- Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA
The t(9;22)(q34;q11) or Philadelphia chromosome creates a BCR–ABL1 fusion gene encoding for a chimeric BCR–ABL1 protein. It is present in 3–4% of pediatric acute lymphoblastic leukemia (Ph+ ALL), and about 25% of adult ALL cases. Prior to the advent of tyrosine kinase inhibitors (TKI), Ph+ ALL was associated with a very poor prognosis despite the use of intensive chemotherapy and frequently hematopoietic stem-cell transplantation (HSCT) in first remission. The development of TKIs revolutionized the therapy of Ph+ ALL. Addition of the first generation ABL1 class TKI imatinib to intensive chemotherapy dramatically increased the survival for children with Ph+ ALL and established that many patients can be cured without HSCT. In parallel, the mechanistic understanding of Ph+ ALL expanded exponentially through careful mapping of pathways downstream of BCR–ABL1, the discovery of mutations in master regulators of B-cell development such as IKZF1 (Ikaros), PAX5, and early B-cell factor (EBF), the recognition of the complex clonal architecture of Ph+ ALL, and the delineation of genomic, epigenetic, and signaling abnormalities contributing to relapse and resistance. Still, many important basic and clinical questions remain unanswered. Current clinical trials are testing second generation TKIs in patients with newly diagnosed Ph+ ALL. Neither the optimal duration of therapy nor the optimal chemotherapy backbone are currently defined. The role of HSCT in first remission and post-transplant TKI therapy also require further study. In addition, it will be crucial to continue to dig deeper into understanding Ph+ ALL at a mechanistic level, and translate findings into complementary targeted approaches. Expanding targeted therapies hold great promise to decrease toxicity and improve survival in this high-risk disease, which provides a paradigm for how targeted therapies can be incorporated into treatment of other high-risk leukemias.
Pharmacologic inhibition of the tyrosine kinase activity of BCR–ABL1 is the poster child for molecularly targeted cancer therapy. The first tyrosine kinase to be targeted, it is still the most effective “novel” therapeutic strategy to date, leading to remissions and possibly cures with single agents in chronic myeloid leukemia (CML) (1). Single-agent tyrosine kinase inhibition has not produced sustained responses in Ph+ ALL, but in combination with standard chemotherapy has revolutionized therapy and outcome for this patient population (2).
The Biology of Philadelphia Chromosome-Positive ALL
BCR–ABL1-Induced Leukemia
BCR–ABL1 translocations are associated with two distinct clinical hematologic malignancies, CML and ALL. For CML, three discrete clinical stages have been defined: chronic phase, accelerated phase, and blast crisis. Genomic instability, the accumulation of additional cytogenetic (trisomy 8, isochromosome 17) and molecular (p53 pathway mutations, loss of p16INK4A/ARF) abnormalities, and BCR–ABL1-independent activation of downstream signaling pathways (LYN, AKT, STAT5) are all associated with – and likely contribute to – the progression to blast crisis (3). In about 30% of the cases, the predominant lineage in blast crisis is B-lymphoid rather than myeloid, speaking to the likely hematopoietic stem-cell origin of the disease. This presumed stem-cell origin may also explain the inability to achieve any durable remissions using conventional chemotherapy. Prior to the advent of tyrosine kinase inhibition, temporary disease stabilization was often achieved using hydroxyurea, low-dose cytarabine, and/or interferon, but the only curative approach was an allogeneic hematopoietic stem-cell transplantation (HSCT).
In addition to CML, BCR–ABL1 translocations are found in a distinct subtype of ALL, called Ph+ ALL. The clinical presentation is indistinguishable from ALL with other cytogenetic abnormalities, and the diagnosis relies on the presence of the BCR–ABL1 translocation (cytogenetics and FISH) and/or fusion transcript (PCR). Outcomes for Ph+ ALL were exceptionally poor when treated with chemotherapy, and HSCT in first remission was usually considered to be the best therapy (4). The frequency of BCR–ABL1 rearrangement in ALL increases with age (Figure 1) (5) and has been reported as high as 50% in the elderly (6). A greater percentage of patients with adverse cytogenetics contributes substantially to the overall worse outcome in adult compared to pediatric ALL (Figure 1).
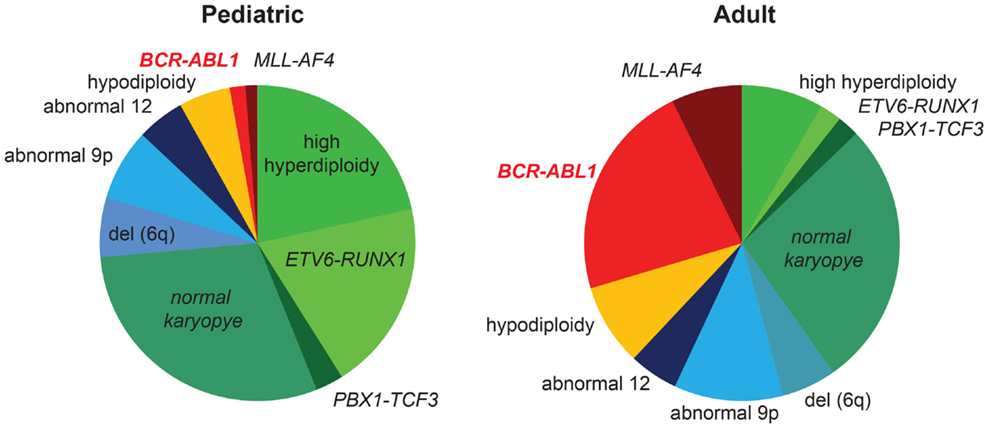
Figure 1. Frequency of BCR/ABL1 rearrangement in pediatric and adult ALL. Cytogenetic abnormalities in pediatric (>1 year) and adult patients with ALL are shown (5). The majority of children <1 year of age carry a rearrangement of the MLL-gene and are not included in this graph. Favorable cytogenetic abnormalities are represented in green, neutral in blue, and unfavorable cytogenetics are represented in yellow/red. Favorable cytogenetics (high hyperdiploidy, ETV6–RUNX1) decrease, while the frequency of BCR/ABL1 rearrangement increases with age. The higher percentage of unfavorable cytogenetics substantially contributes to inferior outcomes in adult versus pediatric ALL.
The first indication that the BCR–ABL1 fusion protein is indeed the crucial driver of CML came from mouse studies showing that expression of BCR–ABL1 in the bone marrow causes a CML-like disease (7–9). Studies that utilized a mutant BCR–ABL1 protein with an inactive tyrosine kinase domain defined the tyrosine kinase activity of ABL1 as absolutely required for transformation (10). This suggested that targeted inhibition of the ABL kinase domain might be an effective therapeutic strategy in BCR–ABL1-driven hematologic malignancies. The pioneering work of Brian Druker spearheaded the clinical development of the first tyrosine kinase inhibitor (TKI), Imatinib (11–13). Imatinib gained FDA approval in 2000 and revolutionized CML therapy, converting a near universally fatal disease requiring HSCT into a chronic condition controlled with monotherapy of a targeted agent (14). In the years since the initial success of imatinib, second [nilotinib, dasatinib, bosutinib (15–17)] and third [ponatinib (18)] generation ABL1 class TKIs have been developed, which are active against multiple imatinib-resistant BCR–ABL1 mutants.
Early studies using imatinib as monotherapy in Ph+ ALL were disappointing, with initial responses rapidly progressing to TKI-resistant disease. However, the integration of TKIs into a high-risk ALL chemotherapy backbone fundamentally changed our approach to Ph+ ALL as well. Overall survival (OS) using this strategy more than doubled compared to chemotherapy-only treated historic controls (2), and HSCT is no longer universally recommended for Ph+ ALL. Despite these advances, the survival of Ph+ ALL still lags behind most other cytogenetic subgroups in pediatric ALL. A better understanding of the biology of Ph+ ALL may help to refine therapy and develop rational combinations of targeted agents that will further improve outcomes for patients with this disease.
The Philadelphia Chromosome and BCR–ABL1 Fusion
Ph+ ALL derives its name from the presence of the Philadelphia (Ph) chromosome, named after the city where it was first described in the leukemia cells of a CML patient by Nowell and Hungerford in 1960 (19). In 1973, Janet Rowley reported that the Philadelphia Chromosome was the der(22) product of the reciprocal t(9;22)(q34;q11.2) translocation (20). The BCR–ABL1 fusion gene is generated by joining almost the entire coding region of the ABL1 tyrosine kinase gene (Abelson murine leukemia virus homolog, exons 2–11, chromosome 9) to the breakpoint cluster region (BCR) gene on chromosome 22 (Figure 2) (21). There are two main regions where breakpoints cluster within the BCR gene. The “CML” breakpoint region lies between exons 12 and 16 in a region called the major breakpoint cluster region (M-BCR). Translocations involving the M-BCR produce the larger p210 BCR–ABL1 protein, which derives its name from its molecular size of 210 kDa. Translocations that occur within the minor “ALL” BCR (m-BCR) yield a smaller p190 gene product that retains only the first exon of BCR. A rare p230 fusion protein (with a “micro”-BCR breakpoint between exons 19 and 20) has also been described. Both p210 and p190 transform primary human and murine bone marrow cells (8, 9, 22). Of the two, the “ALL-type” p190 is the stronger transforming agent (7, 23). About 90% of pediatric Ph+ ALL patients have the classic ALL-type p190 translocation (24) with some variability reported in the literature as CML in B-lymphoid blast crisis can sometimes be hard to distinguish from Ph+ ALL.
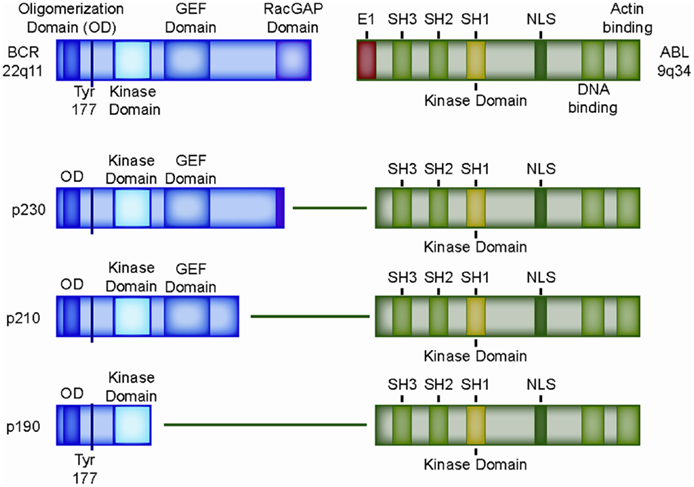
Figure 2. Structure of the most common BCR–ABL1 fusion genes. Domain structure of wild type BCR and wild type ABL1 protein, as well as retained domains in the three most common BCR–ABL1 variants, p230, p210, and p190. OD: oligomerization domain (coiled-coil domain) mediating oligomerization, Tyr177: tyrosine 177, which, when phosphorylated, serves as a docking site for the adaptor protein GRB-2; SH2-domain: SRC homology 2 (binding to phosphorylated tyrosine residues, including BCR exon 1), SH3-domain: SRC homology 2 (binding to proline rich peptides). SH1-domain: SRC homology 1 (ABL1 catalytic domain); GEF-domain: guanine nucleotide exchange factors (G-protein signaling); E1: exon 1 of ABL1, contains the inhibitory N-terminal “cap” that binds the catalytic domain (SH1) of ABL1 and prevents autophosphorylation; NLS: nuclear localization signal.
Wild type ABL1 is a ubiquitously expressed but tightly regulated non-receptor tyrosine kinase that is present throughout hematopoietic development, with declining levels during myeloid maturation. It is predominantly located in the cytoplasm in hematopoietic cells, but can shuttle to the nucleus. In the cytoplasm, ABL1 is found mostly bound to actin, and functions include signaling and modulation of the cytoskeleton. Nuclear ABL1 has been implicated in cell cycle control. The N-terminus of ABL1 negatively regulates ABL1 kinase activity, allowing for tight titration of ABL1 kinase activity under physiologic conditions. Loss of the N-terminus as a result of the BCR–ABL1 translocation results in high constitutive kinase activity. Thus loss of this important regulatory domain is a major contributor to ABL1-mediated leukemogenesis (25) [reviewed in Ref. (26)].
The fusion partner of ABL1, BCR, is a complex locus that is transcribed into two major proteins, both with multiple functional domains implicated in a variety of fundamental biological processes. These include G-protein signaling pathways, cytoskeletal organization, growth, and development. The only exon of BCR that is consistently retained in all fusions is exon 1, which encodes a coiled-coil domain facilitating dimerization and autophosphorylation (amino acids 1–63) (27, 28), a docking site for the adaptor protein GRB-2 (phosphorylated tyrosine 177) (28, 29), and a tyrosine kinase domain (amino acids 298–413) (30). The exact role of the BCR-tyrosine kinase domain is unclear, and in a murine model of CML utilizing retroviral introduction of p210BCR–ABL1 deletion mutants into bone marrow cells, it appeared to be dispensable. On the other hand, the consistent inclusion of the BCR-tyrosine kinase domain in human BCR–ABL1-driven malignancies suggests that it may play a functional role in leukemogenesis (30–32).
Downstream Pathways Activated by BCR–ABL1 Fusion Proteins
The molecular consequence of all BCR–ABL1 fusion proteins is a hyperactive ABL1 kinase domain and aberrant phosphorylation of a variety of targets. Activation results from lack of autoinhibition due to loss of the N-terminal regulatory domain of ABL1, and homodimerization and autophosphorylation of the fusion protein (27). The importance of the homodimerization and autophosphorylation for BCR–ABL1 signaling is underscored by promising in vitro results of peptides and small-molecule inhibitors that cause allosteric inhibition of BCR–ABL1 (33–36). BCR–ABL1 kinase activity leads to direct and indirect activation of multiple pathways (37), including PI3K (38), AKT (39–41), MTOR (42, 43), RAS (39, 44), EGFR, MAP-kinase (40, 43, 45), JNK/SAPK (43), JAK1–3 (46), the SRC-family kinases LYN, HCK, and FGR (47), PTPN11, NF-kB, phospholipase C, and, as a common downstream effector of many of these pathways, STAT5 (Figure 3) (46, 48–50). Most of these pathways have been worked out in CML, but the relevant binding sites or kinase domains are preserved in the p190 fusion protein. Activation of JAK1–3 (50) and STAT1, 3, 5, and 6 (48, 50) has been experimentally confirmed for p190. Work in CML suggests that JAK1–3 activation is mediated through the interaction of BCR–ABL1 with cytokine receptors rather than direct phosphorylation (51). On the other hand, JAK2 appears to directly phosphorylate BCR–ABL1 at the critical tyrosine-177 residue and increase BCR–ABL1 protein stability, thus enhancing BCR–ABL1 signaling (52). Another important downstream pathway that has been confirmed specifically in Ph+ ALL is the PI3K–AKT–MTOR pathway. Deletion of PI3K inhibited leukemogenesis in a murine model of p190 Ph+ ALL. A dual PI3K/MTOR inhibitor was effective on Ph+ ALL patient samples (53) and showed synergy with Imatinib (54). The activation of AKT and MTOR signaling also plays a critical role in steroid resistance in ALL (55, 56), and multiple agents targeting the PI3K/AKT/MTOR axis are currently in clinical trials for pediatric ALL [reviewed in Ref. (57)].
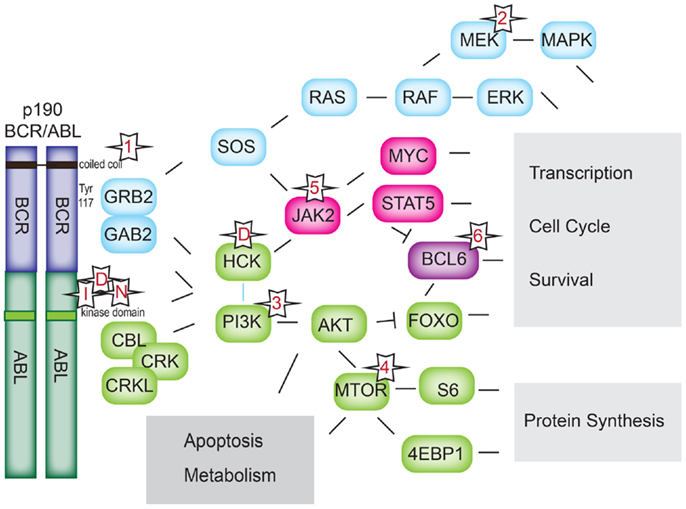
Figure 3. BCR–ABL1 signaling pathways. Downstream signaling pathways activated by BCR–ABL1. Numerical references 1–6 denote classes of inhibitors in Table 1. I: imatinib; D: dasatinib, N: nilotinib.
Of particular importance appears to be the adaptor proteins GRB-2 (58–60) and GAB-2 (60), which interact with, and participate in the activation of nearly all of the signaling pathways cited above. GRB-2 has been shown to bind phosphorylated tyrosine-177 (Figures 2 and 3). The importance of this interaction is demonstrated by the impaired in vivo leukemogenesis in murine models of p210BCR–ABL1 constructs with an engineered inactivating mutation of tyrosine-177 (29, 45, 61). Peptide-inhibition of the SH3 domain of the adaptor protein GRB-2 reduced growth and induced apoptosis in the BCR–ABL1-positive K562 cell line (7). Similarly, genetic inactivation of GAB-2 impairs p210BCR–ABL1-mediated transformation in mice (62).
Recently, overexpression of the epidermal growth factor ERBB was found to be specifically elevated in Ph+ ALL (56 versus 4.8% of Ph− ALL) (63). The molecular details of how this pathway intersects with BCR–ABL1 signaling requires further study; preliminarily, p70S6 kinase (MTOR target) has been implicated. From a translational perspective, the ERBB/HER2/NEU inhibitor lapatinib was synergistic with imatinib and nilotinib (but not dasatinib) on Ph+ ALL cell lines.
The reported activation of the SCR-family kinases LYN, HCK, and FGR by BCR–ABL1 has important implications. BCR–ABL1 has been shown to interact with and activate SRC-family kinases, and inhibition of SRK-family kinases decreased growth and survival of BCR–ABL1-positive myeloid cell lines in vitro (64–71). In addition to being activated by BCR–ABL1, the Src-family kinases Lyn (72) and Hck (73) have been reported to in turn phosphorylate BCR–ABL1 at several sites, including the critical residue for the interaction with the adaptor proteins Grb-2 and Gab-2 (Tyrosine-177). Expression of p210BCR–ABL1 in murine lymphoid progenitors negative for all three kinases (Lyn−/− Hck−/− Fgr−/−) near completely prevented leukemogenesis in a mouse model of Ph+ lymphoid leukemia (47). There was considerable redundancy between Lyn, Hck, and Fgr in this model, and genetic inactivation of at least two kinases was required to protect mice from leukemia. Somewhat surprisingly, given that CML cell lines responded to Src inhibition in vitro (67–71), Lyn, Hck, and Fgr were not required to induce CML in vivo (47). A small-molecule inhibitor of SRC-family kinases improved the survival of mice with lymphoid but not myeloid leukemia. Lack of inhibition of Src-family kinases by imatinib, and dual inhibition of Src-kinases and BCR–ABL1 with dasatinib were proposed to underlie the improved efficacy of dasatinib in the p210 lymphoid leukemia model (74). Under normal physiologic conditions, Lyn−/− Hck−/− FGR−/− mice display defects in B-cell maturation and autoimmune features suggesting a specific role for these kinases in B-cell development, but the early B-cell compartments appear to be preserved (47). Dependency of Src-family kinases may thus be a specific feature of Ph+ ALL. This is highly relevant from a clinical–translational standpoint as it provides a compelling rationale to investigate the dual BCR–ABL1–SRC-family kinase inhibitor dasatinib in Ph+ ALL, and suggests that this agent may be more effective in Ph+ ALL than imatinib or nilotinib.
Many of these downstream pathways – particularly the JAK–STAT pathway, are also targeted by several newly described leukemogenic fusion proteins that induce a disease similar to Ph+ ALL, but without a BCR–ABL1 rearrangement. These “Ph-like” leukemias share with Ph+ ALL a transcriptional signature indicative of kinase activation, co-occurring mutations in the B-cell transcription factor IKZF1, and a poor outcome. Initially identified based solely on transcription profiling, improved molecular techniques have allowed identification of a tyrosine kinase mutation in many of these patients. These include rearrangements of JAK2, ABL1, PDGFRB, CRLF2, and EPOR, deletion of SH2B3 encoding the JAK2-negative regulator LNK, and activating mutations of FLT3 and the IL7 receptor (IL7R) (75, 76). The presence of these mutations opens the door for potential therapeutic impact using targeted inhibitors (77). This fascinating subgroup of ALL is the topic of a dedicated review in this issue.
Differences between p210 and p190 – Lessons from Mouse Models
As mentioned earlier, p190BCR–ABL1 has stronger transforming activity than p210BCR–ABL1, both in fibroblast transforming assays (78) and in mouse models (7, 79). One possible reason may be a higher specific kinase activity and possibly broader substrate range of the p190BCR–ABL1 fusion protein (78). In transgenic animals, p190BCR–ABL1-induced exclusively B-lymphoid leukemia with a short latency, while p210BCR–ABL1 led to development of both lymphoid and myeloid leukemias with a longer latency (79). When introduced into stem cell and progenitor enriched mouse bone marrow, both p210BCR–ABL1 and p190BCR–ABL1 cause a myeloproliferative disease with expansion of granulocytic, myelomonocytic, and lymphoid compartments; however, the disease induced by p190BCR–ABL1 has a significantly shorter latency (7). p190BCR–ABL1 induces stronger STAT1 and STAT5 phosphorylation in Baf3 cells than p210BCR–ABL1 (48), and also induces phosphorylation of STAT6 (50).
Co-Occurring Genetic Abnormalities
Next generation sequencing studies have revealed that many leukemia genomes are remarkably stable – particularly when compared to epithelial cancers. Nevertheless, Ph+ ALL cells have been shown to carry several recurrent mutations that commonly co-occur with BCR–ABL1 fusions and contribute to leukemogenesis. The most frequent co-occurring genetic abnormalities are deletions of the lymphoid-specific transcriptional regulators IKAROS (IKZF1), PAX5 (paired box 5), and EBF1 (early B-cell factor 1). Deletions involving CDKN2A/B are also common. In addition, one of the first examples of “convergent clonal evolution” within the same leukemia was described in Ph+ ALL: one patient’s leukemia contained two cytogenetically distinct subclones that independently acquired a duplication of 8q, corroborating the crucial role of co-occurring mutations (80). Interestingly, GWAS studies have identified genetic polymorphisms of IKZF1 (81–87), PAX5 (88), and CDKN2A/B (81, 89–91) as susceptibility loci that mediate a genetic predisposition to childhood ALL. However, subgroup analysis, when performed, revealed no specific association with Ph+ ALL. This may in part be due to a low number of Ph+ ALL patients in these studies, and a targeted evaluation of IKZF1, PAX5, and CDKN2A/B susceptibility alleles specifically in Ph+ ALL patients may be warranted.
IKZF1 Deletions and Point Mutations in Ph+ ALL
A review of BCR–ABL1 in ALL requires discussion of its most frequent partner in crime, IKZF1 (86). BCR–ABL1 and IKZF1 mutations are strongly linked: about 70–80% of Ph+ ALLs have somatic mutations in IKZF1 (about 90% deletions and 10% point mutations), which is much higher than the rate of IKZF1 mutations in Ph− ALL (92–94). There are three functional types of IKZF1 mutations: haploinsufficiency or near haploinsufficiency (due to monoallelic null mutations such as inactivating point mutations, premature stop codons, and deletions, 55%), complete absence of Ikaros due to bi-allelic deletions (12%) (92, 93, 95), and alterations that create a dominant-negative (DN) form of Ikaros, IK6 (33% of all IKZF1 mutations). The IK6 Ikaros mutant is produced by an in-frame deletion of exons 4–7, which deletes the DNA-binding domain and leads to cytosolic accumulation of the mutant protein (92, 96, 97). The resulting hematopoietic phenotype in a mouse model mimicking this mutation (a smaller deletion that phenocopies the loss of the DNA-binding and nuclear export) is more severe that monoallelic null mutations, as Ik6 associates with the wild type Ikaros and probably traps it in the cytoplasm together with other complex members such as Helios, Aiolos, and Eos (98). The mutations associated with a more profound reduction in Ikaros function (bi-allelic deletion and Ik6) are particularly common in Ph+ ALL (92, 93, 95–97). This underscores the remarkably tight link between Ikaros and Ph+ ALL. In the closely related “Ph-like” ALL subset characterized by a gene-expression profile highly similar to that of Ph+ ALL, but without BCR–ABL1 fusion, IKZF1 mutations are also common. However, the majority of mutations result in a less severe reduction of Ikaros function [i.e., haploinsufficiency in 55–70% of all IKZF1 mutations in non-BCR–ABL1 ALL (95, 99)]. Twin studies and tracking of subclonal populations suggests that BCR–ABL1 fusion is the first hit, and IKZF1 mutations occur later during leukemogenesis (95, 100, 101). There are also reported cases of “convergent” evolution of IKZF1 mutations, with different subclones within the same patient carrying different IKZF1 mutations, underscoring the importance of this locus for Ph+ ALL (72, 80, 101). Much work has been dedicated to understanding the molecular mechanism of loss of Ikaros alone and in the context of Ph+ ALL. Complete loss or expression of DN Ikaros in normal hematopoiesis causes a mild (Ikzf1−/−) to severe (DN) reduction in the number of hematopoietic stem cells (HSC), complete loss of the B-cell and dendritic cell compartments, a skewing toward the T-lymphoid lineage (98, 102–108), and ultimately T-cell malignancies in mice (109). Despite this, IKZF1 mutations are much more common in B-lymphoid than in T-lymphoid malignancies (110). On the surface, the combination of BCR–ABL1 fusion and loss of IKZF1 neatly fits the paradigm proposed by Gilliand for acute myeloid leukemia (AML), which hypothesized that leukemia development requires a combination of class I (signal transduction pathway mutation leading to uncontrolled growth, such as FLT3, or RAS), and class II mutations (aberrant transcription factors resulting in differentiation block, such as PML–RAR, AML–ETO, MLL-translocations, or point mutations in C/EBPα) (111). According to this model, BCR–ABL1 is the class I mutation and IKZF1, the class II mutation. A possible reason for the frequent occurrence of at least the Ik6 mutation in B-cell precursor ALL could lie in the fact that the exons 4 and 7 are flanked by genomic regions that can function as off-target sites for recombination activated gene (RAG) proteins, which mediate VDJ recombination in this cell population (93, 112). Whether the particularly common co-occurrence of BCR–ABL1 and Ik6 is solely a function of the developmental stage of the cell of origin, or whether the presence of the BCR–ABL1 translocation predisposes to aberrant RAG activity is not known. In addition, if and how BCR–ABL1 and mutant Ikaros cooperate on a molecular level is still not fully understood. The normal function of Ikaros suggests that one of its main contributions to B-cell leukemogenesis is a differentiation block in the B-lymphoid lineage at the pro- to pre-B-cell transition. While complete loss of Ikaros results in a complete absence of the entire B-cell compartment, a severely reduced expression of Ikaros allows development up to the Pro-B-cell stage but not beyond (113). However, additional mechanisms likely play a role. Ikaros has been shown to downregulate Myc, thus loss of Ikaros may result in increased Myc activity and increased proliferation (114). Gene-expression profiling suggests that IKAROS mutated B-ALL has a more prominent “stem-cell signature” (99, 115), and a larger leukemia initiating cell pool [LIC, defined by CD34 expression rather than functionally (96)], suggesting that some of the functions of Ikaros in silencing stem-cell programs in HSCs may play a role (105, 116). Finally, it has been suggested that loss of Ikaros may either synergize with or enhance Jak–Stat signaling. This hypothesis is mostly based on the circumstantial evidence that the other main subtype of ALL with frequent IKZF1 mutations are the Ph-like leukemias. Ph-like leukemias share a transcriptional profile with Ph+ ALL and frequently carry mutations that, like BCR–ABL1, activate Stat5 (75, 117). Modulation of this pathway could both provide a competitive advantage at a subclonal level of IKZF1− clones, and provide an escape pathway for BCR–ABL1 inhibition. Loss of Ikaros predicts a poor prognosis even within Ph+ ALL (94, 99).
PAX5 Mutations in Ph+ ALL
Recurrent mutations of PAX5 occur in about one-third of B-ALL cases (99, 118, 119), and in up to 50% of Ph+ ALL (92, 120, 121). PAX 5 is a transcription factor that is expressed specifically during B-cell development, and controls lineage identity and commitment (107, 122). Like loss of Ikaros, loss of Pax5 leads to a differentiation block at the pro- to pre-B-cell stage (122–124). Loss of Pax5 also allows trans-differentiation of already lineage-committed pro-B cells into other lineages, confers a certain degree of self-renewal onto this population (125, 126), and can cause B-cell lymphomas (127). Unlike Ikaros, however, the physiologic expression of PAX5 is limited to B-cell precursor stages, and its loss is not associated with an adverse prognosis (99, 120, 121). It has been speculated that a lack of an effect of PAX5 on hematopoietic stem-cell transcriptional programs may be responsible for the different prognostic implications of PAX5 and IKZF1 mutations (120, 128). In addition to driving B-lymphoid development, Ikaros has been reported to repress hematopoietic stem-cell specific gene-expression programs during early lineage specification, a function not shared with the other two major regulators of B-cell development that are found mutated in Ph+ ALL, PAX5, and EBF1 (Figure 4) (105, 116, 128).
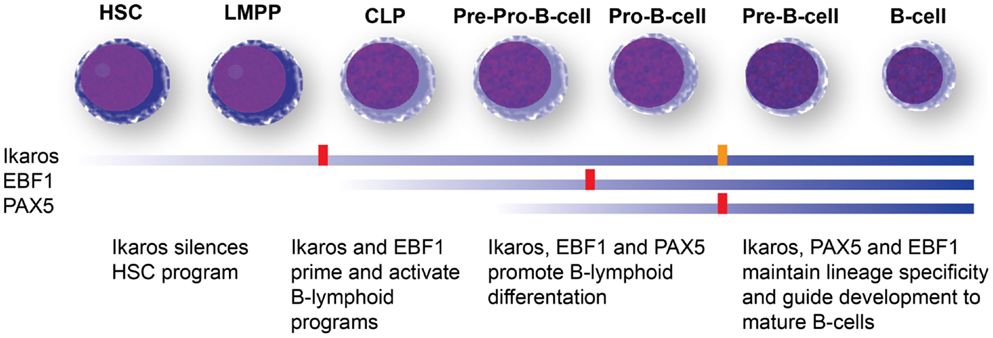
Figure 4. B-cell development and transcription factors mutated in Ph+ ALL. Differentiation stage-dependent expression (blue bars) and function of the three major B-cell developmental regulators mutated in Ph+ ALL, Ikaros, Pax5, and EBF1 (106, 107). Ikaros expression is detected early in hematopoietic development and appears to have a role in shutting down stem-cell programs and nudging cells toward lymphoid development. Ikaros expression is maintained through B-cell development. Complete loss of Ikaros in murine models leads to a differentiation block at the LMPP stage and complete absence of the entire B-cell lineage (red block). A severe reduction allows the development of B-cell progenitors, but maturation is blocked at the Pro-B stage (orange block). EBF1 is turned on in common lymphoid progenitors (CLPs) and controls lineage specification to the B-cell lineage. Loss of EBF1 in murine models leads to a differentiation block at the Pro-B stage (red block). Pax5 is turned on the latest and maintains lineage commitment. Loss of Pax5 causes a differentiation block at the Pro-B-cell stage. Neither Pax5 nor EBF1 appear to have a role in silencing hematopoietic stem-cell programs, which may explain why IKZF1 mutations are associated with a poor prognosis, while PAX5 and EBF1 mutations do not predict adverse outcomes. HSC: hematopoietic stem cells; LMPP: lymphoid-primed multipotent progenitors; CLP: common lymphoid progenitors.
EBF1 Mutations in Ph+ ALL
Early B-cell factor 1 is the predominant transcription factor mediating B-cell lineage commitment (107). It has been shown to co-regulate target genes with PAX5. In mouse models, complete loss of Ebf1 leads to a differentiation block at the pre–pro-B-cell stage (129). In contrast to Ikzf1 and Pax5, Ebf1−/− mice do not develop spontaneous hematologic malignancies (129). However, combining loss of one allele of Ebf1 or Pax5 with a constitutively active Stat5 allele (the downstream effector in both Ph+ and Ph-like ALL) results in B-cell precursor leukemia in all animals (130). EBF1 mutations occur in about 14% of Ph+ ALL.
CDKN2A/B in Ph+ ALL
The CDKN2A/B locus is frequently altered in ALL. The products of the CDKN2A and CDKN2B genes, p16INK4A and p15INK4B, are inhibitors of cyclin-dependent kinases. In addition, transcription of an alternate reading frame of the CDKN2A locus produces p14ARF, which antagonizes the p53 ubiquitin ligase, HDM2. Silencing of the CDKN2A/B locus in HSC has been implicated in HSC self-renewal (131–134). The distribution of CDKN2A/B alterations within cytogenetic subgroups is non-random. CDKN2A/B is rarely deleted in ALL with translocations of E2A [E2A–PBX1 in t(1;19)(q23;p13) and E2A–HLF in t(17;19)(q21–22;p13) ALL] (135). In contrast, increased rates of CDKN2A/B deletions are found in Ph+ ALL with a reported frequency of ~50% (80, 92, 136) compared to around 30% in non-Ph+ B-ALL (137, 138). CDKN2A/B deletions are rare in CML in chronic phase but frequently associated with the transformation to lymphoid blast crisis, suggesting a specific role in B-lymphoid leukemia (139). Similar to IKZF1, the mechanism of deletion of p16 in lymphoid malignancies may involve RAG-mediated recombination (140). Experimental overexpression of BCR–ABL1 induces expression of Arf, which, if unopposed, leads to apoptosis (141). Introduction of p190BCR–ABL1 into Arf -null murine bone marrow decreases the latency and increases resistance to imatinib in the lymphoid malignancy that develops in recipient mice. In most clinical studies that have assessed the prognostic significance of CDKN2A/B loss of function in ALL, CDKN2A/B deletion or hypermethylation do not appear to be associated with changes in outcome for pediatric ALL, while silencing or inactivation of the locus predicts a worse outcome in adults (99, 136, 138, 142–146). In a recent study mapping clonal evolution in adult patient-derived Ph+ ALL cells grown in immunodeficient mice, the loss of CDKN2A/B was associated with increased competitive advantage on a subclonal level, more aggressive growth in xenografts, a higher leukemia initiating frequency, and a trend toward inferior outcome in patients (80). In both children and adults, deletions as well as epigenetic silencing through promoter hypermethylation are found at increased frequencies in relapsed specimens as opposed to those from initial diagnosis, suggesting a role in mediating relapse and resistance to therapy (147–150).
Epigenetic Abnormalities in Ph+ ALL
In addition to genetic abnormalities, Ph+ ALL has a characteristic DNA methylation profile. Ph+ ALL can be distinguished from other subtypes of ALL by hierarchical clustering of DNA methylation profiles. A recent study that quantified differentially methylated regions (DMRs) in all major ALL subtypes (compared to B-cell precursors, i.e., using a developmentally matched control) revealed about 350 DMRs in Ph+ ALL samples (151). This was remarkably different from only about 50 DMRs identified in CRLF2-rearranged ALL samples, many of which were “Ph-like” ALL samples that share a major transcriptional program with Ph+ ALL but are negative for the BCR–ABL1 translocation, and instead commonly have activating Jak1/2 mutations. Whether differential methylation is a consequence of the BCR–ABL1 fusions or co-occurring genetic abnormalities, and whether it plays a role in malignant transformation, resistance or relapse is unknown. However, an active role for DNA methylation in malignant transformation (rather than a mere reflection of the transcriptional landscape) is supported by the dependence of several experimental tumor models on functional DNA methyltransferase Dnmt1 (152, 153), including MLL–AF9 and Myc–Bcl2-driven leukemia (154, 155). Reactivation of silenced CDKN2A/B by demethylating agents may have therapeutic benefit in a subgroup of ALL patients where this locus is intact (148), and demethylating agents are currently in clinical trials for relapsed and refractory ALL. Very little is known about a potential involvement of other epigenetic mechanisms in the biology of Ph+ ALL, such as covalent modifications of histones or nucleosome positioning. A better understanding of Ph+ ALL biology, including associated genetic and epigenetic abnormalities, should facilitate the development of rational synergistic combinations of targeted agents with TKIs.
Mechanisms of Resistance in Ph+ ALL – BCR–ABL1 Mutations
One of the primary mechanisms of resistance and treatment failure in CML is the acquisition of BCR–ABL1 mutations that render the fusion protein completely or relatively unresponsive to TKIs. A plethora of different mutations mediating imatinib-resistance have been described. Most of these mutant BCR–ABL1 proteins are still sensitive to the second generation ABL kinase inhibitors dasatinib and nilotinib. In addition, the recently approved ponatinib is active against the most common mutation that causes resistance to both first and second generation ABL TKIs, the “gatekeeper” T315I mutation (Table 2; Figure 5). Kinase domain mutations develop even more frequently in Ph+ ALL treated with TKI monotherapy despite initial sensitivity (156–162) (161, 163, 164). A rate of BCR–ABL1 kinase domain (TKD) mutations of >80% has been reported in (adult) patients with Ph+ ALL at relapse (165–167), with the most common mutations being T315I, Y253H, and E255K/V (42, 168). There has been considerable debate in the field whether these mutations occur during treatment, or whether TKIs select for pre-existing resistant subclones. Several studies suggest that a substantial percentage of patients harbor subclones with TKD mutations prior to the initiation of therapy (42, 169–171). BCR–ABL1 mutations known to cause resistance have been identified in minor subclones in as many as 40% of Ph+ patients at initial diagnosis (169). Tyrosine kinase domain mutations may be less common in patients treated with a combination of intensive chemotherapy and TKI. In addition, the kinetics of emergence of resistant mutants – when they do develop – is not well-studied. Sequencing of 10 evaluable pediatric patients treated with imatinib and highly intensive chemotherapy on AALL0031 revealed two known resistance causing BCR–ABL1 mutations at relapse, none of which were detected in the initial diagnostic sample (38). Both mutations are responsive to nilotinib or dasatinib (M244V and H396P) (53). In contrast, in the GIMEMA LAL1205 study, the T315I mutation was discovered in four out of seven patients who relapsed after induction with dasatinib + steroids only, followed by intensive consolidation (intensive chemotherapy + TKI + auto HSCT, or allo HSCT) (172). It is possible that the combination of TKIs with an up-front intensive chemotherapy backbone serves to reduce selective pressure on TKI-resistant subclones. However, more in depth analysis will be required to define the respective roles of resistance to TKI and standard chemotherapy, and their interdependence. The fact that BCR–ABL1 has been found mutated at relapse also raises the important question how aggressively patients with Ph+ ALL who receive TKIs on a backbone of standard chemotherapy should be screened for the emergence of TKD mutations. Since patients may still respond to the chemotherapy portion, early warning signs of TKI failure may be missing. If resistant mutants develop or emerge with similar kinetics to what is observed with monotherapy, patients may receive months or years of ineffective TKIs only to ultimately relapse, when early detection of an emergent mutant clone could have prompted switching to another agent active against the mutant BCR–ABL1. A major technical difficulty of such studies is the limit of detection to reliably assess and follow clonal heterogeneity in a minimal residual disease (MRD) setting. The decreased cost of sequencing and novel techniques such as MRD-sort combined with high throughput single cell sequencing may be able to provide answers in the near future. However, the complexity and cost of such an approach would first require a more in depth study of whether BCR–ABL1 mutations are a substantial contributor to resistance and relapse in Ph+ ALL treated with intensive chemotherapy plus TKI.
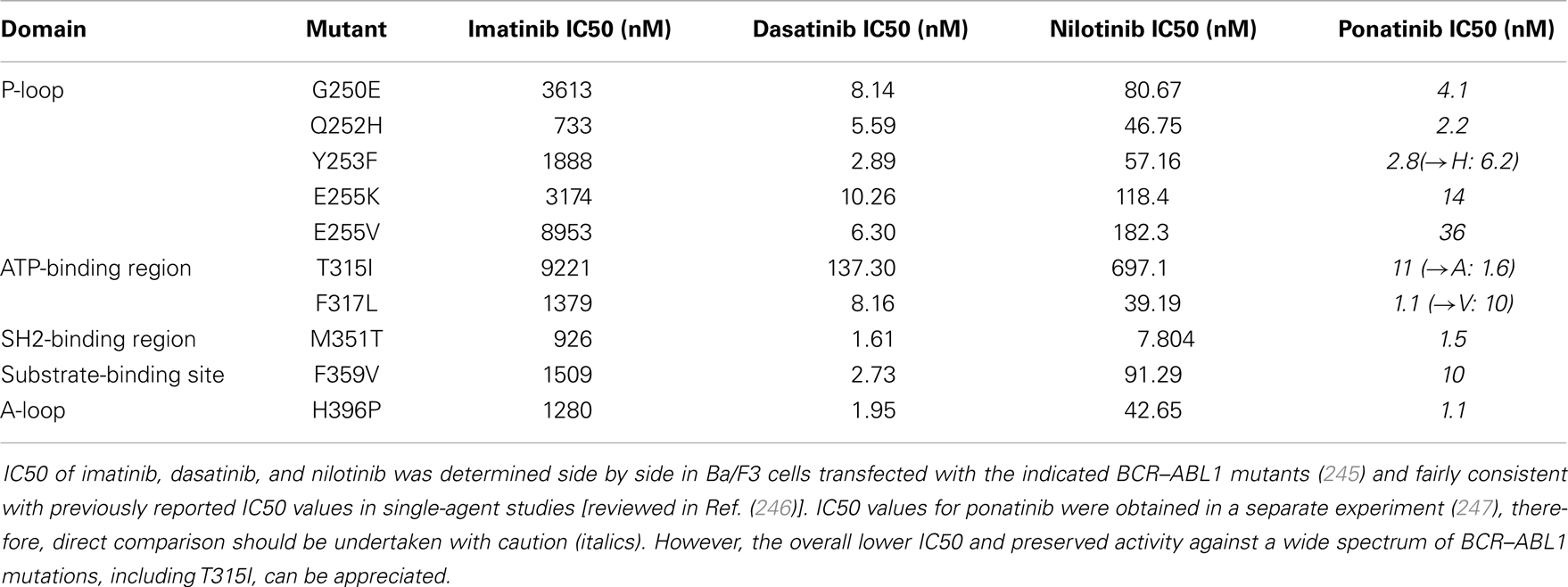
Table 2. Activity (IC50) of imatinib, dasatinib, nilotinib, and ponatinib against selected BCR–ABL1 mutants.
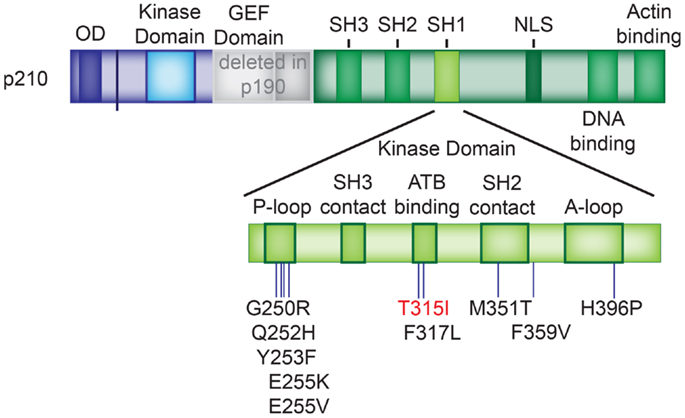
Figure 5. BCR–ABL1 TKD mutations. Location of the BCR–ABL1 tyrosine kinase domain mutations listed in Table 2. The T315I mutation, which causes resistance against imatinib, dasatinib, and nilotinib is depicted in red. P-loop: phosphate-binding loop; A-loop: activation loop.
Mechanisms of Resistance in Ph+ ALL – Increased Intracellular BCR–ABL1
In addition to kinase domain mutations, increasing the amount of BCR–ABL1 protein in the cell can lead to resistance to TKIs. One mechanism includes amplification of the BCR–ABL1 fusion gene or the entire Ph+ chromosome, which have been reported in up to 10% of Ph+ ALL (173, 174). Amplification of p210BCR–ABL1 also appears to be particularly common in lymphoid blast crisis of CML (175).
Mechanisms of Resistance in Ph+ ALL – Overexpression of Drug Exporters
There is also evidence for the involvement of drug exporters such as ABCB1/MDR (176)1/PGP and ABCG2/BCRP in the development of TKI resistance in CML (112, 176–178). Less data is available on the role of drug efflux pumps in Ph+ ALL. One study evaluated the expression levels of LRP, MRP, and ABCB1/MDR1/PGP in Ph+ ALL, and found consistent overexpression of LRP, but not MRP and ABCB1/MDR1/PGP (179). In contrast, promoter CpG methylation of MDR1 (i.e., silencing) in ALL was found to be inversely correlated with the presence of the Philadelphia chromosome, suggesting a role for MDR1 in Ph+ ALL as well (146). Overexpression of ABCB1/MDR1/PGP was also found as a mechanism of resistance to nilotinib in CML cell lines (180).
Mechanisms of Resistance in Ph+ ALL – Upregulation of Parallel Pathways
A large amount of data supports Src-family kinases as a mediator of resistance to imatinib and nilotinib. Src-family kinases are activated downstream of BCR–ABL1, and may play a particularly important role in lymphoid blast crisis and Ph+ ALL (see also “Downstream Pathways Activated by BCR–ABL1 Fusion Proteins”) (47, 64–71, 74). Persistent HCK and LYN activity has been detected in CML patients who failed imatinib despite the absence of a BCR–ABL1 mutation (72, 181, 182). In imatinib-resistant cells, LYN was found in a complex with BCR–ABL1 and GAB-2 (72). LYN and HCK have been shown to be activated by BCR–ABL1 in a non-kinase dependent manner, and are in turn able to phosphorylate BCR–ABL (72, 73). Imatinib-resistant CML cell lines or mouse models overexpressing Src-family kinases were shown to respond to the dual BCR–ABL1 – Src-family kinase inhibitor dasatinib, but not nilotinib or Imatinib (147).
Another parallel pathway with a potential role in TKI resistance is the JAK–STAT pathway. p190BCR–ABL1 activates JAK1, 2, and 3 and STAT1, 3, 5, and 6, likely providing a positive-feedback loop, substantial signal amplification, and redundancy. A dual BCR–ABL1–JAK2 inhibitor was able to overcome TKI resistance in vivo and a K562 mouse model (52, 183). JAK2 inhibitors are currently in clinical trials for myeloproliferative disorders and AML, and are being explored for Ph-like ALL with JAK mutations (184).
An additional escape pathway involves AKT, STAT5, and the B-cell lymphoma 6 (BCL6) transcriptional repressor. BCL6 is a well-established proto-oncogene in lymphoid malignancies that is a common target of activating translocations in diffuse large B-cell lymphoma (DLCBL) (185). STAT5, one of the major downstream targets of the BCR–ABL1 fusion protein, suppresses BCL6 in B-cells (186, 187). In vitro treatment of Ph+ ALL cells was shown to result in about 90-fold upregulation of BCL6 that was dependent on STAT5, to levels similar to those present in DLCBL. In addition, BCL6 is upregulated by FoxO4, which in turn is inactivated by PI3K/AKT signaling. This suggests involvement of both the JAK2/STAT5 and PI3K/AKT signaling pathways in mediating the derepression of BCL6 in response to TKIs. Deletion, expression of a dominant negative, or pharmacologic inhibition of BCL6 synergized with imatinib and nilotinib in syngeneic and primary patient xenograft mouse models (188). The mechanism of BCL6-mediated resistance may include repression of Arf and p53-mediated apoptosis, and the induction of more stem-cell like features such as quiescence and self-renewal, which may specifically enhance the survival and resistance of LICs (189). Induction of cell cycle exit and quiescence is one of the normal functions of BCL6 during B-cell development at the pre-B stage (190).
The RAS-RAF-MEK–MAPK pathway may also contribute to suboptimal efficacy of BCR–ABL1 inhibitors and resistance (43). MAPK activity was found to be increased in a presumed compensatory response to imatinib treatment of human CD34+ cells transduced with p210BCR–ABL1. A MEK-inhibitor reduced proliferation in this model and showed synergy with Imatinib (40). Finally, overexpression of the epidermal growth factor ERBB/HER2/NEU has been linked to drug resistance in ALL (191). ERBB is frequently elevated in Ph+ ALL, and ERBB/HER2/NEU inhibitor lapatinib was synergistic with imatinib and nilotinib (but not dasatinib) on Ph+ ALL cell lines with high ERBB expression (63).
Mechanisms of Resistance in Ph+ ALL – Acquisition of Additional Genetic Abnormalities Interfering with Transcriptional Regulation and B-Cell Development
Mutations of IKZF1 play a fundamental role in Ph+ ALL (86). In addition to being extremely common at diagnosis, IKZF1 mutations are further enriched at relapse. This includes both presumed de novo acquisition of an IKZF1 mutation (192, 193), as well as selection for a subclone with a more severe mutation (Ik6 or homozygous deletion) at relapse that was initially present at a low percentage (95). This pattern suggests a role for Ikaros in mediating drug resistance and relapse. However, how Ikaros would specifically cause drug resistance is unclear. A potential mechanism could lie in the fact that IKZF1 mutated B-ALL has a more “stem-cell like” signature by gene-expression profiling than IKZF1 wild type B-ALL. HSC are highly drug resistant, and expression of a stem-cell program has been associated with drug resistance and poor outcome in other types of leukemia (194–197).
Mechanisms of Resistance in Ph+ ALL – Inhibition of Apoptosis
As discussed above, increased promoter methylation has been described in relapsed ALL specimens when compared to the initial diagnostic specimen. CDKN2A/B appears to be a key locus that influences to drug resistance and is affected by epigenetic silencing or deletion (137, 142, 143, 146, 148, 151, 198). One of the consequences of deleting or silencing the CDKN2A locus is the inability to upregulate p14Arf, which can lead to a loss of functional p53 through increased activity of the p53 E3 ubiquitin ligase HDM2. Another mechanism of inactivating p53 in Ph+ ALL involves the overexpression of BCL6 discussed above (188). In a murine model of Ph+ ALL, imatinib treatment resulted in BCL6 upregulation and downregulation of p53, while genetic inactivation of BCL6 resulted in increased levels of p53 and failure to cause leukemia in mice. Interestingly, BCL6 was also recruited to the CDKN2A locus. Dual inhibition of BCR–ABL1 and BCL6 demonstrated in synergy in xenografts of patient-derived primary Ph+ ALL cells. Efficient inactivation of the p53 pathway by BCR–ABL1 and its downstream targets may explain why genetic mutations or deletions of p53 are rare up-front in Ph+ ALL (199, 200). Acquisition of p53 mutations at relapse has been described, but appears a rather rare event compared to the commonly affected IKZF1, PAX5, and CDKN2A/B loci (150, 201–203).
Mechanisms of Resistance in Ph+ ALL – Resistance to Concomitant Chemotherapeutic Agents
In addition to resistance to TKIs, resistance to standard chemotherapy agents can develop during treatment with chemotherapy plus TKI. Mechanisms of resistance discussed above – mutation, amplification, or upregulation of the molecular target, drug exporters, the expression of stem-cell signatures, upregulation of survival pathways, and inhibition of apoptosis apply to cytotoxic agents as well (204–206). In addition, several recurrent genetic alterations have been found enriched in ALL at relapse that confer resistance to specific cytotoxic agents. These include loss of MSH6 [mediating resistance to thiopurines, alkylating agents, and prednisone (202)], decreased MSH2 protein levels (mediating resistance to purine analogs) (207), and mutations in NR3C1 and CREBBP (mediating glucocorticoid resistance) (55, 193, 203, 208–211). None of these mechanisms appear to be specific to Ph+ ALL. Finally, the contribution of AKT and MTOR – both are activated in response to BCR–ABL1 signaling – to glucocorticoid resistance has been discussed above (55, 56).
Mechanisms of Resistance in Ph+ ALL – Interactions with the Bone Marrow Niche and the Immune System
In addition to cell-autonomous mechanisms of resistance, ALL blasts interact with the bone marrow niche (212) and the immune system. Some of these interactions probably contribute to the emergence of resistance and relapse. At the same time, our improved understanding of leukemic blast–host interactions has led to the development of therapeutic approaches that attempt to disrupt the interaction with the bone marrow niche (213, 214), or aim to break tolerance and effectively engage the patient’s immune system in the eradication of leukemia [reviewed in Ref. (206)]. None of these approaches are specifically targeted at Ph+ ALL.
Current Clinical Concepts in the Treatment of Ph+ ALL
Initial Studies Using Imatinib
Prior to the use of TKI, survival for pediatric Ph+ ALL treated with chemotherapy with or without HSCT was extremely poor. The International Ponte di Legno Childhood ALL Consortium reported 7-year event-free survival (EFS) and OS rates of 25 and 36% for 326 Ph+ ALL patients diagnosed between 1985 and 1996, and 32 and 45% for 610 Ph+ patients diagnosed between 1995 and 2005 and treated without TKI in first remission (4, 215). Although initial white blood cell count, age, and early response to therapy were predictive of outcome, the EFS even for “good-risk” patients was <50% (4). In the pre-TKI era, HSCT in first remission was generally considered to be the best available treatment option, particularly if a matched related donor was available. While HSCT produced slightly better results than chemotherapy alone, relapse was common even after allogeneic HSCT. In the 1995–2005 era, the 5-year EFS and OS rates for patients who achieved a first remission (89%) and went on to HSCT were only 34.2 and 48.3% (4).
Following the landmark reports showing the efficacy of imatinib in CML, several studies had shown transient single-agent efficacy of imatinib in patients with relapsed or refractory Ph+ ALL, and adult trials had shown the feasibility of combining imatinib with chemotherapy, albeit on an intermittent dosing schedule and typically as a bridge to early HSCT (157, 159, 162). In 2002, the Children’s Oncology Group (COG) began the AALL0031 trial that was designed to evaluate the safety and efficacy of combining imatinib (340 mg/m2/day) with a very intensive chemotherapy regimen (2). Imatinib was introduced after 4-weeks of induction therapy were completed and the exposure was successively increased in five cohorts, with expanded accrual in cohort 5 to estimate efficacy. In cohort 5, patients received continuous imatinib therapy starting at day 1 of consolidation therapy, with intermittent (14 days on – 14 days off) dosing during the last year of maintenance therapy. A parallel cohort of Ph− very high-risk ALL patients was treated with the identical chemotherapy regimen, without imatinib. Overall, addition of imatinib to the intensive chemotherapy regimen was tolerated very well, with no significant increase in toxicity of the combined regimen compared to chemotherapy alone. In the initial study report, the 3-year EFS for patients treated in cohort 5 was 80%, as compared to 35% in a historical control group treated in the pre-TKI era (Figure 6) (2). This outstanding outcome has been stable over time with recent analyses showing a 71% 7-year EFS rate for patients in AALL0031 cohort 5 versus 21.4% in the historical control group (Figure 6) (216). AALL0031 also explored the use of HSCT (matched related HSCT for patients on study, the final analysis also included follow-up of patients who went off study for an unrelated-donor HSCT). Neither matched related or unrelated HSCT offered any additional benefit (2, 216). Results of this trial, although based on a relatively small number of patients, revolutionized clinical practice for children and adolescents with Ph+ ALL. Tyrosine kinase inhibition is an integral part of treatment for Ph+ ALL and is now incorporated during induction therapy. HSCT in first remission is no longer routinely recommended for all patients as a default, though it may still be an important treatment for patients that respond poorly to chemotherapy plus TKI.

Figure 6. Survival of children with Ph+ ALL treated with imatinib + chemotherapy on COG AALL0031. (A) Event-free survival (early follow-up) of cohort 5 of AALL0031 treated with the MTD of imatinib in combination with chemotherapy compared to historic controls. Thirteen of the 44 patients went on to receive a matched related HSCT, while the remaining 31 patients received chemotherapy only. Introduction of imatinib onto a backbone of standard chemotherapy dramatically improved the outcome of Ph+ ALL. (B) Disease-free survival (5.2-year median follow-up) of patients treated as per AALL0031 cohort 5 based on transplant status. Chemotherapy only – 24 patients, matched related HSCT on study – 13 patients, unrelated HSCT off study – 6 patients. HSCT did not offer additional benefit compared to chemotherapy + imatinib.
A key pediatric study conducted primarily in Europe by 10 study groups in parallel to COG AALL0031, was the EsPhALL phase 3 trial (91). Patients received induction therapy according to the standard of their respective study group, and were categorized as good (108 patients) or poor risk (70 patients) based on initial response. The study then randomized good-risk patients to chemotherapy with or without post-induction imatinib on the backbone of the intense Associazione Italiana di Ematologia Oncologica Pediatrica – Berlin–Frankfurt–Münster (AIEOP-BFM) ALL 2000 regimen. Poor-risk patients were all assigned to chemotherapy plus imatinib. HSCT was recommended for all patients, and performed in 77% of patients in this trial. The imatinib exposure was intermittent with much lower cumulative exposure than used on AALL0031 (126 versus 616 days on AALL0031 cohort 5). The EsPhALL trial confirmed the superior efficacy of imatinib plus chemotherapy. The 4-year disease-free survival (as treated analysis of 81 patients) was 75.2% for good-risk patients receiving imatinib and 55.9% for those who did not receive imatinib (p = 0.06). Outcomes for the poor-risk population were also encouraging with 4-year EFS of 53.5%, which was significantly better than the outcome of historical control patients treated without imatinib. This is the largest, and the only randomized trial to systematically assess imatinib in combination with intensive multi-agent chemotherapy in children with Ph+ ALL. Given the results of this trial and AALL0031, it is highly unlikely that another trial will ever be conducted for chemotherapy with/without TKI in pediatric Ph+ ALL. The EsPhALL trial was amended to start continuous imatinib treatment at day 15 of induction therapy, and decrease the number of patients who underwent HSCT in first remission. This study will accrue patients through early to mid 2014.
A superior outcome with the addition of imatinib to chemotherapy in pediatric Ph+ ALL was also reported by the Sociedad Española de Hematología y Oncología Pediátricas (217). In the SHOP-2005 trial imatinib was initiated on day 15 of induction, and most patients (15/16) underwent bone marrow transplantation in first remission. The 3-year EFS for patients on this trial was significantly higher than that of historical controls who did not receive imatinib (the rate of BMT in the historical control was 17/27).
Second Generation TKIs (Dasatinib, Nilotinib)
In light of the encouraging but still suboptimal results obtained with chemotherapy plus imatinib in pediatric Ph+ ALL, and the development of second generation BCR–ABL1 inhibitors, it was logical to investigate whether the use of dasatinib or nilotinib in combination with chemotherapy might further improve outcomes (Table 3). Multiple lines of reasoning suggest that this could be the case. Second generation TKIs are more potent inhibitors of BCR–ABL1 in vitro, and lead to earlier and more profound reduction of the leukemic clone burden in CML (15, 16). The CNS is a well-known sanctuary site in ALL and CNS penetration of imatinib is poor. While very little data are available on the CNS penetration of nilotinib, oral administration of dasatinib produces therapeutic levels in the CSF (218). The imatinib-resistant TKD mutations reported to occur in relapsed Ph+ ALL are typically responsive to dasatinib or nilotinib, with the important exception of the gatekeeper T315I mutation that is resistant to all three agents (38, 53). Finally, multiple other kinases are activated by BCR–ABL1, including PI3K/AKT (39, 40), EGFR, MAP-kinase (40, 45), JNK/SAPK, JAK1–3, and the Src-family kinases LYN, HCK, and FGR (47, 67–71). Particularly Src-kinases appears to be an important downstream target of the BCR–ABL1-induced signaling cascade, while also phosphorylating BCR–ABL1 in a positive-feedback loop, and contributing to clinical resistance against TKIs. This pathway may be particularly important in Ph+ lymphoid malignancies (47, 72, 74, 91, 157, 180–182). Because dasatinib targets both BCR–ABL1 and Scr-family kinases, it is a particularly attractive agent to investigate in Ph+ ALL, as it may suppress the BCR–ABL1 signaling cascade at multiple levels. This could result in a more profound inhibition, reduction of the emergence of resistance, and improved clinical outcomes. In contrast to dasatinib, nilotinib does not efficienty inhibit Src-family kinases, and LYN (as well as two other tyrosine kinases that interact with LYN, SYK and AXL) has been implicated in mediating resistance to nilotinib (180, 219).
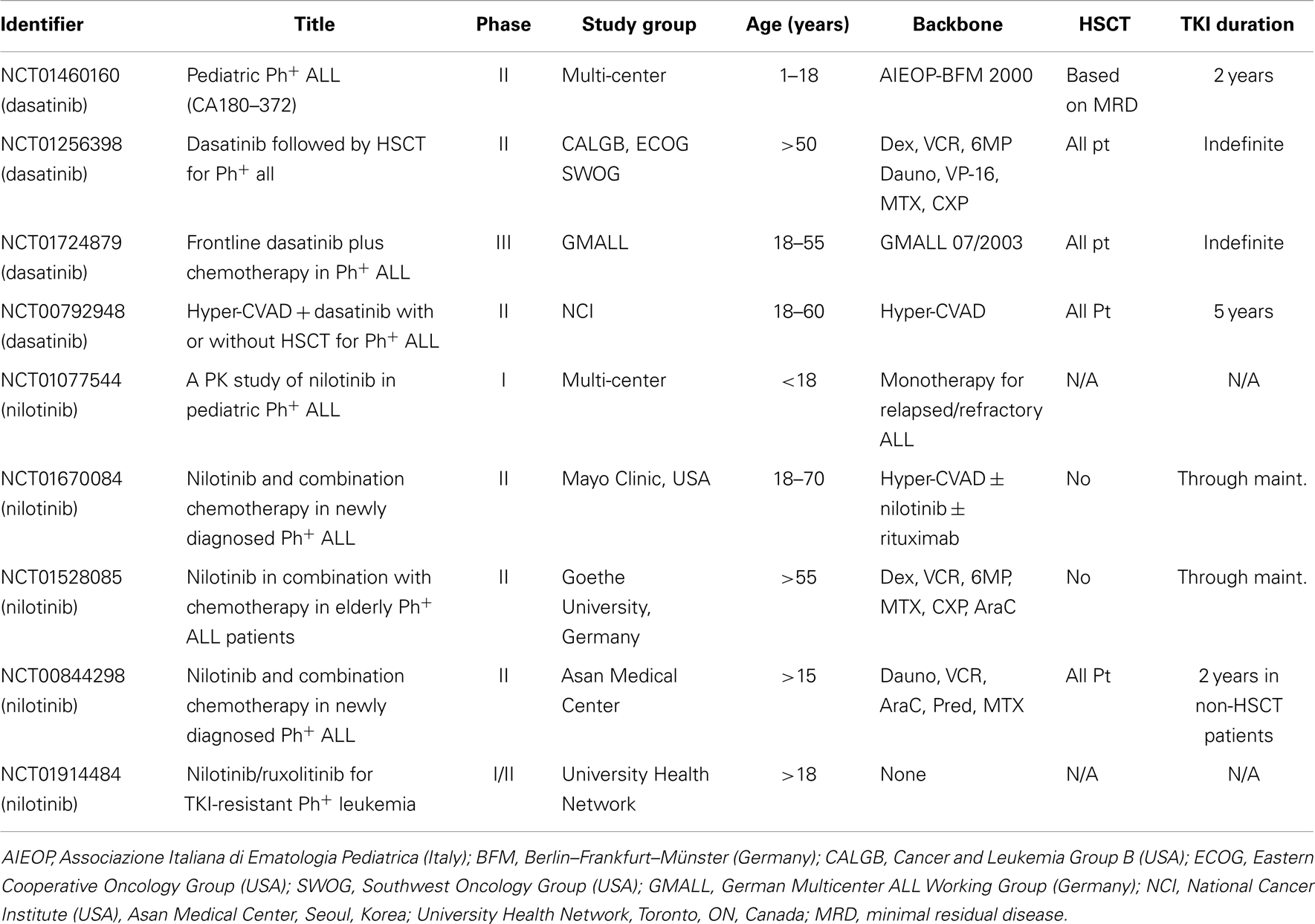
Table 3. Selected open clinical trials (clinicaltrials.gov) investigating dasatinib and nilotinib for Ph+ ALL.
Dasatinib monotherapy demonstrated encouraging efficacy against adult Ph+ ALL in early clinical trials (220, 221). In the GIMEMA LAL1205 study, 55 patients received induction with dasatinib + steroids and IT-MTX, while post-induction consolidation was at the discretion of the treating center. Two patients received no further therapy and 19 patients continued on TKI only (69.6% relapse at 20 months), 14 patients received intensive chemotherapy + TKI (21.7% relapses), 18 went on to allogeneic HSCT (11.1% relapses) (172). This trial demonstrated impressive remission rates for dasatinib and steroids only, but at the same time underscored the importance of adding intensive chemotherapy and/or HSCT to maintain durable remissions. The encouraging early data led to several trials investigating dasatinib in combination with chemotherapy. Incorporation of pulses of twice daily dasatinib with hyper-CVAD, followed by continuous dasatinib during maintenance and indefinitely after completion of chemotherapy resulted in a CR rate of 94% and an estimated 2-year survival of 64% (EFS and OS have not yet been reported) (222). The combination of dasatinib with hyper-CVAD using a similar regimen (but with dasatinib given continuous) in adults with relapsed Ph+ ALL was also recently reported (223). Dasatinib needed to be dose reduced from 100 to 70 mg daily due to prolonged cytopenias. The lower dose was well-tolerated and the CR rate was encouraging at 68%, but only two patients (11%) were alive with at a median follow-up of 52 months.
Two phase I/II pediatric trials showed that dasatinib monotherapy was well-tolerated and safe in children (224, 225). The COG AALL0622 trial tested dasatinib (60 mg/m2/day) in combination with the same chemotherapy regimen used with imatinib in COG AALL0031. In addition to the different TKIs used in the two studies, AALL0622 started dasatinib at day 15 of induction rather than starting after induction therapy was completed as done in AALL0031. AALL0622 has completed accrual and concluded that daily dasatinib therapy was safe with this chemotherapy regimen (226). Although it is too early to report outcome following treatment with chemotherapy plus dasatinib on AALL0622, the early response rates compare favorably to those obtained in AALL0031 with chemotherapy plus imatinib (227). The complete remission and end block 2 consolidation MRD negative rates were 98 and 89% on AALL0622, as compared to 89 and 71% on AALL0031. The COG and the European EsPhALL group are currently collaborating on a study that tests adding dasatinib 60 mg/m2/day to the EsPhALL chemotherapy backbone (COG AALL1122, NCT01460160) and are working together to develop a successor trial that randomizes patients to receive one of two different chemotherapy backbones plus TKI.
The other major second generation TKI is nilotinib, which has similar activity to dasatinib against imatinib-resistant BCR–ABL1 mutants. However, nilotinib does not inhibit the Src-family kinases, which is theoretically important in Ph+ ALL but has not yet been shown to be clinically important (180, 219). It is not clear whether the CNS penetration of nilotinib is as good as that of dasatinib (218). Nevertheless, nilotinib has shown encouraging activity in adult relapsed/refractory Ph+ ALL, with some patients achieving responses that were sustained for months on monotherapy (228, 229). Published studies on nilotinib therapy in newly diagnosed Ph+ ALL are scarce, but a recently presented pilot study of four Ph+ ALL patients who received a combination of nilotinib with intensive chemotherapy reported complete remissions in all patients (230). Early results of combining nilotinib with intensive chemotherapy in the current South Korean cooperative trial have also shown excellent activity (231). Nilotinib was given continuously from induction until completion of chemotherapy (2 years). All patients were eligible for HSCT, and 59/91 of patients underwent HSCT in first remission. The complete remission rate was 90% and the 2-year OS 70%.
Targeting the T315I Mutation: Ponatinib
Dasatinib and nilotinib are effective against the vast majority of reported BCR–ABL1 TKD mutants with one crucial exception: the T315I mutation. For many years, T315I conferred resistance to all available TKIs. Ponatinib is a multi-targeted TKI with strong activity against BCR–ABL and imatinib-resistant mutants including, for the first time, T315I. Ponatinib gained accelerated approval from the FDA in the US in December 2012 based on very promising activity observed in the PACE II clinical trial, including major cytogenetic responses in 70% of patients with T315I mutations (18). Several other studies have shown promising activity in newly diagnosed CML patients (NCT01570868, NCT01641107, and the EPIC trial NCT01650805), but all of these US trials were halted in October 2013, when a high rate of cardiovascular adverse events was reported in post-approval surveillance. Ponatinib was approved for use in the European Union in July 2013, and remains on the market at this time. As of December 2013, the future of this agent, which is the only agent known to be effective for CML and Ph+ ALL patients with T315I mutations, remains uncertain. Concerningly high rates of cardiovascular events were also reported for nilotinib, although they have received less attention in the scientific and lay press (232, 233). This raises the important question how to weigh molecular efficacy against risks and side effects of treatment. The risk–benefit ratio of any given agent will be different in an elderly patient with cardiovascular risk factors or pre-existing disease with newly diagnosed CML, or a child with Ph+ ALL. Irrespective of its potential use in TKI-naïve CML and Ph+ ALL patients, the impressive activity against the T315I BCR–ABL1 will likely make ponatinib a crucial second line agent for patients with this mutation.
Current and Future Clinical Questions
Chemotherapy Backbone
In addition to the choice of TKI to be used in Ph+ ALL, several other critical questions regarding the optimal therapy for pediatric Ph+ ALL remain unresolved. The current highly intense chemotherapy backbones are associated with substantial treatment related morbidity and mortality, as well as significant potential risk of late effects such as infertility and second malignancies. Furthermore, chemotherapy that is too intensive may compromise the ability to deliver optimal TKI therapy or to combine TKIs with other new targeted therapies. As discussed above, COG AALL1122, NCT01460160 is investigating dasatinib on the chemotherapy backbone of the AEIOP-BFM ALL 2000 regimen, which had shown excellent results in the EsPhALL trial. This backbone contains significantly reduced cumulative doses of cyclophosphamide, ifosfamide, etoposide, and high-dose methotrexate compared to AALL0031. Despite the changes, the morbidity and mortality associated with this regimen is still substantial. A successor trial in development proposes to compare the EsPhALL backbone to a less intensive chemotherapy regimen, with identical TKI therapy in the two regimens.
Duration of TKI Inhibition
Another open question is the duration of TKI inhibition. Several current protocols in adult patients propose indefinite tyrosine kinase inhibition after completion of maintenance ALL therapy (Table 1). In contrast, the current pediatric Ph+ ALL trials typically stop TKI therapy when chemotherapy stops at 2–2.5 years post diagnosis. There have been some, but relatively few late events in COG AALL0031 suggesting that this approach is feasible and not associated with excessive relapses. However, it remains uncertain if additional TKI therapy beyond 2–2.5 years is beneficial. In particular, there are concerns about growth and bone mineral density with prolonged TKI therapy in children (234–236). These effects may be compounded by the concomitant use of highly intensive chemotherapy and radiation, both of which have been shown to independently affect longitudinal growth and bone health (237, 238).
The Role of Hematopoietic Stem-Cell Transplant
In AALL0031, HSCT did not improve outcomes for patients treated with imatinib in the final cohort. In contrast, outcomes for patients who did not undergo HSCT on EsPhALL appeared inferior to the transplanted cohort (relapse was reported in three of nine good-risk patients and five of seven poor-risk patients). It is possible that the longer duration of imatinib on AALL0031 (616 versus 126 days on EsPhALL) plays a role. On both trials, the numbers were extremely small, and more data will be required to define the role of HSCT in Ph+ ALL. The current combined follow-up study of AALL0031/AALL0622 and EsPhALL, AALL1122 (NCT01460160), is assessing the effect of earlier, continuous, and longer exposure to dasatinib. Rather than pursuing HSCT in all patients, only patients who fail to meet predefined MRD criteria and have a suitable donor are assigned to undergo HSCT in first remission, which is the approach generally pursued in other pediatric ALL subsets. In adult patients, initial studies were unable to address the question of whether HSCT confers a survival benefit due to small sample sizes, but durable remissions have been observed with the combination of TKI and intensive chemotherapy regimens (157, 159, 162). HSCT in first remission (if possible based on donor availability and clinical status of the recipient) remains the standard of care in younger adults with Ph+ ALL, but outstanding results observed with chemotherapy plus TKI are challenging this consensus. Despite controversy over the role of HSCT in first remission in pediatric Ph+ ALL, there is universal consensus that HSCT in second complete remission is the preferred therapy after relapse.
The Role of Post-Transplant TKI
Another open question centers on whether and for how long TKIs could or should be continued after HSCT. Several studies suggest that post-transplant TKI are beneficial and reduce the rate of relapse (239–242). However, the duration of post-transplant TKI therapy varies widely between studies, and high rates of dose reduction and cessation due to cytopenias and other side effects have been reported. Very few prospective randomized studies are available. A recent GMALL study randomly assigned Ph+ ALL patient to receive imatinib post-HSCT either prophylactically, or triggered by a rise in MRD. Molecular recurrence after HSCT was significantly lower in the prophylactic group (40 versus 69%; P = 0.046), although this did not translate into a difference in survival (243). In a recently reported non-randomized trial of 92 patients with Ph+ ALL, the ability to tolerate post-HSCT imatinib was an independent predictor of DFS and OS (244). This study involved pediatric patients, who had a higher rate of tolerating post-transplant imatinib than adult patients. In the current pediatric AALL1122, NCT01460160 post-transplant imatinib for up to 12 additional months is optional and at the discretion of the treating investigator.
Conclusion
Ph+ ALL is a poster child for the successful integration of targeted small-molecule inhibitors with standard chemotherapy. TKIs have revolutionized the outcomes of this disease. However, survival rates are still inferior to most other types of childhood ALL. Future efforts to improve survival and decrease toxicity need to focus on refining chemotherapy regimens, and optimizing TKI therapy. In addition, increasing the mechanistic understanding of this complex and fascinating disease will facilitate translating new findings into improved targeted therapies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Stephen P. Hunger is the Ergen Family Chair in Pediatric Cancer. This manuscript is dedicated to Janet Rowley, who revolutionized understanding of cancer genetics.
Abbreviations
ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; DFS, disease-free survival; EFS, event-free survival; HSCT, hematopoietic stem-cell transplantation; OS, overall survival; Ph+, Philadelphia chromosome positive; TKD, tyrosine kinase domain; TKI, tyrosine kinase inhibitor.
References
1. Mahon FX. Is going for cure in chronic myeloid leukemia possible and justifiable? Hematology Am Soc Hematol Educ Program (2012) 2012:122–8. doi: 10.1182/asheducation-2012.1.122
2. Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol (2009) 27(31):5175–81. doi:10.1200/JCO.2008.21.2514
3. Ilaria RL Jr. Pathobiology of lymphoid and myeloid blast crisis and management issues. Hematology Am Soc Hematol Educ Program (2005). 188–94. doi:10.1182/asheducation-2005.1.188
4. Arico M, Schrappe M, Hunger SP, Carroll WL, Conter V, Galimberti S, et al. Clinical outcome of children with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia treated between 1995 and 2005. J Clin Oncol (2010) 28(31):4755–61. doi:10.1200/JCO.2010.30.1325
5. Mrozek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematol Oncol Clin North Am (2009) 23(5):991–1010. doi:10.1016/j.hoc.2009.07.001
6. Larson RA. Management of acute lymphoblastic leukemia in older patients. Semin Hematol (2006) 43(2):126–33. doi:10.1053/j.seminhematol.2006.01.007
7. Kelliher M, Knott A, McLaughlin J, Witte ON, Rosenberg N. Differences in oncogenic potency but not target cell specificity distinguish the two forms of the BCR/ABL oncogene. Mol Cell Biol (1991) 11(9):4710–6.
8. Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science (1990) 247(4944):824–30. doi:10.1126/science.2406902
9. Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, Groffen J. Acute leukaemia in bcr/abl transgenic mice. Nature (1990) 344(6263):251–3. doi:10.1038/344251a0
10. Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science (1990) 247(4946):1079–82. doi:10.1126/science.2408149
11. Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med (1996) 2(5):561–6. doi:10.1038/nm0596-561
12. Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med (2001) 344(14):1031–7. doi:10.1056/NEJM200104053441402
13. Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med (2001) 344(14):1038–42. doi:10.1056/NEJM200104053441402
14. O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med (2003) 348(11):994–1004. doi:10.1056/NEJMoa022457
15. Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med (2010) 362(24):2260–70. doi:10.1056/NEJMoa1002315
16. Saglio G, Kim DW, Issaragrisil S, le Coutre P, Etienne G, Lobo C, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med (2010) 362(24):2251–9. doi:10.1056/NEJMoa0912614
17. Puttini M, Coluccia AM, Boschelli F, Cleris L, Marchesi E, Donella-Deana A, et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res (2006) 66(23):11314–22. doi:10.1158/0008-5472.CAN-06-1199
18. Cortes JE, Kim DW, Pinilla-Ibarz J, le Coutre P, Paquette R, Chuah C, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med (2013) 369(19):1783–96. doi:10.1056/NEJMoa1306494
19. Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst (1960) 25:85–109.
20. Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature (1973) 243(5405):290–3. doi:10.1038/243290a0
21. de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature (1982) 300(5894):765–7. doi:10.1038/300765a0
22. Li S, Ilaria RL Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med (1999) 189(9):1399–412. doi:10.1084/jem.189.9.1399
23. McLaughlin J, Chianese E, Witte ON. Alternative forms of the BCR-ABL oncogene have quantitatively different potencies for stimulation of immature lymphoid cells. Mol Cell Biol (1989) 9(5):1866–74.
24. Suryanarayan K, Hunger SP, Kohler S, Carroll AJ, Crist W, Link MP, et al. Consistent involvement of the bcr gene by 9;22 breakpoints in pediatric acute leukemias. Blood (1991) 77(2):324–30.
25. Franz WM, Berger P, Wang JY. Deletion of an N-terminal regulatory domain of the c-abl tyrosine kinase activates its oncogenic potential. EMBO J (1989) 8(1):137–47.
26. Hantschel O. Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes Cancer (2012) 3(5–6):436–46. doi:10.1177/1947601912458584
27. McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol (1993) 13(12):7587–95.
28. He Y, Wertheim JA, Xu L, Miller JP, Karnell FG, Choi JK, et al. The coiled-coil domain and Tyr177 of bcr are required to induce a murine chronic myelogenous leukemia-like disease by bcr/abl. Blood (2002) 99(8):2957–68. doi:10.1182/blood.V99.8.2957
29. Million RP, Van Etten RA. The Grb2 binding site is required for the induction of chronic myeloid leukemia-like disease in mice by the Bcr/Abl tyrosine kinase. Blood (2000) 96(2):664–70.
30. Maru Y, Witte ON. The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell (1991) 67(3):459–68. doi:10.1016/0092-8674(91)90521-Y
31. Muller AJ, Young JC, Pendergast AM, Pondel M, Landau NR, Littman DR, et al. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol Cell Biol (1991) 11(4):1785–92.
32. Pendergast AM, Muller AJ, Havlik MH, Maru Y, Witte ON. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell (1991) 66(1):161–71. doi:10.1016/0092-8674(91)90148-R
33. Beissert T, Hundertmark A, Kaburova V, Travaglini L, Mian AA, Nervi C, et al. Targeting of the N-terminal coiled coil oligomerization interface by a helix-2 peptide inhibits unmutated and imatinib-resistant BCR/ABL. Int J Cancer (2008) 122(12):2744–52. doi:10.1002/ijc.23467
34. Mian AA, Oancea C, Zhao Z, Ottmann OG, Ruthardt M. Oligomerization inhibition, combined with allosteric inhibition, abrogates the transformation potential of T315I-positive BCR/ABL. Leukemia (2009) 23(12):2242–7. doi:10.1038/leu.2009.194
35. Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature (2010) 463(7280):501–6. doi:10.1038/nature08675
36. Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol (2006) 2(2):95–102. doi:10.1038/nchembio760
37. Sattler M, Griffin JD. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin Hematol (2003) 40(2 Suppl 2):4–10. doi:10.1053/shem.2003.50034
38. Chang BH, Willis SG, Stork L, Hunger SP, Carroll WL, Camitta BM, et al. Imatinib resistant BCR-ABL1 mutations at relapse in children with Ph+ ALL: a Children’s Oncology Group (COG) study. Br J Haematol (2012) 157(4):507–10. doi:10.1111/j.1365-2141.2012.09039.x
39. Chu S, Li L, Singh H, Bhatia R. BCR-tyrosine 177 plays an essential role in Ras and Akt activation and in human hematopoietic progenitor transformation in chronic myelogenous leukemia. Cancer Res (2007) 67(14):7045–53. doi:10.1158/0008-5472.CAN-06-4312
40. Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood (2004) 103(8):3167–74. doi:10.1182/blood-2003-04-1271
41. Kharas MG, Janes MR, Scarfone VM, Lilly MB, Knight ZA, Shokat KM, et al. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J Clin Invest (2008) 118(9):3038–50. doi:10.1172/JCI33337
42. Pfeifer H, Lange T, Wystub S, Wassmann B, Maier J, Binckebanck A, et al. Prevalence and dynamics of bcr-abl kinase domain mutations during imatinib treatment differ in patients with newly diagnosed and recurrent bcr-abl positive acute lymphoblastic leukemia. Leukemia (2012) 26(7):1475–81. doi:10.1038/leu.2012.5
43. Redig AJ, Vakana E, Platanias LC. Regulation of mammalian target of rapamycin and mitogen activated protein kinase pathways by BCR-ABL. Leuk Lymphoma (2011) 52(Suppl 1):45–53. doi:10.3109/10428194.2010.546919
44. Goga A, McLaughlin J, Afar DE, Saffran DC, Witte ON. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell (1995) 82(6):981–8. doi:10.1016/0092-8674(95)90277-5
45. Modi H, Li L, Chu S, Rossi J, Yee JK, Bhatia R. Inhibition of Grb2 expression demonstrates an important role in BCR-ABL-mediated MAPK activation and transformation of primary human hematopoietic cells. Leukemia (2011) 25(2):305–12. doi:10.1038/leu.2010.257
46. Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood (2006) 107(12):4898–906. doi:10.1182/blood-2005-09-3596
47. Hu Y, Liu Y, Pelletier S, Buchdunger E, Warmuth M, Fabbro D, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet (2004) 36(5):453–61. doi:10.1038/ng1343
48. Frank DA, Varticovski L. BCR/abl leads to the constitutive activation of Stat proteins, and shares an epitope with tyrosine phosphorylated Stats. Leukemia (1996) 10(11):1724–30.
49. Carlesso N, Frank DA, Griffin JD. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med (1996) 183(3):811–20. doi:10.1084/jem.183.3.811
50. Ilaria RL Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem (1996) 271(49):31704–10. doi:10.1074/jbc.271.49.31704
51. Tao WJ, Lin H, Sun T, Samanta AK, Arlinghaus R. BCR-ABL oncogenic transformation of NIH 3T3 fibroblasts requires the IL-3 receptor. Oncogene (2008) 27(22):3194–200. doi:10.1038/sj.onc.1210979
52. Samanta A, Perazzona B, Chakraborty S, Sun X, Modi H, Bhatia R, et al. Janus kinase 2 regulates Bcr-Abl signaling in chronic myeloid leukemia. Leukemia (2011) 25(3):463–72. doi:10.1038/leu.2010.287
53. O’Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res (2005) 65(11):4500–5. doi:10.1158/0008-5472.CAN-05-0259
54. Yang X, He G, Gong Y, Zheng B, Shi F, Shi R, et al. mTOR inhibitor rapamycin enhances anti-leukemia effect of imatinib on Ph+ acute lymphoblastic leukemia cells. Eur J Haematol (2014) 92(2):111–20. doi:10.1111/ejh.12202
55. Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell (2013) 24(6):766–76. doi:10.1016/j.ccr.2013.10.022
56. Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell (2006) 10(4):331–42. doi:10.1016/j.ccr.2006.09.006
57. Barrett D, Brown VI, Grupp SA, Teachey DT. Targeting the PI3K/AKT/mTOR signaling axis in children with hematologic malignancies. Paediatr Drugs (2012) 14(5):299–316. doi:10.2165/11594740-000000000-00000
58. Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, et al. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell (1998) 95(6):793–803. doi:10.1016/S0092-8674(00)81702-X
59. Kardinal C, Konkol B, Lin H, Eulitz M, Schmidt EK, Estrov Z, et al. Chronic myelogenous leukemia blast cell proliferation is inhibited by peptides that disrupt Grb2-SoS complexes. Blood (2001) 98(6):1773–81. doi:10.1182/blood.V98.6.1773
60. Lewitzky M, Kardinal C, Gehring NH, Schmidt EK, Konkol B, Eulitz M, et al. The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-x-x-P core motif. Oncogene (2001) 20(9):1052–62. doi:10.1038/sj.onc.1204202
61. Pendergast AM, Quilliam LA, Cripe LD, Bassing CH, Dai Z, Li N, et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell (1993) 75(1):175–85. doi:10.1016/0092-8674(93)90689-N
62. Sattler M, Mohi MG, Pride YB, Quinnan LR, Malouf NA, Podar K, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell (2002) 1(5):479–92. doi:10.1016/S1535-6108(02)00074-0
63. Irwin ME, Nelson LD, Santiago-O’Farrill JM, Knouse PD, Miller CP, Palla SL, et al. Small molecule ErbB inhibitors decrease proliferative signaling and promote apoptosis in Philadelphia chromosome-positive acute lymphoblastic leukemia. PLoS One (2013) 8(8):e70608. doi:10.1371/journal.pone.0070608
64. Warmuth M, Bergmann M, Priess A, Hauslmann K, Emmerich B, Hallek M. The Src family kinase Hck interacts with Bcr-Abl by a kinase-independent mechanism and phosphorylates the Grb2-binding site of Bcr. J Biol Chem (1997) 272(52):33260–70. doi:10.1074/jbc.272.52.33260
65. Klejman A, Schreiner SJ, Nieborowska-Skorska M, Slupianek A, Wilson M, Smithgall TE, et al. The Src family kinase Hck couples BCR/ABL to STAT5 activation in myeloid leukemia cells. EMBO J (2002) 21(21):5766–74. doi:10.1093/emboj/cdf562
66. Stanglmaier M, Warmuth M, Kleinlein I, Reis S, Hallek M. The interaction of the Bcr-Abl tyrosine kinase with the Src kinase Hck is mediated by multiple binding domains. Leukemia (2003) 17(2):283–9. doi:10.1038/sj.leu.2402778
67. Danhauser-Riedl S, Warmuth M, Druker BJ, Emmerich B, Hallek M. Activation of Src kinases p53/56lyn and p59hck by p210bcr/abl in myeloid cells. Cancer Res (1996) 56(15):3589–96.
68. Lionberger JM, Wilson MB, Smithgall TE. Transformation of myeloid leukemia cells to cytokine independence by Bcr-Abl is suppressed by kinase-defective Hck. J Biol Chem (2000) 275(24):18581–5. doi:10.1074/jbc.C000126200
69. Wilson MB, Schreiner SJ, Choi HJ, Kamens J, Smithgall TE. Selective pyrrolo-pyrimidine inhibitors reveal a necessary role for Src family kinases in Bcr-Abl signal transduction and oncogenesis. Oncogene (2002) 21(53):8075–88. doi:10.1038/sj.onc.1206008
70. Warmuth M, Simon N, Mitina O, Mathes R, Fabbro D, Manley PW, et al. Dual-specific Src and Abl kinase inhibitors, PP1 and CGP76030, inhibit growth and survival of cells expressing imatinib mesylate-resistant Bcr-Abl kinases. Blood (2003) 101(2):664–72. doi:10.1182/blood-2002-01-0288
71. Pene-Dumitrescu T, Peterson LF, Donato NJ, Smithgall TE. An inhibitor-resistant mutant of Hck protects CML cells against the antiproliferative and apoptotic effects of the broad-spectrum Src family kinase inhibitor A-419259. Oncogene (2008) 27(56):7055–69. doi:10.1038/onc.2008.330
72. Wu J, Meng F, Lu H, Kong L, Bornmann W, Peng Z, et al. Lyn regulates BCR-ABL and Gab2 tyrosine phosphorylation and c-Cbl protein stability in imatinib-resistant chronic myelogenous leukemia cells. Blood (2008) 111(7):3821–9. doi:10.1182/blood-2007-08-109330
73. Pene-Dumitrescu T, Smithgall TE. Expression of a Src family kinase in chronic myelogenous leukemia cells induces resistance to imatinib in a kinase-dependent manner. J Biol Chem (2010) 285(28):21446–57. doi:10.1074/jbc.M109.090043
74. Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci U S A (2006) 103(45):16870–5. doi:10.1073/pnas.0606509103
75. Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell (2012) 22(2):153–66. doi:10.1016/j.ccr.2012.06.005
76. Loh ML, Zhang J, Harvey RC, Roberts K, Payne-Turner D, Kang H, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood (2013) 121(3):485–8. doi:10.1182/blood-2012-04-422691
77. Weston BW, Hayden MA, Roberts KG, Bowyer S, Hsu J, Fedoriw G, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol (2013) 31(25):e413–6. doi:10.1200/JCO.2012.47.6770
78. Lugo TG, Witte ON. The BCR-ABL oncogene transforms Rat-1 cells and cooperates with v-myc. Mol Cell Biol (1989) 9(3):1263–70.
79. Voncken JW, Kaartinen V, Pattengale PK, Germeraad WT, Groffen J, Heisterkamp N. BCR/ABL P210 and P190 cause distinct leukemia in transgenic mice. Blood (1995) 86(12):4603–11.
80. Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature (2011) 469(7330):362–7. doi:10.1038/nature09733
81. Pastorczak A, Gorniak P, Sherborne A, Hosking F, Trelinska J, Lejman M, et al. Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the Polish population. Leuk Res (2011) 35(11):1534–6. doi:10.1016/j.leukres.2011.07.034
82. Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood (2010) 115(9):1765–7. doi:10.1182/blood-2009-09-241513
83. Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet (2009) 41(9):1006–10. doi:10.1038/ng.430
84. Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet (2009) 41(9):1001–5. doi:10.1038/ng.432
85. Linabery AM, Blommer CN, Spector LG, Davies SM, Robison LL, Ross JA. ARID5B and IKZF1 variants, selected demographic factors, and childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Leuk Res (2013) 37(8):936–42. doi:10.1016/j.leukres.2013.04.022
86. Kastner P, Dupuis A, Gaub MP, Herbrecht R, Lutz P, Chan S. Function of Ikaros as a tumor suppressor in B cell acute lymphoblastic leukemia. Am J Blood Res (2013) 3(1):1–13.
87. Lin CY, Li MJ, Chang JG, Liu SC, Weng T, Wu KH, et al. High-resolution melting analyses for genetic variants in ARID5B and IKZF1 with childhood acute lymphoblastic leukemia susceptibility loci in Taiwan. Blood Cells Mol Dis (2014) 52(2–3):140–5. doi:10.1016/j.bcmd.2013.10.003
88. Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet (2013) 45(10):1226–31. doi:10.1038/ng.2754
89. Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet (2010) 42(6):492–4. doi:10.1038/ng.585
90. Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst (2013) 105(10):733–42. doi:10.1093/jnci/djt042
91. Biondi A, Schrappe M, De Lorenzo P, Castor A, Lucchini G, Gandemer V, et al. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol (2012) 13(9):936–45. doi:10.1016/S1470-2045(12)70377-7
92. Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature (2008) 453(7191):110–4. doi:10.1038/nature06866
93. Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood (2009) 114(10):2159–67. doi:10.1182/blood-2008-08-173963
94. Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol (2009) 27(31):5202–7. doi:10.1200/JCO.2008.21.6408
95. Dupuis A, Gaub MP, Legrain M, Drenou B, Mauvieux L, Lutz P, et al. Biclonal and biallelic deletions occur in 20% of B-ALL cases with IKZF1 mutations. Leukemia (2013) 27(2):503–7. doi:10.1038/leu.2012.204
96. Tonnelle C, Imbert MC, Sainty D, Granjeaud S, N’Guyen C, Chabannon C. Overexpression of dominant-negative Ikaros 6 protein is restricted to a subset of B common adult acute lymphoblastic leukemias that express high levels of the CD34 antigen. Hematol J (2003) 4(2):104–9. doi:10.1038/sj.thj.6200235
97. Iacobucci I, Lonetti A, Messa F, Cilloni D, Arruga F, Ottaviani E, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood (2008) 112(9):3847–55. doi:10.1182/blood-2007-09-112631
98. Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell (1994) 79(1):143–56. doi:10.1016/0092-8674(94)90407-3
99. Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med (2009) 360(5):470–80. doi:10.1056/NEJMoa0808253
100. Cazzaniga G, van Delft FW, Lo Nigro L, Ford AM, Score J, Iacobucci I, et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood (2011) 118(20):5559–64. doi:10.1182/blood-2011-07-366542
101. Caye A, Beldjord K, Mass-Malo K, Drunat S, Soulier J, Gandemer V, et al. Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica (2013) 98(4):597–601. doi:10.3324/haematol.2012.073965
102. Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity (1996) 5(6):537–49. doi:10.1016/S1074-7613(00)80269-1
103. Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med (1999) 190(9):1201–14. doi:10.1084/jem.190.9.1201
104. Lopez RA, Schoetz S, DeAngelis K, O’Neill D, Bank A. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc Natl Acad Sci U S A (2002) 99(2):602–7. doi:10.1073/pnas.022412699
105. Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity (2009) 30(4):493–507. doi:10.1016/j.immuni.2009.01.014
106. Malin S, McManus S, Busslinger M. STAT5 in B cell development and leukemia. Curr Opin Immunol (2010) 22(2):168–76. doi:10.1016/j.coi.2010.02.004
107. Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol (2010) 22(2):177–84. doi:10.1016/j.coi.2010.02.003
108. Boggs SS, Trevisan M, Patrene K, Geogopoulos K. Lack of natural killer cell precursors in fetal liver of Ikaros knockout mutant mice. Nat Immun (1998) 16(4):137–45. doi:10.1159/000069438
109. Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell (1995) 83(2):289–99. doi:10.1016/0092-8674(95)90170-1
110. Kastner P, Chan S. Role of Ikaros in T-cell acute lymphoblastic leukemia. World J Biol Chem (2011) 2(6):108–14. doi:10.4331/wjbc.v2.i6.108
111. Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet (2002) 3:179–98. doi:10.1146/annurev.genom.3.032802.115046
112. Ozvegy-Laczka C, Hegedus T, Varady G, Ujhelly O, Schuetz JD, Varadi A, et al. High-affinity interaction of tyrosine kinase inhibitors with the ABCG2 multidrug transporter. Mol Pharmacol (2004) 65(6):1485–95. doi:10.1124/mol.65.6.1485
113. Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol (2002) 32(3):720–30. doi:10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P
114. Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol (2010) 30(17):4149–58. doi:10.1128/MCB.00224-10
115. Iacobucci I, Iraci N, Messina M, Lonetti A, Chiaretti S, Valli E, et al. IKAROS deletions dictate a unique gene expression signature in patients with adult B-cell acute lymphoblastic leukemia. PLoS One (2012) 7(7):e40934. doi:10.1371/journal.pone.0040934
116. Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity (2008) 29(2):217–27. doi:10.1016/j.immuni.2008.05.015
117. Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood (2012) 119(15):3512–22. doi:10.1182/blood-2011-11-394221
118. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature (2007) 446(7137):758–64. doi:10.1038/nature05690
119. Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood (2011) 118(11):3080–7. doi:10.1182/blood-2011-03-341412
120. Iacobucci I, Lonetti A, Paoloni F, Papayannidis C, Ferrari A, Storlazzi CT, et al. The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the GIMEMA Acute Leukemia Working Party. Haematologica (2010) 95(10):1683–90. doi:10.3324/haematol.2009.020792
121. Familiades J, Bousquet M, Lafage-Pochitaloff M, Bene MC, Beldjord K, De Vos J, et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia (2009) 23(11):1989–98. doi:10.1038/leu.2009.135
122. Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol (2007) 8(5):463–70. doi:10.1038/ni1454
123. Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell (1994) 79(5):901–12. doi:10.1016/0092-8674(94)90079-5
124. Nutt SL, Thevenin C, Busslinger M. Essential functions of Pax-5 (BSAP) in pro-B cell development. Immunobiology (1997) 198(1–3):227–35. doi:10.1016/S0171-2985(97)80043-5
125. Schaniel C, Gottar M, Roosnek E, Melchers F, Rolink AG. Extensive in vivo self-renewal, long-term reconstitution capacity, and hematopoietic multipotency of Pax5-deficient precursor B-cell clones. Blood (2002) 99(8):2760–6. doi:10.1182/blood.V99.2.472
126. Schaniel C, Bruno L, Melchers F, Rolink AG. Multiple hematopoietic cell lineages develop in vivo from transplanted Pax5-deficient pre-B I-cell clones. Blood (2002) 99(2):472–8. doi:10.1182/blood.V99.2.472
127. Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature (2007) 449(7161):473–7. doi:10.1038/nature06159
128. Georgopoulos K. Acute lymphoblastic leukemia – on the wings of IKAROS. N Engl J Med (2009) 360(5):524–6. doi:10.1056/NEJMe0809819
129. Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature (1995) 376(6537):263–7. doi:10.1038/376263a0
130. Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med (2011) 208(6):1135–49. doi:10.1084/jem.20101947
131. Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity (2004) 21(6):843–51. doi:10.1016/j.immuni.2004.11.004
132. Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature (1999) 397(6715):164–8. doi:10.1038/16476
133. Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev (2003) 13(1):77–83. doi:10.1016/S0959-437X(02)00013-8
134. Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature (2003) 423(6937):302–5. doi:10.1038/nature01587
135. Maloney KW, McGavran L, Odom LF, Hunger SP. Different patterns of homozygous p16INK4A and p15INK4B deletions in childhood acute lymphoblastic leukemias containing distinct E2A translocations. Leukemia (1998) 12(9):1417–21. doi:10.1038/sj.leu.2401124
136. Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica (2013) 98(7):1081–8. doi:10.3324/haematol.2013.085175
137. Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev (2008) 22(11):1411–5. doi:10.1101/gad.1673908
138. Kim M, Yim SH, Cho NS, Kang SH, Ko DH, Oh B, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet (2009) 195(1):59–65. doi:10.1016/j.cancergencyto.2009.06.013
139. Sill H, Goldman JM, Cross NC. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood (1995) 85(8):2013–6.
140. Kohno T, Yokota J. Molecular processes of chromosome 9p21 deletions causing inactivation of the p16 tumor suppressor gene in human cancer: deduction from structural analysis of breakpoints for deletions. DNA Repair (2006) 5(9–10):1273–81. doi:10.1016/j.dnarep.2006.05.021
141. Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A (2006) 103(17):6688–93. doi:10.1073/pnas.0602030103
142. Gutierrez MI, Siraj AK, Bhargava M, Ozbek U, Banavali S, Chaudhary MA, et al. Concurrent methylation of multiple genes in childhood ALL: correlation with phenotype and molecular subgroup. Leukemia (2003) 17(9):1845–50. doi:10.1038/sj.leu.2403060
143. Nakamura M, Sugita K, Inukai T, Goi K, Iijima K, Tezuka T, et al. p16/MTS1/INK4A gene is frequently inactivated by hypermethylation in childhood acute lymphoblastic leukemia with 11q23 translocation. Leukemia (1999) 13(6):884–90. doi:10.1038/sj.leu.2401437
144. Primo D, Tabernero MD, Perez JJ, Rasillo A, Sayagues JM, Espinosa AB, et al. Genetic heterogeneity of BCR/ABL+ adult B-cell precursor acute lymphoblastic leukemia: impact on the clinical, biological and immunophenotypical disease characteristics. Leukemia (2005) 19(5):713–20. doi:10.1038/sj.leu.2403714
145. Carter TL, Watt PM, Kumar R, Burton PR, Reaman GH, Sather HN, et al. Hemizygous p16(INK4A) deletion in pediatric acute lymphoblastic leukemia predicts independent risk of relapse. Blood (2001) 97(2):572–4. doi:10.1182/blood.V97.2.572
146. Garcia-Manero G, Daniel J, Smith TL, Kornblau SM, Lee MS, Kantarjian HM, et al. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clin Cancer Res (2002) 8(7):2217–24.
147. Maloney KW, McGavran L, Odom LF, Hunger SP. Acquisition of p16(INK4A) and p15(INK4B) gene abnormalities between initial diagnosis and relapse in children with acute lymphoblastic leukemia. Blood (1999) 93(7):2380–5.
148. Bhatla T, Wang J, Morrison DJ, Raetz EA, Burke MJ, Brown P, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood (2012) 119(22):5201–10. doi:10.1182/blood-2012-01-401687
149. Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S, et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res (2011) 17(23):7413–23. doi:10.1158/1078-0432.CCR-11-1227
150. Krentz S, Hof J, Mendioroz A, Vaggopoulou R, Dorge P, Lottaz C, et al. Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia (2013) 27(2):295–304. doi:10.1038/leu.2012.155
151. Figueroa ME, Chen SC, Andersson AK, Phillips LA, Li Y, Sotzen J, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest (2013) 123(7):3099–111. doi:10.1172/JCI66203
152. Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic Mice. Cancer Res (2002) 62(5):1296–9.
153. Trinh BN, Long TI, Nickel AE, Shibata D, Laird PW. DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair. Mol Cell Biol (2002) 22(9):2906–17. doi:10.1128/MCB.22.9.2906-2917.2002
154. Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev (2012) 26(4):344–9. doi:10.1101/gad.184341.111
155. Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet (2009) 41(11):1207–15. doi:10.1038/ng.463
156. Vignetti M, Fazi P, Cimino G, Martinelli G, Di Raimondo F, Ferrara F, et al. Imatinib plus steroids induces complete remissions and prolonged survival in elderly Philadelphia chromosome-positive patients with acute lymphoblastic leukemia without additional chemotherapy: results of the Gruppo Italiano Malattie Ematologiche dell’Adulto (GIMEMA) LAL0201-B protocol. Blood (2007) 109(9):3676–8. doi:10.1182/blood-2006-10-052746
157. Thomas DA, Faderl S, Cortes J, O’Brien S, Giles FJ, Kornblau SM, et al. Treatment of Philadelphia chromosome-positive acute lymphocytic leukemia with hyper-CVAD and imatinib mesylate. Blood (2004) 103(12):4396–407. doi:10.1182/blood-2003-08-2958
158. Lee KH, Lee JH, Choi SJ, Lee JH, Seol M, Lee YS, et al. Clinical effect of imatinib added to intensive combination chemotherapy for newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia (2005) 19(9):1509–16. doi:10.1038/sj.leu.2403886
159. Yanada M, Takeuchi J, Sugiura I, Akiyama H, Usui N, Yagasaki F, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J Clin Oncol (2006) 24(3):460–6. doi:10.1200/JCO.2005.03.2177
160. Wassmann B, Pfeifer H, Goekbuget N, Beelen DW, Beck J, Stelljes M, et al. Alternating versus concurrent schedules of imatinib and chemotherapy as front-line therapy for Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL). Blood (2006) 108(5):1469–77. doi:10.1182/blood-2005-11-4386
161. Druker BJ. Imatinib: a viewpoint by Brian J. Druker. Drugs (2001) 61(12):1775–6. doi:10.2165/00003495-200161120-00009
162. de Labarthe A, Rousselot P, Huguet-Rigal F, Delabesse E, Witz F, Maury S, et al. Imatinib combined with induction or consolidation chemotherapy in patients with de novo Philadelphia chromosome-positive acute lymphoblastic leukemia: results of the GRAAPH-2003 study. Blood (2007) 109(4):1408–13. doi:10.1182/blood-2006-03-011908
163. Delannoy A, Delabesse E, Lheritier V, Castaigne S, Rigal-Huguet F, Raffoux E, et al. Imatinib and methylprednisolone alternated with chemotherapy improve the outcome of elderly patients with Philadelphia-positive acute lymphoblastic leukemia: results of the GRAALL AFR09 study. Leukemia (2006) 20(9):1526–32. doi:10.1038/sj.leu.2404320
164. Wassmann B, Pfeifer H, Scheuring UJ, Binckebanck A, Gokbuget N, Atta J, et al. Early prediction of response in patients with relapsed or refractory Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ALL) treated with imatinib. Blood (2004) 103(4):1495–8. doi:10.1182/blood-2003-01-0154
165. Jones D, Thomas D, Yin CC, O’Brien S, Cortes JE, Jabbour E, et al. Kinase domain point mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia emerge after therapy with BCR-ABL kinase inhibitors. Cancer (2008) 113(5):985–94. doi:10.1002/cncr.23666
166. Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood (2002) 99(9):3472–5. doi:10.1182/blood.V99.9.3472
167. Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res (2006) 12(24):7374–9. doi:10.1158/1078-0432.CCR-06-1516
168. Jones D, Chen SS, Jabbour E, Rios MB, Kantarjian H, Cortes J. Uncommon BCR-ABL kinase domain mutations in kinase inhibitor-resistant chronic myelogenous leukemia and Ph+ acute lymphoblastic leukemia show high rates of regression, suggesting weak selective effects. Blood (2010) 115(26):5428–9. doi:10.1182/blood-2009-11-252155
169. Pfeifer H, Wassmann B, Pavlova A, Wunderle L, Oldenburg J, Binckebanck A, et al. Kinase domain mutations of BCR-ABL frequently precede imatinib-based therapy and give rise to relapse in patients with de novo Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL). Blood (2007) 110(2):727–34. doi:10.1182/blood-2006-11-052373
170. Hofmann WK, Komor M, Wassmann B, Jones LC, Gschaidmeier H, Hoelzer D, et al. Presence of the BCR-ABL mutation Glu255Lys prior to STI571 (imatinib) treatment in patients with Ph+ acute lymphoblastic leukemia. Blood (2003) 102(2):659–61. doi:10.1182/blood-2002-06-1756
171. Soverini S, Vitale A, Poerio A, Gnani A, Colarossi S, Iacobucci I, et al. Philadelphia-positive acute lymphoblastic leukemia patients already harbor BCR-ABL kinase domain mutations at low levels at the time of diagnosis. Haematologica (2011) 96(4):552–7. doi:10.3324/haematol.2010.034173
172. Foa R, Vitale A, Vignetti M, Meloni G, Guarini A, De Propris MS, et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood (2011) 118(25):6521–8. doi:10.1182/blood-2011-05-351403
173. La Starza R, Vitale A, Serra A, Saglio G, Fioritoni G, Falzetti D, et al. Philadelphia-positive acute lymphoblastic leukemia with multiple subclones including duplication of the Philadelphia chromosome and Abelson oncogene. Cancer Genet Cytogenet (2002) 132(1):46–50. doi:10.1016/S0165-4608(01)00507-6
174. Ko BS, Tang JL, Lee FY, Liu MC, Tsai W, Chen YC, et al. Additional chromosomal abnormalities and variability of BCR breakpoints in Philadelphia chromosome/BCR-ABL-positive acute lymphoblastic leukemia in Taiwan. Am J Hematol (2002) 71(4):291–9. doi:10.1002/ajh.10227
175. Jones D, Luthra R, Cortes J, Thomas D, O’Brien S, Bueso-Ramos C, et al. BCR-ABL fusion transcript types and levels and their interaction with secondary genetic changes in determining the phenotype of Philadelphia chromosome-positive leukemias. Blood (2008) 112(13):5190–2. doi:10.1182/blood-2008-04-148791
176. Burger H, van Tol H, Brok M, Wiemer EA, de Bruijn EA, Guetens G, et al. Chronic imatinib mesylate exposure leads to reduced intracellular drug accumulation by induction of the ABCG2 (BCRP) and ABCB1 (MDR1) drug transport pumps. Cancer Biol Ther (2005) 4(7):747–52. doi:10.4161/cbt.4.7.1826
177. Illmer T, Schaich M, Platzbecker U, Freiberg-Richter J, Oelschlagel U, von Bonin M, et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia (2004) 18(3):401–8. doi:10.1038/sj.leu.2403257
178. Burger H, van Tol H, Boersma AW, Brok M, Wiemer EA, Stoter G, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood (2004) 104(9):2940–2. doi:10.1182/blood-2004-04-1398
179. Ramakers-van Woerden NL, Pieters R, Hoelzer D, Slater RM, den Boer ML, Loonen AH, et al. In vitro drug resistance profile of Philadelphia positive acute lymphoblastic leukemia is heterogeneous and related to age: a report of the Dutch and German Leukemia Study Groups. Med Pediatr Oncol (2002) 38(6):379–86. doi:10.1002/mpo.10087
180. Mahon FX, Hayette S, Lagarde V, Belloc F, Turcq B, Nicolini F, et al. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res (2008) 68(23):9809–16. doi:10.1158/0008-5472.CAN-08-1008
181. Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG, et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst (2008) 100(13):926–39. doi:10.1093/jnci/djn188
182. Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood (2003) 101(2):690–8. doi:10.1182/blood.V101.2.690
183. Samanta AK, Chakraborty SN, Wang Y, Schlette E, Reddy EP, Arlinghaus RB. Destabilization of Bcr-Abl/Jak2 network by a Jak2/Abl kinase inhibitor ON044580 overcomes drug resistance in blast crisis chronic myelogenous leukemia (CML). Genes Cancer (2010) 1(4):346–59. doi:10.1177/1947601910372232
184. Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood (2012) 120(17):3510–8. doi:10.1182/blood-2012-03-415448
185. Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell (2007) 12(3):280–92. doi:10.1016/j.ccr.2007.08.011
186. Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene (2007) 26(2):224–33. doi:10.1038/sj.onc.1209775
187. Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol (2004) 24(22):10058–71. doi:10.1128/MCB.24.22.10058-10071.2004
188. Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature (2011) 473(7347):384–8. doi:10.1038/nature09883
189. Hurtz C, Hatzi K, Cerchietti L, Braig M, Park E, Kim YM, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med (2011) 208(11):2163–74. doi:10.1084/jem.20110304
190. Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, et al. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood (2011) 118(15):4174–8. doi:10.1182/blood-2011-01-331181
191. Chevallier P, Robillard N, Wuilleme-Toumi S, Mechinaud F, Harousseau JL, Avet-Loiseau H. Overexpression of Her2/neu is observed in one third of adult acute lymphoblastic leukemia patients and is associated with chemoresistance in these patients. Haematologica (2004) 89(11):1399–401.
192. Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia (2010) 24(7):1258–64. doi:10.1038/leu.2010.87
193. Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science (2008) 322(5906):1377–80. doi:10.1126/science.1164266
194. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med (2011) 17(9):1086–93. doi:10.1038/nm.2415
195. Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA (2010) 304(24):2706–15. doi:10.1001/jama.2010.1862
196. Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med (2004) 350(16):1617–28. doi:10.1056/NEJMoa040465
197. Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJ, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia (2013) 27(4):852–60. doi:10.1038/leu.2012.363
198. Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol (2008) 9(7):810–9. doi:10.1038/ni.1617
199. Lanza C, Gaidano G, Cimino G, Lo Coco F, Basso G, Sainati L, et al. p53 gene inactivation in acute lymphoblastic leukemia of B cell lineage associates with chromosomal breakpoints at 11q23 and 8q24. Leukemia (1995) 9(6):955–9.
200. Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet (2013) 45(3):242–52. doi:10.1038/ng.2532
201. Hof J, Krentz S, van Schewick C, Korner G, Shalapour S, Rhein P, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol (2011) 29(23):3185–93. doi:10.1200/JCO.2011.34.8144
202. Yang JJ, Bhojwani D, Yang W, Cai X, Stocco G, Crews K, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood (2008) 112(10):4178–83. doi:10.1182/blood-2008-06-165027
203. Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood (2011) 118(19):5218–26. doi:10.1182/blood-2011-04-345595
204. Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW, et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood (2006) 108(2):711–7. doi:10.1182/blood-2006-02-002824
205. Rebucci M, Michiels C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem Pharmacol (2013) 85(9):1219–26. doi:10.1016/j.bcp.2013.02.017
206. Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol (2013) 14(6):e205–17. doi:10.1016/S1470-2045(12)70580-6
207. Diouf B, Cheng Q, Krynetskaia NF, Yang W, Cheok M, Pei D, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med (2011) 17(10):1298–303. doi:10.1038/nm.2430
208. Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood (2011) 117(9):2658–67. doi:10.1182/blood-2010-03-275347
209. Fleury I, Primeau M, Doreau A, Costea I, Moghrabi A, Sinnett D, et al. Polymorphisms in genes involved in the corticosteroid response and the outcome of childhood acute lymphoblastic leukemia. Am J Pharmacogenomics (2004) 4(5):331–41. doi:10.2165/00129785-200404050-00006
210. Bokemeyer A, Eckert C, Meyr F, Koerner G, Stackelberg AV, Ullmann R, et al. Copy number genome alterations are associated with treatment response and outcome in relapsed childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Haematologica (2013). doi:10.3324/haematol.2012.072470
211. Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature (2011) 471(7337):235–9. doi:10.1038/nature09727
212. Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood (2013) 121(24):4821–31. doi:10.1182/blood-2012-12-475483
213. Welschinger R, Liedtke F, Basnett J, Dela Pena A, Juarez JG, Bradstock KF, et al. Plerixafor (AMD3100) induces prolonged mobilization of acute lymphoblastic leukemia cells and increases the proportion of cycling cells in the blood in mice. Exp Hematol (2013) 41(3):293.e–302.e. doi:10.1016/j.exphem.2012.11.004
214. Yu M, Gang EJ, Parameswaran R, Stoddart S, Fei F, Schmidhuber S, et al. AMD3100 sensitizes acute lymphoblastic leukemia cells to chemotherapy in vivo. Blood Cancer J (2011) 1(4):e14. doi:10.1038/bcj.2011.13
215. Arico M, Valsecchi MG, Camitta B, Schrappe M, Chessells J, Baruchel A, et al. Outcome of treatment in children with Philadelphia chromosome-positive acute lymphoblastic leukemia. N Engl J Med (2000) 342(14):998–1006. doi:10.1056/NEJM200004063421402
216. Schultz KRCA, Heerema NA, Bowman WP, Aledo A, Slayton WB, Sather H, et al. Long term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group Study AALL0031. Leukemia (2014). doi:10.1038/leu.2014.30
217. Rives S, Estella J, Gomez P, Lopez-Duarte M, de Miguel PG, Verdeguer A, et al. Intermediate dose of imatinib in combination with chemotherapy followed by allogeneic stem cell transplantation improves early outcome in paediatric Philadelphia chromosome-positive acute lymphoblastic leukaemia (ALL): results of the Spanish Cooperative Group SHOP studies ALL-94, ALL-99 and ALL-2005. Br J Haematol (2011) 154(5):600–11. doi:10.1111/j.1365-2141.2011.08783.x
218. Porkka K, Koskenvesa P, Lundan T, Rimpilainen J, Mustjoki S, Smykla R, et al. Dasatinib crosses the blood-brain barrier and is an efficient therapy for central nervous system Philadelphia chromosome-positive leukemia. Blood (2008) 112(4):1005–12. doi:10.1182/blood-2008-02-140665
219. Gioia R, Leroy C, Drullion C, Lagarde V, Etienne G, Dulucq S, et al. Quantitative phosphoproteomics revealed interplay between Syk and Lyn in the resistance to nilotinib in chronic myeloid leukemia cells. Blood (2011) 118(8):2211–21. doi:10.1182/blood-2010-10-313692
220. Ottmann O, Dombret H, Martinelli G, Simonsson B, Guilhot F, Larson RA, et al. Dasatinib induces rapid hematologic and cytogenetic responses in adult patients with Philadelphia chromosome positive acute lymphoblastic leukemia with resistance or intolerance to imatinib: interim results of a phase 2 study. Blood (2007) 110(7):2309–15. doi:10.1182/blood-2007-02-073528
221. Lilly MB, Ottmann OG, Shah NP, Larson RA, Reiffers JJ, Ehninger G, et al. Dasatinib 140 mg once daily versus 70 mg twice daily in patients with Ph-positive acute lymphoblastic leukemia who failed imatinib: results from a phase 3 study. Am J Hematol (2010) 85(3):164–70. doi:10.1002/ajh.21615
222. Ravandi F, O’Brien S, Thomas D, Faderl S, Jones D, Garris R, et al. First report of phase 2 study of dasatinib with hyper-CVAD for the frontline treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia. Blood (2010) 116(12):2070–7. doi:10.1182/blood-2009-12-261586
223. Benjamini O, Dumlao TL, Kantarjian H, O’Brian S, Garcia-Manero G, Faderl S, et al. Phase II trial of HyperCVAD and Dasatinib in patients with relapsed Philadelphia chromosome positive acute lymphoblastic leukemia or blast phase chronic myeloid leukemia. Am J Hematol (2013) 89(3):282–7. doi:10.1002/ajh.23624
224. Zwaan CM, Rizzari C, Mechinaud F, Lancaster DL, Lehrnbecher T, van der Velden VH, et al. Dasatinib in children and adolescents with relapsed or refractory leukemia: results of the CA180-018 phase I dose-escalation study of the Innovative Therapies for Children with Cancer Consortium. J Clin Oncol (2013) 31(19):2460–8. doi:10.1200/JCO.2012.46.8280
225. Aplenc R, Blaney SM, Strauss LC, Balis FM, Shusterman S, Ingle AM, et al. Pediatric phase I trial and pharmacokinetic study of dasatinib: a report from the Children’s Oncology Group Phase I Consortium. J Clin Oncol (2011) 29(7):839–44. doi:10.1200/JCO.2010.30.7231
226. Slayton WB, Schultz KR, Jones T. Continuous dose dasatinib is safe and feasible in combination with intensive chemotherapy in pediatric Philadelphia chromosome positive acute lymphoblastic leukemia (Ph+ ALL): Children’s Oncology Group (COG) Trial AALL0622. Blood (2012) 120:137.
227. Slayton WB, Schultz KR, Jones T, Devidas M, Raetz EA, Borowitz MJ, et al. Outstanding early responses with dasatinib and intensive chemotherapy in pediatric Philadelphia chromosome positive acute lymphoblastic leukemia (Ph+ ALL): Children’s Oncology Group (COG) trial AALL0622. Pediatr Blood Cancer (2012) 59:988.
228. Ottmann OG, Larson RA, Kantarjian HM, le Coutre PD, Baccarani M, Hochhaus A, et al. Phase II study of nilotinib in patients with relapsed or refractory Philadelphia chromosome – positive acute lymphoblastic leukemia. Leukemia (2013) 27(6):1411–3. doi:10.1038/leu.2012.324
229. Usuki K, Tojo A, Maeda Y, Kobayashi Y, Matsuda A, Ohyashiki K, et al. Efficacy and safety of nilotinib in Japanese patients with imatinib-resistant or -intolerant Ph+ CML or relapsed/refractory Ph+ ALL: a 36-month analysis of a phase I and II study. Int J Hematol (2012) 95(4):409–19. doi:10.1007/s12185-012-1026-9
230. Castillo E, Al-Rajabi R, Pandya DM, Varadarajan P, Kelly KR, Swords R, et al. A pilot study of the combination of nilotinib and hyper-CVAD for Philadelphia chromosome positive acute lymphocytic leukemia and lymphoid blast crisis chronic myelogenous leukemia. Blood (2010) 116:885–6.
231. Kim D-Y, Joo YD, Kim S-D, Lee J-H, Lee J-H, Kim D-H, et al. Nilotinib combined with multi-agent chemotherapy for adult patients with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia: final results of prospective multicenter Phase 2 study. Blood (2010) 55.
232. Tefferi A. Nilotinib treatment-associated accelerated atherosclerosis: when is the risk justified? Leukemia (2013) 27(9):1939–40. doi:10.1038/leu.2013.112
233. Coon EA, Zalewski NL, Hoffman EM, Tefferi A, Flemming KD. Nilotinib treatment-associated cerebrovascular disease and stroke. Am J Hematol (2013) 88(6):534–5. doi:10.1002/ajh.23442
234. Berman E, Nicolaides M, Maki RG, Fleisher M, Chanel S, Scheu K, et al. Altered bone and mineral metabolism in patients receiving imatinib mesylate. N Engl J Med (2006) 354(19):2006–13. doi:10.1056/NEJMoa051140
235. Barr RD. Imatinib mesylate in children and adolescents with cancer. Pediatr Blood Cancer (2010) 55(1):18–25. doi:10.1002/pbc.22484
236. Hobernicht SL, Schweiger B, Zeitler P, Wang M, Hunger SP. Acquired growth hormone deficiency in a girl with chronic myelogenous leukemia treated with tyrosine kinase inhibitor therapy. Pediatr Blood Cancer (2011) 56(4):671–3. doi:10.1002/pbc.22945
237. Atkinson SA, Halton JM, Bradley C, Wu B, Barr RD. Bone and mineral abnormalities in childhood acute lymphoblastic leukemia: influence of disease, drugs and nutrition. Int J Cancer Suppl (1998) 11:35–9. doi:10.1002/(SICI)1097-0215(1998)78:11+<35::AID-IJC11>3.3.CO;2-9
238. Sanders JE, Guthrie KA, Hoffmeister PA, Woolfrey AE, Carpenter PA, Appelbaum FR. Final adult height of patients who received hematopoietic cell transplantation in childhood. Blood (2005) 105(3):1348–54. doi:10.1182/blood-2004-07-2528
239. Carpenter PA, Snyder DS, Flowers ME, Sanders JE, Gooley TA, Martin PJ, et al. Prophylactic administration of imatinib after hematopoietic cell transplantation for high-risk Philadelphia chromosome-positive leukemia. Blood (2007) 109(7):2791–3. doi:10.1182/blood-2006-04-019836
240. Burke MJ, Trotz B, Luo X, Weisdorf DJ, Baker KS, Wagner JE, et al. Imatinib use either pre- or post-allogeneic hematopoietic cell transplantation (allo-HCT) does not increase cardiac toxicity in chronic myelogenous leukemia patients. Bone Marrow Transplant (2009) 44(3):169–74. doi:10.1038/bmt.2008.441
241. Burke MJ, Trotz B, Luo X, Baker KS, Weisdorf DJ, Wagner JE, et al. Allo-hematopoietic cell transplantation for Ph chromosome-positive ALL: impact of imatinib on relapse and survival. Bone Marrow Transplant (2009) 43(2):107–13. doi:10.1038/bmt.2008.296
242. Klyuchnikov E, Schafhausen P, Kroger N, Brummendorf TH, Osanmaz O, Asenova S, et al. Second-generation tyrosine kinase inhibitors in the post-transplant period in patients with chronic myeloid leukemia or Philadelphia-positive acute lymphoblastic leukemia. Acta Haematol (2009) 122(1):6–10. doi:10.1159/000228587
243. Pfeifer H, Wassmann B, Bethge W, Dengler J, Bornhauser M, Stadler M, et al. Randomized comparison of prophylactic and minimal residual disease-triggered imatinib after allogeneic stem cell transplantation for BCR-ABL1-positive acute lymphoblastic leukemia. Leukemia (2013) 27(6):1254–62. doi:10.1038/leu.2012.352
244. Chen H, Liu KY, Xu LP, Liu DH, Chen YH, Zhao XY, et al. Administration of imatinib after allogeneic hematopoietic stem cell transplantation may improve disease-free survival for patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. J Hematol Oncol (2012) 5:29. doi:10.1186/1756-8722-5-29
245. Redaelli S, Piazza R, Rostagno R, Magistroni V, Perini P, Marega M, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J Clin Oncol (2009) 27(3):469–71. doi:10.1200/JCO.2008.19.8853
246. O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood (2007) 110(7):2242–9. doi:10.1182/blood-2007-03-066936
247. O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell (2009) 16(5):401–12. doi:10.1016/j.ccr.2009.09.028
248. Ye YB, Lin JY, Chen Q, Liu F, Chen HJ, Li JY, et al. The cytotoxicity of a Grb2-SH3 inhibitor in Bcr-Abl positive K562 cells. Biochem Pharmacol (2008) 75(11):2080–91. doi:10.1016/j.bcp.2007.12.021
249. Sadelain M, Brentjens R, Riviere I. The basic principles of chimeric antigen receptor design. Cancer Discov (2013) 3(4):388–98. doi:10.1158/2159-8290.CD-12-0548
250. Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood (2009) 113(15):3397–405. doi:10.1182/blood-2008-07-168773
Keywords: acute lymphoblastic leukemia, BCR–ABL1, tyrosine kinase inhibition, chemotherapy, hematopoietic stem-cell transplantation
Citation: Bernt KM and Hunger SP (2014) Current concepts in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia. Front. Oncol. 4:54. doi: 10.3389/fonc.2014.00054
Received: 14 January 2014; Accepted: 06 March 2014;
Published online: 25 March 2014.
Edited by:
Mignon Lee-Cheun Loh, University of California San Francisco, USAReviewed by:
Anthony V. Moorman, Newcastle University, UKBill H. Chang, Oregon Health and Science University, USA
Copyright: © 2014 Bernt and Hunger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen P. Hunger, Center for Cancer and Blood Disorders, Children’s Hospital Colorado, 13123 East 16th Avenue, Box B115, Aurora, CO 80045, USA e-mail: stephen.hunger@childrenscolorado.org
 Kathrin M. Bernt
Kathrin M. Bernt Stephen P. Hunger
Stephen P. Hunger