
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 18 February 2016
Sec. Molecular and Cellular Oncology
Volume 6 - 2016 | https://doi.org/10.3389/fonc.2016.00037
This article is part of the Research Topic Glycosylation Changes in Cancer: An Innovative Frontier at the Interface of Cancer and Glycobiology View all 16 articles
Glycosylation is one of the key components influencing several signaling pathways implicated in cell survival and growth. The Notch signaling pathway plays a pivotal role in numerous cell fate specifications during metazoan development. Both Notch and its ligands are repeatedly glycosylated by the addition of sugar moieties, such as O-fucose, O-glucose, or O-xylose, to bring about structural and functional changes. Disruption to glycosylation processes of Notch proteins result in developmental disorders and disease, including cancer. This review summarizes the importance and recent updates on the role of glycosylated Notch proteins in tumorigenesis and tumor metastasis.
It has been 60 years since the discovery of a link between the changes in protein glycosylation and oncogenic transformation (1). Numerous studies in cancer biology have supported this finding and underscored the significance of glycosylation in tumorigenesis and metastasis. Numerous proteins, such as mucins, selectins, gonadrotrophins, with altered glycosylation have been implicated in tumorigenesis (2, 3). Extensive research has been carried out to understand the glycobiology of the cancerous cell (4, 5). Cancer is a complex disease requiring several accumulated mutations, whose progression additionally depends on tumor cell interactions with the surrounding environment (6). Cell-surface proteins and changes associated with them define the course of cell-to-cell interactions. Altered glycosylated cell-surface proteins are one of the unique features of a cancerous cell (7), and specific glycan changes are critical during tumorigenesis and metastasis (8, 9). Several altered glycans serve as biomarkers to identify malignant cells undergoing epithelial–mesenchymal transition (EMT) and metastasis (9–11). Many essential glycosylated proteins, such as Notch, are altered in malignant cells, and the recent finding of GALNT11 as a new molecular marker in Notch-mediated chronic lymphocyte leukemia (CLL) (12) has increased interest in understanding Notch glycosylation.
The Notch signaling pathway facilitates short-range cell–cell communication to play a central role in proliferation and differentiation during animal development (13, 14). Notch signaling regulates a plethora of genes implicated in various cellular processes, and its signaling activity is extremely sensitive to the Notch receptor levels. Therefore, any slight modulation in Notch activity can perturb the regulation of gene expression and thus promoting several disorders, including cancer. Intriguingly, aberrant Notch signaling activity is highly implicated in several forms of leukemia and solid tumor development (11). T-cell acute lymphoblastic (T-ALL) neoplasm is one of the earliest diseases to be associated with the Notch signaling pathway (15). Membrane-bound Notch proteins (Notch receptor and its ligands) undergo rigorous glycosylation to accomplish its activity. Interestingly, deregulation of the components involved in glycosylating Notch proteins are implicated in Notch-induced tumorigenesis (16–19).
There is extremely robust evidence to suggest that aberrant Notch activity (11, 20, 21) or changes in glycosylation (5, 22, 23) can promote EMT and tumor development, despite the unclear role of glycosylated Notch proteins in relation to tumorigenesis. This review discusses recent advances in our understanding of glycosylation of Notch proteins and the impact of altered Notch glycans in promoting tumorigenesis and metastasis.
A century ago, flies with Notch-ed wing phenotype led to identification (24) and characterization of evolutionarily conserved Notch signaling pathway (25, 26). The signaling pathway comprising Notch receptor and Delta/Serrate/LAG-2 (DSL) family of ligands play crucial role in determining cell-fate choices in all animals (13, 27). While Drosophila has one Notch receptor (28), mammalians have four homologs (Notch1–4) (29) with an extracellular domain (ECD) and an intracellular domain (ICD). Both Notch receptor and DSL ligands show a high degree of structural similarities in the ECD (30).
Notch signaling involves receptor activation, Notch ICD (NICD) generation, and target stimulation (Figure 1A). Nascent Notch protein is initially glycosylated in the ER and Golgi apparatus (17, 31–33). In mammals, it is proteolytically cleaved by Furin at site 1 (S1) (this cleavage does not occur in Drosophila) (34). Following this, the mature Notch receptor heterodimer, comprising the ECD and transmembrane-ICD, gets tethered to the cell surface of the signal-receiving cell. Notch receptor interaction with membrane-bound ligands such as Delta/Serrate/LAG-2 (DSL) family proteins in the signal-sending cell initiates two successive proteolytic cleavages at site 2 (S2) and site 3 (S3) mediated by a disintegrin and metalloprotease (ADAM) and presenilin/γ-secretase complex, respectively. The NICD is then released, which translocates into the nucleus and binds to CSL (CBF1/SuH/LAG-1) transcriptional regulators to activate target genes (14, 35). The events that lead to the release of the Notch ICD (and Notch activation) rely on Notch ECD shedding. Evidences indicate presence of a non-canonical mode of signaling without ligand–receptor interaction to release NICD [Figure 1A (36, 37)]. Therefore, factors that influence shedding of the Notch ECD (either positively or negatively) can directly modulate Notch activity. Numerous extra- and intracellular modulators involving glycosylation, and trafficking machineries maintain the cellular pool of Notch in a context-specific manner.
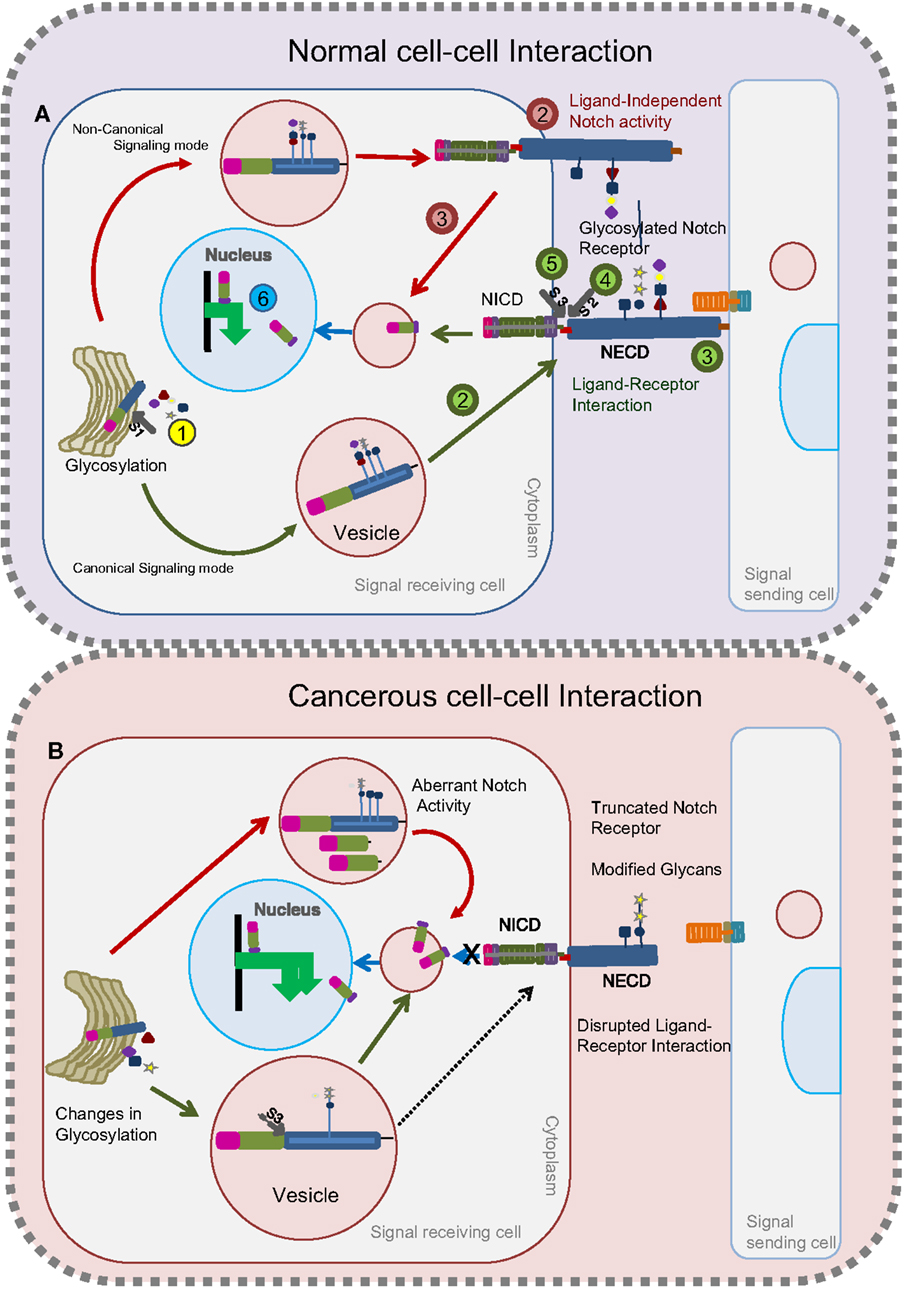
Figure 1. Glycosylated Notch-mediated cell–cell interactions in normal cells and cancerous cells. In normal cells (A), glycosylation on the Notch receptor takes place in the Golgi bodies followed by proteolytic (S1) furin cleavage (Step 1, yellow). Depending on the glycosylation cues, glycosylated Notch receptor takes either a canonical route (as shown by green arrows) or a non-canonical route (as shown by red arrows) to release the NICD fragment. Through the canonical mode (green) of a signal-receiving cell, glycosylated Notch receptor is transferred to the plasma membrane (Step 2) to interact with the ligands on the signal-sending cell (Step 3). Following this, the Notch receptor undergoes two successive proteolytic cleavages (S2) (Step 4) and (S3) (Step 5) to release the NICD that translocates into the nucleus to activate target genes (Step 6). In the non-canonical route (red), the Notch receptor does not interact with ligands (Step 2) but gets proteolytically cleaved inside the vesicles to release the NICD fragment (Step 3) that translocates into the nucleus to activate the target genes (Step 6). In cancerous cells (B), changes in glycosylation lead to the production of Notch receptors with modified glycans or truncated Notch receptors. Notch receptors with modified glycans undergo unusual S3 cleavage in the vesicles that releases NICD to activate Notch targets. Truncated Notch receptors and Notch receptors with modified glycans that reach the plasma membrane cannot interact with ligands on the signal-sending cell to release the NICD.
Notch protein exerts its biological functions by both canonical (ligand-dependent) and non-canonical (ligand-independent) signaling modes (Figure 1A). Contradicting earlier reports that ligand-dependent Notch activity alone is indispensable for the positive regulation of Notch signaling, recent work suggests that ligand-independent Notch signaling also plays a crucial positive role in regulating Notch activity (36, 38). Namely, changes in trafficking can lead to ligand-independent signaling (Figure 1A). Defects in any one of the two signaling modes can lead to tumorigenesis and tumor progression. Strikingly, glycosylation is one such process that modulates ligand–receptor binding and trafficking activities [Figure 1B (39, 40)]. In the following section of this review, we discuss the impact of glycosylation on Notch and its ligands to accomplish its biological function.
Glycosylation is an enzymatic reaction that mediates a chemical linkage of mono- or polysaccharides (glycans) onto other saccharides, proteins, or lipids occurring in Golgi apparatus and endoplasmic reticulum (ER). Nascent Notch proteins enroute the secretory pathway (41) to undergo a rigorous glycosylation on their ECD with 29–36 epidermal growth factors (EGF)-like repeats to emerge as a mature receptor and get localized on the cell surface (42). Predominantly, Notch receptor undergoes O-glycosylation at serine/threonine residues (31), and to a lesser extent, N-glycosylation on AsnXSer/Thr residues of EGF repeats (17).
The EGF repeats are modified by O-fucose, O-glucose, O-GlcNAc, and O-xylose (Figure 2). This short EGF repeat has six conserved Cys residues that form three disulfide bridges, wherein O-fucosylation at C2–X–X–X–S/T–C3 is mediated by O-fucosyltransferase 1 (O-FucT-1) (encoded by Ofut1 gene in Drosophila and Pofut1 in mammals) (39, 43) and elongated by Fringe, an N-acetylglucosaminyl transferase. Fucosylation is one of the prevalent glycosylation types on Notch proteins. Fringe is essential to promote Notch/Delta-binding, in preference to Notch/Serrate, whose interaction is inhibited by this modification (44, 45). Similarly, O-glycosylation (C1–X–S–X–P/A–C2) is mediated by O-glucosyltransferase, Rumi in Drosophila or POGLUT1 in mammals (40, 46–48), and elongated by Shams, a xylosyltransferase. In humans, xylosyltransferase (GXYLT)1 and (GXYLT)2, that add first and second xylose residues to Notch EGF repeats, have been identified (49, 50). Although Rumi is not required for the ligand-binding activity of Notch, it has been suggested that it functions to promote extracellular cleavage. In contrast to the glucose residues, xylosylation negatively regulates Notch signaling. A unique non-nucleocytoplasmic O-GlcNAc is reported to occur on the consensus sequence of Notch C5–X–X–G–X–S/T–G–X–X–C6 by EGF-specific O-GlcNAc-transferase (EOGT) in Drosophila and Eogt1 in mammals that mediate extracellular matrix interactions (51–54). The specific role of O-GlcNAc modifications on Notch activity is still not clear (Figure 2C). Recently, the presence of mucin-type-O-GalNAc glycans on the Notch ECD and N-glycans have been reported (19). The last four amino acids in the spacer between EGF repeats cooperates with calcium-ion binding and plays an important role in enhancing the rigidity and stability of EGF repeats (55). Recent research suggests that a strong crosstalk exists between glycosylating machinery and calcium modulating chaperones in regulating Notch activity.
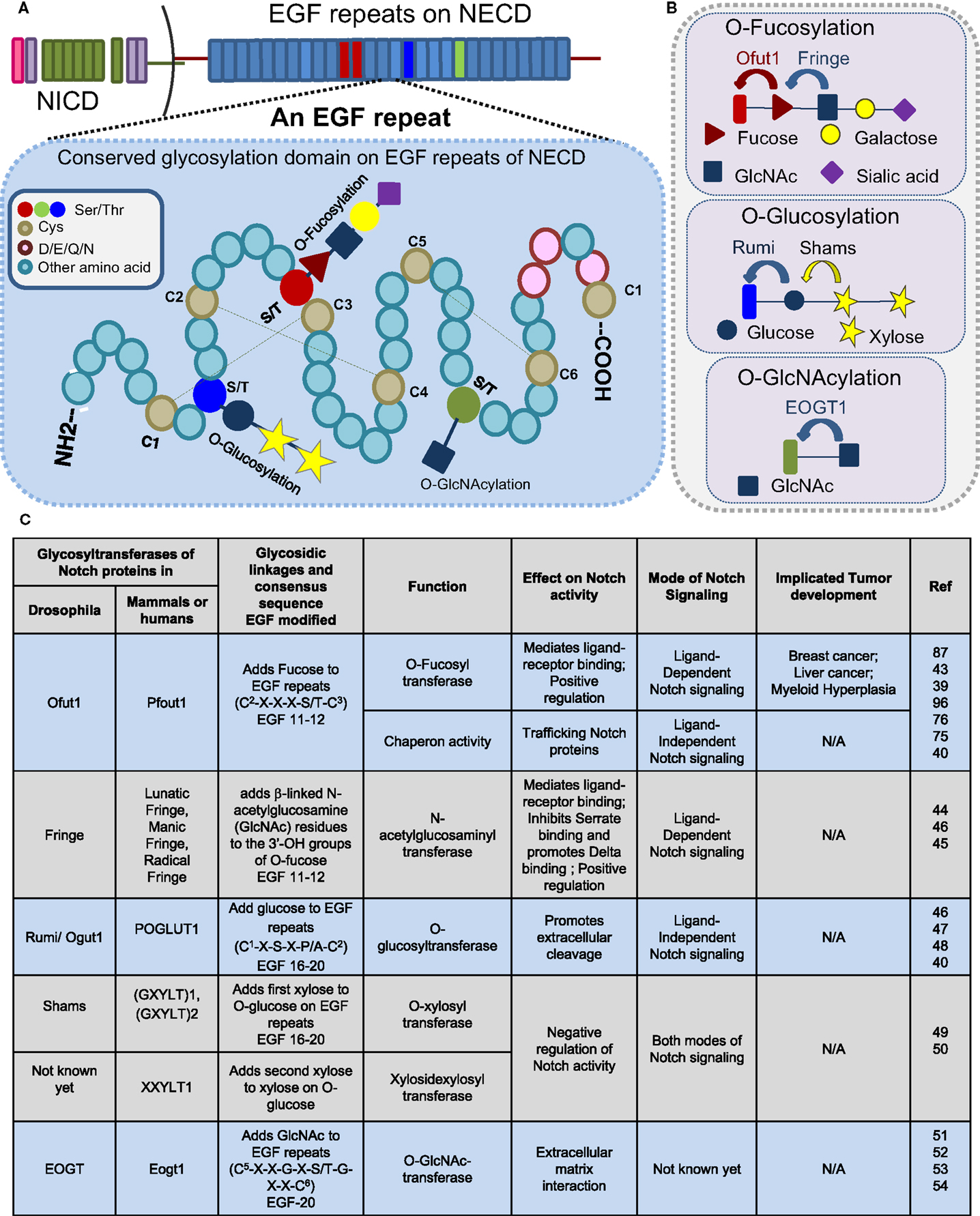
Figure 2. Glycosylation sites present on EGF repeats of the Notch extracellular domain. Consensus sites (A) and glycosyltransferases (B) of O-fucosylation, O-glycosylation, and O-GlcNAcylation that occur on an EGF repeat of NECD. (C) Table displaying a partial list of the glycosyltransferases that modify the Notch receptor.
Recent glycomic studies suggest that common glycosylation changes associated with tumor development are fucosylation, sialylation, O-glycan truncation, and O- and N-linked branching (8), and most of these changes are frequently associated with Notch proteins. Notch pathway cooperates with other mutations or deregulation in other oncogenic or tumor-suppressive genes of other signaling pathways, polarity regulators, and endocytic compartments to potentiate tumor progression (56–58).
Aberrant Notch activity has distinct roles in the development of several solid and hematopoietic tumors, and it has been shown to have either oncogenic or tumor-suppressive roles in a context-specific manner (21). In hematopoietic cancers, for example, T cell active lymphoblastic leukemia (T-ALL), Notch has an oncogenic role (59, 60), while in acute myeloid leukemia (AML), it has a tumor-suppressive role (61, 62). However, in solid tumors, such as hepatocellular carcinoma (HCC) (63) and medulloblastoma (64), Notch may have either an oncogenic or tumor-suppressive role depending on context (65, 66). It has been suggested that the switch between canonical and non-canonical Notch signaling can have a tumor-suppressive role, as demonstrated with Notch1 in the mouse (67). Not only does the glycosylation process of the Notch receptor aid in ligand-dependent signaling activity, but also reports suggest that changes in glycosylation events may lead to ligand-independent Notch activation (37). Most importantly changes in glycosylation could switch from one mode of signaling to the other, leading to deregulated Notch activity.
Notch protein undergoes extensive post-translational modification (68). To a larger extent, the ligand-dependent Notch signaling pathway requires glycosylation of the Notch ECD for signaling activation. It has been demonstrated that altered carbohydrate structure can play a very significant role in modulating ligand-binding activity (31, 69). Glycan modifications on EGF repeats of the Notch receptor indicate that the EGF8 repeat is required for Serrate-specific binding (70), while EGF12 is specifically required for Fringe function to inhibit Serrate and promote Delta-binding (16, 71), which mediates positive regulation on Notch signaling (Figure 2). Biochemical studies suggest that O-fucose addition on EGF14 leads to either dysregulated receptor–ligand activation or truncation effects. Fucosylation, which modulates the receptor–ligand interaction, is an important epitope on the EGF repeat (72–74) and impairment to this process is implicated with various forms of malignancies. Reports indicate that aberrant ligand-dependent Notch activity is highly associated with development of HCC, T-cell leukemia, and breast cancer (23). Interestingly, in humans, upregulation and aberrant gene expression of α-1,6-fucosyltransferase encoded by FUT8 is associated with development of breast cancer (75) and liver cancer (76).
Intriguingly, deletion or truncation of EGF repeats that impair the ligand–receptor interaction has been implicated in tumorigenesis. Such truncation may emerge due to defects in the glycosyl transferases or other factors that cooperate with them to enhance the receptor–ligand interaction. In squamous cell lung carcinoma (SqCC), where the tumor-suppressive role of Notch is impaired, it has been demonstrated that loss of EGF repeats generates truncated receptors that disrupt ligand-binding activity (Figure 1B) (77). Defects in Notch receptor fucosylation by deletion of FX (homolog of human GDP-l-fucose synthase) or O-fucosyltransferase (Pofut1) has been implicated in the development of myeloid hyperplasia (Figure 2) (78). Recently, O-mucin-type glycans have been found on Notch proteins and defects in O-mucin type glycosylation are well-documented in several forms of cancer (3, 19). Defects in ppGALNAcTS (an enzyme involved in initiating O-mucin glycans) (79) and C1GalT1 (chaperones involved in elongation) (80) can lead to the truncation of Notch protein. The association of Notch proteins with ppGALNAcTs and C1GalT1 is yet to be identified. Although the functional defects of glycosyltransferases are correlated to development of cancer, further investigation is required to understand how alterations in glycosylation of Notch contribute to tumor development.
Factors that influence Notch heterodimerization can have a significant impact on receptor–ligand interactions. Interestingly, OFut1 (55, 81) and Rumi (46, 82) are proposed to bind to calcium ions to enhance rigidity and also aid in modulating the thermodynamics of the protein. It has been shown that calcium binds to the EGF 12 repeat (55, 83). Calcium binding on EGF repeats is a highly conserved phenomenon assigning a crucial role to the structure of the protein (84). Calcium binding occurs on certain amino acids on a short linker sequence, N–N–x–N–C1 (where N can be D/E/Q/N, x-any amino acid, and C1 is the first conserved Cys of the EGF) between two EGF repeats (55, 83). From our knowledge of how calcium binding can affect EGF repeats, it has been proposed that depending on the rigidity and flexibility provided, Fringe might facilitate interactions with elongated glycans or inhibit interactions with neighboring regions. Studies indicate that calcium depletion dissociates and activates heterodimeric Notch receptors (85). Reports suggest that crosstalk exists between calcium levels and Notch activity during tumorigenesis. In line with this, it has been demonstrated that calcium/calmodulin-dependent kinase II (CaMKII) regulates Notch1 activity in prostate carcinoma development (86).
In recent years, several reports indicate that ligand-independent Notch signaling is implicated in tumorigenesis. Several endocytic components have been associated with Notch in promoting tumor progression. In spite of this, regulatory mechanisms that initiate ligand-independent Notch signaling activity remain elusive. It is highly logical to think that such events are triggered during early stages of nascent Notch protein production in the Golgi compartment. Glycosylation is not only indispensable for protein folding and protein activity, but it has an unprecedented role in intracellular transport/localization and degradation/half-life of the protein.
Ofut1/Pofut1 has both enzymatic fucosyl activity and fucosyl-independent chaperone activity on Notch proteins (39, 87). In addition to its usual role as O-fucosyltransferase, OFut1 has been implicated in maintaining the Notch pool by recycling cell-surface Notch through endosomes and on to lysosomes in a fucose-dependent manner (39). Similarly, another study has provided evidence of the involvement of OFut1 and fucosylation in localizing Notch to the sub-apical complex/adherens junction of epithelial cells by dynamin dependent transcytosis (88). These interesting data prompt further investigation into the possible mechanisms of the process. Fringe activity follows the Ofut1 reaction on specific EGF repeats of Notch. There is evidence indicating possible glycosylation events on other sites of EGF repeats too. Therefore, Fringe activity on different EGF repeats of Notch proteins, or yet to be identified glycosylation activity, might promote cleavage of Notch that inhibits the localization of processed Notch protein to the plasma membrane, retaining it in the intracellular compartment (Figure 1B). Deregulated function of Fringe or glycosyltransferase like Ofut1 might possibly lead to aberrant Notch activity. Rumi activity has been demonstrated to be required for ligand-independent Notch activation caused by deletion of LNR repeats (47, 48). Mutations in the heterodimerization domain on the EGF repeats may impair S2 cleavage of Notch leading to either ligand-independent activation or ligand-mediated hypersensitivity. A recent report has shown a cooperation of Ofut1 chaperone activity and Rumi in Notch transport (40). In the absence of ligands, preliminary results demonstrating glycosylation-mediated Notch trafficking defects are yet to be linked to tumorigenesis.
Studies to date, in most contexts, demonstrate that the addition of O-glucose positively regulates Notch signaling, while updates from Shams/GXYLT suggest that the addition of O-xylose residues downregulates Notch activity in a context-specific manner. It is proposed that this regulation, by changing the distribution of forms and length of sugar residues, offers a novel paradigm to modulate Notch signaling (48). Report suggests that the addition of Xylose to isolated Ser or Thr residues initiates Glycosaminoglycans (GAG) synthesis (89). As several studies have implicated aberrant GAG synthesis and GAG-conjugated proteins to tumor development and metastasis (90–92), it is highly intriguing to understand the significance of GAGs during Notch-induced tumorigenesis. Current therapeutic developments depend mostly on either modulating ligand–receptor interactions or the proteolytic cleavage of the receptor (93). Recent studies indicate that glycosylating proteins are important auxiliary proteins that modulate Notch activity and could therefore also be potential targets for future therapeutics. Glycan profiling of the modified glycans on Notch proteins may provide a better picture to understand the dramatic “glycome shift” that takes place during tumorigenesis and metastasis.
It is clear from the numerous studies highlighted here that Notch regulation is extremely complex and context dependent. For example, Notch can signal in a ligand-dependent or -independent manner, there are multiple Notch ligands, and the plethora of glycosyl modifications discussed in this review provide a further level of complexity. It is also clear how little we understand about how this regulation affects Notch’s ultimate function, namely, the regulation of gene expression. How does the nature of the ligand (or lack of ligand) affect the combination of genes whose expression will be upregulated or downregulated due to Notch signaling? Do individual or combinations of glycosyl modifications affect Notch’s ability to engage with other DNA-binding proteins and regulate the expression of specific genes? Notch-mediated gene regulation controls multiple cell differentiation processes both during development and adult life, and the complexity of Notch regulation will likely provide the necessary specificity that is required to generate the correct response to Notch signaling in different contexts. Our lack of understanding is tangible and unless we address these questions we cannot begin to understand how Notch regulation leads to specificity of response or the mechanisms by which Notch deregulation can lead to either an oncogenic or tumor-suppressive effect.
SP, AC, UN, and MG wrote and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research in the lab of MG is supported by a Career Establishment Award from Cancer Research UK. We would also like to thank the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India for the Startup grant (YSS/2014/000896) to UN. We apologize for any omissions when citing relevant literature due to space restrictions.
1. Ladenson RP, Schwartz SO, Ivy AC. Incidence of the blood groups and the secretor factor in patients with pernicious anemia and stomach carcinoma. Am J Med Sci (1949) 217:194–7. doi: 10.1097/00000441-194902000-00011
2. Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol (2005) 83:429–39. doi:10.1111/j.1440-1711.2005.01351.x
3. Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep (2006) 7:599–604. doi:10.1038/sj.embor.7400705
4. Kölbl AC, Andergassen U, Jeschke U. The role of glycosylation in breast cancer metastasis and cancer control. Front Oncol (2015) 5:219. doi:10.3389/fonc.2015.00219
5. Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol (2015) 10:473–510. doi:10.1146/annurev-pathol-012414-040438
6. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi:10.1016/j.cell.2011.02.013
7. Häuselmann I, Borsig L. Altered tumor-cell glycosylation promotes metastasis. Front Oncol (2014) 4:28. doi:10.3389/fonc.2014.00028
8. Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics (2014) 14:525–46. doi:10.1002/pmic.201300387
9. Freire-de-Lima L. Sweet and sour: the impact of differential glycosylation in cancer cells undergoing epithelial-mesenchymal transition. Front Oncol (2014) 4:59. doi:10.3389/fonc.2014.00059
10. Tuccillo FM, de Laurentiis A, Palmieri C, Fiume G, Bonelli P, Borrelli A, et al. Aberrant glycosylation as biomarker for cancer: focus on CD43. Biomed Res Int (2014) 2014:742831. doi:10.1155/2014/742831
11. Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell (2014) 25:318–34. doi:10.1016/j.ccr.2014.02.018
12. Libisch MG, Casás M, Chiribao M, Moreno P, Cayota A, Osinaga E, et al. GALNT11 as a new molecular marker in chronic lymphocytic leukemia. Gene (2014) 533:270–9. doi:10.1016/j.gene.2013.09.052
13. Artavanis-Tsakonas S. Notch signaling: cell fate control and signal integration in development. Science (1999) 284:770–6. doi:10.1126/science.284.5415.770
14. Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol (2006) 7:678–89. doi:10.1038/nrm2009
15. Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell (1991) 66:649–61. doi:10.1016/0092-8674(91)90111-B
16. Haines N, Irvine KD. Glycosylation regulates notch signalling. Nat Rev Mol Cell Biol (2003) 4:786–97. doi:10.1038/nrm1228
17. Stanley P. Regulation of notch signaling by glycosylation. Curr Opin Struct Biol (2007) 17:530–5. doi:10.1016/j.sbi.2007.09.007
18. Takeuchi H, Haltiwanger RS. Role of glycosylation of notch in development. Semin Cell Dev Biol (2010) 21:638–45. doi:10.1016/j.semcdb.2010.03.003
19. Takeuchi H, Haltiwanger RS. Significance of glycosylation in notch signaling. Biochem Biophys Res Commun (2014) 453:235–42. doi:10.1016/j.bbrc.2014.05.115
20. Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci (2007) 64:2746–62. doi:10.1007/s00018-007-7164-1
21. Dominguez M. Oncogenic programmes and notch activity: an ‘organized crime’? Semin Cell Dev Biol (2014) 28:78–85. doi:10.1016/j.semcdb.2014.04.012
22. Eklund EA, Freeze HH. Essentials of glycosylation. Semin Pediatr Neurol (2005) 12:134–43. doi:10.1016/j.spen.2005.11.001
23. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer (2015) 15:540–55. doi:10.1038/nrc3982
24. Dexer JS. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am Nat (1914) 48:712–58.
25. Poulson DF. Chromosomal deficiencies and the embryonic development of Drosophila melanogaster. Proc Natl Acad Sci U S A (1937) 23:133–7. doi:10.1073/pnas.23.3.133
26. Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell (1985) 43:567–81. doi:10.1016/0092-8674(85)90229-6
27. Klein T, Arias AM. Interactions among delta, serrate and fringe modulate notch activity during Drosophila wing development. Development (1998) 125:2951–62.
28. Wu JY, Rao Y. Fringe: defining borders by regulating the notch pathway. Curr Opin Neurobiol (1999) 9:537–43. doi:10.1016/S0959-4388(99)00020-3
29. Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian notch. Nature (1995) 377:355–8. doi:10.1038/377355a0
30. Chillakuri CR, Sheppard D, Lea SM, Handford PA. Notch receptor-ligand binding and activation: insights from molecular studies. Semin Cell Dev Biol (2012) 23:421–8. doi:10.1016/j.semcdb.2012.01.009
31. Haltiwanger RS, Stanley P. Modulation of receptor signaling by glycosylation: fringe is an O-fucose-beta1,3-N-acetylglucosaminyltransferase. Biochim Biophys Acta (2002) 1573:328–35. doi:10.1016/S0304-4165(02)00400-2
32. Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem (2004) 73:1019–49. doi:10.1146/annurev.biochem.73.011303.073752
33. Reynders E, Foulquier F, Annaert W, Matthijs G. How golgi glycosylation meets and needs trafficking: the case of the COG complex. Glycobiology (2011) 21:853–63. doi:10.1093/glycob/cwq179
34. Kidd S, Lieber T. Furin cleavage is not a requirement for Drosophila notch function. Mech Dev (2002) 115:41–51. doi:10.1016/S0925-4773(02)00120-X
35. Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical notch signaling: emerging role and mechanism. Trends Cell Biol (2012) 22:257–65. doi:10.1016/j.tcb.2012.02.003
36. Baron M. Endocytic routes to notch activation. Semin Cell Dev Biol (2012) 23:437–42. doi:10.1016/j.semcdb.2012.01.008
37. Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci (2013) 126:2135–40. doi:10.1242/jcs.127308
38. Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and deltex defines a ligand-independent notch signal. J Cell Biol (2011) 195:1005–15. doi:10.1083/jcb.201104146
39. Sasamura T, Ishikawa HO, Sasaki N, Higashi S, Kanai M, Nakao S, et al. The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of notch in Drosophila. Development (2007) 134:1347–56. doi:10.1242/dev.02811
40. Ishio A, Sasamura T, Ayukawa T, Kuroda J, Ishikawa HO, Aoyama N, et al. O-Fucose monosaccharide of Drosophila notch has a temperature-sensitive function and cooperates with O-glucose glycan in notch transport and notch signaling activation. J Biol Chem (2015) 290:505–19. doi:10.1074/jbc.M114.616847
41. Palade G. Intracellular aspects of the process of protein synthesis. Science (1975) 189:347–58. doi:10.1126/science.189.4206.867-b
42. Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol (2011) 21:583–9. doi:10.1016/j.sbi.2011.08.008
43. Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem (2005) 280:11289–94. doi:10.1074/jbc.M414574200
44. Brückner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of fringe modulates notch-delta interactions. Nature (2000) 406:411–5. doi:10.1038/35019075
45. Munro S, Freeman M. The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr Biol (2000) 10:813–20. doi:10.1016/S0960-9822(00)00578-9
46. Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, et al. Mammalian notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem (2000) 275:9604–11. doi:10.1074/jbc.275.13.9604
47. Jafar-Nejad H, Leonardi J, Fernandez-Valdivia R. Role of glycans and glycosyltransferases in the regulation of notch signaling. Glycobiology (2010) 20:931–49. doi:10.1093/glycob/cwq053
48. Lee TV, Sethi MK, Leonardi J, Rana N, Falk FR, Buettner FR, et al. Negative regulation of notch signaling by xylose. PLoS Genet (2013) 9:e1003547. doi:10.1371/journal.pgen.1003547
49. Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J Biol Chem (2010) 285:1582–6. doi:10.1074/jbc.C109.065409
50. Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, et al. Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J Biol Chem (2012) 287:2739–48. doi:10.1074/jbc.M111.302406
51. Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, et al. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem (2008) 283:35486–95. doi:10.1074/jbc.M806202200
52. Sakaidani Y, Furukawa K, Okajima T. O-GlcNAc modification of the extracellular domain of notch receptors. Methods Enzymol (2010) 480:355–73. doi:10.1016/S0076-6879(10)80016-3
53. Sakaidani Y, Ichiyanagi N, Saito C, Nomura T, Ito M, Nishio Y, et al. O-linked-N-acetylglucosamine modification of mammalian notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem Biophys Res Commun (2012) 419:14–9. doi:10.1016/j.bbrc.2012.01.098
54. Müller R, Jenny A, Stanley P. The EGF repeat-specific O-GlcNAc-transferase eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila. PLoS One (2013) 8:e62835. doi:10.1371/journal.pone.0062835
55. Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, et al. Structural and functional properties of the human notch-1 ligand binding region. Structure (2004) 12:2173–83. doi:10.1016/j.str.2004.09.012
56. Djiane A, Krejci A, Bernard F, Fexova S, Katherine Millen K, Bray SJ. Dissecting the mechanisms of notch induced hyperplasia. EMBO J (2013) 32:60–71. doi:10.1038/emboj.2012.326
57. Patel PH, Edgar BA. Tissue design: how Drosophila tumors remodel their neighborhood. Semin Cell Dev Biol (2014) 28:86–95. doi:10.1016/j.semcdb.2014.03.012
58. Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene (2015) 34:939–50. doi:10.1038/onc.2014.59
59. Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Cheryll Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (2004) 306:269–71. doi:10.1126/science.1102160
60. Malyukova A, Dohda T, von der Lehr N, Akhoondi SN, Akhondi S, Corcoran M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for notch signaling. Cancer Res (2007) 67:5611–6. doi:10.1158/0008-5472.CAN-06-4381
61. Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med (2013) 210:321–37. doi:10.1084/jem.20121527
62. Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med (2013) 210:301–19. doi:10.1084/jem.20121484
63. Villanueva A, Alsinet C, Yanger K, Hoshida Y, Zong Y, Toffanin S, et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology (2012) 143:1660.e–9.e. doi:10.1053/j.gastro.2012.09.002
64. Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res (2004) 64:7787–93. doi:10.1158/0008-5472.CAN-04-1446
65. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet (2012) 44:760–4. doi:10.1038/ng.2291
66. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet (2012) 44:694–8. doi:10.1038/ng.2256
67. Blanpain C, William E, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev (2006) 20:3022–35. doi:10.1101/gad.1477606
68. Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell (2009) 16:633–47. doi:10.1016/j.devcel.2009.03.010
69. Chen J, Moloney DJ, Stanley P. Fringe modulation of jagged1-induced notch signaling requires the action of beta 4galactosyltransferase-1. Proc Natl Acad Sci U S A (2001) 98:13716–21. doi:10.1073/pnas.241398098
70. Yamamoto S, Charng W, Rana NA, Kakuda S, Jaiswal M, Bayat V, et al. A mutation in EGF repeat-8 of notch discriminates between serrate/jagged and delta family ligands. Science (2012) 338:1229–32. doi:10.1126/science.1228745
71. Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits notch activation. Development (2003) 130:6411–21. doi:10.1242/dev.00883
72. Dear AE, Medcalf RL. The urokinase-type-plasminogen-activator receptor (CD87) is a pleiotropic molecule. Eur J Biochem (1998) 252:185–93. doi:10.1046/j.1432-1327.1998.2520185.x
73. Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron (1998) 21:63–75. doi:10.1016/S0896-6273(00)80515-2
74. Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem (2001) 276:40338–45. doi:10.1074/jbc.M107849200
75. Blomme B, Van Steenkiste C, Callewaert N, Van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J Hepatol (2009) 50:592–603. doi:10.1016/j.jhep.2008.12.010
76. Potapenko IO, Haakensen VD, Lüders T, Helland A, Bukholm I, Sørlie T, et al. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol Oncol (2010) 4:98–118. doi:10.1016/j.molonc.2009.12.001
77. Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A (2011) 108:17761–6. doi:10.1073/pnas.1114669108
78. Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of notch receptor ligand interactions. Blood (2011) 117:5652–62. doi:10.1182/blood-2010-12-326074
79. Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology (2012) 22:736–56. doi:10.1093/glycob/cwr182
80. Ju T, Cummings RD. A unique molecular chaperone cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A (2002) 99:16613–8. doi:10.1073/pnas.262438199
81. Rao Z, Handford PA, Mayhew M, Knott V, Brownlee GG, Stuart D. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell (1995) 82:131–41. doi:10.1016/0092-8674(95)90059-4
82. Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, et al. Rumi is a CAP10 domain glycosyltransferase that modifies notch and is required for notch signaling. Cell (2008) 132:247–58. doi:10.1016/j.cell.2007.12.016
83. Cordle J, Johnson S, Yan Tay JZ, Roversi P, Wilkin MB, de Madrid BH, et al. A conserved face of the jagged/serrate DSL domain is involved in notch trans-activation and cis-inhibition. Nat Struct Mol Biol (2008) 15:849–57. doi:10.1038/nsmb.1457
84. Xu A, Lei L, Irvine KD. Regions of Drosophila notch that contribute to ligand binding and the modulatory influence of fringe. J Biol Chem (2005) 280:30158–65. doi:10.1074/jbc.M505569200
85. Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol (2000) 20:1825–35. doi:10.1128/MCB.20.5.1825-1835.2000
86. Mamaeva OA, Kim J, Feng G, McDonald JM. Calcium/calmodulin-dependent kinase II regulates notch-1 signaling in prostate cancer cells. J Cell Biochem (2009) 106:25–32. doi:10.1002/jcb.21973
87. Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science (2005) 307:1599–603. doi:10.1126/science.1108995
88. Sasaki N, Sasamura T, Ishikawa HO, Kanai M, Ueda R, Saigo K, et al. Polarized exocytosis and transcytosis of notch during its apical localization in Drosophila epithelial cells. Genes Cells (2007) 12:89–103. doi:10.1111/j.1365-2443.2007.01037.x
89. Esko JD, Kimata K, Lindahl U. Proteoglycans and sulfated glycosaminoglycans. 2nd ed. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology (Chaps. 16), Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (2009). p. 1–20.
90. Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, et al. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J (2012) 279:1177–97. doi:10.1111/j.1742-4658.2012.08529.x
91. Belting M. Glycosaminoglycans in cancer treatment. Thromb Res (2014) 133:S95–101. doi:10.1016/S0049-3848(14)50016-3
92. Nagarajan U, Pakkiriswami S, Pillai AB. Sugar tags and tumorigenesis. Front Cell Dev Biol (2015) 3:69. doi:10.3389/fcell.2015.00069
Keywords: glycosyltransferases, Notch signaling, tumorigenesis, epidermal growth factor
Citation: Pakkiriswami S, Couto A, Nagarajan U and Georgiou M (2016) Glycosylated Notch and Cancer. Front. Oncol. 6:37. doi: 10.3389/fonc.2016.00037
Received: 14 December 2015; Accepted: 05 February 2016;
Published: 18 February 2016
Edited by:
Leonardo Freire-de-Lima, Federal University of Rio de Janeiro, BrazilReviewed by:
Xiaoxia Zhu, University of Miami Miller School of Medicine, USACopyright: © 2016 Pakkiriswami, Couto, Nagarajan and Georgiou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Usha Nagarajan, ushibiotech@gmail.com;
Marios Georgiou, marios.georgiou@nottingham.ac.uk
†Shanmugasundaram Pakkiriswami and Africa Couto contributed equally.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.