- 1Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, Program in Women’s Oncology, Women and Infants Hospital, Providence, RI, United States
- 2Department of Chemistry, Wheaton College, Norton, MA, United States
- 3Center for Genomics and Proteomics, Genomics Core Facility, Brown University, Providence, RI, United States
- 4Department of Obstetrics and Gynecology, Wilmot Cancer Institute, Division of Gynecologic Oncology, University of Rochester Medical Center, Rochester, NY, United States
- 5Roger Williams Medical Center, Department of Surgery, Boston University Medical School, Providence, RI, United States
- 6Department of Biomedical and Pharmaceutical Sciences, University of Rhode Island, Kingston, RI, United States
Human epididymis protein 4 (HE4) has received much attention recently due to its diagnostic and prognostic abilities for epithelial ovarian cancer. Since its inclusion in the Risk of Ovarian Malignancy Algorithm (ROMA), studies have focused on its functional effects in ovarian cancer. Here, we aimed to investigate the role of HE4 in invasion, haptotaxis, and adhesion of ovarian cancer cells. Furthermore, we sought to gain an understanding of relevant transcriptional profiles and protein kinase signaling pathways mediated by this multifunctional protein. Exposure of OVCAR8 ovarian cancer cells to recombinant HE4 (rHE4) promoted invasion, haptotaxis toward a fibronectin substrate, and adhesion onto fibronectin. Overexpression of HE4 or treatment with rHE4 led to upregulation of several transcripts coding for extracellular matrix proteins, including SERPINB2, GREM1, LAMC2, and LAMB3. Gene ontology indicated an enrichment of terms related to extracellular matrix, cell migration, adhesion, growth, and kinase phosphorylation. LAMC2 and LAMB3 protein levels were constitutively elevated in cells overexpressing HE4 and were upregulated in a time-dependent manner in cells exposed to rHE4 in the media. Deposition of laminin-332, the heterotrimer comprising LAMC2 and LAMB3 proteins, was increased in OVCAR8 cells treated with rHE4 or conditioned media from HE4-overexpressing cells. Enzymatic activity of matriptase, a serine protease that cleaves laminin-332 and contributes to its pro-migratory functional activity, was enhanced by rHE4 treatment in vitro. Proteomic analysis revealed activation of focal adhesion kinase signaling in OVCAR8 cells treated with conditioned media from HE4-overexpressing cells. Focal adhesions were increased in cells treated with rHE4 in the presence of fibronectin. These results indicate a direct role for HE4 in mediating malignant properties of ovarian cancer cells and validate the need for HE4-targeted therapies that will suppress activation of oncogenic transcriptional activation and signaling cascades.
Introduction
Epithelial ovarian cancer (EOC) is a highly deadly disease owing to the fact that peritoneal metastasis presents early on in 70% of patients (1, 2). Metastasis most commonly occurs by the transcoelomic route (3), which is a complex process involving epithelial-to-mesenchymal transition (EMT), anoikis resistance, spheroid formation, and homing, attachment, and growth of the malignant cells in a new area. Multiple factors play a role in mediating the above listed steps, including proteases, extracellular matrix components, transmembrane molecules, integrins, chemokines and their receptors, metabolic factors, pro-angiogenic factors, microRNAs, and immune factors (4). Although some progress has been made in identifying potential new treatment targets, novel tumor-enriched targets are desperately needed to allow for more effective treatments with fewer side effects.
Human epididymis protein 4 (HE4) is a secreted glycoprotein and a member of the whey acidic protein (WAP) domain-containing family of anti-proteases (5). It has received much attention in recent years due to its diagnostic and prognostic abilities for EOC. The FDA-approved Risk of Ovarian Malignancy Algorithm (ROMA), which uses HE4 serum levels along with CA125 and menopausal status to detect and monitor ovarian cancer, demonstrates improved sensitivity and specificity over the Risk of Malignancy Index (RMI) that uses CA125, pelvic sonography, and menopausal status (6). Serum HE4 levels predict ovarian cancer with fewer instances of false positives in the case of benign gynecological conditions, as compared to CA125 (7). Since the development of ROMA, studies by us and others have focused on the functional effects of HE4 in ovarian cancer, including its effect on chemotherapy resistance, anti-estrogen resistance, invasion, and migration (8–17).
Several studies have pointed to a role for HE4 in invasion and migration in diverse ovarian cancer cell lines (10, 13, 16), and one study found that HE4 overexpression promoted adhesion of SKOV3 cells onto fibronectin (13). Clinically, HE4 levels were higher in tissues with lymph node metastases (16), and HE4 has also been linked to myometrial invasion in endometrial cancer (18–21). Herein, we expand upon the current literature indicating a role for HE4 in promoting metastatic properties, including invasion, migration, and adhesion. We furthermore present a genome-wide analysis of HE4-mediated transcriptional regulation underlying these effects. Finally, because we have previously shown that HE4 interacts with growth factor signaling cascades (12) and promotes activation of extracellular signal-regulated kinase (ERK) (8), we also sought to elucidate HE4-mediated protein kinase signaling that may contribute to the observed phenotypes.
Materials and Methods
Cell Culture and Treatments
OVCAR8 cells and their derivative lines were cultured in Dulbecco-Modified Eagle’s Medium with 10% fetal bovine serum and 1% penicillin/streptomycin, in a humidified incubator at 37°C/5% CO2. OVCAR8 cells stably expressing null-vector control plasmid (OVCAR8-NV) and HE4 overexpression plasmid (OVCAR8-C5) were established as previously described, with OVCAR8-C5 cells secreting HE4 levels > 800 pM (12). OVCAR8 wild-type (WT) cells were treated with 20 nM human recombinant HE4 (rHE4; MyBioSource, MBS355616) or 50% conditioned media from OVCAR8-C5 cells for various lengths of time, as indicated.
Microarray
Subconfluent OVCAR8-WT cells were treated with rHE4 in triplicate for 6 h, and RNA was isolated by Trizol extraction/LiCl precipitation. Subconfluent OVCAR8-NV and OVCAR8-C5 cells were collected in triplicate and RNA isolated in the same manner. Purity was determined by NanoDrop 2000 (Thermo Scientific), and RNA integrity was measured by Bioanalyzer (Agilent 2100) before microarray analysis (Affymetrix HuGene-1_0-st-v1) of 150 ng starting material at the Brown University Genomics Core Facility. Raw intensity values were converted to CELS files, and Transcriptome Analysis Console (TAC) Software was used to generate fold changes and ANOVA p-values. ANOVA p-values < 0.05 were considered significant. The top 15 genes in either direction were determined for OVCAR8-WT cells (untreated/rHE4-treated) and OVCAR8-NV/OVCAR8-C5 cells. Genes that were changed in both OVCAR8-WT (untreated/rHE4-treated) and OVCAR8-NV/OVCAR8-C5 comparisons with fold change greater than 1.5 in either direction were determined.
Database for Annotation, Visualization, and Integrated Discovery (DAVID) Gene Ontology Analysis
The DAVID v6.7 (22, 23) was used to identify the top four enriched annotation terms among genes differentially expressed (1.5-fold in either direction, p < 0.05) between OVCAR8-WT (untreated/rHE4-treated) and OVCAR8-NV/OVCAR8-C5, as well as among overlapping genes in these two comparisons. Default DAVID parameters were employed as follows:
Kappa Similarity: Similarity Term Overlap—3; Similarity Threshold—0.5
Classification: Initial Group Membership—3; Final Group Membership—3; Multiple Linkage Threshold—0.5
Enrichment Threshold: EASE—1.0
Stringency: Medium
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
RNA was isolated by Trizol extraction/LiCl precipitation. Total RNA (500 ng) was reverse transcribed into cDNA using an iScript cDNA Synthesis Kit (Bio-Rad, 1708890) according to the manufacturer’s protocol. For microarray validation, the same RNA samples were used. 1 µL cDNA reaction, 2 µL each of 5 µM custom forward and reverse primers (Invitrogen, Sino Biological Inc.) or 1 µM forward and reverse-validated primers (http://realtimeprimers.com), 10 µL SYBR Green (Applied Biosciences [ABI], 4367659), and 5 µL RNAse-free water were added to each well of a 96-well plate for qRT-PCR analysis. Plates were run on an ABI 7500 Fast Real-Time PCR System, and data were analyzed using the ΔΔCt method. Relative expression levels were normalized to 18S rRNA to correct for equivalent total RNA levels. Validated LAMC2 and LAMB3 primers were purchased from http://realtimeprimers.com. Validated SERPINB2 primers were purchased from Sino Biological Inc. (HP100614). Custom primer sequences (Invitrogen) are as follows:
GREM1—F-GGGAGCCCTGCATGTGAC
GREM1—R-GAAGCGGTTGATGATGGTG
TNC—F-AAGCGGGGAATGTTGGGATAG
TNC—R-TAGTCTCCTTTCCACCCCTC
18S rRNA—F-CCGCGGTTCTATTTTGTTGG
18S rRNA—R-GGCGCTCCCTCTTAATCATG
Western Blot
Protein was extracted in Cell Lysis Buffer (Cell Signaling, 9803) with 1 mM PMSF, and concentrations were determined by DC Protein Assay (Bio-Rad Laboratories, 5000116). Equal amounts of protein boiled with Novex Sample Reducing Agent (Life Technologies, NP009) and NuPAGE LDS sample buffer (Thermo Fisher Scientific, NP0007) were loaded into a 4–12% gradient NuPAGE Novex Bis-Tris gel [Life Technologies, NP0321BOX (mini) and WG1402BX10 (midi)]. Protein was transferred by semi-dry transfer to methanol-activated 0.2 µm PVDF membranes (Bio-Rad, 162-0177) at 0.12–0.2 A for 1 h. Blocking was performed in 5% milk in phosphate-buffered saline with 0.05% Tween 20 (PBS-T) for 30 min at room temperature. Membranes were incubated in primary antibody in 5% milk in PBS-T overnight at 4°C and then in secondary antibody in 5% milk in PBS-T for 1 h at room temperature, with PBS-T washes in between. HRP-tagged secondary antibodies were detected by Amersham ECL Prime Western Blot Detection System (GE Healthcare, RPN2232). Blots were imaged directly in a Bio-Rad ChemiDoc MP Imaging System. GAPDH was used as a loading control. Original images can be seen in Figure S1 in Supplementary Material. Antibodies and dilutions used are as follows:
LAMC2 (Santa Cruz, sc-28330, 1:200)
LAMB3 (Santa Cruz, sc-135968, 1:200)
GAPDH (Cell Signaling, 2118, 1:2,000)
Densitometry
Densitometry analysis of Western blots was performed using Image J. Blots were analyzed in eight-bit TIFF format with the “analyze gel” function. Band densities were normalized to GAPDH or the appropriate total protein for phosphoproteins. The lowest value was set to 1 for plotted graphs.
Phosphoproteomics
OVCAR8-WT cells were treated with 50% OVCAR8-C5 conditioned media for 48 h or left untreated. Protein was collected using lysis buffer provided in the Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems, ARY003B). The manufacturer’s instructions for the kit were followed, and membranes were developed in a Bio-Rad ChemiDoc MP Imaging System. Image J was used to perform background subtraction and determine spot density.
Invasion Assays
For one replicate of the invasion assays, a Cytoselect 24-Well Cell Invasion Kit (8 μm, Colorimetric, Cell Biolabs, CBA-110) was used according to the manufacturer’s instructions. After overnight starvation, OVCAR8 cells (1 × 105/well) were plated in triplicate in serum-free media in cell culture inserts in the presence or absence of 20 nM rHE4. Media containing 10% FBS were inserted into the lower chamber. After 24 h, media were aspirated from the insert, and the top side of the insert was cleaned with a cotton swab. The insert was then crystal violet stained and washed, and the stained cells were extracted. Extraction solution (150 µL) from each sample was then transferred to a 96-well plate and OD measured at 550 nm. For the following two experimental replicates, 8 µm Transwell Permeable Supports Coated with Cultrex BME (Corning Inc., 3458) were used, with crystal violet staining and acetic acid extraction.
Adhesion Assays
Cytoselect 48-Well Cell Adhesion Assay Kit (Fibronectin-Coated, Colorimetric; Cell Biolabs, CBA-050) was used to determine the effect of 20 nM rHE4 treatment on adhesion of OVCAR8-WT cells. Cells were plated in triplicate in serum-free media at 1 × 105/well with or without 20 nM rHE4 in the assay plate for 2 h. Media were aspirated, and cells were stained, washed, and extracted. Extraction solution (150 µL) from each sample was transferred to a 96-well plate and OD read at 550 nm.
Haptotaxis Assays
Haptotaxis assays were conducted using Transwell plates (6.5 µm thickness, 8 µm pores; Corning Inc., 3422). The lower surfaces of the Transwell membranes were coated by adding 500 µL of serum-free PRF-DMEM/F12 containing 2 µg/mL human fibronectin to the lower reservoir overnight. OVCAR8 cells in serum-free PRF-DMEM/F12 were seeded into the upper reservoirs of the Transwell inserts in the presence or absence of 20 nM HE4 and allowed to migrate overnight. Non-migrated cells were removed from the upper surface of the membrane using a Q-tip, and the cells attached to the lower surface were stained with 0.4% crystal violet in sodium borate buffer, pH 9.2 for 5 min, and then washed 2× in water. Crystal violet was eluted from the cells using acetic acid and measured spectrophotometrically at 550 nm.
Matriptase Assay
A SensoLyte Rh110 Matriptase Activity Kit (AnaSpec, 72241) was used according to the manufacturer’s protocol. Various doses of rHE4 (10, 20, and 40 nM) were tested, and fluorescent readouts (490/520 nm) were performed after 30 min incubation at 37°C. Leupeptin was used as an inhibitor control. Average fluorescence from the substrate control wells was subtracted from the rHE4 test control wells, and the remainder was subtracted from the values obtained for the rHE4-treated wells.
Immunofluorescence
To visualize focal adhesions, OVCAR8 cell plated on glass coverslips were starved in phenol red-free, serum-free media for 48 h. Starved cells were treated with 1 µg/mL rat fibronectin (Millipore, 341668) in the absence or presence of 20 nM HE4 for 2 h at 37°C. Cells were then washed with PBS and fixed in 4% paraformaldehyde for 10 m and permeabilized in 0.1% Triton X-100 for 5 m. Cells were blocked in 4% normal horse serum in PBS for 30 m and then incubated with phosphotyrosine clone 4G10 antibody (Millipore, 05-321) in 4% normal horse serum (1:500) for 1 h at room temperature. Coverslips were washed in PBS, and cell-associated antibodies were detected using DyLight 594 anti-mouse secondary (Vector Laboratories, DI-2594) diluted in 4% normal horse serum (1:1,000) for 45 min at room temperature. After staining, coverslips were washed and mounted on glass slides in Vectashield with DAPI (Vector Laboratories, H-1200). Imaging was performed on a Zeiss Axio Imager M1 using associated AxioVision software. Quantification was performed in Image J by measuring mean gray values of 4–6 fields per replicate after background subtraction (rolling ball radius = 50 pixels, sliding paraboloid). Results are the average of three independent experiments with 1–3 replicates each.
For laminin-332 staining, OVCAR8-WT cells were seeded onto glass coverslips and treated the next day with 20 nM rHE4 for 5 h. Cells were washed with PBS, fixed in 4% paraformaldehyde for 10 min, and permeabilized in 0.1% Triton X-100 for 5 m. Blocking was performed in 4% normal horse serum for 30 min, and the cells were incubated with anti-laminin-332 antibody (Abcam, ab14509) diluted in 4% normal horse serum (1:200) overnight at 4°C. The following day, the coverslips were washed in PBS and incubated in DyLight 488 anti-rabbit secondary (Vector Laboratories, DI-1488) diluted in 4% normal horse serum (1:1,000) for 45 min at room temperature. Coverslips were mounted using Vectashield with DAPI (Vector Laboratories, H-1200). Imaging and quantitative analysis were performed by an experienced technician at the RI Hospital Core Digital Imaging Facility. Results are the average of three independent experiments with three replicates each.
Statistics
Where statistics are shown, n ≥ 3 independent experiments with biological replicates ≥3, and p-values were determined by unpaired one-tailed Student’s t-test. Transcriptome Analysis Console (TAC) software (Affymetrix) was used to generate fold changes and ANOVA p-values for microarray data.
Results
Recombinant HE4 Promotes Invasion, Haptotaxis, and Adhesion of Ovarian Cancer Cells
To evaluate the effect of HE4 on malignant characteristics of OVCAR8 cells, we performed assays to evaluate invasion, adhesion, and migration. When invasion capacity was evaluated, OVCAR8-WT cells treated with rHE4 were found to have a 2.07-fold greater invasion capacity than untreated cells (p = 0.010788). Treatment with rHE4 also increased adhesion of cells to a fibronectin matrix by 1.29-fold (p = 0.002257). Interestingly, adhesion onto other substrates—collagen I and IV, laminin I, and fibrinogen—was not altered (data not shown), revealing the specificity of HE4’s effect on adhesion to fibronectin. We then tested the effect of rHE4 treatment of OVCAR8 cells on haptotaxis toward a fibronectin substrate. Treatment with rHE4 increased haptotaxis of the cells by 1.72-fold (p = 0.000378) (Figure 1). Collectively, our results demonstrate that HE4 has a direct effect on metastatic properties of ovarian cancer cells.
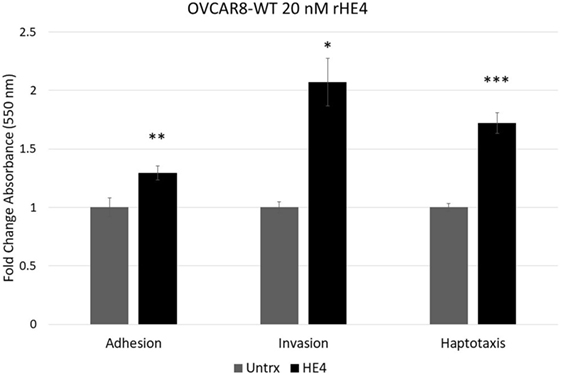
Figure 1. Human epididymis protein 4 (HE4) promotes invasion, adhesion onto a fibronectin substrate, and haptotaxis toward a fibronectin substrate OVCAR8 invasion in response to recombinant HE4 (rHE4) treatment for 24 h was evaluated using Transwell plates. Adhesion in response to 2 h rHE4 treatment was evaluated using fibronectin-coated plates. HE4-mediated haptotaxis toward a fibronectin substrate (24 h treatment) was evaluated using Transwell plates. Error bars indicate standard error of the mean of three biological replicates. *p < 0.05, **p < 0.005, and ***p < 0.0005 versus controls.
HE4 Promotes Expression of Extracellular Matrix-Related Transcripts
To investigate the effect of recombinant HE4 treatment on transcriptional regulation in OVCAR8 ovarian cancer cells, we treated subconfluent cells with 20 nM rHE4 for 6 h. Simultaneously, we collected RNA from OVCAR8 cells stably overexpressing HE4 (C5) or a null vector plasmid (NV). We have previously shown that stable overexpression of HE4 in OVCAR8-C5 cells produces HE4 levels in conditioned media of >800 pM (12). Serial dilution revealed an HE4 concentration of 504 pM in media diluted 1:10 and 51 pM in media diluted 1:100 (data not shown). Total RNA was submitted for microarray analysis comparing OVCAR8-NV to OVCAR8-C5 and OVCAR8-WT untreated cells to OVCAR8-WT rHE4-treated cells. The top 15 annotated, protein-coding genes significantly changed in either direction were identified, as shown in Table 1. The complete results are available through ArrayExpress under the accession number E-MTAB-6366.
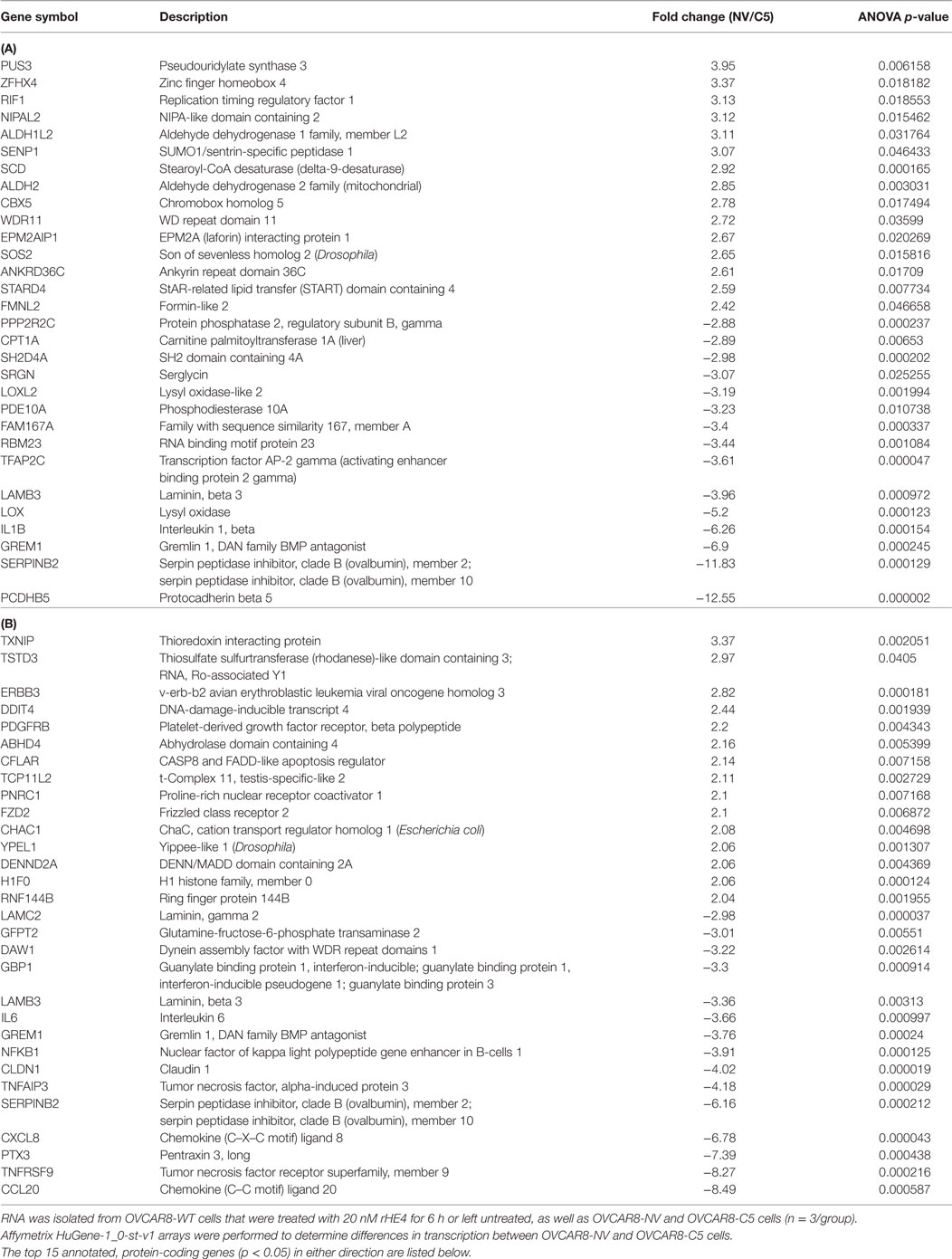
Table 1. Transcripts differentially expressed between OVCAR8-NV/OVCAR8-C5 (A), and OVCAR8-wild-type (WT)-untreated/OVCAR8-WT-recombinant human epididymis protein 4 (rHE4) treated (B) cells.
To narrow down our genes of interest, we then identified all transcripts that were changed ≥1.5-fold in either direction by both HE4 overexpression and rHE4 treatment (Table 2). Six of these genes were regulated in opposite directions (DENN/MADD domain containing 2 A [DENND2A], family with sequence similarity 19, member 1A [FAM19A1], dehydrogenase/reductase member 3 [DHRS3], transgenlin [TAGLN], leucine rich repeat interacting protein 1 [LRRFIP1], and interleukin 6 [IL6]), suggesting that stable overexpression of HE4 has some different effects on transcription than short-term exposure of cells to HE4 protein. While we found a few transcripts that were downregulated by HE4, many upregulated genes were of particular interest because of their involvement with invasion and metastasis in diverse tumor types.
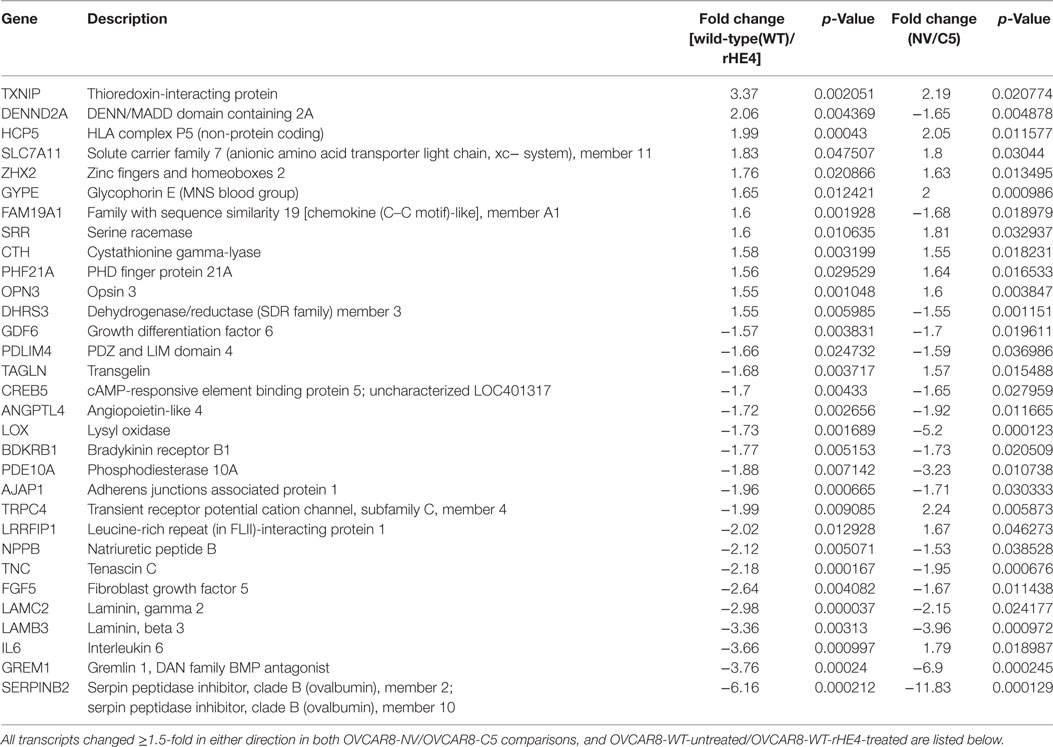
Table 2. Transcripts differentially expressed with both human epididymis protein 4 (HE4) overexpression and recombinant HE4 (rHE4) treatment.
Several genes that were significantly upregulated code for extracellular matrix proteins, such as serpin peptidase inhibitor, member 2 (SERPINB2), gremlin 1 (GREM1), laminin-β3 (LAMB3), laminin-γ2 (LAMC2), fibroblast growth factor 5 (FGF5), tenascin C (TNC), adherens junctions associated protein 1 (AJAP1), and growth and differentiation factor 6 (GDF6). We validated the upregulation of SERPINB2, GREM1, LAMB3, LAMC2, and TNC by qRT-PCR (Figures 2A,B). SERPINB2 was upregulated by 2.13-fold (p = 0.024991) and 3.31-fold (p = 0.015549) by HE4 stable overexpression and rHE4 treatment, respectively; LAMB3 was upregulated by 4.04-fold (p = 0.0004) and 7.02-fold (p = 0.00022), respectively; LAMC2 was upregulated by 4.45-fold (p = 0.002671) and 5.46-fold (p = 0.00079), respectively; GREM1 was upregulated by 3.19-fold (p = 0.015549) and 4.94-fold (p = 0.0000241), respectively; and TNC was upregulated by 3.86-fold (p = 0.028017) and 6.06-fold (p = 0.002942), respectively.
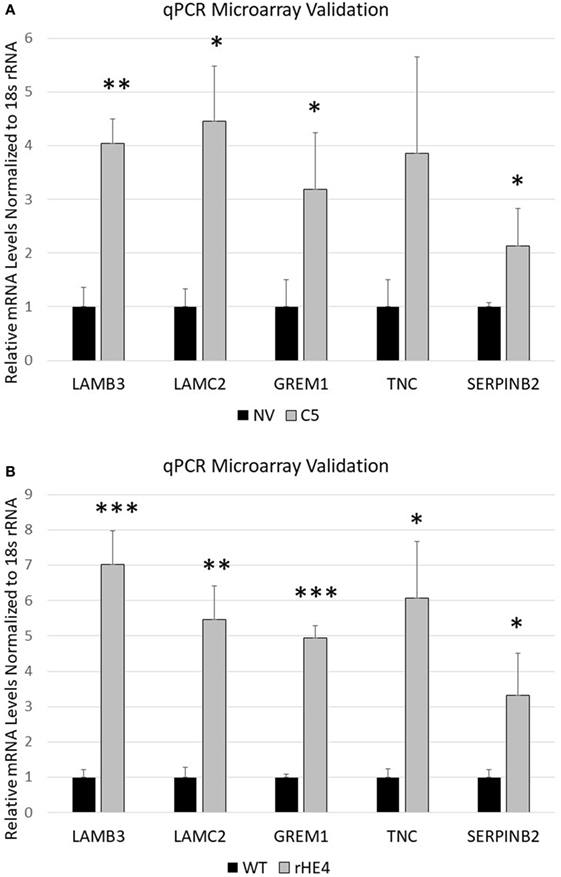
Figure 2. Quantitative reverse transcription polymerase chain reaction validation of microarray SERPINB2, GREM1, LAMC2, LAMB3, and TNC were selected to validate microarray results in OVCAR8-NV versus OVCAR8-C5 (A) and OVCAR8-wild-type (WT) untreated versus recombinant human epididymis protein 4 (rHE4)-treated (B). Error bars represent the SD of three biological replicates, *p < 0.05, **p < 0.005, and ***p < 0.0005 versus controls.
Gene ontology analysis of the differentially expressed transcripts revealed enrichment of terms related to the extracellular matrix, cell migration, adhesion, and growth (Table 3). We furthermore noted that treatment of OVCAR8-WT cells with rHE4 promoted enrichment of gene terms related to phosphorylation/protein kinase activity, suggesting that addition of exogenous HE4 may have an effect on protein kinase cascades.
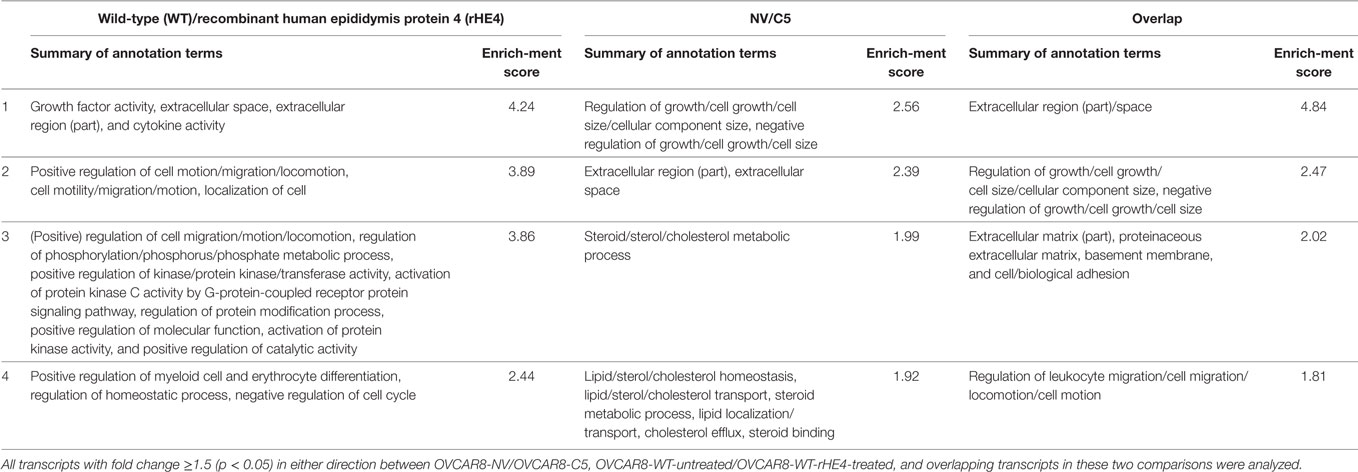
Table 3. Top 4 Database for Annotation, Visualization, and Integrated Discovery annotation clusters of differentially expressed transcripts.
HE4 Upregulates LAMC2 and LAMB3 Protein in a Time-Dependent Manner and Increases Laminin-332 Levels
We chose to further investigate HE4’s regulation of LAMC2 and LAMB3, since these two genes code for chains of laminin-332, a secreted heterotrimer that has been well-described to promote aggression and metastatic properties in diverse cancers (24–31). LAMC2 and LAMB3 protein levels were constitutively elevated in OVCAR8-C5 cells compared to OVCAR8-NV (Figures 3A–C), while their levels peaked between 4 and 24 h after treatment with both rHE4 and conditioned media from OVCAR8-C5 cells (Figures 3D–G). Interestingly, C5 media (which has an approximate HE4 concentration of 5 nM) elicited a stronger response than 20 nM rHE4, suggesting that naturally secreted HE4 may be more bioactive than recombinant protein. Collectively, these results reveal a time-dependent effect of exogenous HE4 on LAMC2 and LAMB3 and indicate that stable overexpression of HE4 leads to constitutively high levels of these two proteins.
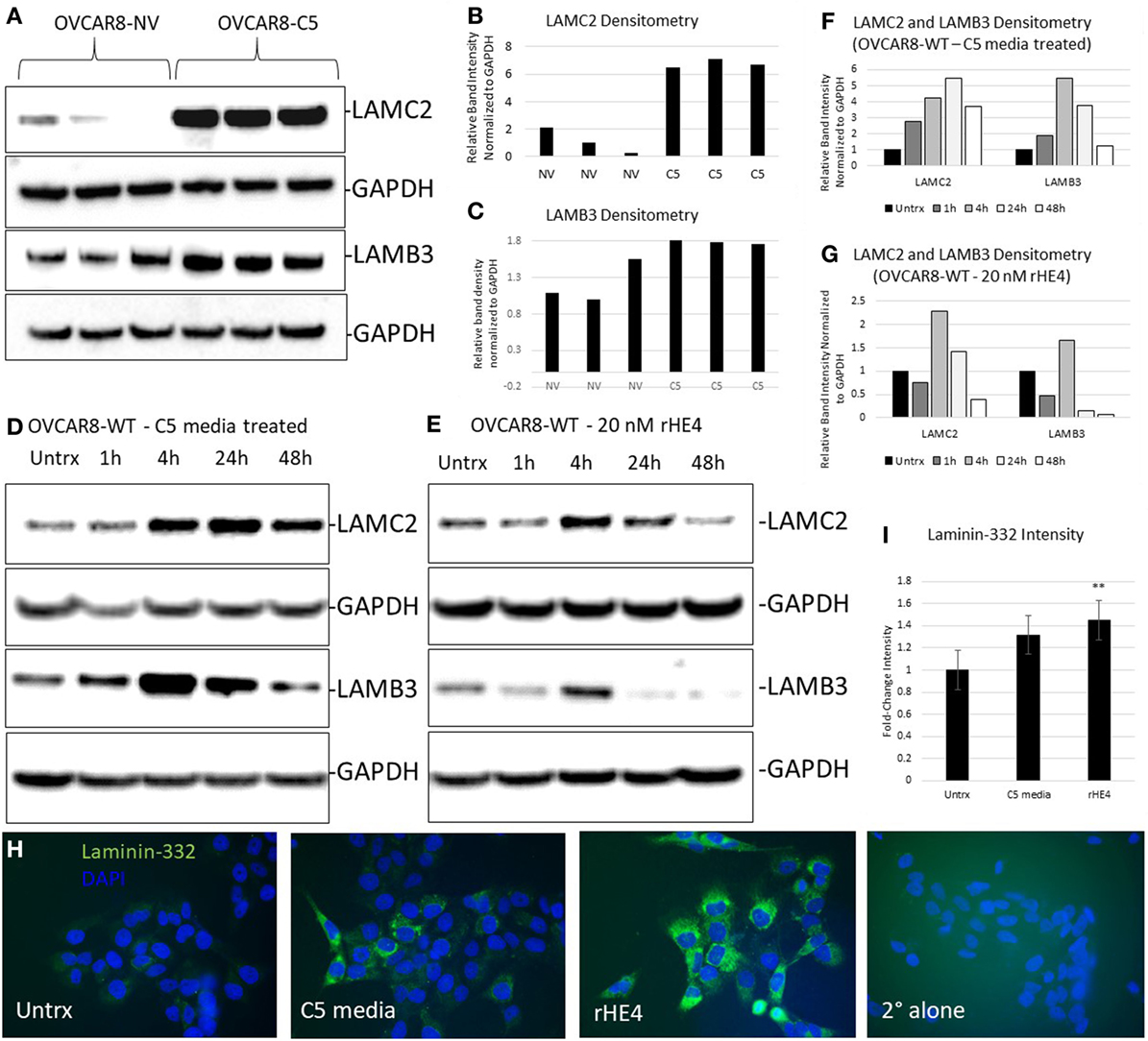
Figure 3. Human epididymis protein 4 (HE4) promotes an increase in laminin-332. (A) Protein levels of LAMC2 and LAMB3 in OVCAR8-NV and OVCAR8-C5 cells were determined by Western blot. GAPDH was used as a loading control. (B,C) Densitometry analysis of LAMC2 and LAMB3 (normalized to GAPDH) from (A). (D,E) Time course analysis of protein levels in OVCAR8-wild-type (WT) cells left untreated or treated with 20 nM recombinant HE4 (rHE4) or 50% conditioned media from OVCAR8-C5 cells. (F,G) Densitometry analysis of LAMC2 and LAMB3 (normalized to GAPDH) from Western blot in (D,E). (H) OVCAR8 cells were treated with rHE4 or conditioned media from OVCAR8-C5 cells for 5 h, and immunofluorescence staining for laminin-332 was performed. Representative images are shown. Green = laminin-332, blue = DAPI, scale = 40×. (I) Intensity was determined for laminin-332. Results are the average of fold change from three independent experiments with three replicates per experiment and four fields for each replicate. Error bars indicate standard error of the mean. **p < 0.005 versus control.
Next, we performed immunofluorescence analysis of the complete laminin-332 heterotrimer to determine if overexpression of its subunits promotes increased levels of the complete heterotrimer. Laminin-332 staining (mean gray value) was increased by 1.36-fold with C5 media treatment (p = 0.069025) and by 1.48-fold with rHE4 treatment (p = 0.002926), indicating that elevated levels of LAMC2 and LAMB3 proteins resulted in increased secretion of the laminin-332 heterotrimer (Figures 3H,I).
HE4 Enhances Enzymatic Activity of Matriptase, a Serine Protease That Cleaves Laminin-332
Since HE4 has been shown to inhibit the activity of multiple proteases (32, 33), we hypothesized that it might also inhibit enzymatic activity of matriptase, a serine protease that is known to proteolytically cleave laminin-332 in its β3 chain. Surprisingly, we observed the opposite to be true. In vitro matriptase activity was enhanced by rHE4 in a dose-dependent manner, while the inhibitor control (leupeptin) almost entirely obliterated matriptase activity (Figure 4). When the results of 3–5 separate experiments per dose were averaged, we saw an average 1.26-fold (±0.534075) increase in activity by 10 nM rHE4 (p = 0.04139), 1.32-fold (±0.489149) by 20 nM rHE4 (p = 0.000798), and 1.63-fold (±0.61337) by 40 nM rHE4 (p = 0.026596) (data not shown). Although these results are in opposition to the presumed role of rHE4 as a protease inhibitor, they indicate that HE4 not only upregulates laminin-332 levels but may also contribute to regulation of its proteolytic processing, which is known to promote migration in prostate cancer cells (34).
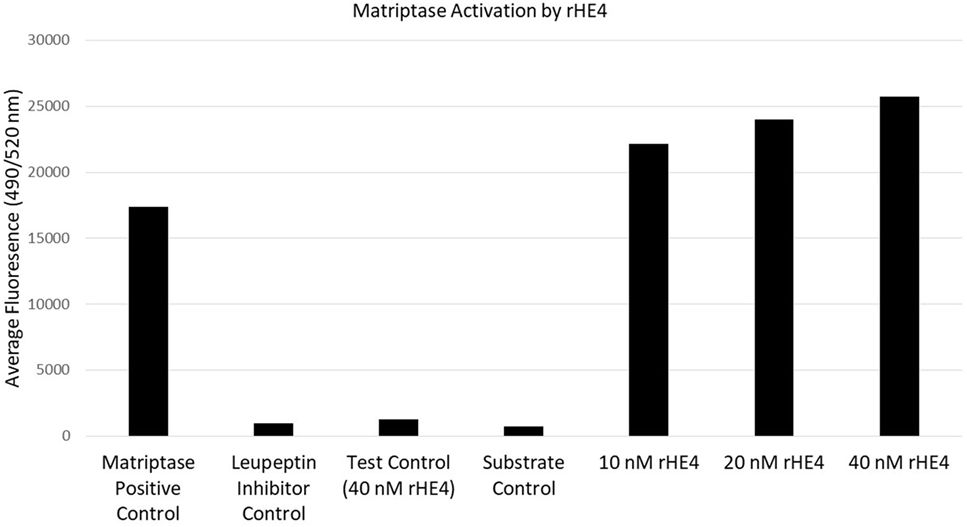
Figure 4. Human epididymis protein 4 (HE4) enhances enzymatic activity of matriptase. The effects of various doses of recombinant HE4 (rHE4) on in vitro matriptase activity were evaluated. The experiment was performed 3–5 times per dose, and the results from one representative experiment including all three doses are shown. Leupeptin = matriptase inhibitor.
HE4 Promotes Activation of Focal Adhesion Kinase (FAK) Signaling
Next, we pursued the hypothesis that HE4 promotes activation of diverse kinase signaling pathways. The activation of MAPK signaling by HE4 has been well documented by us and others (8, 10, 13, 14), but we suspected that activation of other signaling pathways is also stimulated by HE4. We treated OVCAR8-WT cells with conditioned media from OVCAR8-C5 cells for 48 h and analyzed phosphorylation of an array of kinases and their target proteins using a Human Phospho-Kinase Array (R&D Systems; Figure 5A). Upregulation of several phosphoproteins was observed, including phospho-ERK (2.34-fold), as we have previously reported (8). Of note, we also found β-catenin and phospho-FAK (Y397) upregulated by 3.13- and 2.76-fold, respectively. β-Catenin is an intracellular signal transducer of the Wnt-signaling pathway, which has roles in cell–cell adhesion (35), and FAK is a Src-family tyrosine kinase that promotes cell adhesion to the extracellular matrix (36). Furthermore, multiple other members of the Src-family of tyrosine kinases, including phospho-Fgr (Y412), phospho-Fyn (Y420), phospho-Hck (Y411), phospho-Src (Y419), and phospho-Yes (Y426), were all upregulated by around 1.5-fold or more in response to exogenous HE4 exposure.
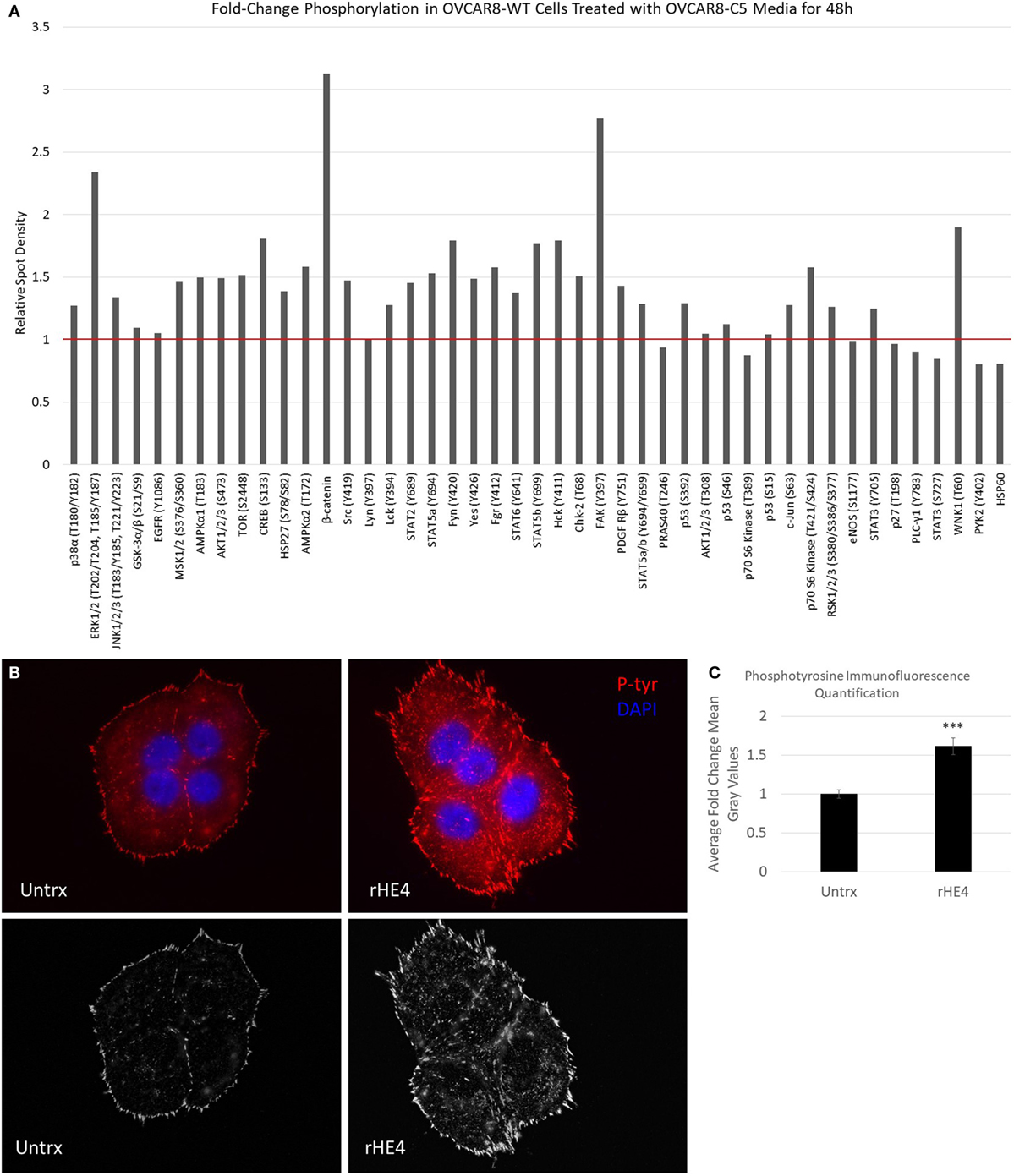
Figure 5. Human epididymis protein 4 (HE4) promotes increased formation of focal adhesions in the presence of fibronectin. (A) Proteome Profiler Human Phospho-Kinase Array (Cell Biolabs) was used to compare levels of protein phosphorylation in OVCAR8-wild-type (WT) cells (untrx) versus OVCAR8-WT cells treated with 50% conditioned media from HE4-overexpressing OVCAR8-C5 cells for 48 h. Red bar delineates proteins with increased phosphorylation. (B) Upper panel: focal adhesions in untreated versus recombinant HE4 (rHE4)-treated OVCAR8-WT cells. Focal adhesions are indicated by phosphotyrosine staining (red). Blue = DAPI. Scale = 100×. (B) Lower panel: focal adhesions are shown with background subtraction performed in Image J (rolling ball radius = 50 pixels). (C) Quantification of phosphotyrosine intensity. Results are the average of fold change from three independent experiments, with 1–3 replicates per experiment and 4–6 fields per replicate. Error bars indicate standard error of the mean. ***p < 0.0005 versus control.
To confirm the effects of HE4-mediated activation of FAK signaling, we stimulated starved OVCAR8-WT cells with rat fibronectin in the presence or absence of rHE4 for 2 h and examined focal adhesions by phosphotyrosine staining. Cells that were treated with rHE4 had 1.61-fold more focal adhesions than untreated cells (p = 0.0000593), indicating that HE4 phosphorylation of FAK at Y397 promotes subsequent formation of focal adhesions (Figures 5B,C).
Discussion
Several relevant gene transcripts were affected by HE4 overexpression or treatment as indicated by our microarray results. SERPINB2 was the foremost upregulated transcript by stable HE4 overexpression and rHE4 treatment. Traditionally, SERPINB2, a secreted glycoprotein that inhibits tPA and urokinase, has been described as a tumor suppressor that inhibits tumor growth and metastasis, whereas SERPINB1 promotes tumor progression (37). This description of SERPINB2 function appears to contradict the effect of HE4 on metastatic properties; thus, the connection between these two proteins and the significance of SERPINB2 upregulation by HE4 is an area for further investigation. The secretion of SERPINB2 protein could be evaluated, since one study found that the unsecreted, intracellular protein had a different role than the secreted protein and protected against TNFα-induced apoptosis (38).
GREM1, which was also significantly upregulated by HE4 stable overexpression or rHE4 treatment, is overexpressed in a wide variety of cancers, including uterine cervical, lung, ovary, kidney, breast, colon, pancreas, and sarcoma (39), and has been shown to promote metastatic properties. GREM1 tissue expression is associated with EMT and coordinates migration at the cancer invasion front in colon cancer (40, 41). Xu et al. also found overexpression of GREM1 in peritoneal metastatic ovarian cancers in comparison to primary tumors (42), indicating the importance of this protein in ovarian cancer progression.
LAMC2, LAMB3, and LAMA2 genes encode for the γ2, β3, and α3 chains that comprise the cross-shaped heterotrimer laminin-332, a secreted glycoprotein that is a major component of the basement membrane of epithelial tissues. Laminin-332 serves as a ligand of various transmembrane receptors including α3β1 and α6β4 integrins. Importantly, abnormal expression and high levels of laminin-332 have been shown to promote invasion in colon, breast, and skin cancers (30). Specific chains of the laminin-332 heterotrimer have been correlated with various aspects of tumor aggression as well. For example, LAMB3 was correlated with chemoresistance and poor prognosis in Stage III colorectal cancer (43). Of particular interest, one study found that expression of another laminin, LAMB2, was mediated by HE4. The authors reported that HE4 promoted invasion and migration in CaOV3 cells via HE4 binding with Annexin-V, and downregulation of the HE4 gene suppressed the expression of MKNK2 and LAMB2. Treatment with exogenous HE4 protein reversed this effect (15). In a recent report, laminin-332 was found to promote EMT and correlate with poor prognosis in lung cancer. The authors found that collagen XVII stabilized laminin-332 and caused activation of the FAK pathway (31), which is relevant to our current results showing upregulation of laminin-332 and FAK activation in response to HE4 exposure.
It is likely that the tyrosine phosphorylation of FAK that we noted in cells treated with exogenous HE4 was a result of rapid upregulation of laminin components following HE4 treatment. Laminin binding to integrin receptors mediates FAK activation (31, 44–46), which in turn activates other Src family kinases to regulate cell survival, apoptosis, proliferation, adhesion, migration, and invasion (36). Activated FAK (Y397) is localized at focal adhesions, which are points of contact between the ECM and the actin cytoskeleton (36). Thus, our results showing increased numbers of focal adhesions with rHE4 treatment are in agreement with the observed upregulation of FAK phosphorylation. Inhibition of this major signaling kinase is a promising cancer therapeutic; however, tumor-specific targeting of FAK remains a barrier to this approach (36). It is possible that inhibiting a regulator of FAK signaling such as HE4, which is selectively upregulated in ovarian tumor tissue, may overcome this problem. Furthermore, evidence thus far suggests that HE4 regulates multiple signaling pathways and aspects of tumor progression, making it a more attractive therapeutic target.
We also wanted to gain insight into possible mechanisms of HE4 regulation of laminin-332. Although we presumed some regulation to occur via signaling and regulation of gene expression of laminin-332 components, we wondered if HE4’s role as a putative protease inhibitor could contribute to its effect on laminin-332 levels. Matriptase is a serine protease that is known to cleave laminin-332 in the β3 chain, thereby promoting the effects of laminin-332 on motility (34). The role of matriptase in ovarian cancer is still being defined, but studies show that its expression is upregulated at early stages of the disease, while it is lost in Stage III/IV tumors (47, 48). While this may seem to indicate that matriptase expression is beneficial in ovarian cancer, this is not a foregone conclusion. One study also found that advanced stage tumors that do express matriptase are less likely to coexpress its inhibitor, HAI-1 (47). Another study found that the high-metastatic human ovarian cancer cell line HO-8910 had higher levels of matriptase than the homologous HO-8910 cell line, and knockdown of matriptase effectively inhibited the cells’ invasion and migration abilities (49). These results suggest that unchecked matriptase activity may play a role in the aggression of a percentage of late-stage tumors. Therefore, if HE4 enhances matriptase activity, this may potentially contribute to increased invasion and migration.
We wondered if HE4 might affect matriptase activity, since it has been shown to act as a cross-class protease inhibitor (32, 33) and belongs to the “four-disulfide core” family that encompasses several extracellular protease inhibitors (50). Our results revealed that recombinant HE4 actually enhances the activity of matriptase in a dose-dependent manner, which shows for the first time that HE4 can promote activity of at least one serine protease. It is possible that the increased matriptase activity may work in concert with upregulation of laminin-332 by HE4 to promote laminin-332 functions affecting migration, invasion, or adhesion. Further exploration of this hypothesis and clarification of how HE4 increases matriptase activity are underway.
Our results give way to many other questions that will need to be investigated in future studies. First, what is the mechanism of the rapid upregulation of LAMC2 and LAMB3 protein expression by HE4? Although it has become clear that HE4 affects many aspects of ovarian cancer progression, the molecular mechanisms behind these effects are not yet clear. It is possible that secreted HE4 binds to cellular receptors to affect intracellular signaling, since we have previously noted HE4 interaction with EGFR (12). However, a specific HE4 receptor has yet to be identified. It is also possible that intracellular HE4 has different functions than extracellular, which is suggested by our data showing differential regulation of some genes in HE4-overexpressing cells and rHE4-treated cells. The possibility of a nuclear role for HE4 may also exist, given our previous results showing nuclear translocation of HE4 upon treatment of cells with various growth factors (12).
Other questions raised by our results remain to be answered as well. Does activation of FAK by HE4 primarily occur through laminin signaling, or does HE4 affect signaling through diverse mechanisms? Finally, the cause of the specificity of HE4 adhesion to fibronectin over other substrates could also be examined. Furthermore, while we tested an array of potential substrates, there are several types of each substrate. It is possible that HE4 may promote adhesion onto one or more of these other forms of extracellular matrix components.
Our results are in agreement with other reported studies on the role of HE4 in promoting metastatic properties, indicating a consistency to the effect of HE4 in diverse cell types. However, it is likely there are also cell-type specific differences in precisely how HE4 promotes invasion, migration, and adhesion. Therefore, it would be interesting to determine the clinical relevance of our findings by determining if laminin proteins or FAK activation correlate with high levels of HE4 in ovarian cancer tissue.
Conclusion
Together, our results support recent research connecting HE4 to increased migratory and invasive capacities, as well as establish a role for HE4 in adhesion to a fibronectin substrate. We have found that HE4 affects ECM protein expression and cell-signaling promoting invasion, haptotaxis, and adhesion of ovarian cancer cells. Furthermore, we have shown for the first time that HE4 enhances the enzymatic activity of the serine protease matriptase, which not only has implications for its role in tumor metastasis but also for the establishment of HE4’s molecular function. Such results firmly establish HE4 as an important protein not just in the clinical diagnosis and management of EOC but also in its mechanistic basis.
Availability of Data Sets
The microarray data set supporting the conclusions of this article is included within the supplementary materials, and also is available through ArrayExpress under accession number E-MTAB-6366.
Author Contributions
JR, HG, RM, PD, and NY designed experiments. JR and HG executed all experiments with assistance from MK, NJ, and MO. NY developed null vector and HE4-overexpressing stable clones. CS performed the microarrays and assisted with data analysis. JR prepared this manuscript. All the authors reviewed and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Research reported in this publication was supported by Alpert Medical School at Brown University Deans’ Emerging Areas of New Science (DEANS) Excellence Award under project number GR40003; the Brown University Genomics Core Facility; the Kilguss Research Core of Women & Infants Hospital of Rhode Island; The Division of Gynecologic Oncology, Program in Women’s Oncology at Women & Infants Hospital of Rhode Island; and The Department of Chemistry at Wheaton College. The Brown Genomics Core Facility received partial support from the National Institutes of Health (NIGMS grant Number P30GM103410, NCRR grant Numbers P30RR031153, P20RR018728 and S10RR02763), National Science Foundation (EPSCoR grant No 0554548), Lifespan Rhode Island Hospital, and the Division of Biology and Medicine at Brown University.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fonc.2017.00332/full#supplementary-material.
Figure S1. Original uncropped Western blot images.
Abbreviations
C5, clone 5; EOC, epithelial ovarian cancer; EGFR, epidermal growth factor receptor; ERK, extracellular signal-regulated kinase; HGSC, high-grade serous ovarian cancer; MAPK, mitogen-activated protein kinase; NV, null vector plasmid; qRT-PCR, quantitative reverse transcription polymerase chain reaction; rHE4, recombinant human epididymis protein 4; RMI, Risk of Malignancy Index; ROMA, Risk of Ovarian Malignancy Algorithm; WAP, whey acidic protein; WT, wild-type.
References
1. Thibault B, Castells M, Delord JP, Couderc B. Ovarian cancer microenvironment: implications for cancer dissemination and chemoresistance acquisition. Cancer Metastasis Rev (2014) 33:17–39. doi:10.1007/s10555-013-9456-2
2. Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet (2009) 374:1371–82. doi:10.1016/S0140-6736(09)61338-6
4. Weidle UH, Birzele F, Kollmorgen G, Rueger R. Mechanisms and targets involved in dissemination of ovarian cancer. Cancer Genomics Proteomics (2016) 13:407–23. doi:10.21873/cgp.20004
5. Bingle L, Singleton V, Bingle CD. The putative ovarian tumour marker gene HE4 (WFDC2), is expressed in normal tissues and undergoes complex alternative splicing to yield multiple protein isoforms. Oncogene (2002) 21:2768–73. doi:10.1038/sj.onc.1205363
6. Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol (2009) 112:40–6. doi:10.1016/j.ygyno.2008.08.031
7. Heliström I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res (2003) 63:3695–700.
8. Ribeiro JR, Schorl C, Yano N, Romano N, Kim KK, Singh RK, et al. HE4 promotes collateral resistance to cisplatin and paclitaxel in ovarian cancer cells. J Ovarian Res (2016) 9:28. doi:10.1186/s13048-016-0240-0
9. Wang H, Zhu L, Gao J, Hu Z, Lin B. Promotive role of recombinant HE4 protein in proliferation and carboplatin resistance in ovarian cancer cells. Oncol Rep (2015) 33:403–12. doi:10.3892/or.2014.3549
10. Zhu YF, Gao GL, Tang SB, Zhang ZD, Huang QS. Effect of WFDC 2 silencing on the proliferation, motility and invasion of human serous ovarian cancer cells in vitro. Asian Pac J Trop Med (2013) 6:265–72. doi:10.1016/S1995-7645(13)60055-3
11. Lokich E, Singh R, Han A, Romano N, Yano N, Kyukwang K, et al. HE4 expression is associated with hormonal elements and mediated by importin-dependent nuclear translocation. Sci Rep (2014) 4:5500. doi:10.1038/srep05500
12. Moore RG, Hill EK, Horan T, Yano N, Kim K, MacLaughlan S, et al. HE4 (WFDC2) gene overexpression promotes ovarian tumor growth. Sci Rep (2014) 4:3574. doi:10.1038/srep03574
13. Lu R, Sun X, Xiao R, Zhou L, Gao X, Guo L. Human epididymis protein 4 (HE4) plays a key role in ovarian cancer cell adhesion and motility. Biochem Biophys Res Commun (2012) 419:274–80. doi:10.1016/j.bbrc.2012.02.008
14. Lee S, Choi S, Lee Y, Chung D, Hong S, Park N. Role of human epididymis protein 4 in chemoresistance and prognosis of epithelial ovarian cancer. J Obstet Gynaecol Res (2017) 43:220–7. doi:10.1111/jog.13181
15. Zhuang H, Tan M, Liu J, Hu Z, Liu D, Gao J, et al. Human epididymis protein 4 in association with Annexin II promotes invasion and metastasis of ovarian cancer cells. Mol Cancer (2014) 13:243. doi:10.1186/1476-4598-13-243
16. Zhuang H, Hu Z, Tan M, Zhu L, Liu J, Liu D, et al. Overexpression of Lewis y antigen promotes human epididymis protein 4-mediated invasion and metastasis of ovarian cancer cells. Biochimie (2014) 105:91–8. doi:10.1016/j.biochi.2014.06.022
17. Zhu L, Guo Q, Jin S, Feng H, Zhuang H, Liu C, et al. Analysis of the gene expression profile in response to human epididymis protein 4 in epithelial ovarian cancer cells. Oncol Rep (2016) 36:1592–604. doi:10.3892/or.2016.4926
18. Moore RG, Miller CM, Brown AK, Robison K, Steinhoff M, Lambert-Messerlian G. Utility of tumor marker HE4 to predict depth of myometrial invasion in endometrioid adenocarcinoma of the uterus. Int J Gynecol Cancer (2011) 21:1185–90. doi:10.1097/igc.0b013e3182229ad8
19. Kalogera E, Scholler N, Powless C, Weaver A, Drapkin R, Li J, et al. Correlation of serum HE4 with tumor size and myometrial invasion in endometrial cancer. Gynecol Oncol (2012) 124:270–5. doi:10.1016/j.ygyno.2011.10.025
20. Zamani N, Modares Gilani M, Zamani F, Zamani MH. Utility of pelvic MRI and tumor markers HE4 and CA125 to predict depth of myometrial invasion and cervical involvement in endometrial cancer. J Family Reprod Health (2015) 9:177–83. doi:10.5812/iranjradiol.5276
21. Prueksaritanond N, Cheanpracha P, Yanaranop M. Association of serum HE4 with primary tumor diameter and depth of myometrial invasion in endometrial cancer patients at Rajavithi Hospital. Asian Pac J Cancer Prev (2016) 17:1489–92. doi:10.7314/APJCP.2016.17.3.1489
22. Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics (2009):1–13. doi:10.1002/0471250953.bi1311s27
23. Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4:44–57. doi:10.1038/nprot.2008.211
24. Oh KH, Choi J, Woo J-S, Baek SK, Jung KY, Koh MJ, et al. Role of laminin 332 in lymph node metastasis of papillary thyroid carcinoma. Auris Nasus Larynx (2017) 44(6):729–34. doi:10.1016/j.anl.2017.01.010
25. Carpenter PM, Sivadas P, Hua SS, Xiao C, Gutierrez AB, Ngo T, et al. Migration of breast cancer cell lines in response to pulmonary laminin 332. Cancer Med (2017) 6:220–34. doi:10.1002/cam4.957
26. Guess CM, Quaranta V. Defining the role of laminin-332 in carcinoma. Matrix Biol (2009) 28(8):445–55. doi:10.1016/j.matbio.2009.07.008
27. Hong S, Baek S, Woo J, Cho J, Jung K, Kwon S. Expression of laminin-5 in papillary thyroid carcinoma; as a biomarker of lymph node metastasis. Thyroid (2014) 24:A15–6.
28. Kim BG, An HJ, Kang S, Choi YP, Gao MQ, Park H, et al. Laminin-332-rich tumor microenvironment for tumor invasion in the interface zone of breast cancer. Am J Pathol (2011) 178:373–81. doi:10.1016/j.ajpath.2010.11.028
29. Baba Y, Iyama KI, Hirashima K, Nagai Y, Yoshida N, Hayashi N, et al. Laminin-332 promotes the invasion of oesophageal squamous cell carcinoma via PI3K activation. Br J Cancer (2008) 98:974–80. doi:10.1038/sj.bjc.6604252
30. Tsuruta D, Kobayashi H, Imanishi H, Sugawara K, Ishii M, Jones JCR. Laminin-332-integrin interaction: a target for cancer therapy? Curr Med Chem (2008) 15:1968–75. doi:10.2174/092986708785132834
31. Liu C-C, Lin J-H, Hsu T-W, Hsu J-W, Chang J-W, Su K, et al. Collagen XVII/laminin-5 activates epithelial-to-mesenchymal transition and is associated with poor prognosis in lung cancer (2016). doi:10.18632/oncotarget.11208
32. LeBleu VS, Teng Y, O’Connell JT, Charytan D, Müller GA, Müller CA, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat Med (2013) 19:227–31. doi:10.1038/nm.2989
33. Chhikara N, Saraswat M, Tomar A, Dey S, Singh S, Yadav S. Human epididymis protein-4 (HE-4): a novel cross-class protease inhibitor. PLoS One (2012) 7:e47672. doi:10.1371/journal.pone.0047672
34. Tripathi M, Potdar AA, Yamashita H, Weidow B, Cummings PT, Kirchhofer D, et al. Laminin-332 cleavage by matriptase alters motility parameters of prostate cancer cells. Prostate (2011) 71:184–96. doi:10.1002/pros.21233
35. Morin PJ. Beta-catenin signaling and cancer. Bioessays (1999) 21:1021–30. doi:10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P
36. Zhao J, Guan J-L. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev (2009) 28:35–49. doi:10.1007/s10555-008-9165-4
37. Croucher DR, Saunders DN, Lobov S, Ranson M. Revisiting the biological roles of PAI2 (SERPINB2) in cancer. Nat Rev Cancer (2008) 8:535–45. doi:10.1038/nrc2400
38. Dickinson J, Bates EJ, Ferrante A, Antalis TM. Plasminogen activator inhibitor type 2 inhibits tumor necrosis factor α-induced apoptosis. J Biol Chem (1995) 270:27894–904. doi:10.1074/jbc.270.46.27894
39. Namkoong H, Shin SM, Kim HK, Ha S-A, Cho GW, Hur SY, et al. The bone morphogenetic protein antagonist gremlin 1 is overexpressed in human cancers and interacts with YWHAH protein. BMC Cancer (2006) 6:74. doi:10.1186/1471-2407-6-74
40. Karagiannis GS, Musrap N, Saraon P, Treacy A, Schaeffer DF, Kirsch R, et al. Bone morphogenetic protein antagonist gremlin-1 regulates colon cancer progression. Biol Chem (2015) 396:163–83. doi:10.1515/hsz-2014-0221
41. Karagiannis GS, Berk A, Dimitromanolakis A, Diamandis EP. Enrichment map profiling of the cancer invasion front suggests regulation of colorectal cancer progression by the bone morphogenetic protein antagonist, gremlin-1. Mol Oncol (2013) 7:826–39. doi:10.1016/j.molonc.2013.04.002
42. Xu H, Ma Y, Zhang Y, Pan Z, Lu Y, Liu P, et al. Identification of cathepsin K in the peritoneal metastasis of ovarian carcinoma using in-silico, gene expression analysis. J Cancer (2016) 7:722–9. doi:10.7150/jca.14277
43. Fukazawa S, Shinto E, Tsuda H, Ueno H, Shikina A, Kajiwara Y, et al. Laminin β3 expression as a prognostic factor and a predictive marker of chemoresistance in colorectal cancer. Jpn J Clin Oncol (2015) 45:533–40. doi:10.1093/jjco/hyv037
44. Delimont D, Dufek BM, Meehan DT, Zallocchi M, Gratton MA, Phillips G, et al. Laminin α2-mediated focal adhesion kinase activation triggers Alport glomerular pathogenesis. PLoS One (2014) 9:e99083. doi:10.1371/journal.pone.0099083
45. Hoshina N, Tezuka T, Yokoyama K, Kozuka-hata H, Oyama M, Yamamoto T. Focal adhesion kinase regulates laminin-induced oligodendroglial process outgrowth. Genes Cells (2007) 12:1245–54. doi:10.1111/j.1365-2443.2007.01130.x
46. Yang XH, Flores LM, Li Q, Zhou P, Xu F, Krop IE, et al. Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res (2010) 70:2256–63. doi:10.1158/0008-5472.CAN-09-4032
47. Oberst MD, Johnson MD, Dickson RB, Lin CY, Singh B, Stewart M, et al. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res (2002) 8:1101–7.
48. Tanimoto H, Shigemasa K, Tian X, Gu L, Beard JB, Sawasaki T, et al. Transmembrane serine protease TADG-15 (ST14/Matriptase/MT-SP1): expression and prognostic value in ovarian cancer. Br J Cancer (2005) 92:278–83. doi:10.1038/sj.bjc.6602320
49. Sun P, Jiang Z, Chen X, Xue L, Mao X, Ruan G, et al. Decreasing the ratio of matriptase/HAI-1 by downregulation of matriptase as a potential adjuvant therapy in ovarian cancer. Mol Med Rep (2016) 14:1465–74. doi:10.3892/mmr.2016.5435
Keywords: human epididymis protein 4, epithelial ovarian cancer, metastasis, invasion, haptotaxis, adhesion, protein kinase signaling, OVCAR8 cells
Citation: Ribeiro JR, Gaudet HM, Khan M, Schorl C, James NE, Oliver MT, DiSilvestro PA, Moore RG and Yano N (2018) Human Epididymis Protein 4 Promotes Events Associated with Metastatic Ovarian Cancer via Regulation of the Extracelluar Matrix. Front. Oncol. 7:332. doi: 10.3389/fonc.2017.00332
Received: 06 September 2017; Accepted: 26 December 2017;
Published: 22 January 2018
Edited by:
Nicolas Wentzensen, National Cancer Institute (NIH), United StatesReviewed by:
Luigi Formisano, Vanderbilt University Medical Center, United StatesRachelle Johnson, Vanderbilt University Medical Center, United States
Copyright: © 2018 Ribeiro, Gaudet, Khan, Schorl, James, Oliver, DiSilvestro, Moore and Yano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer R. Ribeiro, jrribeiro@wihri.org
 Jennifer R. Ribeiro
Jennifer R. Ribeiro Hilary M. Gaudet
Hilary M. Gaudet Mehreen Khan2
Mehreen Khan2 Naohiro Yano
Naohiro Yano