- 1Department of Biophysics, GSI Helmholtzzentrum für Schwerionenforschung, Darmstadt, Germany
- 2Department of Radiation Oncology, Shanghai Proton and Heavy Ion Center, Shanghai, China
- 3Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 4Institute of Molecular Oncology and Experimental Therapeutics, Marienhospital Herne, Ruhr University of Bochum Medical School, Herne, Germany
- 5Institute of Condense Matter Physics, Darmstadt University of Technology, Darmstadt, Germany
Purpose: The purpose of this study is to link both numeric and structural chromosomal aberrations to the effectiveness of radiotherapy in chemotherapy refractory tumor cells.
Materials and methods: Neuroblastoma (LAN-1) and 79HF6 glioblastoma cells derived from patients and their chemoresistant sublines were artificially cultured as neurospheres and irradiated by X-rays and heavy ions sources. All the cell lines were irradiated by Carbon-SIS with LET of 100 keV/μm. However, 79HF6 cells and LAN-1 cells were also irradiated by Carbon-UNILAC with LET of 168 keV/μm and Nickel ions with LET of 174 keV/μm, respectively. The effect of radiation on the survival and proliferation of cells was addressed by standard clonogenic assays. In order to analyze cell karyotype standard Giemsa staining, multicolor fluorescence in situ hybridization (mFISH) and multicolor banding (mBAND) techniques were applied.
Results: Relative biological effectiveness values of heavy ion beams relative to X-rays at the D10 values were found between 2.3 and 2.6 with Carbon-SIS and Nickel for LAN-1 and between 2.5 and 3.4 with Carbon-SIS and Carbon-UNILAC for 79HF6 cells. Chemorefractory LAN-1RETO cells were found more radioresistant than untreated LAN-1WT cells. 79HF6RETO glioblastoma cells were found more radiosensitive than cytostatic sensitive cells 79HF6WT. Sphere formation assay showed that LAN-1RETO cells were able to form spheres in serum-free culture, whereas 79HF6 cells could not. Most of 79HF6WT cells revealed a number of 71–90 chromosomes, whereas 79HF6RETO revealed a number of 52–83 chromosomes. The majority of LAN-1WT cells revealed a number of 40–44 chromosomes. mFISH analysis showed some stable aberrations, especially on chromosome 10 as judged by the impossibility to label this region with specific probes. This was corroborated using mBAND analysis.
Conclusion: Heavy ion irradiation was more effective than X-ray in both cytostatic naive cancer and chemoresistant cell lines. LAN-1RETO chemoresistant neuroblastoma cells were found to be more radioresistant than the cytostatic naive cells (LAN-1WT), whereas this effect was not found in 79HF6 cells.
Introduction
There is convincing evidence that many solid and hematological malignancies are organized hierarchically and contain a small population of cancer stem cells (CSCs) that possess the capacity to self-renew and to cause the heterogeneous lineages of cells that form the tumor (1). Consequently, cell heterogeneity of tumors may play an important role in tumor persistence and metastasis formation. Additionally, there is growing evidence that CSCs are inherently resistant to radiation and perhaps other conventional anticancer treatments, i.e., chemotherapy (2–4). These intrinsic mechanisms of resistance are responsible for a significant number of tumor recurrences (2, 3). Consequently, an effective anticancer treatment can only be achieved if this population is eliminated.
Chemotherapy has the advantage over radiotherapy in fighting the disseminated metastatic situation but at higher costs for the organism as a whole. Contrary to that, radiotherapy is a more localized treatment, but it is less applicable once the cancer has spread to several regions. Contemporaneous studies have consistently shown that CSC phenotypes are triggered after chemotherapy courses with an accompanied radioresistance of cancer cells both in vitro and in vivo probably by preferential activation of the DNA damage response (5). This indicates the urgent necessity for reevaluation of conventional therapies and searching for new ones that focus on CSCs to enhance the efficacy of cancer treatments.
Neuroblastoma is one of the most common extracranial pediatric tumors with a wide spectrum of clinical forms. The long-term survival of children with a high-risk clinical phenotype is <40% (especially those with MYCN amplification) (6). Glioblastoma is the most aggressive brain tumor in adults. In spite of multimodal therapy, the median survival is only around 14 months with early recurrences (and infiltrative events) in the brain (7). The existence (and local spread) of CSCs may be an important reason for the treatment failure due to its resistance to conventional therapy, which leads to a poor prognosis.
Culturing cancer cells in the presence of a low dose of chemotherapeutic agents is one of the approaches to enrich subpopulations with CSC-like phenotypes and related physiology. Etoposide is a topoisomerase inhibitor and causes DNA breaks enforcing apoptosis in dividing cancer cells. It is used as a standard chemotherapy in many tumors, such as neuroblastoma. However, etoposide is also known as an inducing agent of multidrug-resistant cancer phenotypes. In this study, low dose of etoposide was used to enrich CSCs fraction in glioblastoma and neuroblastoma cell lines.
Particle radiotherapy is becoming more widely used because proton and heavy ions have a favorable depth–dose distribution and a higher relative biological effectiveness (RBE) compared with photon. Once cancer cells are exposed to this therapy, they suffer a complex and clustered DNA damage, which is unable to be repaired by cellular mechanisms independent of the reactive oxygen species formed after exposing cells to charged particles. Consequently, malignant cells are less radioresistance because the mechanisms responsible for DNA reparation work less effective (8).
Our works aimed at studying the survival of chemoresistant cells compared with their wild-type parentals after being exposed to X-rays and heavy ions. We also addressed the question if the karyotype and chromosomal number deviations are related to the survival.
Materials and Methods
Cell Lines and Culture Conditions
Two parental and their subtypes highly chemotherapy refractory cell lines LAN-1WT, LAN-1RETO neuroblastoma and 79HF6WT, 79HF6RETO glioblastoma multiforme derived from human tumors were used in this investigation. The LAN-1 cells were isolated from a bone marrow metastasis of a 2-year-old boy with neuroblastoma (clinical Stage IV), and the 79HF6 cells were isolated from a female adult patient. The etoposide-resistant sublines usyed in this work exhibit CSC features among a set of CSC markers, broad spectrum of cross-resistance to several cytostatics, and radioresistance. The phenotype characteristics and the CSC features were published previously (5). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin (all purchased from Biochrom AG, Berlin, Germany), and kept in a humidified atmosphere of 5% CO2 at 37°C. Resistant to ETOposide (RETO) cells were constantly cultured in the medium containing 4 μg/ml etoposide (Teva, Germany). The cell doubling time (tD) was determined in the exponential phase of the growth with the GSI in house program gd (©M. Krämer, 2003).
Clonogenic Assay
Clonogenic assay was performed to determine both clonogenic behavior and cell survival rates after irradiation. Cells were seeded in T25 flask containing around 100 viable colonies after irradiation. LAN-1WT and LAN-1RETO cells were incubated for 9 days. 79HF6WT cells were incubated for 11 days, whereas 79HF6RETO cells were incubated for 25 days. Colonies were fixed and stained with methylene blue. Colonies containing more than 50 cells were defined as survivors.
Sphere Formation Assay
Cells were cultured in serum-free neurobasal A medium (Gibco, Life Technologies, Germany) supplemented with B27 (Gibco, Life Technologies, Germany), 10 ng/ml human fibroblast growth factor-basic (Biochrom, Germany), 20 ng/ml human epidermal growth factor (Biochrom, Germany), and 0.1% bovine serum albumin fraction V (Roche Diagnostics, Germany) to observe the formation of neurospheres (9).
Karyotyping
For chromosome preparations, cells were seeded 48 h in T75 culture flasks with 10 ml medium before the experiment in order to allow stabile attachment. One hundred microliters of colcemid (Roche Deutschland Holding GmbH, Germany) with the concentration of 10 μg/ml were added to the cultures. After 3.5 h of incubation for LAN-1 and 79HF6WT and 4 h for 79HF6RETO, cells were trypsinized and harvested. Cell suspension was pelleted and carefully treated with prewarmed (37°C) 0.075M potassium chloride solution for 8 min and then fixed with 3:1 ratio of MeOH:glacial acetic acid for 30 min at room temperature. After washing, cells were resuspended in proper volume of the mentioned fixative and dropped on wet slides. The slides were then air-dried for 24 h. The slides were stained with 5% Giemsa (Merck, Germany) solution for 10 min, washed with distilled water, and dried overnight.
Multicolor Fluorescence In Situ Hybridization Technique and Multicolor Banding Technique
For multicolor fluorescence in situ hybridization (mFISH) analysis, the slides were hybridized using the 24XCyte mFISH kit (Metasystems, Altlussheim, Germany) according to the protocol recommended by the manufacturer. In brief, the slides were first subjected to a denaturation followed by dehydration. An appropriate volume of DNA denatured probe was incubated in a humidified chamber at 37°C in the dark for 2 days. Afterward, the remaining hybridization probe was washed off. Finally, all DNA material was counterstained using DAPI/antifade (250 ng/ml), and the slide was covered. The chromosomal dispersal was analyzed using fluorescence microscopy Imager Z1 (Zeiss, Germany). Probes labeled with FITC, Orange, Texas Red®, Aqua, CyTM5 (Cy5), and 4′,6-diamidino-2-phenylindol (DAPI) fluorochromes were used to visualize chromosomal segments. Karyotypes were (re)constructed using the Isis/mFISH software (Metasystems). The procedure of multicolor banding (mBAND) is similar to that of mFISH, performed with the mBAND kit (Metasystems, Altlussheim, Germany), as previously described.
Ionizing Irradiation
All the irradiations were performed in GSI. The X-ray irradiation was carried out using an Isovolt DS1 X-ray machine (Seifert, Ahrensberg, Germany), exposing cells to 250 kVp and 16 mA.
Ion irradiation was performed in a synchrotron machine of the GSI. For irradiation at Carbon-SIS facility, cells were cultured in T12.5 culture flasks and were completely filled with culture medium before irradiation. The cells were irradiated with 10 mm spread out Bragg peak (SOBP) with LET of 100 keV/μm. All the cell lines were irradiated by Carbon-SIS. For experiments using a Carbon-UNILAC, 79HF6 cells were cultured in 3 cm Petri dishes and placed into compatible Petri dish magazines for irradiation. The carbon ions had a primary energy of 11.4 MeV/u and the energy decreased to 9.9 MeV/u when stopping on target with the corresponding LET of 168 keV/μm (10). After irradiation, the inner border of Petri dish was cleaned using sterile cotton to remove unirradiated medium accumulated at the bottom of the inner border, because dishes were irradiated in a vertical position. As survival of cells is related to the LET of the beam, we use those two carbon beams with different LETs. LAN-1 cells were also irradiated by Nickel ions with energy of 1 GeV/u and LET of 174 keV/μm, because the beam time is limited in GSI and we got the chance of irradiated by Nickel ions with similar LET to Carbon-UNILAC, which is a higher LET beam compared with Carbon-SIS.
Samples in triplicate were subjected to irradiation sections for each dose with X-ray, heavy ions, and repeated at least three times. The irradiation doses for LAN-1WT were fixed from 0 to 7 Gy for X-ray, from 0 to 2 Gy for Carbon, and from 0 to 2 Gy for Nickel. However, the doses for LAN-1RETO were from 0 to 9 Gy for X-ray, 0 to 3.2 Gy for Carbon, and 0 to 2 Gy for Nickel. The doses for 79HF6WT and 79HF6RETO were applied from 0 to 10 Gy for X-ray, 0 to 5 Gy for Carbon-SIS, and 0 to 2.72 Gy for Carbon-Unilac.
Cell survival curves of X-ray were fitted with the linear-quadratic model (Eq. 1):
Cell survival curves of heavy ions were fitted with a pure exponential equation (Eq. 2):
RBE10 values were calculated at 10% of survival level according to Eq. 3:
All of the fitting was performed with the GSI in house program gd (©M. Krämer, 2003).
Statistical Analysis
Experiments were performed at least in triplicate, and the survival fraction of cells was given as mean ± SD. Karyotype and mBAND figures were descriptive and therefore not statistically analyzed.
Results
Differential Growth Patterns of LAN-1 Neuroblastoma and Glioblastoma 79HF6 Cell Lines
All four cell lines, both wild type and resistant, grew adherently. The growth kinetic for all cells shows differential pattern as jugged by their doubling times. In this regard, the replication of LAN-1WT, LAN-1RETO, 79HF6WT, and 79HF6RETO was observed at 21.7 ± 0.7, 16.9 ± 0.8, 21.6 ± 0.3, and 56.7 ± 5.2 h, respectively. LAN-1WT could form spheres when cultured in serum-free neurobasal A medium, whereas the other tumor cells lines were not able to form neurospheres once cultured under this condition (Figure 1). LAN-1RETO and 79HF6RETO chemotherapy refractory cells are able to stably grow in the medium containing low concentrations of etoposide. The growth kinetic of LAN-1RETO cells showed a faster cell replication than the wild-type parentals. Contrary to that, 79HF6RETO cells grew slower than 79HF6WT. These dissimilar growth patterns are in part a consequence of the development of chemoresistance of both tumor entities, which are biochemically dissimilar.
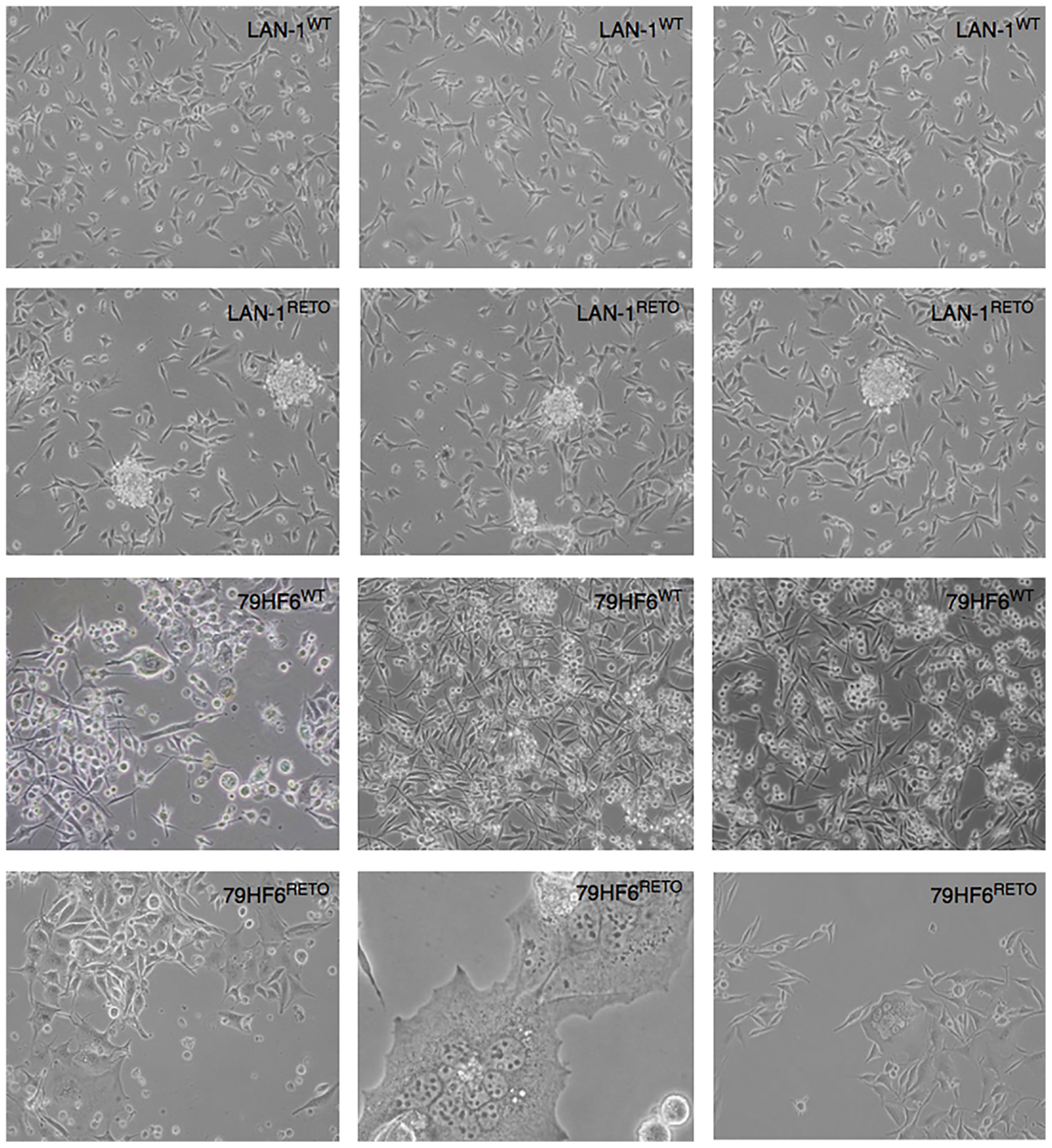
Figure 1. Neurosphere-forming cells. LAN-1WT/RETO neuroblastoma cells and 79HF6WT/RETO glioblastoma cells have shown different morphology in serum-free neurobasal A medium. Resistant LAN-1 subline showed the capacity to form neurospheres, whereas their wild-type parental cells are not able to form spheres. 79HF6WT/RETO cells did not alter their growth pattern once incubated in this defined condition. Magnification: LAN-1, 10×; 79HF6WT, 10×; 79HF6RETO, 10×; central picture, 20×. Pictures are representative for different experiments.
Effectiveness of Heavy Ion Irradiation in Comparison to X-Ray
As previously published by our group and others, resistance to etoposide induces radioresistance in both LAN-1 neuroblastoma and glioblastoma 79HF6 cell lines (5). To explore how heavy ion irradiation has advantages over the conventional X-ray exposures, we monitored the cell survival of these cells exposure to Carbon and Nickel ion irradiation.
The survival curves showed that heavy ion irradiation was more effective than X-ray in all four cell lines (Figures 2 and 3). The RBE values of heavy ions beam relative to X-rays at the D10 values were from 2.3 to 2.6 for LAN-1 cells and 2.5 to 3.4 for 79HF6 cells (Table 1). For LAN-1 cells, the etoposide-resistant subtypes (cultured in the presence of etoposide) were found to be more radioresistant than WT cells (cultured without etoposide) after X-ray and heavy ion irradiation (Figure 2), but for 79HF6 cells, RETO cells are more sensitive than WT cells after X-ray and heavy ion irradiation (Figure 3).
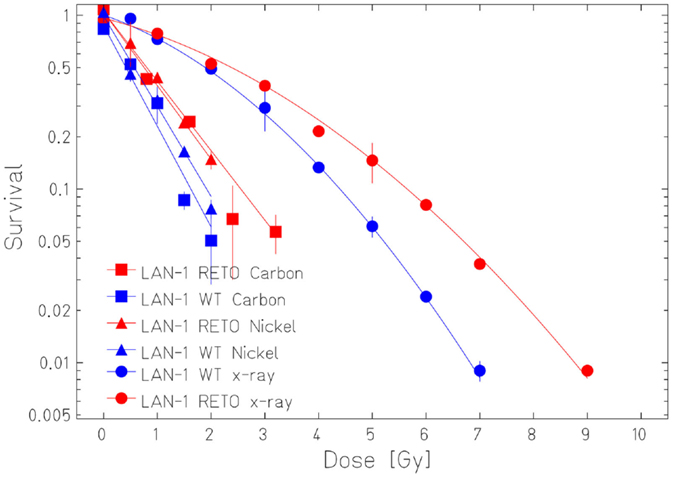
Figure 2. Survival of LAN-1 neuroblastoma cells after ionizing irradiation. LAN-1WT/RETO cells were irradiated with X-ray, Nickel (174 keV/μm), and Carbon-SIS (100 keV/μm). Graphic depicts the results of three independent experiments. Points, the mean survival fractions; bars, SD.
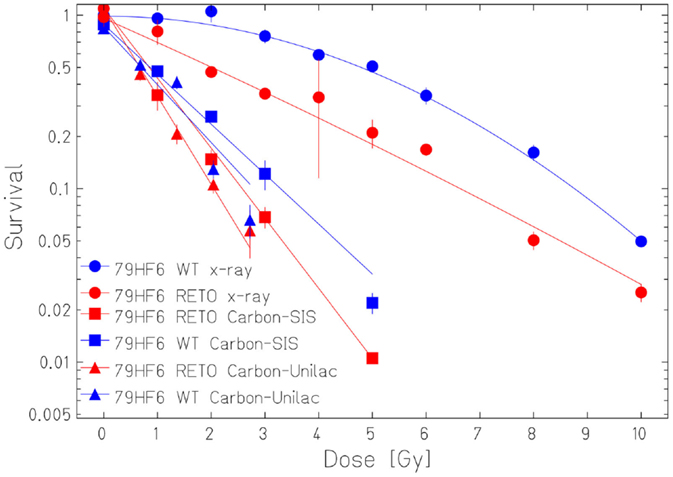
Figure 3. Survival of 79HF6 glioblastoma cells after ionizing irradiation. 79HF6WT/RETO cells were irradiated with X-ray, Carbon-SIS (100 keV/μm), and Carbon-Unilac (168 keV/μm). Graphic depicts the results of three independent experiments. Points, the mean survival fractions; bars, SD.

Table 1. Relative biological effectiveness values of heavy ions in LAN-1 neuroblastoma and 79HF6 glioblastoma cells.
Chromosomal Aberrations Found in LAN-1 Neuroblastoma and Glioblastoma 79HF6 Cell Lines
In order to search for the cause of radioresistance, we analyzed the karyotype of all cells used in our study. Most of LAN− cells had 40–44 chromosomes, and mFISH showed some stable aberrations, especially on chromosome 10 with an unstained region (Figure 4), and mBAND showed the unstained region located on 10p (Figure 5).
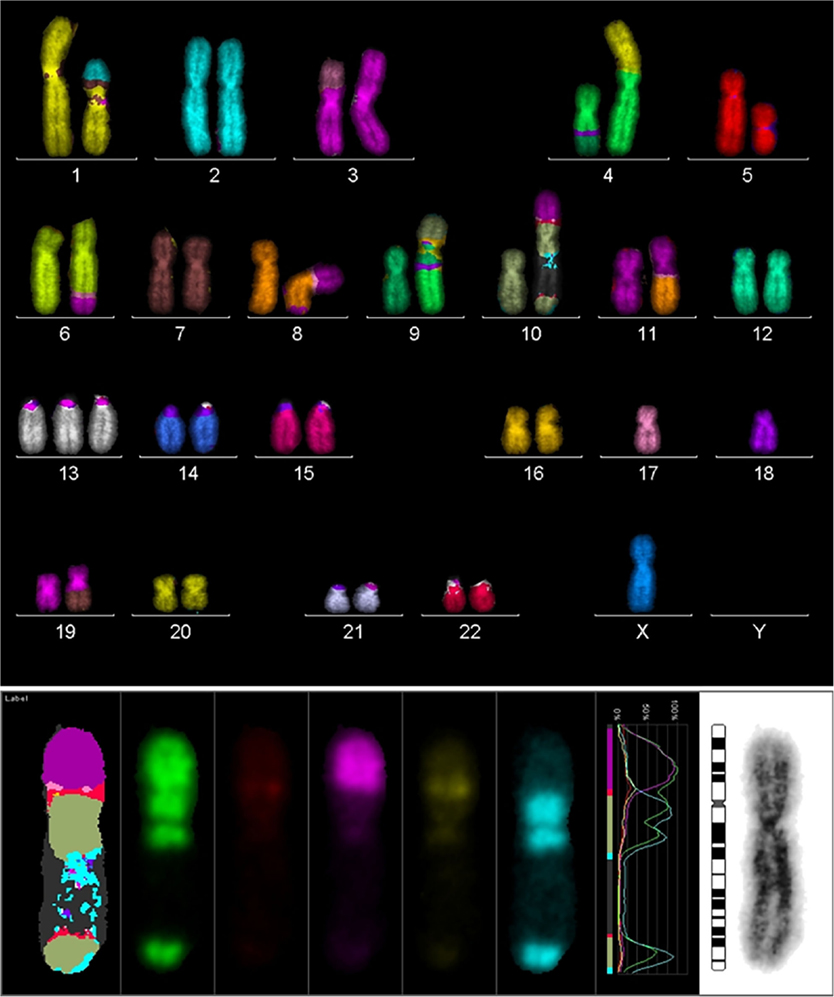
Figure 4. Karyotype of LAN-1 neuroblastoma cells analyzed by mFISH. LAN-1 cells revealed a range of 40–44 chromosomes per cell. Besides the segmental exchange between chromosomes, the most prominent observation in this cell line was an unstained region in chromosome 10. Structurally, chromosome 10 appeared to be intact (lower picture) once stained with Giemsa, but lower arms were not able to hybridize with the corresponding probes. Picture is representative for several cells analyzed under the same conditions. Magnification 100×.
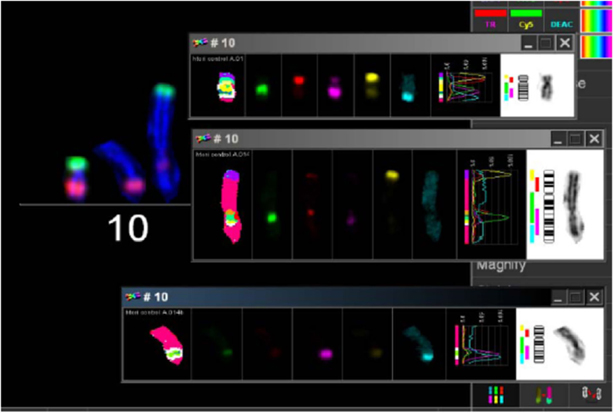
Figure 5. mBAND analysis of chromosome 10 of LAN-1 neuroblastoma cells. As detected in the karyotype, LAN-1 cells showed an unstained region that was localized on 10p. This arm was not able to hybridize with the corresponding probes directed toward this region. Picture is representative for several cells analyzed under the same conditions. Magnification 100×.
The chromosomal number in 79HF6 glioblastoma cells enormously differed in the resistant subline. Most of the 79HF6WT cells revealed a number of 71–90 chromosomes, whereas 79HF6WT cells reflected 52–83 chromosomes. The chromosomal number of 79HF6RETO revealed two peaks (Figure 6). It indicated that when cells were exposed to etoposide, the chromosome had the tendency to decrease to its relative normal ploidy. This phenomenon could be explained as the effort of tumor cells on maintaining gene stability. Diploid chromosomal distribution is more stable compared with polyploidy numbers, especially when tumor cells suffer the injury of chemical agents or other stressors. Tumors cells carrying a polyploidy derived a subgroup with a more stable karyotype in order to maintain the gene stability.
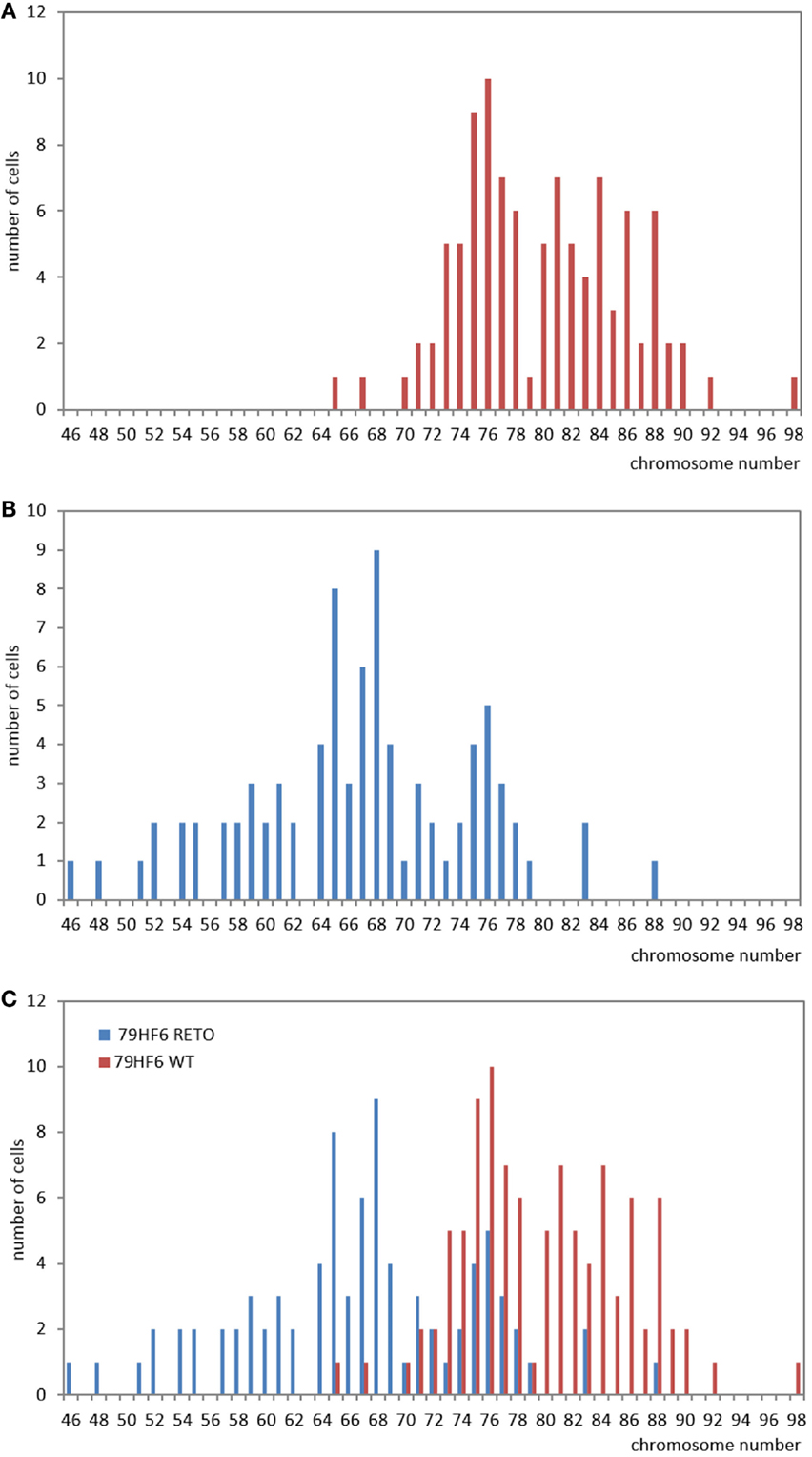
Figure 6. Studies on chromosome number in 79HF6 glioblastoma cells. 79HF6WT/RETO glioblastoma cells revealed strong differences in their chromosome number. Wild-type cells showed polyploidy (A). On the contrary, the resistant subline 79HF6RETO showed less numerical aberrations (B). Both diagrams were merged to provide an overall look of the chromosomal dispersion in both cells types (C). These differences were found to be highly significant once applied a descriptive statistic. Graphics are representative for several experiments with the same experimental conditions.
Discussion
Cancer stem cells show continuous self-renewal, extensive parenchymal migration/infiltration, and potential for full or partial differentiation in all cell types, which constitute a tumor. To explore how LAN-1 neuroblastoma and glioblastoma 79HF6 cell lines growth, we cultured these cells under optimal conditions for propagation in serum-free neurobasal A medium. It is known that under these conditions, cells displayed profound biological differences in growth patterns and were enforced to grow as non-adherent, multicellular spheres, inducing CSC-like populations (9).
Every cell type obviously showed different morphologies, as they were cultured in serum-free medium or serum-contained medium. Sphere formation assay (11, 12) performed to select CSC phenotypes from both cell types revealed that LAN-1RETO cells were able to form neurospheres after culturing them in serum-free medium, instead LAN-1WT, 79HF6WT, and 79HF6RETO cells were not capable to form neurospheres under the same conditions. CSCs may have the competence of durable self-renew, the capacity to develop and maintain tumor-related cell heterogeneity, differentiation, as well as the ability of both radioresistance and chemoresistance. To confirm the existence of CSC features, cells were transplanted to hosts and expected to induce tumor and maintain the features of parental tumor cells (13, 14). Studies in the past (5) have shown that both cell lines have CSC-like features, including chemoresistance and radioresistance to X-ray.
Our studies revealed that heavy ions had higher cell killing efficiency in both neuroblastoma and glioblastoma cell lines, despite its chemoresistance and chromosomal normality status. 79HF6WT cells were very resistant to X-ray. The survival rates of these cells were nearly not affected with 1 or 2 Gy of exposure to X-ray and were still around 5% with 10 Gy. This could explain why glioblastoma is so hard to be treated in clinic with conventional X-ray. However, RBE10 was 2.5–2.9, which indicates that heavy ions had notable advantages in killing radioresistant tumors as glioblastoma.
Our studies also revealed that LAN-1RETO cells were found more radioresistant to X-ray than LAN-1WT. Contrary to that, 79HF6RETO glioblastoma cells were less radioresistant than its wild-type parentals. The probable reason for this difference could be the different growth rates or the chromosomal number (15, 16). 79HF6RETO revealed a high variation in number of chromosomes in comparison to the wild-type cells that are more homogeneous. Thus, cells with a chromosomal abnormality in number are more sensible to ion irradiation. Although LAN-1RETO cells were made more resistant to radiation and were able to form neurospheres after exposure to etoposide, the opposite was true for 79HF6RETO. This may imply the higher level point that general CSC features are enhanced by etoposide in LAN-1 and CSC features may be reduced by etoposide in 79HF6. This difference could be caused by the inherent biological difference of neuroblastoma and glioblastoma. It may also be related to the concentration of etoposide. When the concentration is different, the results could change. Clearly, more work is needed to find the reason.
The biological hallmark of neuroblastomas is the complexity of the genetic abnormalities developed by the tumor cells, which are powerful prognostic markers. The most consistent abnormalities found in this tumor entity include ploidy changes, deletions of chromosome arms, amplification of the MYCN oncogene, and most frequently gains of chromosome arm 17q (6). There was a region in LAN-1 cells, which could not be stained by any fluorescence after mFISH procedure on chromosome no. 10. The analysis of mBAND corroborated that the unstained region was located on chromosome arm 10p. Homogeneously staining regions (HSRs) were localized on chromosome arm 10p and 10q of neuroblastoma cells with G-banding technique (17). HSR was cytogenetic evidence of a probable gene amplification. The unstained region of mFISH and mBAND of LAN-1WT cells was probably caused by repeated gene amplification or very short gene sequences. When gene segments are shorter than the probes of mFISH and mBAND, the fluorescently labeled probes could not properly anneal with the complementary sequences. Because of this, chromosome arm 10p showed an unlabeled region.
Recurrent loss of genetic material is normally found on chromosome arms 3p, 10p, 10q, 16q, and 20q in the hereditary neuroblastomas, in addition to regions usually deleted in sporadic neuroblastomas (1p36 and 11q) (18). These chromosomal sites may harbor tumor suppressor genes. Furthermore, loss of heterozygosis (LOH) at chromosome 10 is found exclusively at 10p11.23–p15.1 and consequently associated with MYCN-amplified Stage IV tumors in neuroblastoma tumors (19). LOH could induce different changes in one pair of alleles on certain gene and loss of part or even whole gene sequence of the allele. Usually, LOH is commonly related to the deficiency of tumor suppressor genes, i.e., p53. In the case where two alleles exist, the tumor suppressor gene will suppress the generation of tumors. In normal cells, when one allele is abnormal or lost, this defective suppression drive cells to immortalization. Thus, chromosome 10p probably encloses certain tumor suppressor genes. In our study, chromosome 17 reflects a deletion. Considering that p53 gene is located on this chromosome, the lack of p53 gene due to LOH induces a defeat of tumor suppressor functions in these cells. Previous studies showed that tumor suppressor genes, i.e., p53 and so on, could regulate cell survival and death (20, 21). Chromosomal aberrations in these cells subsequently affecting tumor suppressor gene expressing could influence the survival of cells.
In summary, heavy ion irradiation is more effective than X-ray for both untreated and chemoresistant tumor cell lines. For LAN-1 cells, the chemoresistant subpopulation LAN-1RETO is definitely more radioresistant than untreated cells (WT), while this effect was not found in 79HF6 cells.
Author Contributions
ZY: substantial contributions to the conception of the work, performing the experiments, analysis, and interpretation of data for the work and drafting the work. CH: substantial contributions to the conception of the work, performing the experiments, analysis. DP: substantial contributions to performing the experiments, analysis. WK-W: substantial contributions to the conception of the work, revising it critically for important intellectual content. G-LJ: substantial contributions to the conception of the work, revising it critically for important intellectual content. DD-C: substantial contributions to the conception of the work, revising it critically for important intellectual content. MD: substantial contributions to the conception of the work, revising it critically for important intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol (2012) 13(2):e83–9. doi: 10.1016/S1470-2045(11)70257-1
2. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene (2010) 29(34):4741–51. doi:10.1038/onc.2010.215
3. Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A (2009) 106(33):13820–5. doi:10.1073/pnas.0905718106
4. Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, et al. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther (2007) 6(2):532–41. doi:10.1158/1535-7163.MCT-06-0462
5. Díaz-Carballo D, Gustmann S, Jastrow H, Acikelli AH, Dammann P, Klein J, et al. Atypical cell populations associated with acquired resistance to cytostatics and cancer stem cell features: the role of mitochondria in nuclear encapsulation. DNA Cell Biol (2014) 33(11):749–74. doi:10.1089/dna.2014.2375
6. Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet (2007) 369(9579):2106–20. doi:10.1016/S0140-6736(07)60983-0
7. Evers P, Lee PP, DeMarco J, Agazaryan N, Sayre JW, Selch M, et al. Irradiation of the potential cancer stem cell niches in the adult brain improves progression-free survival of patients with malignant glioma. BMC Cancer (2010) 10:384. doi:10.1186/1471-2407-10-384
8. Pignalosa D, Durante M. Overcoming resistance of cancer stem cells. Lancet Oncol (2012) 13(5):e187–8. doi:10.1016/S1470-2045(12)70196-1
9. Barrantes-Freer A, Kim E, Bielanska J, Giese A, Mortensen LS, Schulz-Schaeffer WJ, et al. Human glioma-initiating cells show a distinct immature phenotype resembling but not identical to NG2 glia. J Neuropathol Exp Neurol (2013) 72(4):307–24. doi:10.1097/NEN.0b013e31828afdbd
10. Kraft G, Daues H, Fischer B, Kopf U, Liebold HP, Quis D, et al. Irradiation chamber and sample changes for biological samples. Nucl Instrum Methods (1980) 168:175–9. doi:10.1016/0029-554X(80)91249-5
11. Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci (1992) 12(11):4565–74.
12. Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science (1992) 255(5052):1707–10. doi:10.1126/science.1553558
13. Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev (2008) 18:48–53. doi:10.1016/j.gde.2008.01.017
14. Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol (2007) 23:675–99. doi:10.1146/annurev.cellbio.22.010305.104154
15. Yang X, Darling JL, McMillan TJ, Peacock JH, Steel GG. Heterogeneity of radiosensitivity in a human glioma cell line. Int J Radiat Oncol Biol Phys (1992) 22(1):103–8. doi:10.1016/0360-3016(92)90988-T
16. Till JE. Radiosensitivity and chromosome numbers in strain L mouse cells in tissue culture. Radiat Res (1961) 15:400–9. doi:10.2307/3571245
17. Sawyer JR, Miller JP, Roloson GJ. A novel reciprocal translocation (14;15)(q11;q24) in a congenital mesoblastic nephroma. Cancer Genet Cytogenet (1996) 88(1):39–42. doi:10.1016/0165-4608(95)00302-9
18. Altura RA, Maris JM, Li H, Boyett JM, Brodeur GM, Look AT. Novel regions of chromosomal loss in familial neuroblastoma by comparative genomic hybridization. Genes Chromosomes Cancer (1997) 19(3):176–84. doi:10.1002/(SICI)1098-2264(199707)19:3<176::AID-GCC7>3.0.CO;2-V
19. Mora J, Cheung NK, Oplanich S, Chen L, Gerald WL. Novel regions of allelic imbalance identified by genome-wide analysis of neuroblastoma. Cancer Res (2002) 62(6):1761–7.
20. Wee KB, Aguda BD. Akt versus p53 in a network of oncogenes and tumor suppressor genes regulating cell survival and death. Biophys J (2006) 91(3):857–65. doi:10.1529/biophysj.105.077693
21. Tarunina M, Alger L, Chu G, Munger K, Gudkov A, Jat PS. Functional genetic screen for genes involved in senescence: role of Tid1, a homologue of the Drosophila tumor suppressor l(2)tid, in senescence and cell survival. Mol Cell Biol (2004) 24(24):10792–801. doi:10.1128/MCB.24.24.10792-10801.2004
Keywords: chemoresistance, X-ray and heavy ion irradiation, relative biological effectiveness, neuroblastoma, glioblastoma
Citation: Yu Z, Hartel C, Pignalosa D, Kraft-Weyrather W, Jiang G-L, Diaz-Carballo D and Durante M (2016) The Effect of X-Ray and Heavy Ions Radiations on Chemotherapy Refractory Tumor Cells. Front. Oncol. 6:64. doi: 10.3389/fonc.2016.00064
Received: 30 September 2015; Accepted: 07 March 2016;
Published: 29 March 2016
Edited by:
John Varlotto, University of Massachusetts Medical Center, USAReviewed by:
Wenyin Shi, Thomas Jefferson University, USAEvagelia C. Laiakis, Georgetown University, USA
John Eley, University of Maryland School of Medicine, USA
Copyright: © 2016 Yu, Hartel, Pignalosa, Kraft-Weyrather, Jiang, Diaz-Carballo and Durante. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhan Yu, zhan.yu@sphic.org.cn
 Zhan Yu
Zhan Yu Carola Hartel
Carola Hartel Diana Pignalosa1
Diana Pignalosa1 David Diaz-Carballo
David Diaz-Carballo Marco Durante
Marco Durante