Insm1a Is Required for Zebrafish Posterior Lateral Line Development
- 1ENT Institute and Otorhinolaryngology Department of Affiliated Eye and ENT Hospital, State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai, China
- 2Key Laboratory of Hearing Medicine of NHFPC, Shanghai, China
- 3Key Laboratory for Developmental Genes and Human Disease, Ministry of Education, Institute of Life Sciences, Southeast University, Nanjing, China
- 4Jiangsu Key Laboratory of Neuroregeneration, Co-innovation Center of Neuroregeneration, Nantong University, Nantong, China
- 5Research Institute of Otolaryngology, Nanjing, China
- 6Institutes of Biomedical Sciences, Fudan University, Shanghai, China
- 7Shanghai Engineering Research Centre of Cochlear Implant, Shanghai, China
- 8The Institutes of Brain Science and the Collaborative Innovation Center for Brain Science, Fudan University, Shanghai, China
Insulinoma-associated 1 (Insm1), a zinc-finger transcription factor, is widely expressed in the developing nervous system and plays important roles in cell cycle progression and cell fate specification. However, the functions of Insm1 in the embryonic development of the sensory system and its underlying molecular mechanisms remain largely unexplored. Here, through whole-mount in situ hybridization, we found that the zebrafish insm1a gene was expressed in the posterior lateral line (pLL) system, including both the migrating pLL primordium and the deposited neuromast cells. In order to decipher the specific roles of insm1a in zebrafish pLL development, we inhibited insm1a expression by using a morpholino knockdown strategy. The insm1a morphants exhibited primordium migration defects that resulted in reduced numbers of neuromasts. The inactivation of insm1a reduced the numbers of hair cells in neuromasts, and this defect could be a secondary consequence of disrupting rosette formation in the pLL primordium. Additionally, we showed that insm1a knockdown decreased the proliferation of pLL primordium cells, which likely contributed to these pLL defects. Furthermore, we showed that loss of insm1a resulted in elevated Wnt/β-catenin signaling and downregulation of Fgf target genes in the primordium. Insm1a knockdown also perturbed the expression patterns of chemokine signaling genes. Taken together, this study reveals a pivotal role for Insm1a in regulating pLL development during zebrafish embryogenesis.
Introduction
The lateral line of zebrafish is a mechanosensory system that is implicated in several behaviors, including water movement detection and localization, schooling, predator avoidance, prey detection and capture, and so on. This system is composed of the anterior lateral line, located on the head, jaw, and opercle, and the posterior lateral line (pLL) that is distributed in a stereotypical pattern over the surface of the trunk and tail. The pLL develops from the pLL primordium (pLLp), which consists of ~100 cells and arises at the cranial placode just posterior to the otic vesicle. During early embryonic stages, the pLLp migrates caudally along the horizontal myoseptum of the embryo between 22 and 48 h post-fertilization (hpf), depositing the sensory organs called neuromasts at regular intervals and ultimately generating five or six neuromasts on the trunk and two or three terminal neuromasts at the tail (Ghysen and Dambly-Chaudiere, 2004, 2007). Each neuromast is composed of mechanosensory hair cells in the center and supporting cells at the periphery (Dambly-Chaudière et al., 2003; Ghysen and Dambly-Chaudiere, 2004). The neuromast hair cells are functionally and morphologically related to the hair cells in the vertebrate inner ear (Whitfield, 2002; Nicolson, 2005).
Morphogenesis of the pLL is a complicated and highly regulated set of processes, including pLLp directional migration, proliferation, neuromast deposition, and hair cell differentiation. All of these processes are tightly controlled by a network of signaling pathways, including chemokine, canonical Wnt/β-catenin, and fibroblast growth factor (Fgf) signaling (Aman and Piotrowski, 2008, 2009, 2011; Ma and Raible, 2009). Canonical Wnt/β-catenin and Fgf signaling are essential for pLL morphogenesis during embryonic development, and misregulation of these signaling pathways through genetic or pharmacological modulation results in multiple pLL malformations due to defects in the processes of cell proliferation, cell fate determination, and cell differentiation (Aman and Piotrowski, 2008; Lecaudey et al., 2008; Aman et al., 2011; McGraw et al., 2011). Additionally, knockdown of cxcl12a or either of its receptors cxcr4b or cxcr7b, which are expressed in the leading or trailing part of the pLLp, respectively, leads to strong migration defects (Li et al., 2004; Haas and Gilmour, 2006; Dambly-Chaudiere et al., 2007). Accumulating evidence has shown that much of the signaling regulation is accomplished by the sequential expression of cascades of transcription factors in which the activities of one set of transcription factors control some signaling pathways as well as the expression of other sets of transcription factors that in turn regulate signaling pathways. However, the specific transcription factors involved in pLL development are still largely uncharacterized.
Insulinoma-associated protein 1 (Insm1 or IA-1) is a conserved zinc-finger transcription factor that was originally cloned from a human insulinoma subtraction library (Goto et al., 1992). Insm1 mRNA is abundantly expressed in the developing central and peripheral nervous systems, in the olfactory epithelium, and in the endocrine cells (Gierl et al., 2006; Farkas et al., 2008; Wildner et al., 2008; Rosenbaum et al., 2011), and several lines of evidence indicate that insm1 expression can be controlled by the basic helix-loop-helix transcription factors (Breslin et al., 2003; Mellitzer et al., 2006). Insm1 is an important transcriptional repressor that performs multiple functions in early embryonic neurogenesis (Breslin et al., 2002). Deletion of the insm1 gene in the cortex and olfactory epithelium leads to fewer basal progenitors and consequently to fewer neurons (Farkas et al., 2008; Rosenbaum et al., 2011), and global insm1 deletion in mice results in early embryonic lethality (Gierl et al., 2006). In the zebrafish sensory system, insm1a is necessary for photoreceptor differentiation, and it regulates cell cycle progression in retinoblasts and functions upstream of the bHLH transcription factors during retinal development (Forbes-Osborne et al., 2013). In the embryonic mouse ear, insm1 is expressed in delaminating progenitor cells and in nascent spiral and vestibular ganglia (SVG) neurons (Lorenzen et al., 2015). Insm1 is also expressed in nascent outer hair cells of the embryonic mouse cochlea, but not in inner hair cells, and insm1 ablation in mice results in fewer SVG neurons and reduced proliferation of delaminated progenitors (Lorenzen et al., 2015). Despite knowledge of its specific roles in some contexts, the role of insm1 in zebrafish pLL development has not yet been studied.
Here, we show that the zebrafish insm1a gene is expressed in the developing pLL system. By knocking down insm1a with its specific morpholino, we demonstrate that disrupting the expression of insm1a results in a disorganized pLL pattern with fewer numbers of neuromasts along the body and a lack of hair cells within the neuromasts. Loss of insm1a decreased the normal proliferation of pLLp cells, and the transcription of lef1 and axin2, which are known Wnt-activated transcription factors that are important for pLL morphogenesis, were upregulated in insm1a-deficient primordia. Importantly, our results also suggest that chemokine signaling in the pLL might be regulated by Insm1a. Taken together, this work represents the first report on the function of Insm1a in regulating some signaling pathways that control pLL formation during zebrafish embryogenesis.
Materials and Methods
Zebrafish Lines and Maintenance
All zebrafish animal procedures were carried out following the institutional guidelines approved by the Institutional Animal Care and Use Committee of Fudan University, Shanghai. The pLLp and hair cells were visualized using the tg(cldnb:lynGFP) line and the tg(Brn3c:GFP) line, respectively. Embryos were obtained by natural spawning and developed at 28.5°C on a 14 h light:10 h dark cycle. Embryos were staged as previously described (Kimmel et al., 1995).
Micro Injections of Morpholinos and mRNA
For morpholino oligonucleotide (MO) knockdowns, embryos were injected with MOs at the one-cell to two-cell stage. All MOs were synthesized by GeneTools, LLC (Philomath, OR, USA). The MO injections were 6 ng of insm1a-MO (5′-AAATCCTCTGGGCATCTTCGCCAGC-3′) or 6 ng of the standard control-MO, (5′-CCTCTTACCTCAGTTACAATTTATA-3′). To avoid off-target effects, 9 ng of antisense p53-MO (5′-GCGCCATTGCTTTGCAAGAATTG-3′) was co-injected with insm1a-MO. Insm1a mRNA was transcribed with the mMESSAGE Machine Sp6 Kit (Ambion, Austin, TX, US) according to the manufacturer's instructions. For mRNA injections, 200 pg of insm1a mRNA was injected into one-cell-stage embryos and incubated at 28°C until the desired stages.
Immunohistochemistry
Embryos were fixed overnight at 4°C in 4% paraformaldehyde (PFA) and were washed with PBT-2 (PBS containing 0.5% Triton X-100) three times followed by incubation in blocking solution for 1 h at room temperature. Primary antibodies were then added to this blocking solution and incubated overnight at 4°C with rocking. The following antibodies were used as primary antibodies: anti-GFP (1:1000 dilution; Abcam, Cambridge, UK), anti-cleaved caspase-3 (1:500 dilution; Cell Signaling Technology Inc., Danvers, MA, USA), and anti-E-cadherin (1:200 dilution; BD Biosciences,USA). After three washes with PBT-2, Alexa Fluor 488– and 594–conjugated secondary antibodies (Jackson Immuno Research, West Grove, PA, USA) were all used at 1:500 dilution and incubated overnight at 4°C with rocking. Nuclei were labeled with 4,6-diamidino-2-phenylindole (DAPI; 1:1000 dilution; Invitrogen, Carlsbad, CA, USA) for 20 min at room temperature. For image collection, Z-sections were taken at 1 μm intervals through the depth of the primordium/neuromast. Maximum-intensity projections were generated for analysis, and images were processed using Photoshop software (Adobe). Cell counts were performed at the time of imaging by viewing the images under a fluorescence microscope (Eclipse; Nikon Instruments) using a 40× objective.
BrdU Injection and Immunohistochemistry
For proliferation analysis, control and insm1a morphant embryos were dechorionated and incubated with a 15 mM solution of BrdU (Sigma-Aldrich, St. Louis, MO, USA). The embryos were returned to 28°C and collected at 1 h post treatment. The embryos were anesthetized with ethyl 3-aminobenzoate methanesulfonate salt (MS-222, Tricaine, Sigma-Aldrich). The embryos were immunostained as described above, with the addition of 2N HCl for 0.5 h at 37°C prior to blocking in 10% normal goat serum. Following incubation with the monoclonal anti-BrdU primary antibody, embryos were washed three times with PBT-2 and then incubated with the secondary antibody for 1 h at 37°C. Fluorescently labeled embryos were imaged with a Leica confocal fluorescence microscope (TCS SP8; Leica, Wetzlar, Germany). Maximum-intensity projections were generated for analyses, and images were processed using Photoshop software (Adobe).
Whole-Mount In situ Hybridization
Regular whole-mount in situ hybridization (WISH) of zebrafish embryos was performed as previously described (Thisse and Thisse, 2008). Briefly, the embryos were depigmented with 1-phenyl-2-thiourea (Sigma-Aldrich), euthanized in MS-222, and fixed in 4% PFA at 4°C overnight. Fixed embryos were then washed in PBST (PBS with 0.1% Tween-20) and stored in 100% methanol at −20°C for dehydration. For in situ hybridization, embryos were rehydrated in a graded methanol series and washed three times with PBST. To permeabilize the embryos, proteinase K (20 μg/ml in PBST) was added, and the embryos were re-fixed in 4% PFA for 20 min. After washing in PBST, the embryos were prehybridized at 65°C for ≥2 h in hybridization buffer. The labeled probes were added to the hybridization buffer at 65°C overnight. Embryos were washed through a graded SSC series at 65°C before blocking for a minimum of 1 h in blocking buffer (Roche). Embryos were incubated overnight at 4°C with an anti-digoxigenin-AP Fab fragment (Roche) diluted 1:4000 in blocking buffer. The following day, the embryos were washed 4 × 30 min with 2 mg/mL BSA in PBST and equilibrated in NTMT buffer (0.1M Tris pH 9.5, 0.05M MgCl2, 0.1M NaCl, and 0.1% Tween-20). The embryos were then stained with BM purple AP substrate (Roche) in the dark. The color reaction was stopped by washing with PBST, and the embryos were then re-fixed in PFA, cleared in 75% glycerol/PBS, and imaged on a bright-field microscope (Eclipse Ti-U; Nikon Instruments, Melville, NY, USA). Sites of binding were identified as blue-black dots.
TUNEL Staining
The TUNEL (Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) cell death assay was performed as described previously (Cai et al., 2016) using the In Situ Cell Death Detection Kit, Fluorescein (Roche, Nutlet, NJ, USA; cat. no. 11684795910).
Western Blot Analysis
Embryos collected at the indicated times were lysed in RIPA buffer. The proteins were separated by SDS-PAGE and then transferred onto PVDF membranes (Immobilon-P; Millipore, Bedford, MA, USA). The membranes were then blocked with 5% nonfat dried milk in TBST (20 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 0.1% Tween-20) for 1 h at room temperature and subsequently incubated with the corresponding primary antibody overnight at 4°C. The next day, the blots were washed three times with TBST (5 min/wash) and subsequently blotted with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The reactions were detected using ECL Prime Western Blotting Detection Reagent (GE Healthcare, Wauwatosa, WI, USA).
Statistical Analysis
All data were analyzed using GraphPad Prism (version 6). Statistical analyses were performed by two-tailed Student's t-test (see figure legends for details), and a p-value < 0.05 was considered statistically significant. All data are presented as the mean ± s.e.m.
Results
Insm1a Is Expressed in the Zebrafish Lateral Line System
The insm1a gene is widely expressed in the developing nervous system during the early stages of embryonic development (Breslin et al., 2003; Forbes-Osborne et al., 2013). WISH experiments in wild-type larvae showed that insm1a was strongly expressed in the brain at 24 hpf and could be detected in the pLLp (Figure 1A). From 28 to 48 hpf, zebrafish insm1a mRNA was expressed in the pLLp throughout its migration along the body and was expressed in the deposited neuromast cells (Figures 1B,C). Immunohistochemistry for E-cadherin and WISH for insm1a confirmed that insm1a was specifically expressed in the center of the pLLp of the zebrafish embryos (Figure 1B). At 48 hpf, insm1a expression was still present in the pLL system but was beginning to decrease in the neuromasts (Figure 1C). At 72 hpf, insm1a mRNA continued to be expressed in the terminal neuromasts with reduced or no expression in the neuromasts along the trunk (Figure 1D). These results suggest that insm1a might have an important role in the regulation of embryonic zebrafish pLL development.
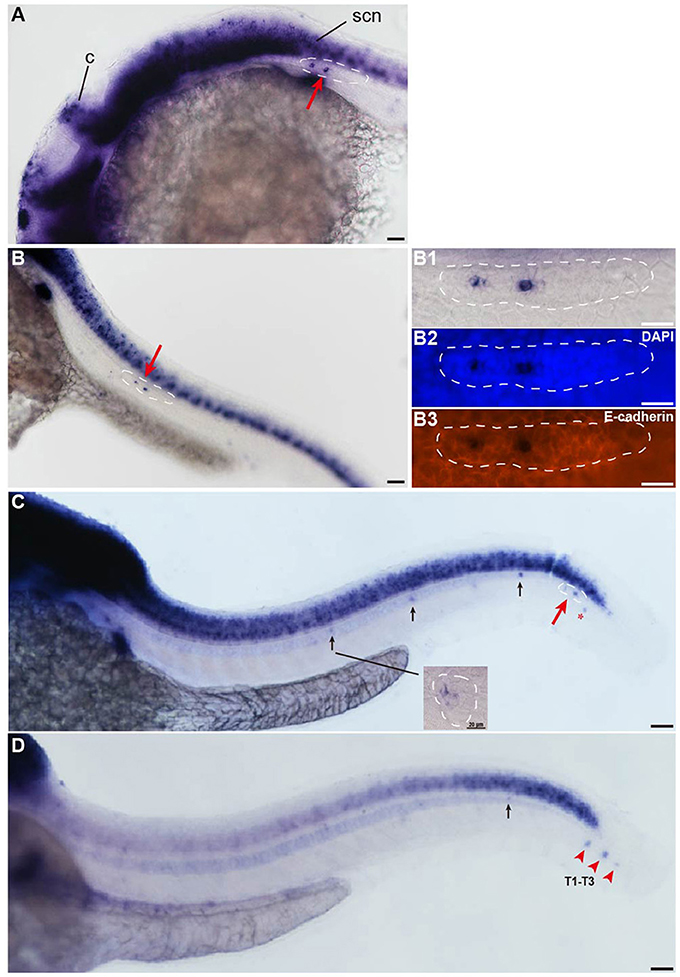
Figure 1. Insm1a is expressed in the developing pLL system. (A–D) Expression of insm1a detected by whole-mount in situ hybridization (WISH) in embryos at 24 hpf (A), 30 hpf (B), 48 hpf (C), and 72 hpf (D). Insm1a is expressed in the pLL system (Lateral view). (B1–B3) The right column of (B) shows the pLLp at higher magnification. The primordia are outlined with dotted lines and indicated by red arrows, and the neuromasts are indicated by black arrows. The red arrowheads in (D) indicate the terminal neuromasts (T1-T3), and the red asterisk in (C) indicates the primordium on the other side of the zebrafish. c, cerebellum; scn, spinal cord neuron. In all figures, scale bars are 50 μm, except the higher magnification image in (C), where the scale bar is 20 μm.
Insm1a Is Required for Embryonic pLL Morphogenesis
To investigate the function of insm1a in pLL morphogenesis,we used a translational antisense morpholino knockdown strategy to inhibit insm1a.We first assessed the efficiency of the knockdown by performing western blot on protein extracted from embryos injected with control-MO or insm1a-MO. There was a marked reduction of Insm1a protein in embryos injected with insm1a-MO compared to control-MO (Figure S1A). To exclude the general nonspecific effects of morpholinos linked to p53 activation, we co-injected embryos with p53-MO and insm1a-MO in all experiments in this study. Co-injection with control-MO and p53-MO resulted in embryos that were indistinguishable from control-MO–injected embryos, suggesting that pLL development was unaffected by loss of p53 (Figures S1B,D), thus control embryos only received control-MO in the following experiments. At 72 hpf, the majority of the insm1a morphants showed no obvious differences in terms of overall morphology (Figures S1E,F), although some insm1a morphants had slightly smaller heads, slightly smaller eyes, and slight cardiac edema. To evaluate the pLL phenotypes, tg(cldnb:lynGFP) transgenic embryos that express GFP in the pLLp and the neuromasts were injected with control-MO and insm1a-MO. At 48 hpf, the pLL system in control-MO–injected embryos contained an average of 5.48 neuromasts (n = 25) in a periodic pattern over the trunk and two or three terminal neuromasts at the tip of the tail (Figures 2A,E). However, the neuromast numbers in the pLL system were significantly reduced in insm1a-MO–injected embryos (2.35 ± 0.08 neuromasts; n = 121; Figures 2B,C,E). Because pLLp migration was usually slightly delayed at 48 hpf in insm1a morphants, we examined the number of neuromasts formed at 72 hpf. Almost all of the primordia had reached the tail-tip where the pLLp normally stops migrating, but neuromast numbers were still drastically reduced (Figure S2).
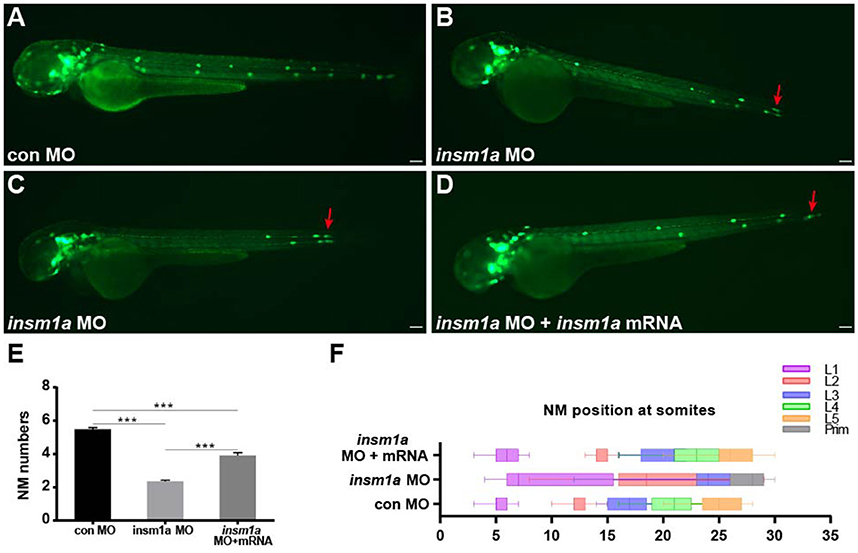
Figure 2. Insm1a is required for proper migration of the pLLp and neuromast deposition. (A–D) Fluorescent images of cldnb:lynGFP embryos at 48 hpf injected with control-MO (A), insm1a-MO (B,C), and insm1a-MO + insm1a mRNA (D). Red arrows indicate the position of the primordium. Scale bars: 100 μm. (E) Quantification of the number of neuromasts (NM) along the body at 48 hpf in control-MO (n = 25), insm1a-MO (n = 121), and insm1a-MO + insm1a mRNA (n = 24). ***p < 0.001. (F) The distribution of deposited neuromasts L1 to L5 in control-MO, insm1a-MO, and insm1a-MO + insm1a mRNA at 48 hpf.
We next focused on the positioning of neuromasts in the insm1a morphants. As illustrated in Figures 2B,C, loss of insm1a severely altered the neuromast distribution resulting in an erratic pattern compared with the regularly spaced neuromasts along the trunk of control embryos (Figure 2F) at 48 hpf. In about 55% (n = 66/121) of the morphants, the first neuromast (L1) was deposited normally, whereas the following neuromast (L2) was found at a more posterior position with the last two or three neuromasts (L3/L4–L5) often absent (Figures 2B,F). In the remaining morphants (n = 55), the L1 position was displaced very posteriorly and the last few neuromasts were often lost (Figures 2C,F). To further validate the phenotype specificity, we co-injected insm1a mRNA along with insm1a-MO. Co-injection of insm1a mRNA partially rescued the pLL defects, with the number of trunk neuromasts recovering to 3.9 (n = 24) and the neuromast positioning appearing normal (Figures 2D–F). Taken together, these results suggest that loss of insm1a disrupts proper pLL morphogenesis during early zebrafish development.
Insm1a Regulates the Development of Hair Cells
To determine whether the loss of insm1a influences hair cell formation, tg(Brn3c:GFP) transgenic embryos were injected with control-MO or insm1a-MO, and anti-GFP antibody was used to label the hair cells in the neuromasts. At 72 hpf, insm1a morphants exhibited decreased numbers of GFP-labeled hair cells compared with controls (3.2 ± 0.13 hair cells, n = 28 vs. 6.58 ± 0.16 hair cells, n = 26, p < 0.001; Figures 3A–C). To further investigate the effect of insm1a on hair cell precursors in neuromasts, we injected insm1a-MO into the wild-type embryos at the one-cell to two-cell stage. At 30 hpf, insm1a morphants were collected and subjected to WISH analysis using the probe for the proneural gene atoh1a, which is a marker for cells fated to become neuromast hair cells (Itoh and Chitnis, 2001). We observed a reduction in signal for the migrating pLLp and a reduction in the number of deposited neuromasts in insm1a morphants (Figures 3E1,E2) compared to controls (Figures 3D1,D2). These data suggest that insm1a can regulate atoh1a expression and thus promote hair cell formation.
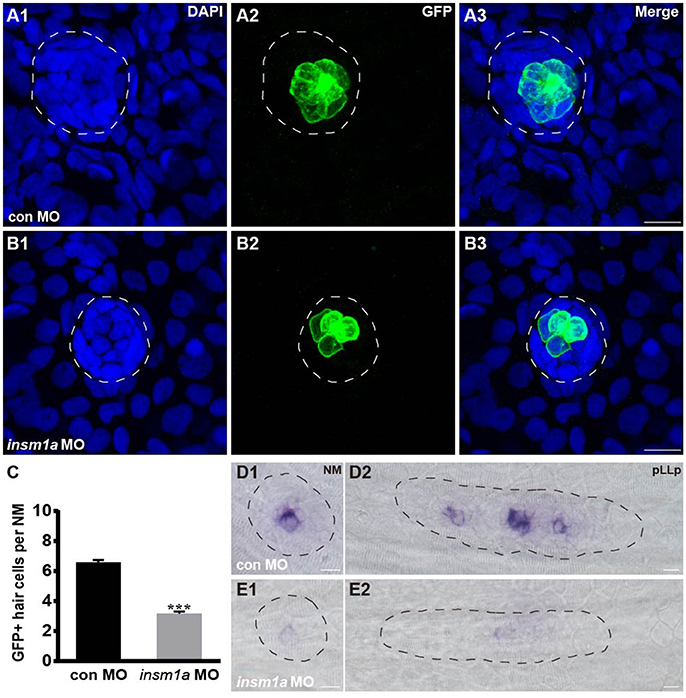
Figure 3. Insm1a regulates hair cell formation in the neuromasts. (A,B) Representative images of the GFP-expressing hair cells in 72 hpf Brn3c:GFP control embryos (A) and insm1a morphants (B). (C) Bar graph of the quantification of the average GFP-positive hair cell numbers per neuromast (NM) for each group. Data are expressed as means ± s.e.m. ***p < 0.001. (D,E) Lateral views of neuromasts and primordia stained by WISH with an atoh1a probe at 30 hpf. The primordium and neuromast are outlined with dotted lines. In all figures, scale bars are 10 μm.
Insm1a Regulates Cell Proliferation in the Primordium
The number of epithelial rosettes was significantly reduced in insm1a morphants at 32 hpf when compared with control primordia, which normally contained two to four rosettes (white arrows in Figures 4A–C). This suggests that insm1a is required for rosette assembly in the migrating pLLp. Loss of insm1a decreased the size of the pLLp throughout the migration process, and cell counts showed a significant decrease in cell numbers in insm1a morphant pLL primordia compared to control pLL primordia (76.5 ± 3.7 cells, n = 10 vs. 94.6 ± 3.66 cells, n = 14, p = 0.0026; Figure 4D). Because previous studies showed that Insm1 is involved in cell proliferation by interacting with cell cycle regulatory proteins (Liu et al., 2006; Zhang et al., 2009), we asked whether reduced cell proliferation is the cause of the reduced cell counts in the primordia of insm1a morphants. We performed a bromodeoxyuridine (BrdU) incorporation assay to determine the proliferation status of the pLLp in insm1a morphants and control embryos at 32 hpf. There was a significant decrease in the BrdU index in the insm1a pLLp compared to controls (28.06 ± 1.92%, n = 10 vs. 41.2 ± 2.02%, n = 14, p = 0.0002; Figures 4A,B,E). To determine if insm1a knockdown induces cell death, insm1a morphants were allowed to develop until 32 hpf and were analyzed for apoptosis with the TUNEL assay and caspase-3 immunolabeling. No significant apoptosis was detected either in control primordia or in insm1a primordia (Figure S3). Taken together, our results indicated that insm1a is likely an important regulator of cell proliferation during pLL development.
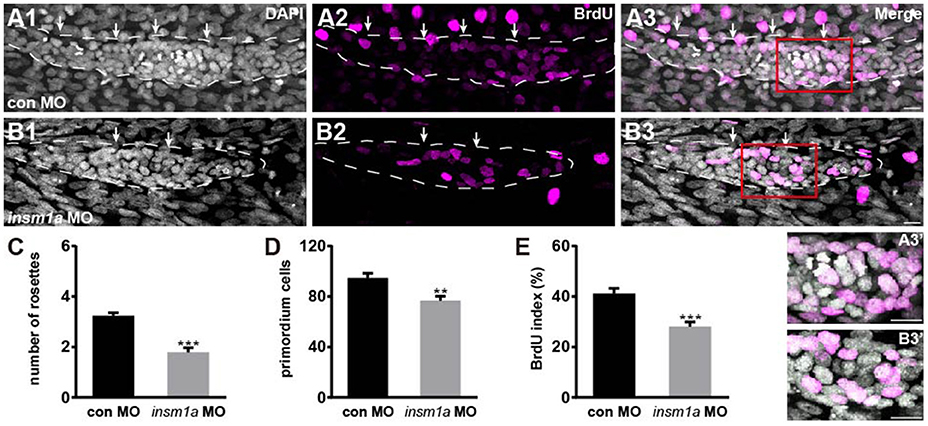
Figure 4. Insm1a is required for cell proliferation in the migrating pLLp. (A,B) Representative images of the BrdU immunostaining for proliferating cells in the pLLp in controls (A1–A3) and insm1a morphants (B1–B3) at 32 hpf showing that knocking down of insm1a impairs rosette assembly and cell proliferation. The primordium is outlined with a dotted line. (A3′–B3′) BrdU-positive cells in the rosette at high magnification. White arrows indicate rosettes. Scale bars: 10 μm. (C–E) Quantification of rosette numbers (C), cell numbers (D), and BrdU+ primordium cells (BrdU index) (E) in controls and insm1a morphants. Data are expressed as mean ± s.e.m. ***p < 0.001; **p < 0.01.
Insm1a Is Required for cxcr12a and cxcr4b Expression in the Primordium
In zebrafish, chemokine signaling pathways play central roles in the directional migration of the pLLp (Haas and Gilmour, 2006). The chemokine ligand cxcl12a is expressed along the horizontal myoseptum of zebrafish, and the receptors cxcr4b and cxcr7b are normally polarized to the leading and trailing zone of the migrating primordium, respectively (Valentin et al., 2007). Given the severe primordium migration defects in insm1a morphants, we asked whether Insm1a functions in the control of pLL morphogenesis by regulating chemokine signaling. We examined the expression of a suite of region-specific chemokine markers during the early developmental stage, and WISH analysis showed that the expression of the myoseptum-expressed cxcl12a was altered in insm1a morphants compared to controls (Figures 5A,B). The insm1a-MO embryos showed a broad expansion of cxcr4b expression (Figures 5C–E), whereas the expression domains of cxcr7b were reduced in the primordia of these embryos at 30–32 hpf (Figures 5F–H). Thus, we speculated that Insm1a might regulate pLL morphogenesis in part by modulating chemokine gene expression and thus controlling primordium migration.
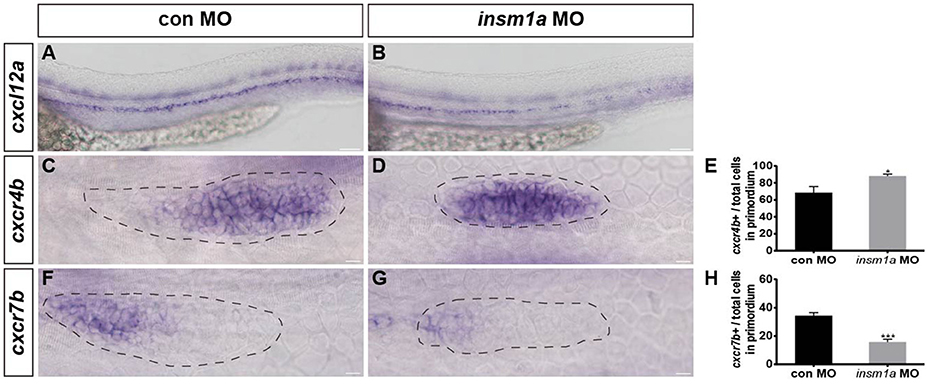
Figure 5. Insm1a regulates the expression of chemokine signaling components. WISH was used to detect the expression of cxcl12a (A,B), cxcr4b (C,D), and cxcr7b (F,G) in control and insm1a morphants at 30–32 hpf. (E,H) The ratio of cxcr4b-positive cells (E) or cxcr7b-positive cells (H) to total cells in the primordia of controls (black bars) and insm1a morphants (gray bars). Data are expressed as mean ± s.e.m. *p < 0.05; ***p < 0.001. The cxcl12a expression is reduced in insm1a morphants, while cxcr4b expression is up-regulated and cxcr7b expression is down-regulated in insm1a morphants compared to controls. The primordium is outlined with a dotted line, and the anterior is to the left. Scale bar: 50 μm (A,B) and 10 μm (C–F).
Loss of insm1a Affects Wnt and Fgf Signaling in the Primordium
Wnt/β-catenin plays central roles in regulating the migration and proliferation of the primordium during early pLL development (Aman and Piotrowski, 2008; Aman et al., 2011), and Wnt/β-catenin signaling is thought to restrict the localized expression pattern of the chemokine receptors cxcr4b and cxcr7b to the leading and trailing edges of the primordium, respectively (Aman and Piotrowski, 2008). To determine whether Insm1a functions in the control of pLL morphogenesis through regulation of the Wnt/β-catenin signaling pathway, we first examined the expression of lef1, a direct Wnt target gene, whose expression is normally restricted to the leading-edge cells in the migrating primordium, as shown in control embryos (Figure 6A). In contrast, in insm1a morphants, the expression of lef1 significantly expanded, although it was still restricted to the leading-edge cells of the primordium compared to controls (Figures 6B,C). Likewise, the expression area of axin2 was also expanded in the primordium of insm1a morphants as compared to controls (Figures 6D–F). These findings indicate that the insm1a gene is a negative regulator of Wnt/β-catenin signaling in the control of pLL development in zebrafish. Because the correct patterning and migration of the pLLp depends on the interaction between Wnt/β-catenin and Fgf signaling, we next asked whether the pLL phenotypes observed in insm1a morphants were also associated with the Fgf signaling pathway. WISH showed that loss of insm1a had no significant effect on the expression levels of fgf3 (Figure 6H) or fgf10a (Figure 6J) compared to controls (Figures 6G,I). In contrast, embryos injected with insm1a-MO showed markedly reduced expression of pea3, a Fgf target gene, in the primordium (Figures 6K,L).
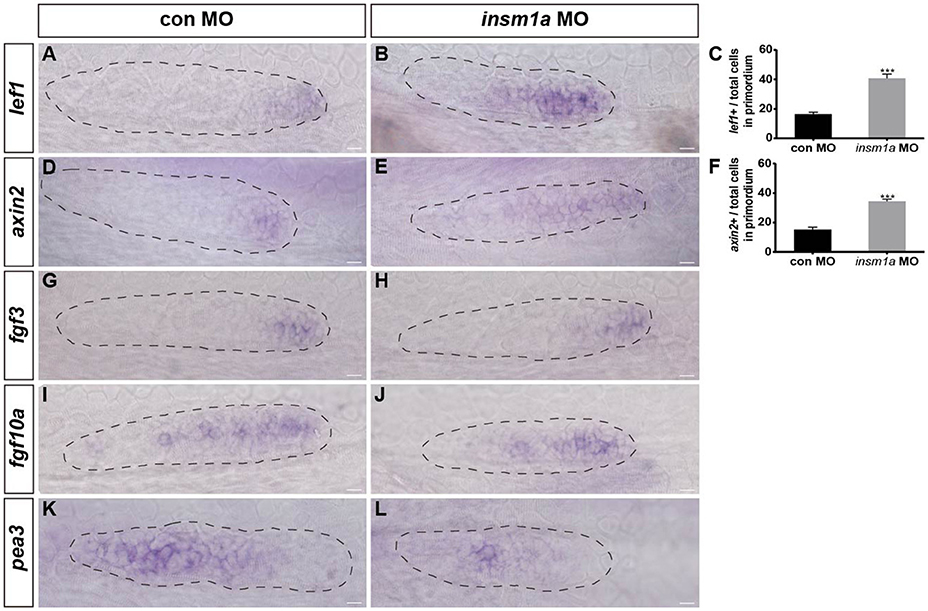
Figure 6. Insm1a regulates the expression of Wnt and Fgf signaling components. Expression of the Wnt and Fgf target genes lef1 (A,B), axin2 (D,E), fgf3 (G,H), fgf10a (I,J), and pea3 (K,L) in control embryos and in insm1a morphants at 30–32 hpf. (C,F) The ratio of lef1-positive cells (C) or axin2-positive cells (F) to total cells in the primordia of controls (black bars) and insm1a morphants (gray bars). Data are expressed as mean ± s.e.m. ***p < 0.001. In insm1a primordia, the expression domains of lef1 and axin2 expand, whereas fgf3 and fgf10a expression is normal but pea3 is reduced. The primordium is outlined with a dotted line, and the anterior is to the left. Scale bar: 10 μm.
Discussion
In the developing mouse ear, insm1 is expressed in the delaminating and delaminated neuronal progenitors and promotes their proliferation along with SVG neurogenesis beginning at embryonic day (E)10.5 (Lorenzen et al., 2015). Insm1 mutant mice have fewer cell divisions and thus produce fewer neurons within the otic ganglia at developmental stages after E10.5. However, whether insm1 is expressed in the auditory organs of other model organisms, and whether it plays a role in driving the specific differentiation of these sensory organs, are not known. In this study, we identify the importance of Insm1a function in normal pLL development in zebrafish. We showed that insm1a was expressed in the embryonic pLLp and deposited neuromasts, implying that insm1a might play important roles in many aspects of pLL development. Indeed, using morpholino antisense technology, our findings indicated that insm1a not only regulated pLL formation but also contributed to the differentiation of hair cells. Furthermore, we showed that insm1a is required for the normal expression patterning of chemokine, Wnt/β-catenin, and Fgf signaling in the primordium. Thus, our findings identify transcription factor Insm1a as a novel regulator in pLL development.
In zebrafish, insm1a is expressed throughout the developing nervous system, including the forebrain, midbrain, hindbrain, olfactory placode, cerebellum, and retina, and it is an important player in early embryonic neurogenesis (Lukowski et al., 2006). Our WISH analysis demonstrated that zebrafish insm1a mRNA was transiently expressed in the embryonic pLL system, including the migrating primordium and the newly deposited neuromasts, providing new lines of evidence for a role for insm1a in normal sensory organ development. The timing of peak insm1a expression in the zebrafish pLL system corresponds to the period of primary pLL formation. The signal for insm1a was significantly reduced in the neuromasts spaced along the trunk later in development when these neuromasts mature into mechanosensory neuromasts. Our findings are consistent with reports that the insm1 transcript is transiently expressed in the neuronal progenitors and nascent neurons throughout the developing nervous system and is dramatically decreased after birth (Duggan et al., 2008; Farkas et al., 2008).
In order to investigate the role of insm1a in pLL development, we used a translation-blocking antisense morpholino to knockdown insm1a expression in zebrafish. Our results demonstrated that insm1a gene knockdown resulted in severe pLL developmental defects. Because insm1a morphants did not exhibit significant morphological abnormalities during early development, the pLL defects in the morphants are probably due to deficiency of insm1a. In insm1a-deficient embryos, we observed fewer cells in the migrating primordium when compared with controls. We performed a TUNEL assay and caspase-3 immunohistochemistry to assess the level of cell death in control embryos and insm1a morphants. No significant changes were observed in the insm1a morphants, demonstrating that the decrease in cell numbers is unlikely to be the result of increased apoptosis. Another potential explanation for the reduced cell numbers in insm1a morphants is that loss of insm1a might alter cell cycle progression. We next examined the proliferation level in the pLLp by using the BrdU incorporation assay for labeling S-phase cells. We found fewer proliferating cells in the insm1a primordia compared to controls. Our results suggest that Insm1a is involved in cell cycle progression. As a transcription factor, Insm1a has been shown to be a critical regulator in the transcriptional network that mediates various cellular processes such as cell proliferation, cell specification, and cell fates (Liu et al., 2006; Farkas et al., 2008; Wildner et al., 2008; Zhang et al., 2009). For example, Insm1a has been implicated in regulating the transcription of cell cycle genes in the mouse neocortex (Farkas et al., 2008), the secondary sympathetic ganglia (Wildner et al., 2008), and the developing and regenerating zebrafish retina (Ramachandran et al., 2012; Forbes-Osborne et al., 2013). Previous reports have shown that Insm1 is involved in modulating transcriptional activity through recruitment of cyclin D1 and histone deacetylases (Liu et al., 2006). Cyclin D1 plays an important role in transcriptional regulation through its association with CDK4 (Sherr, 1996), and cyclin D1-deficient mice have reduced body size and suffer from neurological impairment (Sicinski et al., 1995). Whether the molecular mechanism of insm1 transcriptional activity in cell cycle functions is due to the recruitment of cyclin D1 or other unknown major components remains unclear and will be investigated in future studies.
The process of pLLp migration is mediated by a network of signaling pathways, including chemokine, canonical Wnt/β-catenin, and Fgf signaling. Among these, chemokine signaling is important in guiding the directed collective migration of the pLLp (David et al., 2002; Li et al., 2004; Dambly-Chaudiere et al., 2007). The chemokine receptors cxcr4b and cxcr7b are expressed in the leading and trailing zones of the primordium, respectively, and their ligand cxcl12a is expressed along the horizontal myoseptum of the zebrafish (David et al., 2002; Haas and Gilmour, 2006; Valentin et al., 2007). Knockdown of cxcl12a or its receptor cxcr4b or cxcr7b leads to strong defects in primordium migration (David et al., 2002; Li et al., 2004; Haas and Gilmour, 2006; Dambly-Chaudiere et al., 2007; Valentin et al., 2007). To understand the effects of the loss of insm1a on the chemokine signaling pathway, we examined the expression of cxcl12a, cxcr4b, and cxcr7b. We found that insm1a knockdown led to reduced cxcl12a expression along the myoseptum and led to an expansion of cxcr4b expression into the trailing part of the pLLp. It also led to a loss of cxcr7b expression, although this was not completely abolished compared with control embryos. Our findings show that insm1a is involved in the regulation of chemokine signaling in the primordium during development. It has been shown previously that inactivation of Cxcl12a signaling by cxcl12a-MO results in a significant expansion of cxcr7b expression (Gamba et al., 2010a). However, the expanded expression of cxcr7b never extended to the anteriormost cells of the primordium, suggesting that other factors can regulate cxcr7b independently of Cxcl12a/cxcr4 signaling (Gamba et al., 2010a). A marked decrease in the expression of cxcr4b in the absence of cxcl12 was also observed (Gamba et al., 2010a). Interestingly, cxcr4b still exists in the leading domain, even if the pLLp migrates away from its normal migratory path in cxcl12a morphants, suggesting that expression of cxcr4b might be under the direct control of other important signaling pathways, perhaps Wnt/β-catenin signaling. The disruption of cxcr4b and cxcr7b expression in insm1a-MO embryos might reflect a direct requirement for insm1a in Cxcl12a/cxcr4 signaling or might be a consequence of some other ectopic signaling expression in the primordium. It would be important to verify these proposals in the future by genetic approaches (e.g., inactivation of cxcl12a, cxcr4b, or cxcr7b) in combination with other approaches such as next-generation sequencing and bioinformatics analysis.
Proper pLLp migration also depends on the Wnt/β-catenin and Fgf signaling feedback system (Aman and Piotrowski, 2008, 2009, 2011). Wnt/β-catenin signaling is active in the leading region of the primordium, where it controls the expression of the Wnt targets lef1 and axin2 and regulates multiple aspects of pLL development, including primordium migration, proneuromast formation, and cell proliferation in the primordium (Aman and Piotrowski, 2008; Gamba et al., 2010b; Aman et al., 2011; McGraw et al., 2011; Valdivia et al., 2011). Wnt/β-catenin signaling controls primordium migration by coordinating the localized expression of chemokine receptors cxcr4b and cxcr7. Wnt activity confines cxcr7b expression to the trailing part of the pLLp and is required for maintenance of normal cxcr4b expression in the leading part of the pLLp (Aman and Piotrowski, 2008; McGraw et al., 2011; Breau et al., 2013). Activation of the Fgf signaling pathway in the trailing zone of the primordium triggers morphogenesis of the apically constricted rosettes within the primordium and restricts the activation of Wnt activity to the leading zone. Wnt activation, in turn, is necessary for the expression of the Fgf ligands fgf3 and fgf10a in the leading zone of the primordium (Lecaudey et al., 2008; Nechiporuk and Raible, 2008). Activation of the Wnt pathway and/or loss of Fgf signaling results in cxcr7b downregulation and cxcr4b expansion, which contribute to aberrant pLL development, including disruptions in rosette organization and proneuromast deposition (Aman and Piotrowski, 2008; Nechiporuk and Raible, 2008). It is informative to compare these results with the expression and function of Insm1a. We investigated the effects of loss of insm1a expression on Wnt/Fgf signaling and found that the expression of lef1 and axin2 were markedly increased in the insm1a morphant primordia. To investigate the effects of insm1a expression on Fgf signaling, we analyzed the expression of fgf3, fgf10a, and pea3, which are necessary for proper patterning and migration of the primordium and for neuromast formation and deposition (Aman and Piotrowski, 2008; Lecaudey et al., 2008; Nechiporuk and Raible, 2008). We found that the expression of both the fgf3 and fgf10a genes appeared unchanged in insm1a morphants, but there was a marked reduction in pea3 expression. However, it is still unclear whether insm1a is directly or indirectly involved in regulating Wnt/Fgf signaling in the pLLp, and the detailed mechanisms await further investigation.
We have shown here that zebrafish insm1a mRNA was transiently expressed during the entire pLLp migration process. Because E-cadherin immunohistochemistry and WISH with an insm1a antisense probe showed that insm1a was not localized to the leading or trailing cells but was specifically present in the central part of the primordium of the zebrafish embryos, we hypothesized that Insm1a is required for rosette assembly and regulates hair cell formation during pLL development. To test this hypothesis, we knocked down insm1a expression and showed that loss of insm1a significantly reduced the numbers of rosettes in the primordium of insm1a morphants compared to controls. As a consequence, the final pattern of pLL was aberrant with very few neuromasts being deposited from the primordia and those that were deposited lacking organized rosettes. Neuromast maturation is another important aspect of the development of the pLL system, and in control embryos the numbers of hair cells in the pLL neuromast increased over time. When we injected insm1a-MO into the Brn3c:GFP transgenic line, which express GFP in the neuromast hair cells, we observed a significant decrease in the number of hair cells. Our findings also support a recent study documenting the function of Insm1 in delaminating progenitors, nascent neurons, and differentiating outer hair cells of the embryonic mouse inner ear (Lorenzen et al., 2015). Deletion of insm1 resulted in fewer SVG neurons during otic neurogenesis, suggesting that Insm1 plays a functional role in the differentiation of the nascent neurons. However, that study did not report any obvious abnormalities in the differentiation of hair cells upon the loss of insm1. This might be partly due to the embryonic lethality of the insm1-null mouse. In our study, because the insm1a transcript was expressed transiently in neuromasts during the early developmental stages in zebrafish, we hypothesized that the effect of insm1a knockdown on hair cell formation is probably a secondary consequence of the absence of organized rosettes in the primordium. Previous studies have shown that atoh1a is typically restricted to central cells in the pLLp, which gives central cell clusters the ability to develop into sensory hair cells (Lecaudey et al., 2008; Nechiporuk and Raible, 2008). In our study, we observed that atoh1a expression was lost in some hair cell clusters of both the migrating pLLp and neuromasts in insm1a morphants, suggesting that insm1a might regulate hair cell formation by regulating atoh1a expression.
In summary, our study suggests an important role for the transcription factor Insm1a in zebrafish pLL development. We show that Insm1a is required for the proliferation of pLLp cells and the development of hair cells. We further reveal the association between Insm1a and Wnt signaling in regulating pLL formation. Further analysis is needed to determine whether Insm1a is also involved in other signaling pathways and whether it regulates the expression of other transcription factors involved in primordium migration and neuromast differentiation. Identifying the direct targets of Insm1a during pLL development will provide helpful insights into the underlying mechanisms of hearing development and hearing-related diseases.
Author Contributions
YH, RC, HL, and DL conceived and designed the work. YH and RC wrote the manuscript. YH, XL, and FQ performed the zebrafish experiments. HL, XL, FQ, and DL performed data analyses. All authors discussed the data, and all authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Min Yu for their technical assistance and Yalin Huang for help with the confocal microscope. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0103900, 2015CB965000), the National Natural Science Foundation of China (Nos. 81620108005, 81500784, 81230019, 81470687, 81622013, 31500852, 81470692), the Jiangsu Province Natural Science Foundation (BK20150022, BK20140620, BK20160125), the Biomedical Program of Shanghai Committee of Science and Technology (14DJ1400203), the National Science and Technology Major Project (2016YFC0905200), the Boehringer Ingelheim Pharma GmbH, the Yingdong Huo Education Foundation, and the Fundamental Research Funds for the Central Universities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fnmol.2017.00241/full#supplementary-material
Figure S1. (A) Western blot detection of Insm1a protein in embryos injected with 6 ng insm1a antisense morpholino or 6 ng control morpholino showing a marked reduction of Insm1a protein in the insm1a morphants. The experiment was repeated two times. (B,C) Fluorescent images of cldnb:lynGFP embryos at 48 hpf injected with control-MO (con MO) (B) and con MO + p53 MO (C). Scale bars: 100 μm. (D) Quantification of the number of neuromasts along the body at 48 hpf in con MO (n = 20) and con MO + p53 MO (n = 20). There was no significant difference (p > 0.05). (E,F) Transmitted light images of 72 hpf control morpholino-injected embryo (con MO) (E) or insm1a morphants (insm1a MO) (F). The overall morphology of the insm1a morphants was indistinguishable from controls. Scale bars: 100 μm.
Figure S2. Insm1a knockdown shows a reduction in the number of pLL neuromasts. (A,B) Fluorescent images of cldnb:lynGFP embryos at 72 hpf injected with control-MO and insm1a-MO. Scale bars: 100 μm. (C) Quantification of the number of neuromasts along the body at 72 hpf in control-MO (n = 20) and insm1a-MO (n = 38). ***p < 0.001.
Figure S3. Loss of insm1a has no significant effect on cell death. (A,B) TUNEL assay on sections from a control (A) and insm1a morphant (B) at 32 hpf. (C,D) Representative images of cleaved caspase-3 immunohistochemistry in primordia from a control (C) and insm1a morphant (D). No significant difference in the number of apoptotic cells in the insm1a morphant was detected compared to controls. Scale bar: 100 μm (A,B) and 10 μm (C,D).
References
Aman, A., and Piotrowski, T. (2008). Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev. Cell 15, 749–761. doi: 10.1016/j.devcel.2008.10.002
Aman, A., and Piotrowski, T. (2009). Multiple signaling interactions coordinate collective cell migration of the posterior lateral line primordium. Cell Adh. Migr. 3, 365–368. doi: 10.4161/cam.3.4.9548
Aman, A., and Piotrowski, T. (2011). Cell-cell signaling interactions coordinate multiple cell behaviors that drive morphogenesis of the lateral line. Cell Adh. Migr. 5, 499–508. doi: 10.4161/cam.5.6.19113
Aman, A., Nguyen, M., and Piotrowski, T. (2011). Wnt/beta-catenin dependent cell proliferation underlies segmented lateral line morphogenesis. Dev. Biol. 349, 470–482. doi: 10.1016/j.ydbio.2010.10.022
Breau, M. A., Wilkinson, D. G., and Xu, Q. (2013). A hox gene controls lateral line cell migration by regulating chemokine receptor expression downstream of Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 16892–16897. doi: 10.1073/pnas.1306282110
Breslin, M. B., Zhu, M., and Lan, M. S. (2003). NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J. Biol. Chem. 278, 38991–38997. doi: 10.1074/jbc.M306795200
Breslin, M. B., Zhu, M., Notkins, A. L., and Lan, M. S. (2002). Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 30, 1038–1045. doi: 10.1093/nar/30.4.1038
Cai, C., Lin, J., Sun, S., and He, Y. (2016). JNK inhibition inhibits lateral line neuromast hair cell development. Front. Cell. Neurosci. 10:19. doi: 10.3389/fncel.2016.00019
Dambly-Chaudiere, C., Cubedo, N., and Ghysen, A. (2007). Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev. Biol. 7:23. doi: 10.1186/1471-213X-7-23
Dambly-Chaudière, C., Sapède, D., Soubiran, F., Decorde, K., Gompel, N., and Ghysen, A. (2003). The lateral line of zebrafish: a model system for the analysis of morphogenesis and neural development in vertebrates. Biol. Cell 95, 579–587. doi: 10.1016/j.biolcel.2003.10.005
David, N. B., Sapede, D., Saint-Etienne, L., Thisse, C., Thisse, B., Dambly-Chaudière, C., et al. (2002). Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc. Natl. Acad. Sci. U.S.A. 99, 16297–16302. doi: 10.1073/pnas.252339399
Duggan, A., Madathany, T., de Castro, S. C., Gerrelli, D., Guddati, K., García-Añoveros, J., et al. (2008). Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J. Comp. Neurol. 507, 1497–1520. doi: 10.1002/cne.21629
Farkas, L. M., Haffner, C., Giger, T., Khaitovich, P., Nowick, K., Birchmeier, C., et al. (2008). Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55. doi: 10.1016/j.neuron.2008.09.020
Forbes-Osborne, M. A., Wilson, S. G., and Morris, A. C. (2013). Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Dev. Biol. 380, 157–171. doi: 10.1016/j.ydbio.2013.05.021
Gamba, L., Cubedo, N., Ghysen, A., Lutfalla, G., and Dambly-Chaudière, C. (2010a). Estrogen receptor ESR1 controls cell migration by repressing chemokine receptor CXCR4 in the zebrafish posterior lateral line system. Proc. Natl. Acad. Sci. U.S.A. 107, 6358–6363. doi: 10.1073/pnas.0909998107
Gamba, L., Cubedo, N., Lutfalla, G., Ghysen, A., and Dambly-Chaudiere, C. (2010b). Lef1 controls patterning and proliferation in the posterior lateral line system of zebrafish. Dev. Dyn. 239, 3163–3171. doi: 10.1002/dvdy.22469
Ghysen, A., and Dambly-Chaudiere, C. (2004). Development of the zebrafish lateral line. Curr. Opin. Neurobiol. 14, 67–73. doi: 10.1016/j.conb.2004.01.012
Ghysen, A., and Dambly-Chaudiere, C. (2007). The lateral line microcosmos. Genes Dev. 21, 2118–2130. doi: 10.1101/gad.1568407
Gierl, M. S., Karoulias, N., Wende, H., Strehle, M., and Birchmeier, C. (2006). The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 20, 2465–2478. doi: 10.1101/gad.381806
Goto, Y., De Silva, M. G., Toscani, A., Prabhakar, B. S., Notkins, A. L., and Lan, M. S. (1992). A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J. Biol. Chem. 267, 15252–15257.
Haas, P., and Gilmour, D. (2006). Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev. Cell 10, 673–680. doi: 10.1016/j.devcel.2006.02.019
Itoh, M., and Chitnis, A. B. (2001). Expression of proneural and neurogenic genes in the zebrafish lateral line primordium correlates with selection of hair cell fate in neuromasts. Mech. Dev. 102, 263–266. doi: 10.1016/S0925-4773(01)00308-2
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Lecaudey, V., Cakan-Akdogan, G., Norton, W. H., and Gilmour, D. (2008). Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development 135, 2695–2705. doi: 10.1242/dev.025981
Li, Q., Shirabe, K., and Kuwada, J. Y. (2004). Chemokine signaling regulates sensory cell migration in zebrafish. Dev. Biol. 269, 123–136. doi: 10.1016/j.ydbio.2004.01.020
Liu, W. D., Wang, H. W., Muguira, M., Breslin, M. B., and Lan, M. S. (2006). INSM1 functions as a transcriptional repressor of the neuroD/beta2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem. J. 397, 169–177. doi: 10.1042/BJ20051669
Lorenzen, S. M., Duggan, A., Osipovich, A. B., Magnuson, M. A., and García-Añoveros, J. (2015). Insm1 promotes neurogenic proliferation in delaminated otic progenitors. Mech. Dev. 138, 233–245. doi: 10.1016/j.mod.2015.11.001
Lukowski, C. M., Ritzel, R. G., and Waskiewicz, A. J. (2006). Expression of two insm1-like genes in the developing zebrafish nervous system. Gene Expr. Patterns 6, 711–718. doi: 10.1016/j.modgep.2005.12.008
Ma, E. Y., and Raible, D. W. (2009). Signaling pathways regulating zebrafish lateral line development. Curr. Biol. 19, R381–R386. doi: 10.1016/j.cub.2009.03.057
McGraw, H. F., Drerup, C. M., Culbertson, M. D., Linbo, T., Raible, D. W., and Nechiporuk, A. V. (2011). Lef1 is required for progenitor cell identity in the zebrafish lateral line primordium. Development 138, 3921–3930. doi: 10.1242/dev.062554
Mellitzer, G., Bonne, S., Luco, R. F., Van De Casteele, M., Lenne-Samuel, N., Collombat, P., et al. (2006). IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 25, 1344–1352. doi: 10.1038/sj.emboj.7601011
Nechiporuk, A., and Raible, D. W. (2008). FGF-dependent mechanosensory organ patterning in zebrafish. Science 320, 1774–1777. doi: 10.1126/science.1156547
Nicolson, T. (2005). The genetics of hearing and balance in zebrafish. Annu. Rev. Genet. 39, 9–22. doi: 10.1146/annurev.genet.39.073003.105049
Ramachandran, R., Zhao, X. F., and Goldman, D. (2012). Insm1a-mediated gene repression is essential for the formation and differentiation of Muller glia-derived progenitors in the injured retina. Nat. Cell Biol. 14, 1013–1023. doi: 10.1038/ncb2586
Rosenbaum, J. N., Duggan, A., and Garcia-Anoveros, J. (2011). Insm1 promotes the transition of olfactory progenitors from apical and proliferative to basal, terminally dividing and neuronogenic. Neural Dev. 6:6. doi: 10.1186/1749-8104-6-6
Sicinski, P., Donaher, J. L., Parker, S. B., Li, T., Fazeli, A., Gardner, H., et al. (1995). Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82, 621–630. doi: 10.1016/0092-8674(95)90034-9
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Valdivia, L. E., Young, R. M., Hawkins, T. A., Stickney, H. L., Cavodeassi, F., Schwarz, Q., et al. (2011). Lef1-dependent Wnt/beta-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development. Development 138, 3931–3941. doi: 10.1242/dev.062695
Valentin, G., Haas, P., and Gilmour, D. (2007). The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr. Biol. 17, 1026–1031. doi: 10.1016/j.cub.2007.05.020
Whitfield, T. T. (2002). Zebrafish as a model for hearing and deafness. J. Neurobiol. 53, 157–171. doi: 10.1002/neu.10123
Wildner, H., Gierl, M. S., Strehle, M., Pla, P., and Birchmeier, C. (2008). Insm1 (IA-1) is a crucial component of the transcriptional network that controls differentiation of the sympatho-adrenal lineage. Development 135, 473–481. doi: 10.1242/dev.011783
Keywords: insulinoma-associated 1, zebrafish, posterior lateral line primordium, hair cell, Wnt/β-catenin signaling
Citation: He Y, Lu X, Qian F, Liu D, Chai R and Li H (2017) Insm1a Is Required for Zebrafish Posterior Lateral Line Development. Front. Mol. Neurosci. 10:241. doi: 10.3389/fnmol.2017.00241
Received: 22 May 2017; Accepted: 17 July 2017;
Published: 02 August 2017.
Edited by:
Hong-Bo Zhao, University of Kentucky, United StatesReviewed by:
Angelika Doetzlhofer, Johns Hopkins University, United StatesPhilip Forsyth Copenhaver, Oregon Health & Science University, United States
Copyright © 2017 He, Lu, Qian, Liu, Chai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renjie Chai, renjiec@seu.edu.cn
Huawei Li, hwli@shmu.edu.cn
†These authors have contributed equally to this work.
 Yingzi He
Yingzi He Xiaoling Lu
Xiaoling Lu Fuping Qian
Fuping Qian Dong Liu
Dong Liu Renjie Chai
Renjie Chai Huawei Li
Huawei Li