Pluripotent stem cells for the study of CNS development
- 1 Department of Psychiatry, Weill Cornell Medical College, New York, NY, USA
- 2 Program in Neuroscience, Weill Cornell Medical College, New York, NY, USA
The mammalian central nervous system is a complex neuronal network consisting of a diverse array of cellular subtypes generated in a precise spatial and temporal pattern throughout development. Achieving a greater understanding of the molecular and genetic mechanisms that direct a relatively uniform population of neuroepithelial progenitors into diverse neuronal subtypes remains a significant challenge. The advent of pluripotent stem cell (PSC) technology allows researchers to generate diverse neural populations in vitro. Although the primary focus of PSC-derived neural cells has been their therapeutic potential, utilizing PSCs to study neurodevelopment is another frequently overlooked and equally important application. In this review, we explore the potential for utilizing PSCs to study neural development. We introduce the types of neurodevelopmental questions that PSCs can help to address, and we discuss the different strategies and technologies that researchers use to generate diverse subtypes of PSC-derived neurons. Additionally, we highlight the derivation of several thoroughly characterized neural subtypes; spinal motoneurons, midbrain dopaminergic neurons and cortical neurons. We hope that this review encourages researchers to develop innovative strategies for using PSCs for the study of mammalian, and specifically human, neurodevelopment.
Introduction
How the highly organized human nervous system arises from a relatively uniform population of neural progenitors remains one of the most fascinating and challenging questions in neurobiology. Progress has been made toward understanding many of the key signaling pathways responsible for patterning the developing mammalian nervous system. However, the molecular and genetic fate-determining mechanisms that give rise to the diversity of different neural cell types are poorly understood. The ability to manipulate gene expression in lower vertebrates is an invaluable tool for examining mammalian neurodevelopment, but studying the inimitable process of human neurodevelopment is significantly more complicated. This problem persists because obtaining human embryonic tissue is difficult and animal models seldom completely recapitulate human developmental programs. A more comprehensive technique for studying development of the mammalian (and in particular human) nervous system would lead to a greater understanding of both normal neurodevelopment and provide insight into the pathophysiologies of numerous neurodevelopmental diseases.
Over the last decade, pluripotent stem cells (PSCs) have emerged as powerful tools with the potential to further illuminate key mechanisms underlying neuronal development. The advent of embryonic stem cell (ESC) and induced pluripotent stem cell (iPSC) technologies has facilitated the analysis of the developing nervous system on a cellular and molecular level that was previously unattainable. Several recent reviews have focused on stem cell pluripotency and cellular reprogramming (Hochedlinger and Plath, 2009; Hanna et al., 2010; Stadtfeld and Hochedlinger, 2010) and the potential for PSC- and iPSC-derived neural cells to treat neurological disease (Kim and de Vellis, 2009; Marchetto et al., 2010; Aboody et al., 2011; Cundiff and Anderson, 2011; Gaspard and Vanderhaeghen, 2011). In this review, we focus the recent advances in the application of PSCs as model systems for studying normal brain development. We will examine the types of questions that PSCs can address, and the advantages and disadvantages of using PSCs for studying neurodevelopment. Guided by the developing embryo, we will summarize the different PSC differentiation techniques and genetic manipulations that are used to generate distinct neural subtypes, and highlight several in more detail.
Types of Pluripotent Stem Cells
The fusion of sperm and egg produces totipotent stem cells that can differentiate into all known cell types. As development proceeds, cells progress from a totipotent state to more restricted lineages such as PSCs (the ability to differentiate into any of the three germ layers), multipotent stem cells (more restricted cell fates), and unipotent cells (can only produce one cell type). PSCs are capable of self-renewal, meaning that given the appropriate conditions, they can proliferate while maintaining identical fate potential to the parent cell. The first PSCs to be isolated were ESCs from the inner cell mass of the mouse blastula (mESCs; Figure 1A; Evans and Kaufman, 1981; Martin, 1981), which differentiated into all three germ layers upon transplantation. In addition to mESCs, stem cells have also been generated from the mouse epiblast after implantation, referred to as mEpiSCs (Brons et al., 2007; Tesar et al., 2007). Although mEpiSCs display many characteristics similar to mESCs, their molecular profile represents that of a more specified cell type, and their potency is more restricted. In the late 1990s the first human ESCs (hESCs) were generated (Thomson et al., 1998). Interestingly hESCs have different gene expression profiles compared to mESCs (Ginis et al., 2004), and appear to be functionally more similar to mEpiSCs than mESCs (Tesar et al., 2007). Since studying human neurodevelopment at the cellular and molecular level presents significant technical challenges, hESCs open the door to many experimental possibilities that were previously impossible.
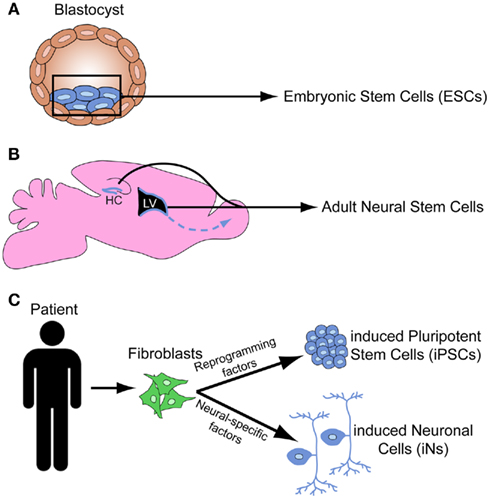
Figure 1. Schematic depicts the origin of embryonic stem cells from the inner cell mass of the blastocyst (A), the stem cell niches in the subventricular zone and the hippocampus in the adult mouse brain (B), and the induced pluripotent stem cells and induced neural cells derived from human fibroblasts (C).
In addition to ESCs, the maintenance and repair of adult tissue is achieved by niches of adult stem cells. In the brain, adult neural stem cells (NSCs) can give rise to neurons, astrocytes, and oligodendrocytes. There are two active zones of neurogenesis in the adult brain: the subventricular zone (SVZ) of the lateral ventricle gives rise to neurons that populate the olfactory bulb, and the subgranular zone (SGZ) of the dentate gyrus gives rise granule cells that integrate into the hippocampal circuitry (Figure 1B; Zhao et al., 2008). While adult stem cells may provide important insights into aspects of neurodevelopment, ESCs are the primary stem cell tool for studying neurodevelopment.
More recently, researchers have discovered how to reprogram both mouse (Takahashi and Yamanaka, 2006) and human (Takahashi et al., 2007; Yu et al., 2007) somatic cells into iPSCs (Figure 1C). This was initially achieved by introducing retroviruses engineered to express four genes that mediate the reprogramming to pluripotency. Improved techniques for somatic cell reprogramming to PSCs include the genetic excision of virally introduced programming factors, small molecules that reactivate quiescent transcription factors to induce pluripotency, utilization of transposons to reprogram cells, synthetic modified mRNAs that do not alter the cell’s genome, and microRNAs (miRNAs; Miyoshi et al., 2011; Sidhu, 2011). iPSCs have several advantages over ESCs, such as the ability to create patient-specific human iPSC lines and reduced ethical concerns since their generation does not involve the destruction of human blastocyst stage embryos. iPSCs and ESCs have a well-defined core pluripotency genetic network in common. However, several recent studies found that iPSCs contain increased somatic mutations (Gore et al., 2011), increased copy number variations (Hussein et al., 2011), and significant variability in DNA methylation and epigenomic events (Lister et al., 2011) as compared to both their fibroblast origin cells and hESCs. These issues have raised concerns regarding the potential limitations of iPSCs to study disease. However, many different iPSC lines can be differentiated into motor neurons (MNs) using a standard differentiation protocol (Boulting et al., 2011), providing hope that iPSC lines with significant genetic and epigenetic variability may display similar differentiation potential. Thus, iPSCs have tremendous potential for both therapeutics and for the elucidation of developmental processes, but the safety of iPSCs and the mechanisms of reprogramming require further investigation.
To circumvent this reprogramming step, numerous groups have bypassed the PSC stage and successfully converted mouse and human fibroblasts directly into functional neurons, called induced neural cells (iNs; Figure 1C). Initial studies identified ∼3–5 critical transcription factors that could be overexpressed to directly convert fibroblasts to neurons (Vierbuchen et al., 2010; Pang et al., 2011), whereas others used combinations of miRNAs and transcription factors (Ambasudhan et al., 2011; Yoo et al., 2011). The ability to derive specific neural subtypes from iNs remains unclear, although several groups have generated dopaminergic iNs (Caiazzo et al., 2011; Pfisterer et al., 2011) and motor neuron iNs (Son et al., 2011). An alternate strategy is to convert fibroblasts directly to neuronal progenitors (rather than PSCs or mature neurons), which could retain the ability to differentiate into multiple subtypes (Kim et al., 2011a). Additionally, patient-specific iNs could be generated to enhance the study of developmental disorders and other neurological diseases (Qiang et al., 2011). Thus, the significant decrease in time and resources to derive neurons directly from somatic cells justifies further investigation into this strategy.
Structure and Derivation of the Developing Nervous System
Organization of the Developing Nervous System
In order to direct PSCs toward a neuronal fate in vitro, one must understand the basic cellular organization and patterning factors within the developing embryonic nervous system (for review see Martin, 2003). The gastrula is a tri-layered structure comprised of three germinal layers: endoderm (primordial viscera), mesoderm (primordial bones/muscles), and ectoderm (primordial nervous system and skin). The neural plate is a flat sheet of ectodermal cells (the neuroepithelium) that evaginates over time and fuses dorsally to form a cylinder of cells known as the neural tube. Throughout its rostro-caudal axis, the neural tube contains multipotent NSCs that generate both neurons and glia of the central nervous system (CNS). The peripheral nervous system (PNS) is derived from the neural crest, a population of cells that emerges from the dorsal neural tube and migrates peripherally. Subsequently, three primary vesicles develop in the rostral portion of the neural tube: the prosencephalon (forebrain), mesencephalon (midbrain), and rhombencephalon (hindbrain). The caudal neural tube remains undifferentiated and will form the mature spinal cord. Secondary vesicles emerging from the prosencephalon give rise to the telencephalon (cerebral hemispheres) and diencephalon (thalamus and hypothalamus).
Neural Induction and Patterning the Neuraxis
How does the relatively simple gastrula initiate neural induction and give rise to many distinct regions of the CNS, which then produce a vast array of neural subtypes? The first molecules that play a role in specifying neural fates during gastrulation were discovered in a series of studies in Xenopus in the 1990s (reviewed in Munoz-Sanjuan and Brivanlou, 2002; Stern, 2005; Levine and Brivanlou, 2007). Bone morphogenetic proteins (BMPs) and Wnts are secreted factors that suppress the neural induction of ectoderm and promote an epidermal lineage. The gastrula organizer is a group of cells that express BMP inhibitors to suppress epidermal differentiation and initiate neural induction. Thus, neural induction primarily requires inhibition of non-neural fates by blocking neural inhibitors (a double negative), raising the hypothesis that neural fates are a default mechanism that must be overcome by progenitor cells to differentiate into other tissue types (Munoz-Sanjuan and Brivanlou, 2002).
The initial patterning of the CNS is achieved using a relatively small number of evolutionarily conserved signaling factor families that are expressed as gradients along the rostro-caudal (R-C, also called the anterior–posterior or A–P axis) and dorsal–ventral (D–V) axis (Figure 2). The same signaling molecule can induce different effects depending upon its concentration, temporal and spatial expression pattern, and the presence of various receptor subfamilies and modulating factors. For example, although BMPs and Wnts initially inhibit neural induction, BMPs, and Wnts are expressed throughout the dorsal portion of the neuraxis and are required for the formation of dorsal neural tissue (Liu and Niswander, 2005; Ulloa and Marti, 2010). Ventralization of the neuraxis is primarily directed by the morphogen Sonic hedgehog (Shh), which is secreted at the ventral midline of the neural tube by a specialized structure known as the floor plate (and the underlying non-neuronal notochord) that spans the entire R-C neuraxis. In addition to Shh, much of the ventral neuraxis also expresses BMP inhibitors that suppress BMP action.
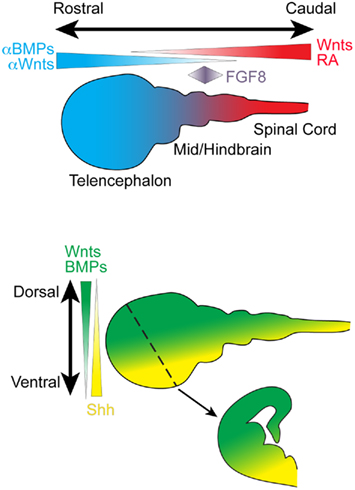
Figure 2. Schematic depicts important secreted signaling factors that pattern the rostro-caudal and dorso-ventral neuraxis during embryonic development. A coronal section through the developing telencephalon is depicted (dotted line, arrow). BMP, bone morphogenetic protein; FGF, fibroblast growth factor; RA, retinoic acid; Shh, Sonic hedgehog
One of the most important caudalizing molecules is retinoic acid (RA). RA is present at high levels in the primitive spinal cord and, in combination with fibroblast growth factors (FGFs), is required for patterning the different spinal cord segments (Maden, 2006). Wnts are also expressed in the caudal portion of the neuraxis, and Wnt antagonists are expressed by cells along the rostral axis. Additionally, a patch of cells located at the midbrain–hindbrain boundary (the isthmic organizer) secretes FGF8, an important signal for regulating development of midbrain and hindbrain structure (Partanen, 2007).
Thus, the fate of an individual neuron depends upon its spatial position along the neuraxis, temporal birthdate, genetic profile, and the patterning factors expressed by neighboring cells. It is therefore logical that PSCs can be differentiated toward many different neural subtypes by modulating exogenous levels of (or intrinsic responses to) BMP, Wnt, Shh, FGF, and other relevant signaling pathways (Figure 3).
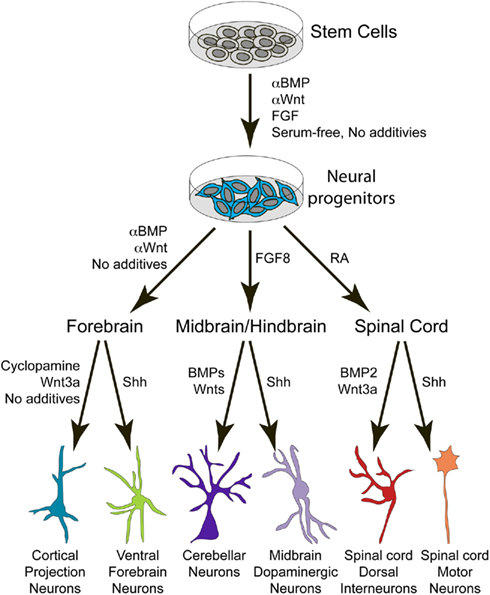
Figure 3. Schematic depicts the general procedure for deriving different neuronal subtypes from PSCs by utilizing secreted patterning factors identified in the developing embryo. BMP, bone morphogenetic protein; FGF, fibroblast growth factor; RA, retinoic acid; Shh, Sonic hedgehog.
Neurodevelopmental Questions that may be Addressed Using PSCs
As the PSC field is still in its infancy, many studies focus on developing efficient protocols for optimizing the derivation of distinct neuronal cell types (Table 1). The challenge is to harness this potential to address specific questions of neurodevelopment.
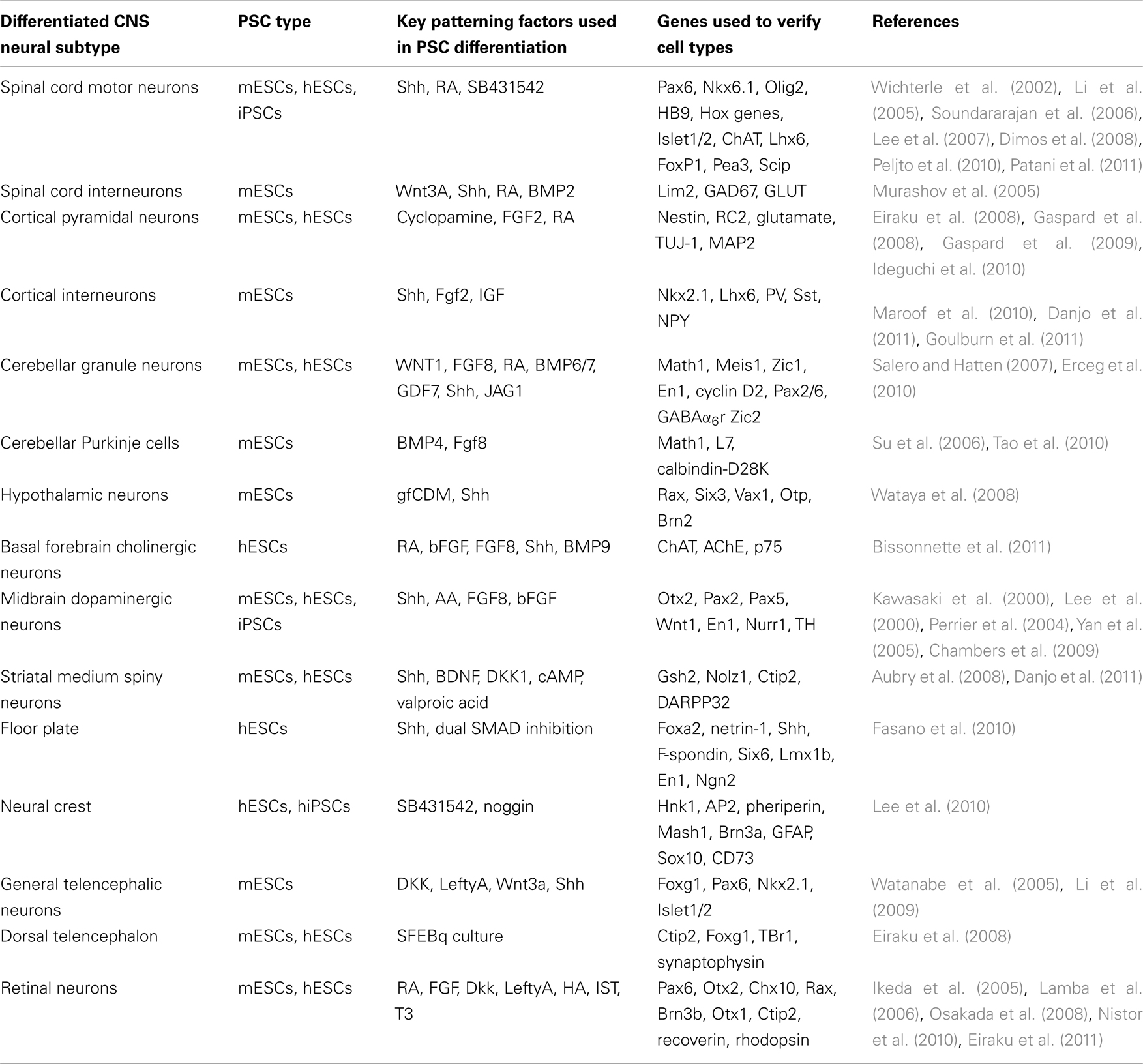
Table 1. Table details the many different neural cells that have been derived from PSCs to date, specifying the type of PSC, the key exogenous signaling factors, the markers used to verify cell type specificity, and the corresponding references.
Neurogenesis as a Default Mechanism
As mentioned above, evidence from the developing embryo supports a model whereby ectodermal tissue becomes specified to a neural identity by default (Munoz-Sanjuan and Brivanlou, 2002). ESC differentiations in vitro are an ideal tool to more closely examine this theory, but the variety of distinct culture techniques can produce different results. Culturing mESCs in a chemically defined serum-free medium as low-density aggregates is sufficient to induce differentiation into neural cells without exogenous factors (Tropepe et al., 2001; Smukler et al., 2006; Gaspard et al., 2008). When grown as adherent monolayers, mESCs required the addition of FGF for neural induction (Ying et al., 2003). Another study developed a protocol in which mESCs cultured in the absence of any extrinsic factors produced ∼80% Sox1+ cells, a marker of neural progenitors, but inhibiting Wnt and Nodal signaling increased it to ∼90% (Watanabe et al., 2005). More detailed analysis revealed that the lack of extrinsic cues appears to drive mESCs to an anterior/forebrain fate (Watanabe et al., 2005; Gaspard et al., 2008), in agreement with the lack of instructive cues to rostralize the neural tube during embryonic development (Figure 2). However, removal of KSR and insulin from the culture medium may promote a hypothalamic cell fate (Wataya et al., 2008). Additionally, in one instance, MNs were derived from mESCs grown at a low-density in the absence of added factors (Peljto et al., 2010), highlighting that subtle changes in medium composition and culture techniques can have significant effects on fate and differentiation efficiency.
Similar findings have been demonstrated for hESCs. hESCs grown as embryoid body-like aggregates in the absence of feeder cells and exogenous cues produced ∼35% Pax6+ cells (a dorsal, “pallial” telencephalic marker when co-expressed with FoxG1), but addition of Wnt, Nodal, and BMP inhibitors boosted this percentage to ∼75% Pax6+ cells (Watanabe et al., 2007). Multiple studies found that hESCs and hiPSCs are driven toward a neural fate when cultured in the presence of inhibitors of SMAD signaling (noggin and SB431542), a downstream transducer of TGFβ activity (Pera et al., 2004; Smith et al., 2008; Chambers et al., 2009). Thus, these data generally support the “default model”; ESCs undergo neural induction in vitro without instruction from extrinsic factors, and simply blocking inhibitors of neural fate can further enhance neurogenesis.
Temporal Specification of Neural Cell Types
Pluripotent stem cell-derived neurons can be used to study the temporal specification or differentiation process of neuronal subtypes. Neural specification is often determined by differential responses of receptor cells to a gradient of a given signaling molecule. Thus, subtle changes in the concentration or timing of exogenous factors can have a dramatic effect on cell type specification. For example, in hESCs using identical differentiation protocols, early Shh exposure produces floor plate tissue while later Shh exposure induces telencephalic progenitors (Fasano et al., 2010). While this is a dramatic change in cell fate, one could examine more subtle fate effects. In one protocol, a moderate increase in Shh concentration (from 10 to 30 nM) can shift ESCs from and LGE-like to an medial ganglionic eminence (MGE)-like fate (Danjo et al., 2011).
Neurogenesis in the developing cortex occurs in a predictable temporal manner with the sequential generation of specific neural subtypes: preplate neurons such as Cajal–Retzius cells are born first, and then deep cortical layers followed by more superficial layers. There are specific temporal markers for each of these cell types, allowing for the characterization of maturing PSC-derived neurons in a temporal fashion. It was previously shown that individual cortical progenitors plated in culture undergo repeated asymmetric divisions to produce a variety of neuronal cell types that mimic the normal differentiation schedule in vivo (Shen et al., 2006). This pattern also holds true for mESC differentiations in vitro. When mESCs are cultured in the absence of serum or extrinsic factors (except for a Shh inhibitor), they sequentially generate cortical neurons that grossly recapitulate the temporal pattern of normal development (Eiraku et al., 2008; Gaspard et al., 2008). A similar finding was demonstrated with mESC-derived retinal cells. Amazingly, floating EBs can develop optic cup-like structures that give rise to different retinal cells in a temporal manner that mimics their developmental time course in vivo (Eiraku et al., 2011). Thus, mESCs (and potentially hESCs) may provide a simplified, reductionist approach for studying the temporal patterning of cortical development.
Identification of Novel Genes and Molecules that Specify Neuronal Fate Determination
The fact that distinct CNS regions, and individual neuronal progenitors within these regions, can give rise to a wide variety of cell types in vivo presents a challenge for understanding how individual genes determine neuronal fate specification. The ability to generate and isolate a homogeneous population of neurons in vitro can better address this question. One can purify a specific PSC-derived population of cells and perform microarrays or transcriptome sequencing at different time points to identify novel genes and relevant signaling pathways that are important for fate specification, as was done for hESC-derived floor plate cells (Fasano et al., 2010). Additionally, small molecule screens can be conducted to identify compounds that enhance the derivation of distinct cell types. Improvements in the specificity and efficiency of differentiation protocols should lead to a significant increase in these types of studies.
Post-Mitotic Development
Pluripotent stem cell-derived neurons are an excellent tool for studying aspects of post-mitotic neuronal maturation, such as cell migration, axiogenesis, dendritogenesis, target recognition, synaptogenesis, and proper integration into neuronal circuitry. A common technique is to purify fluorescently labeled PSC-derived neurons and transplant them into host environments to verify that they form synapses (Maroof et al., 2010; Danjo et al., 2011). In addition to being a crucial aspect of protocol verification, this procedure provides a mechanism to study various developmental aspects of a neuron in its endogenous environment. One can visualize transplanted cells in real time, compare gene expression in fluorescently labeled cells with their endogenous neighbors, and evaluate their synaptogenesis and integration into the cortical circuitry using electrophysiological recordings. In addition to transplantation studies, PSC-derived neurons can also be plated on specific feeder cells to study synaptogenesis in vitro (Brennand et al., 2011; Kim et al., 2011b). The ability to manipulate gene expression in PSCs allows one to characterize a role for specific genes in distinct processes of neuronal maturation. Put simply, generating an unlimited source of PSC-derived neural subtypes can open the door to many possible experiments examining neuronal maturation.
Effect of Exogenous Factors on Neural Development
Another less appreciated use of PSCs is the derivation of neural progenitors from PSCs to study the effects of exogenous factors on neural development. For example, ethanol consumption during pregnancy can have wide ranging effects on many aspects of brain development, making it a difficult to determine the exact mechanism of ethanol’s effect. A recent study found that hESCs exposed to ethanol during neuronal differentiation impaired the survival, morphology, gene expression, and differentiation capabilities of neural progenitors (Talens-Visconti et al., 2011). There is a growing field of developmental toxicology studies that use PSC-derived cells to examine the effects of hazardous agents on cell survival, proliferation, and differentiation (Wobus and Loser, 2011). For instance, one study examined the effect of botulinum toxin on mESC-derived MN development (Kiris et al., 2011). These detailed findings would be challenging to uncover from more traditional methods, and one can envision these types of experiments being applied to a wide range of candidate compounds that perturb normal neuronal development.
Specific Insights into Human Development
Human ESCs and hiPSCs allow researchers to specifically investigate aspects of human development that were previously unattainable due to the technical and ethical concerns of studying human fetuses. While mice (and mESCs) are good models for studying neural development, there are clearly significant differences between mice and humans, and thus discoveries in mice (and mESCs) may not be directly applicable to human development. mESCs generally differentiate toward a given neural cell type faster than hESCs under equivalent conditions, which may be due to differences in intrinsic kinetics or differentiation competence. There is ample evidence that mESCs and hESCs have significantly different gene expression profiles (Ginis et al., 2004; Tesar et al., 2007). For example, the transcription factor Pax6 is a requisite fate determinant for neural induction of hESCs, whereas neither loss nor gain of Pax6 function affects neural specification in mESCs (Zhang et al., 2010). Additionally, some protocols that are efficient at neural induction in hESCs and hiPSCs (dual SMAD inhibition; Chambers et al., 2009) do not efficiently generate neurons from mESCs (J. Tyson and S. Anderson, unpublished data).
While hPSCs hold more promise for studying human development than mPSC-derived cells, a challenge is to develop assays to study synaptogenesis and functional connectivity in these cells. Long-term cultures in vitro or xenograft transplants in vivo are required to thoroughly analyze the development of hPSC-derived neurons. Additionally, since relatively little is known about the molecular nature of human neurodevelopment, it can be difficult to verify whether discoveries made with hPSCs represent real insights into human neurodevelopment or are instead artifacts of the culture system. The Allen Institute for Brain Science has recently launched BrainSpan, an atlas for the developing human brain consisting of RNA sequencing, microarray analysis, and in situ hybridizations spanning multiple brain regions at 13 different developmental stages (http://developinghumanbrain.org/). The combination of the developing mouse and human brain atlases are invaluable resources for the study of human brain development.
Genetic Techniques for Enhancing PSC-Derived Neural Cell Production, Specification, and Isolation
Even with well-defined differentiation protocols, cultures contain a heterogeneous population of different cell types, so having a marker for labeling and isolating the desired cell type is crucial. Some strategies for cell type specific purification rely on cell surface markers to sort neuronal progenitors (Pruszak et al., 2007) For example, the forebrain surface antigen-1 (FORSE1) has been used to isolate hESC-derived neuronal progenitors (Elkabetz et al., 2008). The use of cell type selective promoter elements to direct expression of fluorescent markers is another powerful tool for efficiently identifying and purifying distinct PSC-derived neurons via fluorescence-activated cell sorting (FACS).
Traditional techniques for gene transfer in PSCs utilize site-directed targeting or non-targeted insertion of promoter-driven fluorescent markers. These transgenes can be introduced into the PSC genome via electroporation, nucleofection, or viral-mediated delivery systems. Site-directed targeting requires homologous recombination and should faithfully recapitulate the targeted gene by integrating into the endogenous locus, but this process can be labor intensive and often results in a heterozygotic phenotype. Random insertion leaves both endogenous copies of the gene of interest intact, but requires identification of promoter or enhancer structures to drive protein expression. Additionally, randomly inserted transgenes are often dysregulated or unstable and can disrupt expression of other genes depending on their integration site. Several recent technologies described below show promise for enhancing the derivation of neural cells from PSCs.
Bacterial artificial chromosomes (BACs) can be used to introduce large pieces of genomic DNA (∼100–300 Kb) into PSCs via homologous recombination. BACs are advantageous because their large size likely retains critical regulatory regions for gene expression and limits silencing due to unwanted chromatin modifications. BACs can be modified to express fluorescent proteins under the control of specific genes. BAC transgenesis into mESCs (Tomishima et al., 2007) and hESCs (Placantonakis et al., 2009) has facilitated the production of many fluorescent-labeled ESC lines, with a tremendous potential to modify other genes. Some potential drawbacks to BAC transgenesis include inefficient BAC integration, disruption of endogenous gene expression at the integration site, and unwanted gene dosage effects since BACs often contain multiple genes.
Transposons are DNA sequences that can move from one location to another within the genome of a single cell, with the piggyBac transposon being most active in mammalian cells (Ding et al., 2005). The transposon system consists of a DNA sequence flanked by two terminal repeats, and a transposase enzyme that catalyzes mobilization of the transposon to another genomic site. Importantly, transposons can be seamlessly removed by re-expressing the transposase, eliminating any unwanted genomic footprint. In addition to providing a viral-free mechanism for deriving iPSCs from fibroblasts (Sidhu, 2011), transposons can also be used to inducibly express pro-neural genes that efficiently drive hESCs to differentiate toward a neuronal fate (Lacoste et al., 2009). Combined with the improved efficiency of piggyBac transposase in mammalian cells (Yusa et al., 2011), transposons represent a highly efficient technique with minimal genomic manipulation for enhancing PSC-derived neural cells in the future.
Finally, zinc finger nucleases (ZFNs) can induce site-specific double-strand DNA breaks to facilitate homologous recombination for genetic manipulation in PSCs. Several zinc finger motifs that specify a DNA recognition site are fused to a non-specific nuclease that cleaves double stranded DNA and induces homology-directed repair, allowing for the incorporation of exogenous sequences into at the cleavage site. Successful demonstration of ZFN in hESCs was recently achieved (Lombardo et al., 2007), and another study successfully targeted the PITX3 gene in hESCs (Hockemeyer et al., 2009), a known marker for dopaminergic neurons. While this approach significantly improves the efficiency for making fluorescent reporter lines, it remains to be seen if ZFNs can be used to enhance gene expression to promote neural differentiation.
In addition to the above strategies for labeling PSCs, it bears mentioning that a variety of genetic manipulation techniques can be utilized to direct fate specification of PSC-derived cells. One group transfected two transcription factors into hESC-derived neural progenitors to enhance generation of basal forebrain cholinergic neurons (Bissonnette et al., 2011). Another promising tool is to inducibly express fate-determining genes during PSC cell differentiations. For example, one can utilize a mESC line modified with the reverse tetracycline transactivator system (rtTA, “doxy-on”) system to inducibly and reversibly express genes of interest during the differentiation protocol (Ting et al., 2005). Other strategies that can be used to manipulate gene expression in PSCs include using the LoxP-Cre, CreER, or FRT-Flp systems, which provide inducible control of recombination.
The ongoing progress towards identifying novel genes expressed in specific neuronal subtypes combined with improved techniques for genetically modifying PSCs will greatly advance the use of PSC-derived neurons for the study of neurodevelopment.
Generation of Specific Neural Subtypes from PSCs
A wide variety of protocols have been developed to generate many PSC-derived neural subtypes that span the entire neural tube (Table 1). In this section, we will briefly summarize the PSC derivation of three of the more thoroughly investigated and clinically relevant neuronal subtypes (Figure 4). (See the Neurogenesis as a Default Mechanism? section for a summary of general neural induction techniques). It is important to note that there are significant differences in protocol development, such as the media composition, the timing and concentration of exogenous signaling factors, cell density at plating, the adherent and feeder environment, and the physical dimensions of the culture system. While we highlight the most important signaling factors that are used to generate each type of neuron, the medium often contains other growth factors and nutrients not mentioned. We recommend that readers refer to the original studies for a more detailed description of the protocols.
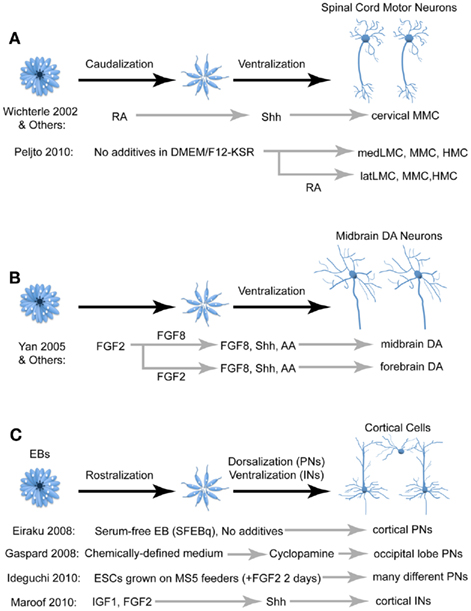
Figure 4. Comparison of different techniques for deriving three of the more studied neural cell types: spinal motor neurons (A), midbrain dopaminergic neurons (B), and cortical cells (C). FGF, fibroblast growth factor; RA, retinoic acid; Shh, Sonic hedgehog; IGF, insulin growth factor; AA, arachidonic acid; PN, projection neurons; IN, interneurons; MMC, median motor neurons; LMC, lateral motor neurons; HMC, hypaxial motor neurons; DA, dopaminergic.
Spinal Motor Neurons
In the developing embryo, MNs are generated from neural progenitors in the ventral portion of the spinal cord. A combinatorial code involving the Hox family of transcription factors establishes the identity of multiple MN pools located throughout the longitudinal axis of the spinal cord (Dasen and Jessell, 2009). Mature MNs in vivo adopt one of three columnar fates; either the median (MMC), hypaxial (HMC), or lateral (LMC) motor column. These three motor columns can be further divided into subgroups categorized by the individual cells’ spatial position, molecular identity, and musculature innervations. Optimizing protocols to generate different types of PSC-derived MNs will be beneficial for studying both normal neural development and therapeutic applications for MN diseases.
Since the derivation of MNs from PSC requires caudalization and ventralization of neural progenitors, the two likely candidates for inducing this identity are RA and Shh, respectively (Figure 4A; Wichterle et al., 2002). After the caudalizing action of RA, Shh signaling promotes MN fate defined by the expression of several MN markers including HB9, which is selectively expressed by MNs in vivo. Upon transplantation into chick spinal cord, HB9::GFP mESC-derived MNs project axons along endogenous MN axon pathways and form synapses on the corresponding muscle targets (Wichterle et al., 2002). Using this strategy, most mESC-derived MNs express the cervical MMC marker Lhx3 (Wichterle et al., 2002; Soundararajan et al., 2006). While most studies utilize RA and Shh to generate MNs, efforts to generate other MN fates have utilized retinoid-independent techniques (Patani et al., 2011). A recent study derived LMC MNs by eliminating exogenous RA and Shh from the protocol and instead relying on endogenous signaling hubs within EBs (Peljto et al., 2010). Furthermore, late addition of exogenous RA induced post-mitotic MNs to a lateral LMC fate. Thus, although growing PSCs in serum-free, morphogen-free conditions tends to promote anterior neural fates, this is an example where slight modifications in the medium and culture conditions can give rise to different cell fates and induction efficiencies. Further protocol optimization will likely improve our ability to generate homogenous populations of different specific MN subtypes.
More recently, protocols utilizing RA and Shh have been developed to generate MNs from hESCs (Li et al., 2005; Di Giorgio et al., 2008) and iPSCs (Dimos et al., 2008). The iPSC study used fibroblasts taken from patients with amyotrophic lateral sclerosis (ALS) and healthy controls, and demonstrated that the ALS iPSCs could differentiate into MNs in vitro, highlighting the potential for using iPSCs to study neurological diseases. Taken together, these findings suggest that one can recapitulate fundamental biological principles of MN development and maturation in vitro, reinforcing the notion that PSCs provide a system to analyze mechanisms of normal and abnormal human neurodevelopment.
Midbrain Dopaminergic Neurons
Dopaminergic (DA) neurons are found in many brain regions, but the most prominent groups of DA neurons occupy the substantia nigra and ventral tegmental area of the midbrain. Degeneration of midbrain DA neurons is one of the hallmarks of Parkinson’s disease, and thus developing methods to derive DA neurons from PSCs has significant therapeutic potential. Much like PSC-derived MNs, the successful conversion of PSCs into midbrain DA neurons in vitro is dependent on the signaling factors used to induce genes expressed in endogenously developing DA neurons. As mentioned above, FGF8 is an important patterning factor for the midbrain and hindbrain regions (Partanen, 2007). The ventral location of midbrain DA neurons implies that Shh may also play a prominent role in their derivation.
In 2000, two separate studies utilizing distinct protocols successfully derived DA cells from mESCs. One study treated dissociated mESCs with Shh, FGF8, and ascorbic acid to generate cells that expressed a variety of midbrain DA markers and confirmed that these cells released dopamine into the medium (Lee et al., 2000). A second study generated DA neurons simply by growing mESCs on stromal cells in the absence of any patterning factors, and these cells survived upon transplantation into the mouse striatum, indicating that they are more likely forebrain rather than midbrain DA neurons (Kawasaki et al., 2000). Strikingly, these two drastically different protocols both reported that ∼30% of Tuj-1+ cells (a neuronal marker) expressed the dopaminergic marker tyrosine hydroxylase (TH). A later study utilized Shh and FGF8 to produce hESC-derived midbrain DA neurons, noting that early FGF8 treatment produced midbrain DA neurons whereas later FGF8 treatment resulted in cells that were more likely forebrain DA neurons (Figure 4B; Yan et al., 2005). Additionally, Shh, FGF8, and ascorbic acid were utilized to drive hESCs and iPSCs to midbrain DA neurons using the feeder-free, dual SMAD inhibition protocol mentioned above (Chambers et al., 2009). Transplantation of both mESC-derived (Kim et al., 2002) and hESC-derived (Roy et al., 2006) midbrain DA neurons have shown promise in ameliorating some of the motor defects in rodent models of Parkinson’s, although significant work is required to ensure the long-term efficacy and safety of this technique.
Cortical Neurons
As mentioned above, PSCs that are differentiated either as EBs, adherent monolayers, or co-cultures with stromal cells and then expanded in serum-free media without exogenous patterning factors predominantly generate forebrain progenitors expressing the telencephalic marker FoxG1 (also known as BF1). Notably, many of these progenitors give rise to cortical pyramidal neurons, which can be partially attributed to endogenous Wnt signaling that represses Shh signaling (Li et al., 2009). Generation of cortical pyramidal neurons is further enriched when the ventralizing action of the morphogen Shh is inhibited (Figure 4C; Gaspard et al., 2008). mESC- and hESC-derived forebrain progenitors are capable of forming 3D structures with an apical–basal polarity and organized cellular layers reminiscent of cortical development in vivo (Eiraku et al., 2008). These cells are similar to their in vivo equivalents in morphology, neurotransmitter profile, expression of cortical layer markers and precise axonal projections upon transplantation in vivo.
Cortical pyramidal neurons have a wide variety of cortical and subcortical targets depending on their location and functional modality, and there are both intrinsic and extrinsic mechanisms that regulate this organization (O’Leary et al., 2007). Initial studies found that many ESC-derived neural progenitors would develop into cortical pyramidal cells and integrate into the cortical circuitry when transplanted into the lateral ventricles of mice and rats (Wernig et al., 2004; Muotri et al., 2005). More detailed studies characterized the specific connectivity of these cells. In one instance, ESC-derived cortical pyramidal cells transplanted into the frontal cortex adopted projection patterns resembling that of visual and limbic occipital cortical projection neurons (and expressed the occipital cortical markers CoupTFI and CoupTFII), indicating that fates of these cells were determined in vitro (Gaspard et al., 2008). Conversely, another study found that ESC-derived pyramidal cells developed dendritic and axonal connectivity that were appropriate for their transplanted location into the somatosensory, visual, or motor cortices, implying that the fate of these cells was still influenced by environmental factors (Ideguchi et al., 2010).
GABAergic interneurons comprise ∼20% of all cortical cells and are important modulators of cortical activity. While cortical pyramidal cells originate from the dorsal telencephalon, cortical interneurons are born in the ventral telencephalon, specifically the MGE and caudal ganglionic eminence (CGE), and migrate dorsally into the cortical plate. Two recent studies have developed protocols to differentiate mESCs into cortical interneurons by adding the ventralizing agent Shh to the culture medium upon neural induction (Figure 4C; Maroof et al., 2010; Danjo et al., 2011). These cells express characteristic interneuronal neurochemical and electrophysiological properties when transplanted into postnatal cortices (Maroof et al., 2010). Modulation of FGF signaling can alter the balance between MGE- and CGE-derived interneuron subgroups (Danjo et al., 2011).
The details of how exactly intrinsic expression profiles act in combination with extrinsic functional inputs to produce the diverse array of cortical pyramidal and interneuron subgroups remains unclear. Therefore using PSC-based strategies to define the intrinsic genetic programs and corresponding temporal patterning signals in maturing progenitors in vitro could help uncover signaling pathways involved in human cortical development in vivo.
Conclusion
The ability to generate PSC-derived neural cells that faithfully recapitulate normal embryonic development enables the study of mammalian neural development, and in particular human, in a more accessible manner then has previously been achieved. Rapid progress is being made in manipulating gene expression in PSCs and improving techniques for generating, isolating and studying diverse populations of neural subtypes. For example, efforts are underway for PSC-derived neural cells to more faithfully recapitulate in vivo development using bioreactors and 3D-scaffolds to improve organized cell migration and axon extension in the dish, as well improved reproducibility over conditions such as temperature, pH and oxygen levels, nutrient replacement, and flow rate of the media (Ghasemi-Mobarakeh et al., 2008; Dalton and Mey, 2009). These types of advances will aid in defining molecular mechanisms controlling the establishment of the complex and precise neuronal connectivity during CNS development. Like all emerging scientific endeavors, significant hurdles remain in expanding the use of PSCs to study neural development.
First, there is considerable diversity and nuances within PSC culture techniques, making it unclear how reproducible differentiation protocols are between labs. For example, differentiation techniques that are efficient for floating embryoid bodies might not translate well to adherent dissociated PSC cultures. This lack of uniformity is one of the driving forces for developing novel strategies for neuronal differentiation, but it is also a challenge for determining which systems best recapitulate the in vivo scenario. If there are eight diverse protocols for generating MNs with different efficiencies and purity, should researchers adopt the most efficient technique to maximize conformity and verification of results? Or does each differentiation technique result in subtle differences that provide distinct insights into some aspect of neurodevelopment? In addition to differences in cell culture techniques, there is also significant variability of how researchers present efficiency values in papers. Some calculate efficiencies based on a sampling method that estimates the total number of cells in a culture, some on the number of neuronal progenitors (i.e., Sox1+ or Nestin+ cells), and some reports are extremely vague about how (or if) they calculate efficiencies or purity. The use of different cell type specific markers for quantification of derivation and efficiency also complicates comparison of inter-laboratory techniques. These are challenging questions for the field of stem cells in general, but in particular for neuroscience because of the remarkable diversity of different cell types in the CNS.
Another issue is how best to study the maturity and functionality of PSC-derived neural cells. For certain questions, analyzing developmental aspects of PSC-derived fate-restricted neural progenitors may be adequate. However, to fully characterize a given neural subtype, one needs to verify that it can mature and integrate into the circuitry of a host environment. Simply demonstrating that cells express cell type specific (or enriched) genes does not verify that the cells display similar characteristics to their endogenous counterparts. This daunting task requires several important considerations: (1) when to isolate cells in the differentiation process, (2) how, when and where to transplant the cells and/or plate on adherent substrates or feeder cells (i.e., prepare slices after transplant and record from labeled cells, etc.). Again, the lack of conformity due to different laboratories having distinct areas of expertise makes this a challenging task. However, there is some push to address these conformity and reproducibility issues using many different cell lines across multiple laboratories (Boulting et al., 2011).
The use of PSC-derived neural cells to recapitulate and study normal and pathological neurodevelopment is clearly a “work-in progress,” but one of tremendous promise for the field of developmental neurobiology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Adam Goulburn and Dr. Takeshi Sakurai for comments on the manuscript. We apologize to all of our colleagues whose work we did not discuss due to space limitations. This work was supported by NIH grants R01 MH066912 and RC1 MH089690 (Stewart A. Anderson).
References
Aboody, K., Capela, A., Niazi, N., Stern, J. H., and Temple, S. (2011). Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a rosetta stone. Neuron 70, 597–613.
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., and Ding, S. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118.
Aubry, L., Bugi, A., Lefort, N., Rousseau, F., Peschanski, M., and Perrier, A. L. (2008). Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc. Natl. Acad. Sci. U.S.A. 105, 16707–16712.
Bissonnette, C. J., Lyass, L., Bhattacharyya, B. J., Belmadani, A., Miller, R. J., and Kessler, J. A. (2011). The controlled generation of functional Basal forebrain cholinergic neurons from human embryonic stem cells. Stem Cells 29, 802–811.
Boulting, G. L., Kiskinis, E., Croft, G. F., Amoroso, M. W., Oakley, D. H., Wainger, B. J., Williams, D. J., Kahler, D. J., Yamaki, M., Davidow, L., Rodolfa, C. T., Dimos, J. T., Mikkilineni, S., Macdermott, A. B., Woolf, C. J., Henderson, C. E., Wichterle, H., and Eggan, K. (2011). A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 29, 279–286.
Brennand, K. J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., Li, Y., Mu, Y., Chen, G., Yu, D., Mccarthy, S., Sebat, J., and Gage, F. H. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225.
Brons, I. G., Smithers, L. E., Trotter, M. W., Rugg-Gunn, P., Sun, B., Chuva De Sousa Lopes, S. M., Howlett, S. K., Clarkson, A., Ahrlund-Richter, L., Pedersen, R. A., and Vallier, L. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195.
Caiazzo, M., Dell’anno, M. T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., Sotnikova, T. D., Menegon, A., Roncaglia, P., Colciago, G., Russo, G., Carninci, P., Pezzoli, G., Gainetdinov, R. R., Gustincich, S., Dityatev, A., and Broccoli, V. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227.
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280.
Cundiff, P. E., and Anderson, S. A. (2011). Impact of induced pluripotent stem cells on the study of central nervous system disease. Curr. Opin. Genet. Dev.
Danjo, T., Eiraku, M., Muguruma, K., Watanabe, K., Kawada, M., Yanagawa, Y., Rubenstein, J. L., and Sasai, Y. (2011). Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J. Neurosci. 31, 1919–1933.
Dasen, J. S., and Jessell, T. M. (2009). Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 88, 169–200.
Di Giorgio, F. P., Boulting, G. L., Bobrowicz, S., and Eggan, K. C. (2008). Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3, 637–648.
Dimos, J. T., Rodolfa, K. T., Niakan, K. K., Weisenthal, L. M., Mitsumoto, H., Chung, W., Croft, G. F., Saphier, G., Leibel, R., Goland, R., Wichterle, H., Henderson, C. E., and Eggan, K. (2008). Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221.
Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y., and Xu, T. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483.
Eiraku, M., Takata, N., Ishibashi, H., Kawada, M., Sakakura, E., Okuda, S., Sekiguchi, K., Adachi, T., and Sasai, Y. (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56.
Eiraku, M., Watanabe, K., Matsuo-Takasaki, M., Kawada, M., Yonemura, S., Matsumura, M., Wataya, T., Nishiyama, A., Muguruma, K., and Sasai, Y. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532.
Elkabetz, Y., Panagiotakos, G., Al Shamy, G., Socci, N. D., Tabar, V., and Studer, L. (2008). Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22, 152–165.
Erceg, S., Ronaghi, M., Zipancic, I., Lainez, S., Rosello, M. G., Xiong, C., Moreno-Manzano, V., Rodriguez-Jimenez, F. J., Planells, R., Alvarez-Dolado, M., Bhattacharya, S. S., and Stojkovic, M. (2010). Efficient differentiation of human embryonic stem cells into functional cerebellar-like cells. Stem Cells Dev. 19, 1745–1756.
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156.
Fasano, C. A., Chambers, S. M., Lee, G., Tomishima, M. J., and Studer, L. (2010). Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell 6, 336–347.
Gaspard, N., Bouschet, T., Herpoel, A., Naeije, G., Van Den Ameele, J., and Vanderhaeghen, P. (2009). Generation of cortical neurons from mouse embryonic stem cells. Nat. Protoc. 4, 1454–1463.
Gaspard, N., Bouschet, T., Hourez, R., Dimidschstein, J., Naeije, G., Van Den Ameele, J., Espuny-Camacho, I., Herpoel, A., Passante, L., Schiffmann, S. N., Gaillard, A., and Vanderhaeghen, P. (2008). An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357.
Gaspard, N., and Vanderhaeghen, P. (2011). From stem cells to neural networks: recent advances and perspectives for neurodevelopmental disorders. Dev. Med. Child Neurol. 53, 13–17.
Ghasemi-Mobarakeh, L., Prabhakaran, M. P., Morshed, M., Nasr-Esfahani, M. H., and Ramakrishna, S. (2008). Electrospun poly(epsilon-caprolactone)/gelatin nanofibrous scaffolds for nerve tissue engineering. Biomaterials 29, 4532–4539.
Ginis, I., Luo, Y., Miura, T., Thies, S., Brandenberger, R., Gerecht-Nir, S., Amit, M., Hoke, A., Carpenter, M. K., Itskovitz-Eldor, J., and Rao, M. S. (2004). Differences between human and mouse embryonic stem cells. Dev. Biol. 269, 360–380.
Gore, A., Li, Z., Fung, H. L., Young, J. E., Agarwal, S., Antosiewicz-Bourget, J., Canto, I., Giorgetti, A., Israel, M. A., Kiskinis, E., Lee, J. H., Loh, Y. H., Manos, P. D., Montserrat, N., Panopoulos, A. D., Ruiz, S., Wilbert, M. L., Yu, J., Kirkness, E. F., Izpisua Belmonte, J. C., Rossi, D. J., Thomson, J. A., Eggan, K., Daley, G. Q., Goldstein, L. S., and Zhang, K. (2011). Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67.
Goulburn, A. L., Alden, D., Davis, R. P., Micallef, S. J., Ng, E. S., Yu, Q. C., Lim, S. M., Soh, C. L., Elliott, D. A., Hatzistavrou, T., Bourke, J., Watmuff, B., Lang, R. J., Haynes, J. M., Pouton, C. W., Giudice, A., Trounson, A. O., Anderson, S. A., Stanley, E. G., and Elefanty, A. G. (2011). A targeted NKX2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells 29, 462–473.
Hanna, J. H., Saha, K., and Jaenisch, R. (2010). Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143, 508–525.
Hochedlinger, K., and Plath, K. (2009). Epigenetic reprogramming and induced pluripotency. Development 136, 509–523.
Hockemeyer, D., Soldner, F., Beard, C., Gao, Q., Mitalipova, M., Dekelver, R. C., Katibah, G. E., Amora, R., Boydston, E. A., Zeitler, B., Meng, X., Miller, J. C., Zhang, L., Rebar, E. J., Gregory, P. D., Urnov, F. D., and Jaenisch, R. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857.
Hussein, S. M., Batada, N. N., Vuoristo, S., Ching, R. W., Autio, R., Narva, E., Ng, S., Sourour, M., Hamalainen, R., Olsson, C., Lundin, K., Mikkola, M., Trokovic, R., Peitz, M., Brustle, O., Bazett-Jones, D. P., Alitalo, K., Lahesmaa, R., Nagy, A., and Otonkoski, T. (2011). Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58–62.
Ideguchi, M., Palmer, T. D., Recht, L. D., and Weimann, J. M. (2010). Murine embryonic stem cell-derived pyramidal neurons integrate into the cerebral cortex and appropriately project axons to subcortical targets. J. Neurosci. 30, 894–904.
Ikeda, H., Osakada, F., Watanabe, K., Mizuseki, K., Haraguchi, T., Miyoshi, H., Kamiya, D., Honda, Y., Sasai, N., Yoshimura, N., Takahashi, M., and Sasai, Y. (2005). Generation of Rx+/Pax6+ neural retinal precursors from embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 102, 11331–11336.
Kawasaki, H., Mizuseki, K., Nishikawa, S., Kaneko, S., Kuwana, Y., Nakanishi, S., Nishikawa, S. I., and Sasai, Y. (2000). Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron 28, 31–40.
Kim, J., Efe, J. A., Zhu, S., Talantova, M., Yuan, X., Wang, S., Lipton, S. A., Zhang, K., and Ding, S. (2011a). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843.
Kim, J. E., O’sullivan, M. L., Sanchez, C. A., Hwang, M., Israel, M. A., Brennand, K., Deerinck, T. J., Goldstein, L. S., Gage, F. H., Ellisman, M. H., and Ghosh, A. (2011b). Investigating synapse formation and function using human pluripotent stem cell-derived neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 3005–3010.
Kim, J. H., Auerbach, J. M., Rodriguez-Gomez, J. A., Velasco, I., Gavin, D., Lumelsky, N., Lee, S. H., Nguyen, J., Sanchez-Pernaute, R., Bankiewicz, K., and Mckay, R. (2002). Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature 418, 50–56.
Kim, S. U., and de Vellis, J. (2009). Stem cell-based cell therapy in neurological diseases: a review. J. Neurosci. Res. 87, 2183–2200.
Kiris, E., Nuss, J. E., Burnett, J. C., Kota, K. P., Koh, D. C., Wanner, L. M., Torres-Melendez, E., Gussio, R., Tessarollo, L., and Bavari, S. (2011). Embryonic stem cell-derived motoneurons provide a highly sensitive cell culture model for botulinum neurotoxin studies, with implications for high-throughput drug discovery. Stem Cell Res 6, 195–205.
Lacoste, A., Berenshteyn, F., and Brivanlou, A. H. (2009). An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell 5, 332–342.
Lamba, D. A., Karl, M. O., Ware, C. B., and Reh, T. A. (2006). Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 12769–12774.
Lee, G., Chambers, S. M., Tomishima, M. J., and Studer, L. (2010). Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 5, 688–701.
Lee, H., Shamy, G. A., Elkabetz, Y., Schofield, C. M., Harrsion, N. L., Panagiotakos, G., Socci, N. D., Tabar, V., and Studer, L. (2007). Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells 25, 1931–1939.
Lee, S. H., Lumelsky, N., Studer, L., Auerbach, J. M., and Mckay, R. D. (2000). Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat. Biotechnol. 18, 675–679.
Levine, A. J., and Brivanlou, A. H. (2007). Proposal of a model of mammalian neural induction. Dev. Biol. 308, 247–256.
Li, X. J., Du, Z. W., Zarnowska, E. D., Pankratz, M., Hansen, L. O., Pearce, R. A., and Zhang, S. C. (2005). Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221.
Li, X. J., Zhang, X., Johnson, M. A., Wang, Z. B., Lavaute, T., and Zhang, S. C. (2009). Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development 136, 4055–4063.
Lister, R., Pelizzola, M., Kida, Y. S., Hawkins, R. D., Nery, J. R., Hon, G., Antosiewicz-Bourget, J., O’malley, R., Castanon, R., Klugman, S., Downes, M., Yu, R., Stewart, R., Ren, B., Thomson, J. A., Evans, R. M., and Ecker, J. R. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73.
Liu, A., and Niswander, L. A. (2005). Bone morphogenetic protein signalling and vertebrate nervous system development. Nat. Rev. Neurosci. 6, 945–954.
Lombardo, A., Genovese, P., Beausejour, C. M., Colleoni, S., Lee, Y. L., Kim, K. A., Ando, D., Urnov, F. D., Galli, C., Gregory, P. D., Holmes, M. C., and Naldini, L. (2007). Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306.
Marchetto, M. C., Winner, B., and Gage, F. H. (2010). Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Hum. Mol. Genet. 19, R71–R76.
Maroof, A. M., Brown, K., Shi, S. H., Studer, L., and Anderson, S. A. (2010). Prospective isolation of cortical interneuron precursors from mouse embryonic stem cells. J. Neurosci. 30, 4667–4675.
Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638.
Miyoshi, N., Ishii, H., Nagano, H., Haraguchi, N., Dewi, D. L., Kano, Y., Nishikawa, S., Tanemura, M., Mimori, K., Tanaka, F., Saito, T., Nishimura, J., Takemasa, I., Mizushima, T., Ikeda, M., Yamamoto, H., Sekimoto, M., Doki, Y., and Mori, M. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638.
Munoz-Sanjuan, I., and Brivanlou, A. H. (2002). Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3, 271–280.
Muotri, A. R., Nakashima, K., Toni, N., Sandler, V. M., and Gage, F. H. (2005). Development of functional human embryonic stem cell-derived neurons in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 102, 18644–18648.
Murashov, A. K., Pak, E. S., Hendricks, W. A., Owensby, J. P., Sierpinski, P. L., Tatko, L. M., and Fletcher, P. L. (2005). Directed differentiation of embryonic stem cells into dorsal interneurons. FASEB J. 19, 252–254.
Nistor, G., Seiler, M. J., Yan, F., Ferguson, D., and Keirstead, H. S. (2010). Three-dimensional early retinal progenitor 3D tissue constructs derived from human embryonic stem cells. J. Neurosci. Methods 190, 63–70.
O’Leary, D. D., Chou, S. J., and Sahara, S. (2007). Area patterning of the mammalian cortex. Neuron 56, 252–269.
Osakada, F., Ikeda, H., Mandai, M., Wataya, T., Watanabe, K., Yoshimura, N., Akaike, A., Sasai, Y., and Takahashi, M. (2008). Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat. Biotechnol. 26, 215–224.
Pang, Z. P., Yang, N., Vierbuchen, T., Ostermeier, A., Fuentes, D. R., Yang, T. Q., Citri, A., Sebastiano, V., Marro, S., Sudhof, T. C., and Wernig, M. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223.
Partanen, J. (2007). FGF signalling pathways in development of the midbrain and anterior hindbrain. J. Neurochem. 101, 1185–1193.
Patani, R., Hollins, A. J., Wishart, T. M., Puddifoot, C. A., Alvarez, S., De Lera, A. R., Wyllie, D. J., Compston, D. A., Pedersen, R. A., Gillingwater, T. H., Hardingham, G. E., Allen, N. D., and Chandran, S. (2011). Retinoid-independent motor neurogenesis from human embryonic stem cells reveals a medial columnar ground state. Nat. Commun. 2, 214.
Peljto, M., Dasen, J. S., Mazzoni, E. O., Jessell, T. M., and Wichterle, H. (2010). Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 7, 355–366.
Pera, M. F., Andrade, J., Houssami, S., Reubinoff, B., Trounson, A., Stanley, E. G., Ward-Van Oostwaard, D., and Mummery, C. (2004). Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell. Sci. 117, 1269–1280.
Perrier, A. L., Tabar, V., Barberi, T., Rubio, M. E., Bruses, J., Topf, N., Harrison, N. L., and Studer, L. (2004). Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 101, 12543–12548.
Pfisterer, U., Kirkeby, A., Torper, O., Wood, J., Nelander, J., Dufour, A., Bjorklund, A., Lindvall, O., Jakobsson, J., and Parmar, M. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 10343–10348.
Placantonakis, D. G., Tomishima, M. J., Lafaille, F., Desbordes, S. C., Jia, F., Socci, N. D., Viale, A., Lee, H., Harrison, N., Tabar, V., and Studer, L. (2009). BAC transgenesis in human embryonic stem cells as a novel tool to define the human neural lineage. Stem Cells 27, 521–532.
Pruszak, J., Sonntag, K. C., Aung, M. H., Sanchez-Pernaute, R., and Isacson, O. (2007). Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells 25, 2257–2268.
Qiang, L., Fujita, R., Yamashita, T., Angulo, S., Rhinn, H., Rhee, D., Doege, C., Chau, L., Aubry, L., Vanti, W. B., Moreno, H., and Abeliovich, A. (2011). Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell 146, 359–371.
Roy, N. S., Cleren, C., Singh, S. K., Yang, L., Beal, M. F., and Goldman, S. A. (2006). Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat. Med. 12, 1259–1268.
Salero, E., and Hatten, M. E. (2007). Differentiation of ES cells into cerebellar neurons. Proc. Natl. Acad. Sci. U.S.A. 104, 2997–3002.
Shen, L., Nam, H. S., Song, P., Moore, H., and Anderson, S. A. (2006). FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus 16, 875–890.
Sidhu, K. S. (2011). New approaches for the generation of induced pluripotent stem cells. Expert Opin. Biol. Ther. 11, 569–579.
Smith, J. R., Vallier, L., Lupo, G., Alexander, M., Harris, W. A., and Pedersen, R. A. (2008). Inhibition of activin/nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol. 313, 107–117.
Smukler, S. R., Runciman, S. B., Xu, S., and Van Der Kooy, D. (2006). Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J. Cell Biol. 172, 79–90.
Son, E. Y., Ichida, J. K., Wainger, B. J., Toma, J. S., Rafuse, V. F., Woolf, C. J., and Eggan, K. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218.
Soundararajan, P., Miles, G. B., Rubin, L. L., Brownstone, R. M., and Rafuse, V. F. (2006). Motoneurons derived from embryonic stem cells express transcription factors and develop phenotypes characteristic of medial motor column neurons. J. Neurosci. 26, 3256–3268.
Stadtfeld, M., and Hochedlinger, K. (2010). Induced pluripotency: history, mechanisms, and applications. Genes Dev. 24, 2239–2263.
Stern, C. D. (2005). Neural induction: old problem, new findings, yet more questions. Development 132, 2007–2021.
Su, H. L., Muguruma, K., Matsuo-Takasaki, M., Kengaku, M., Watanabe, K., and Sasai, Y. (2006). Generation of cerebellar neuron precursors from embryonic stem cells. Dev. Biol. 290, 287–296.
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872.
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676.
Talens-Visconti, R., Sanchez-Vera, I., Kostic, J., Perez-Arago, M. A., Erceg, S., Stojkovic, M., and Guerri, C. (2011). Neural differentiation from human embryonic stem cells as a tool to study early brain development and the neuroteratogenic effects of ethanol. Stem Cells Dev. 20, 327–339.
Tao, O., Shimazaki, T., Okada, Y., Naka, H., Kohda, K., Yuzaki, M., Mizusawa, H., and Okano, H. (2010). Efficient generation of mature cerebellar Purkinje cells from mouse embryonic stem cells. J. Neurosci. Res. 88, 234–247.
Tesar, P. J., Chenoweth, J. G., Brook, F. A., Davies, T. J., Evans, E. P., Mack, D. L., Gardner, R. L., and Mckay, R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199.
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147.
Ting, D. T., Kyba, M., and Daley, G. Q. (2005). “Inducible transgene expression in mouse stem cells,” in Methods in Molecular Medicine: Developmental Hematopoiesis Methods and Protocols, ed. M. H. Baron (Totowa, NJ: Humana Press, Inc.), 23–46.
Tomishima, M. J., Hadjantonakis, A. K., Gong, S., and Studer, L. (2007). Production of green fluorescent protein transgenic embryonic stem cells using the GENSAT bacterial artificial chromosome library. Stem Cells 25, 39–45.
Tropepe, V., Hitoshi, S., Sirard, C., Mak, T. W., Rossant, J., and Van Der Kooy, D. (2001). Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron 30, 65–78.
Ulloa, F., and Marti, E. (2010). Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 239, 69–76.
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Sudhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041.
Watanabe, K., Kamiya, D., Nishiyama, A., Katayama, T., Nozaki, S., Kawasaki, H., Watanabe, Y., Mizuseki, K., and Sasai, Y. (2005). Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 8, 288–296.
Watanabe, K., Ueno, M., Kamiya, D., Nishiyama, A., Matsumura, M., Wataya, T., Takahashi, J. B., Nishikawa, S., Muguruma, K., and Sasai, Y. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686.
Wataya, T., Ando, S., Muguruma, K., Ikeda, H., Watanabe, K., Eiraku, M., Kawada, M., Takahashi, J., Hashimoto, N., and Sasai, Y. (2008). Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 11796–11801.
Wernig, M., Benninger, F., Schmandt, T., Rade, M., Tucker, K. L., Bussow, H., Beck, H., and Brustle, O. (2004). Functional integration of embryonic stem cell-derived neurons in vivo. J. Neurosci. 24, 5258–5268.
Wichterle, H., Lieberam, I., Porter, J. A., and Jessell, T. M. (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397.
Wobus, A. M., and Loser, P. (2011). Present state and future perspectives of using pluripotent stem cells in toxicology research. Arch. Toxicol. 85, 79–117.
Yan, Y., Yang, D., Zarnowska, E. D., Du, Z., Werbel, B., Valliere, C., Pearce, R. A., Thomson, J. A., and Zhang, S. C. (2005). Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells 23, 781–790.
Ying, Q. L., Stavridis, M., Griffiths, D., Li, M., and Smith, A. (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186.
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., Lee-Messer, C., Dolmetsch, R. E., Tsien, R. W., and Crabtree, G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I., and Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920.
Yusa, K., Zhou, L., Li, M. A., Bradley, A., and Craig, N. L. (2011). A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536.
Zhang, X., Huang, C. T., Chen, J., Pankratz, M. T., Xi, J., Li, J., Yang, Y., Lavaute, T. M., Li, X. J., Ayala, M., Bondarenko, G. I., Du, Z. W., Jin, Y., Golos, T. G., and Zhang, S. C. (2010). Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100.
Keywords: stem cells, pluripotent, derivation, neurons, nervous system, development, embryonic
Citation: Petros TJ, Tyson JA and Anderson SA (2011) Pluripotent stem cells for the study of CNS development. Front. Mol. Neurosci. 4:30. doi: 10.3389/fnmol.2011.00030
Received: 05 July 2011;
Accepted: 23 September 2011;
Published online: 12 October 2011.
Edited by:
Alistair N. Garratt, Max Delbrück Center for Molecular Medicine, GermanyReviewed by:
Ye He, University of California San Francisco, USASmita Jha, Baylor College of Medicine, USA
Copyright: © 2011 Petros, Tyson and Anderson. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Stewart A. Anderson, Department of Psychiatry, Weill Cornell Medical College, Box 244, 1300 York Avenue, New York, NY 10065, USA. e-mail: saa2007@med.cornell.edu