The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice
- 1State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China
- 3School of Life Sciences, University of Science and Technology of China, Hefei, China
- 4The State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 5Department of Orthopaedic Surgery, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 6The Solomon H. Snyder Department of Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Histone demethylase UTX mediates removal of repressive trimethylation of histone H3 lysine 27 (H3K27me3) to establish a mechanistic switch to activate large sets of genes. Mutation of Utx has recently been shown to be associated with Kabuki syndrome, a rare congenital anomaly syndrome with dementia. However, its biological function in the brain is largely unknown. Here, we observe that deletion of Utx results in increased anxiety-like behaviors and impaired spatial learning and memory in mice. Loss of Utx in the hippocampus leads to reduced long-term potentiation and amplitude of miniature excitatory postsynaptic current, aberrant dendrite development and defective synapse formation. Transcriptional profiling reveals that Utx regulates a subset of genes that are involved in the regulation of dendritic morphology, synaptic transmission, and cognition. Specifically, Utx deletion disrupts expression of neurotransmitter 5-hydroxytryptamine receptor 5B (Htr5b). Restoration of Htr5b expression in newborn hippocampal neurons rescues the defects of neuronal morphology by Utx ablation. Therefore, we provide evidence that Utx plays a critical role in modulating synaptic transmission and cognitive behaviors. Utx cKO mouse models like ours provide a valuable means to study the underlying mechanisms of the etiology of Kabuki syndrome.
Introduction
Accumulating evidence suggests that epigenetic regulations play critical roles in neurological disorders (Tsankova et al., 2007; Ma et al., 2010). Trimethylation at Lysine 27 of histone H3 (H3K27me3) establishes a repressive chromatin state in silencing an array of crucial genes, which contributes to important biological processes including X-inactivation, genomic imprinting, stem cell maintenance, circadian rhythms, and cancer (Plath et al., 2003; Etchegaray et al., 2006; Sparmann and van Lohuizen, 2006; Van der Meulen et al., 2014). In mammals, the dynamic steady-state levels of dimethylation and trimethylation of histone H3 lysine 27 (H3K27me2/3) are mainly maintained through balance between methyltransferase Polycomb Repressor Complex 2 (PRC2) and the demethylases UTX (also known as KDM6A) and Jumonji D3 (JMJD3, also known as KDM6B) (Agger et al., 2007; Lee et al., 2007; Van der Meulen et al., 2014). H3K27me3 is involved not only in the balance between self-renewal and differentiation of neural stem cells (NSCs) (Pereira et al., 2010; Zhang et al., 2014), but also in the development of neurodegenerative diseases (Li et al., 2013; von Schimmelmann et al., 2016).
The Utx gene is encoded on the X chromosome but escapes X inactivation in females and is ubiquitously expressed (Greenfield et al., 1998). Earlier studies have demonstrated a critical role of Utx in cell reprogramming, cell differentiation, embryonic development, muscle regeneration, circadian rhythm, and senescence (Agger et al., 2007; Mansour et al., 2012). Utx mutation has been found in a variety of human cancers, including multiple myeloma, esophageal, renal cancer, bladder cancer, and T-cell acute lymphoblastic leukemia (van Haaften et al., 2009; Van der Meulen et al., 2014). Interestingly, recent studies have shown that de novo deletion and point mutations of Utx are associated with Kabuki syndrome (Lederer et al., 2012; Miyake et al., 2013), a rare congenital anomaly syndrome with mild to severe intellectual disability, growth retardation, and a variety of visceral malformations. Recent clinical data suggests that UTX and UTX-mediated H3K27me2/3 demethylation may play a critical role in the brain development (Miyake et al., 2013). Although Utx is highly expressed in most of the brain regions (Xu et al., 2008), its functional role in the central nervous system (CNS) is largely unknown. More importantly, we still do not know whether deletion of UTX in brain could replicate moderate-to-severe congenital anomaly/mental retardation observed from clinical Kabuki syndrome patients, and the pathomechanisms of Kabuki syndrome as well as the roles of Utx in the brain are largely unknown.
To investigate the function of Utx in CNS, we generated forebrain specific Utx deletion mice (cKO). Here we show that Utx cKO mice exhibited anxiety-like behaviors, learning and memory impairments. At the physiological and cellular levels, these cKO mice displayed abnormal synaptic transmission and long-term potentiation (LTP) accompanied with the abnormal neuronal morphology. Bioinformatics analysis revealed that UTX mediated-H3K27me3 demethylation suppressed expression of a subset of genes that are involved in the regulation of neuronal morphology and synaptic activity. The neurotransmitter 5-hydroxytryptamine receptor 5B (Htr5b) is a downstream target of Utx. Overexpression of Htr5b can rescue neuronal morphological impairment induced by Utx deficiency. The results from the present study provide evidence for the first time that Utx plays important roles in neuroplasticity and behaviors. Our work also suggests that Utx deficiency lead to cognition deficits underlying intellectual disability in Kabuki syndrome.
Materials and Methods
Mice
All experiments involving animals were performed in accordance with the animal protocol approved by the Institutional Animal Care and Use Committee at the Institute of Zoology, Chinese Academy of Sciences. Mice were housed in groups of 3–5 animals in a 12 h light/12 dark cycle, with standard mouse chow and water ad libitum. Utxf/f (stock number 021926), transgenic Nestin-Cre (stock number 003771), and Emx1-Cre transgenic mice (stock number 005628) were bought from Jackson Lab. The conditional Utx knockout mice were generated by breeding Utxf/f mice with either Nestin-Cre or Emx1-Cre transgenic mice.
Behavioral Tests
All behavioral tests were performed during the light cycle between 07:00 and 19:00. Male mice at 2–3 months of age were used for all the behavioral tests. All the videos were analyzed by the Smart software (Pan Lab, Harvard Apparatus).
Open Field Test
The open field test was conducted in a large box (72 × 72 × 36 cm) in a brightly lit room. Mice were placed in the center of the maze and were monitored from above by a video camera. The number of entries in the center zone (18 × 18 cm) of the maze was recorded over a 5-min trial to evaluate anxiety effects.
Light-Dark Box Test
Light-dark box test was performed as previously described (Costall et al., 1993). An apparatus (45 × 27 × 27 cm) consisting of two chambers, a black chamber (18 × 27 cm) and a light chamber (27 × 27 cm), was used for the light/dark exploration test. Mice were placed into the dark box and allowed to move between the light box and dark box for 5 min. The total number of transitions and the time spent in each box were analyzed.
Elevated Plus Maze
The elevated plus maze is a plus-shaped apparatus with four arms (two open, 62 × 8.5 cm; two closed, 62 × 8.5 × 30 cm), elevated 70 cm from the floor. Mice were placed at the junction of the four arms of the maze, and allowed to freely explore the maze for 5 min. The number of entries and duration in each arm were analyzed (Rodgers and Dalvi, 1997).
Morris Water Maze
Morris water maze test was performed as described previously (Vorhees and Williams, 2006). A 120 cm diameter, 45 cm deep Morris water maze was filled with water to a depth of 25 cm. Target escape platform (diameter 13 cm) was hidden 1.5 cm beneath the surface of the water at the center of a given quadrant of the water tank. Four extra-maze cues, in different shapes, colors, and sizes, were uniformly located on the wall surrounding the water tank. The water temperature was adjusted to 21 ± 1°C. During training trials, mice were trained in four trials per day starting from different sites. The mice were allowed to swim for up to 1 min to locate the platform. If it failed to locate the platform within that time, escape was assisted. Mice were introduced gently to the hidden platform and allowed to rest on the platform for 15 s. For the probe trial, 24 h after the final training day the platform was removed and time spent and entry in each of the four quadrants were recorded.
Electrophysiology
Acute Hippocampal Slice Preparation
Acute hippocampal slices were prepared from 8-week-old cKO mice and their WT littermates. Briefly, mice were deeply anesthetized with isoflurane and decapitated. The brain was rapidly removed and transferred into ice-cooled artificial cerebrospinal fluid (aCSF) (in mM: 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 D-Glucose, 2 CaCl2, and 1.5 MgCl2 saturated with 95% O2 and 5% CO2 to pH: 7.4). A filter paper was placed on the bottom of a petri dish in advance. The brain was transferred into the petri dish containing ice-cold aCSF. Coronal slices with 300 μm thickness containing hippocampus were cut with a vibratome (Leica, VT 1000 S, Germany), which was filled with the cold aCSF. The prepared slices were incubated in oxygenated aCSF at room temperature at least for 1 h, and then individual slices were transferred to a recording chamber, which was bubbled with oxygenated aCSF. The temperature of the aCSF in the recording chamber was 31 ± 1°C. Individual pyramidal neurons visualized with an Olympus microscope (Olympus BX50-WI, Olympus, Japan) fitted with a 40x long-working distance objective (NA 0.8).
Electrophysiological Recordings
Whole-cell patch-clamp recordings were made using an Axopatch 700B amplifier and the pClamp10.6 software was used for data acquisition and analysis. Patch pipettes (4–6 MΩ) were pulled from borosilicate glass capillaries (GB 150F-8P) with a micropipette puller (Sutter instrument, USA). The internal pipette solution contained: (in mM) 135 K-gluconate, 10 HEPES, 2 MgCl2, 10 EGTA, 0.3 MgGTP, 0.5 Na2ATP (pH 7.3 with KOH). Spontaneous miniature excitatory postsynaptic currents (mEPSCs) were recorded under the whole-cell configuration of voltage clamp. The membrane potential was held at −70 mV. Series resistances and cell capacitance compensation were carried out prior to recording. The recordings were included only in those with high resistance seal (>1 GΩ) and a series resistance <25 MΩ. To isolate AMPA receptor-mediated mEPSCs, 10 nM glycine, 10 μM bicuculline (the GABAA receptor antagonist), and D-AP-5(NMDA receptor antagonist) were added to the aCSF. In addition, TTX (0.5 μM) was included in the extracellular solution.
Field excitatory postsynaptic potentials (fEPSPs) were recorded in the CA1 region of hippocampus. A bipolar concentric stimulating electrode (FHC Inc., Bowdoin, ME) was placed at the Schaffer collaterals to deliver stimuli. A glass recording electrode (3–4 MΩ) filled with aCSF was positioned in the stratum radiatum of the CA1 area. fEPSPs in CA1 were induced by stimulus at 0.033 Hz with an intensity that elicited a fEPSP amplitude of 40–50% of the maximum. After establishment of stable baseline recordings for at least 15 min, LTP was induced by a high-frequency stimulation (HFS) consisting of one train of 100 Hz stimulation for 1 s at baseline stimulation intensity. The fEPSP signals were digitized using Digidata1440A interface board. The data were sampled at 10 kHz and filtered at 2 kHz. Recordings were analyzed using the Clampfit 10.6 (Axon Instruments, Foster City, CA).
Neuronal Culture and Transfection
Primary hippocampal neurons (1 × 104 cells per well in a 24-well plate) were cultured from P0 Utx cKO and WT mice on plates coated with poly-D-lysine (100 μg/ml). The dissected hippocampus tissue was digested with trypsin-EDTA for 10 min at 37°C. The tissue was then washed three times with MEM plus 10% FBS and dissociated with the culture medium. Then neurons were grown in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen) and 2 mM GlutaMAX (Invitrogen) and penicillin/streptomycin. Primary hippocampal neurons were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Neuronal Morphology Analysis
In vivo Dendritic and Spine Density Analysis
We analyzed in vivo dendrites and spine density by using Golgi Stain Kit following the manufacturer's instructions. Generally, freshly 8-week-old mouse brains were incubated in the dark in Golgi solution A+B (FD Rapid Golgi Stain Kit, PK401, FD NeuroTechnologies) for 2 weeks. After incubation, brains were transferred into Solution C and were stored at room temperature in the dark for 3 days. Coronal sections (200 μm) were cut with a Leica CM1950 cryostat and mounted on 3% gelatin-coated slides. Staining procedures were followed according to the manufacturer's protocol (FD NeuroTechnologies), and slides were dehydrated in ethanol and mounted with Permount for microscopy. Images of the dendrites and the second segment apical dendrite spine were acquired on LSM 710 confocal microscope with 20 × and 100 × oil lense, respectively. Dendritic branches were traced, and their lengths were calculated using the Simple Neurite Tracer plugin of Fiji.
In vitro Dendritic and Spine Analysis
For dendrites analysis, cultured neurons were fixed with 4% paraformaldehyde and then were washed with PBS at 7 day in vitro (DIV-7). Fixed neurons were blocked by 2% normal goat serum and 0.1% Triton X-100 in TBS for 1 h at room temperature. Neurons were incubated in primary antibodies (Map2, Mab3418, Millipore, 1:1,000) overnight at 4°C, and then incubated with secondary antibodies. Images of dendrites were acquired on Zeiss LSM710 confocal microscope with a 20 × lense. For spine analysis, cultures were used for immunostaining at DIV-19. Images of spines were acquired on Zeiss LSM710 confocal microscope with a 63 × oil lense. The secondary dendritic spines were analyzed.
Stereotactic Injection
Stereotactic injections into the hippocampus (stereotaxic coordinates from Bregma: 2.0 mm caudal, 1.2 mm lateral, 2.0 mm ventral; 2.8 mm caudal, 2.0 mm lateral, 1.7 mm ventral) were performed as follows: 8-week-old Utxflox/flox were bilaterally injected with 1 μl Adeno-associated virus (AAV) 2/8 (Heyuan, China; Titer: Control virus:6.24 × 1012 V.G./ml; Cre virus: 5.04 × 1012 V.G./ml) at a rate of 0.125 μl/min.
RNA Isolation and RT-qPCR
Total RNA were extracted from hippocampus tissues or cultured cells according to procedures using Trizol reagent (Invitrogen). Two micrograms of total RNA was reverse transcribed using either oligo (dT) primers or specific primers by using a Transcriptor First Strand cDNA Synthesis Kit (Roche). For real-time PCR analysis, according to the manufacturer's instructions, using a SYBR mix from Roche. 25 ng of cDNA and 0.5 mM primers were used in a final volume of 20 μl. The PCR steps were performed 30 s pre-denaturation at 95°C, followed by 45 cycles of 10 s denaturation at 94°C, 30 s annealing at 60°C, 30 s extension at 72°C. The analysis of RT-qPCR used the 2−ΔΔCT method. Each reaction was run in triplicate and analyzed following the ΔΔCt method using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalization control. All primers are listed in Table 1.
Protein Quantification
Hippocampal tissues and cultured neurons were lysed in buffer containing 25 mM HEPES at pH7.9, 150 mM NaCl, 1 mM PMSF, 20 mM NaF, 1 mM DTT, 0.1% NP40, and proteinase inhibitor cocktails (Roche). Protein concentrations were determined by Folin phenol method with bovine serum albumin as standard. Twenty micrograms of the protein was separated on 8–12% SDS-PAGE gels (Bio-Rad) and transferred to PVDF membranes (Millipore). The membranes were blocked in 5% BSA in TBS-T with 0.05% Tween-20 and incubated with primary antibodies at 4°C overnight. Dilutions of primary antibodies were 1:1000 for UTX (E409, Millipore), H3K27me3 (1:1,000, 07449, Millipore), PSD95 (ab2723, Abcam), Synapsin (ab8049, Abcam), and 1:10,000 for β-actin antibody (Sigma). As for the secondary antibodies, we used HRP-linked goat anti-mouse or HRP-Linked goat anti-rabbit at 1:500. Enhanced chemoluminescence (ECL, Pierce) was used for detection. Quantification of the blots was determined with Quantity One Ver.4.4.0 (BioRad, USA).
Immunohistochemistry
Adult mice were anesthetized, perfused with 4% PFA. Brain tissue was dissected out, equilibrated in 30% sucrose, and sectioned into 40 μm-thick serial sections. The brain sections were washed in PBS for 15 min three times, and then blocked in blocking solution (3% BSA+0.3%Triton X-100+0.2% sodium azide) at room temperature for 1 h. The primary antibodies we used are as follows: anti-H3K27me3 (1:1,000, 07449, Millipore), anti-Map2 (1:1,000, Mab3418, Millipore), anti-GFP (1:1,000, A10262, Life technology), anti-NeuN (1:1,000, millipore), anti-Doublecortin (1:500, millipore). After incubation in primary antibody solution at 4°C overnight, the brain sections were washed with TBS for 30 min three times and then incubated with the secondary antibodies conjugated to Alexa Fluor 488 or 594 with a concentration of 1:500 at room temperature. The sections were finally stained with DAPI and mounted using adhesion anti-fade medium.
Bioinformatics Analyses
Only transcripts that showed more than 1.5-fold differential expression compared to control were subjected to relevance network analysis. GO analysis was performed by Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7) (Huang et al., 2009). Mouse phenotype and gene enrichment were analyzed by WEB-based Toolkit (https://toppgene.cchmc.org/).
Statistical Analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) with one-way or two-way analysis of variance as specified in legend of each figure. Prior to all statistical analyses, data were examined for normality of variance using the Kolmogorov-Smirnov test. All data were presented as mean ± SEM, and statistically significant was defined as *p < 0.05; **p < 0.01; ***p < 0.001.
Results
Utx cKO Mice Display Anxiety-Like Behaviors and Spatial Learning and Memory Deficits
To evaluate the function of Utx in the brain, we used Utx conditional knockout (cKO) mice that lack Utx expression in NSCs during development (Figure 1A). Utx cKO male mice could survive to adult, however, almost all Utx cKO female mice died within 3 weeks after birth. Utx deletion was then confirmed at both mRNA and protein levels in cKO mice (Figures 1B,D), and we did not observe any change of its Y chromosome homolog Uty at mRNA level in the cKO brain (Figure 1C). Since Utx is a histone demethylase and functions in removal of repressive trimethylation of H3K27 (Agger et al., 2007), we next examined the histone marks H3K27me3 in cKO mice. Consistent with our expectations, Utx deletion significantly resulted in increased expression of H3K27me3 in the hippocampus compared to that in the WT littermates (Figures 1E,F).
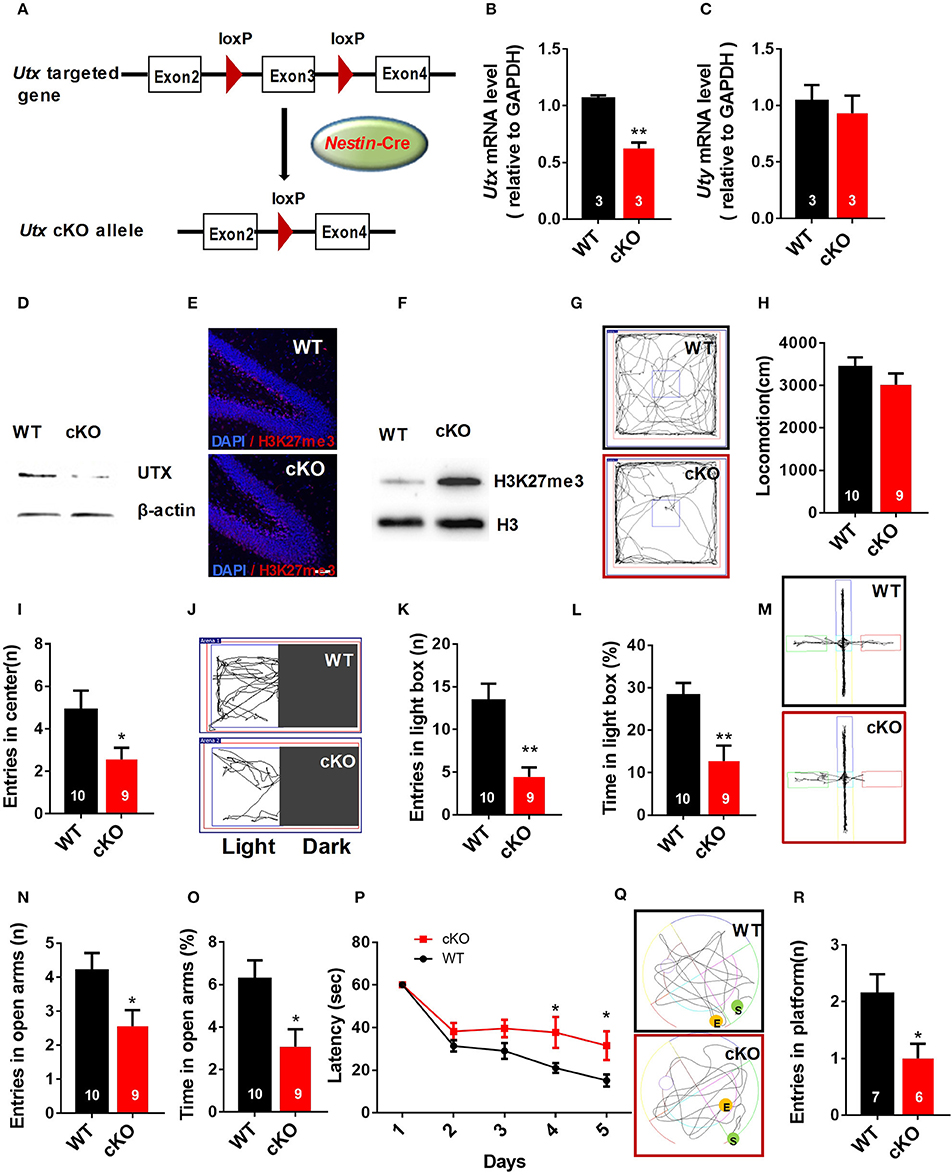
Figure 1. Nestin-Cre Utx cKO mice display anxiety-like behaviors and spatial learning and memory deficits. (A) Strategy for the generation of Nestin-Cre Utx cKO mice. (B) Real-time PCR analysis showing that Utx mRNA is decreased in the hippocampus of cKO mice (n = 3, p < 0.01). (C) Real-time PCR analysis showing that Uty mRNA had no change in the hippocampus of cKO mice (n = 3, p = 0.6287). (D) Western blotting analysis confirmed knockout of Utx in the hippocampus of cKO mice. (E) Representative images of fluorescent immunohistochemistry showing an increase of H3K27me3 in the dentate gyrus of hippocampus in cKO mice. Scale bars, 50 μm. (F) Protein levels of H3K27me3 are increased in the hippocampus from cKO mice. (G) Representative trajectory maps in an open field test. (H) Utx cKO mice had comparable locomotivity to WT littermate mice in open field test over a 5-min period [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 1.181, P = 0.2539]. (I) cKO mice showed decreased entry into the center zone during 5-min open field test [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 2.157; P = 0.0456]. (J) Representative trajectory maps of the light-dark box test. (K) cKO mice had decreased entry into the light box during 5-min light-dark box test [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 3.855, P = 0.0013]. (L) cKO mice spent less time in the light box over a 5-min light-dark box test [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 3.423, P = 0.0032]. (M) Representative trajectory maps of WT and Utx CKO mice in the elevated plus maze test. (N) cKO mice showed decreased entry into the open arms during 5-min elevated plus maze test [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 2.338, P = 0.0318]. (O) cKO mice spent less time significantly in the open arms over a 5-min elevated plus maze compared to WT littermates [WT, n = 10; cKO, n = 9; two-tailed t-test: t(17) = 2.659, P = 0.0165]. P cKO mice spent more time in reaching the platform during 5-day training in Morris water maze test [WT, n = 7; cKO, n = 6; repeated measures analysis of variance (ANOVA), followed by Turkey post-hoc test; Group effect: F(1, 11) = 8.977, P = 0.012; Day4: t(11) = 2.337, P = 0.039; Day5: t(11) = 2.366, P = 0.037]. (Q) Representative trajectory maps of WT and Utx CKO mice in water maze test. (R) cKO mice showed less platform crossing in Morris water maze test [WT, n = 7; cKO, n = 6; two-tailed t-test: t(11) = 2.602, P = 0.0246]. (P), repeated measures analysis of variance (ANOVA), followed by Turkey post-hoc test; others, unpaired t-test. n, number of mice. *P < 0.05; **P < 0.01. Error bars, s.e.m.
Male Utx cKO mice and their WT littermates were subjected to a battery of behavioral tests including tests for anxiety-like behaviors as well as learning and memory. We firstly conducted open field test and found that Utx cKO mice had smaller entries into the center of the arena (Figures 1G,I), whereas total distance traveled was comparable to that of WT littermates (Figure 1H). Increased anxiety-like behaviors of cKO mice were then confirmed in the light-dark box test and the elevated plus maze test. In the light-dark box test, Utx cKO mice showed a substantially decreased entries (Figures 1J,K) and time spent (Figure 1L) in the light box. Similarly, Utx cKO mice displayed decreased entries (Figures 1M,N) and spent less time in the open arms when compared with the WT littermates in the elevated plus maze test (Figure 1O).
Next, we determined whether Utx cKO mice have deficits in spatial learning and memory using the Morris water maze test. We found that cKO mice take a longer time to locate the hidden platform in the training trials (Figure 1P) and spend less time in the platform zone during the probe test (Figures 1Q,R), indicating that Utx cKO mice have deficits in hippocampus-dependent spatial learning and memory.
To further confirm the function of Utx in the brain, we crossed homozygous Utxf/f mice with transgenic Emx1-Cre mice, in which the endogenous Emx1 locus directs expression of Cre recombinase specifically in the neocortex and hippocampus, to generate Emx1-Cre Utx cKO mice with Utx deletion in the forebrain (Figure 2A). Consistent with the observations in Nestin-Cre Utx cKO mice, these Emx1-Cre Utx cKO mice also displayed anxiety-like behaviors as indicated by decreased entry into the center zone in the open field test (Figures 2B–D), decreased entries (Figures 2E,F), and time spent (Figure 2G) in the light box in the light-dark box test. In the Morris water maze test, Emx1-Cre Utx cKO mice took prolonged time to locate the hidden platform during both training (Figure 2H) and probe trials (Figures 2I,J), indicating that impaired spatial learning and memory also exists in Emx1-Cre Utx cKO mice. These results suggest that Utx specific ablation in the forebrain phenocopies the behavioral deficits observed in Nestin-Cre Utx-cKO mice, further strengthening the critical role of Utx in regulation of cognitive behaviors.
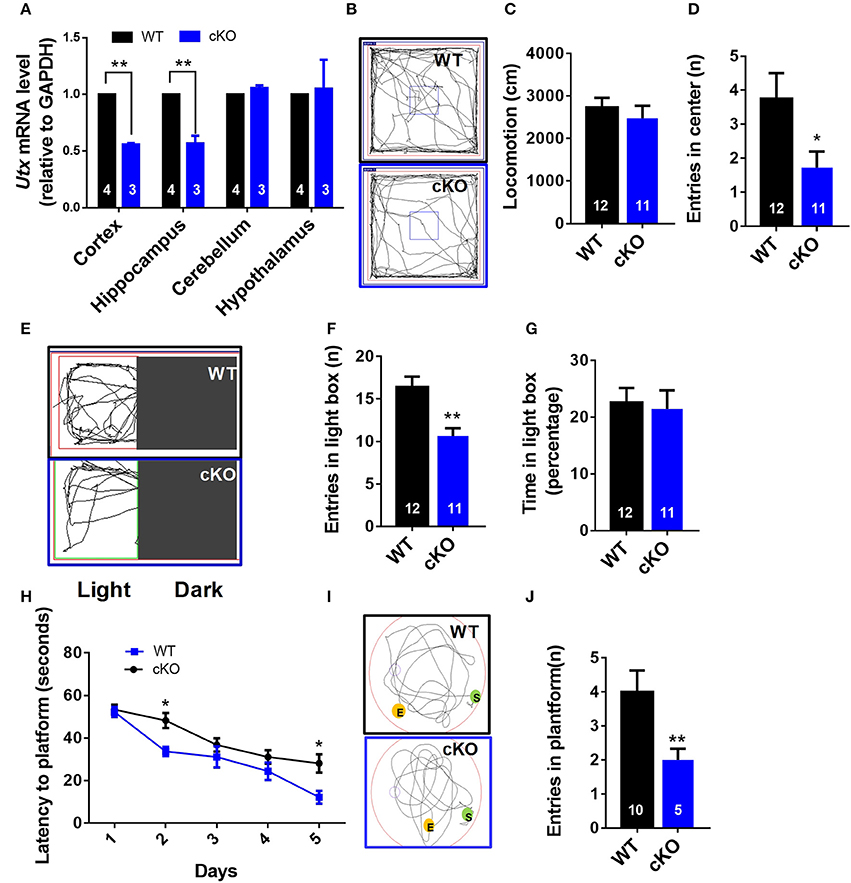
Figure 2. Emx1-cre Utx cKO mice display anxiety-like behaviors and congenital abnormities. (A) Real-time PCR analysis showing that Utx mRNA are decreased in the cortex and hippocampus in Emx1-Cre Utx cKO mice [WT, n = 4; cKO, n = 3; two-tailed t-test: t(5) = 40.531, P < 0.001] and hippocampus [WT, n = 4; cKO, n = 3; two-tailed t-test: t(5) = 7.610, P = 0.001]. (B) Representative trajectory maps of Utx cKO and WT mice in the open field test. (C) Utx cKO and WT mice displayed similar locomotivity in the open field test over a 5-min period [WT n = 12, cKO n = 11; two-tailed t-test; t(21) = 0.7064; P = 0.4877 for WT vs. cKO]. (D) Utx cKO mice had decreased entries into the center zone during 5-min open field test [WT n = 12, cKO n = 11; two-tailed t-test; t(21) = 2.238; P = 0.0362 for WT vs. cKO]. (E) Representative trajectory maps in the light-dark box test. (F) Utx cKO mice showed decreased entries into the light box during 5-min light-dark test [WT, n = 12; cKO, n = 11; two-tailed t-test: t(21) = 3.730, P = 0.0012]. (G) Utx cKO mice spent comparable time in the light box over a 5-min light-dark test [WT, n = 12; cKO, n = 11; two-tailed t-test: t(21) = 0.270, P = 0.7845]. (H) Utx cKO mice spent significantly longer time in locating the platform during 5-day training [WT n = 5, cKO n = 10; repeated measures analysis of variance (ANOVA), followed by Turkey post-hoc test; Group effect: F(1, 13) = 7.016, P = 0.02; Day2: t(13) = 2.738; P = 0.0169; Day5: t(13) = 2.632; P = 0.0207 for WT vs. cKO]. (I) Representative trajectory maps in the Morris water maze test. (J) Utx cKO mice showed significant decreased number of platform crossing in the Morris water maze test [WT, n = 5; cKO, n = 10; two-tailed t-test: t(13) = 3.103, P = 0.0084]. *P < 0.05; **P < 0.01. Error bars, s.e.m.
Down-Regulation of Utx in Adult Hippocampus Phenocopies the Behavioral Deficits As in Utx cKO Mice
The hippocampus is an important region in the brain in the information processing, consolidation, and storage, and responsible for cognitive ability and memory retention. It is also involved in emotional and cognitional functions (Eichenbaum, 2004). We then asked the question as to whether deletion of Utx in the hippocampus of adults could phenocopy the cognitive deficits exhibited in Utx cKO mice. To test this idea, we stereotactically injected AAV 2/8 virus expressing either GFP or Cre into the hippocampus of 8-week-old Utxf/f mice (Figure 3A). As shown in Figure 3B, expression of Utx at the protein level was significantly downregulated along with Cre recombinase after virus injection. In particular, we observed that deletion of Utx in the hippocampus of adult Utxf/f mice exhibited normal locomotion (Figure 3D), but showed fewer entries into the center zone (Figures 3C,E) in the open field test. This is consistent with the anxiety-like behavior and cognitional deficits in Utx cKO mice. Similarly, mice with Utx down-regulation in the hippocampus also showed decreased entries in the light chamber in the light-dark box test (Figures 3F,G,H), and fewer entries (Figures 3I,J), and time spent (Figures 3K) in the open arms in the elevated plus maze test. In the Morris water maze test, mice with Utx down-regulation in the hippocampus displayed the lower ability to locate the platform (Figures 3L,M,N), indicating impaired spatial learning and memory abilities. Taken together, these results indicate that deletion of Utx in the adult hippocampus indeed replicates the behavioral deficits observed in cKO mice in which deletion of Utx occurs during development, further supporting the idea that Utx plays critical roles in regulation of emotion and cognition in adults.
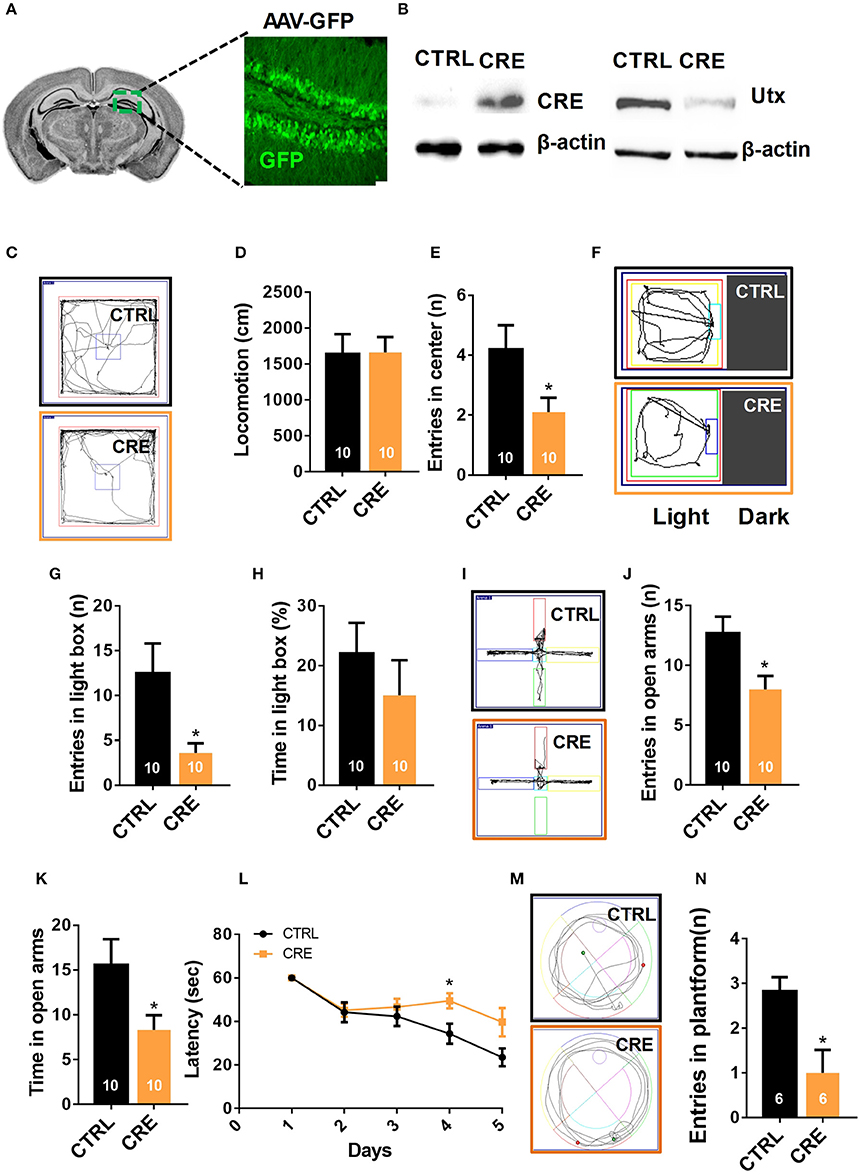
Figure 3. Knocking-down of Utx expression in adult hippocampus phenocopies the behavioral deficits observed in Utx cKO mice. (A) Schematic illustrating injection of AAV-GFP-CRE and AAV-GFP control viruses into the adult Utx f/f hippocampus (left), representative images showing GFP expression in the hippocampus 2 months after viral injection (right). (B) UTX protein level was significantly reduced in the Utx f/f hippocampus 2 months after AAV2/8-cre viral injection. (C) Representative trajectory maps of WT and Utx cKO mice in the open field test. (D) Utx knockdown had no effect on locomotivity [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 0.05696, P = 0.9552]. (E) Utx knockdown resulted in less entries into the center zone during the open field test [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 2.249, P = 0.0373]. (F) Representative trajectory maps of WT and Utx cKO mice in the light-dark field test. (G) Mice with Utx knockdown had decreased entries into the light box during 5-min light-dark box test [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 2.561, P = 0.0196]. (H) Mice with Utx knockdown showed no significant changes of time in the light box compared to their controls over a 5-min light-dark box test [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 0.9038, P = 0.3780]. (I) Representative trajectory maps of WT and Utx cKO mice in elevated plus maze test. (J) Mice with Utx knockdown displayed decreased entries into the open arms during 5-min elevated plus maze test [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 2.655, P = 0.0161]. (K) Mice with Utx knockdown spent less time in the open arms over a 5-min elevated plus maze [CTRL, n = 10; CRE, n = 10; two-tailed t-test: t(18) = 2.655, P = 0.0161]. (L) Mice with Utx knockdown spent more time in reaching the platform during 5-day training in Morris water maze test [CTRL, n = 7; CRE, n = 8; repeated measures analysis of variance (ANOVA), followed by Turkey post-hoc test; Group effect: F(1, 13) = 6.038, P = 0.029; Day4: t(13) = 2.663, P = 0.020]. (M) Representative trajectory maps in a water maze test. (N) Mice with Utx knockdown showed significant changes of target crossing in Morris water maze test [CTRL, n = 6; CRE, n = 6; two-tailed t-test: t(10) = 3.051, P = 0.0122]. *P < 0.05. Error bars, s.e.m.
Utx cKO Mice Display Abnormalities of LTP and Basal Synaptic Transmission in the Hippocampus
To explore the role of Utx loss-of-function in hippocampal synaptic transmission and plasticity, we recorded hippocampal LTP, a cellular model for learning and memory, in acute hippocampal slices prepared from 6 to 7 week-old Utx cKO and WT mice. As shown in Figures 4B,C, LTP at Schaffer collateral synapses in the CA1 region (Figure 4A) was significantly impaired in cKO mice when compared with that in WT littermates. There was a significant difference in the amplitude of LTP at 55–60 min after induction between cKO and WT mice (Figures 4B,C). These results indicated that Utx deficiency impairs hippocampal long-term synaptic plasticity, which may underlie the behavioral deficits displayed by cKO mice.
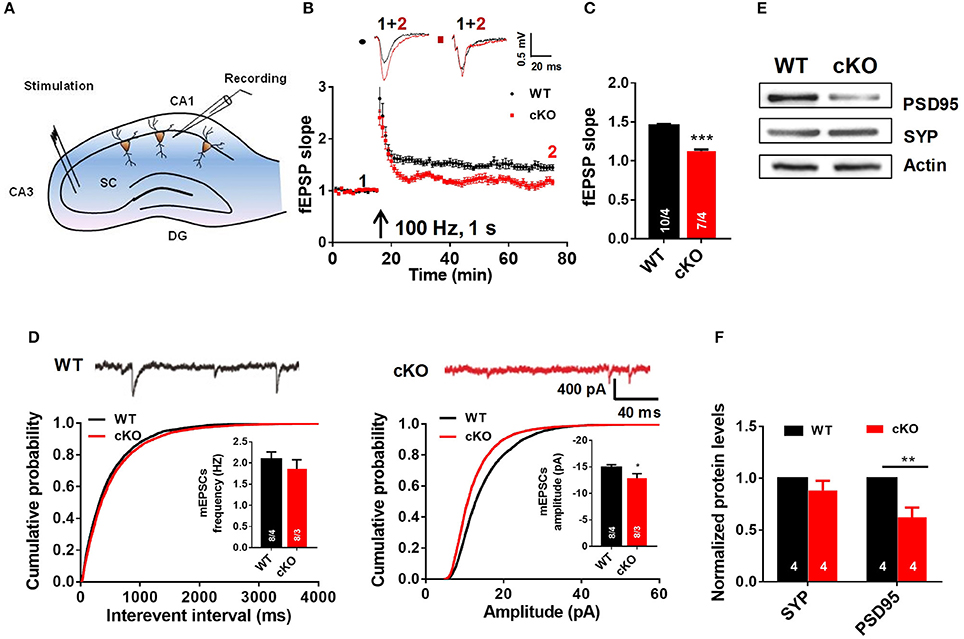
Figure 4. Utx cKO mice display abnormalities of LTP and basal synaptic transmission in CA1 of the hippocampus. (A) Schematic overview of the electrophysiological protocol for acute hippocampal slice recording. (B) Typical experiment showing time course of CA1 LTP for a single recording from WT and cKO mice. fEPSP traces before (1, black) and after (2, red) are shown in the inset above. (C) cKO mice showed lower LTP amplitude measured at 55–60 min post-induction [WT, 10 slices from 4 animals; cKO, 7 slices from 4 animals; two-tailed t-test: t(338) = 12.07, P < 0.0001]. (D) Amplitudes [WT, 8 slices from 4 animals; cKO, 8 slices from 3 animals; two-tailed t-test: t(14) = 2.211, P = 0.0442] but not frequencies [WT, 8 slices from 4 animals; cKO, 8 slices from 3 animals; two-tailed t-test: t(14) = 0.8486, P = 0.4104] of spontaneous mEPSCs (monitored in 0.5 μM tetrodotoxin) was impaired in cKO CA1 neurons [top, representative traces; bottom, cumulative plots and summary graphs of the mEPSC (left) frequency and amplitude (right)]. (E) Representative western blotting for PSD95 and Synapsin (SYP) in total hippocampus extracts of WT and cKO mice. (F) Quantification of immunoreacitivity revealed a significant decrease of PSD95 expression in cKO mice. SYP protein levels were similar in cKO mice to WT mice [WT, n = 4; Cko, n = 4; two-tailed t-test: t(6) = 3.951, P = 0.0075]. SYP protein levels were similar in cKO mice to WT mice [WT, n = 4; cKO, n = 4; two-tailed t-test: t(6) = 1.262, P = 0.2538]. *P < 0.01; **P < 0.01; ***P < 0.001. Error bars, s.e.m.
To further determine the cause of the impaired hippocampal synaptic plasticity in Utx cKO mice, we recorded AMPA receptor (AMPAR)-mediated miniature excitatory postsynaptic currents (mEPSCs) in CA1 pyramidal cells using the whole-cell patchclamp technique. AMPAR mediated mEPSCs were pharmacologically isolated by bath application of the GABAA and NMDA receptor antagonists bicuculline (10 μM) and D-AP5 (25 μM) in the presence of TTX (0.5 μM). At a holding potential of −70 mV, mEPSCs displayed fast inward currents and sensitive to 20 μM DNQX, indicating AMPAR-mediated currents. As shown in Figure 4D, the amplitude, but not the frequency, of mEPSCs was significant decreased in slices from cKO mice when compared with that from WT mice. The results from electrophysiological recordings further support our speculation that Utx deletion impairs hippocampal synaptic transmission.
Impaired LTP and decreased amplitude of mEPSCs are most likely due to the deficits in postsynaptic function and/or reduction of functional synapses (Liao et al., 1995; Xiao et al., 2007). To test this assumption, we examined the immunoreactivity of synaptic-related proteins in the hippocampus of both WT and cKO mice. As shown in Figures 4E,F, expression of postsynaptic density protein PSD-95, a postsynaptic marker, was significantly decreased, while synapsin, a presynaptic marker, remained unchanged in cKO mice when compared to that in WT littermates. These results suggest that decreased expression of PSD-95 may lead to a deficit of postsynaptic plasticity in Utx cKO mice.
Morphological Abnormalities of Hippocampal Neurons in Utx cKO Mice In vivo
Neuronal signal integration as well as synaptic transmission and plasticity highly depend on the morphology of dendrites and their spines (Hering and Sheng, 2001). The impairment of hippocampal synaptic plasticity and the reduced PSD95 expression in Utx cKO mice prompted us to postulate that Utx deficiency may cause abnormal neuronal morphology. To determine whether disruption of Utx affects neuronal morphology, we traced Golgi-stained hippocampal neurons in the CA1 and dentate gyrus (DG). Utx cKO mice exhibited a significant reduction in dendritic length and number of branches in neurons in both the CA1 (Figures 5A–D) and DG regions (Figures 5E–G). Moreover, a reduction of spine density in hippocampal CA1 pyramidal neurons was also found in Utx deletion mice (Figures 5H,I). These data, together with the reduction in the postsynaptic PSD95 protein (Figure 4E), suggest an essential role for UTX in synaptic formation and dendritic development in hippocampal neurons, which in turn affect spatial learning and memory.
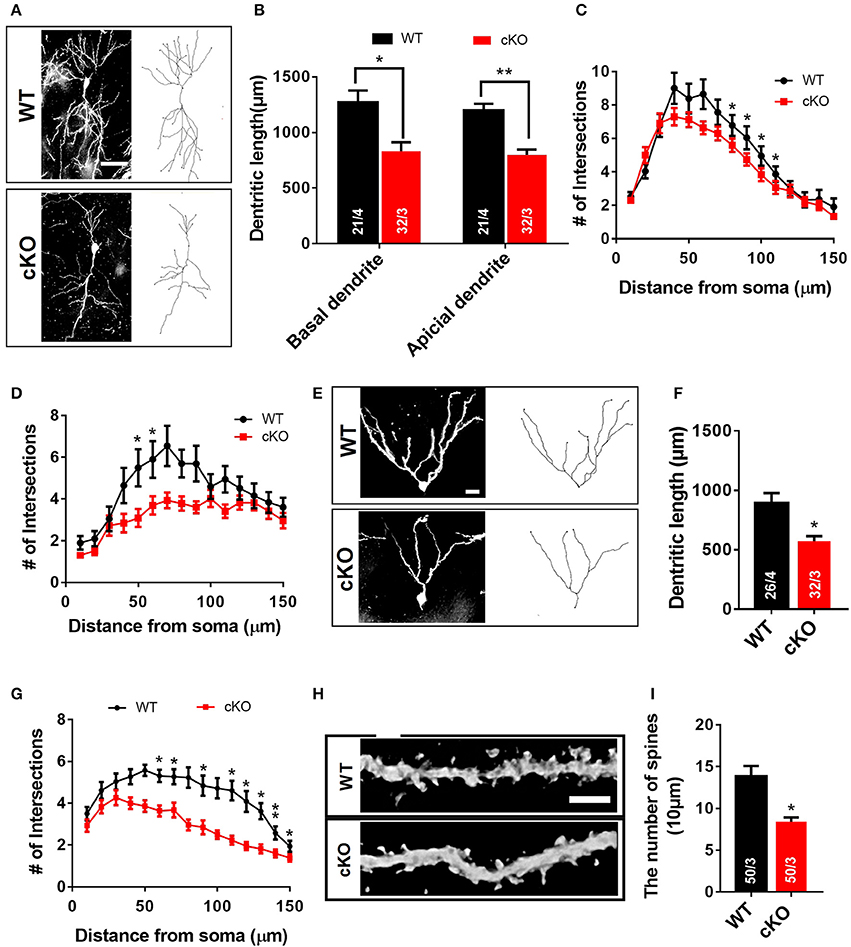
Figure 5. Morphological abnormalities of hippocampal neurons in Utx cKO mice. (A) Representative projection images of Golgi-stained pyramidal neurons in the hippocampal CA1 region. Scale bar, 50 μm. (B) Quantification of dendritic length of CA1 neurons [WT, 21 neurons from 4 mice; cKO, 32 neurons from 3 mice; two-tailed t-test: basal dendrite, t(5) = 3.122, P = 0.0262; apical dendrite, t(5) = 5.137, P = 0.0037]. (C) Quantification of basal dendritic number of CA1 neurons [80 μm, t(5) = 3.090, P = 0.027; 90 μm, t(5) = 2.988, P = 0.031; 100 μm, t(5) = 3.313, P = 0.021; 110 μm, t(5) = 2.988, P = 0.031]. (D) Reduced branching of apical dendrites was found in Utx deletion CA1 neurons [50 μm, t(5) = 3.828, P = 0.012; 60 μm, t(5) = 4.781, P = 0.005]. (E) Representative images of dendritic spines of Golgi-stained secondary dendrites of pyramidal neurons in the hippocampal CA1 region. Scale bars, 2 μm. (F) Utx cKO mice decreased spine density in the CA1 neurons compared to WT littermate [WT, 50 neurons in 3 mice; cKO, 50 neurons in 3 mice; two-tailed t-test: t(4) = 4.082, P = 0.0151]. (G) Representative projection images of Golgi-stained neurons in the dentate gyrus. Scale bar, 50 μm. (H) Quantification of dendritic length of neurons in the dentate gyrus [WT, 26 neurons from 4 mice; cKO, 32 neurons from 3 mice; two-tailed t-test: t(5) = 3.086, P = 0.0273]. (I) Quantification results showed that Utx deletion decreased dendritic branching in the dentate gyrus [60 μm, t(5) = 3.140, P = 0.026; 70 μm, t(5) = 2.869, P = 0.035; 90 μm, t(5) = 3.042, P = 0.029; 110 μm, t(5) = 2.587, P = 0.049; 120 μm, t(5) = 3.886, P = 0.012; 130 μm, t(5) = 3.054, P = 0.028; 140 μm, t(5) = 6.444, P = 0.001; 150 μm, t(5) = 3.095, P = 0.027). *P < 0.05; **P < 0.01. Error bars, s.e.m.
Utx Deletion in Hippocampal Neurons Results in Morphological Defects In vitro
We next used a well-established in vitro primary neuron culture system to examine the effects of UTX deletion on the morphology of hippocampal neurons. Indeed, Utx cKO hippocampus neurons displayed a significant reduced in total dendritic length and dendritic complexity compared with control neurons isolated from littermate newborn pups (Figures 6A–C) at Day 7 in vitro (DIV7). At DIV19, cKO hippocampal neurons showed significant reductions in spine density (Figures 6D,E), which is consistent with the Golgi staining observations in the CA1 pyramidal neurons in vivo (Figures 5H,I). To further determine whether UTX regulates neuronal morphology, we acutely manipulated UTX expression by infecting in newborn hippocampus neurons isolated from Utxf/f pups with lenti-cre viruses to delete UTX expression. As expected, we found that acute knockdown of UTX in hippocampus neurons also led to a significant decrease in total dendritic length and dendritic complexity (Figures 6F–H). These loss-of-function data in primary neurons further support our in vivo observation that deletion of UTX inhibits neuronal dendritic development.
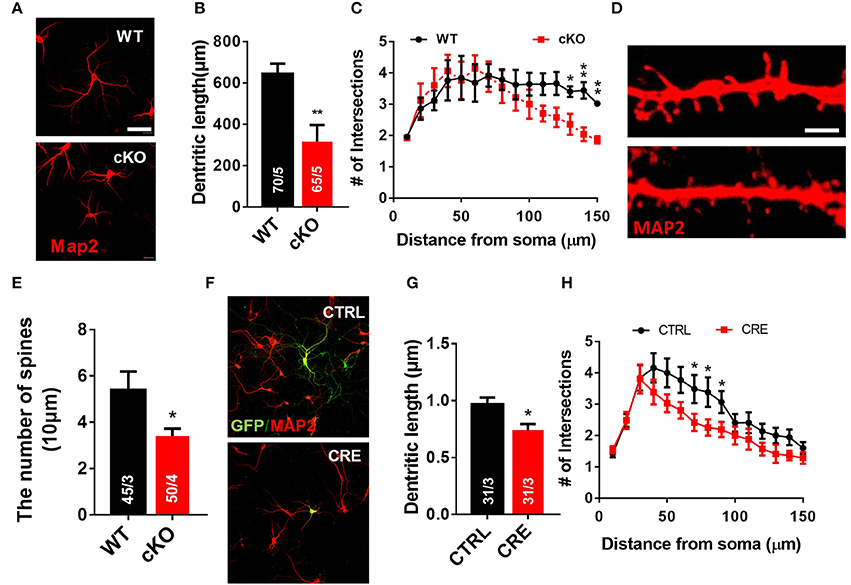
Figure 6. Utx deletion results in abnormal dendritic development in cultured hippocampus neurons. (A) Neurons from the hippocampus of Utx cKO and WT littermate newborn mice were cultured for 7 days in vitro (DIV-7), fixed and stained for expression of Map2 (red). Scale bars, 50 μm. (B) Quantification of dendritic length of WT and cKO P0 hippocampal neurons. (C) Quantification of the branching of P0 hippocampal neurons (DIV-7) [WT, 70 neurons from 5 mice; cKO, 65 neurons from 5 mice; two-tailed t-test: t(8) = 3.494, P = 0.0082]. (D) Representative images of spines from Map2 stained secondary dendrites of cultured neurons (DIV-19) from P0 WT and cKO hippocampus [WT, 45 neurons from 3 mice, Cko, 50 neurons from 4 mice; two-tailed t-test: t(8) = 2.309, P = 0.0497]. Scale bars, 2 μm. (E) Spine density from Map2 stained secondary dendrites of cultured neurons (DIV-19) was reduced in cKO. (F) Representative images of Utxf/f hippocampal neurons infected with Cre-expressing lenti-virus (DIV7). GFP (green), Map2 (red). Scale bars, 50 μm. (G) Cre-mediated Utx deletion resulted in shorter dendritic length in cultured hippocampal neurons [CTRL, 31 neurons from 3 mice; CRE, 31 neurons from 3 mice; two-tailed t-test: t(60) = 2.872; P = 0.0056]. (H) The dendrite complexity was decreased in cultured Utxf/f hippocampal neurons that infected with Cre-expressing lenti-virus [CTRL, 31 neurons from 3 mice; CRE, 31 neurons from 3 mice; 70 μm: t(60) = 2.024, P = 0.047; 80 μm: t(60) = 2.106, P = 0.039; 90 μm: t(60) = 2.012, P = 0.019]. *P < 0.05, **P < 0.01. Error bars, s.e.m.
Altered Expression of Synaptic Plasticity and Cognition Associated Genes in the Hippocampus of Utx cKO Mice
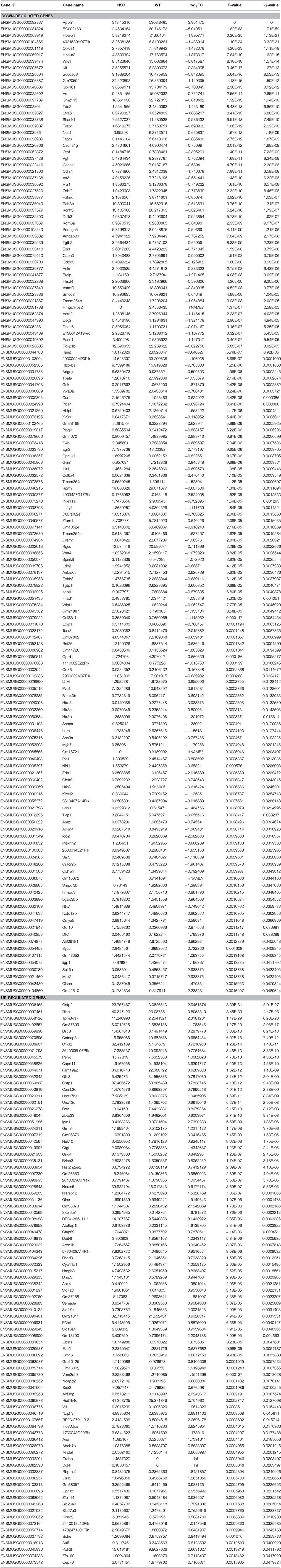
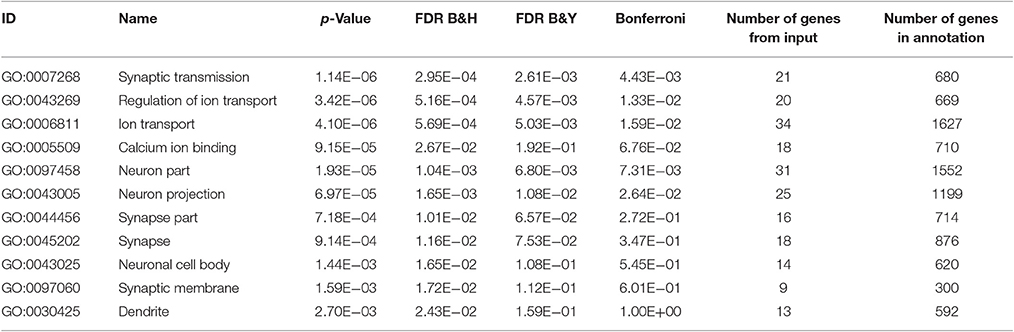
Next, we investigated the effects of Utx loss-of-function on hippocampal gene expression, in order to identify potential mechanisms that might be associated with the synaptic plasticity and cognitive dysfunction in Utx cKO mice. To delineate the molecular pathways regulated by Utx, we conducted gene expression profiling (RNA-seq) using total RNA from the hippocampus of WT and cKO mice. One hundred forty seven down-regulated and 87 up-regulated genes were identified in cKO mice when compared with WT mice (Figures 7A–C and Table 2). To uncover the genes involved in the phenotypes of Utx loss-of-function, we performed Gene Ontology (GO) enrichment analysis of dysregulated genes in Utx cKO mice and found several enriched GO terms for biological processes, including cognition, learning and memory, anxiety, and synaptic transmission (Figure 7C). We then focused on the 147 genes with decreased expression in cKO mice, as their downregulation likely resulted from a loss of Utx. Most of those down-regulated genes are involved in regulation of Ion transport, calcium ion binding, neurite elongation, dendritic, and synaptic formation (Table 3), suggesting an essential role of Utx in Cognition, Learning and Memory, Anxiety, Synaptic Formation, and Function (Figures 7C,D and Table 3).
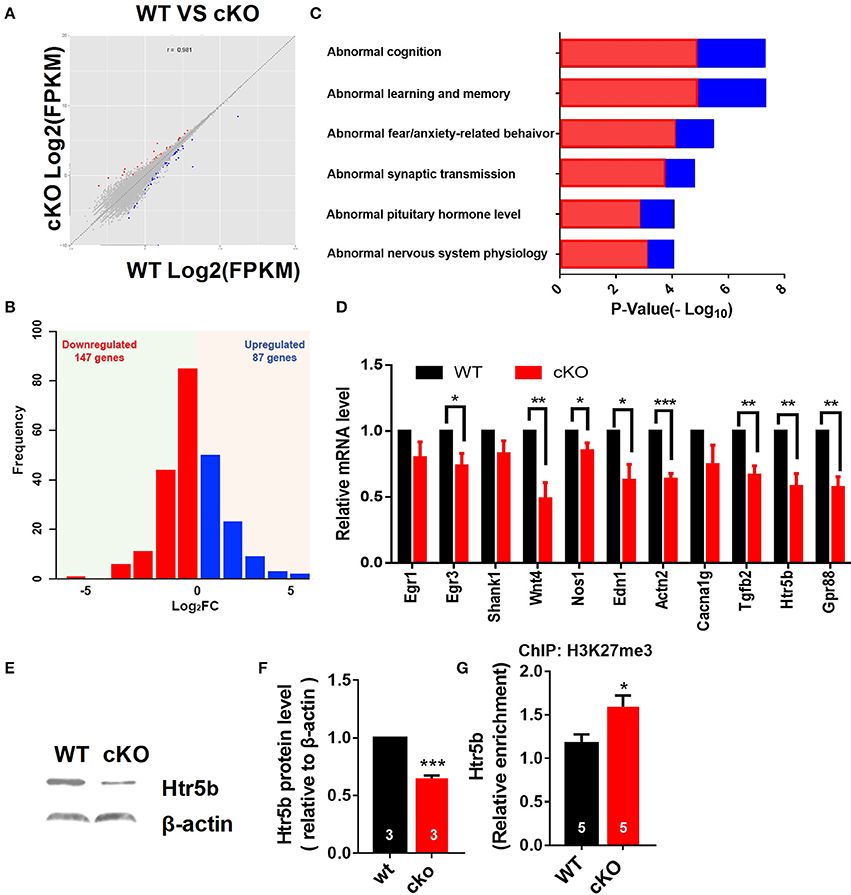
Figure 7. Altered expression of synaptic plasticity and cognition associated genes in the hippocampus of Utx cKO mice. (A) Correlation analysis of differential genes between cKO and WT samples. (B) Histograms of differentially expressed genes in the hippocampus of cKO mice compared to the littermate WT mice. (C) Gene ontology (GO) analysis of differentially expressed genes. Red represents down-regulated genes; Blue represents up-regulated genes. (D) Validation of down-regulated genes in cKO hippocampus by RT-qPCR. (D), paired t-test; *P < 0.05, **P < 0.01. Error bars, s.e.m. (E) Representative image of western blotting analysis on Htr5b. (F) Quantification of Htr5b protein levels in the hippocampus of cKO and littermate WT mice [WT, n = 3; cKO, n = 3; two-tailed t-test: t(4) = 14.79, P = 0.0001]. (G) ChIP assay showed that H3K27me3 was enriched at the promoter of Htr5b gene (n = 5, P = 0.0105). *P < 0.05, ***P < 0.001. Error bars, s.e.m.
We then further examined some of those down-regulated genes relevant for synaptic plasticity and cognition found to be altered by RNA-seq. Using RT-qPCR, we verified that several genes critical for synaptic plasticity, and/or dendrite development including nitric oxide synthase 1 (Nos1), actin filament cross-linker α-Actinin-2 (Actn2), zinc finger transcription factor Egr3, transforming growth factor-β2 (Tgfb2), and Wnt4 were down-regulated in the hippocampus of Utx cKO mice (Figure 7D). Several other essential genes (Gpr88 and Edn1) involved in mental disorder, learning, and memory were also suppressed in the hippocampus of Utx cKO mice (Figure 7D). Amongst the down-regulated genes, we were particularly interested in Htr5b, a functionally unknown serotonin receptor. Serotonin and its receptors have been implicated in dendrite morphology, synaptic transmission and cognition (Wirth et al., 2016). To validate the results from RNA-seq analysis, we conducted qPCR and western blotting analyses and observed that Utx deletion significantly decreased expression of Htr5b in the hippocampus of cKO mice (Figures 7D–F). To further verify whether H3K27me3 enriches on the promoter of Htr5b, we performed ChIP analysis and found that H3K27me3 was enriched in the promoter region of Htr5b (Figure 7G), suggesting that Utx mediated H3K27me3 demethylase activity is required for the alteration in Htr5b in the hippocampus.
Htr5b Modulates Neuronal Morphology, and Its Gain-of-Function Rescues the Impairment of Neuronal Morphology in Utx cKO neurons
The serotonergic system is involved in many aspects of neural development, including neurite outgrowth, somatic morphology regulation, synaptogenesis, and control of dendritic spine shape and density (Wirth et al., 2016). To examine a functional relationship between Utx and Htr5b in mediating neural development, newborn hippocampal neurons from WT mice were transfected with lentivirus expressing both GFP and Htr5b shRNA sequences to suppress Htr5b at both mRNA and protein levels (Figures 8A,B). Knockdown of Htr5b resulted in a significant reduction in total dendritic length (Figures 8C,D) and spine density (Figures 8H,I). Given the fact that the expression level of Htr5b is decreased in Utx cKO mice that displayed neuronal morphological abnormalities (Figures 7E,F), we reasoned whether Htr5b gain-of-function could rescue neural development deficits in Utx cKO neurons. To test this assumption, exogenous Htr5b was expressed in Utx cKO newborn hippocampal neurons. Interestingly, Htr5b gain-of-function in cKO neurons was sufficient to restore the dendritic length (Figures 8E–G) and spine density (Figures 8J,K). These results suggest that Htr5b is a functional downstream target of Utx in modulating neuronal growth and morphology.
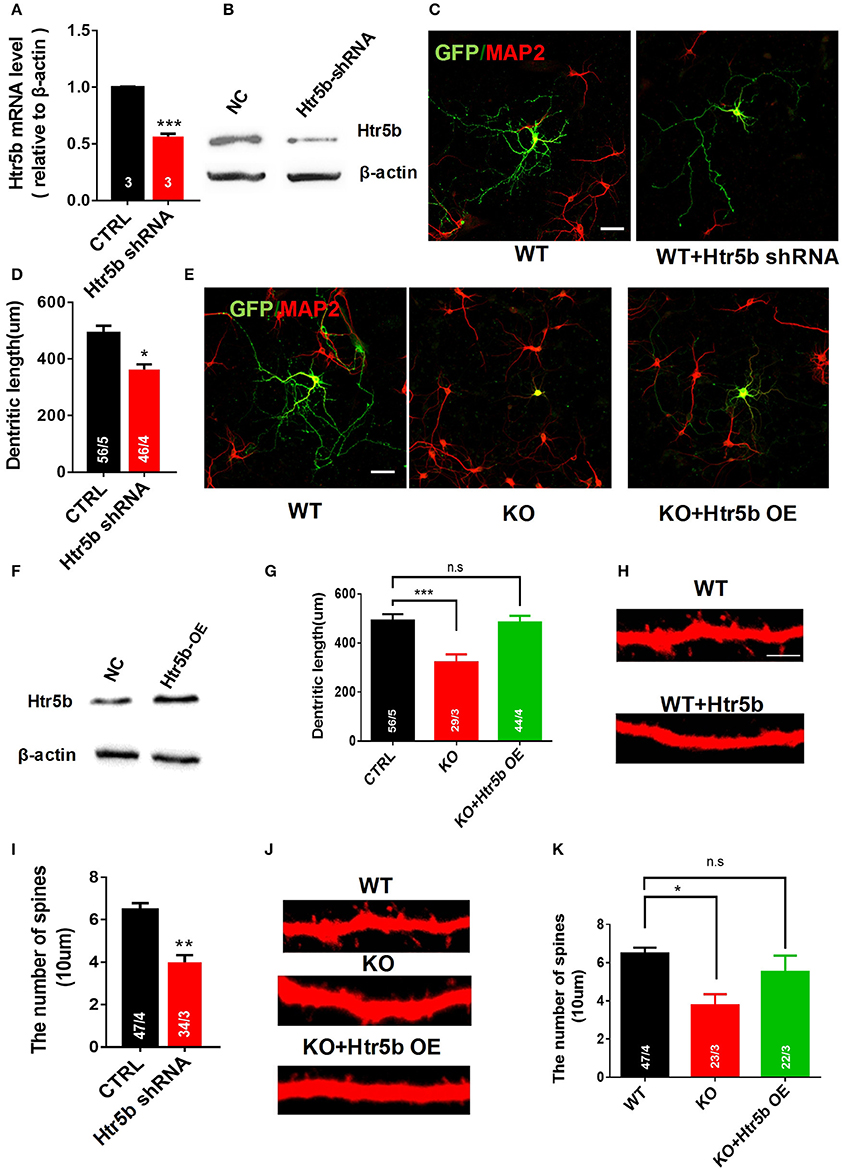
Figure 8. Restoring Htr5b in the hippocampal neurons of Utx cKO mice rescues morphological impairment. (A) Htr5b mRNA was decreased in newborn hippocampal neurons infected with Lenti-virus expressing Htr5b shRNA [Three independent experiments; two-tailed t-test: t(4) = 18.03, P < 0.001]. (B) Western blotting for testing Htr5b protein levels in newborn hippocampal neurons infected with Lenti-virus expressing Htr5b shRNA. (C) Representative images of GFP (Green) and Map2 (Red) double positive neurons expressing either GFP or Htr5b shRNA in WT newborn hippocampal neurons (DIV7). Scale bars, 50 μm. (D) Quantification of dendritic length after Htr5b knockdown in WT newborn hippocampal neurons (DIV7) [WT, 56 neurons from 5 mice, cKO, 46 neurons from 4 mice; two-tailed t-test: t(7) = 2.487, P = 0.0418]. (E) Representative images of GFP (Green) and Map2 (Red) double positive neurons expressing either GFP or Htr5b in WT or cKO newborn hippocampal neurons (DIV7). Scale bars, 50 μm. (F) Western blotting for testing Htr5b protein levels in newborn hippocampal neurons infected with Lenti-Htr5b overexpression virus. (G) Htr5b overexpression rescued the deficits of the dendritic length in cKO newborn hippocampal neurons (DIV7) [WT, 56 neurons from 5 mice; cKO, 29 neurons from 3 mice; KO+Htr5b, 44 neurons from 4 mice; One-way ANOVA: F(2, 9) = 14.17, P = 0.0017]. (H) Representative images of spines from Map2 stained secondary dendrites of WT and cKO P0 hippocampal neurons (DIV19) expressing either GFP or Htr5b protein. Scale bars, 2 μm. (I) Htr5b knockdown resulted in the lower dendritic spine density in newborn hippocampal neurons (DIV7). (J) Representative images of spines from Map2 stained secondary dendrites of WT and cKO hippocampal neurons (DIV19) expressing either GFP or exogenous Htr5b protein. Scale bars, 2 μm. (K) Htr5b overexpression reversed the lower dendritic spine density in cKO neurons to similar levels of WT neurons (DIV19). Scale bars, 2 μm. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars, s.e.m.
Discussion
Recent human genetic findings indicate that epigenetic modifications, including histone methylation, are important regulators of neural development (Portela and Esteller, 2010). However, the underlying mechanisms remain largely unknown. Histone H3 lysine 27 demethylase Utx has been identified as a Kabuki syndrome-risk gene in patients with intellectual disability (Lederer et al., 2012; Miyake et al., 2013). Here, we demonstrate for the first time that forebrain deletion of Utx in mice leads to anxiety-like behaviors, spatial learning, and memory impairments. We further demonstrated that Utx deficiency impairs neuronal morphology and hippocampal synaptic transmission, providing novel evidence for the involvement of Utx in neural development and cognitive behaviors.
Regulation of neuronal development, such as dendrite and spine development, in the brain is critically important in a variety of physiological and pathological conditions (Hering and Sheng, 2001; de la Torre-Ubieta and Bonni, 2011). For instance, mental disorders and alterations in learning and memory are frequently accompanied by analogous modifications of dendrite and spine (Beique et al., 2006). In the present study, we provided evidence for the first time both in vivo and in vitro that UTX is involved in the regulation of neuronal morphogenesis, including spine density and dendrite complexity. In particular, specific deletion of Utx in embryonic nervous system or down-regulation of Utx in adult hippocampus caused abnormal behavioral phenotypes, including anxiety-like behaviors and impairments in learning and memory, which may underscore congenital anomalies in Kabuki syndrome patients. Our study might shed new light on the understanding of epigenetic regulation, especially histone modification in neural development and cognitive behaviors. Since Utx is also expressed in other cell types (Smith et al., 2014), it would be interesting to explore if loss of Utx demethylase activity in non-neuronal cells also contributes to impairments in mood and cognition, as observed in Utx cKO mice.
LTP is widely considered to represent a cellular mechanism for the formation of specific types of anxiety-related behaviors, learning, and memory (Lisman and Raghavachari, 2006). Impaired LTP has been found in several anxiety animal models (Bannerman et al., 2014). In agreement with our behavioral results, we found that deletion of Utx significantly impairs LTP at CA3-CA1 synapses in the hippocampus, further confirming that Utx is an important modulator for neuronal behaviors. Cognitive abnormalities are closely correlated with altered synaptic transmission and plasticity in the hippocampus (Bannerman et al., 2014). AMPA-mediated mEPSCs are widely used to determine pre- and/or postsynaptic contribution to synaptic transmission and plasticity (Bekkers and Stevens, 1990). Utx cKO mice showed a decrease in the amplitude, but not the frequency, of mEPSCs, suggesting a postsynaptic defect in hippocampus (Xiao et al., 2007). Consistent with this finding, we observed a significant reduction of PSD-95, a key player in postsynaptic transmission (Beique et al., 2006). Our results suggest that impaired LTP and reduced amplitude of mEPSCs after Utx deletion lead to synaptic dysfunction, which, in turn, causes cognitive deficits in Utx cKO mice.
We observed that a null mutation of Utx caused mid-gestational lethality, which is consistent with the previous reports by others (Shpargel et al., 2012; Welstead et al., 2012). Male Utx-null mice can escape embryonic lethality, suggesting that this is likely due to the expression of Uty, a paralog that lacks H3K27 demethylase activity (Shpargel et al., 2012; Wang et al., 2012). However, the behavioral abnormalities observed in male Utx-null mice appeared not to be associated with Uty since we did not find any changes in Uty expression after Utx deletion in the brain, suggesting that Uty may not functionally compensate for Utx during neural development. In consistent with our observations, a recent study also showed that UTY cannot compensate for certain demethylase-dependent activities of UTX in T cell acute lymphoblastic leukemia (T-ALL) (Van der Meulen et al., 2015). Further studies in molecular and behavioral levels will be required to fully understand why male Utx-null mice can escape embryonic lethality and what functional role of Uty plays in the brain.
The involvement of the serotonin system in neural development has been well-established (Sparta and Stuber, 2014). Most serotonin receptor family members are known for their roles in mediating morphogenic signaling in neurons (Wirth et al., 2016) and cognition (Meneses, 1999). For example, several studies showed that Htr3 promotes neurite outgrowth in thalamic neurons as well as in PC12 cells (Homma et al., 2006). Serotonin 3A receptor is involved in learning, cognition, and emotion (Gatt et al., 2010). Our transcriptional profiling RT-PCR results revealed that expression of Htr5b, a calcium signaling related gene encoding serotonin 5B receptor, is decreased in the hippocampus of Utx cKO mice. While Htr5b is highly expressed in CNS (Meneses, 1999), the signaling pathways regulated by Htr5b in vivo are still not well-understood. This is largely due to the lack of proper receptor ligands (Wirth et al., 2016). More recently, there is evidence showing that Htr5b is a downstream target for transcription factor ATF-7, which mediates abnormal behaviors in mice (Maekawa et al., 2010). In that study, authors observed that ATF-7-deficient mice exhibit abnormal behaviors and increased Htr5b mRNA levels in the dorsal raphe nuclei (Maekawa et al., 2010). In the present study, we observed that knockout of Utx led to down-regulation of Htr5b and displaying anxiety-like behaviors and spatial learning and memory disability, suggesting that a balance in Htr5b expression levels would be critically important for maintaining the functional normality in the nervous system.
In addition to the observations made in vivo, we also found that down-regulation of Htr5b in cultured hippocampal neurons resulted in reduced dendritic length and spine density. Our results reveal a novel functional role of Htr5b in regulation of dendrite morphology development. Specifically, we observed that the deletion of Utx results in a decreased Htr5b associated with an increased H3K27me3 modification. ChIP assay showed high enrichment of H3K27me3 in the promoter region of Htr5b. These results suggest that regulation of Utx in Htr5b expression is likely mediated through demethylation of H3K27me3. Functionally, overexpression of Htr5b in cultured neurons could rescue the impairments in dendritic length and spine density induced by Utx deletion, suggesting that Htr5b is involved in UTX-mediated H3K27me3 de-methylation in regulation of neuronal morphology. Our findings provide a novel mechanism underlying the role of Utx in neural development, synaptic transmission, and cognition. Assessment of behavioral performance by modulating H3k27me3 levels in genetically animal models together with UTX inhibitors would provide more insights into the mechanisms underlying H3K27me3 modification in cognitive function, which remains to be studied.
In summary, we show here that H3K27 demethylase enzyme Utx loss-of-function contributes to impairments in neuronal development and synaptic plasticity, which are responsible for mood and cognitive deficits in Utx cKO mice. Given the fact that behavior changes in Utx loss-of-function mice replicate some symptoms in human Kabuki patients, Utx cKO mouse models like ours therefore provide a valuable means to study the underlying mechanisms of the etiology of Kabuki syndrome, and to develop novel clinical implications.
Significance Statement
Trimethylation of histone H3 lysine 27 (H3K27me3) establishes a repressive chromatin state in silencing gene expression. The demethylases UTX mediates the removal of H3K27me2/3 to establish a mechanistic switch to activate large sets of genes. Moreover, defects in UTX cause Kabuki syndrome characterized by congenital anomaly and mental retardation. We discovered that deletion of UTX in the brain results in increased anxiety-like behaviors, impaired spatial learning and memory, and neuronal morphology deficiency in mice. UTX regulates a subset of genes that are associated with dendritic morphology, synaptic transmission, and cognition. This study enhances our understanding the cognitive and developmental deficits in Kabuki syndrome.
Author Contributions
GT, ZT, and CL designed research; GT, YZ, and PL performed the majority of the experiments; TM and LY performed the electrophysiological recordings; SZ, SD, QT, YX, HY, and HD contributed to collection and assembly of data; GT, ZT, and CL wrote the paper; CL, FZ, and ZT supervised the research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Chu Chen at Louisiana State University Health Sciences Center for editing the manuscript. This work was supported by grants from the National Science Foundation of China (grant nos. 31571043 to CL. and 81571212 to ZT), the National Key Research and Development Program of China Project (grant no. 2016YFA0101402 to CL), State Key Laboratory of Stem Cell and Reproductive Biology, and the Hundred Talents Program of CAS.
References
Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734. doi: 10.1038/nature06145
Bannerman, D. M., Sprengel, R., Sanderson, D. J., McHugh, S. B., Rawlins, J. N., Monyer, H., et al. (2014). Hippocampal synaptic plasticity, spatial memory and anxiety. Nat. Rev. Neurosci. 15, 181–192. doi: 10.1038/nrn3677
Beique, J. C., Lin, D. T., Kang, M. G., Aizawa, H., Takamiya, K., and Huganir, R. L. (2006). Synapse-specific regulation of AMPA receptor function by PSD-95. Proc. Natl. Acad. Sci. U.S.A. 103, 19535–19540. doi: 10.1073/pnas.0608492103
Bekkers, J. M., and Stevens, C. F. (1990). Presynaptic mechanism for long-term potentiation in the hippocampus. Nature 346, 724–729. doi: 10.1038/346724a0
Costall, B., Domeney, A. M., Kelly, M. E., Tomkins, D. M., Naylor, R. J., Wong, E. H., et al. (1993). The effect of the 5-HT3 receptor antagonist, RS-42358-197, in animal models of anxiety. Eur. J. Pharmacol. 234, 91–99. doi: 10.1016/0014-2999(93)90710-Y
de la Torre-Ubieta, L., and Bonni, A. (2011). Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron 72, 22–40. doi: 10.1016/j.neuron.2011.09.018
Eichenbaum, H. (2004). Hippocampus: cognitive processes and neural representations that underlie declarative memory. Neuron 44, 109–120. doi: 10.1016/j.neuron.2004.08.028
Etchegaray, J. P., Yang, X. M., DeBruyne, J. P., Peters, A. H. F. M., Weaver, D. R., Jenuwein, T., et al. (2006). The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 281, 21209–21215. doi: 10.1074/jbc.M603722200
Gatt, J. M., Nemeroff, C. B., Schofield, P. R., Paul, R. H., Clark, C. R., Gordon, E., et al. (2010). Early life stress combined with serotonin 3A receptor and brain-derived neurotrophic factor valine 66 to methionine genotypes impacts emotional brain and arousal correlates of risk for depression. Biol. Psychiatry 68, 818–824. doi: 10.1016/j.biopsych.2010.06.025
Greenfield, A., Carrel, L., Pennisi, D., Philippe, C., Quaderi, N., Siggers, P., et al. (1998). The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7, 737–742. doi: 10.1093/hmg/7.4.737
Hering, H., and Sheng, M. (2001). Dendritic spines: structure, dynamics and regulation. Nat. Rev. Neurosci. 2, 880–888. doi: 10.1038/35104061
Homma, K., Kitamura, Y., Ogawa, H., and Oka, K. (2006). Serotonin induces the increase in intracellular Ca2+ that enhances neurite outgrowth in PC12 cells via activation of 5-HT3 receptors and voltage-gated calcium channels. J. Neurosci. Res. 84, 316–325. doi: 10.1002/jnr.20894
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Lederer, D., Grisart, B., Digilio, M. C., Benoit, V., Crespin, M., Ghariani, S. C., et al. (2012). Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 90, 119–124. doi: 10.1016/j.ajhg.2011.11.021
Lee, M. G., Villa, R., Trojer, P., Norman, J., Yan, K. P., Reinberg, D., et al. (2007). Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450. doi: 10.1126/science.1149042
Li, J., Hart, R. P., Mallimo, E. M., Swerdel, M. R., Kusnecov, A. W., and Herrup, K. (2013). EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat. Neurosci. 16, 1745–1753. doi: 10.1038/nn.3564
Liao, D., Hessler, N. A., and Malinow, R. (1995). Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature 375, 400–404. doi: 10.1038/375400a0
Lisman, J., and Raghavachari, S. (2006). A unified model of the presynaptic and postsynaptic changes during LTP at CA1 synapses. Sci. STKE 2006:re11. doi: 10.1126/stke.3562006re11
Ma, D. K., Marchetto, M. C., Guo, J. U., Ming, G.-L., Gage, F. H., and Song, H. (2010). Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 13, 1338–1344. doi: 10.1038/nn.2672
Maekawa, T., Kim, S., Nakai, D., Makino, C., Takagi, T., Ogura, H., et al. (2010). Social isolation stress induces ATF-7 phosphorylation and impairs silencing of the 5-HT 5B receptor gene. EMBO J. 29, 184–195. doi: 10.1038/emboj.2009.318
Mansour, A. A., Gafni, O., Weinberger, L., Zviran, A., Ayyash, M., Rais, Y., et al. (2012). The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488, 409–413. doi: 10.1038/nature11272
Meneses, A. (1999). 5-HT system and cognition. Neurosci. Biobehav. Rev. 23, 1111–1125. doi: 10.1016/S0149-7634(99)00067-6
Miyake, N., Mizuno, S., Okamoto, N., Ohashi, H., Shiina, M., Ogata, K., et al. (2013). KDM6A point mutations cause Kabuki syndrome. Hum. Mutat. 34, 108–110. doi: 10.1002/humu.22229
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M.-W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. doi: 10.1073/pnas.1002530107
Plath, K., Fang, J., Mlynarczyk-Evans, S. K., Cao, R., Worringer, K. A., Wang, H. B., et al. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135. doi: 10.1126/science.1084274
Portela, A., and Esteller, M. (2010). Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068. doi: 10.1038/nbt.1685
Rodgers, R. J., and Dalvi, A. (1997). Anxiety, defence and the elevated plus-maze. Neurosci. Biobehav. Rev. 21, 801–810. doi: 10.1016/S0149-7634(96)00058-9
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8:e1002964. doi: 10.1371/journal.pgen.1002964
Smith, S. M., Kimyon, R. S., and Watters, J. J. (2014). Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method. ASN Neuro 6, 193–207. doi: 10.1042/AN20130050
Sparmann, A., and van Lohuizen, M. (2006). Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856. doi: 10.1038/nrc1991
Sparta, D. R., and Stuber, G. D. (2014). Cartography of serotonergic circuits. Neuron 83, 513–515. doi: 10.1016/j.neuron.2014.07.030
Tsankova, N., Renthal, W., Kumar, A., and Nestler, E. J. (2007). Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 8, 355–367. doi: 10.1038/nrn2132
Van der Meulen, J., Sanghvi, V., Mavrakis, K., Durinck, K., Fang, F., Matthijssens, F., et al. (2015). The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21. doi: 10.1182/blood-2014-05-577270
Van der Meulen, J., Speleman, F., and Van Vlierberghe, P. (2014). The H3K27me3 demethylase UTX in normal development and disease. Epigenetics 9, 658–668. doi: 10.4161/epi.28298
van Haaften, G., Dalgliesh, G. L., Davies, H., Chen, L., Bignell, G., Greenman, C., et al. (2009). Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41, 521–523. doi: 10.1038/ng.349
von Schimmelmann, M., Feinberg, P. A., Sullivan, J. M., Ku, S. M., Badimon, A., Duff, M. K., et al. (2016). Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 19, 1321–1330. doi: 10.1038/nn.4360
Vorhees, C. V., and Williams, M. T. (2006). Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858. doi: 10.1038/nprot.2006.116
Wang, C. C., Lee, J. E., Cho, Y. W., Xiao, Y., Jin, Q. H., Liu, C. Y., et al. (2012). UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 109, 15324–15329. doi: 10.1073/pnas.1204166109
Welstead, G. G., Creyghton, M. P., Bilodeau, S., Cheng, A. W., Markoulaki, S., Young, R. A., et al. (2012). X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. U.S.A. 109, 13004–13009. doi: 10.1073/pnas.1210787109
Wirth, A., Holst, K., and Ponimaskin, E. (2016). How serotonin receptors regulate morphogenic signalling in neurons. Prog. Neurobiol. 151, 35–56. doi: 10.1016/j.pneurobio.2016.03.007
Xiao, M., Xu, L., Laezza, F., Yamada, K., Feng, S., and Ornitz, D. M. (2007). Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol. Cell. Neurosci. 34, 366–377. doi: 10.1016/j.mcn.2006.11.020
Xu, J., Deng, X. X., Watkins, R., and Disteche, C. M. (2008). Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 28, 4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008
Keywords: Utx, H3K27me3, synaptic transmission, cognition
Citation: Tang G-B, Zeng Y-Q, Liu P-P, Mi T-W, Zhang S-F, Dai S-K, Tang Q-Y, Yang L, Xu Y-J, Yan H-L, Du H-Z, Teng Z-Q, Zhou F-Q and Liu C-M (2017) The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice. Front. Mol. Neurosci. 10:267. doi: 10.3389/fnmol.2017.00267
Received: 16 May 2017; Accepted: 07 August 2017;
Published: 24 August 2017.
Edited by:
Oliver Stork, Otto von Guericke University of Magdeburg, GermanyReviewed by:
Jaewon Ko, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaChiara Verpelli, Istituto di Neuroscienze (CNR), Italy
Valentin Stein, University of Bonn, Germany
Copyright © 2017 Tang, Zeng, Liu, Mi, Zhang, Dai, Tang, Yang, Xu, Yan, Du, Teng, Zhou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao-Qian Teng, tengzq@ioz.ac.cn
Feng-Quan Zhou, fzhou4@jhmi.edu
Chang-Mei Liu, liuchm@ioz.ac.cn
†These authors have contributed equally to this work.
 Gang-Bin Tang1†
Gang-Bin Tang1†  Pei-Pei Liu
Pei-Pei Liu Ting-Wei Mi
Ting-Wei Mi Zhao-Qian Teng
Zhao-Qian Teng Feng-Quan Zhou
Feng-Quan Zhou Chang-Mei Liu
Chang-Mei Liu