
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 11 September 2017
Sec. Molecular and Cellular Oncology
Volume 7 - 2017 | https://doi.org/10.3389/fonc.2017.00203
This article is part of the Research Topic Cancer Models View all 14 articles
 Manuela Gaebler1†
Manuela Gaebler1† Alessandra Silvestri2†
Alessandra Silvestri2† Johannes Haybaeck3,4
Johannes Haybaeck3,4 Peter Reichardt1
Peter Reichardt1 Caitlin D. Lowery5
Caitlin D. Lowery5 Louis F. Stancato5
Louis F. Stancato5 Gabriele Zybarth2
Gabriele Zybarth2 Christian R. A. Regenbrecht2*
Christian R. A. Regenbrecht2*
Over the past decade, the development of new targeted therapeutics directed against specific molecular pathways involved in tumor cell proliferation and survival has allowed an essential improvement in carcinoma treatment. Unfortunately, the scenario is different for sarcomas, a group of malignant neoplasms originating from mesenchymal cells, for which the main therapeutic approach still consists in the combination of surgery, chemotherapy, and radiation therapy. The lack of innovative approaches in sarcoma treatment stems from the high degree of heterogeneity of this tumor type, with more that 70 different histopathological subtypes, and the limited knowledge of the molecular drivers of tumor development and progression. Currently, molecular therapies are available mainly for the treatment of gastrointestinal stromal tumor, a soft-tissue malignancy characterized by an activating mutation of the tyrosine kinase KIT. Since the first application of this approach, a strong effort has been made to understand sarcoma molecular alterations that can be potential targets for therapy. The low incidence combined with the high level of histopathological heterogeneity makes the development of clinical trials for sarcomas very challenging. For this reason, preclinical studies are needed to better understand tumor biology with the aim to develop new targeted therapeutics. Currently, these studies are mainly based on in vitro testing, since cell lines, and in particular patient-derived models, represent a reliable and easy to handle tool for investigation. In the present review, we summarize the most important models currently available in the field, focusing in particular on the three-dimensional spheroid/organoid model. This innovative approach for studying tumor biology better represents tissue architecture and cell–cell as well as cell–microenvironment crosstalk, which are fundamental steps for tumor cell proliferation and survival.
Cancer is a group of diseases with a multitude of genomic aberrations typically classified by the cell of origin. Solid malignant neoplasms are predominantly carcinomas, which derive from epithelial cells, while a far less frequent group of solid neoplasms originates from mesenchymal cells. Normal mesenchymal cells form the soft and connective tissues as well as the bones. Tumors stemming from these cells are called sarcomas. They are malignant in most cases, and while their incidence in adults ranges from 1 to 2% (1–4), they account for up to 15% of all childhood and adolescence cancers (2, 3). Two main groups can be subdivided: soft-tissue sarcomas (STS) are more common in adults and represent 87% of all sarcomas, while sarcomas of the bone [osteosarcomas, Ewing sarcomas (EWS), and chondrosarcomas] occur more often below the age of 20 years (4, 5). Currently, the American cancer registry reports 4.2 cases per 100,000 for STS and 1.0 per 100,000 for sarcomas of the bone (6). Similar incidence rates have been reported for Europe (5, 7–9). Based on these numbers and according common definitions (10), sarcomas meet the criteria of rare diseases.
As for any rare disease, diagnostics and treatment should take place in specialized centers (7–9). Despite increased survival resulting from numerous multidisciplinary curative and palliative treatment options including surgery, monodrug or multidrug chemotherapy and/or targeted therapy, radiation therapy, hyperthermia, and isolated limb perfusion in a neoadjuvant or adjuvant setting (7–9), the disease outcome is often fatal. Currently, the 5-year relative survival rate for a patient with sarcoma considering the type, stage, localization, and age is about 60% (5) but dramatically dropping to 10% when only patients with advanced stages are considered (11). Due to the limited availability of tumor tissue for research and the complexity of the disease, progress in clinical management of sarcomas is lagging behind that of carcinomas. Since the lack of effective treatment options contributes to the low survival rate, the need for improving the treatment is evident.
Sarcomas could stem from virtually any mesenchymal cell in the body, and new pathological and molecular methods used for tumor classification currently allow for the distinction of more than 70 histopathological subtypes (1, 2, 12, 13). This high degree of heterogeneity combined with low incidence makes systematic research of sarcomas scientifically challenging.
A large group of sarcomas develop spontaneously, but environmental and predisposing genomic factors have been found to increasing the risk of contracting this kind of tumor. For example, Kaposi sarcomas are known to be HIV or human herpes virus 8 induced (14). Common risk factors known to be causative for many malignancies such as exposure to certain environmental pollutants and chemicals, ionizing radiation (often in form of a previous radiotherapy), and inherited genetic aberrations are also confirmed to play a role in sarcomas (Table 1). Sarcomas can be classified based on their genomics into genetically simple and genetically complex sarcomas (15, 16). Sarcomas of the genetically simple category (hypomutated) are characterized by only one disease-specific “driver” aberration such as a translocation or mutation (Table 2) and are more common in younger patients. Most of the known translocations result in fusion genes which code for transcription or growth factors (15). Identifying these translocations is of great value to the pathologist, as they allow for a confirmed diagnosis where simple histopathology alone is not definite. For example, detecting the amplification of MDM2 helps to confirm the diagnosis of a well-differentiated or dedifferentiated liposarcoma (17, 18). The genetically complex group (hypermutated) is made up by more or less chaotic karyotypes with high mutation frequencies in key oncogenes and tumor suppressor genes like TP53 or RB1 (15, 16). These complex genomic aberrations are commonly found in adult patients and/or as secondary lesions after radiation exposure (15) (Table 2).
Moreover, there are ongoing discussions about other potential risk factors for sarcoma development. Congenital or acquired immunodeficiency and, interestingly, also hernias seem to have suggestive evidence (3, 4). While often a trauma is reported in the patients’ medical history, publication showed that there is no such causative link between injury and sarcoma development, except for fibrosarcoma, dermatofibrosarcoma, and for patients with Gardner’s syndrome who underwent surgery (19).
Gastrointestinal stromal tumors represent approximately 18% of all sarcomas and are the most common mesenchymal neoplasms of the gastrointestinal tract. Historically, GISTs have a poor prognosis with tumor recurrence within 5 years after complete resection in up to 50% of patients. An important improvement in the management of this neoplasm was achieved in 1998 due to the discovery of oncogenic mutations in the tyrosine kinase KIT (20). The subsequent development and exploitation of kinase inhibitors that specifically downregulate this aberrant signal transduction pathway improved GIST patient outcome (21, 22) and made this approach a model for treating sarcoma by targeting altered intracellular signaling molecules. In some cases, a specific treatment can now be selected to target a mutation in a molecular pathway if a drug targeting this pathway is available, even if this drug was originally approved for a different tumor type. An important example is imatinib, a kinase inhibitor originally approved for the treatment of patients with BCR-ABL-positive chronic myeloid leukemia, which is also a very effective inhibitor of KIT and thus showed increased efficacy in KIT-mutated GIST. However, further investigation of mutation status in GIST has revealed a specific mutation (PDGFRA D842V) that according to current guidelines mostly prohibits the use of imatinib (9) as patients with this mutation harbor a primary resistance to this drug (21).
Irrespective of the clinical characteristics and in contrast to carcinomas, which arise from epithelial cells and are well defined by their tissue of origin, sarcomas are a group of highly heterogeneous tumors and evidence suggest that they develop directly from mesenchymal stem cells (MSCs) (23). MSCs are multipotent precursor cells of mesenchymal tissues such as bone, cartilage, fat, and muscle; several studies indicate their involvement in sarcomagenesis. Based on the wide variety of sarcoma subtypes, the origin of these tumors can be explained by two different hypotheses: the development of malignant alterations in a committed cell, distinct for every sarcoma subtype, or the presence of a common multipotent cell of origin that after transformation can differentiate into specific lineages (Figure 1).
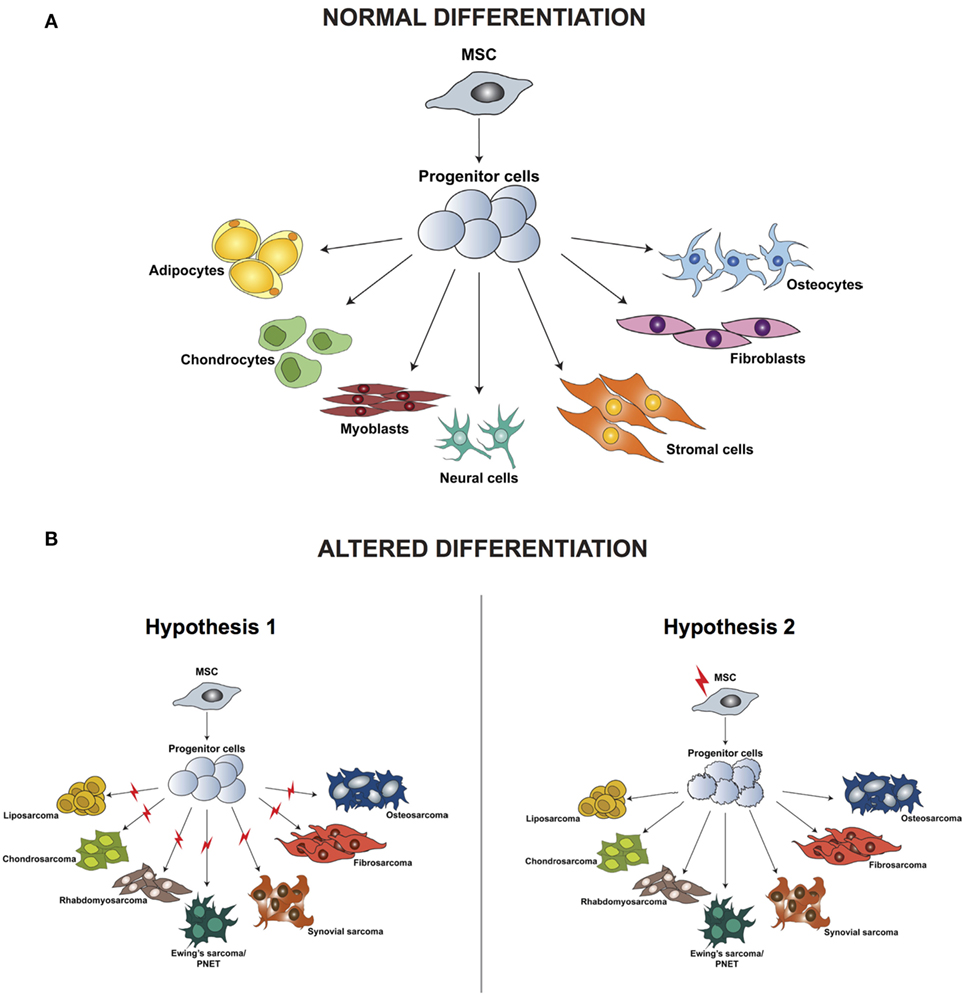
Figure 1. Differentiation of normal mesenchymal stem cells (A) and altered differentiation (B). (B) The difference between the two hypotheses, whereby the initiating aberration occurs either at a later stage of differentiation (hypothesis 1) or hits the stem cell (hypothesis 2). Modified from the study by Teicher (24).
According to the first hypothesis, tumors with a distinct phenotype and grade develop based on the basis of the lineage and of the differentiation stage when the initiating mutation occurs. This hypothesis is supported by studies in which the comparison between gene expression profiles of sarcomas and tissue-specific differentiation stages of MSCs showed a signature overlap in tumor and normal tissue according to their lineage of differentiation (25–29). One of the limitations of these analyses is the fact that they are based on in vitro cell culturing, which is known to induce alterations in the gene expression profile, thereby introducing a bias in the results. Moreover, it has been demonstrated that cells of a specific sarcoma subtype can differentiate into multiple lineages in vitro when specific inducing factors are added, thus indicating that not only the cell of origin but also the tumor microenvironment is fundamental for determining the final tumor phenotype.
Increasing evidence indicates that sarcomagenesis might be initiated by an aberration in a multipotent cell, and this hypothesis is currently favored by most researchers in the field. Several studies have demonstrated that mouse and human transformed MSCs can give rise to sarcomas after transplantation into mice. Miura and coworkers (30) showed that murine bone marrow-derived mesenchymal stem cells (BMMSCs) undergo spontaneous malignant transformation after prolonged culture (passage 29–54). Moreover, when injected in mice, these cells can form fibrosarcomas.
To test if MSCs are able not only to develop tumors when injected in mice after transformation but also to directly transform in vivo, Li et al. (31) transplanted bone marrow or MSC from male C57BL/6J mice into transgenic mice expressing a non-mammalian beta-gal enzyme (ROSA), chicken h-actin-enhanced GFP, and into WT littermates with bone marrow or MSC from male C57BL/6J mice as a control. After 18–24 months from transplantation, fibrosarcomas were the most common tumor detected, and immunohistochemistry analysis demonstrated that these tumors were derived from the transplanted bone marrow.
Compared to murine cells, human BMMSCs showed senescence without immortalization indicating that human MSCs cannot spontaneously transform (30); therefore, to translate the results obtained in mouse models, transformation of human MSC prior to inoculation is required. Genetic approaches aimed to knock out tumor suppressor genes and overexpress specific oncogenes have been used to induce MSCs transformation. The most common way to transform normal cells into malignant counterparts is the endogenous expression of human telomerase reverse transcriptase, simian virus 40 large T antigen (SV40-LT), and oncogenic H-RAS (32–34). Li et al. (35) applied this approach to study the origin of osteosarcoma. By using hMSC, they established cell lines by serially introducing these genetic alterations, and the effect of this manipulation on cellular phenotype, gene expression profiles, karyotype, and multilineage differentiation capacity was compared to osteosarcoma. They showed that two distinct genotypic and phenotypic sarcoma cell lines developed from these genetic events and that the transformed cells were characterized by increased motility. Moreover, transformed cells could be induced, so that osteogenic, adipogenic, and chondrogenic differentiation occurred, demonstrating that their multilineage differentiation potential was maintained.
Other groups studied MSC as cell of origin of osteosarcoma. Mohseny et al. (36) deeply characterized murine MSCs, transformed MSCs, and derived osteosarcoma cells lines genetically, phenotypically, and functionally, as well as for mRNA and protein expression. They identified aneuploidization, translocation, and homozygous loss of the cdkn2 region as the key mediators of MSC transformation. In a cohort of 88 osteosarcoma patients, they showed a correlation between CDKN2A/p16 protein expression and prognosis, thus linking murine MSC model to human osteosarcoma. The genetic alterations that were found in both the in vitro cultured tumorigenic MSCs and the derived mouse tumors demonstrated that osteosarcoma could originate from MSCs. Interestingly, the fact that these cells could differentiate in vitro to chondrocytes and adipocytes but were prone to form osteosarcomas in vivo reveals the importance of the tumor microenvironment in determining the final tumor phenotype.
Finally, it has been shown that a subgroup of these multipotent cells express not only mesenchymal markers but also stem cell markers such as OCT3/4, NANOG, and SOX (37, 38) and that they are associated with drug resistance and metastasis development (39, 40). Taken together, these data suggest that MSCs might be not only the sarcoma initiating cells but also, due to their stemness, the cells responsible for maintaining tumor growth.
Besides earlier detection by novel imaging techniques, the overall survival of sarcoma patients has not improved in the last 30 years. This is mainly due to the lack of understanding of the biological consequences of the genomic alterations involved in sarcomagenesis. Therefore, it is clear that a better understanding of human sarcoma tumorigenesis and metastasis is pivotal to improve the management of sarcoma patients in terms of new therapeutic targets and approaches. Since each sarcoma subtype is characterized by a low incidence, the development of clinical trials is challenging, and the results are often biased by the limited number of patients involved (16). These limitations related to the nature of sarcomas make interdisciplinary approaches indispensable and the development of reliable preclinical models for molecular analysis and research of potential targetable nodes a priority. Even with technologies such as next-generation sequencing finding their way into the field of pathology, only the detailed understanding of the biology of sarcomas will foster new insights and as consequence translate to more effective therapeutic regimens in the clinic.
Recently, the efficacy of molecular methods in improving sarcoma diagnosis was tested in a multicenter, prospective study. For this study, the diagnosis of 384 patients from 32 French sarcoma centers using histopathology exclusively versus a combination of histopathology plus molecular characterization was reevaluated. The authors reported that for 53 of the patients considered an improvement was obtained when the diagnosis made by an expert pathologist was revised according to molecular genetic testing (41). This underlines the importance of sarcoma molecular characterization and demonstrates that molecular testing could significantly increase diagnostic accuracy.
In recent years, it has been extensively demonstrated that malignant tumors are characterized by varying degrees of heterogeneity where not only the primary tumor but also the corresponding distant metastasis have distinct genetic profiles (42, 43). Considering this heterogeneity, searching for actionable mutations using only next-generation sequencing techniques may be very challenging, and the treatment of tumor cells with specific mutations by targeted therapy could select for specific subpopulations resistant to the initial therapy (44–46), making the combination of multiple drugs with different targets the most promising approach, aiming at the inhibition of tumor growth at multiple levels. For example, Patwardhan et al. reported that a selective c-Fms/KIT inhibitor in combination with an mTORC1 inhibitor could be more effective than the c-Fms/KIT inhibitor alone in reducing tumor growth in malignant peripheral nerve sheath tumors in cell lines and xenograft in vivo models (47). These recent findings in sarcoma biology have encouraged the sarcoma research community into developing new predictive models for improving sarcoma treatment.
Preclinical and translational studies of tumor mutations and aberrations as well as validation of therapeutic targets are based mainly on in vitro testing. Currently, the number of sarcoma models available for functional testing is still very limited, with only 2% of commercially available cell lines derived from STS (48). Moreover, the cell lines available do not represent the diversity of sarcomas, but are limited to the most common groups like osteosarcoma, leiomyosarcoma, and rhabdomyosarcoma with a total lack of more rare subtypes such as alveolar soft part sarcoma.
Due to these limitations, several groups focused on the establishment of new sarcoma cell lines. More than three decades ago, Bruland and coworkers isolated primary cells from 11 primary and metastatic human sarcoma specimens by enzymatic dissociation (49). Since the general success rate of sarcoma cell isolation was limited, they developed an alternative procedure using a non-adherent cell cultivation method (cellular spheroids) to the classical monolayer culture. With this approach, they produced stable monolayer cultures in 5 of the 11 samples used. These cells formed colonies in clonogenic soft-agar assays and developed tumors upon subcutaneous injection into nude mice. In 2002, additional cell lines were established, the majority from lung metastatic specimens derived from different sarcoma subtypes (50). In this study, all 11 cell lines analyzed expressed VEGF and basic-FGF, and they grew in anchorage-independent conditions. Moreover, when injected intramuscularly, six of the cell lines tested formed tumors and five of these spontaneously developed lung metastases, thus demonstrating the retention of tumorigenic and metastatic potential of the original tumor.
Recently, Salawa and coworkers established (48) primary cell cultures from fresh soft-tissue sarcoma samples with a success rate of 70%. For the seven long-term cell cultures that remained proliferative for at least 3 years and for more that 60 passages, they confirmed that the genomic and phenotypic characteristics were comparable to the original tumors. Since it is well known that long-term culture affects cell molecular characteristics, they analyzed the loss of heterozygosity (LOH) highlighting an increase of LOH after ~40 passages, thus demonstrating the presence of a genomic evolution commonly observed in in vitro cell cultures. Interestingly, three of the seven cell lines isolated were derived from undifferentiated pleomorphic sarcomas (UPSs) and the other four were derived from high-grade subtypes, suggesting a correlation between aggressive clinical course and the potential of in vitro growth.
Since studies based on the use of cancer cell lines often showed conflicting results, the characterization of the in vitro models used is very important, so that the results achieved by different laboratories can be compared. In 2010, the EuroBoNeT consortium characterized a set of 36 commonly used bone tumor cell lines (51), including osteosarcoma, EWS, and chondrosarcoma. After DNA fingerprint analysis to exclude cross-contamination of tumor cell lines, they showed that clones derived from the same original cell line (in this case, HOS) showed some differences from the parental line, suggesting a genomic evolution of the clones used in the study. Moreover, they highlighted a discrepancy between CDKN2A homozygous deletions in osteosarcoma and EWS cell lines (42 and 36%, respectively) compared to primary sarcoma samples, in which the frequency of this deletion is expected to be lower. This observation suggests that the cell line panel analyzed may be enriched in more aggressive tumors that easily grow in vitro. Finally, they analyzed the expression of TP53, a marker accepted for response to chemotherapy. They reported that 7 of the 10 TP53wt osteosarcoma cell lines showed low levels of TP53 mRNA transcripts and only weak or no staining for the corresponding protein. Moreover, this downregulation was present only in osteosarcoma cell lines but not in the other seven TP53wt bone tumor lines analyzed.
To select cell lines that are more representative of human osteosarcoma, the same research group further characterized the 19 osteosarcoma cell lines available in that study (52) by analyzing their ability to differentiate in vitro and their tumorigenic potential in nude mice. While the differentiation capacity toward osteoblasts, adipocytes, or chondrocytes was maintained in all cell lines with some cell lines able to differentiate in more than one lineage, only eight cell lines developed tumors after subcutaneous and intramuscular injection into nude mice. In mice injected with HOS-143B cell line, multiple lung metastasis was detected during autopsy, demonstrating the metastatic potential of these cells. Interestingly, the availability of the non-tumorigenic HOS parental line and the corresponding non-metastatic HOS-MNNG makes these lines an excellent model for studying osteosarcoma progression.
Since 2D in vitro models are inexpensive and relatively easy to generate and maintain, they have been broadly used in preclinical research. However, these models do not accurately recapitulate the three-dimensional (3D) structure of tumor tissues and the complex crosstalk between tumor cells and microenvironment.
Forcing cells to grow in 2D induces alterations in cell morphology that in turn translates in changes of the gene and protein expression, as well as cell behavior compared to the tissue of origin (53–55). These limitations are partially overcome by 3D cell cultures that represent the donor-tissues’ architecture including cell–cell and cell–matrix interactions and are thus valuable tools for investigating the influence of the microenvironment and gradients of nutrients and oxygen on the interplay of cells within a tumor and their response to drug treatment (56). Since little is known about the molecular biology of sarcomas including unknown contextual cross talk between signaling pathways and other components presumably including epigenetic modifications and regulatory RNA sequences, patient-derived sarcoma tumor models are desirable tools to fulfill the promises of personalized medicine.
Several reports mainly aimed at the study of the presence of cells with cancer stem cell characteristics and their role in sarcoma tumorigenesis, local relapse, metastasis, and therapy resistance were published demonstrating that sarcoma cells can grow in non-adherent conditions, forming 3D structures called spheroids (49, 57–59). In 2009, Fujii et al. showed that commercially available human sarcoma cell lines such as MG63 (osteosarcoma), HTB166 (EWS), and HT1080 (fibrosarcoma) are characterized by the ability to form sarcospheres with stem-like properties. Moreover, they showed that cells grown as spheroids were resistant to doxorubicin and cisplatin, drugs frequently used for sarcoma treatment (39).
In additional to immortalized cell lines, primary cells can also form sarcospheres when grown in non-adherent and serum-starved conditions. Salerno and coworkers demonstrated that isolated tumor spheres were tumorigenic after transplantation into mice and that the tumors formed recapitulated the corresponding human disease (58). In addition, they showed that by modification of cell culture conditions, it was possible to influence the growth of the sarcospheres. Mimicking the tumor microenvironment by reducing O2 conditions to 1% induced a significant increase in the number and the size of the spheres demonstrating that 3D sarcoma models are useful tools for studying sarcoma development due to their flexibility.
To better model morphology, growth kinetics, and protein expression profiles of human tumors, Fong et al. (59) established an ex vivo 3D model of EWS by culturing TC-71 cells in porous 3D electrospun poly(ε-caprolactone) scaffolds. After a 20-day culture, a well-differentiated EWS-like phenotype was preserved in this in vitro 3D model as verified by expression of diagnostic markers such as CD99, keratin, and smooth muscle actin. Considering that one of the most promising new treatment strategies in EWS is the inhibition of the IGF-1R/mTOR pathway, they showed that the activation of IGF-1R/mTOR signal was higher in the 3D model, compared to the 2D counterpart, suggesting that the 3D microenvironment has a more physiological effect on the intracellular signaling cascade. Finally, they tested the sensitivity of this 3D model to doxorubicin, a cytotoxic chemotherapeutic agent used in EWS treatment. Similar to the lower sensitivity observed in xenografts, an increased resistance was observed in the 3D model compared to 2D cells. Taken together, these data demonstrated that EWS 3D models are useful and reliable tools for evaluating new IGF-1R antagonists not only as single agents but also in combined strategies. Moreover, since they better mimic the tumor microenvironment, they provide important information for identifying new signaling nodes that can represent potential targets for therapeutic intervention.
One of the main characteristics of EWS cells is the recruitment and activation of osteoclasts, leading to the destruction of bone tissue by osteoclast-mediated osteolysis. As this process is crucial, several groups focused on the development of in vitro models of bone osteolysis by coculturing tumor cells with osteoclasts and osteoblasts. Recently, Villasante et al. (60) engineered a healthy bone tissue by co-culturing osteoblasts derived from hMSC and osteoclasts derived from monocytes isolated from blood samples. First, hMSC were seeded within a decellularized bone scaffold and differentiated toward osteoblasts. CD14+ monocytes were then cocultured with osteoblast and differentiated in osteoclasts. EWS aggregates were finally infused into the tissue-engineered bone and maintained in culture. The analysis of the bone microenvironment highlighted a decrease in bone density, connectivity, and matrix deposition in the presence of EWS cells. Moreover, the treatment with antiosteolytic drugs inhibited this process limiting osteoclast-mediated bone resorption. Taken together these data highlight the feasibility of developing bone-mimicking models for the study of bone tumors and bone metastasis development. The possibility of using patient-derived induced pluripotent stem cells for developing the bone niche suggests the potential of using this model for creating personalized models useful for precision medicine.
In the last decade, several 3D cell culture models have been developed to study different aspects of tumor biology and to test the efficacy of new anticancer molecules. While these approaches have mainly been established using carcinoma cells, with little effort, they can be also applied to the study of sarcoma biology. The least complex and therefore most frequently used models are based on spontaneous cell aggregation where the reduction of the adhesive forces to the surface of the culture plate allows cells to adhere spontaneously to each other forming cellular spheroids (Figure 2A). These models can be maintained either by using non-adhesive surfaces or spinner flasks and gyratory rotators, the use of hanging drop cultures, embedding of tumor cells in hydrogel matrices, or by the use of microcarrier beads and scaffolds. To avoid cell adhesion to the substrate surface, non-adhesive surfaces (Figure 3A) can be generated by using coatings such as agarose, polyHEMA, positively charged polystyrene, or proteoglycans (61–63). More recently, culture plates with modified surface chemistry have been developed allowing for “out-of-the-box”-ready technology and more reproducible growth of cellular spheroids. Besides the biological limitations, the main technical limitation of this method is the formation of spheroids with variable size and the inability to process upscaling.
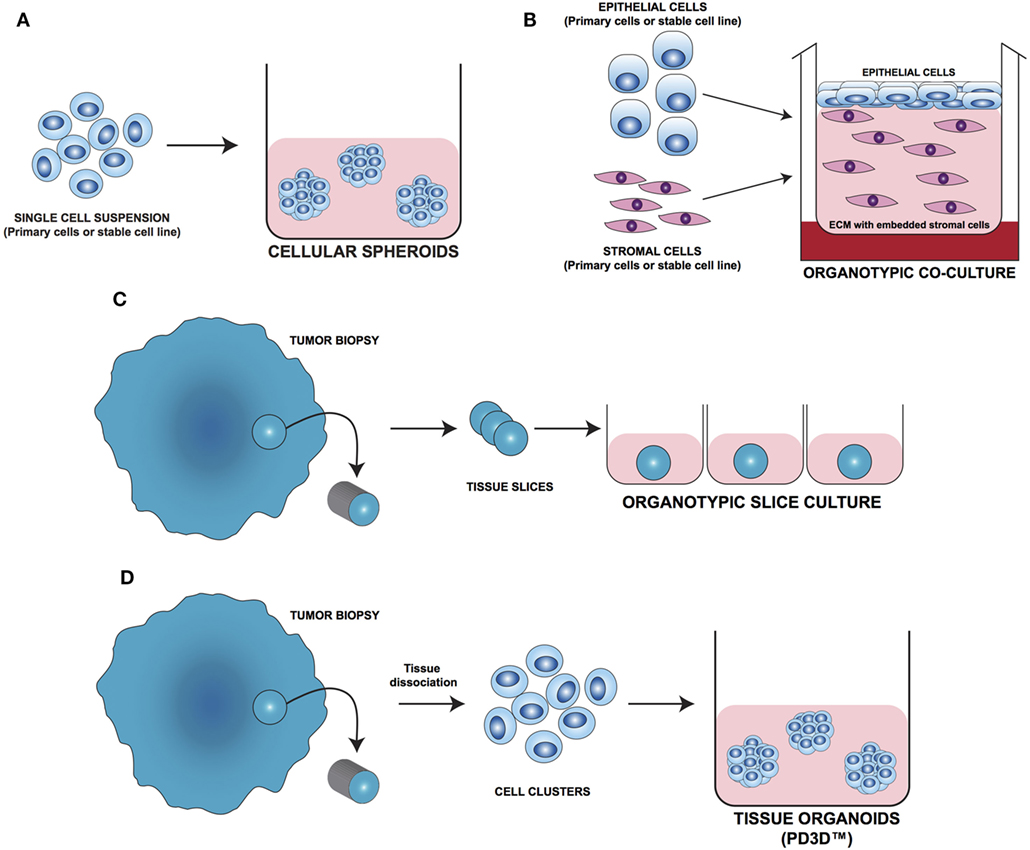
Figure 2. Different approaches for 3D cell culture model development. (A) Cellular spheroids: single cells from primary or stable cell lines aggregate together forming 3D structures. (B) Organotypic coculture: epithelial cells are cocultured with stroma cells embedded in a supporting matrix. (C) Organotypic slice culture: tissue slices obtained from the whole organ or from fragments of it are directly cultivated ex vivo. (D) Tissue organoids (PD3D™): primary cells isolated from fresh tissue without prior cell enrichment are grown as 3D multicellular structures [Modified from Silvestri et al. (64)].
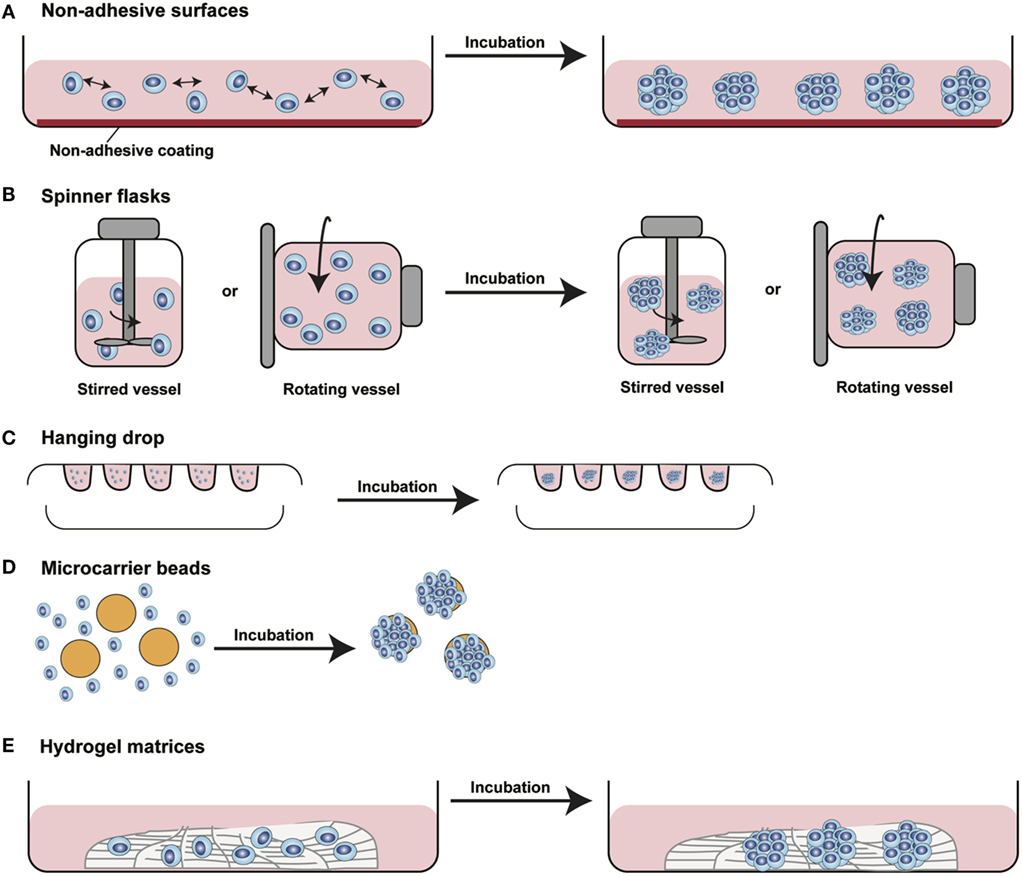
Figure 3. Different methods for 3D spheroids development and growth. (A) Non-adhesive surfaces: culture plates with modified surfaces to reduce cell adhesion stimulate cell aggregation and formation of 3D structures. (B) Spinner flasks: stirred or rotating vessels are used to prevent cell adhesion to the surface of the plate allowing 3D spheroids formation. (C) Hanging drop: cells seeded in small drops of medium form cellular aggregates at the tip of the drop due to gravity forces. (D) Microcarrier beads: cells adhere to and proliferate on the surface of natural or synthetic solid beads forming 3D structures. (E) Hydrogel matrices: cells are seeded into matrices of natural or synthetic origin forming 3D structures by single cells aggregation or by monoclonal cell growth. [Modified from Silvestri et al. (64)].
The aforementioned limitations are partially overcome using spinner flasks or gyratory rotator (Figure 3B) systems, bioreactors that allow continuous mixing of medium or a constant rotatory movement of the flask, which prevents cell adhesion (65). These methods allow massive production of spheroids, therefore representing the method of choice for growing high amounts of homogeneous spheroids for downstream applications.
A technique often used is the so-called hanging drop method (Figure 3C) that makes use of gravity to stimulate cell aggregation. Cells in suspension are plated in small drops onto the underside of a plate lid that is then carefully inverted. Due to gravity, the cells accumulate in the tip of the drop, forming spheroidal aggregates (66). For those cells that do not spontaneously aggregate, systems that facilitate cell to cell interaction have been developed. Microcarrier beads (Figure 3D) are characterized by differences in size and composition. Surface coating allows adhesion and proliferation of cells consequently forming minispheroids that in turn aggregate one to each other, thus forming bigger spheroids (67). Another system to facilitate cell aggregation is the use of solid scaffolds with different porosity composed by natural or synthetic materials such as collagen, chitosan, or d,d,-l,l-polylactic acid. After seeding, cells can migrate along the surface, aggregate, and create 3D structures (68).
As tumor cells do not exist as isolated entities but rather are part of a complex microenvironment, natural or synthetic hydrogel matrices (Figure 3E) that mimic the in vivo tissue architecture can be used to grow tumor cells in 3D structures. The choice of a naturally or synthetically composed gel can be based on the aim of the analysis, ranging from single component hydrogels, i.e., laminin, collagen, and fibronectin to more complex ones such as Matrigel™ or Puramatrix™ (69, 70). Tumor cells can also be grown together with other tissue components such as stroma and epithelial cells in organotypic cocultures (Figure 2B). This more complex model allows to study the influence of tumor microenvironment on tumor development and progression as well as on drug sensitivity (71–74).
One of the most important tools in medical research is the model able to mimic the physiological situation in the closest way possible. Since they are commercially available and easy to handle, most of the basic and preclinical research in the oncological field was done using immortalized cell lines. On the downside, long-term in vivo culturing and the immortalization process often cause alterations in the molecular and phenotypic characteristics of these cells that can strongly differ from the cell of origin. To overcome these limitations, fresh tissue directly obtained during surgery has been used for isolating and cultivating tumor cells. One of the most straight forward methods is to cultivate fragments/slices of the tumor tissue as so-called organotypic slice cultures (Figure 2C). Several research groups use this method to study drug uptake, proliferation, and cell death, as well as for molecular characterization before and after treatment (75–77). The main advantage of this system is that the original tissue architecture is preserved, allowing the immediate study of the normal/altered physiology. Using tissue slices that can be maintained in culture for a short period of time only and without the opportunity of further expansion strongly limits the use of these models.
The currently most innovative and promising approach for in vitro model is represented by tumor tissue organoids (Figures 2D and 4). Organoids are multicellular structures directly isolated from primary tissue and grown in well-defined conditions. 3D organoids maintain the complex architecture of their tissue of origin and self-organize by reproducing their unique architecture and marker expression. This innovative tool has been used by several research groups for studying tumor development and progression and for testing drug efficacy (78–81). Interestingly, it has been recently demonstrated that this tumor model can be easily applied to high-throughput drug screening (82) and to correlate patient’s tumor molecular profiles to drug sensitivity (83).
In 2010, Barretina and colleagues (84) performed an integrative system analysis of DNA sequence, copy number, and mRNA expression on 207 soft-tissue sarcoma samples including 7 major subtypes to identify novel subtype-specific genomic alterations representing potential therapeutic targets. They first studied genomic alterations in 47 tumor/normal DNA pairs highlighting 21 totally modified genes. These results were then validated in a second study-set of 160 tumors confirming the presence of subtype-specific mutations in several genes such as PIK3CA in myxoid/round cell liposarcoma, TP53 in pleomorphic liposarcoma, and NF1 in myxofibrosarcoma and pleomorphic liposarcoma. The data obtained are of high clinical potential since they helped identify tumors that might be responsive to PI3K or mTOR inhibitors, since NF1 loss causes mTOR pathway activation.
Recently, panel sequencing of 194 cancer-related genes in 25 STS was performed to identify actionable mutations (85). This analysis revealed the presence of different mutational profiles. In particular, in 60% of the cases targetable mutations for which clinical trials are available were highlighted while for another 28% of cases mutations which are currently not targetable were present. This study demonstrates the versatility of next-generation sequencing both in patient stratification for treatment with currently available therapeutics and in the identification of potential targets for developing new molecular treatments.
In the recent years, strong efforts have been made to find new biomarkers for selecting the best treatment based on specific tumor molecular profiles. With the goal of developing an efficient approach for patient stratification for treatment, Hanes and colleagues (86) combined tumor genomic characterization with drug testing in vitro in patient-derived cell lines. Three metastases from a patient with high-grade dedifferentiated liposarcoma previously treated with different chemotherapeutic agents were used for the study. Tumor tissues were analyzed by exome and transcriptome sequencing as well as DNA copy number analysis to highlight genomic aberrations that could represent potential targets for treatment. The data obtained were then used for selecting those drugs that can directly affect the altered gene or the corresponding signaling pathway in a cell line derived from the metastatic tissue. Among the altered genes observed in the tumor sample, an amplification of FRS2, the gene coding for fibroblast growth factor receptor substrate 2, was revealed. Based on this molecular alteration, they tested the in vitro efficacy of NVP-BGJ398 (infigratinib), a pan-FGFR inhibitor showing promising inhibition of proliferation induced by cell cycle arrest. Taken together, these data demonstrate the benefit of combining tumor genomic profiling with in vitro testing for a better selection of treatment in sarcoma patients and for selecting new promising treatments for improving sarcoma survival.
Even if specific mutations have been associated with certain sarcoma subtypes, their etiology remain largely unknown. An equally important approach in biomarker discovery is the analysis of the proteome. Since the proteome is a functional translation of the genome, the information provided by its in-depth analysis may be a key in understanding sarcoma progression and therapy failure. Several research groups focused on differential expression of proteins in tumor tissue compared to the normal counterpart using diverse technologies such as Digiwest (87), 2D-PAGE (86, 87), mass spectrometry (87), and array technology (88).
Developing new sarcoma diagnostic biomarkers, Suehara and colleagues used 2D difference electrophoresis (2D-DIGE) analysis performing global protein expression analysis in different histological subtypes of soft-tissue sarcoma. Profiling data highlighted a set of 67 proteins distinguishing the 80 sarcoma samples based on their histological classification. Moreover, a signature of five proteins was able to differentiate at time of that publication known as grade III malignant fibrous histocytomas (today classified as UPS) and leiomyosarcomas into low- and high-risk groups characterized by significantly different survival rates (88).
The same research group applied a combined 2D-DIGE and mass spectrometry approach for profiling patients with GIST characterized by good and poor clinical prognosis (89), demonstrating the potential of this marker in GIST clinical management. This analysis highlighted 43 proteins (spots) differentially expressed and corresponding to 25 distinct gene products. Among these proteins, the authors focused on pfetin, a potassium channel protein, since 8 of the 43 spots that were found derived from this protein, and 4 of these had discriminative power between the two groups. Pfetin expression and its correlation with tumor metastasis was confirmed by real-time PCR and western blot. Moreover, the authors demonstrated that pfetin expression and 5-year metastasis-free survival rate were directly correlating.
In another recent study, 59 rhabdomyosarcoma samples were microdissected to enrich tumor cell content and analyzed by reverse phase protein microarrays (90), an antibody-based technology useful in studying the level of expression of selected total and phosphoproteins. This study showed that the phosphorylation of several components of the Akt/mTOR pathway was increased in tumors from patients with short-term survival. Moreover, an altered relationship between insulin receptor substrate 1, and this pathway was highlighted in patients with poor survival. The significance of these results was demonstrated by treating mouse xenografts with CCI-779, an mTOR inhibitor, that compared to controls greatly reduced the growth of two different rhabdomyosarcoma cell lines. These data showed the utility of phosphoproteomic pathway mapping for the study of functional drivers of sarcoma progression and for selecting patients for anti-mTOR/IRS therapy. These and other proteomic studies in different sarcoma subtypes were extensively reviewed by Kondo and colleagues (91).
Genomic and proteomic approaches can be synergistically applied for a deeper understanding of tumor biology at molecular level. Integrating these profiling systems, it is possible to correlate the presence of tumor-specific mutations to functional alterations in intracellular pathways. The information obtained from a multiomics approach may help in both designing new targeted therapies and selecting the best treatment option for a specific patient.
Since no innovative therapeutic approaches are available for most sarcoma subtypes, several research groups focused on the discovery of new targets for sarcoma treatment by screening of compound libraries mainly on immortalized 2D cell lines and studying their effect on sarcoma cell biology.
As mentioned before, several sarcoma types are characterized by the presence of chromosomal translocations that cause the production of altered transcription factors. About 85% of EWSs express the EWS/FLI1 fusion protein, known to cause alterations in transcriptional regulation and RNA processing. EWS/FLI1 represents a very attractive drug target since it is specifically expressed by the tumor cells, but it is absent in the healthy tissue. Since, currently, no drugs targeting transcription factors are available, one approach is to directly or indirectly inhibit the players of the altered connected pathway. To this aim, Grohar and coworkers (92) screened more than 50,000 compounds in TC32 EWS cells for the ability of altering the expression level of the EWS/FLI1 downstream target NR0B1, that was prior transfected with a luciferase construct. The 200 compounds that showed activity in primary screening were further validated by multiplex PCR assay with the aim of selecting those hits that best inhibited the expression of a predetermined set of EWS/FLI1 downstream target genes. With this approach, they selected mithramycin as lead compound able to inhibit EWS/FLI1 activity. This effect was validated and further characterized in in vitro experiments and in in vivo xenograft models. Taken together, these data demonstrate the potential efficacy of this compound in treating EWS and the utility of applying high-throughput screening approaches for selecting new potential drug targets and new sarcoma therapies.
A similar study screened a small-molecule compound library containing FDA-approved drugs modulating the expression of EWS/FLI1 target genes on a panel of six EWS cell lines (93). To determine compound efficacy, the expression levels of few, well-characterized EWS/FLI1 target genes was measured. Among the 10 hits with the highest efficacy, several know therapeutic agents and fenretinide, currently in clinical trials for Ewing’s sarcoma, have been highlighted demonstrating the robustness of this screening approach. Moreover, midostaurin, a pan-kinase inhibitor, resulted in one of the most promising novel compounds. Interestingly, the efficacy of this drug was already shown in rhabdomyosarcoma, another pediatric sarcoma type. Moreover, midostaurin is currently undergoing phase II clinical trials for leukemia in adults and children, with a low toxicity in the pediatric population that make this drug a promising candidate to be tested in pediatric sarcoma patients.
The determination of new drug targets and efficient therapeutics requires even more the investigation of sarcoma subtypes that, unlike EWS, are not characterized by a known driver molecular alteration. Several groups tested available compound libraries to better characterize the drug sensitivity of different sarcoma subtypes and to correlate the response to specific compounds with the molecular characteristics of the tumor. Teicher and coworkers (94) screened the response of 63 sarcoma cell lines to 100 FDA-approved anticancer drugs and to a library of 345 investigational oncology agents. Moreover, they correlated treatment response with cell molecular profiles obtained by exon and microRNA arrays. The authors highlighted important correlations between cell characteristics such as sarcoma subtype and gene/miRNA expression, demonstrating that this screening approach is useful in studying the efficacy of FDA-approved drugs in specific sarcoma subtypes, in defining new potential therapeutic agents and for correlating sarcoma molecular profiles with drug sensitivity.
With such screening platforms available, it will become possible to investigate combination therapies for “vertical inhibition” of a single pathway or inhibition spanning multiple pathways.
Ever since the sequencing of the first human genome, hopes were high that knowledge of the cancer genome landscape would bring an end to cancer and other diseases. Yet, sequencing alone has proven to be “remarkably unhelpful,” and the belief that sequencing your DNA is going to extend your life is “a cruel illusion” as James Watson put it in a recent interview with the New York Times (95). Today, genome researchers still struggle to be able to sufficiently support clinical decision-making with meaningful sequence data, and to compensate for this deficit, they propose that “more is more” (96). Yet, these genome centrics are neither feasible in the clinical setting nor payable by the majority of patients and insurance companies. Using PD3D cell cultures and exploiting their phenomics by combining multiple layers of evidence is expected to soon become the state-of-the-art approach. All current reports share the assumption that short-term PD3D cell cultures have already proven their superior predictive value in the preclinical arena, ousting other in vitro models in the development of new drugs. Pauli et al. have reported that they can successfully establish 3D cell cultures from surgically removed specimen within weeks (97). Therefore, leading comprehensive cancer centers around the world have started including patient-specific cell culture data in their infrastructure, as detailed by Shraddha Chakrandhar in Nature Medicine (98). Drug screenings in an automated setup, as described in 2016 by Boehnke et al. (82), take about 1 week once enough cells are available. In parallel, ultra-deep targeted sequencing can be performed, focusing on only those mutations that are relevant for the clinical decision-making.
Of course, the panels of target genes to be sequenced has to be updated with latest clinically relevant information to ensure that oncologists can stay focused on the immediate needs of the patients. Protein extracts from before and after the drug screening can be used for methods like Digiwest, a bead-based, multiplexed western blot (87). With this method, a selection of up to 200 (phospho-)proteins can be quantified at once, providing differential information not only on expression levels but also on the activation of key signaling kinases, such as those along them TOR, WNT, MAPK, or PI3K axes. Taking into consideration the time frame in which additional information can be used to support the decision-making process and the nature of information that becomes available from measuring cellular phenomics for the discussion in tumor boards, PD3D models may indeed become an integral part in clinical oncology of the 21st century.
Recent research efforts in sarcoma has enabled important improvements in the knowledge of sarcoma histopathology that in turn defined sarcomas not as a single tumor entity but rather as different tumor subtypes with histology-specific molecular characteristics. The recent development of targeted therapies significantly contributed to the improved treatment options for sarcoma patients. Considering that this approach, first developed in carcinomas, showed efficacy in GIST, the next step in sarcoma research is to focus on the molecular characterization of the different subtypes to highlight new potential targets for therapy.
Since the availability of in vitro models that reliably represent the physiological tumor behavior is a prerequisite for successful sarcoma preclinical and translational research, several models have been developed for discovering new potential targets for therapeutic intervention. In particular, patient-derived cell lines and more recently 3D organoids represent innovative tools for studying molecular pathways promoting sarcoma development and tumor progression and for drug efficacy screenings. Moreover, these models could directly impact clinical decisions if and when used as a tool for precision medicine. 3D cultures directly isolated from a patient’s sarcoma could be used for testing the efficacy of drugs currently available, thus supporting clinicians in the selection of the most efficacious and promising treatment.
MG, AS, and CR drafted the original manuscript. CL provided the figures. JH, PR, LS, and GZ edited the draft and supported MG, AS, CR, and CL in finalizing the manuscript.
No aspect of the submitted work has received payment or services from a third party. No entity has influenced the content of the submitted work. CR is founder and CEO of cpo. AS and GZ are employed at cpo. cpo is a contract research company working as a service provider in the field of 3D in vitro models and drug screening. CL and LS work in research at Eli Lilly and Company. MG and PR work at the Sarcoma Center at HELIOS Kliniken Berlin-Buch. HELIOS operates multiple hospitals in Germany and Europe. PR reports grants and personal fees from Novartis, personal fees from Pfizer, personal fees from Bayer, personal fees from PharmaMar, personal fees from Amgen, personal fees from GlaxoSmthKline, personal fees from Clinigen, personal fees from Lilly, and personal fees from AstraZeneca. None of them has been influencing the submitted work. All other authors declare no conflict of interest.
This study was supported by Berliner Krebsgesellschaft e.V. grant to CR (grant #201402).
1. Mastrangelo G, Coindre J-M, Ducimetière F, Dei Tos AP, Fadda E, Blay J-Y, et al. Incidence of soft tissue sarcoma and beyond: a population-based prospective study in 3 European regions. Cancer (2012) 118:5339–48. doi:10.1002/cncr.27555
2. Ryan CW, Desai J. The past, present, and future of cytotoxic chemotherapy and pathway-directed targeted agents for soft tissue sarcoma. Am Soc Clin Oncol Educ Book (2013) 33:e386–93. doi:10.1200/EdBook_AM.2013.33.e386
3. Patel SR, Benjamin RS. Weichgewebe- und Knochensarkome und Knochenmetastasen. In: Suttorp N, Dietel M, editors. Harrison’s Principles of Internal Medicine (Harrisons Innere Medizin). Berlin: ABWWissenschaftsverlag GmbH, The McGraw-Hill Companies, Inc. (2013). p. 876–81.
4. Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res (2012) 2:14. doi:10.1186/2045-3329-2-14
5. Stiller CA, Trama A, Serraino D, Rossi S, Navarro C, Chirlaque MD, et al. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur J Cancer (2013) 49:684–95. doi:10.1016/j.ejca.2012.09.011
6. American Cancer Society. Special Section: Rare Cancers in Adults. Atlanta: American Cancer Society (2017).
7. The ESMO/European Sarcoma Network Working Group. Soft tissue and visceral sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol (2014) 25:iii102–12. doi:10.1093/annonc/mdu254
8. The ESMO/European Sarcoma Network Working Group. Bone sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol (2014) 25:iii113–23. doi:10.1093/annonc/mdu256
9. The ESMO/European Sarcoma Network Working Group. Gastrointestinal stromal tumours: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol (2014) 25:iii21–6. doi:10.1093/annonc/mdu255
10. Gatta G, van der Zwan JM, Casali PG, Siesling S, Dei Tos AP, Kunkler I, et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur J Cancer (2011) 47:2493–511. doi:10.1016/j.ejca.2011.08.008
11. Schütte J, Bauer S, Brodowicz T, Grünwald V, Hofer S, Hohenberger P, et al. Weichgewebssarkome des Erwachsenen. (2017). Available from: www.dgho.de
12. Fletcher CDM, Krishnan Unni K, Mertens F, editors. WHO Classification of Tumours, Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon (2002). p. 1–415.
13. Jo VY, Fletcher CDM. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology (2014) 46:95–104. doi:10.1097/PAT.0000000000000050
14. Curtiss P, Strazzulla LC, Friedman-Kien AE. An update on Kaposi’s sarcoma: epidemiology, pathogenesis and treatment. Dermatol Ther (Heidelb) (2016) 6:465–70. doi:10.1007/s13555-016-0152-3
15. Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer (2003) 3:685–94. doi:10.1038/nrc1168
16. Dancsok AR, Asleh-Aburaya K, Nielsen TO. Advances in sarcoma diagnostics and treatment. Oncotarget (2017) 8:7068–93. doi:10.18632/oncotarget.12548
17. Binh MBN, Sastre-Garau X, Guillou L, de Pinieux G, Terrier P, Lagacé R, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol (2005) 29:1340–7. doi:10.1097/01.pas.0000170343.09562.39
18. Italiano A, Bianchini L, Keslair F, Bonnafous S, Cardot-Leccia N, Coindre J-M, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer (2008) 122:2233–41. doi:10.1002/ijc.23380
19. Morrison BA. Soft tissue sarcomas of the extremities. Proc (Bayl Univ Med Cent) (2003) 16:285–90.
20. Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science (1998) 279:577–80. doi:10.1126/science.279.5350.577
21. Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer (2011) 11:865–78. doi:10.1038/nrc3143
22. Kurtovic-Kozaric A, Kugic A, Hasic A, Beslija S, Ceric T, Pasic A, et al. Long-term outcome of GIST patients treated with delayed imatinib therapy. Eur J Cancer (2017) 78:118–21. doi:10.1016/j.ejca.2017.03.024
23. Mohseny AB, Hogendoorn PCW. Concise review: mesenchymal tumors: when stem cells go mad. Stem Cells (2011) 29:397–403. doi:10.1002/stem.596
24. Teicher BA. Searching for molecular targets in sarcoma. Biochem Pharmacol (2012) 84:1–10. doi:10.1016/j.bcp.2012.02.009
25. Danielson LS, Menendez S, Attolini CS-O, Guijarro MV, Bisogna M, Wei J, et al. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol (2010) 177:908–17. doi:10.2353/ajpath.2010.091150
26. Matushansky I, Hernando E, Socci ND, Matos T, Mills J, Edgar MA, et al. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol (2008) 172:1069–80. doi:10.2353/ajpath.2008.070284
27. Cleton-Jansen A-M, Anninga JK, Briaire-de Bruijn IH, Romeo S, Oosting J, Egeler RM, et al. Profiling of high-grade central osteosarcoma and its putative progenitor cells identifies tumourigenic pathways. Br J Cancer (2009) 101:2064–2064. doi:10.1038/sj.bjc.6605482
28. Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet (2002) 359:1301–7. doi:10.1016/S0140-6736(02)08270-3
29. Boeuf S, Kunz P, Hennig T, Lehner B, Hogendoorn P, Bovée J, et al. A chondrogenic gene expression signature in mesenchymal stem cells is a classifier of conventional central chondrosarcoma. J Pathol (2008) 216:158–66. doi:10.1002/path.2389
30. Miura M, Miura Y, Padilla-Nash HM, Molinolo AA, Fu B, Patel V, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells (2006) 24:1095–103. doi:10.1634/stemcells.2005-0403
31. Li H, Fan X, Kovi RC, Jo Y, Moquin B, Konz R, et al. Spontaneous expression of embryonic factors and p53 point mutations in aged mesenchymal stem cells: a model of age-related tumorigenesis in mice. Cancer Res (2007) 67:10889–98. doi:10.1158/0008-5472.CAN-07-2665
32. Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature (1999) 400:464–8. doi:10.1038/22780
33. Rich JN, Guo C, McLendon RE, Bigner DD, Wang XF, Counter CM. A genetically tractable model of human glioma formation. Cancer Res (2001) 61:3556–60.
34. Shima Y, Okamoto T, Aoyama T, Yasura K, Ishibe T, Nishijo K, et al. In vitro transformation of mesenchymal stem cells by oncogenic H-rasVal12. Biochem Biophys Res Commun (2007) 353:60–6. doi:10.1016/j.bbrc.2006.11.137
35. Li N, Yang R, Zhang W, Dorfman H, Rao P, Gorlick R. Genetically transforming human mesenchymal stem cells to sarcomas: changes in cellular phenotype and multilineage differentiation potential. Cancer (2009) 115:4795–806. doi:10.1002/cncr.24519
36. Mohseny AB, Szuhai K, Romeo S, Buddingh EP, Briaire-de Bruijn I, de Jong D, et al. Osteosarcoma originates from mesenchymal stem cells in consequence of aneuploidization and genomic loss of Cdkn2. J Pathol (2009) 219:294–305. doi:10.1002/path.2603
37. Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, et al. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res (2010) 70:4602–12. doi:10.1158/0008-5472.CAN-09-3463
38. Levings PP, McGarry SV, Currie TP, Nickerson DM, McClellan S, Ghivizzani SC, et al. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res (2009) 69:5648–55. doi:10.1158/0008-5472.CAN-08-3580
39. Fujii H, Honoki K, Tsujiuchi T, Kido A, Yoshitani K, Takakura Y. Sphere-forming stem-like cell populations with drug resistance in human sarcoma cell lines. Int J Oncol (2009) 34:1381–6. doi:10.3892/ijo_00000265
40. Honoki K. Do stem-like cells play a role in drug resistance of sarcomas? Expert Rev Anticancer Ther (2010) 10:261–70. doi:10.1586/era.09.184
41. Italiano A, Di Mauro I, Rapp J, Pierron G, Auger N, Alberti L, et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol (2016) 17:532–8. doi:10.1016/S1470-2045(15)00583-5
42. Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet (2014) 46:225–33. doi:10.1038/ng.2891
43. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med (2012) 366:883–92. doi:10.1056/NEJMoa1113205
44. Rye IH, Helland Å, Saetersdal A, Naume B, Almendro V, Polyak K, et al. Intra-tumor heterogeneity as a predictor of therapy response in HER2 positive breast cancer. Cancer Res (2012) 72. doi:10.1158/0008-5472.SABCS12-P3-05-04
45. Tougeron D, Lecomte T, Pages JC, Villalva C, Collin C, Ferru A, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol (2013) 24:1267–73. doi:10.1093/annonc/mds620
46. Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q, Duan J, et al. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One (2013) 8:e54170. doi:10.1371/journal.pone.0054170
47. Patwardhan PP, Surriga O, Beckman MJ, de Stanchina E, Dematteo RP, Tap WD, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clin Cancer Res (2014) 20:3146–58. doi:10.1158/1078-0432.CCR-13-2576
48. Salawu A, Fernando M, Hughes D, Reed MWR, Woll P, Greaves C, et al. Establishment and molecular characterisation of seven novel soft-tissue sarcoma cell lines. Br J Cancer (2016) 115:1058–68. doi:10.1038/bjc.2016.259
49. Bruland O, Fodstad O, Pihl A. The use of multicellular spheroids in establishing human sarcoma cell lines in vitro. Int J Cancer (1985) 35:793–8. doi:10.1002/ijc.2910350616
50. Hu M, Nicolson GL, Trent JC, Yu D, Zhang L, Lang A, et al. Characterization of 11 human sarcoma cell strains. Cancer (2002) 95:1569–76. doi:10.1002/cncr.10879
51. Ottaviano L, Schaefer K-L, Gajewski M, Huckenbeck W, Baldus S, Rogel U, et al. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer (2010) 49:40–51. doi:10.1002/gcc.20717
52. Mohseny AB, Machado I, Cai Y, Schaefer K-L, Serra M, Hogendoorn PCW, et al. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest (2011) 91:1195–205. doi:10.1038/labinvest.2011.72
53. Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol (2007) 1:84–96. doi:10.1016/j.molonc.2007.02.004
54. Silberstein GB. Tumour-stromal interactions. Role of the stroma in mammary development. Breast Cancer Res (2001) 3:218–23. doi:10.1186/bcr299
55. Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci (2003) 116:2377–88. doi:10.1242/jcs.00503
56. Akkerman N, Defize LHK. Dawn of the organoid era: 3D tissue and organ cultures revolutionize the study of development, disease, and regeneration. Bioessays (2017) 39:1600244. doi:10.1002/bies.201600244
57. Bai C, Yang M, Fan Z, Li S, Gao T, Fang Z. Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J Exp Clin Cancer Res (2015) 34:58. doi:10.1186/s13046-015-0175-0
58. Salerno M, Avnet S, Bonuccelli G, Eramo A, De Maria R, Gambarotti M, et al. Sphere-forming cell subsets with cancer stem cell properties in human musculoskeletal sarcomas. Int J Oncol (2013) 43:95–102. doi:10.3892/ijo.2013.1927
59. Fong ELS, Lamhamedi-Cherradi S-E, Burdett E, Ramamoorthy V, Lazar AJ, Kasper FK, et al. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc Natl Acad Sci U S A (2013) 110:6500–5. doi:10.1073/pnas.1221403110
60. Villasante A, Marturano-Kruik A, Robinson ST, Liu Z, Guo XE, Vunjak-Novakovic G. Tissue-engineered model of human osteolytic bone tumor. Tissue Eng Part C Methods (2017) 23:98–107. doi:10.1089/ten.TEC.2016.0371
61. Yuhas JM, Li AP, Martinez AO, Ladman AJ. A simplified method for production and growth of multicellular tumor spheroids. Cancer Res (1977) 37:3639–43.
62. Bae SI, Kang GH, Kim YI, Lee BL, Kleinman HK, Kim WH. Development of intracytoplasmic lumens in a colon cancer cell line cultured on a non-adhesive surface. Cancer Biochem Biophys (1999) 17:35–47. doi:10.1016/S0940-2993(99)80053-0
63. Lin R-Z, Lin R-Z, Chang H-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J (2008) 3:1172–84. doi:10.1002/biot.200700228
64. Silvestri A, Schumacher D, Silvestrov M, Schäfer R, Reinhard C, Hoffmann J, et al. In vitro three-dimensional cell cultures as tool for precision medicine. In: Haybaeck J, editor. Mechanisms of Molecular Carcinogenesis. (Vol. 2), Cham: Springer International Publishing (2017). p. 281–313.
65. Sutherland RM, Inch WR, McCredie JA, Kruuv J. A multi-component radiation survival curve using an in vitro tumour model. Int J Radiat Biol Relat Stud Phys Chem Med (1970) 18:491–5. doi:10.1080/09553007014551401
66. Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol Bioeng (2003) 83:173–80. doi:10.1002/bit.10655
67. Johns RA, Tichotsky A, Muro M, Spaeth JP, Le Cras TD, Rengasamy A. Halothane and isoflurane inhibit endothelium-derived relaxing factor-dependent cyclic guanosine monophosphate accumulation in endothelial cell-vascular smooth muscle co-cultures independent of an effect on guanylyl cyclase activation. Anesthesiology (1995) 83:823–34. doi:10.1097/00000542-199510000-00023
68. Bell E. Strategy for the selection of scaffolds for tissue engineering. Tissue Eng (1995) 1:163–79. doi:10.1089/ten.1995.1.163
69. Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics (2010) 10:1886–90. doi:10.1002/pmic.200900758
70. Abu-Yousif AO, Rizvi I, Evans CL, Celli JP, Hasan T. PuraMatrix encapsulation of cancer cells. J Vis Exp (2009) (34):e1692. doi:10.3791/1692
71. Mackenzie IC, Fusenig NE. Regeneration of organized epithelial structure. J Invest Dermatol (1983) 81:S189–94. doi:10.1111/1523-1747.ep12541093
72. Stark HJ, Baur M, Breitkreutz D, Mirancea N, Fusenig NE. Organotypic keratinocyte cocultures in defined medium with regular epidermal morphogenesis and differentiation. J Invest Dermatol (1999) 112:681–91. doi:10.1046/j.1523-1747.1999.00573.x
73. Barton CE, Johnson KN, Mays DM, Boehnke K, Shyr Y, Boukamp P, et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Dis (2010) 1:e74. doi:10.1038/cddis.2010.49
74. Boehnke K, Mirancea N, Pavesio A, Fusenig NE, Boukamp P, Stark H-J. Effects of fibroblasts and microenvironment on epidermal regeneration and tissue function in long-term skin equivalents. Eur J Cell Biol (2007) 86:731–46. doi:10.1016/j.ejcb.2006.12.005
75. van der Kuip H, Mürdter TE, Sonnenberg M, McClellan M, Gutzeit S, Gerteis A, et al. Short term culture of breast cancer tissues to study the activity of the anticancer drug taxol in an intact tumor environment. BMC Cancer (2006) 6:86. doi:10.1186/1471-2407-6-86
76. Estes JM, Oliver PG, Straughn JM, Zhou T, Wang W, Grizzle WE, et al. Efficacy of anti-death receptor 5 (DR5) antibody (TRA-8) against primary human ovarian carcinoma using a novel ex vivo tissue slice model. Gynecol Oncol (2007) 105:291–8. doi:10.1016/j.ygyno.2006.12.033
77. Kiviharju-af Hällström TM, Jäämaa S, Mönkkönen M, Peltonen K, Andersson LC, Medema RH, et al. Human prostate epithelium lacks Wee1A-mediated DNA damage-induced checkpoint enforcement. Proc Natl Acad Sci U S A (2007) 104:7211–6. doi:10.1073/pnas.0609299104
78. Kondo J, Endo H, Okuyama H, Ishikawa O, Iishi H, Tsujii M, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A (2011) 108:6235–40. doi:10.1073/pnas.1015938108
79. Endo H, Okami J, Okuyama H, Kumagai T, Uchida J, Kondo J, et al. Spheroid culture of primary lung cancer cells with neuregulin 1/HER3 pathway activation. J Thorac Oncol (2013) 8:131–9. doi:10.1097/JTO.0b013e3182779ccf
80. Ashley N, Jones M, Ouaret D, Wilding J, Bodmer WF. Rapidly derived colorectal cancer cultures recapitulate parental cancer characteristics and enable personalized therapeutic assays. J Pathol (2014) 234:34–45. doi:10.1002/path.4371
81. Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev (2014) 24:68–73. doi:10.1016/j.gde.2013.11.012
82. Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, et al. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J Biomol Screen (2016) 21:931–41. doi:10.1177/1087057116650965
83. Schütte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M, et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun (2017) 8:14262. doi:10.1038/ncomms14262
84. Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, DeCarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet (2010) 42:715–21. doi:10.1038/ng.619
85. Jour G, Scarborough JD, Jones RL, Loggers E, Pollack SM, Pritchard CC, et al. Molecular profiling of soft tissue sarcomas using next-generation sequencing: a pilot study toward precision therapeutics. Hum Pathol (2014) 45:1563–71. doi:10.1016/j.humpath.2014.04.012
86. Hanes R, Grad I, Lorenz S, Stratford EW, Munthe E, Reddy CCS, et al. Preclinical evaluation of potential therapeutic targets in dedifferentiated liposarcoma. Oncotarget (2016) 7:54583–95. doi:10.18632/oncotarget.10518
87. Treindl F, Ruprecht B, Beiter Y, Schultz S, Döttinger A, Staebler A, et al. A bead-based western for high-throughput cellular signal transduction analyses. Nat Commun (2016) 7:12852. doi:10.1038/ncomms12852
88. Suehara Y, Kondo T, Fujii K, Hasegawa T, Kawai A, Seki K, et al. Proteomic signatures corresponding to histological classification and grading of soft-tissue sarcomas. Proteomics (2006) 6:4402–9. doi:10.1002/pmic.200600196
89. Suehara Y, Kondo T, Seki K, Shibata T, Fujii K, Gotoh M, et al. Pfetin as a prognostic biomarker of gastrointestinal stromal tumors revealed by proteomics. Clin Cancer Res (2008) 14:1707–17. doi:10.1158/1078-0432.CCR-07-1478
90. Petricoin EF, Espina V, Araujo RP, Midura B, Yeung C, Wan X, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res (2007) 67:3431–40. doi:10.1158/0008-5472.CAN-06-1344
91. Kondo T, Kawai A. A proteomics approach for the development of sarcoma biomarkers. EuPA Open Proteom (2014) 4:121–8. doi:10.1016/j.euprot.2014.06.004
92. Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen Q-R, Yeung C, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst (2011) 103:962–78. doi:10.1093/jnci/djr156
93. Boro A, Prêtre K, Rechfeld F, Thalhammer V, Oesch S, Wachtel M, et al. Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing’s sarcoma. Int J Cancer (2012) 131:2153–64. doi:10.1002/ijc.27472
94. Teicher BA, Polley E, Kunkel M, Evans D, Silvers T, Delosh R, et al. Sarcoma cell line screen of oncology drugs and investigational agents identifies patterns associated with gene and microRNA expression. Mol Cancer Ther (2015) 14:2452–62. doi:10.1158/1535-7163.MCT-15-0074
96. Schütte M, Ogilvie LA, Rieke DT, Lange BMH, Yaspo M-L, Lehrach H. Cancer precision medicine: why more is more and DNA is not enough. Public Health Genomics (2017). doi:10.1159/000477157
97. Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov (2017) 7:462–77. doi:10.1158/2159-8290.CD-16-1154
Keywords: sarcoma, preclinical model, in vitro organoid culture, patient-derived in vitro model, drug screening, sarcoma treatment, personalized medicine
Citation: Gaebler M, Silvestri A, Haybaeck J, Reichardt P, Lowery CD, Stancato LF, Zybarth G and Regenbrecht CRA (2017) Three-Dimensional Patient-Derived In Vitro Sarcoma Models: Promising Tools for Improving Clinical Tumor Management. Front. Oncol. 7:203. doi: 10.3389/fonc.2017.00203
Received: 12 June 2017; Accepted: 21 August 2017;
Published: 11 September 2017
Edited by:
Paolo Pinton, University of Ferrara, ItalyReviewed by:
Ritva Tikkanen, Justus Liebig Universität Gießen, GermanyCopyright: © 2017 Gaebler, Silvestri, Haybaeck, Reichardt, Lowery, Stancato, Zybarth and Regenbrecht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian R. A. Regenbrecht, christian.regenbrecht@cellphenomics.com
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.