the figure depicts a map of the different cortical parcels that are parts of the region of interest and the corresponding identifiers. The third column represents (in blue and green) the results summarized in columns 2 (CQ1 query) and 3 (CQ2 query) of Table 5, respectively. The fourth column of the figure represents (in blue and green) the results summarized in columns 3 and 4 of Table 5, respectively. The cortical parcels that were found to meet the criteria of several competency questions were represented in orange color.
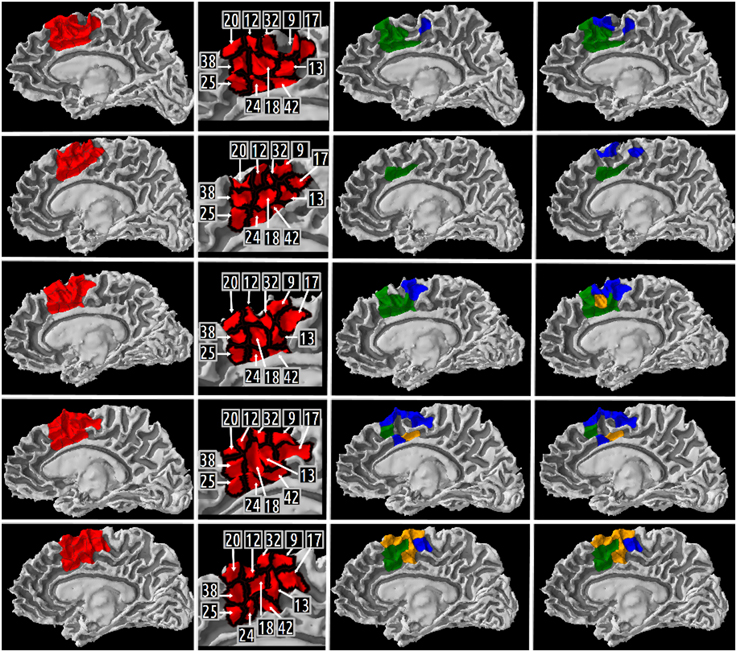
Figure 8. Medial view of the right gray/white interface of the five subjects. Column 1 represents in red the medial Brodmann area 6 region of interest for subject 01, 02, 03, 04, 05, respectively. Column 2 depicts a map of the different high resolution cortical parcels that were parts of the region of interest and the corresponding identifiers. The two last columns represent the results of three different queries corresponding to some of our Competency Questions (CQ) that were translated into terms of the Human Connectomics Ontology (HCO) and submitted to the FaCT++ reasoning engine (cf. Table 3). Column 3 represents in blue (resp. green) the cortical parcels matching the query 1 (resp. 2) criteria (cf. Table 5). When a parcel was the result of both queries, it was represented in orange. Column 4 represents in blue the cortical parcels matching the query 3 criteria (cf. Table 5). On column 3, the same color code was kept for the green and orange cortical parcels.
4. Discussion
The aim of this study was to design an ontology for in vivo human connectomics, i.e., suitable to describe connectomics data revealed by MRI, thus facilitating their retrieval, sharing and comparison with other neuroscience knowledge resources. A new ontology was created, called the “Human Connectomics Ontology” (HCO), that models brain regions and connectivity relationships assessed by diffusion tractography, using a three step methodology. First, the domain of discourse was specified using a set of competency questions grounded on the paradigmatic medial BA6 case study. Then, the HCO was based on a neuroanatomical ontology module called “NEURO-DL-FMA” in order to represent gross anatomy of the brain (i.e., gray matter regions, white matter fiber bundles). Finally, a set of entities was explicitly defined in the HCO to represent some aspects of the connectivity that could be observed through diffusion MRI. Moreover, an experimental work was achieved in order to show how the HCO could be effectively used to express complex queries and process them using a DL reasoning engine.
4.1. A Neuroanatomical Ontology for in vivo Human Connectomics
The medial BA6 case study provided an interesting use case for expressing competency questions for the specification of the HCO. In comparing macaque to human brain, Johansen-Berg et al. showed how the medial BA6 could be subdivided into two major anatomo-functional regions—Supplementary Motor Area (SMA) and pre-SMA—using distinct long-range connectivity patterns assessed by diffusion tractography (Johansen-Berg et al., 2004). As these connectivity patterns were concerned with rich neuroanatomical concepts denoting regions at different levels of resolution (e.g., “part of superior frontal gyrus,” “part of medial parietal cortex,” etc.), our set of competency questions (Neuhaus and Vizedom, 2013), inspired by this study, involved multi-level analysis and rich neuroanatomical expressivity.
NEURO-DL-FMA, defined as a neuroanatomical ontology of the gross-anatomy of the brain, was first extracted as a view from the FMA reference ontology in OWL Full and then translated into OWL DL. Different studies tried to convert the entire FMA (Protege frames version) into different OWL versions and to use reasoning engines (Golbreich et al., 2006; Golbreich et al., 2013). Our strategy was to extract from the OWL Full version of FMA a “view” of the brain constituted only of entities which were parts of the neuraxis, following (Turner et al., 2010; Shaw et al., 2011). Though the latter study achieved brain image analysis using the DXBrain software (Detwiler et al., 2009), no DL reasoning engine was used, however.
The HCO was designed in taking into account both the reference ontology in neuroanatomy FMA and the conceptual framework of structural connectivity FMC. If NEURO-DL-FMA was clearly grounded on a subset of FMA, however no concept of the BAMS18 ontology was used in the HCO. Indeed, structural connectivity addressed in BAMS primarily concerns pathway-tracing experiments in animals, whereas we were focusing on connectivity as observed in diffusion MRI. Nevertheless, the FMC was a useful source of inspiration. Some terms (i.e., gray-matter-region19, node20, route21, connection22) of the FMC thesaurus were instrumental in the definition of some new HCO entities (i.e., hco:Gray_matter_part, hco:MR_Node, hco:MR_Route, hco:mr_connection) dedicated to MRI connectomics.
4.2. Experimental Work
The experimental work was achieved in order to illustrate how the semantic annotation and the reasoning about MRI connectomics datasets could enhance the analysis of connectivity patterns present in this data. Connectivity was assessed using a probabilistic tractography method in the living human brain. It is worth saying that tractography results should be interpreted with care (Jones et al., 2013). Indeed, anatomical connectivity denotes the white matter fibers which physically connect brain regions, whereas connectivity assessed by diffusion tractography relies on water diffusion as an indirect probe of axon geometry. In fact, tractography infers fiber bundles pathways through the diffusion field in assuming that the direction of least hindered diffusion is aligned with axons (Jbabdi and Johansen-Berg, 2011). If this hypothesis seems reasonable at the axon level (microscopic scale), it has several practical consequences at the imaging level (macroscopic scale). For example, complex microscopic architectures of white matter fibers are often oversimplified by local models of axons-diffusion mapping. Moreover, tractography algorithms cannot determine with accuracy the origin and the termination of connections in the cortex (Jbabdi and Johansen-Berg, 2011). Thus, these different ambiguities combined with imaging noise generate spurious connections between brain regions. This is why it is so important to describe such connections using conceptual entities that allow distinguishing them from connections observed using tracer-based methods, e.g., collated in the BAMS database. Furthermore, although probabilistic tractography methods do not estimate the connection strength between two regions (as tracer-based methods actually do), they allow assessing the confidence in the pathway of least hindrance to diffusion. This is a major advantage of probabilistic tractography over deterministic tractography, since the latter cannot provide such confidence cues. So, although tractography is limited by several biases, it is currently the only available tool that gives us the opportunity to investigate anatomical connectivity non invasively and in the living human brain.
Our automatic annotation of brain images was based on brain segmentations achieved thanks to the use of atlases such as the Freesurfer (Fischl et al., 2004; Desikan et al., 2006) or JHU white matter tractography atlas (Wakana et al., 2007; Hua et al., 2008). Such atlases should be used with care in case of brain pathology, however. Another approach for automatic probabilistic reconstruction of in vivo white matter bundles based on global tractography called “Tracula” seems more robust in presence of pathology (Yendiki et al., 2011). However, the JHU white matter tractography atlas was preferred in our experimental work, because FMA provided a one-to-one mapping with the terminology of this atlas. As this atlas provided a probability that a particular voxel belonged to a white matter bundle, some white matter voxels could be mislabelled particularly in case of two close white matter bundles. As an illustration, Table 5 column 4 gives the names of the anatomical white matter fiber bundles reconstructed by diffusion tractography which connected some gray matter parts of the right superior frontal gyrus to some gray matter parts of the right temporal lobe. Two different anatomical bundles were found in our data: the right superior longitudinal fasciculus (found in subjects 01, 02, 03, and 05) and the right inferior longitudinal fasciculus (found in subject 05). If the right superior longitudinal fasciculus was found anatomically relevant in the MRI atlas of human white matter (Mori et al., 2005), the right inferior longitudinal fasciculus was not, however. This spurious annotation may have resulted from some mislabelled white matter voxels, since superior and inferior longitudinal fasciculi appeared close to one another particularly in the occipital lobe of the brain.
The HCO ontology aimed at representing brain regions and connectivity relationships assessed by diffusion tractography in the living brain. This was achieved by creating the corresponding instances of the ontology classes in the annotation file. If the seed of the tractography was located in a segmented white matter bundle, then the instance representing the pathway generated by the tractography algorithm was related to the instance representing this anatomical white matter bundle using the fma:regional_part_of object property (cf. Figure 4). This is based on the assumption that the whole pathway of the tractography was located within the segmented white matter bundle, which would need to be verified.
4.3. Automatic Inferences on Brain Connectivity
Automatic annotation of brain images with terms of an ontology and subsequent analysis using reasoning engines enable powerful information retrieval thanks to the high level representation of the image content embedded in the ontology. As an illustration, when an investigator queries through the HCO all cortical parts of the orbitobasal segment of the right frontal lobe which are connected to some medial BA6 parts via fiber bundles assessed by diffusion tractography, the reasoning engine takes advantage of both class-level knowledge (what are the gyri included in the orbitobasal segment of the right frontal lobe cortex?) and instance-level facts derived from image evidence (which data instantiate some regional parts of these gyri classes?).
Different initiatives used automatic inferences based on structured knowledge in order to represent cerebral connectivity: (1) Neurolex and (2) KEfED (Knowledge Engineering from Experimental Design) approach. (1) Neurolex is a semantic wiki-based website and knowledge management system dedicated to neurobiology whose primary goals are to assist neuroscientists in reviewing anatomical features, linking them to other neuroscience resources, and stimulating discussion with other scientists especially about controversial or missing features (Larson and Martone, 2013). Due to the fact that the semantic MediaWiki platform (on which Neurolex was built) did not support many of the first-order logic features that are needed to achieve OWL DL reasoning, the RDF version of Neurolex was deployed into an instance of the OWL-IM semantic repository (http://www.ontotext.com/owlim) providing SPARQL 1.1 querying capabilities. In Larson and Martone (2013), the authors demonstrated how a SPARQL query could retrieve from Neurolex “all brain regions that send projections into the cerebellum or any of its parts via mossy fibers.” In order to search recursively all subclasses which were regional parts of the cerebellum, the authors used the “property paths”23 feature of SPARQL 1.1. (2) Another initiative was based on a KEfED approach. First an experimental design was modeled as a workflow using a set of KEfED models which aimed at representing the experiment using structured information. Secondly, interpretations of the experimental observations were achieved using a domain-specific reasoning. In Russ et al. (2011), the authors illustrated the relevance of a KEfED approach through a neural connectivity use case based on tract-tracing experiments in animal subjects. Tract-tracing experiments consist of injecting in a site a chemical tracer which is then transported along neurons' axonal fibers. Interpretation of such tract-tracing experiments aims at describing connections between different brain regions. Spatial reasoning was used especially to process the part-whole and the overlaps relationships between regions. Basic geometric features were imported from the BAMS neuroanatomical ontology of the rat into PowerLoom, a first-order logic knowledge representation and closed-world reasoning system. Thus, the authors demonstrated how connectivity matrices could be inferred through the use of spatial reasoning and the modeling of the tract-tracing experiments using a KEfED approach. HCO and its NEURO-DL-FMA module differ from these approaches because they were expressed in the W3C standard OWL language and used the OWL DL description logics sublanguage. Moreover, the FaCT++ reasoning engine was used both to ensure the satisfiability of the ontologies, and to infer new axioms using transitive part-whole, spatial relationships, and connectivity relationships assessed by diffusion tractography. As a result, complex queries (cf. Table 3) could be formulated directly via the “DL Query” tab of the Protégé ontology editor, in a more expressive way than using the SPARQL language.
An interesting initiative dedicated to diffusion tractography called the “White Matter Query Language” (Wassermann et al., 2013) used a textual language in order to express anatomical descriptions of white matter tracts. In selecting streamlines from a whole brain deterministic tractography using both anatomical structure terms describing where streamlines end or pass through, relative position terms of streamlines from other anatomical structures and finally logical operations terms, Wassermann et al. used different expressions to define some association, projection and commissural tracts (Wassermann et al., 2013). Although the latter approach explicitly defined some white matter tracts using a near-to-English syntax, it did not provide an ontology in order to annotate results of in vivo human connectomics based on diffusion tractography nor reasoning capabilities in order to infer part-whole, spatial or connectivity relationships at different level of granularity.
5. Conclusion and Perspectives
In this article we have described a neuroanatomical ontology dedicated to human connectomics called the Human Connectomics Ontology (HCO) that could represent brain regions and connectivity assessed by diffusion tractography in the living human brain. Moreover, an experimental work was achieved in order to show how the HCO could be effectively used within an information system to express complex queries concerning MRI connectomics datasets and process them using a DL reasoning engine. This approach can facilitate comparison of data across scales, modalities and species.
Future work will consist in the development of a visualization module in order to display macro-connectome at different levels of granularity in a matrix or a network form. This module could leverage the reasoning engine to retrieve connections assessed by diffusion tractography between different gray matter regions. To finish, a long-term goal will consist in facilitating the consistent querying of in vivo MRI connectomics data and tracer-based observations made in multiple species. This would be of major interest for assessing the validity of putative connections highlighted in human MR connectomics.
Author Contributions
TM and BG designed the work, analyzed data, drafted the work, approved the final version to be published and agreed to be accountable for all aspects of the work ensuring questions related to the accuracy or integrity of any part of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank C. Poupon for the brain datasets used in this paper which were part of the NMR public database (Poupon et al., 2006). We would like to thank Pr. X. Morandi and R. Seizeur, professors in anatomy and neurosurgery, for their assistance in interpreting anatomy.
Footnotes
1. ^OWL, http://www.w3.org/TR/owl-ref/
2. ^OWL DL, http://www.w3.org/TR/owl-ref/#OWLDL
3. ^FaCT++, http://owl.man.ac.uk/factplusplus/
4. ^Freesurfer, http://surfer.nmr.mgh.harvard.edu/fswiki
5. ^FSL, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL
6. ^HCO, https://medicis.univ-rennes1.fr/activities/theme3/projects/semantic-annotation/index
7. ^University of Washington's Structural Informatics Group, http://sig.biostr.washington.edu/
8. ^MySQL, https://www.mysql.com/
9. ^Jena, https://jena.apache.org/
10. ^RDF, http://www.w3.org/TR/2014/REC-rdf11-concepts-20140225/
11. ^OWL API, http://owlapi.sourceforge.net/
12. ^Gray-matter-region: http://brancusi1.usc.edu/thesaurus/definition/gray-matter-region/
13. ^Node: http://brancusi1.usc.edu/thesaurus/definition/node/
14. ^Route: http://brancusi1.usc.edu/thesaurus/definition/route/
15. ^Connection: http://brancusi1.usc.edu/thesaurus/definition/connection/
16. ^CMTK, http://www.cmtk.org/
17. ^FSL, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL
18. ^BAMS, http://brancusi1.usc.edu/ontology/
19. ^Gray-matter-region, http://brancusi1.usc.edu/thesaurus/definition/gray-matter-region/
20. ^Node, http://brancusi1.usc.edu/thesaurus/definition/node/
21. ^Route, http://brancusi1.usc.edu/thesaurus/definition/route/
22. ^Connection, http://brancusi1.usc.edu/thesaurus/definition/connection/
23. ^Property paths, http://www.w3.org/TR/sparql11-query/#propertypaths
References
Anwander, A., Tittgemeyer, M., von Cramon, D. Y., Friederici, A. D., and Knösche, T. R. (2007). Connectivity-based parcellation of broca's area. Cereb. Cortex 17, 816–825. doi: 10.1093/cercor/bhk034
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Baader, F., Calvanese, D., McGuinness, D., Nardi, D., and Patel-Schneider, P. (2003). The Description Logic Handbook: Theory, Implementation, and Applications. New York, NY: Cambridge University.
Basser, P. J., Pajevic, S., Pierpaoli, C., Duda, J., and Aldroubi, A. (2000). In vivo fiber tractography using dt-mri data. Magn. Reson. Med. 44, 625–632. doi: 10.1002/1522-2594(200010)44:4<625::AID-MRM17>3.0.CO;2-O
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Behrens, T. E. J., Berg, H. J., Jbabdi, S., Rushworth, M. F. S., and Woolrich, M. W. (2007). Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 34, 144–155. doi: 10.1016/j.neuroimage.2006.09.018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Behrens, T. E. J., and Sporns, O. (2012). Human connectomics. Curr. Opin. Neurobiol. 22, 144–153. doi: 10.1016/j.conb.2011.08.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bota, M., Talpalaru, S., Hintiryan, H., Dong, H.-W., and Swanson, L. W. (2014). Bams2 workspace: a comprehensive and versatile neuroinformatic platform for collating and processing neuroanatomical connections. J. Comp. Neurol. 522, 3160–3176. doi: 10.1002/cne.23592
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Caspers, S., Eickhoff, S. B., Zilles, K., and Amunts, K. (2013). Microstructural grey matter parcellation and its relevance for connectome analyses. Neuroimage 80, 18–26. doi: 10.1016/j.neuroimage.2013.04.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Craddock, R. C., Jbabdi, S., Yan, C.-G., Vogelstein, J. T., Castellanos, F. X., Martino, A. D., et al. (2013). Imaging human connectomes at the macroscale. Nat. Methods 10, 524–539. doi: 10.1038/nmeth.2482
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Daducci, A., Gerhard, S., Griffa, A., Lemkaddem, A., Cammoun, L., Gigandet, X., et al. (2012). The connectome mapper: an open-source processing pipeline to map connectomes with mri. PLoS ONE 7:e48121. doi: 10.1371/journal.pone.0048121
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dale, A. M., Fischl, B., and Sereno, M. I. (1999). Cortical surface-based analysis. i. segmentation and surface reconstruction. Neuroimage 9, 179–194. doi: 10.1006/nimg.1998.0395
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dameron, O., Rubin, D. L., and Musen, M. A. (2005). Challenges in converting frame-based ontology into owl: the foundational model of anatomy case-study. AMIA Annu. Symp. Proc. 181–185.
Desikan, R. S., Segonne, F., Fischl, B., Quinn, B. T., Dickerson, B. C., Blacker, D., et al. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. doi: 10.1016/j.neuroimage.2006.01.021
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Detwiler, L. T., Suciu, D., Franklin, J. D., Moore, E. B., Poliakov, A. V., Lee, E. S., et al. (2009). Distributed xquery-based integration and visualization of multimodality brain mapping data. Front. Neuroinform. 3:2. doi: 10.3389/neuro.11.002.2009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Essen, D. C. V., and Ugurbil, K. (2012). The future of the human connectome. Neuroimage 62, 1299–1310. doi: 10.1016/j.neuroimage.2012.01.032
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fischl, B., van der Kouwe, A., Destrieux, C., Halgren, E., Sgonne, F., Salat, D. H., et al. (2004). Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22. doi: 10.1093/cercor/bhg087
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fortino, A., Zalesky, A., and Breakspear, M. (2013). Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage 80, 426–444. doi: 10.1016/j.neuroimage.2013.04.087
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Golbreich, C., Grosjean, J., and Darmoni, S. J. (2013). The foundational model of anatomy in owl 2 and its use. Artif. Intell. Med. 57, 119–132. doi: 10.1016/j.artmed.2012.11.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Golbreich, C., Zhang, S., and Bodenreider, O. (2006). The foundational model of anatomy in owl: experience and perspectives. Web Semant. 4, 181–195. doi: 10.1016/j.websem.2006.05.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gruber, T. R. (1995). Toward principles for the design of ontologies used for knowledge sharing? Int. J. Hum. Comput. Stud. 43, 907–928.
Hagmann, P., Cammoun, L., Gigandet, X., Meuli, R., Honey, C. J., Wedeen, V. J., et al. (2008). Mapping the structural core of human cerebral cortex. PLoS Biol. 6:e159. doi: 10.1371/journal.pbio.0060159
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hagmann, P., Kurant, M., Gigandet, X., Thiran, P., Wedeen, V. J., Meuli, R., et al. (2007). Mapping human whole-brain structural networks with diffusion mri. PLoS ONE 2:e597. doi: 10.1371/journal.pone.0000597
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Honey, C. J., Thivierge, J.-P., and Sporns, O. (2010). Can structure predict function in the human brain? Neuroimage 52, 766–776. doi: 10.1016/j.neuroimage.2010.01.071
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hua, K., Zhang, J., Wakana, S., Jiang, H., Li, X., Reich, D. S., et al. (2008). Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage 39, 336–347. doi: 10.1016/j.neuroimage.2007.07.053
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jbabdi, S., and Johansen-Berg, H. (2011). Tractography: where do we go from here? Brain Conn. 1, 169–183. doi: 10.1089/brain.2011.0033
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jbabdi, S., Woolrich, M. W., and Behrens, T. E. J. (2009). Multiple-subjects connectivity-based parcellation using hierarchical dirichlet process mixture models. Neuroimage 44, 373–384. doi: 10.1016/j.neuroimage.2008.08.044
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Johansen-Berg, H., Behrens, T. E. J., Robson, M. D., Drobnjak, I., Rushworth, M. F. S., Brady, J. M., et al. (2004). Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc. Natl. Acad. Sci. U.S.A. 101, 13335–13340. doi: 10.1073/pnas.0403743101
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones, D. K., Knösche, T. R., and Turner, R. (2013). White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion mri. Neuroimage 73, 239–254. doi: 10.1016/j.neuroimage.2012.06.081
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larson, S. D., and Martone, M. E. (2013). Neurolex.org: an online framework for neuroscience knowledge. Front. Neuroinform. 7:18. doi: 10.3389/fninf.2013.00018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leergaard, T. B., Hilgetag, C. C., and Sporns, O. (2012). Mapping the connectome: multi-level analysis of brain connectivity. Front. Neuroinform. 6:14. doi: 10.3389/fninf.2012.00014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mechouche, A., Morandi, X., Golbreich, C., and Gibaud, B. (2009). A hybrid system using symbolic and numeric knowledge for the semantic annotation of sulco-gyral anatomy in brain mri images. IEEE Trans. Med. Imaging 28, 1165–1178. doi: 10.1109/TMI.2009.2026746
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mori, S., Wakana, S., van Zijl, P. C. M., and Nagae-Poetscher, L. M. (2005). MRI Atlas of Human White Matter (Amsterdam: Elsevier).
Neuhaus, F., and Vizedom, A. (2013). Towards ontology evaluation across the life cycle: The Ontology Summit 2013. Appl. Ontol. 8, 179–194. doi: 10.3233/AO-130125
Nichols, B. N., Mejino, J. L., Detwiler, L. T., Nilsen, T. T., Martone, M. E., Turner, J. A., et al. (2014). Neuroanatomical domain of the foundational model of anatomy ontology. J. Biomed. Semant. 5:1. doi: 10.1186/2041-1480-5-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Noy, N. F., and Rubin, D. L. (2008). Translating the foundational model of anatomy into owl. Web Semant. 6, 133–136. doi: 10.1016/j.websem.2007.12.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poupon, C., Poupon, F., Allirol, L., and Mangin, J.-F. (2006). “A database dedicated to anatomo-functional study of human brain connectivity,” in Twelfth Annual Meeting of the Organization for Human Brain Mapping (HBM) (Florence), 1–8.
Reese, T. G., Heid, O., Weisskoff, R. M., and Wedeen, V. J. (2003). Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn. Reson. Med. 49, 177–182. doi: 10.1002/mrm.10308
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rosse, C., and Mejino, J. L. V. (2003). A reference ontology for biomedical informatics: the foundational model of anatomy. J. Biomed. Inform. 36, 478–500. doi: 10.1016/j.jbi.2003.11.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rubin, D. L., Noy, N. F., and Musen, M. A. (2007). Protg: a tool for managing and using terminology in radiology applications. J. Digit. Imaging 20(Suppl. 1), 34–46. doi: 10.1007/s10278-007-9065-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Russ, T. A., Ramakrishnan, C., Hovy, E. H., Bota, M., and Burns, G. A. P. C. (2011). Knowledge engineering tools for reasoning with scientific observations and interpretations: a neural connectivity use case. BMC Bioinform. 12:351. doi: 10.1186/1471-2105-12-351
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shaw, M., Detwiler, L. T., Noy, N., Brinkley, J., and Suciu, D. (2011). vsparql: a view definition language for the semantic web. J. Biomed. Inform. 44, 102–117. doi: 10.1016/j.jbi.2010.08.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smith, B., Ceusters, W., Klagges, B., Khler, J., Kumar, A., Lomax, J., et al. (2005). Relations in biomedical ontologies. Genome Biol. 6:R46. doi: 10.1186/gb-2005-6-5-r46
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smith, S. M., Miller, K. L., Salimi-Khorshidi, G., Webster, M., Beckmann, C. F., Nichols, T. E., et al. (2011). Network modelling methods for fmri. Neuroimage 54, 875–891. doi: 10.1016/j.neuroimage.2010.08.063
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sporns, O. (2013). The human connectome: origins and challenges. Neuroimage 80, 53–61. doi: 10.1016/j.neuroimage.2013.03.023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sporns, O., Tononi, G., and Ktter, R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1:e42. doi: 10.1371/journal.pcbi.0010042
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Swanson, L. W., and Bota, M. (2010). Foundational model of structural connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc. Natl. Acad. Sci. U.S.A. 107, 20610–20617. doi: 10.1073/pnas.1015128107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Talairach, J., and Tournoux, P. (1988). Co-Planar Stereotaxic Atlas of the Human Brain. New York, NY: Thieme medical publishers.
Turner, J. A., Mejino, J. L. V., Brinkley, J. F., Detwiler, L. T., Lee, H. J., Martone, M. E., et al. (2010). Application of neuroanatomical ontologies for neuroimaging data annotation. Front. Neuroinform. 4:10. doi: 10.3389/fninf.2010.00010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., et al. (2002). Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. Neuroimage 15, 273–289. doi: 10.1006/nimg.2001.0978
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wakana, S., Caprihan, A., Panzenboeck, M. M., Fallon, J. H., Perry, M., Gollub, R. L., et al. (2007). Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 36, 630–644. doi: 10.1016/j.neuroimage.2007.02.049
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wassermann, D., Makris, N., Rathi, Y., Shenton, M., Kikinis, R., Kubicki, M., et al. (2013). On describing human white matter anatomy: the white matter query language. Med. Image Comput. Comput. Assist. Interv. 16(Pt 1), 647–654. doi: 10.1007/978-3-642-40811-3/81
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yendiki, A., Panneck, P., Srinivasan, P., Stevens, A., Zllei, L., Augustinack, J., et al. (2011). Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front. Neuroinform. 5:23. doi: 10.3389/fninf.2011.00023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zalesky, A., Fornito, A., Seal, M. L., Cocchi, L., Westin, C.-F., Bullmore, E. T., et al. (2011). Disrupted axonal fiber connectivity in schizophrenia. Biol. Psychiatry 69, 80–89. doi: 10.1016/j.biopsych.2010.08.022
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zilles, K., and Amunts, K. (2010). Centenary of brodmann's map–conception and fate. Nat. Rev. Neurosci. 11, 139–145. doi: 10.1038/nrn2776
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: ontology, connectome, data sharing, neuroanatomy, neuroimaging, tractography, semantic web, MRI
Citation: Moreau T and Gibaud B (2015) Ontology-based approach for in vivo human connectomics: the medial Brodmann area 6 case study. Front. Neuroinform. 9:9. doi: 10.3389/fninf.2015.00009
Received: 30 September 2014; Accepted: 24 March 2015;
Published: 10 April 2015.
Edited by:
Maryann E. Martone, University of California San Diego, USAReviewed by:
Mihail Bota, University of Southern California, USAJose L. V. Mejino, University of Washington, USA
Copyright © 2015 Moreau and Gibaud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tristan Moreau, UMR 1099 LTSI, INSERM, Universite de Rennes 1, 2 avenue du Pr. Leon Bernard, Rennes F-35000, France tristan.soyyo@gmail.com
 Tristan Moreau
Tristan Moreau Bernard Gibaud
Bernard Gibaud