- 1Department of Molecular and Cellular Pharmacology, Pharmacogenomics and Pharmacoinformatics, Mie University Graduate School of Medicine, Tsu, Japan
- 2Mie University Medical Zebrafish Research Center, Tsu, Japan
- 3Department of Systems Pharmacology, Mie University Graduate School of Medicine, Tsu, Japan
- 4Department of Omics Medicine, Mie University Industrial Technology Innovation Institute, Tsu, Japan
- 5Department of Bioinformatics, Mie University Life Science Research Center, Tsu, Japan
- 6Department of Anesthesiology and Critical Care Medicine, Mie University Graduate School of Medicine, Tsu, Japan
- 7Department of Pediatrics, Mie University Graduate School of Medicine, Tsu, Japan
Pulmonary arterial hypertension (PAH) is a heterogeneous disorder associated with a progressive increase in pulmonary artery resistance and pressure. Although various therapies have been developed, the 5-year survival rate of PAH patients remains low. There is thus an important need to identify novel genes that are commonly dysregulated in PAH of various etiologies and could be used as biomarkers and/or therapeutic targets. In this study, we performed comparative transcriptome analysis of five mammalian PAH datasets downloaded from a public database. We identified 228 differentially expressed genes (DEGs) from a rat PAH model caused by inhibition of vascular endothelial growth factor receptor under hypoxic conditions, 379 DEGs from a mouse PAH model associated with systemic sclerosis, 850 DEGs from a mouse PAH model associated with schistosomiasis, 1598 DEGs from one cohort of human PAH patients, and 4260 DEGs from a second cohort of human PAH patients. Gene-by-gene comparison identified four genes that were differentially upregulated or downregulated in parallel in all five sets of DEGs. Expression of coiled-coil domain containing 80 (CCDC80) and anterior gradient two genes was significantly increased in the five datasets, whereas expression of SMAD family member six and granzyme A was significantly decreased. Weighted gene co-expression network analysis revealed a connection between CCDC80 and collagen type I alpha 1 (COL1A1) expression. To validate the function of CCDC80 in vivo, we knocked out ccdc80 in zebrafish using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system. In vivo imaging of zebrafish expressing a fluorescent protein in endothelial cells showed that ccdc80 deletion significantly increased the diameter of the ventral artery, a vessel supplying blood to the gills. We also demonstrated that expression of col1a1 and endothelin-1 mRNA was significantly decreased in the ccdc80-knockout zebrafish. Finally, we examined Ccdc80 immunoreactivity in a rat PAHmodel and found increased expression in the hypertrophied media and adventitia of the pre-acinar pulmonary arteries (PAs) and in the thickened intima, media, and adventitia of the obstructed intra-acinar PAs. These results suggest that increased expression of CCDC80 may be involved in the pathogenesis of PAH, potentially by modulating the expression of endothelin-1 and COL1A1.
Introduction
Pulmonary arterial hypertension is a progressive disease characterized by increased pulmonary vascular resistance due to vasoconstriction and remodeling (reviewed in Pullamsetti et al., 2014; McLaughlin et al., 2015). The pathologic abnormalities in vascular lesions include intimal hyperplasia, medial thickness, plexiform lesions, and thrombosis in situ (Iwashita et al., 2014; Otsuki et al., 2015; Shinohara et al., 2015), which are caused by increased migration and proliferation of smooth muscle cells and adventitial fibroblasts, abnormal endothelial cell proliferation, and impaired apoptosis. Although several treatment options have become available and have significantly improved morbidity and mortality, the 5-year survival rate for PAH patients remains at ~60% (Korsholm et al., 2015). Early diagnosis and accurate prognostic stratification of patients at baseline and during follow-up are important to ensure optimal therapeutic strategies (Pezzuto et al., 2015). Thus, finding novel genes involved in the pathogenesis of PAH could provide a better understanding of the pathophysiological mechanisms and suggest novel therapeutic approaches for the disease (Guignabert et al., 2015; Machado et al., 2015).
Transcriptome analysis could represent a new frontier in the search for novel biomarkers and/or therapeutic targets in various diseases, because it facilitates the identification of panels of genes specifically dysregulated in affected tissues (Nishimura et al., 2007, 2015b; Oldham et al., 2008; Oka et al., 2010; Sasagawa et al., 2016). A number of transcriptome analyses of PAH patients and PAH animal models have been performed and the data have been deposited in a public database (Barrett et al., 2009). These include data derived from two cohorts of human patients (Mura et al., 2012; Zhao Y. et al., 2014; Zhao Y.D. et al., 2014); a rat PAH model caused by treatment with the vascular endothelial growth factor receptor inhibitor SU5416 under conditions of hypoxia (Moreno-Vinasco et al., 2008); a mouse PAH model caused by overexpression of Fra-2 (Biasin et al., 2014), a causative gene for systemic sclerosis; a mouse PAH model caused by schistosomiasis (Graham et al., 2013); a rat model caused by left heart disease (Hoffmann et al., 2011); a rat model caused by Pneumocystis infection (Swain et al., 2014); and a mouse PAH model caused by deletion of cavin-1 (Swärd et al., 2013). In this study, we sought to identify genes commonly dysregulated in PAH in both human and rodent models. Therefore, we selected for analysis both cohorts of human PAH patients (Mura et al., 2012; Zhao Y. et al., 2014; Zhao Y.D. et al., 2014); two mouse models caused by schistosomiasis (Graham et al., 2013) and Fra-2 overexpression (Biasin et al., 2014), which were selected because schistosomiasis and connective tissue diseases such as systemic sclerosis are major causes of PAH (Simonneau et al., 2013); and a rat PAH model caused by SU5416 and hypoxia (Moreno-Vinasco et al., 2008), which we included in this study because we have successfully employed this PAH model (Otsuki et al., 2015; Shinohara et al., 2015). We acknowledg that our transcriptome analysis of these datasets may not detect genes involved in other common causes of PAH, such as left heart and/or lung diseases.
We performed a comparative transcriptome analysis of the two human and three rodent PAH datasets and found that coiled-coil domain containing 80 (CCDC80) may be a novel biomarker and therapeutic target in PAH. We validated the function of CCDC80 as it relates to PAH using zebrafish. Many transgenic zebrafish lines expressing fluorescent proteins in various cells have been developed and are available through public resources, enabling multiple approaches to in vivo imaging of the relevant target cells in zebrafish larvae (Nishimura et al., 2015a). Specific genes can also be knocked out using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 systems (Auer and Del Bene, 2014). Thus, when used in combination with advanced technologies for genetic manipulation, in vivo imaging of the zebrafish ventral artery, which supplies blood to the gill arches where gas exchange occurs by diffusion (Jonz and Nurse, 2008), may be a useful tool to characterize the function of genes related to PAH. Our in vivo validation using the zebrafish model revealed that ccdc80 knockout increased the diameter of the ventral artery. Finally, we examined Ccdc80 expression in a rat PAH model and found increased staining in the hypertrophied media and adventitia of the pre-acinar pulmonary arteries (PAs) and in the thickened intima, media, and adventitia of the obstructed intra-acinar PAs.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with Japanese law [The Humane Treatment and Management of Animals (2014), Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain (2013), and the Guidelines for Proper Conduct of Animal Experiments (2006) (Science Council of Japan, 2006; Ministry of the Environment Japan, 2013, 2014)]. All efforts were made to minimize animal suffering. Mie University Institutional Animal Care and Use Committee guidelines state that no approval is required for experiments using zebrafish.
Compounds
Sodium nitroprusside (SNP) was obtained from Millipore (Billerica, MA, USA) and dissolved in 0.3x Danieau’s solution (19.3 mM NaCl, 0.23 mM KCl, 0.13 mM MgSO4, 0.2 mM Ca(NO3)2, 1.7 mM HEPES, pH 7.2) immediately before use in experiments. KT5823 was obtained from Tocris (Bristol, UK). Stock solutions of KT5823 were prepared by dissolving in dimethyl sulfoxide (Nacalai Tesque, Kyoto, Japan). L-NAME was obtained from Dojin Chemicals (Kumamoto, Japan). 2-Phenoxyethanol was obtained from Wako Chemicals (Osaka, Japan).
Comparative Transcriptome Analysis
To compare DEGs in human PAH and rodent models of PAH of different etiologies, we analyzed five lung transcriptome datasets. GSE8078 was derived from a rat PAH model caused by treatment with the VEGF receptor inhibitor SU5416 under conditions of hypoxia (Moreno-Vinasco et al., 2008). Adult male rats received a single subcutaneous injection of SU-5416 and were housed under hypoxic condition for 3.5 weeks. mRNA was isolated from whole lung tissue of the PAH rats (n = 3) and the control rats (n = 4). GSE51222 was derived from a mouse PAH model caused by overexpression of Fra-2 (Biasin et al., 2014), and mRNA was isolated from the lung homogenates of 8-week-old transgenic mice (n = 3) and wild-type littermate controls (n = 3). GSE48936 was derived from a mouse PAH model caused by schistosomiasis (Graham et al., 2013). Female mice received an intraperitoneal injection of Schistosoma mansoni eggs followed 2 weeks later by challenge with S. mansoni eggs injected via the tail vein. One week later, lung mRNA was isolated from injected (n = 3) and uninjected (n = 3) mice. GSE24988 and GSE53408 were derived from human PAH patients (Mura et al., 2012; Zhao Y. et al., 2014; Zhao Y.D. et al., 2014). For our study, we compared the transcriptomes of patients with pulmonary fibrosis without PAH (n = 22) and with intermittent PAH (n = 45) in GSE24988 and controls (n = 11) and patients with PAH (n = 12) in GSE53408.
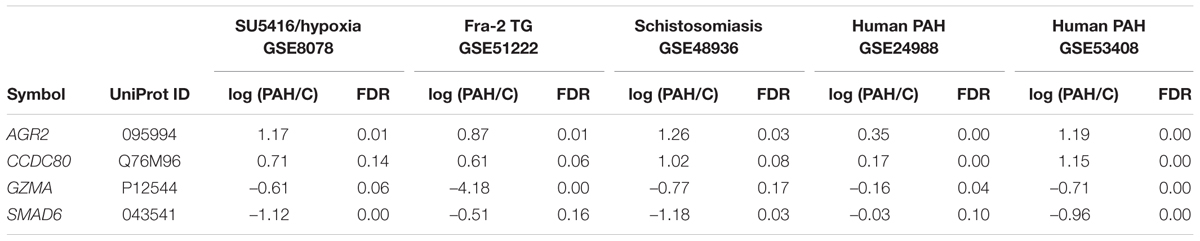
The raw data were normalized using the packages “affy” (Gautier et al., 2004) for GSE8078, “limma” (Ritchie et al., 2015) for GSE51222, and “oligo” (Carvalho and Irizarry, 2010) for GSE48936, GSE24988, and GSE53408 in Bioconductor (Gentleman et al., 2004). Probes with reliable signals were selected and subjected to RankProd (Breitling et al., 2004) analysis to identify DEGs in the PAH groups compared with control groups in each dataset. False discovery rates were set at 20% for the rodent PAH models, 10% for one human cohort (GSE24988), and 5% for the second human cohort (GSE53408). The gene symbols of the DEGs were converted to the human orthologs using Life Science Knowledge Bank (World Fusion, Tokyo, Japan), and UniProt IDs were added using the ID mapping tool (UniProt Consortium, 2015). The lists of DEGs are shown in Supplementary Tables S1-1–S1-5. The DEGs common to all five PAH transcriptome datasets are shown in Table 1.
Weighted Gene Co-Expression Network Analysis
To identify molecular interactions between genes potentially related to PAH, we performed weighted gene co-expression network analysis (WGCNA). WGCNA can organize transcriptomic data into networks based on underlying expression relationships, such as correlations, to elucidate novel connections between genes (Oldham et al., 2008). Expression data for 40 genes commonly dysregulated in the five PAH transcriptome datasets (Supplementary Table S2) were subjected to WGCNA (Langfelder and Horvath, 2008) in Bioconductor (Gentleman et al., 2004). The 40 genes were clustered into three modules of correlated genes: the first contained CCDC80, the second contained both SMAD6 and GZMA, and the third contained AGR2. The gene networks in modules 1, 2, and 3 were identified using thresholds of 0.05, 0.25, and 0.02, respectively.
Zebrafish Strains
We used two lines of zebrafish: a Tg (fli1:EGFP) line (Lawson and Weinstein, 2002) obtained from Zebrafish International Resource Center (Eugene, OR, USA) (Varga, 2011) and an albino line (Kelsh et al., 1996) obtained from the Max Planck Institute for Developmental Biology (Tübingen, Germany). Zebrafish were bred and maintained according to previously described methods (Westerfield, 2007; Nishimura et al., 2016). Briefly, zebrafish were raised at 28.5°C ± 0.5°C with a 14 h/10 h light/dark cycle. Embryos were obtained by natural mating and cultured in 0.3x Danieau’s solution until 6 dpf, at which time they were used for the in vivo imaging analyses or were processed for quantitative polymerase chain reaction (qPCR).
Knockout of ccdc80 in Zebrafish
Knockout of ccdc80 in zebrafish was performed by the ready-to-use CRISPR/Cas9 method (Kotani et al., 2015). CRISPR RNA (crRNA) targeting a 5′-AAGCAGCCCGACCGATAAAC-3′ sequence in the ccdc80 genome and trans-activating crRNA (tracrRNA; Kotani et al., 2015) were obtained from FASMAC (Kanagawa, Japan). Recombinant Cas9 protein was obtained from Toolgen (Seoul, South Korea). In brief, crRNA, tracrRNA, and Cas9 protein were dissolved in sterilized water at concentrations of 250, 1000, and 1000 ng/μl, respectively, and stored at -80°C until required. For microinjection, the crRNA, tracrRNA, Cas9 protein, and a lissamine-labeled control morpholino with no known target gene (Gene Tools, Philomath, OR, USA) were mixed in Yamamoto’s Ringer’s solution (0.75% NaCl, 0.02% KCl, 0.02% CaCl2, 0.002% NaHCO3) to final concentrations of 100 ng/μl, 100 ng/μl, 400 ng/μl, and 50 nM, respectively. The solution was injected into 1–4-cell-stage zebrafish embryos derived from the Tg (fli1: EGFP) line or albino lines. At 1 dpf, the embryos exhibiting bright lissamine fluorescence were selected and maintained until 6 dpf. At 6 dpf, the selected zebrafish were used for in vivo imaging of the ventral artery or were processed for qPCR. After completion of the in vivo imaging experiments, genomic DNA was extracted from the zebrafish by incubation in 50 μl of lysis buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 0.2% Triton X-100, 200 μg/ml proteinase K) at 55°C overnight, followed by incubation at 99°C for 10 min. The solution was then placed at 4°C and used as the template for PCR. To detect the crRNA-induced mutations, a short fragment of the ccdc80 gene encompassing the crRNA target sites was amplified from the genomic DNA using ccdc80_gF1 and ccdc80_gR1 primers and QuickTaq (Toyobo, Osaka, Japan). PCR cycling conditions were: 94°C for 2 min followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 30 s. The PCR products were electrophoresed on 10% poly-acrylamide gel (Wako Chemicals) and visualized by ethidium bromide staining. The crRNA, tracrRNA, and PCR primer sequences are shown in Supplementary Table S3.
In Vivo Imaging of the Ventral Artery of Tg (fli1:EGFP) Zebrafish
Zebrafish at 6 dpf were incubated with or without SNP (1–10 mM) and/or L-NAME (10 mM) in the presence or absence of the protein kinase G (PKG) inhibitor KT5823 (1 μM). Zebrafish were exposed to the chemicals for 1 h for experiments examining the SNP dose-response and the effect of KT5823, and for 2 h for the experiments examining the effect of L-NAME with or without SNP. The zebrafish were then transferred onto glass slides, placed on their backs, and immobilized by covering with a few drops of 3% low-melting point agarose solution. The ventral artery was visualized using an epifluorescence microscope (SMZ25, Nikon, Tokyo, Japan) with GFP-B filters, and images were recorded at 100 frames/s for 5 s. Quantitative assessment of the ventral artery diameter was performed using Volocity (Perkin Elmer, Cambridge, MA, USA). Briefly, the time-lapse images were used to generate M-mode tracings at the level immediately below the first branches of the ventral artery. The ventral artery diameters were then measured manually. The lengths of ventral artery (from the level immediately below the bifurcation to 1st afferent branchial artery to the level immediately above arterial bulb) were also measured manually.
qPCR Analysis
Total RNA was extracted from control or ccdc80-KO albino zebrafish at 6 dpf using an RNAqueous Micro Kit (Takara, Kyoto, Japan) according to the manufacturer’s protocol. RNA concentrations were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA), and aliquots were reverse transcribed using an iScript Select cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qPCR was performed using an ABI Prism 7300 (Life Technologies, Carlsbad, CA, USA) with SYBR Green Realtime PCR Master Mix Plus (Toyobo). The thermal cycling conditions were: 95°C for 1 min followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 45 s. We measured the expression of collagen type I alpha 1 (col1a1), secreted frizzled-related protein 2 (sfrp2), endothelin-1 (edn1), and glyceraldehyde-3-phosphate dehydrogenase (gapdh) mRNA. The data were normalized to gapdh mRNA levels to correct for variability in the initial template concentration and the conversion efficiency of the reverse transcription reaction. The primer sequences are shown in Supplementary Table S3.
Immunohistochemistry
Immunohistochemistry was performed as described previously on lungs excised from control rats or rats with PAH caused by SU5416/hypoxia (Otsuki et al., 2015; Shinohara et al., 2015). Briefly, sections of paraffin-embedded lung tissue were deparaffinized and hydrated. Epitope retrieval was performed by boiling the sections in citrate buffer (0.01 M, pH 6.0), and endogenous peroxidase activity was quenched with 0.3% hydrogen peroxide in methanol. Sections were then blocked in 5% normal goat serum and incubated overnight at 4°C with a rabbit polyclonal antibody to Ccdc80 (ab75881, Abcam, Cambridge, UK) or a mouse monoclonal antibody to α-smooth muscle actin (α-SMA, 1A4; Sigma, St. Louis, MO, USA). Antibody binding was amplified with streptavidin-biotin (LSAB2 kit, Dako, Kyoto, Japan), and sections were incubated with 3,3′-diaminobenzidine substrate and counterstained with hematoxylin. For the negative controls, sections were incubated with rabbit or mouse immunoglobulin instead of the primary antibody (Figures 5D,H). The localization and intensity of staining were assessed by two independent examiners (HS and EZ) who were blinded to the rat treatment group.
Statistical Analysis
Statistical analysis was performed using Prism 6 (GraphPad, La Jolla, CA, USA). Group means were compared by unpaired t-test for two groups and by analysis of variance (ANOVA) for four groups. Alpha was set at 0.05, and Dunnett’s multiple comparisons test was used for post hoc analysis when significant effects were found by ANOVA. Data are shown as the mean ± standard error (SEM).
Results
Identification of DEGs Common to the Five Mammalian PAH Transcriptome Datasets
We performed comparative transcriptome analysis to identify genes dysregulated in the three rodent PAH models and two cohorts of human PAH patients. We identified 228, 379, 850, 1598, and 4260 DEGs in PAH caused by SU5416/hypoxia, Fra-2 TG, schistosomiasis, human PAH cohort 1, and human PAH cohort 2, respectively, compared with the relevant controls (Supplementary Tables S1-1–S1-5). A Venn diagram showing unique and shared DEGs is shown in Figure 1. Four DEGs were either upregulated or downregulated in all five datasets (Table 1). Expression of coiled-coil domain containing 80 (CCDC80) and anterior gradient 2 (AGR2) was significantly increased in the five PAH transcriptome datasets, whereas SMAD family member 6 (SMAD6) and granzyme A (GZMA) were significantly decreased in all datasets. These results suggest that the four DEGs may be novel biomarkers in PAH.
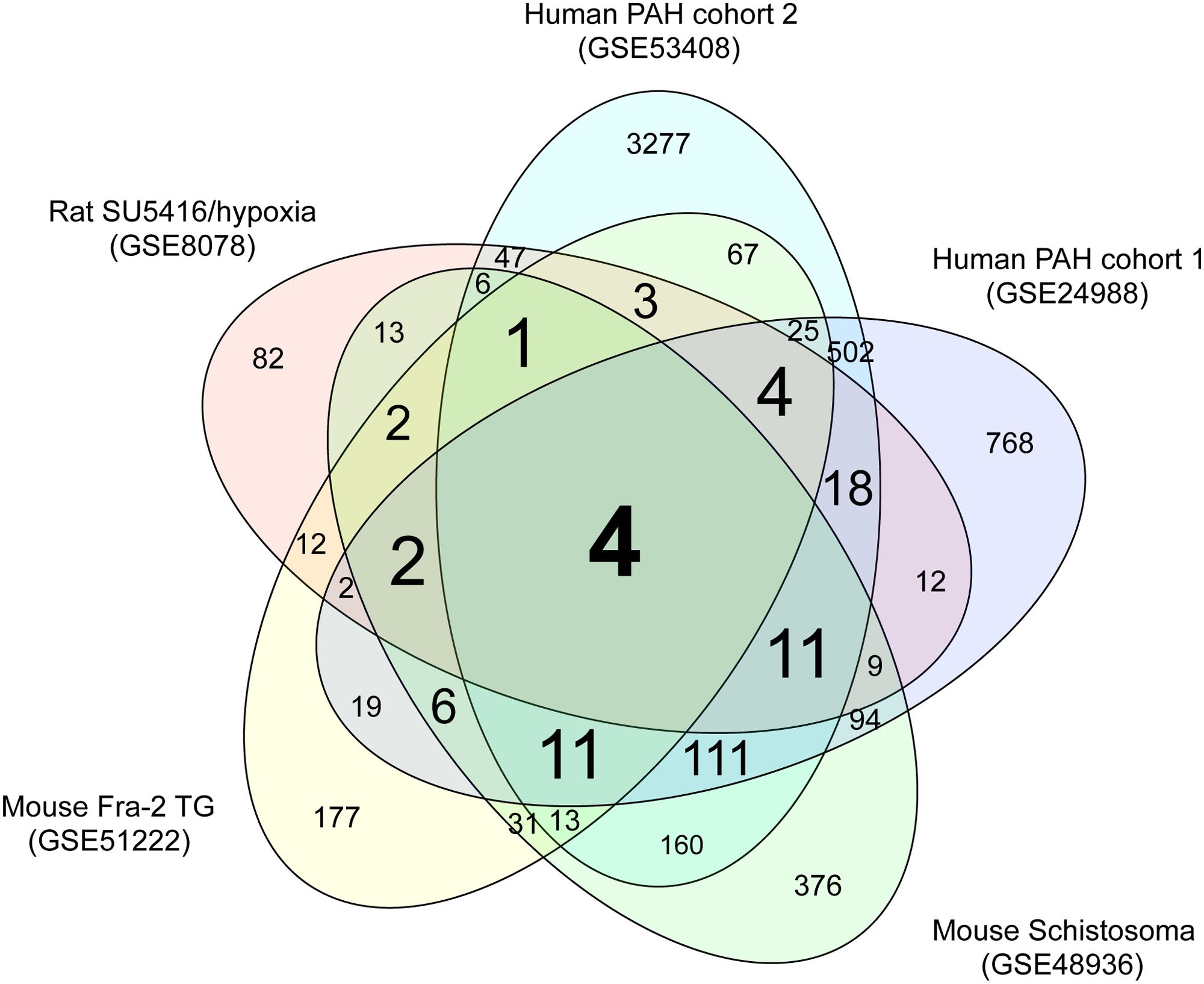
FIGURE 1. Venn diagram of DEGs in the five PAH transcriptome datasets. Transcriptome data from a rat PAH model caused by treatment with the VEGFR inhibitor SU5416 and hypoxia (GSE8078); a mouse PAH model caused by overexpression of Fra-2, a causative gene for systemic sclerosis (GSE51222); a mouse PAH model caused by schistosomiasis (GSE48936); and two cohorts of human PAH patients (GSE24988 and GSE53408) were downloaded from a public database (GEO). Genes that were differentially expressed in the PAH and control groups of each dataset were identified as described in the methods. The numbers of DEGss unique to each transcriptome dataset and shared between datasets are shown.
Identification of Gene Networks Dysregulated in PAH
To identify molecular networks common to the genes dysregulated in PAH, we used WGCNA. We calculated the coefficient of variation (CV) of the normalized probe intensity for each gene in each transcriptome dataset and then sorted the genes in descending order by CV. The top 3000 genes in each PAH transcriptome dataset were selected (Supplementary Tables S1-1–S1-5). A total of 40 genes were common to the five sets of 3000 genes (Supplementary Tables S2-3–S2-6) and were subjected to WGCNA. Figure 2 shows the networks assigned to the genes dysregulated in PAH by WGCNA. The four genes identified in Table 1 (CCDC80, AGR2, SMAD6, and GZMA) are included in the three networks identified by WGCNA (Figure 2). CCDC80 is connected to collagen type I alpha 1 (COL1A1), secreted frizzled-related protein 2 (SFRP2), and insulin-like growth factor 1 (IGF1). SMAD6 is connected to endothelin 1 (EDN1) and GZMA. COL1A1, IGF1, and EDN1 have all been associated with the pathophysiology of PAH (Ooi et al., 2010; Pullamsetti et al., 2014; Madonna et al., 2015; McLaughlin et al., 2015), supporting our hypothesis that CCDC80, SMAD6, and GZMA may be involved in the disease pathophysiology.
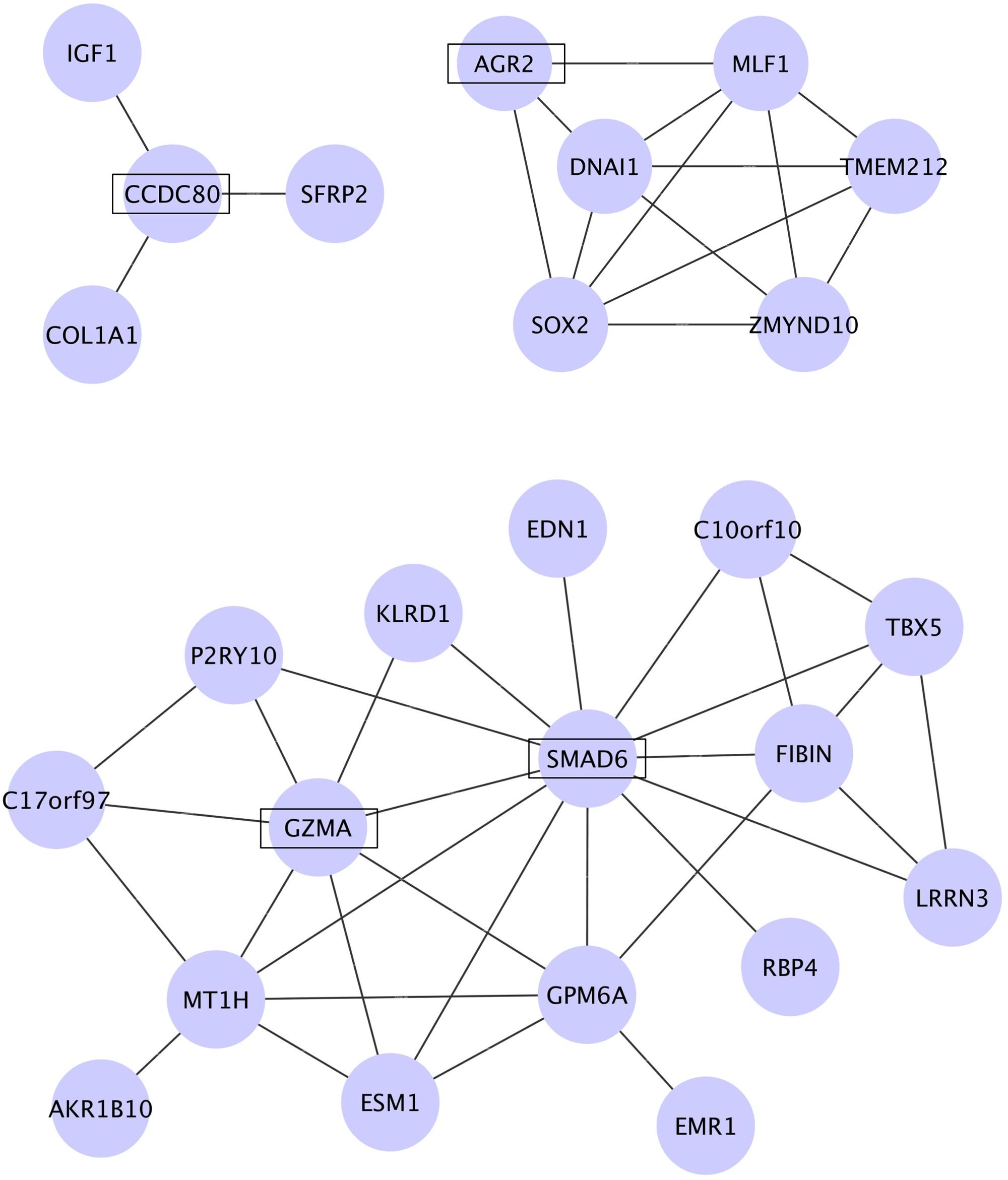
FIGURE 2. Networks among the genes dysregulated in PAH identified by WGCNA. The coefficient of variation (CV) of the normalized probe intensity for each gene in each transcriptome dataset was calculated, and the genes were sorted in descending order by CV. Of the top 3000 genes in each dataset, 40 genes were common to all five datasets and were subjected to WGCNA. The genes were clustered into three modules and the networks identified in these modules are shown. CCDC80, AGR2, SMAD6, and GZMA, which were significantly dysregulated in all five PAH transcriptome datasets, are shown in boxes.
Nitric Oxide Induces Ventral Artery Dilation Through Activation of PKG in Zebrafish
Although comparative transcriptome analysis identified CCDC80 as a novel gene in PAH, the function of CCDC80 in the vasculature is largely unknown. To analyze the function of ccdc80 in vivo, we first established a functionally relevant PAH model using Tg (fli1:EGFP) zebrafish, which express EGFP in vascular endothelial cells (Lawson and Weinstein, 2002). For this, we examined the effect of nitric oxide (NO) on the ventral artery. Treatment of zebrafish with the NO donor SNP dose-dependently dilated the ventral artery (Supplementary Figure S1A), and this was significantly attenuated by co-treatment with KT5823, a specific inhibitor of PKG (Supplementary Figure S1B), indicating that the effects of NO on the ventral artery diameter were regulated though PKG. Inhibition of NO synthase with the specific inhibitor L-NAME significantly decreased the ventral artery diameter when added alone and reversed the artery dilation caused by SNP treatment (Figure 3). The length of ventral artery was not significantly different between the groups. These results indicate that, like the pulmonary artery in mammals, the ventral artery in zebrafish is regulated through NO–PKG signaling.
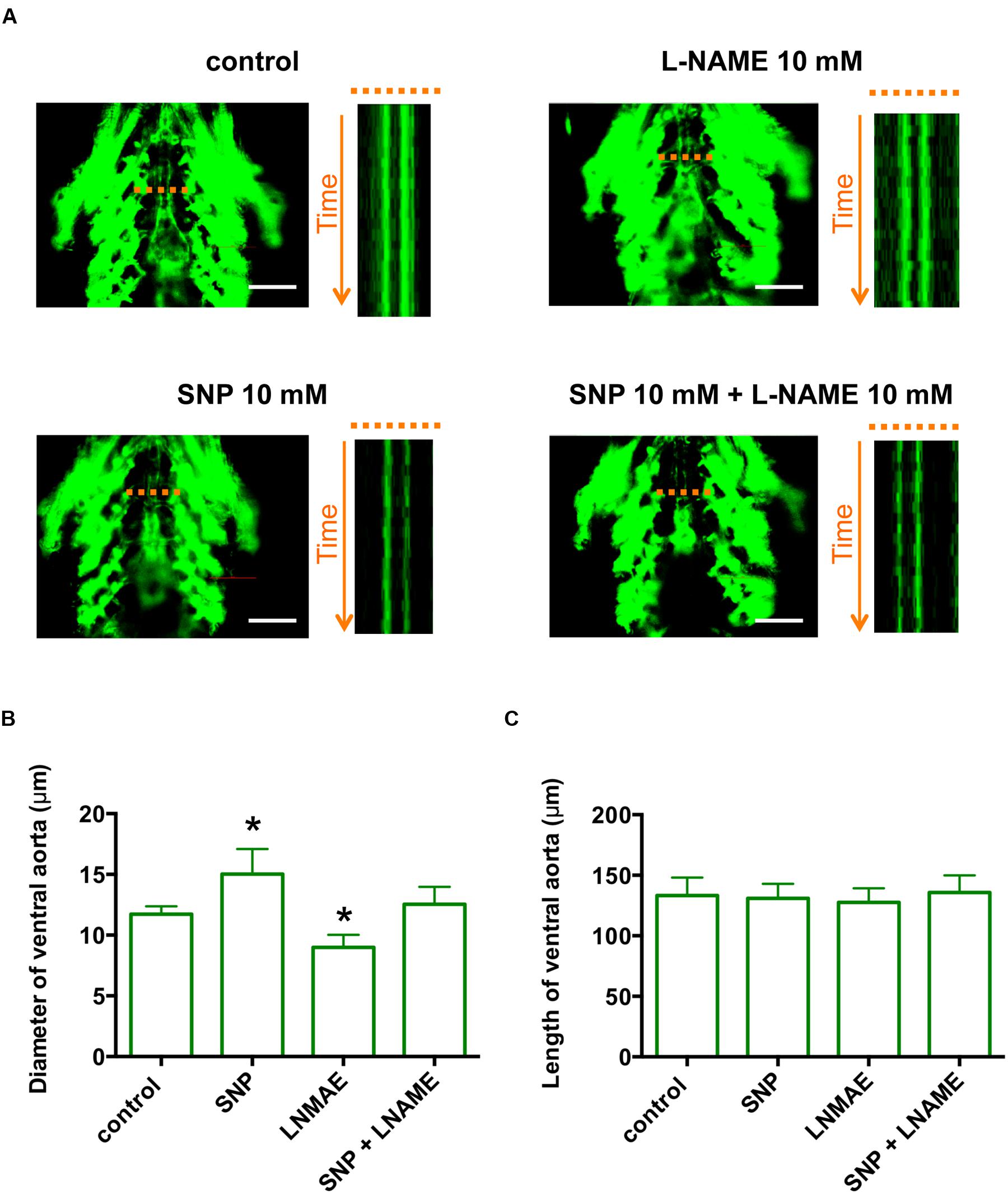
FIGURE 3. The zebrafish ventral artery diameter is increased by a NO donor and decreased by a NO synthase inhibitor. (A) In vivo imaging of the ventral artery of Tg (fli1:EGFP) zebrafish at 6 dpf. Zebrafish were incubated with or without 10 mM Sodium nitroprusside (SNP) and/or 10 mM L-NAME for 2 h and then placed on microscope slides on their backs. The ventral artery was imaged under a fluorescent microscope at 100 frames/s for 5 s. The image stack projections and time-lapse imaging traces at the level of the ventral artery (dotted lines) are shown. Scale bars, 100 μm. (B) The diameter of the ventral artery was increased and decreased by zebrafish treatment with SNP and L-NAME, respectively, relative to untreated zebrafish. The ventral artery diameter of zebrafish co-treated with SNP and L-NAME was not significantly different from that of control zebrafish. The ventral artery diameters were measured using time-lapse imaging. N = 5 for each group. *p < 0.05 vs control. (C) The length of ventral artery in zebrafish treated with SNP and/or L-NAME was not significantly different compared to that in untreated zebrafish. N = 5 for each group.
ccdc80 Knockout Increases Ventral Artery Diameter and Decreases Expression of col1a1 and edn1 in Zebrafish
We next determined whether ccdc80 plays a role in controlling ventral artery diameter in zebrafish. Previous reports have demonstrated that ccdc80 is expressed in the cardiovascular system in zebrafish (Brusegan et al., 2012; Della Noce et al., 2015). Using the CRISPR/Cas9 system (Aida et al., 2015; Kotani et al., 2015), we knocked out ccdc80 in Tg (fli1:EGFP) zebrafish (Supplementary Figure S2). When these animals were examined by fluorescence microscopy, we observed that the ventral artery diameter in ccdc80-KO zebrafish was significantly larger than that of control zebrafish (Figure 4A). The length of ventral artery was not significantly different between the groups, verifying that the wider diameter of the ventral artery in ccdc80-KO zebrafish was not related to the length of the ventral artery in the KO group (Figure 4A).
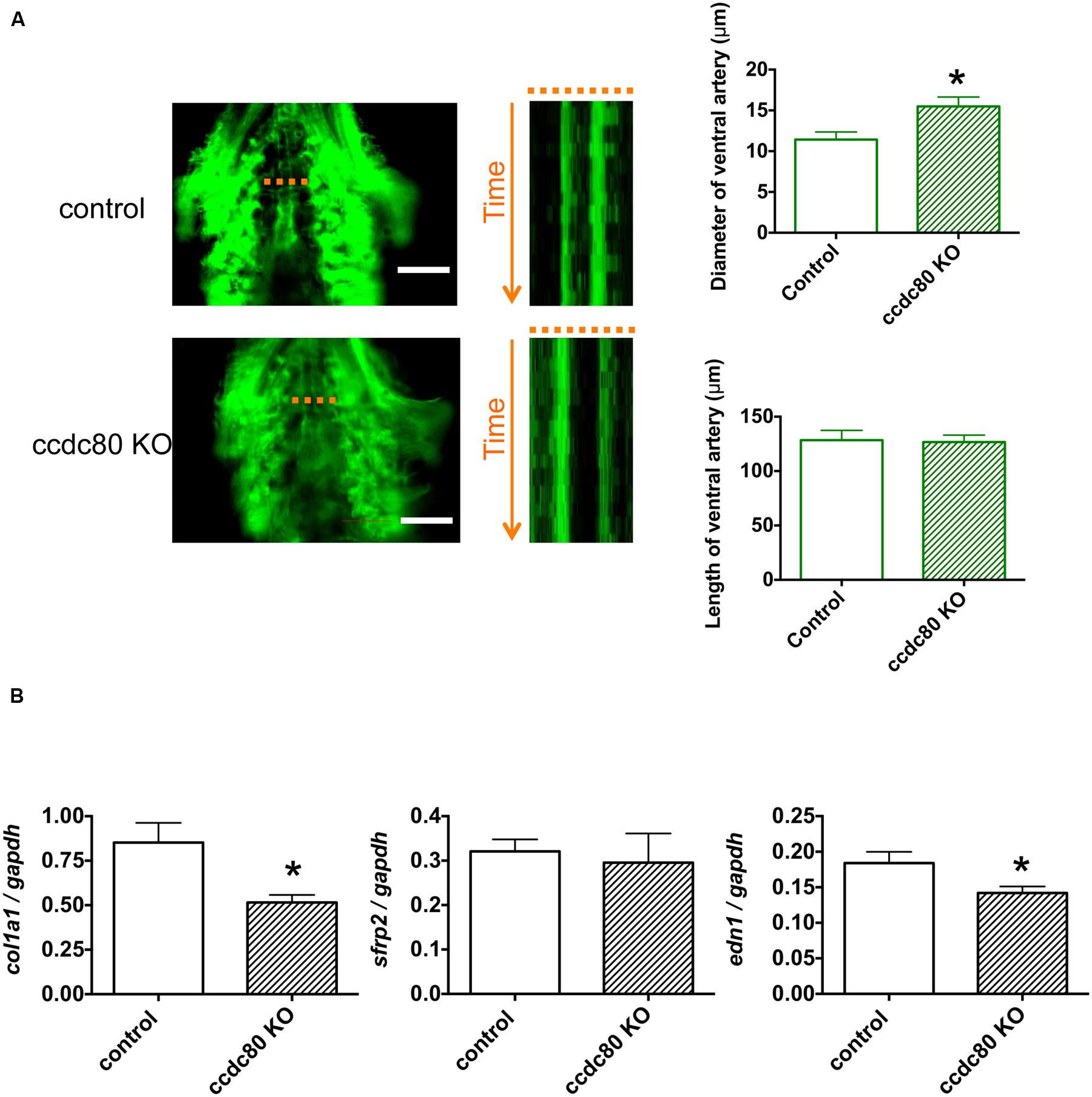
FIGURE 4. Knockout of ccdc80 causes ventral artery dilation and decreases col1a1 and edn1 expression in zebrafish. (A) In vivo imaging of the ventral artery of control and ccdc80 knockout Tg (fli1:EGFP) zebrafish at 6 dpf. Zebrafish were placed on slides on their backs, and the ventral artery was imaged under a fluorescence microscope at 100 frames/s for 5 s. Image stack projections and time-lapse imaging traces at the level of the ventral artery (dotted lines) are shown. Bar, 100 μm. The ventral artery diameter of the ccdc80 knockout zebrafish was significantly larger than that of control, whereas the length of ventral artery was not significantly different between groups. The upper right bar graph shows quantification of ventral artery diameters measured using time-lapse imaging. The lower right bar graph shows quantification of ventral artery lengths. N = 6 or 9 for the control and ccdc80 knockout groups, respectively. *p < 0.05 vs. control. (B) qPCR analysis of col1a1, sfrp2, and edn1 mRNA levels in control and ccdc80 knockout zebrafish at 6 dpf. Expression of col1a1, sfrp2, and edn1 was normalized to that of gapdh. N = 8 per group. *p < 0.05 vs. control groups.
Because WGCNA identified COL1A1 and SFRP2 in the CCDC80 network (Figure 2), we compared the whole-body expression of col1a1 and sfrp2 mRNA in control and ccdc80-KO zebrafish. As shown in Figure 4B, the expression of col1a1 mRNA was significantly lower in ccdc80-KO animals compared with the controls, whereas srfp2 mRNA levels were unchanged. COL1A1 expression is increased by overexpression of EDN1 (Hirt et al., 2012) and decreased by inhibition of EDN1 synthesis (Cowling, 2015); therefore, we also measured the effect of ccdc80 deletion on edn1 expression. We found that edn1 mRNA levels were significantly lower in ccdc80-KO zebrafish compared with the control animals (Figure 4B). These results suggest that CCDC80 may regulate the pulmonary artery tone by modulating endothelin-1-induced collagen expression.
Ccdc80 Expression Is Increased in the Pulmonary Vascular Lesion in a Rat PAH Model
To establish the significance of our findings for PAH pathology, we performed immunohistochemical staining of Ccdc80 protein in the lung tissues of control rats and rats with PAH caused by SU5416 and hypoxia. Ccdc80 was not readily detected in the PAs of control rats (Figures 5A,E), but there was intense immunoreactivity in the hypertrophied media and adventitia of the pre-acinar PAs (Figure 5B) and in the thickened intima, media, and adventitia of the obstructed intra-acinar PAs (Figure 5F) in the PAH rat. The Ccdc80 immunoreactivity was localized in the α-SMA–positive cells in the PA media but not in the intima or adventitia (Figures 5C,G).
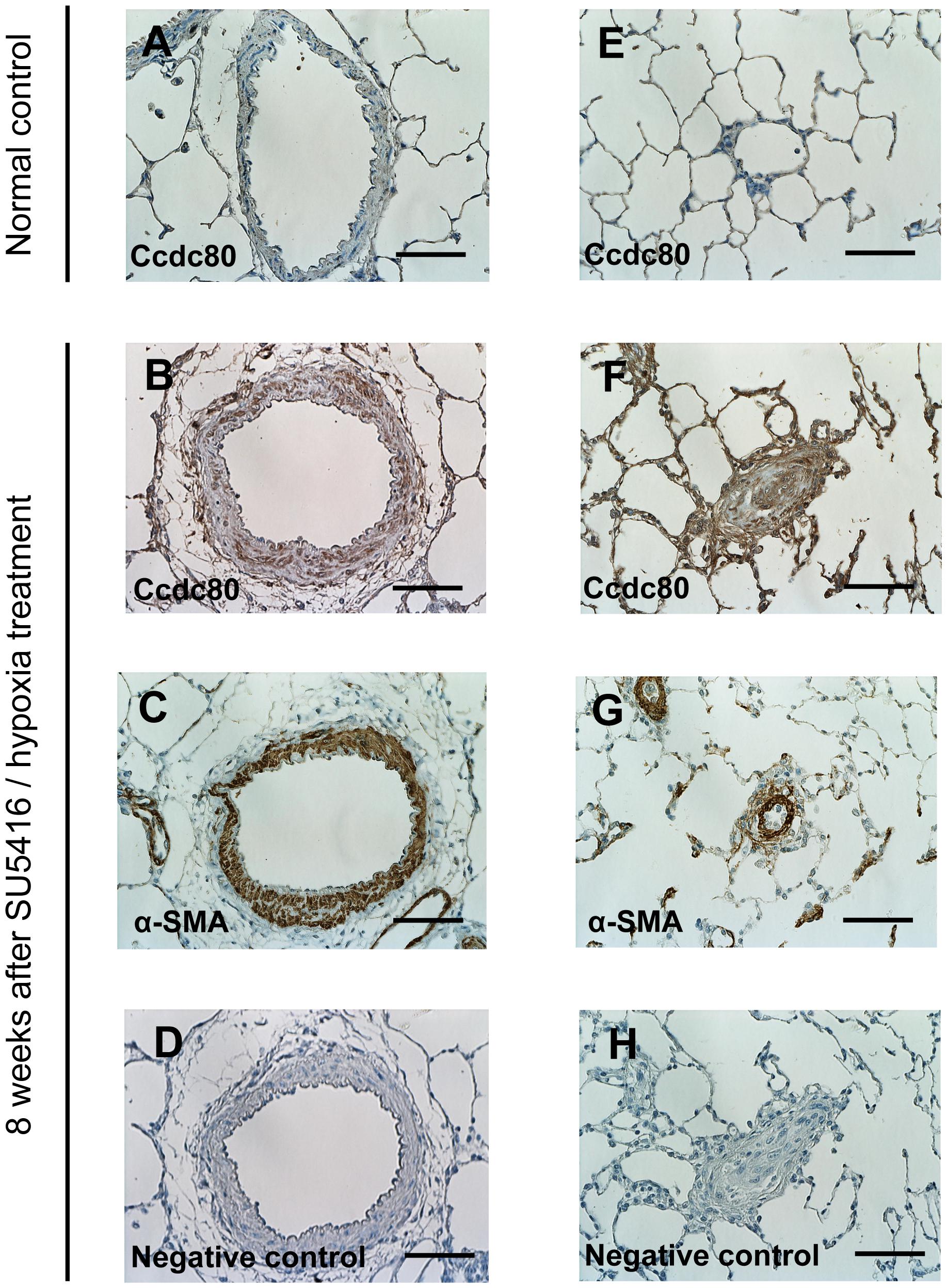
FIGURE 5. Expression of Ccdc80 is increased in the lungs of rats with PAH. (A–H) Representative photomicrographs of pulmonary arteries (PAs) from control rats (A and E) or rats with PAH induced by SU5416 /hypoxia (B–D and F–H). Serial cross-sectional views of pre-acinar PA (A–D) and intra-acinar PA (E–H) immunostained for Ccdc80 and α-smooth muscle actin (α-SMA). Scale bars, 50 μm. Original magnification (×400).
Discussion
Involvement of CCDC80, SMAD6, AGR2, and GZMA in PAH
We demonstrated that expression of CCDC80, SMAD6, AGR2, and GZMA was significantly dysregulated in two cohorts of human PAH patients (Mura et al., 2012; Zhao Y. et al., 2014; Zhao Y.D. et al., 2014) and in three rodent PAH models caused by: (i) treatment with a VEGF receptor inhibitor under conditions of hypoxia (Moreno-Vinasco et al., 2008); (ii) overexpression of Fra-2, a causative gene for systemic sclerosis (Biasin et al., 2014); and (iii) schistosomiasis (Graham et al., 2013).
CCDC80, which was significantly upregulated in the five PAH transcriptome datasets, is a 950-amino acid secreted protein that binds to ECM proteins, including glycosaminoglycans, and promotes cell adhesion (Manabe et al., 2008). Interestingly, human steroid-sensitive gene 1 (SSG1), which is identical to the 530-amino acid amino-terminal sequence of human CCDC80 protein, is highly expressed in cardiovascular systems and is phosphorylated by PKG (Wang et al., 2013). These studies suggest that CCDC80 may regulate vascular function via the ECM and PKG signaling. Consistent with this, we demonstrated that knockout of ccdc80 in zebrafish caused dilation of the ventral artery and decreased the expression of col1a1. We also found that pharmacological inhibition of PKG significantly attenuated the effect of ccdc80 knockout on the zebrafish ventral artery (data not shown). Collectively, these findings point to a possible role for CCDC80 in the pathogenesis of PAH (discussed later).
SMAD6, a member of the SMAD family of TGFβ signaling regulators (Li, 2015), was also downregulated in all five PAH transcriptome datasets. SMAD6 has been shown to negatively regulate TGFβ signaling through the TGFβ-activated kinase (TAK1)–mitogen-activated protein kinase (MAPK) pathway (Jung et al., 2013). Heterozygous mutations in the type II receptor for bone morphogenetic protein (BMPR2), which underlie the majority of the inherited and familial forms of PAH (Guignabert et al., 2015), stimulate the TGFβ–TAK1–MAPK pathway (Nasim et al., 2012). These studies suggest that reduced expression of SMAD6 may increase TGFβ–TAK1–MAPK pathway activity, similar to the effects of BMPR2 mutation. Because inhibition of the TGFβ–TAK1–MAPK pathway rescues abnormal proliferation and apoptosis of pulmonary artery smooth muscle cells isolated from BMPR2 mutant mice (Nasim et al., 2012), normalizing the decrease in SMAD6 expression may be therapeutic in PAH.
The expression of AGR2 was significantly increased in the five PAH transcriptome datasets. AGR2 has been proposed to be involved in development, tissue regeneration, and cancer metastasis (Brychtova et al., 2011). AGR2 can bind to α-dystroglycan, which regulates the ECM and interaction with integrins (Edeleva and Shcherbata, 2013). Thus, the increase in AGR2 expression may be related to the dysregulated ECM observed in PAH.
GZMA was significantly downregulated in all five PAH transcriptome datasets. GZMA is a member of the granzyme family of proteins, which are important mediators of cell death induced by immune cells (Hendel et al., 2010). Previous work has shown that TGFβ decreases GZMA expression and inhibits its function in cytotoxic T cells (Thomas and Massague, 2005). Dysregulation of cytotoxic T cells has been reported in PAH (Ulrich et al., 2008; Savai et al., 2012), raising the possibility that the decrease in GZMA expression in PAH may be related to the dysregulation of cytotoxic T cells.
In addition to these observations, the expression levels and/or activities of CCDC80, AGR2, and GZMA in human plasma have been related to various pathological states (Accardo-Palumbo et al., 2010; Kani et al., 2013; Menni et al., 2015). It may therefore be worthwhile to perform similar measurements of these proteins in the plasma of human PAH patients and determine whether their levels and/or activities correlate with the clinical stages of PAH.
CCDC80 May Regulate Vascular Tone by Modulating Endothelin-1–induced Collagen Expression
In this study, we demonstrated that knockout of ccdc80 in zebrafish caused dilation of the ventral artery and decreased the expression of both edn1 and col1a1. We also showed that Ccdc80 immunoreactivity was increased in the hypertrophied media and adventitia of the pre-acinar PAs and in the thickened intima, media, and adventitia of the obstructed intra-acinar PAs in a rat PAH model.
The expression of CCDC80 was correlated with fibrillin-1 whose expression is induced by TGFβ (Summers et al., 2010), suggesting that it may be induced by TGFβ signaling. TGFβ signaling is activated in familial PAH caused by mutation of BMPR2 and in syndromic PAH caused by systemic sclerosis or schistosomiasis (Varga and Pasche, 2009; Aschner and Downey, 2016). In various models, PAH is ameliorated by blockade of TGFβ signaling through diverse mechanisms, including administration of neutralizing antibodies, antisense nucleotides, or TGFβ receptor kinase inhibitors, and by gene transfer of inhibitory SMAD (Varga and Pasche, 2009; Aschner and Downey, 2016). TGFβ signaling regulates vascular tone, including that of PAs, by regulating the expression of vasodilators such as NO and vasoconstrictors such as endothelin-1 (Pardali and ten Dijke, 2012). These studies suggest that expression of CCDC80 may positively correlate with TGFβ signaling, leading to decreased NO and increased endothelin-1 levels. Further studies using both in vitro and in vivo approaches are required to fully elucidate the functions of CCDC80 in the pathophysiology of PAH and to examine its potential as a marker and/or a therapeutic target in PAH.
Author Contributions
YN conceived the study, performed the bioinformatics analyses, and wrote the manuscript. SS performed the analysis of zebrafish. HS and EZ developed the rat PAH model, performed the immunohistochemical analysis of Ccdc80 expression, and wrote the manuscript. SO, SM, YA, MY, KK, and RK performed the experiments. YM and KM developed the rat PAH model and wrote the manuscript. TT conceived the study and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by JSPS KAKENHI (25670127, 15K15051, 24510069), JST A-step (AS262Z00004Q), the Long-range Research Initiative of the Japan Chemical Industrial Association (13_PT01-01). We thank Junko Koiwa, Hiroko Nakayama, Yuka Hayakawa, Yuka Takahashi, Chizuru Hirota, and Michiko Ariyoshi for assistance with the experiments, and Rie Ikeyama and Yuka Mizutani for administrative support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphar.2016.00142
Abbreviation
DEGs, differentially expressed genes; dpf, days post-fertilization; ECM, extracellular matrix.; L-NAME, N-omega-nitro-L-arginine methyl ester; PAH, pulmonary artery hypertension; PKG, cGMP-dependent protein kinase; SNP, sodium nitroprusside.
References
Accardo-Palumbo, A., D’amelio, L., Pileri, D., D’arpa, N., Mogavero, R., Amato, G., et al. (2010). Reduction of plasma granzyme A correlates with severity of sepsis in burn patients. Burns 36, 811–818. doi: 10.1016/j.burns.2009.11.009
Aida, T., Chiyo, K., Usami, T., Ishikubo, H., Imahashi, R., Wada, Y., et al. (2015). Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 16:87. doi: 10.1186/s13059-015-0653-x
Aschner, Y., and Downey, G. P. (2016). Transforming growth factor-beta: master regulator of the respiratory system in health and disease. Am. J. Respir. Cell Mol. Biol. 54, 647–655. doi: 10.1165/rcmb.2015-0391TR
Auer, T. O., and Del Bene, F. (2014). CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods 69, 142–150. doi: 10.1016/j.ymeth.2014.03.027
Barrett, T., Troup, D. B., Wilhite, S. E., Ledoux, P., Rudnev, D., Evangelista, C., et al. (2009). NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37, D885–D890. doi: 10.1093/nar/gkn764
Biasin, V., Marsh, L. M., Egemnazarov, B., Wilhelm, J., Ghanim, B., Klepetko, W., et al. (2014). Meprin beta, a novel mediator of vascular remodelling underlying pulmonary hypertension. J. Pathol. 233, 7–17. doi: 10.1002/path.4303
Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92. doi: 10.1016/j.febslet.2004.07.055
Brusegan, C., Pistocchi, A., Frassine, A., Della Noce, I., Schepis, F., and Cotelli, F. (2012). Ccdc80-l1 Is involved in axon pathfinding of zebrafish motoneurons. PLoS ONE 7:e31851. doi: 10.1371/journal.pone.0031851
Brychtova, V., Vojtesek, B., and Hrstka, R. (2011). Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett. 304, 1–7. doi: 10.1016/j.canlet.2010.12.023
Carvalho, B. S., and Irizarry, R. A. (2010). A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367. doi: 10.1093/bioinformatics/btq431
Cowling, R. T. (2015). The aging heart, endothelin-1 and the senescent cardiac fibroblast. J. Mol. Cell Cardiol. 81, 12–14. doi: 10.1016/j.yjmcc.2015.01.018
Della Noce, I., Carra, S., Brusegan, C., Critelli, R., Frassine, A., De Lorenzo, C., et al. (2015). The Coiled-Coil Domain Containing 80 (ccdc80) gene regulates gadd45beta2 expression in the developing somites of zebrafish as a new player of the hedgehog pathway. J. Cell. Physiol. 230, 821–830. doi: 10.1002/jcp.24810
Edeleva, E. V., and Shcherbata, H. R. (2013). Stress-induced ECM alteration modulates cellular microRNAs that feedback to readjust the extracellular environment and cell behavior. Front. Genet. 4:305. doi: 10.3389/fgene.2013.00305
Gautier, L., Cope, L., Bolstad, B. M., and Irizarry, R. A. (2004). affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. doi: 10.1093/bioinformatics/btg405
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. doi: 10.1186/gb-2004-5-10-r80
Graham, B. B., Chabon, J., Kumar, R., Kolosionek, E., Gebreab, L., Debella, E., et al. (2013). Protective role of IL-6 in vascular remodeling in Schistosoma pulmonary hypertension. Am. J. Respir. Cell Mol. Biol. 49, 951–959. doi: 10.1165/rcmb.2012-0532OC
Guignabert, C., Tu, L., Girerd, B., Ricard, N., Huertas, A., Montani, D., et al. (2015). New molecular targets of pulmonary vascular remodeling in pulmonary arterial hypertension: importance of endothelial communication. Chest 147, 529–537. doi: 10.1378/chest.14-0862
Hendel, A., Hiebert, P. R., Boivin, W. A., Williams, S. J., and Granville, D. J. (2010). Granzymes in age-related cardiovascular and pulmonary diseases. Cell Death Differ. 17, 596–606. doi: 10.1038/cdd.2010.5
Hirt, M. N., Sörensen, N. A., Bartholdt, L. M., Boeddinghaus, J., Schaaf, S., Eder, A., et al. (2012). Increased afterload induces pathological cardiac hypertrophy: a new in vitro model. Basic Res. Cardiol. 107:307. doi: 10.1007/s00395-012-0307-z
Hoffmann, J., Yin, J., Kukucka, M., Yin, N., Saarikko, I., Sterner-Kock, A., et al. (2011). Mast cells promote lung vascular remodelling in pulmonary hypertension. Eur. Respir. J. 37, 1400–1410. doi: 10.1183/09031936.00043310
Iwashita, Y., Zhang, E., Maruyama, J., Yokochi, A., Yamada, Y., Sawada, H., et al. (2014). Thrombomodulin protects against lung damage created by high level of oxygen with large tidal volume mechanical ventilation in rats. J. Intensive Care 2:57. doi: 10.1186/s40560-014-0057-0
Jonz, M. G., and Nurse, C. A. (2008). New developments on gill innervation: insights from a model vertebrate. J. Exp. Biol. 211, 2371–2378. doi: 10.1242/jeb.010587
Jung, S. M., Lee, J. H., Park, J., Oh, Y. S., Lee, S. K., Park, J. S., et al. (2013). Smad6 inhibits non-canonical TGF-beta1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat. Commun. 4:2562. doi: 10.1038/ncomms3562
Kani, K., Malihi, P. D., Jiang, Y., Wang, H., Wang, Y., Ruderman, D. L., et al. (2013). Anterior gradient 2 (AGR2): blood-based biomarker elevated in metastatic prostate cancer associated with the neuroendocrine phenotype. Prostate 73, 306–315. doi: 10.1002/pros.22569
Kelsh, R. N., Brand, M., Jiang, Y. J., Heisenberg, C. P., Lin, S., Haffter, P., et al. (1996). Zebrafish pigmentation mutations and the processes of neural crest development. Development 123, 369–389.
Korsholm, K., Andersen, A., Kirkfeldt, R. E., Hansen, K. N., Mellemkjaer, S., and Nielsen-Kudsk, J. E. (2015). Survival in an incident cohort of patients with pulmonary arterial hypertension in Denmark. Pulm. Circ. 5, 364–369. doi: 10.1086/681270
Kotani, H., Taimatsu, K., Ohga, R., Ota, S., and Kawahara, A. (2015). Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 Protein Complex in Zebrafish. PLoS ONE 10:e0128319. doi: 10.1371/journal.pone.0128319
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lawson, N. D., and Weinstein, B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318. doi: 10.1006/dbio.2002.0711
Li, Q. (2015). Inhibitory SMADs: potential regulators of ovarian function. Biol. Reprod. 92:50. doi: 10.1095/biolreprod.114.125203
Machado, R. D., Southgate, L., Eichstaedt, C. A., Aldred, M. A., Austin, E. D., Best, D. H., et al. (2015). Pulmonary arterial hypertension: a current perspective on established and emerging molecular genetic defects. Hum. Mutat. 36, 1113–1127. doi: 10.1002/humu.22904
Madonna, R., De Caterina, R., and Geng, Y. J. (2015). Epigenetic regulation of insulin-like growth factor signaling: a novel insight into the pathophysiology of neonatal pulmonary hypertension. Vascul. Pharmacol. 73, 4–7. doi: 10.1016/j.vph.2015.08.002
Manabe, R., Tsutsui, K., Yamada, T., Kimura, M., Nakano, I., Shimono, C., et al. (2008). Transcriptome-based systematic identification of extracellular matrix proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 12849–12854. doi: 10.1073/pnas.0803640105
McLaughlin, V. V., Shah, S. J., Souza, R., and Humbert, M. (2015). Management of pulmonary arterial hypertension. J. Am. Coll. Cardiol. 65, 1976–1997. doi: 10.1016/j.jacc.2015.03.540
Menni, C., Kiddle, S. J., Mangino, M., Vinuela, A., Psatha, M., Steves, C., et al. (2015). Circulating proteomic signatures of chronological age. J. Gerontol. A Biol. Sci. Med. Sci. 70, 809–816. doi: 10.1093/gerona/glu121
Ministry of the Environment Japan (2013). Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain. Notice No.88 [Accessed on April 28, 2006].
Ministry of the Environment Japan (2014). The Law for the Humane Treatment and Management of Animals, Law No. 105 [accessed on October 1, 1973].
Moreno-Vinasco, L., Gomberg-Maitland, M., Maitland, M. L., Desai, A. A., Singleton, P. A., Sammani, S., et al. (2008). Genomic assessment of a multikinase inhibitor, sorafenib, in a rodent model of pulmonary hypertension. Physiol. Genomics 33, 278–291. doi: 10.1152/physiolgenomics.00169.2007
Mura, M., Anraku, M., Yun, Z., Mcrae, K., Liu, M., Waddell, T. K., et al. (2012). Gene expression profiling in the lungs of patients with pulmonary hypertension associated with pulmonary fibrosis. Chest 141, 661–673. doi: 10.1378/chest.11-0449
Nasim, M. T., Ogo, T., Chowdhury, H. M., Zhao, L., Chen, C. N., Rhodes, C., et al. (2012). BMPR-II deficiency elicits pro-proliferative and anti-apoptotic responses through the activation of TGFbeta-TAK1-MAPK pathways in PAH. Hum. Mol. Genet. 21, 2548–2558. doi: 10.1093/hmg/dds073
Nishimura, Y., Inoue, A., Sasagawa, S., Koiwa, J., Kawaguchi, K., Kawase, R., et al. (2016). Using zebrafish in systems toxicology for developmental toxicity testing. Congenit. Anom. (Kyoto) 56, 18–27. doi: 10.1111/cga.12142
Nishimura, Y., Martin, C. L., Vazquez-Lopez, A., Spence, S. J., Alvarez-Retuerto, A. I., Sigman, M., et al. (2007). Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 16, 1682–1698. doi: 10.1093/hmg/ddm116
Nishimura, Y., Murakami, S., Ashikawa, Y., Sasagawa, S., Umemoto, N., Shimada, Y., et al. (2015a). Zebrafish as a systems toxicology model for developmental neurotoxicity testing. Congenit. Anom. (Kyoto) 55, 1–16. doi: 10.1111/cga.12079
Nishimura, Y., Sasagawa, S., Ariyoshi, M., Ichikawa, S., Shimada, Y., Kawaguchi, K., et al. (2015b). Systems pharmacology of adiposity reveals inhibition of EP300 as a common therapeutic mechanism of caloric restriction and resveratrol for obesity. Front. Pharmacol. 6:199. doi: 10.3389/fphar.2015.00199
Oka, T., Nishimura, Y., Zang, L., Hirano, M., Shimada, Y., Wang, Z., et al. (2010). Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 10:21. doi: 10.1186/1472-6793-10-21
Oldham, M. C., Konopka, G., Iwamoto, K., Langfelder, P., Kato, T., Horvath, S., et al. (2008). Functional organization of the transcriptome in human brain. Nat. Neurosci. 11, 1271–1282. doi: 10.1038/nn.2207
Ooi, C. Y., Wang, Z., Tabima, D. M., Eickhoff, J. C., and Chesler, N. C. (2010). The role of collagen in extralobar pulmonary artery stiffening in response to hypoxia-induced pulmonary hypertension. Am. J. Physiol. Heart Circ. Physiol. 299, H1823–H1831. doi: 10.1152/ajpheart.00493.2009
Otsuki, S., Sawada, H., Yodoya, N., Shinohara, T., Kato, T., Ohashi, H., et al. (2015). Potential contribution of phenotypically modulated smooth muscle cells and related inflammation in the development of experimental obstructive pulmonary vasculopathy in rats. PLoS ONE 10:e0118655. doi: 10.1371/journal.pone.0118655
Pardali, E., and ten Dijke, P. (2012). TGFβ Signaling and Cardiovascular Diseases. Int. J. Biol. Sci. 8, 195–213. doi: 10.7150/ijbs.3805
Pezzuto, B., Badagliacca, R., Poscia, R., Ghio, S., D’alto, M., Vitulo, P., et al. (2015). Circulating biomarkers in pulmonary arterial hypertension: update and future direction. J. Heart Lung Transplant. 34, 282–305. doi: 10.1016/j.healun.2014.12.005
Pullamsetti, S. S., Schermuly, R., Ghofrani, A., Weissmann, N., Grimminger, F., and Seeger, W. (2014). Novel and emerging therapies for pulmonary hypertension. Am. J. Respir. Crit. Care Med. 189, 394–400. doi: 10.1164/rccm.201308-1543PP
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Sasagawa, S., Nishimura, Y., Hayakawa, Y., Murakami, S., Ashikawa, Y., Yuge, M., et al. (2016). E2F4 promotes neuronal regeneration and functional recovery after spinal cord injury in zebrafish. Front. Pharmacol. 7:119. doi: 10.3389/fphar.2016.00119
Savai, R., Pullamsetti, S. S., Kolbe, J., Bieniek, E., Voswinckel, R., Fink, L., et al. (2012). Immune and inflammatory cell involvement in the pathology of idiopathic pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 186, 897–908. doi: 10.1164/rccm.201202-0335OC
Science Council of Japan (2006). Guidelines for Proper Conduct of Animal Experiments. Tokyo: Science Council of Japan.
Shinohara, T., Sawada, H., Otsuki, S., Yodoya, N., Kato, T., Ohashi, H., et al. (2015). Macitentan reverses early obstructive pulmonary vasculopathy in rats: early intervention in overcoming the survivin-mediated resistance to apoptosis. Am. J. Physiol. Lung. Cell Mol. Physiol. 308, L523–L538. doi: 10.1152/ajplung.00129.2014
Simonneau, G., Gatzoulis, M. A., Adatia, I., Celermajer, D., Denton, C., Ghofrani, A., et al. (2013). Updated clinical classification of pulmonary hypertension. J. Am. Coll. Cardiol. 62, D34–D41. doi: 10.1016/j.jacc.2013.10.029
Summers, K. M., Raza, S., Van Nimwegen, E., Freeman, T. C., and Hume, D. A. (2010). Co-expression of FBN1 with mesenchyme-specific genes in mouse cell lines: implications for phenotypic variability in Marfan syndrome. Eur. J. Hum. Genet. 18, 1209–1215. doi: 10.1038/ejhg.2010.91
Swain, S. D., Siemsen, D. W., Pullen, R. R., and Han, S. (2014). CD4+ T cells and IFN-gamma are required for the development of Pneumocystis-associated pulmonary hypertension. Am. J. Pathol. 184, 483–493. doi: 10.1016/j.ajpath.2013.10.027
Swärd, K., Sadegh, M. K., Mori, M., Erjefält, J. S., and Rippe, C. (2013). Elevated pulmonary arterial pressure and altered expression of Ddah1 and Arg1 in mice lacking cavin-1/PTRF. Physiol. Rep. 1:e00008. doi: 10.1002/PHY2.8
Thomas, D. A., and Massague, J. (2005). TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380. doi: 10.1016/j.ccr.2005.10.012
Ulrich, S., Nicolls, M. R., Taraseviciene, L., Speich, R., and Voelkel, N. (2008). Increased regulatory and decreased CD8+ cytotoxic T cells in the blood of patients with idiopathic pulmonary arterial hypertension. Respiration 75, 272–280. doi: 10.1159/000111548
UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Varga, J., and Pasche, B. (2009). Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat. Rev. Rheumatol. 5, 200–206. doi: 10.1038/nrrheum.2009.26
Varga, Z. M. (2011). Aquaculture and husbandry at the zebrafish international resource center. Methods Cell Biol. 104, 453–478. doi: 10.1016/B978-0-12-374814-0.00024-0
Wang, G. R., Surks, H. K., Tang, K. M., Zhu, Y., Mendelsohn, M. E., and Blanton, R. M. (2013). Steroid-sensitive gene 1 is a novel cyclic GMP-dependent protein kinase I substrate in vascular smooth muscle cells. J. Biol. Chem. 288, 24972–24983. doi: 10.1074/jbc.M113.456244
Westerfield, M. (2007). A Guide for the Laboratory use of Zebrafish (Danio Rerio). Eugene: University of Oregon Press.
Zhao, Y., Peng, J., Lu, C., Hsin, M., Mura, M., Wu, L., et al. (2014). Metabolomic heterogeneity of pulmonary arterial hypertension. PLoS ONE 9:e88727. doi: 10.1371/journal.pone.0088727
Keywords: pulmonary arterial hypertension, comparative transcriptome analysis, weighted gene co-expression network analysis, CCDC80, EDN1, COL1A1, systems pharmacology
Citation: Sasagawa S, Nishimura Y, Sawada H, Zhang E, Okabe S, Murakami S, Ashikawa Y, Yuge M, Kawaguchi K, Kawase R, Mitani Y, Maruyama K and Tanaka T (2016) Comparative Transcriptome Analysis Identifies CCDC80 as a Novel Gene Associated with Pulmonary Arterial Hypertension. Front. Pharmacol. 7:142. doi: 10.3389/fphar.2016.00142
Received: 09 March 2016; Accepted: 16 May 2016;
Published: 07 June 2016.
Edited by:
Adel Giaid Schwertani, McGill University, CanadaReviewed by:
James S. K. Sham, Johns Hopkins University School of Medicine, USAPascal Bernatchez, University of British Columbia, Canada
Copyright © 2016 Sasagawa, Nishimura, Sawada, Zhang, Okabe, Murakami, Ashikawa, Yuge, Kawaguchi, Kawase, Mitani, Maruyama and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshio Tanaka, tanaka@doc.medic.mie-u.ac.jp
†These authors have contributed equally to this work.
 Shota Sasagawa1†
Shota Sasagawa1† Yuhei Nishimura
Yuhei Nishimura Hirofumi Sawada
Hirofumi Sawada Toshio Tanaka
Toshio Tanaka