- Department of Pathology, Sir Mortimer B. Davis-Jewish General Hospital, McGill University, Montreal, QC, Canada
Biomarker testing has become standard of care for patients diagnosed with non-small cell lung carcinoma (NSCLC). Although, it can be successfully performed in circulating tumor cells, at present, the vast majority of investigations are carried out using direct tumor sampling, either through aspiration methods, which render most often isolated cells, or tissue sampling, that could range from minute biopsies to large resections. Consequently, pathologists play a central role in this process. Recent evidence suggests that refining NSCLC diagnosis might be clinically significant, particularly in cases of lung adenocarcinomas (ADC), which in turn, has prompted a new proposal for the histologic classification of such pulmonary neoplasms. These changes, in conjunction with the mandatory incorporation of biomarker testing in routine NSCLC tissue processing, have directly affected the pathologist’s role in lung cancer work-up. This new role pathologists must play is complex and demanding, and requires a close interaction with surgeons, oncologists, radiologists, and molecular pathologists. Pathologists often find themselves as the central figure in the coordination of a process, that involves assuring that the tumor samples are properly fixed, but without disruption of the DNA structure, obtaining the proper diagnosis with a minimum of tissue waste, providing pre-analytical evaluation of tumor samples selected for biomarker testing, which includes assessment of the proportion of tumor to normal tissues, as well as cell viability, and assuring that this entire process happens in a timely fashion. Therefore, it is part of the pathologist’s responsibilities to assure that the samples received in their laboratories, be processed in a manner that allows for optimal biomarker testing. This article goal is to discuss the essential role pathologists must play in NSCLC biomarker testing, as well as to provide a summarized review of the main NSCLC biomarkers of clinical interest.
Introduction
In Canada, lung cancer represents the second most common cancer in both males and females (14 and 13%, respectively), and it is the leading cause of cancer death for both sexes (1). In fact, lung cancer, with 27.2 and 26.3% mortality rate in males and females, respectively, is responsible for more deaths among Canadians than the other two leading organ-specific cancers combined [colorectal (12.7%) and prostate (10.0%) in males, and breast (13.9%) and colorectal (11,6%) in females] (1). In the United States, approximately 84% of new lung cancer cases are classified as non-small cell lung carcinomas (NSCLC), and 15% as small cell carcinomas (SCC) (2), with the majority of patients being diagnosed at advanced-stage (56%) (3). The prognosis is poor, with the overall 5-year survival rate of 6.1% for SCC and 17.1% for NSCLC (2).
Implementation of personalized targeted therapies has become a reality for a group of lung cancer patients, but this therapeutic option is usually reserved for those patients whom tumor samples have been screened for specific biomarkers. A multitude of potentially useful biomarkers have recently emerged and this list continues to grow. It has become increasingly difficult for pathologists and oncologists to define which biomarkers should be routinely tested. An expert panel in pathology and oncology, assembled by the College of American Pathologists (CAP) with representatives from the International Association for the Study of Lung Cancer (IASLC) and Association for Molecular Pathology (AMP), has recently met in an attempt to address questions regarding biomarker testing in lung cancer. The conclusions have been published in the format of testing guidelines, which presently recommends investigations of abnormalities involving only two genes: the epidermal growth factor receptor (EGFR) and the anaplastic lymphoma kinase (ALK) (4).
This review will focus on the role of the pathologist as an essential figure in the NSCLC biomarker testing process.
Tissue/Cytological Diagnosis and Biomarker Testing
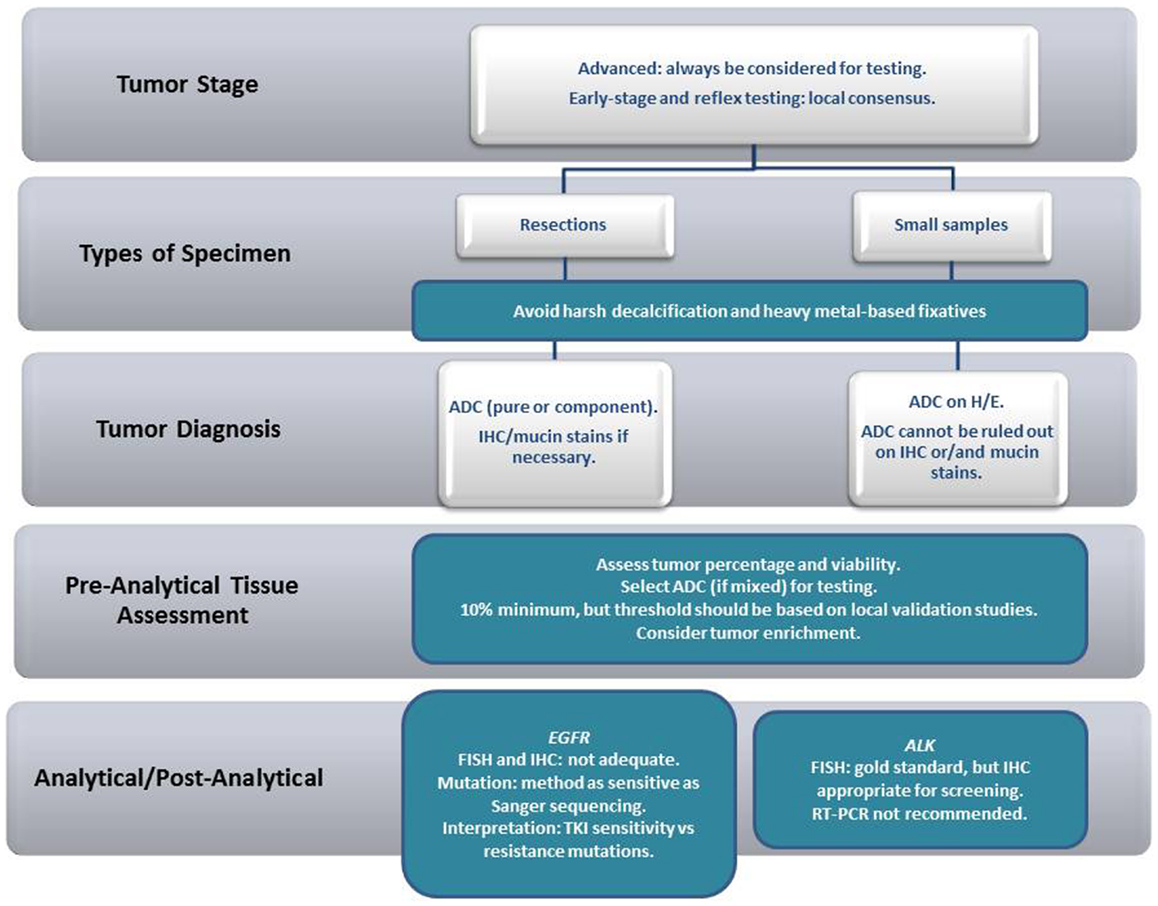
NSCLC, as a standing alone diagnosis, in either tissue or cytological samples, should be avoided whenever possible. In some situations (when the tumor sample is restricted to a smear from a bronchial brushing of a poorly differentiated carcinoma, for example), further characterization might be impossible. However, in our experience, further characterization, particularly with the help of special histochemical stains for the detection of mucin (often with the use of PAS-D or mucicarmin), and/or immunocytochemistry, can be achieved in the majority of cases. From a practical point of view, samples containing adenocarcinoma (ADC) either pure or mixed should undergo biomarker testing. In small samples, the recommendations are less stringent, and, as long as an ADC component cannot be excluded, the tissue should undergo biomarker testing (irrespective of the main tumor component identified). In resections, however, when the pathologist has an opportunity to examine the lesion in its entirety, “pure” tumors [large cell carcinoma (LGC), squamous cell carcinoma (SqCC) or others] should not be tested (Figure 1).
Numerous immunomarkers are available in order to help in the sub classification of NSCLC. The most commonly used are TTF-1, Napsin-A, p63, CK 5/6, and p40 (5–13). Although, it is true that in most cases the pathologist will be reasonably at ease to sub classify NSCLC’s, in some cases, sub classification might be rather difficult. It is our understanding that if the pathologist is uncertain about the specific sub classification, then the sample should be submitted for biomarker testing.
Despite the emphasis placed on focusing on ADC for biomarker testing, it is important to highlight that there are, however, isolated reports in the literature of the detection of either EGFR mutations or ALK rearrangements in tumors classified as SqCC (14–17).
Interestingly, some genetic aberrations can be generally associated with specific NSCLC subtypes and/or clinical profile (i.e., smokers versus non-smokers) (18). ADC is the predominant histologic type associated with both EGFR-mutated, as well as in ALK-rearranged cases. However, EGFR mutations are particularly prevalent in those cases containing non-mucinous bronchioloalveolar (lepidic) pattern (19), while in ALK-rearranged ADC, the most striking correlation is made with the presence of a signet-ring component (Table 1) (20, 21).
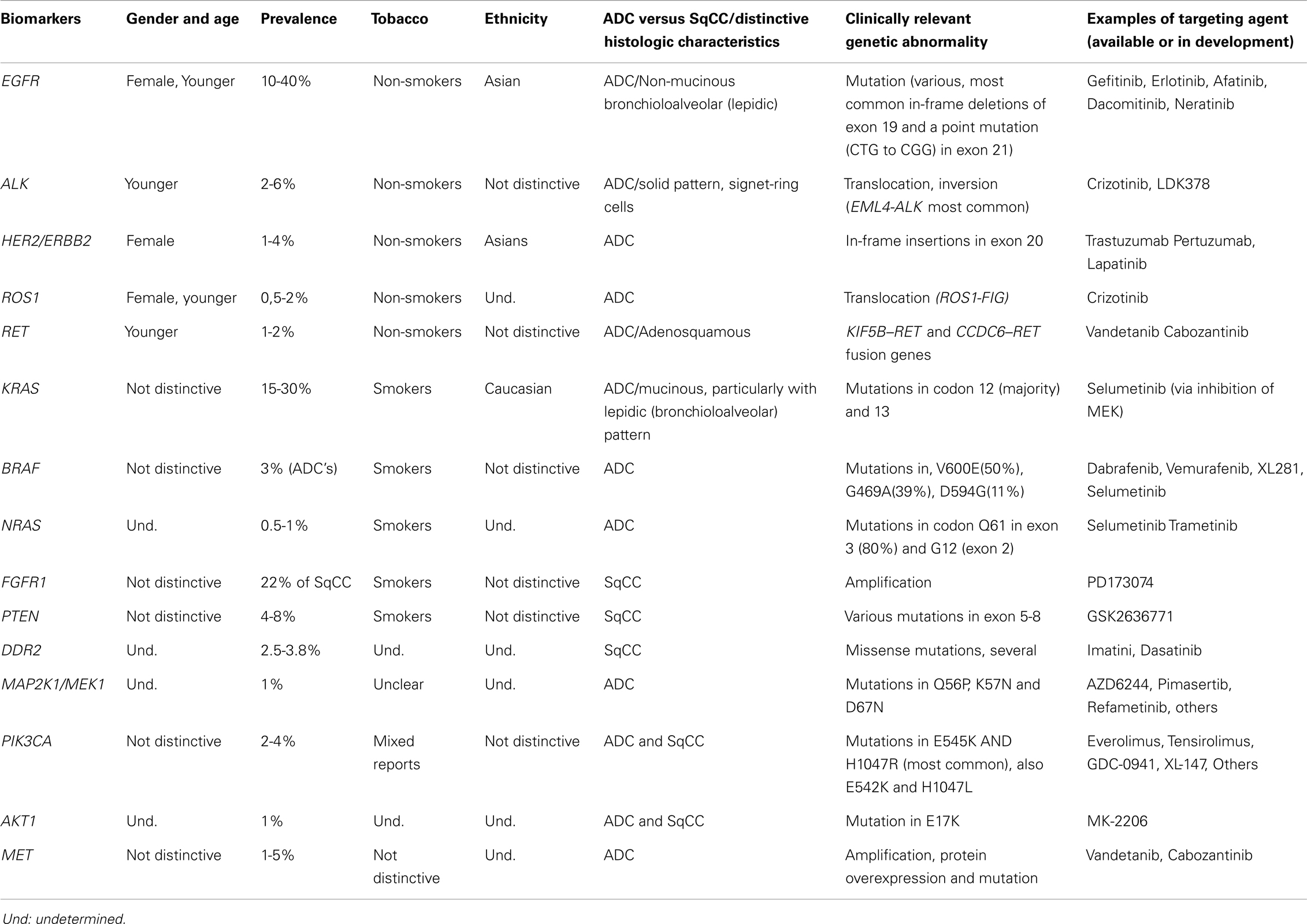
Table 1. Summary of the clinical characteristics, common genetic abnormalities and respective targeting agents of the main NSCLC biomarkers.
An important aspect that affects biomarker testing is the amount of available tumor present in a determined sample. This is a rather difficult topic to address, since the test sensitivity varies significantly according to the employed technique, particularly when searching for EGFR mutations, where normal DNA might interfere with test sensitivity (22). Nevertheless, the pathologist should provide an estimation of the percentage of tumor present in the sample, as well as, the viability of the tumor cells. It is recommended that testing sensitivity, as well as determination of limiting factors that might influence optimal results (fixative choice for example), should be defined locally, through proper validation methods. Of note, samples collected from aspiration biopsy methods, including direct lesion sampling (transbronchial needle aspiration biopsies), as well as the drainage of effusions, should be considered for biomarker testing (23–25).
Epidermal Growth Factor Receptor
Epidermal growth factor receptor (also known as HER-1 or Erb1) is a member of the ErbB receptor tyrosine kinase family, which also includes HER-2/neu (ErbB2), HER-3 (ErbB3), and HER-4 (ErbB4). EGFR activation is associated with cancer cell growth, invasion, proliferation, apoptosis, tumor angiogenesis, and metastatic spread. Therefore, it plays an important role in carcinogenesis and tumor progression by activation mechanisms, including overexpression, mutation, and autocrine ligand production. These actions are accomplished through activation of the RAS/RAF/MEK/MAPK and the PI3K/AKT/mTOR pathways (26).
The two most common EGFR activating mutations that confer sensitivity to tyrosine kinase inhibitors (TKI) are short in-frame deletions of exon 19, and a point mutation (CTG to CGG) in exon 21 at nucleotide 2573, that results in substitution of leucine by arginine at codon 858 (L858R) (27). Despite the fact that these two mutations might represent approximately 90% of all known clinically significant EGFR mutations, the consensus recommendations are that all EGFR mutations that account for at least 1% should be tested (4). It is important to emphasize that among the tested mutations, exon 20 T790M, as well as most exon 20 insertions are associated with resistance to first-generation TKI’s (28).
Epidermal growth factor receptor mutations occur at a higher frequency in tumors from East Asians than from non-Asians (30 versus 8%), from women than from men (59 versus 26%), from never smokers than from ever smokers (66 versus 22%), and in ADC’s compared with other NSCLC histologies (49 versus 2%) (29). In the United States, it is estimated that activating EGFR mutations are found in 15% of patients with primary lung ADC (Table 1) (30).
Turn-around-time (TAT) might be a very important factor for advanced-stage patients, whom might benefit from early institution of targeted therapy. The consensus recommends a maximum of 10 working days as an acceptable TAT from the date the laboratory receives the sample to be tested (4).
Anaplastic Lymphoma Kinase
Translocations involving ALK have previously been identified in anaplastic large cell lymphomas (ALCL), and in a rare mesenchymal neoplasm known as inflammatory myofibroblastic tumor or inflammatory pseudotumor (31, 32). In lung carcinomas, ALK rearrangement was first demonstrated in 2007 by Soda et al. (33) when ALK fusion transcripts were found in 6.7% (5 out of 75) of NSCLC samples. However, the prevalence of ALK rearrangement in lung carcinomas varies significantly (34–36).
ALK rearrangements tend to be mutually exclusive with other known driver mutations in NSCLC (18). However, it has rarely been described together with EGFR and PI3K mutations (36–38).
ALK-rearranged NSCLC patients, when compared to ALK-non-rearranged, are more frequently non- or light-smokers, younger, and present with advanced clinical stage. Histologically, the tumors demonstrate most frequently ADC with solid pattern and signet-ring cells (20, 21, 39).
Although fluorescence in situ hybridization (FISH) is currently the gold standard method for detecting ALK rearrangements according to the United States Food and Drug Administration (FDA) (40), the CAP consensus accepts that, if carefully validated, immunohistochemistry can be considered as a screening method (4). This proposition is in concert with the literature, which has shown in several different articles that immunohistochemistry can be very effective in the detection of ALK rearrangement in lung carcinomas (41–43).
Other Biomarkers
Currently, in over 50% of NSCLC’s, a driver oncogene can be identified (18). In addition to the previously discussed ALK and EGFR genes, several other potential targets have been uncovered in NSCLC’s, including the V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), the human epidermal growth factor receptor 2 (HER2), reactive oxygen species 1 (ROS1), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), phosphoinositide-3-kinase catalytic alpha polypeptide (PIK3CA), c-mesenchymal-epithelial transition mitogen (c-MET), activated protein kinase (MAP2K1), fibroblast growth factor receptor (FGFR), discoidin domain receptor 2 (DDR2), phosphatase and tensin homolog (PTEN), protein kinase B (AKT), rearranged during transfection (RET), and the neuroblastoma RAS viral oncogene homolog (NRAS). It is beyond the scope of this review to discuss each in detail. Current general knowledge of the characteristics of lung cancers carrying abnormalities in these genes has been summarized in Table 1 (18, 20, 21, 28, 31, 33, 36, 37, 41, 44–72).
In conclusion, targeted therapy is already a reality for many patients and it is certain that several other components will soon follow to become valid options in the therapeutic arsenal of oncologists. In view of the overwhelming amount of information being constantly generated into the molecular derangements associated with the development of lung cancer, it is not farfetched to expect that the current consensus guidelines will soon become obsolete. As a pathologist, I witness on a daily basis a continuous and inexorable change in our practice: our job no longer ends with the histological diagnosis. In fact, molecular profiling has become an integral part of the surgical pathology report. It is crucial that us pathologists embrace this new format of oncologic surgical pathology practice, and question ourselves, after each new malignant diagnosis: “what should I do now that might translate into a potential treatment alternative for this patient?”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
NSCLC, non-small cell lung carcinoma; SCC, small cell carcinomas; EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; ADC, adenocarcinoma; LGC, large cell carcinoma; SqCC, squamous cell carcinoma; TKI, tyrosine kinase inhibitors; TAT, turn-around-time; ALCL, anaplastic large cell lymphomas; FISH, fluorescence in situ hybridization.
References
1. Canadian Cancer Society’s Advisory Committee on Cancer Statistics. Canadian Cancer Statistics 2013. Toronto, ON: CCS (2013).
2. Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin (2012) 62(4):220–41. doi: 10.3322/caac.21149
3. Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, et al. SEER Cancer Statistics Review, 1975-2010. Bethesda, MD: National Cancer Institute (2013).
4. Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med (2013) 137(6):828–60. doi:10.5858/arpa.2012-0720-OA
5. Bishop JA, Teruya-Feldstein J, Westra WH, Pelosi G, Travis WD, Rekhtman N. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol (2012) 25(3):405–15. doi:10.1038/modpathol.2011.173
6. Mukhopadhyay S, Katzenstein AL. Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. Am J Surg Pathol (2011) 35(1):15–25. doi:10.1097/PAS.0b013e3182036d05
7. Nobre AR, Albergaria A, Schmitt F. p40: a p63 isoform useful for lung cancer diagnosis - a review of the physiological and pathological role of p63. Acta Cytol (2013) 57(1):1–8. doi:10.1159/000345245
8. Nonaka D. A study of DeltaNp63 expression in lung non-small cell carcinomas. Am J Surg Pathol (2012) 36(6):895–9. doi:10.1097/PAS.0b013e3182498f2b
9. Pelosi G, Fabbri A, Bianchi F, Maisonneuve P, Rossi G, Barbareschi M, et al. DeltaNp63 (p40) and thyroid transcription factor-1 immunoreactivity on small biopsies or cellblocks for typing non-small cell lung cancer: a novel two-hit, sparing-material approach. J Thorac Oncol (2012) 7(2):281–90. doi:10.1097/JTO.0b013e31823815d3
10. Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol (2011) 24(10):1348–59. doi:10.1038/modpathol.2011.92
11. Righi L, Graziano P, Fornari A, Rossi G, Barbareschi M, Cavazza A, et al. Immunohistochemical subtyping of nonsmall cell lung cancer not otherwise specified in fine-needle aspiration cytology: a retrospective study of 103 cases with surgical correlation. Cancer (2011) 117(15):3416–23. doi:10.1002/cncr.25830
12. Ring BZ, Seitz RS, Beck RA, Shasteen WJ, Soltermann A, Arbogast S, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol (2009) 22(8):1032–43. doi:10.1038/modpathol.2009.60
13. Stoll LM, Johnson MW, Gabrielson E, Askin F, Clark DP, Li QK. The utility of napsin-A in the identification of primary and metastatic lung adenocarcinoma among cytologically poorly differentiated carcinomas. Cancer Cytopathol (2010) 118(6):441–9. doi:10.1002/cncy.20108
14. Caliò A, Nottegar A, Gilioli E, Bria E, Pilotto S, Peretti U, et al. ALK/EML4 Fusion Gene May Be Found in Pure Squamous Carcinoma of the Lung. J Thorac Oncol (2014) 9(5):729–32. doi:10.1097/JTO.0000000000000109
15. Miyamae Y, Shimizu K, Hirato J, Araki T, Tanaka K, Ogawa H, et al. Significance of epidermal growth factor receptor gene mutations in squamous cell lung carcinoma. Oncol Rep (2011) 25(4):921–8. doi:10.3892/or.2011.1182
16. Park SH, Ha SY, Lee JI, Lee H, Sim H, Kim YS, et al. Epidermal growth factor receptor mutations and the clinical outcome in male smokers with squamous cell carcinoma of lung. J Korean Med Sci (2009) 24(3):448–52. doi:10.3346/jkms.2009.24.3.448
17. Tanaka K, Hata A, Kida Y, Kaji R, Fujita S, Katakami N, et al. Gefitinib for a poor performance status patient with squamous cell carcinoma of the lung harboring EGFR mutation. Intern Med (2012) 51(6):659–61. doi:10.2169/internalmedicine.51.6386
18. Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol (2011) 22(12):2616–24. doi:10.1093/annonc/mdr489
19. Blons H, Côté JF, Le Corre D, Riquet M, Fabre-Guilevin E, Laurent-Puig P, et al. Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol (2006) 30(10):1309–15. doi:10.1097/01.pas.0000213285.65907.31
20. Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, Barletta JA, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res (2009) 15(16):5216–23. doi:10.1158/1078-0432.CCR-09-0802
21. Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol (2009) 27(26):4247–53. doi:10.1200/JCO.2009.22.6993
22. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res (2007) 13(17):4954–5. doi:10.1158/1078-0432.CCR-07-1387
23. Aisner DL, Deshpande C, Baloch Z, Watt CD, Litzky LA, Malhotra B, et al. Evaluation of EGFR mutation status in cytology specimens: an institutional experience. Diagn Cytopathol (2013) 41(4):316–23. doi:10.1002/dc.21851
24. Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: molecular testing by using cytology specimens. Cancer Cytopathol (2011) 119(2):111–7. doi:10.1002/cncy.20151
25. Malapelle U, Bellevicine C, Zeppa P, Palombini L, Troncone G. Cytology- based gene mutation tests to predict response to anti-epidermal growth factor receptor therapy: a review. Diagn Cytopathol (2011) 39(9):703–10. doi:10.1002/dc.21512
26. Salomon DS, Brandt R, Ciardiello F, Normanno N, et al. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol (1995) 19(3):183–232. doi:10.1016/1040-8428(94)00144-I
27. Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol (2008) 21(Suppl 2):S16–22. doi:10.1038/modpathol.3801018
28. Ludovini V, Bianconi F, Pistola L, Pistola V, Chiari R, Colella R, et al. Optimization of patient selection for EGFR-TKIs in advanced non-small cell lung cancer by combined analysis of KRAS, PIK3CA, MET, and non-sensitizing EGFR mutations. Cancer Chemother Pharmacol (2012) 69(5):1289–99. doi:10.1007/s00280-012-1829-7
29. Bell DW, Brannigan BW, Matsuo K, Finkelstein DM, Sordella R, Settleman J, et al. Increased prevalence of EGFR-mutant lung cancer in women and in East Asian populations: analysis of estrogen-related polymorphisms. Clin Cancer Res (2008) 14(13):4079–84. doi:10.1158/1078-0432.CCR-07-5030
30. Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol (2011) 29(15):2121–7. doi:10.1200/JCO.2010.31.8923
31. Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol (2000) 157(2):377–84. doi:10.1016/S0002-9440(10)64550-6
32. Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science (1994) 263(5151):1281–4. doi:10.1126/science.8122112
33. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature (2007) 448(7153):561–6. doi:10.1038/nature05945
34. Boland JM, Erdogan S, Vasmatzis G, Yang P, Tillmans LS, Johnson MR, et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol (2009) 40(8):1152–8. doi:10.1016/j.humpath.2009.01.012
35. Camidge DR, Kono SA, Flacco A, Tan AC, Doebele RC, Zhou Q, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res (2010) 16(22):5581–90. doi:10.1158/1078-0432.CCR-10-0851
36. Wang Z, Zhang X, Bai H, Zhao J, Zhuo M, An T, et al. EML4-ALK rearrangement and its clinical significance in Chinese patients with advanced non-small cell lung cancer. Oncology (2012) 83(5):248–56. doi:10.1159/000341381
37. Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther (2012) 11(2):485–91. doi:10.1158/1535-7163.MCT-11-0692
38. Wallander ML, Geiersbach KB, Tripp SR, Layfield LJ. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med (2012) 136(7):796–803. doi:10.5858/arpa.2011-0321-OA
39. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res (2008) 14(13):4275–83. doi:10.1158/1078-0432.CCR-08-0168
40. FDA. CRIZOTINIB (2011). Available from: http://www.fda.gov/aboutfda/centersoffices/officeofmedicalproductsandtobacco/cder/ucm270058.htm
41. McLeer-Florin A, Moro-Sibilot D, Melis A, Salameire D, Lefebvre C, Ceccaldi F, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol (2012) 7(2):348–54. doi:10.1097/JTO.0b013e3182381535
42. Paik JH, Choe G, Kim H, Choe JY, Lee HJ, Lee CT, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol (2011) 6(3):466–72. doi:10.1097/JTO.0b013e31820b82e8
43. Selinger CI, Rogers TM, Russell PA, O’Toole S, Yip P, Wright GM, et al. Testing for ALK rearrangement in lung adenocarcinoma: a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol (2013) 26(12):1545–53. doi:10.1038/modpathol.2013.87
44. Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol (2012) 30(8):863–70. doi:10.1200/JCO.2011.35.6345
45. Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol (2009) 27(10):1667–74. doi:10.1200/JCO.2008.19.1635
47. Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov (2013) 3(6):630–5. doi:10.1158/2159-8290.CD-13-0035
48. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science (2007) 316(5827):1039–43. doi:10.1126/science.1141478
49. Go H, Kim DW, Kim D, Keam B, Kim TM, Lee SH, et al. Clinicopathologic analysis of ROS1-rearranged non-small-cell lung cancer and proposal of a diagnostic algorithm. J Thorac Oncol (2013) 8(11):1445–50. doi:10.1097/JTO.0b013e3182a4dd6e
50. Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov (2011) 1(1):78–89. doi:10.1158/2159-8274.CD-11-0005
51. Huang L, An SJ, Chen ZH, Su J, Yan HH, Wu YL. MET expression plays differing roles in non-small-cell lung cancer patients with or without EGFR mutation. J Thorac Oncol (2014) 9(5):725–8. doi:10.1097/JTO.0000000000000105
52. Iida M, Brand TM, Campbell DA, Starr MM, Luthar N, Traynor AM, et al. Targeting AKT with the allosteric AKT inhibitor MK-2206 in non-small cell lung cancer cells with acquired resistance to cetuximab. Cancer Biol Ther (2013) 14(6):481–91. doi:10.4161/cbt.24342
53. Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol (2013) 14(1):38–47. doi:10.1016/S1470-2045(12)70489-8
54. Jin G, Kim MJ, Jeon HS, Choi JE, Kim DS, Lee EB, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer (2010) 69(3):279–83. doi:10.1016/j.lungcan.2009.11.012
55. Kawano O, Sasaki H, Endo K, Suzuki E, Haneda H, Yukiue H, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer (2006) 54(2):209–15. doi:10.1016/j.lungcan.2006.07.006
56. Kohno T, Tsuta K, Tsuchihara K, Nakaoku T, Yoh K, Goto K. RET fusion gene: translation to personalized lung cancer therapy. Cancer Sci (2013) 104(11):1396–400. doi:10.1111/cas.12275
57. Li C, Sun Y, Fang R, Han X, Luo X, Wang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol (2012) 7(1):85–9. doi:10.1097/JTO.0b013e318234f0a2
58. Malanga D, Scrima M, De Marco C, Fabiani F, De Rosa N, De Gisi S, et al. Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle (2008) 7(5):665–9. doi:10.4161/cc.7.5.5485
59. Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res (2008) 68(14):5524–8. doi:10.1158/0008-5472.CAN-08-0099
60. Ohashi K, Sequist LV, Arcila ME, Lovly CM, Chen X, Rudin CM, et al. Characteristics of lung cancers harboring NRAS mutations. Clin Cancer Res (2013) 19(9):2584–91. doi:10.1158/1078-0432.CCR-12-3173
61. Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol (2009) 4(1):5–11. doi:10.1097/JTO.0b013e3181913e0e
62. Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol (2011) 29(15):2046–51. doi:10.1200/JCO.2010.33.1280
63. Pan Y, Zhang Y, Li Y, Hu H, Wang L, Li H, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer (2014) 84(2):121–6. doi:10.1016/j.lungcan.2014.02.007
64. Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol (2011) 12(2):175–80. doi:10.1016/S1470-2045(10)70087-5
65. Reungwetwattana T, Dy GK. Targeted therapies in development for non-small cell lung cancer. J Carcinog (2013) 12:22. doi:10.4103/1477-3163.123972
66. Reungwetwattana T, Weroha SJ, Molina JR. Oncogenic pathways, molecularly targeted therapies, and highlighted clinical trials in non-small-cell lung cancer (NSCLC). Clin Lung Cancer (2012) 13(4):252–66. doi:10.1016/j.cllc.2011.09.004
67. Serizawa M, Koh Y, Kenmotsu H, Isaka M, Murakami H, Akamatsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single-institute study. Cancer (2014) 120(10):1471–81. doi:10.1002/cncr.28604
68. Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res (2005) 65(5):1642–6. doi:10.1158/0008-5472.CAN-04-4235
69. Tsuta K, Kohno T, Yoshida A, Shimada Y, Asamura H, Furuta K, et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer (2014) 110(6):1571–8. doi:10.1038/bjc.2014.36
70. Villaruz LC, Socinski MA, Cunningham DE, Chiosea SI, Burns TF, Siegfried JM, et al. The prognostic and predictive value of KRAS oncogene substitutions in lung adenocarcinoma. Cancer (2013) 119(12):2268–74. doi:10.1002/cncr.28039
71. Warth A, Muley T, Dienemann H, Goeppert B, Stenzinger A, Schnabel PA, et al. ROS1 expression and translocations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology (2014). doi:10.1111/his.12379
Keywords: ALK, EGFR, NSCLC, adenocarcinoma, biomarker, histology, lung
Citation: Brega E and Brandao G (2014) Non-small cell lung carcinoma biomarker testing: the pathologist’s perspective. Front. Oncol. 4:182. doi: 10.3389/fonc.2014.00182
Received: 01 May 2014; Accepted: 27 June 2014;
Published online: 16 July 2014.
Edited by:
Vera Hirsh, McGill University Health Centre, CanadaReviewed by:
Joachim Diebold, Lucerne Cantonal Hospital, SwitzerlandK. Shilo, The Ohio State University, USA
Copyright: © 2014 Brega and Brandao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guilherme Brandao, Department of Pathology, Sir Mortimer B. Davis-Jewish General Hospital, Pavilion G, Suite 120.1, 3755 Côte Ste-Catherine Road, Montreal, QC H3T 1E2, Canada e-mail: guilherme.brandao@mcgill.ca
 Elisa Brega
Elisa Brega Guilherme Brandao
Guilherme Brandao