- 1Key Laboratory of Receptor-Mediated Gene Regulation and Drug Discovery, School of Medicine, Henan University, Kaifeng, China
- 2State Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China
Aging is associated with age-related diseases and an increase susceptibility of cancer. Dissecting the molecular mechanisms that underlie aging and longevity would contribute to implications for preventing and treating the age-dependent diseases or cancers. Multiple signaling pathways such as the insulin/IGF-1 signaling pathway, TOR signaling, AMPK pathway, JNK pathway and germline signaling have been found to be involved in aging and longevity. And DAF-16/FOXO, as a key transcription factor, could integrate different signals from these pathways to modulate aging, and longevity via shuttling from cytoplasm to nucleus. Hence, understanding how DAF-16/FOXO functions will be pivotal to illustrate the processes of aging and longevity. Here, we summarized how DAF-16/FOXO receives signals from these pathways to affect aging and longevity. We also briefly discussed the transcriptional regulation and posttranslational modifications of DAF-16/FOXO, its co-factors as well as its potential downstream targets participating in lifespan according to the published data in C. elegans and in mammals, and in most cases, we may focus on the studies in C. elegans which has been considered to be a very good animal model for longevity research.
Introduction
Aging is an inevitable process, commonly defined as gradually functional decline in the time-dependent manner of most living organisms. Characterized by a progressive loss of physiological integrity, it is always accompanied by the risks of many human age-related diseases such as neurodegenerative disorders, cardiovascular diseases, type 2 diabetes and various cancers (Sun et al., 2015). In addition, the world is becoming older as about 20% of the globe will be over 60 years in the near future, which will causes much higher health-care costs as well as more burdens on society (Hansen and Kennedy, 2016). Therefore, how to delay the process of aging and eliminate the potential risk factors for the age-related diseases seem to be urgently required.
As a biological process, aging is not so easily measured because it contains dynamic changes in cells to tissues and organs over time as well as an increased probability of death (Tissenbaum, 2012). Human longevity and healthy aging are complex phenotypes, as they are not only controlled by the heritably genetic factors but also are modulated by environments including living conditions, diet, physical activity, health habits, and psychological factors as well as social interaction. Environments are so rapidly changing that it may cause outer or inner stress conditions for individuals, subsequently requiring the gene regulation work coordinately, which declines during the process of aging, leading to more and more cellular damage. And it has been considered that accumulation of cellular damage is the general cause of aging (Vijg and Campisi, 2008; Gems and Partridge, 2013). Hence, it is not surprising that aging is regarded as the outcome of a balance between damage and repair (Haigis and Yankner, 2010). Lopez-Otin et al. proposed nine relatively comprehensive hallmarks to determine common denominators of aging in different organisms especially for mammalian aging: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. Pathological dysfunctions of these nine processes are considered to accelerate aging in mammals while the factors involved in regulation of these hallmarks may contribute to aging (Lopez-Otin et al., 2013; Martins et al., 2016).
The genetic pathways and biochemical processes that modulate aging and longevity are well conserved from budding yeast to the nematode worm Caenorhabdites elegans and mammals (Fontana et al., 2010b; Kenyon C. J., 2010). The forkhead transcription factor FOXO as the key downstream regulator that integrates different signals from these pathways plays a crucial role in aging and longevity. Taken incomparable advantages into account, the roundworm C. elegans has been considered to be an excellent system for studying molecular mechanisms in regulating animal aging and longevity. Here we discuss the evidence for the role of DAF-16/FOXO in aging and longevity, especially the data in C. elegans, which could give clues to the further studies for human aging and longevity.
General Information about DAF-16/FOXO
FOXOs belong to the class O of the Forkhead transcription factors, which is featured by a conserved DNA-binding domain as the Forkhead box or FOX that participates a wide range of important cellular processes such as cell cycle arrest, apoptosis, and metabolism besides its function in stress resistance and longevity (Accili and Arden, 2004).
There are four FOXO genes in mammals: FOXO1 (FKHR), FOXO3 (FKHRL1), FOXO4 (AFX), and FOXO6 sharing high similarity in their structure and function as well as regulation with each other, while invertebrates have only one FOXO gene, named daf-16 in C. elegans, based on the initially isolated dauer defective phenotype when mutated (Albert et al., 1981). daf-16 is predicted to encode eight distinct transcripts from daf-16a to daf-16h, of which daf-16a and daf-16d/f/h are indicated to be the major isoforms involved in dauer arrest and longevity. Human FOXO1 (FKHR), FOXO3 (FKHRL1), FOXO4 proteins share highly similar sequences to DAF-16A and DAF-16D/F/H, especially in Forkhead binding domain. DAF-16A also has the same RxRxxS/T phosphotylation motif with the three FOXOs at their amino-termini, whereas DAF-16D/F/H contains the amino-terminal QxRxxS motif (Kwon et al., 2010; Murphy and Hu, 2013).
As the transcription factor, DAF-16/FOXO contains the DNA binding domain that recognizes a core consensus TTGTTTAC sequence known as the DBE (DAF-16 Binding Element), through which numerous downstream genes called class 1 DAF-16/FOXO targets were discovered (Murphy et al., 2003; Schuster et al., 2010). Besides, DAF-16/FOXO has also been reported to contain DAE (DAf-16 Associated Element) with the GATA site that reverses to CTTATCA sequence over-present in the promoter of the potential target genes, which is responsible for the determination of class 2 DAF-16/FOXO targets as well as some co-transcription factors (McElwee et al., 2004; Tepper et al., 2013).
DAF-16/FOXO Integrates Signals of Different Pathways
The Classic IIS Pathway
“Deregulated nutrient sensing” as one of the aging hallmarks to be firstly described to influence longevity, is mainly regulated by the insulin and IGF-1 signaling (IIS) pathway. And this pathway is so highly conserved to modulate aging and longevity across a great evolutionary distance from invertebrates to mammals that the components in every step found in C. elegans could be corresponded to the homologs in mice or human (Figure 1) (Fontana et al., 2010b; Kenyon C. J., 2010). The IIS pathway is a signal transduction cascades that consists of insulin-like peptides (ILPs), an insulin/IGF-1 receptor (DAF-2), a phosphoinositide 3-kinase (AGE-1/AAP-1/PI3K), serine/threonine kinases (PDK-1, AKT-1 and AKT-2) and the pivotal downstream Forkhead Box O transcription factor (DAF-16) in C. elegans.
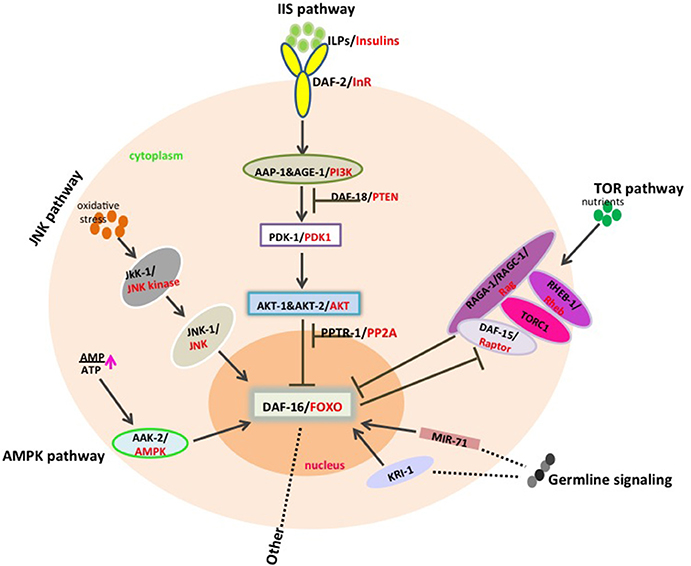
Figure 1. DAF-16/FOXO integrates signals from different pathways to modulate aging and longevity. Insulin-like molecules bind to DAF-2 receptor to lead to the activation of PI3P pathway composed of AGE-1/AAP-1, PDK-1 as well as AKT-1/2, which inhibits DAF-16/FOXO translocation into nucleus by phosphorylation. DAF-18/PTEN and PPTR-1/PP2A negatively regulate the IIS pathway through antagonizing AGE-1/AAP-1 and AKT kinases respectively. Additionally, JNK activity increases under oxidative stress and AMPK is activated upon high AMP/ATP ratios, and both kinases subsequently phosphorylate DAF-16 to promote its activity. Signals from germline, especially in the absence of germline, DAF-16/FOXO would be activated by KRI-1 or by mir-71 in a cell non-autonomous way. TOR pathway is partially dependent on DAF-16 in the complex of TORC1•DAF-15/Raptor together with Rag GTPases such as RAGA-1/RAGC-1, RHEB-1/Rheb, and in turn, DAF-16/FOXO could also inhibit the expression of the TORC1 coactivator daf-15/Raptor.
DAF-16/FOXO receives phosphorylation from the direct upstream AKT kinases mediated signal transduction response to insulin or IGF and is subsequently sequestered in the cytoplasm by 14-3-3 proteins, which antagonizes FoxO and negatively regulates longevity (Brunet et al., 1999). Therefore, any mutations in the pathway genes including the upstream insulin receptor daf-2/IGFR, the signal transducers age-1, pdk-1 as well as akt-1, and akt-2 all show long-lived phenotypes in the corresponding mutants compared with wild type. And the mechanism is dependent on DAF-16 activity, as daf-16 null mutations could fully suppress the life span extension phenotypes of the above mutants. Hence, it is not surprising that the negative regulators of the IIS pathway DAF-18/PTEN and PPTR-1/PP2A mutations exhibit short longevity, because DAF-18/PTEN as a lipid phosphatase antagonizes PI3Ks while PPTR-1/PP2A dephosphorylates ATK kinases, which finally affects the distribution of DAF-16/FOXO (Solari et al., 2005; Padmanabhan et al., 2009). Other genes that function upstream of the IIS pathway, such as unc-31, unc-64, unc-18, or unc-13 also modulate lifespan in a daf-16-dependent manner (Mukhopadhyay et al., 2006). Any tense conditions that cause inner stress to block the IIS pathway like in the presence of the food restriction or the signals failed to be transduced to DAF-16/FOXO would increase the transcriptional activity of DAF-16/FOXO by inducing the translocation of DAF-16/FOXO to nucleus, which could subsequently promote or repress the expression of downstream targets to trigger the resistance to different kinds of stress and prolong the lifespan of the organisms.
TOR Pathway
Another pathway correlated with nutrition affecting longevity is TOR (target of rapamycin) pathway, which was firstly described in C. elegans and was proved evolutionarily conserved later in other organisms (Vellai et al., 2003; Fontana et al., 2010b). Various dietary interventions such as caloric restriction (CR) or dietary restriction (DR) may inactivate TOR pathway to promote lifespan extension. The TOR kinase exists in two distinct complexes, TORC1 containing the coactivator DAF-15/Raptor and TORC2 including RICT-1/Rictor, which function differently in C. elegans and in mammalian cells as well (Zoncu et al., 2011). TORC1-mediated longevity via the GTPases RAGA-1/RAGC-1, RHEB-1/Rheb, and DAF-15/Raptor is dependent on DAF-16/FOXO and is also possibly regulated by another transcription factor SKN-1/Nrf in a feedback manner, whereas TORC2/Rict-1 modulates lifespan mainly through SKN-1/Nrf (Lapierre and Hansen, 2012; Robida-Stubbs et al., 2012). It has also been reported that the long-lived phenotype caused by depletion of LET-363/TOR activity through RNAi interference could not be suppressed by daf-16 mutation, suggesting that the function of TOR may be independent of DAF-16, although let-363 RNAi enhanced dauer formation in daf-2(e1370) mutant worms (Vellai et al., 2003). Therefore, it still needs more details to reveal the correlation between TOR-mediated longevity and DAF-16. Additionally, DAF-16/FOXO negatively regulates the expression of the TORC1 coactivator daf-15/Raptor (Jia et al., 2004).
TORC1 and IIS have distinct effect on DAF-16 as well as its downstream target genes. IIS inhibits and sequesters DAF-16 in cytoplasm via phosphorylation, so that multiple DAF-16 isoforms accumulate in nuclei once IIS is reduced. In contrast, genetic inhibition of TORC1 increases the daf-16 mRNA level and leads to only one isoform (DAF-16f) translocate in intestinal nuclei (Robida-Stubbs et al., 2012). In addition, TORC1 may also affect longevity by phosphorylation S6K (S6 kinase), a crucial regulator of mRNA translation that is involved in longevity in C. elegans (Kapahi et al., 2010).
AMP-Activated Protein Kinase (AMPK) Pathway
AMPK pathway as an energy-sensing signaling pathway responses to stimuli of decreased energy as well as reduced glucose or leptin levels (Greer et al., 2009). And it is the theoretical basis of dietary restriction regimen that is considered to extend both the mean and maximal lifespan in a wide range of species. AMPK is composed of three subunits including the catalytic α subunit and two β, γ regulatory subunits. In mammalian cells, through binding to AMP, ADP or ATP, the γ subunit could induce a conformational change that allosterically influence the activity of the α subunit which could be activated through phosphorylation by the upstream kinase LKB-1 and CAMKKβ (Solari et al., 2005; Woods et al., 2005). In C. elegans, α subunit is encoded by aak-1 and aak-2, while aakb-1 and aakb-2 is for β subunit and five isoforms of γ subunits AAKG-1~5. DAF-16 is necessary for AMPK function in oxidative stress resistance and longevity, as the increased longevity caused by overexpression constitutively active (CA) AMPK was reverted when DAF-16 was inhibited (Greer et al., 2007a). Moreover, the mRNA level of sod-3, one known DAF-16 target gene involved in both stress resistance and longevity, was highly increased in the CA form of AMPK while it was significantly decreased in the aak-2 mutant worms, of which the mechanism is probably that AMPK activates the DAF-16 transcription activity by phosphorylation as AMPK could directly phosphorylates DAF-16 in vitro via the residues different from the consensus motif phosphorylated by AKT kinases (Greer et al., 2007a). And it is also the same with mammalian FOXO3, indicating it may be conserved throughout evolution (Greer et al., 2007b).
It seems that there is a crosstalk between the IIS pathway and AMPK pathway: previous studies showed that the extension lifespan caused by daf-2(lf) could be suppressed by aak-2 mutation, and one potential explanation is that DAF-16 promotes longevity by stimulating expression of genes encoding AMPK in IIS mutants as DAF-16 could activate expression of aak-2, aakb-1, aakg-4, and aakg-5 according to the previous data, which also suggests the mutual activators between DAF-16, and AMPK (Greer et al., 2007a; Tullet et al., 2014).
JNK Signaling Pathway
The JNK (Jun N-terminal kinase) family, a subgroup of MAPK (mitogen-activated protein kinase) superfamily, as a part of signal transduction cascade that is activated by cytokines such as TNF and IL-1, serves as a molecular sensor for various stresses including UV irradiation, ROS (reactive oxygen species), DNA damage, heat, and inflammatory cytokines (Davis, 2000). In C. elegans, overexpression JNK or in the vhp-1 mutant worms that increases JNK activity due to loss of phosphatase activity, showed extension lifespan and resistance to heavy metal toxicity, which may function through phosphorylation DAF-16. Moreover, JNK-1 also promotes the translocation of DAF-16 into nucleus upon heat stress (Mizuno et al., 2004; Oh et al., 2005). Mammalian JNK can directly phosphorylate FOXO4 to enhance its activity (Essers et al., 2004), and JNK may also facilitate FOXO into nucleus by releasing its binding partner 14-3-3 protein via phosphorylation (Sunayama et al., 2005; Yoshida et al., 2005).
JNK pathway has been regarded to act in parallel with the IIS pathway to regulate lifespan before converging onto DAF-16 in C. elegans (Oh et al., 2005). Mammalian components of the JNK signaling pathway also interact with the insulin receptor substrate 1 (IRS-1) and the AKT protein kinase. According to the previous studies, JNK inhibited insulin signal transduction through phosphorylation IRS-1 and activated AKT1 via the scaffold protein JIP1 (JNK-interacting protein) that organizes members of the JNK pathway together (Aguirre et al., 2000; Kim et al., 2003).
Collectively, JNK signaling antagonizes IIS pathway to regulate DAF-16/FOXO, although there is a crosstalk between them. JNK directly phosphorylates DAF-16/FOXO to promote its nuclear localization whereas phosphorylated DAF-16/FOXO by IIS pathway AKT inactively retains in the cytoplasm.
Germline Signaling
Reproductive system that may integrate nutrient signaling and communicate with other tissues through germline to affect aging has been observed in C. elegans, flies as well as in mice, indicating a conserved regulation mechanism across different organisms (Kenyon C., 2010). And it has been reported that the lifespan could be extended by 40–60% if the germline precursor cells were removed or the germline stem cell division were prevented in C. elegans (Hsin and Kenyon, 1999; Arantes-Oliveira et al., 2002). A steroid hormone pathway that includes the key components DAF-36/NVD, DAF-9/CYP27 as well as DAF-12/NHR is required for lifespan extension in response to germline loss, and DAF-12/NHR and DAF-9/CYP27 probably form complex with DAF-16/FOXO to function, although the detailed mechanisms remain to be further determined (Dowell et al., 2003; Hansen et al., 2013). In ablated-germline worms, transcription factor DAF-16/FOXO was activated and primarily translocated to the intestinal nucleus, which was found to be regulated by an intestinal ankyrin repeat protein KRI-1 or by microRNAs such as mir-71 in a cell non-autonomous way (Berman and Kenyon, 2006; Boulias and Horvitz, 2012). Once DAF-16/FOXO was translocated into nucleus, it would activates some downstream targets that may be involved in fat metabolism such as the lipases lipl-4 and lips-17, the direct regulator for longevity in germline-less animals (Wang et al., 2008; McCormick et al., 2012). In addition, DAF-16/FOXO also modulates numerous genes that take part in steroid hormone metabolism such as the steroid hormone dehydrogenases cytochrome P450s (Hansen et al., 2013), suggesting an indirect regulation manner.
Several components of the IIS pathway including the negative regulator DAF-18/PTEN (Berman and Kenyon, 2006), the cofactor SMK-1/SMEK-1 (Wolff et al., 2006) and the transcription factor HSF-1 (Hansen et al., 2005) are also responsible for lifespan extension upon germline loss besides that germline ablation further extends lifespan of long-lived daf-2 mutants, indicating a connection between IIS pathway, and germline signaling. However, the distribution of DAF-16/FOXO in tissues shows different under the regulation of the two pathways. The IIS pathway mainly affects DAF-16/FOXO in both neuronal and intestinal cells, whereas germline ablation leads to DAF-16/FOXO translocation to the nucleus primarily in intestinal cells (Lapierre and Hansen, 2012).
There also exist other components that function dependent on DAF-16/FOXO. Example, in C. elegans, there is a special developmental larval stage called dauer, and the worms arrest at the dauer diapause to live longer upon tense conditions, which is mainly regulated by IIS pathway as well as TGF-β like signaling pathway composed of TGF-β-like ligand DAF-7, the Type 1 and 2 receptors DAF-1 and DAF-4, and the downstream DAF-3 Smad and DAF-5 Sno/Ski (Fielenbach and Antebi, 2008; Gumienny and Savage-Dunn, 2013). According to the genetics epistasis analysis, these two pathways may function in parallel. The TGF-β like signaling pathway could also regulate DAF-16 localization and the DAF-16 target gene sod-3 transcription (Shaw et al., 2007). In addition, PDP-1, genetically function at the level of the R-SMAD proteins DAF-16, and DAF-8 in the TGF-β like signaling pathway, also promotes DAF-16 nucleus localization and transcriptional activity, therefore, it was considered to link TGF-β, and IIS pathways to modulate longevity and development (Narasimhan et al., 2011).
Regulations of DAF-16/FOXO
Temporal regulation of DAF-16/FOXO expression is conserved. FOXO3 and FOXO4 transcript are undetectable at very young stage but increased in the duodenum in elder rats (Huang et al., 2011), and the human FOXO1 mRNA level shows significantly enriched in old individual muscles as well (Buford et al., 2011), while C. elegans daf-16d/f transcription expression also exhibits dramatically increased during the young adult stage, and this upregulation of daf-16d/f expression is responsible for longevity. So far, elt-2 (GATA transcription factor) and swsn-1 (core subunit of SWI/SNF complex) have been identified to modulate daf-16d/f mRNA level besides that TORC1 negatively regulate the daf-16 gene transcription expression (Bansal et al., 2014). However, the crucial roles of DAF-16/FOXO in aging and longevity as well as other cellular processes are mainly through post-translational modifications including phosphorylation, acetylation, methylation and ubiquitination.
DAF-16/FOXO is prone to be phosphorylated by a group of protein kinases at different sites in response to external or internal stimuli, which leads to the alteration of the subcellular localization, protein stability, DNA-binding properties and transcriptional activity. As the primary substrate of AKT/PKB, the phosphoacceptor sites of C. elegans DAF-16 are conserved in mammals, and phosphorylated DAF-16/FOXO by ATK/PKB shows inhibitory transcription activity with retention in cytoplasm (Paradis and Ruvkun, 1998; Brunet et al., 1999; Kwon et al., 2010). In contrast, another C. elegans AGC family serine-threonine kinase SGK-1 exhibits opposite effect to influence longevity as well as stress resistance in a DAF-16 -dependent manner without affecting DAF-16 subcellular localization (Chen et al., 2013), which is different in mammalian cells (Brunet et al., 2001). DAF-16/FOXO also undergoes inhibitory phosphorylation by protein kinases such as AMPK, ERK, CDK2, GSK3, CK1, and IKK when exposed to stimuli (Hu et al., 2004; Huang et al., 2006; Greer et al., 2007a; Yang et al., 2008; Huo et al., 2014), whereas DAF-16/FOXO is usually activated and translocated in nucleus via phosphorylation in the presence of CDK1, JNK, MST1 as well as CAMKII (Essers et al., 2004; Oh et al., 2005; Lehtinen et al., 2006; Yuan et al., 2008; Tao et al., 2013).
DAF-16/FOXO also undergoes acetylation to mediate numerous biological processes besides aging and longevity under the control of histone acetyltransferase CBP/P300 (van der Heide and Smidt, 2005). And acetylated DAF-16/FOXO is more likely to localize to cytoplasm with the abolished DNA-binding capacity (Matsuzaki et al., 2005). On the other hand, Histone deacetylases (HDACs) SIR2/SIRT1 has been demonstrated to be required for lifespan extension possibly through its deacetylase activity and modulation the downstream targets of DAF-16/FOXO (Bordone and Guarente, 2005; Guarente and Picard, 2005; Wang and Tissenbaum, 2006).
The protein stability of DAF-16/FOXO is determined by the ubiquitin-proteasome pathway. Several ubiquitin E3 ligases such as MDM2, SKP2, COP1, and CHIP are responsible for the ubiquitination and degradation of FOXOs (Huang et al., 2005; Kato et al., 2008; Yang et al., 2008; Li et al., 2009). Additionally, Phosphorylation by ERK or IKK also contributes to the FOXOs degradation (Hu et al., 2004; Yang et al., 2008).
DAF-16/FOXO could also be methylated by the protein arginine methyltransferase PRMT1. By blocking the phosphorylation of DAF-16 via AKT, C. elegans PRMT1 plays an important role in longevity and stress tolerance through direct methylation DAF-16 (Takahashi et al., 2011), which is similar to what has been discovered in mammalian cells (Yamagata et al., 2008).
Targets of DAF-16/FOXO
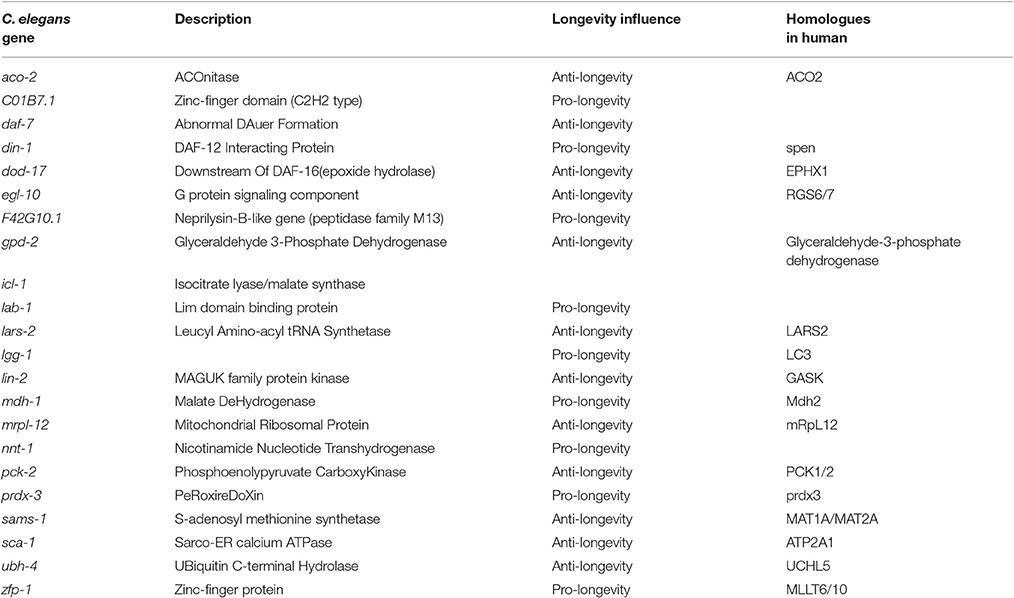
For the crucial role in aging and longevity, it is essential to determine the targets of DAF-16/FOXO transcription factor that might presumably function by activating or inhibiting its downstream genes. By using various high throughout techniques including microarray (McElwee et al., 2003; Murphy et al., 2003), proteomics (Dong et al., 2007), and DamID (DNA adenine methyltransferase identification) (Schuster et al., 2010), numerous direct or indirect targets were identified, and about 109 DAF-16 direct targets in C. elegans were indicated (Li and Zhang, 2016). Since DAF-16 is involved in multiple biological processes besides the regulation in aging and longevity, so do these downstream targets. Here, we just summarized the potential targets in the list that have been reported to influence aging and longevity (see in Table 1) (Oh et al., 2006; Tullet, 2015; Li and Zhang, 2016), and the homologs in human are illustrated based on the descriptions in Wormbase.
Conclusions and Perspectives
Future exploration needs unravel the precise roles of DAF-16/FOXO in aging and longevity so that it would provide more implications to delay or prevent aging and age-dependent diseases.
Author Contributions
XS wrote the manuscript. YW and WC reviewed and revised the manuscript.
Funding
This study was conducted for a project in the Strategic Program of the National Institute for Public Health and the Environment (RIVM: S132001/Personalised Medicine).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. This work is supported by the National Natural Science Foundation of China (Grant No. 31601091) to XS, the National Natural Science Foundation of China (Grant No. 81472232 and Grant No. 81270522) to WC, the National Natural Science Foundation of China (Grant No. 81370537 and No. 81672433) to YW, Program for Science and Technology Innovation Talents in Universities of Henan Province (HASTIT, Grant No. 13HASTIT024) and Plan for Scientific Innovation Talent of Henan Province to WC, and the Fundamental Research Funds for the Central Universities (Grant No. PYBZ1706 and Grant No. YS1407) to YW.
References
Accili, D., and Arden, K. C. (2004). FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421–426. doi: 10.1016/S0092-8674(04)00452-0
Aguirre, V., Uchida, T., Yenush, L., Davis, R., and White, M. F. (2000). The c-Jun NH-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser (307). J. Biol. Chem. 275, 9047–9054. doi: 10.1074/jbc.275.12.9047
Albert, P. S., Brown, S. J., and Riddle, D. L. (1981). Sensory control of dauer larva formation in Caenorhabditis elegans. J. Comp. Neurol. 198, 435–451. doi: 10.1002/cne.901980305
Arantes-Oliveira, N., Apfeld, J., Dillin, A., and Kenyon, C. (2002). Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505. doi: 10.1126/science.1065768
Bansal, A., Kwon, E. S., Conte, D. Jr., Liu, H., Gilchrist, M. J., MacNeil, L. T., et al. (2014). Transcriptional regulation of Caenorhabditis elegans FOXO/DAF-16 modulates lifespan. Longev. Healthspan. 3:5. doi: 10.1186/2046-2395-3-5
Berman, J. R., and Kenyon, C. (2006). Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124, 1055–1068. doi: 10.1016/j.cell.2006.01.039
Bordone, L., and Guarente, L. (2005). Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 6, 298–305. doi: 10.1038/nrm1616
Boulias, K., and Horvitz, H. R. (2012). The, C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15, 439–450. doi: 10.1016/j.cmet.2012.02.014
Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., et al. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868. doi: 10.1016/S0092-8674(00)80595-4
Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A., and Greenberg, M. E. (2001). Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21, 952–965. doi: 10.1128/MCB.21.3.952-965.2001
Buford, T. W., Cooke, M. B., Shelmadine, B. D., Hudson, G. M., Redd, L. L., and Willoughby, D. S. (2011). Differential gene expression of FoxO1, ID1, and ID3 between young and older men and associations with muscle mass and function. Aging Clin. Exp. Res. 23, 170–174. doi: 10.1007/BF03324957
Chen, A. T., Guo, C., Dumas, K. J., Ashrafi, K., and Hu, P. J. (2013). Effects of Caenorhabditis elegans sgk-1 mutations on lifespan, stress resistance, and DAF-16/FoxO regulation. Aging Cell. 12, 932–940. doi: 10.1111/acel.12120
Davis, R. J. (2000). Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252. doi: 10.1016/S0092-8674(00)00116-1
Dong, M. Q., Venable, J. D., Au, N., Xu, T., Park, S. K., Cociorva, D., et al. (2007). Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663. doi: 10.1126/science.1139952
Dowell, P., Otto, T. C., Adi, S., and Lane, M. D. (2003). Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J. Biol. Chem. 278, 45485–45491. doi: 10.1074/jbc.M309069200
Essers, M. A., Weijzen, S., de Vries-Smits, A. M., Saarloos, I., de Ruiter, N. D., Bos, J. L., et al. (2004). FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812. doi: 10.1038/sj.emboj.7600476
Fielenbach, N., and Antebi, A. (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22, 2149–2165. doi: 10.1101/gad.1701508
Fontana, L., Partridge, L., and Longo, V. D. (2010b). Extending healthy life span–from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
Gems, D., and Partridge, L. (2013). Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644. doi: 10.1146/annurev-physiol-030212-183712
Greer, E. L., Banko, M. R., and Brunet, A. (2009). AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann. N. Y. Acad. Sci. 1170, 688–692. doi: 10.1111/j.1749-6632.2009.04019.x
Greer, E. L., Dowlatshahi, D., Banko, M. R., Villen, J., Hoang, K., Blanchard, D., et al. (2007a). An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656. doi: 10.1016/j.cub.2007.08.047
Greer, E. L., Oskoui, P. R., Banko, M. R., Maniar, J. M., Gygi, M. P., Gygi, S. P., et al. (2007b). The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119. doi: 10.1074/jbc.M705325200
Guarente, L., and Picard, F. (2005). Calorie restriction–the SIR2 connection. Cell 120, 473–482. doi: 10.1016/j.cell.2005.01.029
Gumienny, T. L., and Savage-Dunn, C. (2013). TGF-β signaling in C. elegans. WormBook. doi: 10.1895/wormbook.1.22.2
Haigis, M. C., and Yankner, B. A. (2010). The aging stress response. Mol. Cell. 40, 333–344. doi: 10.1016/j.molcel.2010.10.002
Hansen, M., and Kennedy, B. K. (2016). Does longer lifespan mean longer healthspan? Trends Cell Biol. 26, 565–568. doi: 10.1016/j.tcb.2016.05.002
Hansen, M., Flatt, T., and Aguilaniu, H. (2013). Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 17, 10–19. doi: 10.1016/j.cmet.2012.12.003
Hansen, M., Hsu, A. L., Dillin, A., and Kenyon, C. (2005). New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1:10017. doi: 10.1371/journal.pgen.0010017
Hsin, H., and Kenyon, C. (1999). Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366. doi: 10.1038/20694
Hu, M. C., Lee, D. F., Xia, W., Golfman, L. S., Ou-Yang, F., Yang, J. Y., et al. (2004). IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117, 225–237. doi: 10.1016/S0092-8674(04)00302-2
Huang, H., Regan, K. M., Lou, Z., Chen, J., and Tindall, D. J. (2006). CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314, 294–297. doi: 10.1126/science.1130512
Huang, H., Regan, K. M., Wang, F., Wang, D., Smith, D. I., van Deursen, J. M., et al. (2005). Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 102, 1649–1654. doi: 10.1073/pnas.0406789102
Huang, P., Zhou, Z. Q., Huang, R. H., Zhou, B., Wei, Q. W., and Shi, F. X. (2011). Age-dependent expression of forkhead box O proteins in the duodenum of rats. J. Zhejiang Univ. Sci. B. 12, 730–735. doi: 10.1631/jzus.B1000298
Huo, X., Liu, S., Shao, T., Hua, H., Kong, Q., Wang, J., et al. (2014). GSK3 protein positively regulates type I insulin-like growth factor receptor through forkhead transcription factors FOXO1/3/4. J. Biol. Chem. 289, 24759–24770. doi: 10.1074/jbc.M114.580738
Jia, K., Chen, D., and Riddle, D. L. (2004). The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906. doi: 10.1242/dev.01255
Kapahi, P., Chen, D., Rogers, A. N., Katewa, S. D., Li, P. W., Thomas, E. L., et al. (2010). R., less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465. doi: 10.1016/j.cmet.2010.05.001
Kato, S., Ding, J., Pisck, E., Jhala, U. S., and Du, K. (2008). COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J. Biol. Chem. 283, 35464–35473. doi: 10.1074/jbc.M801011200
Kenyon, C. (2010). A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann. N.Y. Acad. Sci. 1204, 156–162. doi: 10.1111/j.1749-6632.2010.05640.x
Kim, A. H., Sasaki, T., and Chao, M. V. (2003). JNK-interacting protein 1 promotes Akt1 activation. J. Biol. Chem. 278, 29830–29836. doi: 10.1074/jbc.M305349200
Kwon, E. S., Narasimhan, S. D., Yen, K., and Tissenbaum, H. A. (2010). A new DAF-16 isoform regulates longevity. Nature 466, 498–502. doi: 10.1038/nature09184
Lapierre, L. R., and Hansen, M. (2012). Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 23, 637–644. doi: 10.1016/j.tem.2012.07.007
Lehtinen, M. K., Yuan, Z., Boag, P. R., Yang, Y., Villen, J., Becker, E. B., et al. (2006). A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125, 987–1001. doi: 10.1016/j.cell.2006.03.046
Li, F., Xie, P., Fan, Y., Zhang, H., Zheng, L., Gu, D., et al. (2009). C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J. Biol. Chem. 284, 20090–20098. doi: 10.1074/jbc.M109.017046
Li, Y. H., and Zhang, G. G. (2016). Towards understanding the lifespan extension by reduced insulin signaling: bioinformatics analysis of DAF-16/FOXO direct targets in Caenorhabditis elegans. Oncotarget 7, 19185–19192. doi: 10.18632/oncotarget.8313
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153, 1194–1217. doi: 10.1016/j.cell.2013.05.039
Martins, R., Lithgow, G. J., and Link, W. (2016). Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 15, 196–207. doi: 10.1111/acel.12427
Matsuzaki, H., Daitoku, H., Hatta, M., Aoyama, H., Yoshimochi, K., and Fukamizu, A. (2005). Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 102, 11278–11283. doi: 10.1073/pnas.0502738102
McCormick, M., Chen, K., Ramaswamy, P., and Kenyon, C. (2012). New genes that extend Caenorhabditis elegans' lifespan in response to reproductive signals. Aging Cell. 11, 192–202. doi: 10.1111/j.1474-9726.2011.00768.x
McElwee, J. J., Schuster, E., Blanc, E., Thomas, J. H., and Gems, D. (2004). Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279, 44533–44543. doi: 10.1074/jbc.M406207200
McElwee, J., Bubb, K., and Thomas, J. H. (2003). Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2, 111–121. doi: 10.1046/j.1474-9728.2003.00043.x
Mizuno, T., Hisamoto, N., Terada, T., Kondo, T., Adachi, M., Nishida, E., et al. (2004). The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 23, 2226–2234. doi: 10.1038/sj.emboj.7600226
Mukhopadhyay, A., Oh, S. W., and Tissenbaum, H. A. (2006). Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 41, 928–934. doi: 10.1016/j.exger.2006.05.020
Murphy, C. T., and Hu, P. J. (2013). Insulin/insulin-like growth factor signaling in C. elegans. WormBook. doi: 10.1895/wormbook.1.164.1
Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., et al. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283. doi: 10.1038/nature01789
Narasimhan, S. D., Yen, K., Bansal, A., Kwon, E. S., Padmanabhan, S., and Tissenbaum, H. A. (2011). PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet. 7:e1001377. doi: 10.1371/journal.pgen.1001377
Oh, S. W., Mukhopadhyay, A., Dixit, B. L., Raha, T., Green, M. R., and Tissenbaum, H. A. (2006). Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38, 251–257. doi: 10.1038/ng1723
Oh, S. W., Mukhopadhyay, A., Svrzikapa, N., Jiang, F., Davis, R. J., and Tissenbaum, H. A. (2005). JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. U.S.A. 102, 4494–4499. doi: 10.1073/pnas.0500749102
Padmanabhan, S., Mukhopadhyay, A., Narasimhan, S. D., Tesz, G., Czech, M. P., and Tissenbaum, H. A. (2009). A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell 136, 939–951. doi: 10.1016/j.cell.2009.01.025
Paradis, S., and Ruvkun, G. (1998). Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12, 2488–2498. doi: 10.1101/gad.12.16.2488
Robida-Stubbs, S., Glover-Cutter, K., Lamming, D. W., Mizunuma, M., Narasimhan, S. D., Neumann-Haefelin, E., et al. (2012). TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724. doi: 10.1016/j.cmet.2012.04.007
Schuster, E., McElwee, J. J., Tullet, J. M., Doonan, R., Matthijssens, F., Reece-Hoyes, J. S., et al. (2010). DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 6:399. doi: 10.1038/msb.2010.54
Shaw, W. M., Luo, S., Landis, J., Ashraf, J., and Murphy, C. T. (2007). The, C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr. Biol. 17, 1635–1645. doi: 10.1016/j.cub.2007.08.058
Solari, F., Bourbon-Piffaut, A., Masse, I., Payrastre, B., Chan, A. M., and Billaud, M. (2005). The human tumour suppressor PTEN regulates longevity and dauer formation in Caenorhabditis elegans. Oncogene 24, 20–27. doi: 10.1038/sj.onc.1207978
Sun, X., Chen, W. D., and Wang, Y. D. (2015). beta-Amyloid: the key peptide in the pathogenesis of Alzheimer's disease. Front. Pharmacol. 6:221. doi: 10.3389/fphar.2015.00221
Sunayama, J., Tsuruta, F., Masuyama, N., and Gotoh, Y. (2005). JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 170, 295–304. doi: 10.1083/jcb.200409117
Takahashi, Y., Daitoku, H., Hirota, K., Tamiya, H., Yokoyama, A., Kako, K., et al. (2011). Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 13, 505–516. doi: 10.1016/j.cmet.2011.03.017
Tao, L., Xie, Q., Ding, Y. H., Li, S. T., Peng, S., Zhang, Y. P., et al. (2013). CAMKII and calcineurin regulate the lifespan of Caenorhabditis elegans through the FOXO transcription factor DAF-16. Elife 2:e00518. doi: 10.7554/eLife.00518
Tepper, R. G., Ashraf, J., Kaletsky, R., Kleemann, G., Murphy, C. T., and Bussemaker, H. J. (2013). PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154, 676–690. doi: 10.1016/j.cell.2013.07.006
Tissenbaum, H. A. (2012). Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 67, 503–510. doi: 10.1093/gerona/gls088
Tullet, J. M. (2015). DAF-16 target identification in C. elegans: past, present and future. Biogerontology 16, 221–234. doi: 10.1007/s10522-014-9527-y
Tullet, J. M., Araiz, C., Sanders, M. J., Au, C., Benedetto, A., Papatheodorou, I., et al. (2014). DAF-16/FoxO directly regulates an atypical AMP-activated protein kinase gamma isoform to mediate the effects of insulin/IGF-1 signaling on aging in Caenorhabditis elegans. PLoS Genet. 10:e1004109. doi: 10.1371/journal.pgen.1004109
van der Heide, L. P., and Smidt, M. P. (2005). Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 30, 81–86. doi: 10.1016/j.tibs.2004.12.002
Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A. L., Orosz, L., and Muller, F. (2003). Genetics, influence of TOR kinase on lifespan in C. elegans. Nature 426:620. doi: 10.1038/426620a
Vijg, J., and Campisi, J. (2008). Puzzles, promises and a cure for ageing. Nature 454, 1065–1071. doi: 10.1038/nature07216
Wang, M. C., O'Rourke, E. J., and Ruvkun, G. (2008). Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960. doi: 10.1126/science.1162011
Wang, Y., and Tissenbaum, H. A. (2006). Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Aging Dev. 127, 48–56. doi: 10.1016/j.mad.2005.09.005
Wolff, S., Ma, H., Burch, D., Maciel, G. A., Hunter, T., and Dillin, A. (2006). SMK-1, an essential regulator of DAF-16-mediated longevity. Cell 124, 1039–1053. doi: 10.1016/j.cell.2005.12.042
Woods, A., Dickerson, K., Heath, R., Hong, S. P., Momcilovic, M., Johnstone, S. R., et al. (2005). Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2, 21–33. doi: 10.1016/j.cmet.2005.06.005
Yamagata, K., Daitoku, H., Takahashi, Y., Namiki, K., Hisatake, K., Kako, K., et al. (2008). Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell. 32, 221–231. doi: 10.1016/j.molcel.2008.09.013
Yang, J. Y., Zong, C. S., Xia, W., Yamaguchi, H., Ding, Q., Xie, X., et al. (2008). ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 10, 138–148. doi: 10.1038/ncb1676
Yoshida, K., Yamaguchi, T., Natsume, T., Kufe, D., and Miki, Y. (2005). JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 7, 278–285. doi: 10.1038/ncb1228
Yuan, Z., Becker, E. B., Merlo, P., Yamada, T., DiBacco, S., Konishi, Y., et al. (2008). Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319, 1665–1668. doi: 10.1126/science.1152337
Keywords: DAF-16/FOXO, aging, longevity, lifespan, C. elegans
Citation: Sun X, Chen W-D and Wang Y-D (2017) DAF-16/FOXO Transcription Factor in Aging and Longevity. Front. Pharmacol. 8:548. doi: 10.3389/fphar.2017.00548
Received: 06 June 2017; Accepted: 04 August 2017;
Published: 23 August 2017.
Edited by:
José das Neves, Instituto de Investigação e Inovação em Saúde, PortugalReviewed by:
Alexey Moskalev, Institute of Biology of Komi Science Center of Ural Division of RAS, RussiaEun-soo Kwon, Korea Research Institute of Bioscience and Biotechnology, South Korea
Copyright © 2017 Sun, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Dong Chen, wdchen666@163.com
Yan-Dong Wang, ydwangbuct2009@163.com
 Xiaojuan Sun
Xiaojuan Sun Wei-Dong Chen
Wei-Dong Chen Yan-Dong Wang
Yan-Dong Wang