- Molecular Cell Biology, Groningen Biomolecular Sciences and Biotechnology Institute, Kluyver Centre for Genomics of Industrial Fermentation, University of Groningen, Groningen, Netherlands
Peroxisomes are ubiquitous eukaryotic organelles, which perform a plethora of functions including hydrogen peroxide metabolism and β-oxidation of fatty acids. Reactive oxygen species produced by peroxisomes are a major contributing factor to cellular oxidative stress, which is supposed to significantly accelerate aging and cell death according to the free radical theory of aging. However, relative to mitochondria, the role of the other oxidative organelles, the peroxisomes, in these degenerative pathways has not been extensively investigated. In this contribution we discuss our current knowledge on the role of peroxisomes in aging and cell death, with focus on studies performed in yeast.
Introduction
The production and accumulation of reactive oxygen species (ROS) is a profound stress factor in living cells due to the fact that ROS can oxidize and therefore damage vital macromolecules such as nucleic acids, proteins, and lipids. Intracellular accumulation of these damaged components leads to aging – a process defined as the deterioration of cells in time which is accompanied by gradual loss of cell viability. Until recently, mitochondria were considered as the main players in ROS production and hence in aging of eukaryotic cells. However, recent reports proposed that peroxisomes also produce significant amounts of ROS. Therefore, like mitochondrial dysfunction, peroxisomal dysfunction may also contribute to cell death and aging. The role of mitochondria in cell death pathways such as apoptosis and necrosis is well established. However, the importance of peroxisomes in these processes is much less understood. In this contribution we summarize findings on the role of peroxisomes in aging and cell death processes. We focus on data obtained in yeast and discuss the relevance of these findings for higher eukaryotes including man.
Peroxisomes: Structure and Function
Peroxisomes are highly dynamic organelles: their morphology, abundance, and function depending on species, developmental stage, and external stimuli (Oku and Sakai, 2010; Schrader et al., 2011). The predominant feature of peroxisomes is the presence of H2O2 producing oxidases and the antioxidant enzyme catalase to detoxify this ROS species. In yeast, peroxisomes are predominantly involved in the primary metabolism of unusual carbon sources, such as oleic acid in Saccharomyces cerevisiae and methanol in methylotrophic yeasts. In man, peroxisomes are involved in the α- and β-oxidation of very long chain fatty acids, biosynthesis of ether phospholipids and bile acids (Wanders and Waterham, 2006). Recently also novel, non-metabolic functions have been identified for mammalian peroxisomes, among which anti-viral innate immunity and anti-viral signaling (Dixit et al., 2010).
The Peroxisome Life Cycle and Implication for Aging
Like mitochondria, peroxisomes can multiply by division of pre-existing ones (Figure 1). However, so far no evidence has been obtained that peroxisomes fuse. The initial stage of peroxisome fission is organelle elongation mediated by the peroxisomal membrane protein Pex11p. Dynamin-related proteins (DRPs) are responsible for the final scission event. In S. cerevisiae the DRPs Vps1p and Dnm1p are involved in peroxisome fission (Hoepfner et al., 2001; Kuravi et al., 2006), whereas in the yeast Hansenula polymorpha peroxisome fission entirely depends on Dnm1p (Nagotu et al., 2008). Recruitment of Dnm1p to the peroxisomal membrane is mediated by the peroxisomal membrane protein Fis1p. Interestingly, Fis1p also recruits Dnm1p to mitochondria for mitochondrial fission (Mozdy et al., 2000). Similarly, mammalian Fis1 and Drp1 are both involved in peroxisome and mitochondrial fission. Hence, peroxisomes and mitochondria share key components of their fission machineries.
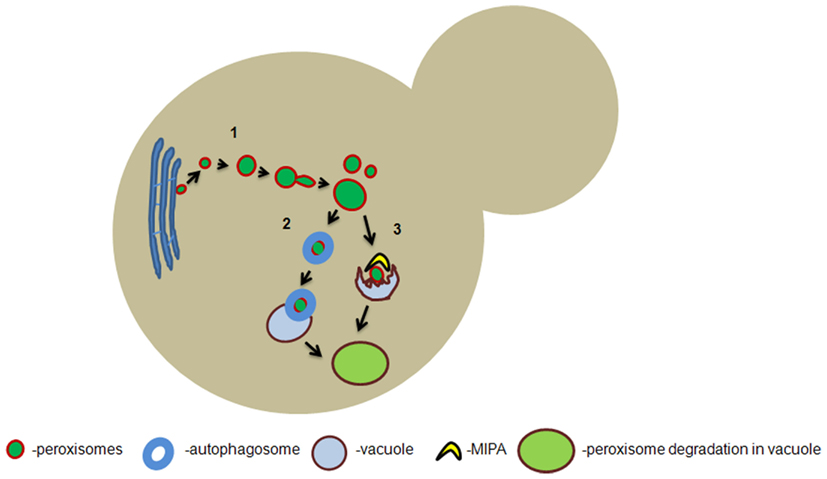
Figure 1. Hypothetical model of peroxisome formation and degradation in yeast. De novo formation of peroxisomes from the endoplasmic reticulum takes place in yeast cells which are devoid of pre-existing peroxisomes (1). In wild-type yeast cells, the predominant mode of peroxisome proliferation occurs via fission of the pre-existing organelles which involves Pex11p dependent elongation and dynamin-related protein (DRP) dependent scission of the peroxisomal membrane. When peroxisomes are targeted for macropexophagy they are engulfed by autophagosomes which ultimately fuse with the vacuolar membrane to deliver the organelle for degradation (2). Peroxisomes can also be targeted for micropexophagy in which the protrusion of the vacuolar membrane engulfs the organelle and they are subsequently degraded (3). MIPA: micropexophagy-specific membrane apparatus.
In two fungal model systems for aging, the filamentous ascomycete Podospora anserina and baker’s yeast down-regulation of mitochondrial fission by deletion of the DNM1 gene leads to a robust increase in replicative lifespan (Scheckhuber et al., 2007, 2008). Moreover, deletion of DNM1 also has a positive effect on chronological aging in baker’s yeast (Palermo et al., 2007). These beneficial effects might be based on improved content mixing of mitochondria so that molecular damage to proteins, lipids, and mtDNA can be ameliorated more efficiently. However, it has to be stressed that the effect of DNM1 deletion on peroxisome fission was not investigated in these studies. Hence, the observed effects may also be partially due to defects in peroxisomal fission.
Several data suggest that peroxisomes divide asymmetrically, resulting in larger mature organelles and small, nascent ones (Koch et al., 2003, 2010; Cepinska et al., 2011; Huber et al., 2012). As a consequence cells contain a heterologous population of peroxisomes, ranging from relatively young and vital nascent organelles to relatively old, mature ones, in which dysfunctional components accumulate in time due to damage caused by products of peroxisomal metabolism.
In addition to fission, peroxisomes can also be formed de novo, a process that is most prominent in yeast pex3 or pex mutants that lack pre-existing peroxisomes, upon reintroduction of the corresponding genes. Several data suggested that these new organelles originate from the endoplasmic reticulum (ER; Hoepfner et al., 2005; Zipor et al., 2009).
Data in yeast, data in yeast indicate that the major pathway of peroxisome proliferation is fission. However, possibly in other species the formation of peroxisomes from the ER is a more prominent process (Geuze et al., 2003; Tabak et al., 2003; Kim et al., 2006; Yonekawa et al., 2011).
Autophagy can result in a reduction in the number of peroxisomes per cell. Autophagy is the pivotal cellular housekeeping process that can eliminate redundant or unwanted components or entire organelles from the cell. Selective degradation of peroxisomes by autophagy is designated “pexophagy,” a phenomenon mainly studied in the methylotrophic yeast species H. polymorpha and Pichia pastoris.
In H. polymorpha, macropexophagy is induced when methanol-grown cells are shifted to glucose (Figure 1). Under these conditions, the key peroxisomal enzymes of methanol metabolism become redundant for growth. The surplus in organelles is then selectively degraded by macropexophagy (Monastryska et al., 2004), a process that involves sequestration of individual peroxisomes from the cytosol and subsequent fusion with the vacuole for degradation (Veenhuis et al., 1983).
When methanol-grown cells of P. pastoris are shifted to glucose, peroxisomes are degraded by micropexophagy, which involves the formation of finger-like protrusions by the vacuole and subsequent engulfment of clusters of peroxisomes from the cytosol with ultimate degradation in the vacuole. Before this takes place, a double membrane flattened sac-like structure termed micropexophagy-specific membrane apparatus (MIPA) is synthesized at the peroxisome surface to complete the sequestration process (Tuttle and Dunn, 1995; Mukaiyama et al., 2004).
In addition, data obtained with H. polymorpha revealed a constant removal of peroxisomes by autophagy (Aksam et al., 2007). This process most likely prevents the accumulation of damaged peroxisomal components, which would be potentially hazardous for the cells. Hence, at peroxisome-inducing conditions, organelle proliferation and degradation occur simultaneously to maintain tightly controlled organelle homeostasis. This mechanism is consistent with the view that timely rejuvenation of peroxisomes is vital for cell viability and survival. Most likely the oldest, mature organelles are most susceptible for autophagic degradation. A central question that offers interesting perspectives for future research is how these peroxisomes are recognized by the pexophagy machinery. The peroxisomal membrane proteins Pex3p and Pex14p have been shown to play a role in the early stage of pexophagy (Bellu et al., 2001, 2002; van Zutphen et al., 2008) and may play a role in this recognition process. Indeed Pex3p was shown to interact with Atg30p, a phosphoprotein that is necessary for the presentation of the compromised peroxisome to the vacuole for its subsequent degradation (Farre et al., 2008).
Peroxisomal Damage and Repair Mechanisms
Peroxisomal ROS and antioxidant enzymes
Peroxisomes counteract the compromising effects of ROS by antioxidant enzymes. Among these are peroxisomal catalase, glutathione peroxidase, peroxiredoxin I and Pmp20p to degrade hydrogen peroxide and CuZnSOD and MnSOD to detoxify superoxide anions (Bonekamp et al., 2009).
Saccharomyces cerevisiae contains two catalases, namely peroxisomal Cta1p and cytosolic Ctt1p. Unexpectedly, inactivation of both catalases in this yeast was shown to result in an increase of the chronological lifespan (Mesquita et al., 2010). This surprising finding could be explained by the activation of superoxide dismutases by the elevated hydrogen peroxide levels. In this view elevated levels of hydrogen peroxide in catalase-deficient S. cerevisiae cells prolong chronological lifespan.
However, earlier studies indicated that deletion of the gene encoding the peroxisomal catalase in S. cerevisiae resulted in a shortened lifespan (Petriv and Rachubinski, 2004). These seemingly contradictory results may be possibly related to differences in the experimental procedures to determine the chronological lifespan (i.e., use of different cultivation conditions such as media composition and growth temperatures; Mesquita et al., 2010: SC medium, 26°C; Petriv and Rachubinski, 2004: YNBD medium, 30°C). The same authors also reported that in Caenorhabditis elegans the deletion of peroxisomal catalase (Ctl-2) in the genetic background of the long lived Δclk-1 mutant shortens the maximum lifespan as well. The shortened lifespan was accompanied by altered peroxisome morphology that might point to compromised peroxisomal function with increased production of ROS (Petriv and Rachubinski, 2004).
Similar to catalase, the peroxisomal peroxiredoxin Pmp20p is also involved in degradation of H2O2. Peroxiredoxins are thiol-specific evolutionary conserved antioxidant enzymes. In addition to H2O2 breakdown they are also involved in degradation of organic hydroperoxides (ROOH) and therefore important for maintaining the integrity of lipid membranes (Yamashita et al., 1999).
Studies in the methylotrophic yeasts H. polymorpha and C. boidinii demonstrated that Pmp20p is important for cell viability during growth on methanol due to its capability to repair ROS generated damages (i.e., lipid peroxidation) at the peroxisomal membrane surface (Horiguchi et al., 2001; Aksam et al., 2007). In the fission yeast Schizosaccharomyces pombe it was shown that Pmp20p (in addition to thioredoxin peroxidase and glutathione peroxidase) inhibited thermal aggregation of citrate synthase at a high temperature (43°C) in vitro. This suggests that at least in S. pombe peroxisomal Pmp20p may have a second function as a molecular chaperone which could be important for organelle quality control (Kim et al., 2010).
In man catalase gene mutations have been identified and linked to detrimental conditions like diabetes, hypertension, macular degeneration, cataracts, cancer, and skin pigmentation disorders (Goth et al., 2004). A severe decrease of catalase activity is found in clinical cases of hypo- or acatalasemia and has been predominantly identified in Japan and certain European countries (e.g., Hungary, Switzerland). Clinical symptoms are severe (among them hemolytic anemia), illustrating the importance of functional catalase for health. Re-direction of functional catalase to peroxisomes in catalase-deficient cell lines led to increased detoxification of H2O2 and also restoration of cellular plasmalogen and fatty acid levels (Sheikh et al., 1998).
Reactive oxygen species are usually associated with their potent damaging capabilities but they are also involved in crucial cellular signaling processes. With regard to aging, low levels of ROS might induce a hormesis response which increases chances of cellular survival by up-regulating pathways dedicated to high-stress adaptation like the retrograde response (Jazwinski, 2012) and the target of rapamycin (TOR) and adenosine mono phosphate kinase (AMPK) pathways (Gems and Partridge, 2008). Although it is clear that mitochondria are supposedly the main players in this regard, it is likely that also peroxisomes integrate into the signaling network as important mediators of aging processes.
Peroxisomal Lon protease
So far one conserved peroxisomal protease is known that plays a role in peroxisomal protein quality control, namely the peroxisomal Lon protease, Pln (Kikuchi et al., 2004; Aksam et al., 2007). Pln belongs to the ATPases associated with diverse cellular activities (AAA) protein family which is supposed to be involved in degradation of unfolded and non-assembled peroxisomal matrix proteins (Aksam et al., 2007; Ngo and Davies, 2007). Studies in H. polymorpha showed that the deletion of the gene encoding Pln leads to a pronounced decrease in cell viability accompanied by enhanced ROS production (Aksam et al., 2009). One possible explanation could be that the accumulation of unfolded proteins leads to the formation of protein aggregates, resulting in a disturbance of ROS homeostasis which ultimately leads to cell death. Indeed, electron dense aggregates are often observed in peroxisomes of H. polymorpha cells lacking Pln (Aksam et al., 2007). However, the precise molecular mechanism and interplay between protein aggregation, ROS production, and cell death still remains unclear and needs to be studied in more detail.
Several studies have investigated the function of Pln in mammalian cells after the identification of this enzyme in rat liver peroxisomes (Kikuchi et al., 2004). In mammals Pln also seems to play a role in peroxisomal matrix protein import as catalase import is compromised when Pln is overproduced (Omi et al., 2008).
The Role of Peroxisomes in Yeast Aging
Mechanistic insights into the role of peroxisomes in aging have been gained mainly from studies conducted in S. cerevisiae. When this yeast is grown in glucose rich media, neutral lipids (e.g., triacylglycerols) are produced in the ER and incorporated in lipid bodies (Olofsson et al., 2009). When these storage lipids need to be mobilized, peroxisomes and lipid bodies come into close contact to allow uptake of the neutral lipids for subsequent oxidation of the non-esterified fatty acids (free fatty acids; Binns et al., 2006). An intriguing model has been put forward linking life span determination and the synthesis and degradation of lipids in the ER, lipid bodies, and peroxisomes (Goldberg et al., 2009a,b; Titorenko and Terlecky, 2011). According to this model, repression of essential components of fatty acid β-oxidation by ethanol (produced as a side-product of glucose fermentation) leads to the accumulation of free fatty acids. This challenges the cell with detrimental effects, i.e., stimulation of necrotic cell death (Aksam et al., 2008; Jungwirth et al., 2008) and lipoapoptosis (shown in the fission yeast S. pombe; Low et al., 2005). Moreover, as a result of impaired β-oxidation in peroxisomes diacylglycerol accumulates in the ER and mediates the induction of protein kinase C-dependent signaling which ultimately affects cellular pathways involved in various stress responses (as demonstrated in the nematode C. elegans; Feng et al., 2007).
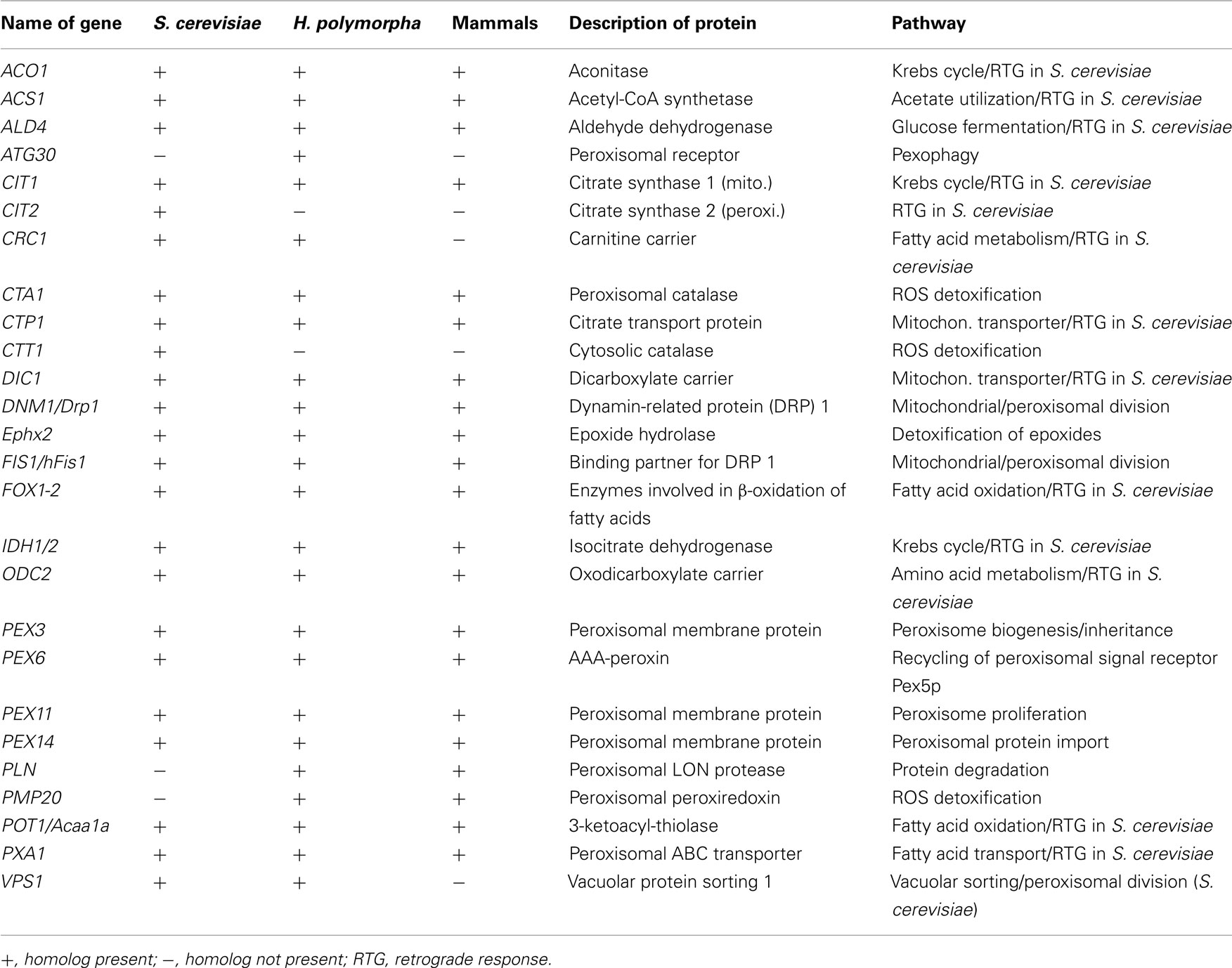
Peroxisomes and peroxisomal enzymes also play a vital role in the phenomenon of retrograde response (RTG) in baker’s yeast (Butow and Avadhani, 2004; Jazwinski, 2012). RTG is activated in yeast cells that are confronted with mitochondrial respiratory dysfunction. Interestingly, induction of RTG leads to an increased replicative lifespan (Kirchman et al., 1999). The more pronounced RTG induction is, the larger the beneficial effect on aging. RTG leads to the induction of transcription of several nuclear genes that help to promote the survival of the cell despite mitochondrial dysfunction (Table 1). Genes encoding Cit1p, Aco1p, Idh1/2p (first enzymes of the citric acid cycle), Ald4p and Acs1p (enzymes for cytosolic biosynthesis of acetyl-CoA), Crc1p, Ctp1p, Dic1p, and Odc2p (membrane transporters for shuttling metabolites between mitochondria, peroxisomes, and the cytosol), and Pex11p, Pxa1p, Cit2p, Fox1p, Fox2p, Pot1p (peroxisomal proteins) belong to this group. Collectively, these (and further) proteins enable yeast cells to enhance oxidation of fatty acids and to synthesize essential metabolic intermediates of the Krebs cycle that otherwise would not be available.
The Role of Peroxisomes in Aging in Mammals
In rat hepatocytes subjection to conditions of hyperinsulinemia leads to an inhibition of β-oxidation and a concomitant acceleration of aging in these animals (Xu et al., 1995). Clearly, the activity of the peroxisomal marker enzyme catalase decreases by 30–40% in liver samples isolated from old mice and rats (Haining and Legan, 1973; Semsei et al., 1989; Perichon and Bourre, 1995, 1996; Xia et al., 1995). Decreased levels/activity of this H2O2-decomposing protein is contributing to the phenomenon of peroxisome senescence. This process leads to diminished regulation of organelle maturation and division, protein import and overall dysfunction (Legakis et al., 2002; Koepke et al., 2008). A comparative study with the goal to identify differences between liver subproteomes from young and old mice revealed several up-regulated peroxisomal proteins. One of the up-regulated enzymes was epoxide hydrolase 2 (Ephx2), which detoxifies epoxides and converts these to excretable dihydrodiols (Amelina et al., 2011). This may constitute a counteracting mechanism in old animals. Another up-regulated enzyme, peroxisomal 3-ketoacyl-thiolase A [Acaa1a, Pot1p (Fox3p) in baker’s yeast], was correlated to increased cholesterol levels in old animals (Amelina et al., 2011).
Recently, an inter-organelle crosstalk between mitochondria and peroxisomes regarding the production of ROS was described (Ivashchenko et al., 2011). Enhanced formation of these compounds in peroxisomes profoundly disturbs the redox balance within mitochondria which results in fragmentation of these organelles. This finding proves that peroxisome dysfunction can have a pronounced effect on mitochondrial structure and function. It is thus conceivable that certain scenarios of mitochondrial dysfunction involving elevated cellular ROS levels can also be linked to peroxisomes.
Peroxisomes and Cell Death in Yeast and Mammals
Cellular death has many different faces. Two of the most common ones, apoptosis and necrosis, have been studied in much detail over the past few years (Golstein and Kroemer, 2007; Taylor et al., 2008). Apoptosis is a ubiquitous mode of programmed cell death, strictly regulated and conserved in eukaryotes (Madeo et al., 2004). This highly organized process is manifested by condensation and cleavage of nuclear DNA, release of pro-apoptotic proteins from mitochondria and, at later stages, “blebbing” of the plasma membrane. Mitochondria are key factors when it comes to the execution of apoptosis. Necrosis, on the other hand, is accompanied by rupture of organelles and the plasma membrane. Recent evidence demonstrates that necrosis can be also tightly controlled, similar to apoptosis (Golstein and Kroemer, 2007). Peroxisomes are involved in regulation of necrosis. For example, S. cerevisiae PEX6 deletion cells display hallmarks of necrosis and strongly elevated formation of ROS (Jungwirth et al., 2008). Moreover, in the methylotrophic yeast H. polymorpha deletion of PMP20 leads to pronounced induction of necrosis when the cells are grown on methanol-containing medium (Aksam et al., 2008). Interestingly, matrix proteins of the peroxisomes were found to be leaking into the cytosol of PMP20 deletion cells during necrosis. This is reminiscent of protein release by mitochondria during apoptosis and constitutes an interesting parallel between mitochondria and apoptosis on the one hand and peroxisomes and necrosis on the other hand (Eisenberg et al., 2010).
Perspectives
Research on the role peroxisomes play during cellular degeneration is supposed to unravel exciting new mechanisms and will likely integrate these fascinating organelles into the network of cellular pathways mediating aging. Some of the interesting lines of research that will yield promising insights into the role of peroxisomes might comprise (i) identification and characterization of a peroxisomal unfolded protein response (UPR) as a mechanism to signal organelle dysfunction (and subsequent degradation), (ii) studying selective inheritance of peroxisomes during replicative aging so that the daughter cells receive “healthy” peroxisomes (Cepinska et al., 2011), and (iii) investigating the role of de novo formation of peroxisomes vs. fission in maintaining a functional peroxisomal population during chronological and replicative aging.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. Christian Quintus Scheckhuber is supported by a fellowship from the Deutsche Forschungsgemeinschaft (SCHE 1686/2-1).
References
Aksam, E. B., de Vries, B., van der Klei, I. J., and Kiel, J. A. (2009). Preserving organelle vitality: peroxisomal quality control mechanisms in yeast. FEMS Yeast Res. 9, 808–820.
Aksam, E. B., Jungwirth, H., Kohlwein, S. D., Ring, J., Madeo, F., Veenhuis, M., and van der Klei, I. J. (2008). Absence of the peroxiredoxin Pmp20 causes peroxisomal protein leakage and necrotic cell death. Free Radic. Biol. Med. 45, 1115–1124.
Aksam, E. B., Koek, A., Kiel, J. A., Jourdan, S., Veenhuis, M., and van der Klei, I. J. (2007). A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3, 96–105.
Amelina, H., Sjodin, M. O., Bergquist, J., and Cristobal, S. (2011). Quantitative subproteomic analysis of age-related changes in mouse liver peroxisomes by iTRAQ LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 3393–3400.
Bellu, A. R., Komori, M., van der Klei, I. J., Kiel, J. A., and Veenhuis, M. (2001). Peroxisome biogenesis and selective degradation converge at Pex14p. J. Biol. Chem. 276, 44570–44574.
Bellu, A. R., Salomons, F. A., Kiel, J. A., Veenhuis, M., and van der Klei, I. J. (2002). Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 277, 42875–42880.
Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., Gilpin, C., Chapman, K. D., Anderson, R. G., and Goodman, J. M. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731.
Bonekamp, N. A., Volkl, A., Fahimi, H. D., and Schrader, M. (2009). Reactive oxygen species and peroxisomes: struggling for balance. Biofactors 35, 346–355.
Butow, R. A., and Avadhani, N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15.
Cepinska, M. N., Veenhuis, M., van der Klei, I. J., and Nagotu, S. (2011). Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic 12, 925–937.
Dixit, E., Boulant, S., Zhang, Y., Lee, A. S., Odendall, C., Shum, B., Hacohen, N., Chen, Z. J., Whelan, S. P., Fransen, M., Nibert, M. L., Superti-Furga, G., and Kagan, J. C. (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681.
Eisenberg, T., Carmona-Gutierrez, D., Büttner, S., Tavernarakis, N., and Madeo, F. (2010). Necrosis in yeast. Apoptosis 15, 257–268.
Farre, J. C., Manjithaya, R., Mathewson, R. D., and Subramani, S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376.
Feng, H., Ren, M., Chen, L., and Rubin, C. S. (2007). Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J. Biol. Chem. 282, 31273–31288.
Gems, D., and Partridge, L. (2008). Stress-response hormesis and aging: “that which does not kill us makes us stronger.” Cell Metab. 7, 200–203.
Geuze, H. J., Murk, J. L., Stroobants, A. K., Griffith, J. M., Kleijmeer, M. J., Koster, A. J., Verkleij, A. J., Distel, B., and Tabak, H. F. (2003). Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell 14, 2900–2907.
Goldberg, A. A., Bourque, S. D., Kyryakov, P., Boukh-Viner, T., Gregg, C., Beach, A., Burstein, M. T., Machkalyan, G., Richard, V., Rampersad, S., and Titorenko, V. I. (2009a). A novel function of lipid droplets in regulating longevity. Biochem. Soc. Trans. 37, 1050–1055.
Goldberg, A. A., Bourque, S. D., Kyryakov, P., Gregg, C., Boukh-Viner, T., Beach, A., Burstein, M. T., Machkalyan, G., Richard, V., Rampersad, S., Cyr, D., Milijevic, S., and Titorenko, V. I. (2009b). Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp. Gerontol. 44, 555–571.
Golstein, P., and Kroemer, G. (2007). Cell death by necrosis: towards a molecular definition. Trends Biochem. Sci. 32, 37–43.
Goth, L., Rass, P., and Pay, A. (2004). Catalase enzyme mutations and their association with diseases. Mol. Diagn. 8, 141–149.
Haining, J. L., and Legan, J. S. (1973). Catalase turnover in rat liver and kidney as a function of age. Exp. Gerontol. 8, 85–91.
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95.
Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F., and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990.
Horiguchi, H., Yurimoto, H., Kato, N., and Sakai, Y. (2001). Antioxidant system within yeast peroxisome. Biochemical and physiological characterization of CbPmp20 in the methylotrophic yeast Candida boidinii. J. Biol. Chem. 276, 14279–14288.
Huber, A., Koch, J., Kragler, F., Brocard, C., and Hartig, A. (2012). A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 13, 157–167.
Ivashchenko, O., van Veldhoven, P. P., Brees, C., Ho, Y. S., Terlecky, S. R., and Fransen, M. (2011). Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol. Biol. Cell 22, 1440–1451.
Jazwinski, S. M. (2012). The retrograde response and other pathways of interorganelle communication in yeast replicative aging. Subcell. Biochem. 57, 79–100.
Jungwirth, H., Ring, J., Mayer, T., Schauer, A., Büttner, S., Eisenberg, T., Carmona-Gutierrez, D., Kuchler, K., and Madeo, F. (2008). Loss of peroxisome function triggers necrosis. FEBS Lett. 582, 2882–2886.
Kikuchi, M., Hatano, N., Yokota, S., Shimozawa, N., Imanaka, T., and Taniguchi, H. (2004). Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 279, 421–428.
Kim, J. S., Bang, M. A., Lee, S., Chae, H. Z., and Kim, K. (2010). Distinct functional roles of peroxiredoxin isozymes and glutathione peroxidase from fission yeast, Schizosaccharomyces pombe. BMB Rep. 43, 170–175.
Kim, P. K., Mullen, R. T., Schumann, U., and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521–532.
Kirchman, P. A., Kim, S., Lai, C. Y., and Jazwinski, S. M. (1999). Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152, 179–190.
Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M. A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605.
Koch, J., Pranjic, K., Huber, A., Ellinger, A., Hartig, A., Kragler, F., and Brocard, C. (2010). PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123, 3389–3400.
Koepke, J. I., Wood, C. S., Terlecky, L. J., Walton, P. A., and Terlecky, S. R. (2008). Progeric effects of catalase inactivation in human cells. Toxicol. Appl. Pharmacol. 232, 99–108.
Kuravi, K., Nagotu, S., Krikken, A. M., Sjollema, K., Deckers, M., Erdmann, R., Veenhuis, M., and van der Klei, I. J. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994–4001.
Legakis, J. E., Koepke, J. I., Jedeszko, C., Barlaskar, F., Terlecky, L. J., Edwards, H. J., Walton, P. A., and Terlecky, S. R. (2002). Peroxisome senescence in human fibroblasts. Mol. Biol. Cell 13, 4243–4255.
Low, C. P., Liew, L. P., Pervaiz, S., and Yang, H. (2005). Apoptosis and lipoapoptosis in the fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 5, 1199–1206.
Madeo, F., Herker, E., Wissing, S., Jungwirth, H., Eisenberg, T., and Fröhlich, K. U. (2004). Apoptosis in yeast. Curr. Opin. Microbiol. 7, 655–660.
Mesquita, A., Weinberger, M., Silva, A., Sampaio-Marques, B., Almeida, B., Leao, C., Costa, V., Rodrigues, F., Burhans, W. C., and Ludovico, P. (2010). Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc. Natl. Acad. Sci. U.S.A. 107, 15123–15128.
Monastryska, I., Sjollema, K., van der Klei, I. J., Kiel, J. A., and Veenhuis, M. (2004). Microautophagy and macropexophagy may occur simultaneously in Hansenula polymorpha. FEBS Lett. 568, 135–138.
Mozdy, A. D., McCaffery, J. M., and Shaw, J. M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380.
Mukaiyama, H., Baba, M., Osumi, M., Aoyagi, S., Kato, N., Ohsumi, Y., and Sakai, Y. (2004). Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70.
Nagotu, S., Saraya, R., Otzen, M., Veenhuis, M., and van der Klei, I. J. (2008). Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim. Biophys. Acta 1783, 760–769.
Ngo, J. K., and Davies, K. J. (2007). Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann. N. Y. Acad. Sci. 1119, 78–87.
Oku, M., and Sakai, Y. (2010). Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 277, 3289–3294.
Olofsson, S. O., Bostrom, P., Andersson, L., Rutberg, M., Perman, J., and Boren, J. (2009). Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta 1791, 448–458.
Omi, S., Nakata, R., Okamura-Ikeda, K., Konishi, H., and Taniguchi, H. (2008). Contribution of peroxisome-specific isoform of Lon protease in sorting PTS1 proteins to peroxisomes. J. Biochem. 143, 649–660.
Palermo, V., Falcone, C., and Mazzoni, C. (2007). Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia Microbiol. (Praha) 52, 479–483.
Perichon, R., and Bourre, J. M. (1995). Peroxisomal beta-oxidation activity and catalase activity during development and aging in mouse liver. Biochimie 77, 288–293.
Perichon, R., and Bourre, J. M. (1996). Aging-related decrease in liver peroxisomal fatty acid oxidation in control and clofibrate-treated mice. A biochemical study and mechanistic approach. Mech. Ageing Dev. 87, 115–126.
Petriv, O. I., and Rachubinski, R. A. (2004). Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. J. Biol. Chem. 279, 19996–20001.
Scheckhuber, C. Q., Erjavec, N., Tinazli, A., Hamann, A., Nyström, T., and Osiewacz, H. D. (2007). Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9, 99–105.
Scheckhuber, C. Q., Rödel, E., and Wüstehube, J. (2008). Regulation of mitochondrial dynamics-characterization of fusion and fission genes in the ascomycete Podospora anserina. Biotechnol. J. 3, 781–790.
Schrader, M., Bonekamp, N. A., and Islinger, M. (2011). Fission and proliferation of peroxisomes. Biochim. Biophys. Acta
Semsei, I., Rao, G., and Richardson, A. (1989). Changes in the expression of superoxide dismutase and catalase as a function of age and dietary restriction. Biochem. Biophys. Res. Commun. 164, 620–625.
Sheikh, F. G., Pahan, K., Khan, M., Barbosa, E., and Singh, I. (1998). Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc. Natl. Acad. Sci. U.S.A. 95, 2961–2966.
Tabak, H. F., Murk, J. L., Braakman, I., and Geuze, H. J. (2003). Peroxisomes start their life in the endoplasmic reticulum. Traffic 4, 512–518.
Taylor, R. C., Cullen, S. P., and Martin, S. J. (2008). Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241.
Titorenko, V. I., and Terlecky, S. R. (2011). Peroxisome metabolism and cellular aging. Traffic 12, 252–259.
Tuttle, D. L., and Dunn, W. A. Jr (1995). Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25–35.
van Zutphen, T., Veenhuis, M., and van der Klei, I. J. (2008). Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 4, 63–66.
Veenhuis, M., Douma, A., Harder, W., and Osumi, M. (1983). Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 134, 193–203.
Wanders, R. J., and Waterham, H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332.
Xia, E., Rao, G., Van, R. H., Heydari, A. R., and Richardson, A. (1995). Activities of antioxidant enzymes in various tissues of male Fischer 344 rats are altered by food restriction. J. Nutr. 125, 195–201.
Xu, L., Ash, M., Abdel-aleem, S., Lowe, J. E., and Badr, M. (1995). Hyperinsulinemia inhibits hepatic peroxisomal beta-oxidation in rats. Horm. Metab. Res. 27, 76–78.
Yamashita, H., Avraham, S., Jiang, S., London, R., van Veldhoven, P. P., Subramani, S., Rogers, R. A., and Avraham, H. (1999). Characterization of human and murine PMP20 peroxisomal proteins that exhibit antioxidant activity in vitro. J. Biol. Chem. 274, 29897–29904.
Yonekawa, S., Furuno, A., Baba, T., Fujiki, Y., Ogasawara, Y., Yamamoto, A., Tagaya, M., and Tani, K. (2011). Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 12746–12751.
Keywords: aging, autophagy, cell death, peroxisome, yeast
Citation: Manivannan S, Scheckhuber CQ, Veenhuis M and van der Klei IJ (2012) The impact of peroxisomes on cellular aging and death. Front. Oncol. 2:50. doi: 10.3389/fonc.2012.00050
Received: 26 March 2012; Paper pending published: 11 April 2012;
Accepted: 01 May 2012; Published online: 21 May 2012.
Edited by:
Manuela Côrte-Real, Universidade do Minho, PortugalReviewed by:
Hua Yan, New York University School of Medicine, USAKai-Uwe Fröhlich, University of Graz, Austria
Copyright: © 2012 Manivannan, Scheckhuber, Veenhuis and van der Klei. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ida Johanna van der Klei, Molecular Cell Biology, Groningen Biomolecular Sciences and Biotechnology Institute, Kluyver Centre for Genomics of Industrial Fermentation, University of Groningen, Linnaeusborg, Nijenborgh 7, 9747 AG Groningen, Netherlands. e-mail: I.J.van.der.Klei@rug.nl
†Selvambigai Manivannan and Christian Quintus Scheckhuber have contributed equally to this work.