Maintenance of Positional Identity of Neural Progenitors in the Embryonic and Postnatal Telencephalon
- 1Department of Neurological Surgery, University of California, San Francisco, San Francisco, CA, United States
- 2Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, University of California, San Francisco, San Francisco, CA, United States
- 3Biomedical Sciences Program, University of California, San Francisco, San Francisco, CA, United States
- 4Medical Scientist Training Program, University of California, San Francisco, San Francisco, CA, United States
- 5San Francisco Veterans Affairs Medical Center, San Francisco, CA, United States
Throughout embryonic development and into postnatal life, regionally distinct populations of neural progenitor cells (NPCs) collectively generate the many different types of neurons that underlie the complex structure and function of the adult mammalian brain. At very early stages of telencephalic development, NPCs become organized into regional domains that each produce different subsets of neurons. This positional identity of NPCs relates to the regional expression of specific, fate-determining homeodomain transcription factors. As development progresses, the brain undergoes vast changes in both size and shape, yet important aspects of NPC positional identity persist even into the postnatal brain. How can NPC positional identity, which is established so early in brain development, endure the many dynamic, large-scale and complex changes that occur over a relatively long period of time? In this Perspective article, we review data and concepts derived from studies in Drosophila regarding the function of homeobox (Hox) genes, Polycomb group (PcG) and trithorax group (trxG) chromatin regulators. We then discuss how this knowledge may contribute to our understanding of the maintenance of positional identity of NPCs in the mammalian telencephalon. Similar to the axial body plan of Drosophila larvae, there is a segmental nature to NPC positional identity, with loss of specific homeodomain transcription factors causing homeotic-like shifts in brain development. Finally, we speculate about the role of mammalian PcG and trxG factors in the long-term maintenance of NPC positional identity and certain neurodevelopmental disorders.
Positional Identity as a Determinant of Cell Fate
Cell fate determination is a fundamental aspect of metazoan development. At very early stages of mammalian embryogenesis, differences in cell position begin to correspond to distinct developmental fates. For instance, after totipotent blastomeres undergo a process known as compaction (Johnson and McConnell, 2004), cells that are located more superficially give rise to the placenta, while those positioned deeper in the embryo generate the pluripotent progenitors of the inner cell mass (Tarkowski and Wróblewska, 1967; Balakier and Pedersen, 1982; Pedersen et al., 1986; Dyce et al., 1987). As the major axes of the body plan are elaborated, progenitor cells attain more refined positional identities that further correspond to their cell fate decisions (Gilbert, 2010).
In the fruit fly, Drosophila melanogaster, the establishment of body plan positional identity can be conceptually divided into two major processes—segmentation and specification. Shortly after gastrulation, the expression of segmentation genes patterns the embryonic ectoderm along the anterior-posterior (AP) axis (Figure 1A; Martinez-Arias and Lawrence, 1985; Akam, 1987). Early segmentation genes (known as gap genes) are responsible for large-scale aspects of the AP axis, while those that are expressed later (pair-rule genes) refine the pattern (Scott and Carroll, 1987). Mutations in segmentation genes result in the loss of segments. For instance, mutations in the gap gene kruppel result in a larva missing all thoracic segments and the first five abdominal segments. Pair-rule gene even-skipped functions later in segmentation, and its mutation results in the absence of even-numbered segments throughout the length of the AP axis.
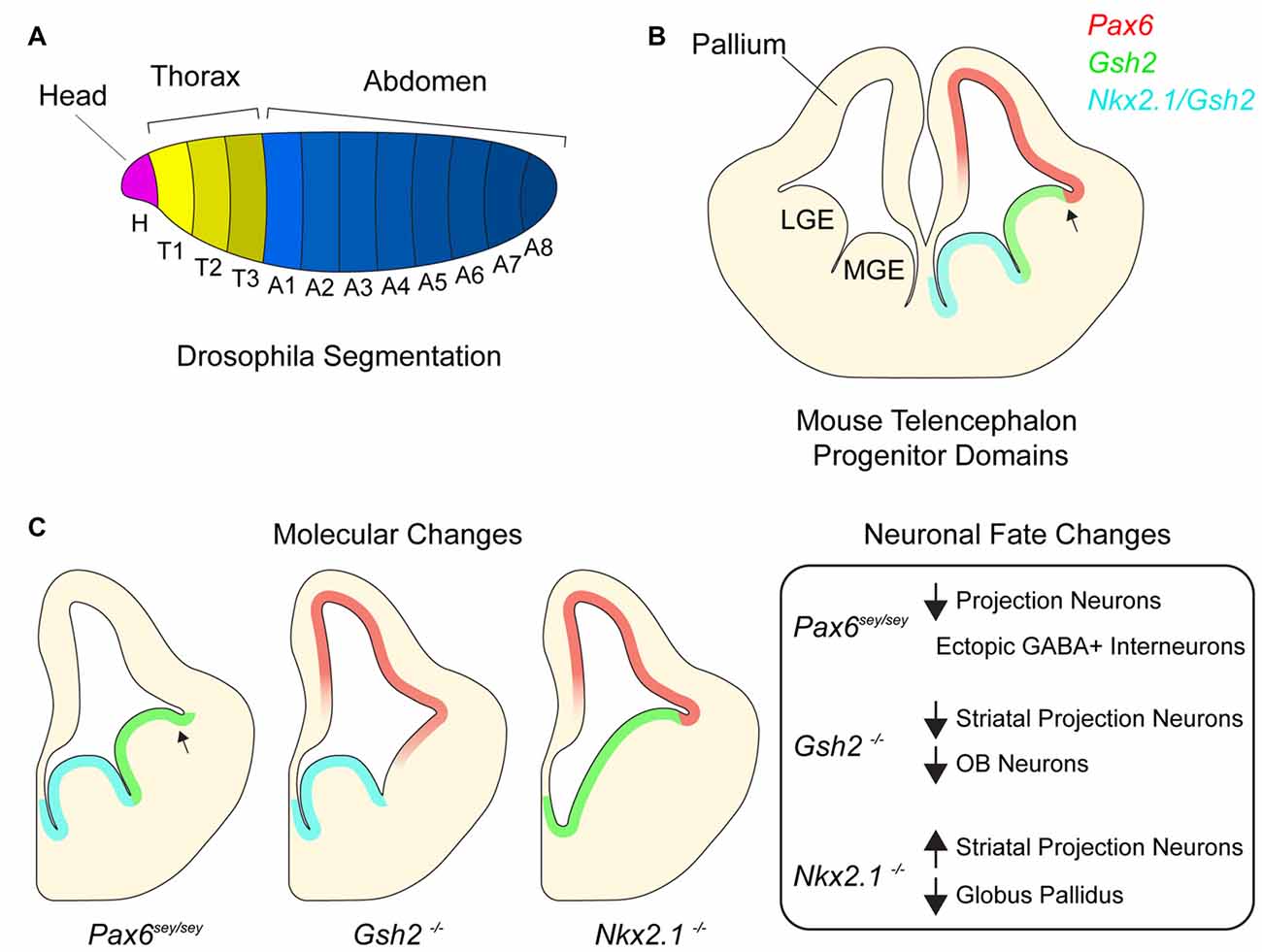
Figure 1. Homeotic transcription factors pattern the developing telencephalon. (A) Drosophila embryo with different body segments. Different colors of each segment signify different combinatorial expression of homeotic transcription factors. (B) Germinal domains and transcription factor patterning in embryonic mouse telencephalon. Medial ganglionic eminence (MGE), lateral ganglionic eminence (LGE). Arrow denotes pallial-subpallial boundary (PSB). (C) Schematic depicting molecular changes in various transcription factor mutants and table listing corresponding neuronal fate changes.
Segmentation genes induce the expression of Drosophila homeobox (Hox) transcription factors, which in turn specify the identity of each segment (White and Lehmann, 1986; McGinnis and Krumlauf, 1992; Lawrence and Morata, 1994). Mutations in Hox genes do not result in the loss of segment number but rather cause a change in segment identity. For example, the Hox gene Ubx is expressed in the third thoracic segment where it is required to generate an appendage called a haltere (Ouweneel and van der Meer, 1973; Lawrence and Morata, 1983). In the absence of Ubx, the third thoracic segment fails to generate halters and instead makes an additional wing, an appendage that normally develops from an adjacent segment. This change in segment identity is known as a “homeotic shift” and results from the ectopic expression of Antp, a Hox gene responsible for wing development. Such ectopic expression of Hox genes that are normally expressed in neighboring regions is a common transcriptional phenotype of Hox gene mutations. Similar findings have been made in studies of mammalian Hox genes (Pearson et al., 2005) as well as the larger set of homeodomain-containing transcription factors, including those that regulate telencephalic development (Hebert and Fishell, 2008), as we later discuss.
Polycomb and Trithorax Group Chromatin Regulators and The Maintenance of Positional Identity
The local structure of chromatin—the dynamic polymer of DNA and histone proteins—can influence whether a locus is expressed or silenced. Thus, changes to chromatin structure can engage and maintain particular genetic programs, helping determine cellular identity. The Polycomb group (PcG) and trithorax group (trxG) gene products—which were initially discovered in Drosophila—comprise an evolutionarily conserved set of chromatin regulators that appear to serve as a transcriptional “memory” system (Geisler and Paro, 2015; Schuettengruber et al., 2017). By assembling into large multiprotein complexes that modify chromatin structure, PcG and trxG factors help organize the genome regionally into transcriptionally silent or active states, respectively (for a review of PcG and trxG protein molecular mechanisms, please see Steffen and Ringrose, 2014; Geisler and Paro, 2015; Schuettengruber et al., 2017).
In Drosophila, shortly after the establishment of Hox gene expression, gap and pair-rule genes are downregulated, and PcG and trxG genes are required to maintain normal patterns of Hox gene expression. For instance, trxG genes are required to maintain the appropriate regional expression of Ubx (Kassis et al., 2017). Reminiscent of the homeotic shift observed in Ubx mutants, the loss of trxG function results in the development of a second set of wing tissue in place or the normal halters (Breen, 1999). Conversely, PcG genes are required to repress Ubx expression in the tissue anterior to the Ubx expression domain (Kassis et al., 2017). Of note, Hox gene expression is properly induced in both PcG and trxG mutants but is lost over time (Lewis, 1978; Struhl and Akam, 1985; Yu et al., 1998; Ernst et al., 2004). Thus, in the absence of proper PcG and trxG function, the expression of certain Hox genes is not maintained, resulting in homeotic shifts (Lewis, 1978).
PcG and trxG proteins are also required for the maintenance of Hox gene expression in mice (Schuettengruber et al., 2017). The prototypical trxG gene trithorax (trx) and its mammalian homolog Mixed-lineage leukemia 1 (Mll1) are both required to positively maintain Hox gene expression (Ingham and Whittle, 1980; Yu et al., 1998). Similar to phenotypes observed in Drosophila, Hox gene expression is established normally in Mll1-null mice but is not properly maintained, resulting in homeotic transformations of their axial skeleton (Yu et al., 1995, 1998). Homeotic skeletal transformations are also observed in mice null for Bmi1, a PcG gene (van der Lugt et al., 1994). Thus, PcG and trxG genes are key components of a “cellular memory system” that maintains the positional identity of progenitor cells in mammalian development.
Positional Identity in The Embryonic Telencephalon
Similar to regional patterning that occurs in the early embryo, progenitors throughout the developing mammalian central nervous system are organized into distinct domains with different positional identities that are in part defined by the expression of homeodomain transcription factors (Shimamura et al., 1995; Flames et al., 2007; Dasen and Jessell, 2009; Narita and Rijli, 2009; Tümpel et al., 2009). In the embryonic telencephalon, excitatory neurons are born dorsally in the pallium while most inhibitory neurons are born ventrally in the subpallium (Anderson et al., 1997; Puelles et al., 2000; Molyneaux et al., 2007; Kepecs and Fishell, 2014). Pallial progenitors express PAX6 and generate cortical projection neurons which migrate radially and give rise to the six-layered neocortex (Custo Greig et al., 2013). The juxtaposition of PAX6+ progenitors and GSH2+ progenitors of the subpallium forms the pallial-subpallial boundary (PSB; Toresson et al., 2000; Corbin et al., 2003). The subpallium is further subdivided into several subdomains including the lateral and medial ganglionic eminences (LGE and MGE), which generate different subtypes of inhibitory neurons. The LGE is located immediately ventral to the pallium and is dorsal to the MGE. LGE progenitors give rise to a large number of olfactory bulb (OB) interneurons and striatal projection neurons while the MGE generates cortical interneurons and the globus pallidus as well as a small number of OB interneurons (Butt et al., 2008; Xu et al., 2008; Flandin et al., 2010). Nkx2.1 is expressed throughout the MGE and not detected in the LGE. Together, the pallium, LGE and MGE form three molecularly distinct domains along the dorsoventral axis of the embryonic telencephalon (Figure 1B). In addition to their expression defining the location and regional boundaries of these domains, Pax6, Gsh2 and Nkx2.1 are also required to specify the developmental potential of neural progenitor cells (NPCs) in their respective domains, as we discuss below.
Homeotic-Like Shifts in The Developing Brain
Small eye (Sey) is a naturally occurring Pax6 allele that is a loss-of-function nonsense mutation (Hill et al., 1991). While the developing neocortex of Pax6Sey/Sey mice maintains the expression of certain pallial genes such as Tbr1 and Math2, some NPCs gradually become mis-specified, adopting a subpallial-like identity (Figure 1C; Manuel et al., 2015), with Gsh2 and subpallial genes Ascl1 and Dlx2 becoming expressed dorsally across the PSB (Toresson et al., 2000). Furthermore, by E15.5, pallial NPCs in Pax6Sey/Sey mice begin generating GABAergic interneurons with an LGE-like identity (Kroll and O’Leary, 2005). These data suggest that as the neocortex develops, sustained Pax6 expression is required to repress ventral telencephalic gene expression and associated neuronal fates.
In the subpallium, Gsh2 is critical to the specification of LGE positional identity. In the LGE of E12.5 Gsh2−/− mice, the expression of Ascl1 and Dlx2 is nearly undetectable (Szucsik et al., 1997; Corbin et al., 2000; Toresson et al., 2000; Yun et al., 2001). Furthermore, expression of Pax6 as well other pallial genes (e.g., Tbr2 and Ngn2) extends ventrally past the PSB into the dorsal LGE. This early absence of LGE identity and ectopic expression of pallial genes correlates with the development of a smaller striatum, and embryonic OB neurogenesis is also impaired. Thus, without Gsh2, the dorsal LGE initially takes on a pallial-like NPC identity, and the genesis of LGE-lineage neuronal subpopulations is defective (Figure 1C; Corbin et al., 2000; Yun et al., 2001).
Nkx2.1 plays key roles in the positional identity of the MGE. In the absence of Nkx2.1 expression, the MGE adopts an LGE-like identity and fails to generate MGE-specific neuron populations (Figure 1C; Sussel et al., 1999; Butt et al., 2008; Nóbrega-Pereira et al., 2008). For instance, conditional deletion of Nkx2.1 at E10.5 from the subpallium decreases the production of MGE-derived cortical interneurons (Butt et al., 2008), and in Nkx2.1−/− mice, development of the globus pallidus is severely impaired (Sussel et al., 1999). Without Nkx2.1, the mutant MGE appears to become partially dorsalized, having ectopic expression of the normally LGE-specific transcription factors Isl1, SCIP and GOLF. Consistent with this LGE-like transcriptional character, the Nkx2.1-null MGE generates striatal neurons. Furthermore, conditional deletion of Nkx2.1 at E16 results in the loss of chandelier cells (Taniguchi et al., 2013), a later-born population of cortical interneurons (Inan et al., 2012). Taken together, these data indicate that Nkx2.1 is required for the maintenance of MGE identity and proper developmental potential of this population of ventral NPCs in both the early and late embryonic brain.
NPCs in The Postnatal Brain: Maintenance of Embryonic Positional Identity
The postnatal mammalian brain harbors NPCs in the ventricular-subventricular zone (V-SVZ), a layer of cells found along the walls of the cerebral ventricles (Lim and Alvarez-Buylla, 2016). In the adult mouse brain, V-SVZ NPCs—known as B1 cells—give rise to neuroblasts that migrate to the OB where they differentiate into several types of interneurons. The two main categories of OB interneurons are granule cells (GCs) and periglomerular cells (PGCs), both of which can be further divided into additional subtypes (Price and Powell, 1970; Kosaka et al., 1997). GC interneurons can be categorized as superficial or deep depending on their location within the GC layer. PGCs can be divided into three mutually exclusive groups by their expression of Calbinden (CalB), Calretinin (CalR), or tyrosine hydroxylase (TH).
Similar to embryonic NPCs, B1 cells have distinct positional identities that give rise to different subtypes of PGCs and GCs. For instance, while B1 cells in the dorsal V-SVZ produce superficial GCs and TH+ PGCs, ventral B1 cells generate deep GCs and CalB+ PGCs (Figure 2A; Merkle et al., 2007; Alvarez-Buylla et al., 2008; Rushing and Ihrie, 2016). Interestingly, such regional differences in the developmental potential of V-SVZ NPCs are retained even when serially passaged in vitro and transplanted into different locations of this postnatal germinal zone. For example, ventrally derived NPCs transplanted to the dorsal V-SVZ still produce deep granule neurons but not TH-positive PGCs (Merkle et al., 2007). Thus, regional differences in V-SVZ NPCs appear to be in large part cell-intrinsic and stable through serial cell divisions.
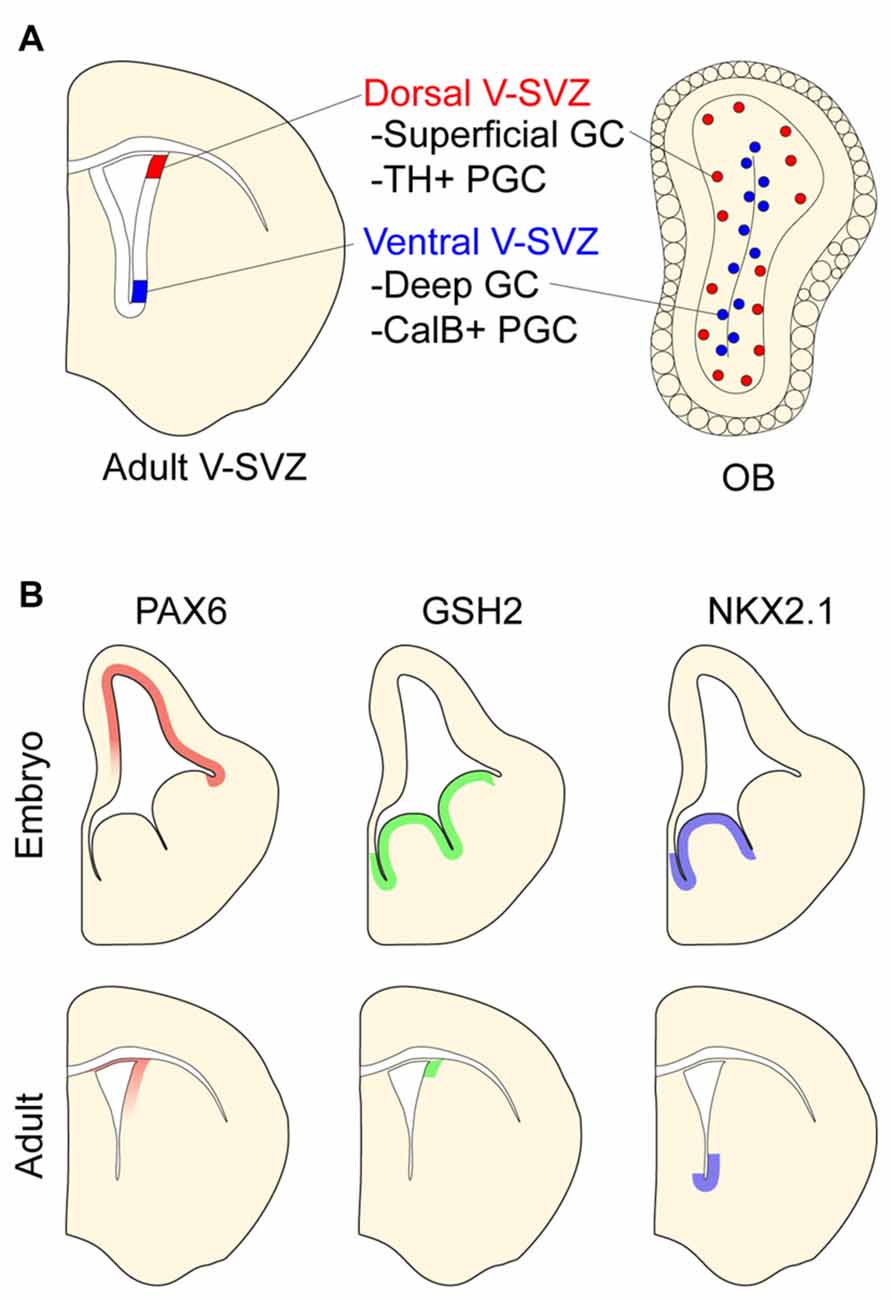
Figure 2. Positional identity in the postnatal ventricular-subventricular zone (V-SVZ). (A) Schematic of postnatal V-SVZ and corresponding olfactory bulb (OB) section depicting positional identity of V-SVZ neural stem cells. Deep (blue) and superficial (red) granule cells (GCs) depicted in OB. (B) Regional transcription factor patterning in embryonic and postnatal V-SVZ germinal zones. Boundaries of protein expression depicted in both embryonic and postnatal germinal zones.
Postnatal B1 cells arise from embryonic NPCs (Merkle et al., 2004; Young et al., 2007; Delgado and Lim, 2015). Clonal analysis with a “barcoded” retroviral library demonstrates that B1 cells share a common embryonic NPC with those that generate neurons for the cortex, striatum and septum (Fuentealba et al., 2015). When NPCs are transduced with retroviral vectors at E12.5 and brains analyzed after ~5 weeks, approximately 35% of the clones contain postnatally-born OB neurons as well as embryonically-born forebrain cells. Further analysis of such clones suggests that B1 cells retain positional information of the shared embryonic NPC. For example, clones containing cortical projection neurons (indicating their birth from pallial NPCs) include superficial OB GCs (which arise from B1 cells close to the pallium). Thus, the positional identity of V-SVZ NPCs appears to be established during embryogenesis and persists throughout development and into postnatal life.
Like the embryonic germinal zones, the postnatal V-SVZ exhibits regional patterns of transcription factor expression that correspond to the developmental potential of local NPCs (Figure 2B and reviewed in Alvarez-Buylla et al., 2008). For instance, the dorsal V-SVZ expresses Emx1, and consistent with the developmental potential of B1 cells in this region (Merkle et al., 2007), Emx1-lineage cells predominantly generate TH+ and CalR+ PGCs in adulthood (Fuentealba et al., 2015). In the most ventral aspect of the V-SVZ, B1 cells express Nkx2.1. Administering tamoxifen to adult Cre-reporter mice carrying the Nkx2.1-CreER “knock-in” allele labels ventral B1 cells that produce deep OB GCs, which is coherent with results from stereotactic methods of labeling ventral B1 cells (Merkle et al., 2007). Several other regional V-SVZ subdomains defined by the expression of specific transcription factors have been similarly defined and found to generate region-appropriate OB subtypes (Merkle et al., 2014).
The pattern of regional transcription factor expression in the V-SVZ is similar to that observed in embryonic development (Alvarez-Buylla et al., 2008). Interestingly, for some of these genes, it appears that such regional expression is maintained in NPCs throughout embryonic development and into adulthood. For example, pallial NPCs labeled with Emx1-CreER at E10.5 give rise to cells that populate the dorsal V-SVZ (Young et al., 2007) where Emx1 continues to be expressed postnatally. Similarly, MGE NPCs labeled at E12.5 with Nkx2.1-CreER give rise to ventral V-SVZ cells including local B1 cells (Delgado and Lim, 2015). Of note, virtually all V-SVZ cells expressing the Cre-reporter are also immunopositive for NKX2.1 protein in adult mice. Even when serially passaged in culture, ventral V-SVZ NPCs retain the expression of Nkx2.1 (Delgado et al., 2016). These data suggest that key transcriptional differences that define the positional identity of B1 cells relate to the “retention” of region-specific gene expression that was established very early in brain development.
Potential Mechanisms Underlying The Maintenance of NPC Positional Identity
The persistence of regionally discrete gene expression along the developmental continuum of NPCs from the early embryo to the adult is remarkable not only in terms of duration, but also because of the tremendous increase in size and anatomic complexity of the brain over this period of time. How is NPC positional identity maintained in the face of these challenges? Below, we touch upon the potential roles of morphogens, transcriptional feedback mechanisms, non-coding RNAs (ncRNAs) and the mammalian PcG/trxG chromatin regulators. To simplify the context of this brief discussion, we focus on the population of Nkx2.1+ NPCs.
The establishment of regional transcription factor expression in the embryonic telencephalon requires the actions of morphogens such as Sonic hedgehog (Shh; Wilson and Rubenstein, 2000; Monuki and Walsh, 2001; Hebert and Fishell, 2008). Shh is required for the early induction of Nkx2.1 in the ventral neural tube, and at E12.5, Shh appears to maintain Nkx2.1 expression in the MGE (Xu et al., 2005; Gulacsi and Anderson, 2006). Given that genes downstream of Shh signaling are expressed in the postnatal V-SVZ (Ihrie et al., 2011), it is possible that Shh is also required to maintain Nkx2.1 in ventral B1 cells. Deletion of Shh in adulthood reduces the production of ventrally derived OB interneurons (Ihrie et al., 2011), suggestive of a loss of ventral NPC identity. However, Shh is also a mitogen for NPCs (Palma et al., 2005), and it remains to be determined if changes in the proliferation of V-SVZ cells contribute to these findings. Can morphogens alone be expected to maintain regionally discrete gene expression as the brain grows in size and anatomic complexity? Though certainly conjecture, we suggest that biological mechanisms other than Shh signaling are required to maintain Nkx2.1 expression at some point along the developmental continuum of early embryonic to postnatal NPCs.
Transcriptional autoregulation is an important mechanism underlying the maintenance of homeotic gene expression during embryogenesis (Lou et al., 1995; Packer et al., 1998). The Nkx2.1 promoter region contains conserved NKX2.1-binding sites to which NKX2.1 can bind and positively regulate transcription (Oguchi and Kimura, 1998; Das et al., 2011). Thus, it is possible that NKX2.1 uses a positive feedback loop for transcriptional maintenance, which could also contribute to the “discreteness” of Nkx2.1 expression. While transcription of the Nkx2.1 locus does not require functional NKX2.1 protein (mutant transcripts are detected in Nkx2.1−/− NPCs; Sussel et al., 1999; Toresson et al., 2000), whether expression of the mutant alleles diminishes over time has not been reported. In any case, transcriptional autoregulation (as well as additional regulatory mechanisms such as the activation or repression of other genes) likely requires the function of chromatin regulatory factors.
The mammalian genome transcribes a large number and diversity of ncRNAs, and specific ncRNAs can regulate the expression of Hox transcription factors. Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that do not code for protein, and it is now clear that certain lncRNAs have important cellular function and interact with PcG/trxG factors (Davidovich and Cech, 2015; Engreitz et al., 2016). In the developing brain, many lncRNAs are highly cell-type specific (Liu et al., 2016), and some lncRNAs play key roles in neurodevelopment (Andersen and Lim, 2017). In the mouse genome, the lncRNA NANCI is located adjacent to Nkx2.1 and is co-expressed with Nkx2.1 in the lungs where it positively regulate Nkx2.1 transcription (Herriges et al., 2014, 2017). In the forebrain including the MGE, NANCI is also co-expressed with Nkx2.1 (Herriges et al., 2014), but whether NANCI helps maintain Nkx2.1 expression in a stable and heritable manner throughout development has not been reported.
microRNAs (miRNAs) are an important class of ncRNAs that downregulate gene expression post-transcriptionally via base-paring with complementary sequences within the target mRNA transcript (Bartel, 2009). Some miRNAs target the mRNAs of Hox transcription factors and may thus play roles in NPC positional identity. For instance, miR-7a is expressed in a ventral-to-dorsal gradient in the mouse V-SVZ, and this miR-7a gradient contributes to the regional expression of PAX6 protein (de Chevigny et al., 2012). While miR-365 has been shown to negatively regulate NKX2.1 in lung cancer cell lines (Kang et al., 2013) its potential role in regulating Nkx2.1 expression in the forebrain has not been reported. Importantly, given that miRNAs can regulate the expression of PcG genes such as EZH2 (Szulwach et al., 2010; Neo et al., 2014), and that some miRNAs are embedded within the Hox gene clusters (Mansfield et al., 2004; Tehler et al., 2011), it will be important to consider the role of miRNAs in NPC positional identity.
Most studies investigating the role of mammalian PcG and trxG chromatin regulators in neural development have focused on the general processes of NPC self-renewal, neuronal differentiation and gliogenesis (reviewed in Hirabayashi and Gotoh, 2010; Lim and Alvarez-Buylla, 2014). However, given that PcG/trxG proteins play key roles in the maintenance of Hox gene expression and positional identity during development of the axial body plan, it seems reasonable to consider the possibility that these factors also maintain the positional identity of NPCs during brain development. For instance, similar to the requirement for Mll1 in the maintenance of Hox gene expression of the early mouse embryo (Yu et al., 1995), does the continuous expression of Nkx2.1 and related ventral identity of NPCs also depend on the action of mammalian trxG proteins? More generally, would deficiencies of trxG or PcG activity lead to homeotic-like shifts in telencephalic NPCs, changing the proportions of neuronal subtypes that are produced from each region?
The Importance of Understanding How NPCs Maintain Positional Identity
Current concepts regarding NPC positional identity in the mammal primarily involve morphogens (or other inductive signals) and transcription factors. Based on data and concepts described in sections above, we suggest that certain chromatin regulators are integral to the maintenance of NPC positional identity. That is, while morphogen gradients indeed establish regional patterns of NPCs in the early telencephalon, and transcription factor regulatory networks are likely critical to such positional identity, certain chromatin regulators may be required for NPCs to “remember” key region-specific aspects of their transcriptome as they continue to proliferate throughout development. The notion that chromatin regulators are critical to NPC positional identity (and thus their developmental potential) may have important implications for our understanding of human disease as well as the culture of NPCs for human therapeutics and drug development.
The maintenance of NPC positional identity is likely crucial to proper brain development, as loss of such developmental information might be expected to cause homeotic-like shifts that result in abnormal neuroanatomy. Recent studies have implicated mutations in many chromatin regulators as causes of human neurodevelopmental and psychiatric disorders (Ronan et al., 2013; De Rubeis et al., 2014). For instance, EZH2 and MLL1 are mutated in Weaver syndrome (Tatton-Brown et al., 2011) and Weidemann-Steiner syndrome (Jones et al., 2012; Strom et al., 2014), respectively, both of which are associated with intellectual disability. Understanding the potential role that these PcG and trxG factors play in NPC positional identity may provide important insights into the pathology of certain neurodevelopmental disorders.
The ability to produce specific neural cell types from cultured NPCs is broadly useful for research into human neurological disease and therapeutic development. The types of neurons generated from NPCs relates to their positional identity, which is generally induced by the application of morphogens. Discovering the mechanisms by which proliferating NPCs retain specific transcriptional programs for long periods of time may thus enhance our ability to durably propagate specific types of NPCs in vitro for such translational purposes. Furthermore, such research may inform methods by which we can “erase” and re-establish NPC identity.
Concluding Remarks
It is now clear that the positional identity of neural progenitors is an important contributor to neuronal diversity. As discussed above, the processes regulating NPC positional identity are reminiscent of those that regulate segment identity in the developing Drosophila larvae. As for progenitor cells of Drosophila larval segments, the positional identity of telencephalic NPCs requires the sustained expression of homeodomain transcription factors. Loss of such region-specific fate regulators causes embryonic NPCs to become misspecified and adopt the identity of the adjacent region, which results in abnormal anatomy. The mechanisms required for the maintenance of NPC positional identity are poorly understood at this time, but our understanding of chromatin regulators such as PcG and trxG factors can be incorporated into current concepts related to morphogen signaling and transcription factor networks. Given the particularly long duration of NPC proliferation in human brain development, the maintenance of NPC positional identity will likely have important implications to our understanding of certain neurodevelopmental disorders.
Author Contributions
RND and DAL wrote the perspective article. Some parts of this manuscript have been adopted from RND’s Doctoral Thesis.
Funding
This work was supported by National Institutes of Health, award no. R01NS091544 and U.S. Department of Veterans Affairs, award no. I01BX000252 to DAL; California Institute of Regeneration Medicine, award no. TG2-01153 to RND.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Akam, M. (1987). The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1–22.
Alvarez-Buylla, A., Kohwi, M., Nguyen, T. M., and Merkle, F. T. (2008). The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harb. Symp. Quant. Biol. 73, 357–365. doi: 10.1101/sqb.2008.73.019
Anderson, S. A., Eisenstat, D. D., Shi, L., and Rubenstein, J. L. (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476. doi: 10.1126/science.278.5337.474
Andersen, R. E., and Lim, D. A. (2017). Forging our understanding of lncRNAs in the brain. Cell Tissue Res. doi: 10.1007/s00441-017-2711-z [Epub ahead of print].
Balakier, H., and Pedersen, R. A. (1982). Allocation of cells to inner cell mass and trophectoderm lineages in preimplantation mouse embryos. Dev. Biol. 90, 352–362. doi: 10.1016/0012-1606(82)90384-0
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Breen, T. R. (1999). Mutant alleles of the Drosophila trithorax gene produce common and unusual homeotic and other developmental phenotypes. Genetics 152, 319–344.
Butt, S. J., Sousa, V. H., Fuccillo, M. V., Hjerling-Leffler, J., Miyoshi, G., Kimura, S., et al. (2008). The requirement of Nkx2–1 in the temporal specification of cortical interneuron subtypes. Neuron 59, 722–732. doi: 10.1016/j.neuron.2008.07.031
Corbin, J. G., Gaiano, N., Machold, R. P., Langston, A., and Fishell, G. (2000). The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development 127, 5007–5020.
Corbin, J. G., Rutlin, M., Gaiano, N., and Fishell, G. (2003). Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development 130, 4895–4906. doi: 10.1242/dev.00717
Custo Greig, L. F., Woodworth, M. B., Galazo, M. J., Padmanabhan, H., and Macklis, J. D. (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769. doi: 10.1038/nrn3586
Das, A., Acharya, S., Gottipati, K. R., McKnight, J. B., Chandru, H., Alcorn, J. L., et al. (2011). Thyroid transcription factor-1 (TTF-1) gene: identification of ZBP-89, Sp1, and TTF-1 sites in the promoter and regulation by TNF-α in lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 301, L427–L440. doi: 10.1152/ajplung.00090.2011
Dasen, J. S., and Jessell, T. M. (2009). Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 88, 169–200. doi: 10.1016/S0070-2153(09)88006-X
Davidovich, C., and Cech, T. R. (2015). The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022. doi: 10.1261/rna.053918.115
de Chevigny, A., Coré, N., Follert, P., Gaudin, M., Barbry, P., Béclin, C., et al. (2012). miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat. Neurosci. 15, 1120–1126. doi: 10.1038/nn.3142
De Rubeis, S., He, X., Goldberg, A. P., Poultney, C. S., Samocha, K., Cicek, A. E., et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. doi: 10.1038/nature13772
Delgado, R. N., and Lim, D. A. (2015). Embryonic Nkx2.1-expressing neural precursor cells contribute to the regional heterogeneity of adult V-SVZ neural stem cells. Dev. Biol. 407, 265–274. doi: 10.1016/j.ydbio.2015.09.008
Delgado, R. N., Lu, C., and Lim, D. A. (2016). Maintenance of neural stem cell regional identity in culture. Neurogenesis 3:e1187321. doi: 10.1080/23262133.2016.1187321
Dyce, J., George, M., Goodall, H., and Fleming, T. P. (1987). Do trophectoderm and inner cell mass cells in the mouse blastocyst maintain discrete lineages? Development 100, 685–698.
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi: 10.1038/nrm.2016.126
Ernst, P., Mabon, M., Davidson, A. J., Zon, L. I., and Korsmeyer, S. J. (2004). An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 14, 2063–2069. doi: 10.1016/j.cub.2004.11.012
Flames, N., Pla, R., Gelman, D. M., Rubenstein, J. L., Puelles, L., and Marin, O. (2007). Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007
Flandin, P., Kimura, S., and Rubenstein, J. L. (2010). The progenitor zone of the ventral medial ganglionic eminence requires Nkx2–1 to generate most of the globus pallidus but few neocortical interneurons. J. Neurosci. 30, 2812–2823. doi: 10.1523/JNEUROSCI.4228-09.2010
Fuentealba, L. C., Rompani, S. B., Parraguez, J. I., Obernier, K., Romero, R., Cepko, C. L., et al. (2015). Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655. doi: 10.1016/j.cell.2015.05.041
Geisler, S. J., and Paro, R. (2015). Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development 142, 2876–2887. doi: 10.1242/dev.120030
Gulacsi, A., and Anderson, S. A. (2006). Shh maintains Nkx2.1 in the MGE by a Gli3-independent mechanism. Cereb. Cortex 16, i89–i95. doi: 10.1093/cercor/bhk018
Hebert, J. M., and Fishell, G. (2008). The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 9, 678–685. doi: 10.1038/nrn2463
Herriges, M. J., Swarr, D. T., Morley, M. P., Rathi, K. S., Peng, T., Stewart, K. M., et al. (2014). Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 28, 1363–1379. doi: 10.1101/gad.238782.114
Herriges, M. J., Tischfield, D. J., Cui, Z., Morley, M. P., Han, Y., Babu, A., et al. (2017). The NANCI-Nkx2.1 gene duplex buffers Nkx2.1 expression to maintain lung development and homeostasis. Genes Dev. 31, 889–903. doi: 10.1101/gad.298018.117
Hill, R. E., Favor, J., Hogan, B. L., Ton, C. C., Saunders, G. F., Hanson, I. M., et al. (1991). Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525. doi: 10.1038/354522a0
Hirabayashi, Y., and Gotoh, Y. (2010). Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11, 377–388. doi: 10.1038/nrn2810
Ihrie, R. A., Shah, J. K., Harwell, C. C., Levine, J. H., Guinto, C. D., Lezameta, M., et al. (2011). Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71, 250–262. doi: 10.1016/j.neuron.2011.05.018
Inan, M., Welagen, J., and Anderson, S. A. (2012). Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb. Cortex 22, 820–827. doi: 10.1093/cercor/bhr148
Ingham, P., and Whittle, R. (1980). Trithorax: a new homoeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol. Gen. Genet. 179, 607–614. doi: 10.1007/bf00271751
Johnson, M. H., and McConnell, J. M. (2004). Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev. Biol. 15, 583–597. doi: 10.1016/j.semcdb.2004.04.002
Jones, W. D., Dafou, D., McEntagart, M., Woollard, W. J., Elmslie, F. V., Holder-Espinasse, M., et al. (2012). De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91, 358–364. doi: 10.1016/j.ajhg.2012.06.008
Kang, S. M., Lee, H. J., and Cho, J. Y. (2013). MicroRNA-365 regulates NKX2–1, a key mediator of lung cancer. Cancer Lett. 335, 487–494. doi: 10.1016/j.canlet.2013.03.006
Kassis, J. A., Kennison, J. A., and Tamkun, J. W. (2017). Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699–1725. doi: 10.1534/genetics.115.185116
Kepecs, A., and Fishell, G. (2014). Interneuron cell types are fit to function. Nature 505, 318–326. doi: 10.1038/nature12983
Kosaka, K., Toida, K., Margolis, F. L., and Kosaka, T. (1997). Chemically defined neuron groups and their subpopulations in the glomerular layer of the rat main olfactory bulb—II. Prominent differences in the intraglomerular dendritic arborization and their relationship to olfactory nerve terminals. Neuroscience 76, 775–786. doi: 10.1016/s0306-4522(96)00308-9
Kroll, T. T., and O’Leary, D. D. (2005). Ventralized dorsal telencephalic progenitors in Pax6 mutant mice generate GABA interneurons of a lateral ganglionic eminence fate. Proc. Natl. Acad. Sci. U S A 102, 7374–7379. doi: 10.1073/pnas.0500819102
Lawrence, P. A., and Morata, G. (1983). The elements of the bithorax complex. Cell 35, 595–601. doi: 10.1016/0092-8674(83)90091-0
Lawrence, P. A., and Morata, G. (1994). Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell 78, 181–189. doi: 10.1016/0092-8674(94)90289-5
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi: 10.1038/276565a0
Lim, D. A., and Alvarez-Buylla, A. (2014). Adult neural stem cells stake their ground. Trends Neurosci. 37, 563–571. doi: 10.1016/j.tins.2014.08.006
Lim, D. A., and Alvarez-Buylla, A. (2016). The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb. Perspect. Biol. 8:a018820. doi: 10.1101/cshperspect.a018820
Liu, S. J., Nowakowski, T. J., Pollen, A. A., Lui, J. H., Horlbeck, M. A., Attenello, F. J., et al. (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17:67. doi: 10.1186/s13059-016-0932-1
Lou, L., Bergson, C., and McGinnis, W. (1995). Deformed expression in the Drosophila central nervous system is controlled by an autoactivated intronic enhancer. Nucleic Acids Res. 23, 3481–3487. doi: 10.1093/nar/23.17.3481
Mansfield, J. H., Harfe, B. D., Nissen, R., Obenauer, J., Srineel, J., Chaudhuri, A., et al. (2004). MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 36, 1079–1083. doi: 10.1038/ng1421
Manuel, M. N., Mi, D., Mason, J. O., and Price, D. J. (2015). Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 9:70. doi: 10.3389/fncel.2015.00070
Martinez-Arias, A., and Lawrence, P. A. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313, 639–642. doi: 10.1038/313639a0
McGinnis, W., and Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi: 10.1016/0092-8674(92)90471-n
Merkle, F. T., Fuentealba, L. C., Sanders, T. A., Magno, L., Kessaris, N., and Alvarez-Buylla, A. (2014). Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat. Neurosci. 17, 207–214. doi: 10.1038/nn.3610
Merkle, F. T., Mirzadeh, Z., and Alvarez-Buylla, A. (2007). Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384. doi: 10.1126/science.1144914
Merkle, F. T., Tramontin, A. D., García-Verdugo, J. M., and Alvarez-Buylla, A. (2004). Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. U S A 101, 17528–17532. doi: 10.1073/pnas.0407893101
Molyneaux, B. J., Arlotta, P., Menezes, J. R., and Macklis, J. D. (2007). Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437. doi: 10.1038/nrn2151
Monuki, E. S., and Walsh, C. A. (2001). Mechanisms of cerebral cortical patterning in mice and humans. Nat. Neurosci. 4, 1199–1206. doi: 10.1038/nn752
Narita, Y., and Rijli, F. M. (2009). Hox genes in neural patterning and circuit formation in the mouse hindbrain. Curr. Top. Dev. Biol. 88, 139–167. doi: 10.1016/s0070-2153(09)88005-8
Neo, W. H., Yap, K., Lee, S. H., Looi, L. S., Khandelia, P., Neo, S. X., et al. (2014). MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J. Biol. Chem. 289, 20788–20801. doi: 10.1074/jbc.m113.525493
Nóbrega-Pereira, S., Kessaris, N., Du, T., Kimura, S., Anderson, S. A., and Marín, O. (2008). Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron 59, 733–745. doi: 10.1016/j.neuron.2008.07.024
Oguchi, H., and Kimura, S. (1998). Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP)/thyroid-specific transcription factor-1 (TTF-1) gene: evidence of autoregulation. Endocrinology 139, 1999–2006. doi: 10.1210/endo.139.4.5933
Ouweneel, W. J., and van der Meer, J. M. (1973). Differentiation capacities of the dorsal metathoracic (haltere) disc of Drosophila melanogaster: 1. Normal organ map. Wilhelm Roux Arch. Entwickl. Mech. Org. 172, 149–161. doi: 10.1007/bf00576938
Packer, A. I., Crotty, D. A., Elwell, V. A., and Wolgemuth, D. J. (1998). Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development 125, 1991–1998.
Palma, V., Lim, D. A., Dahmane, N., Sánchez, P., Brionne, T. C., Herzberg, C. D., et al. (2005). Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132, 335–344. doi: 10.1242/dev.01567
Pearson, J. C., Lemons, D., and McGinnis, W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904. doi: 10.1038/nrg1726
Pedersen, R. A., Wu, K., and Balakier, H. (1986). Origin of the inner cell mass in mouse embryos: cell lineage analysis by microinjection. Dev. Biol. 117, 581–595. doi: 10.1016/0012-1606(86)90327-1
Price, J. L., and Powell, T. P. (1970). The synaptology of the granule cells of the olfactory bulb. J. Cell Sci. 7, 125–155.
Puelles, L., Kuwana, E., Puelles, E., Bulfone, A., Shimamura, K., Keleher, J., et al. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol. 424, 409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7
Ronan, J. L., Wu, W., and Crabtree, G. R. (2013). From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359. doi: 10.1038/nrg3413
Rushing, G., and Ihrie, R. A. (2016). Neural stem cell heterogeneity through time and space in the ventricular-subventricular zone. Front. Biol. 11, 261–284. doi: 10.1007/s11515-016-1407-1
Schuettengruber, B., Bourbon, H.-M., Di Croce, L., and Cavalli, G. (2017). Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171, 34–57. doi: 10.1016/j.cell.2017.08.002
Scott, M. P., and Carroll, S. B. (1987). The segmentation and homeotic gene network in early Drosophila development. Cell 51, 689–698. doi: 10.1016/0092-8674(87)90092-4
Shimamura, K., Hartigan, D. J., Martinez, S., Puelles, L., and Rubenstein, J. L. (1995). Longitudinal organization of the anterior neural plate and neural tube. Development 121, 3923–3933.
Steffen, P. A., and Ringrose, L. (2014). What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15, 340–356. doi: 10.1038/nrm3789
Strom, S. P., Lozano, R., Lee, H., Dorrani, N., Mann, J., O’Lague, P. F., et al. (2014). De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med. Genet. 15:49. doi: 10.1186/1471-2350-15-49
Struhl, G., and Akam, M. (1985). Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4, 3259–3264.
Sussel, L., Marin, O., Kimura, S., and Rubenstein, J. L. (1999). Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370.
Szucsik, J. C., Witte, D. P., Li, H., Pixley, S. K., Small, K. M., and Potter, S. S. (1997). Altered forebrain and hindbrain development in mice mutant for the Gsh-2 homeobox gene. Dev. Biol. 191, 230–242. doi: 10.1006/dbio.1997.8733
Szulwach, K. E., Li, X., Smrt, R. D., Li, Y., Luo, Y., Lin, L., et al. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141. doi: 10.1083/jcb.200908151
Taniguchi, H., Lu, J., and Huang, Z. J. (2013). The spatial and temporal origin of chandelier cells in mouse neocortex. Science 339, 70–74. doi: 10.1126/science.1227622
Tarkowski, A. K., and Wróblewska, J. (1967). Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155–180.
Tatton-Brown, K., Hanks, S., Ruark, E., Zachariou, A., Duarte Sdel, V., Ramsay, E., et al. (2011). Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2, 1127–1133. doi: 10.18632/oncotarget.385
Tehler, D., Høyland-Kroghsbo, N. M., and Lund, A. H. (2011). The miR-10 microRNA precursor family. RNA Biol. 8, 728–734. doi: 10.4161/rna.8.5.16324
Toresson, H., Potter, S. S., and Campbell, K. (2000). Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development 127, 4361–4371.
Tümpel, S., Wiedemann, L. M., and Krumlauf, R. (2009). Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103–137. doi: 10.1016/s0070-2153(09)88004-6
van der Lugt, N. M., Domen, J., Linders, K., van Roon, M., Robanus-Maandag, E., te Riele, H., et al. (1994). Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8, 757–769. doi: 10.1101/gad.8.7.757
White, R. A., and Lehmann, R. (1986). A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell 47, 311–321. doi: 10.1016/0092-8674(86)90453-8
Wilson, S. W., and Rubenstein, J. L. (2000). Induction and dorsoventral patterning of the telencephalon. Neuron 28, 641–651. doi: 10.1016/s0896-6273(00)00171-9
Xu, Q., Tam, M., and Anderson, S. A. (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29. doi: 10.1002/cne.21529
Xu, Q., Wonders, C. P., and Anderson, S. A. (2005). Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development 132, 4987–4998. doi: 10.1242/dev.02090
Young, K. M., Fogarty, M., Kessaris, N., and Richardson, W. D. (2007). Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 27, 8286–8296. doi: 10.1523/jneurosci.0476-07.2007
Yu, B. D., Hanson, R. D., Hess, J. L., Horning, S. E., and Korsmeyer, S. J. (1998). MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. U S A 95, 10632–10636. doi: 10.1073/pnas.95.18.10632
Yu, B. D., Hess, J. L., Horning, S. E., Brown, G. A., and Korsmeyer, S. J. (1995). Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508. doi: 10.1038/378505a0
Keywords: positional identity, neural development, chromatin regulator, trithorax, polycomb, EZH2, MLL1
Citation: Delgado RN and Lim DA (2017) Maintenance of Positional Identity of Neural Progenitors in the Embryonic and Postnatal Telencephalon. Front. Mol. Neurosci. 10:373. doi: 10.3389/fnmol.2017.00373
Received: 08 August 2017; Accepted: 26 October 2017;
Published: 13 November 2017.
Edited by:
M. Albert Basson, King’s College London, United KingdomReviewed by:
Yonghe Wu, Deutsches Krebsforschungszentrum (DKFZ), GermanyDavide De Pietri Tonelli, Fondazione Istituto Italiano di Technologia, Italy
Copyright © 2017 Delgado and Lim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel A. Lim, daniel.lim@ucsf.edu
 Ryan N. Delgado
Ryan N. Delgado Daniel A. Lim
Daniel A. Lim